ABSTRACT.
The global burden of sepsis is concentrated in sub-Saharan Africa (SSA), where epidemic HIV and unique pathogen diversity challenge the effective management of severe infections. In this context, patient stratification based on biomarkers of a dysregulated host response may identify subgroups more likely to respond to targeted immunomodulatory therapeutics. In a prospective cohort of adults hospitalized with suspected sepsis in Uganda, we applied machine learning methods to develop a prediction model for 30-day mortality that integrates physiology-based risk scores with soluble biomarkers reflective of key domains of sepsis immunopathology. After model evaluation and internal validation, whole-blood RNA sequencing data were analyzed to compare biological pathway enrichment and inferred immune cell profiles between patients assigned differential model-based risks of mortality. Of 260 eligible adults (median age, 32 years; interquartile range, 26–43 years; 59.2% female, 53.9% living with HIV), 62 (23.8%) died by 30 days after hospital discharge. Among 14 biomarkers, soluble tumor necrosis factor receptor 1 (sTNFR1) and angiopoietin 2 (Ang-2) demonstrated the greatest importance for mortality prediction in machine learning models. A clinicomolecular model integrating sTNFR1 and Ang-2 with the Universal Vital Assessment (UVA) risk score optimized 30-day mortality prediction across multiple performance metrics. Patients assigned to the high-risk, UVA-based clinicomolecular subgroup exhibited a transcriptional profile defined by proinflammatory innate immune and necroptotic pathway activation, T-cell exhaustion, and expansion of key immune cell subsets including regulatory and gamma-delta T cells. Clinicomolecular stratification of adults with suspected sepsis in Uganda enhanced 30-day mortality prediction and identified a high-risk subgroup with a therapeutically targetable immunological profile. Further studies are needed to advance pathobiologically informed sepsis management in SSA.
INTRODUCTION
The global burden of sepsis, a heterogeneous syndrome of acute organ dysfunction resulting from a complex host response to infection, is concentrated in sub-Saharan Africa (SSA).1,2 In contrast to high-income countries (HICs), where sepsis typically affects older adults with severe bacterial infections, sepsis in SSA disproportionately affects young adults with HIV hospitalized with severe tuberculosis (TB), malaria, and other infections highly divergent from those prevalent in HICs.3,4 In this unique context, predictive performance of physiology-driven prognostic scores for sepsis-related mortality are suboptimal.5 Moreover, recent clinical trials of sepsis treatment strategies, developed in HICs and deployed agnostic of locally relevant immunopathology, have shown harm when implemented in the region.6–8 In the search for effective sepsis treatment strategies relevant to SSA, patient stratification based on a combination of molecular and physiological markers may identify patient subgroups more likely to respond to targeted therapeutics.
In this pilot study, we examined the prognostic and pathobiological implications of a clinicomolecular approach to sepsis triage in SSA. In a prospective cohort of adults hospitalized with suspected sepsis in Uganda, we developed a risk model for mortality prediction at 30 days after hospital discharge that integrates soluble immune biomarkers and bedside physiological parameters. After model evaluation and internal validation, we used whole-blood RNA sequencing to compare biological pathway enrichment and inferred immune cell profiles between patients assigned differential model-based risks of mortality.
MATERIALS AND METHODS
Study setting, participants, and outcomes.
In this study, we analyzed data and blood samples from a prospective observational cohort, Research in the Epidemiology of Severe and Emerging Infections in Uganda (RESERVE-U), of adults (age, ≥ 18 years) hospitalized with severe, undifferentiated infection (suspected sepsis) at Entebbe General Referral Hospital in central Uganda from April 2017 to August 2019.9 Entebbe General Referral Hospital is a 200-bed public district referral hospital with a catchment area of approximately 3 million persons. In the primary catchment area, HIV prevalence is approximately 6% and malaria is endemic.9 The RESERVE-U study enrollment occurred within 24 hours of hospital admission. The primary outcome of the RESERVE-U study was vital status at 30 days after hospital discharge (obtained via telephone from patients or their surrogates). Further details of RESERVE-U study protocols, clinical data and sample collection, and study site capacity have been published and are summarized in the supplemental materials.9,10
Soluble immune biomarkers.
From cryopreserved serum samples collected at the time of study enrollment, we quantified, using custom Luminex 200 system kits (Luminex, Austin, TX) from MilliporeSigma (Burlington, MA) and R&D Systems (Minneapolis, MN), 14 soluble biomarkers reflective of putative domains of sepsis immunopathology (innate/adaptive immune activation, endothelial dysfunction, fibrinolysis). These included interleukin (IL)-6, IL-8, IL-10, interferon (IFN)-γ, IFN-γ-induced protein-10/C-X-C motif chemokine 10 (IP-10/CXCL10), macrophage inflammatory protein-1-alpha/chemokine (C-C motif) ligand 3 (MIP-1α/CCL3), macrophage inflammatory protein-1-beta/chemokine (C-C motif) ligand 4 (MIP-1β/CCL4), tumor necrosis factor-alpha (TNF-α), angiopoietin-1 (Ang-1), angiopoietin-2 (Ang-2), macrophage migration inhibitory factor (MIF), plasminogen activator inhibitor-1 (PAI-1), soluble TNF-receptor type 1 (sTNFR1), and soluble IL-2 receptor alpha/soluble CD25 (sIL-2RA/sCD25). Further details are in the supplemental materials.
Whole-blood RNA isolation, library preparation, and sequencing.
From cryopreserved whole-blood samples collected in PAXgene blood RNA tubes (PreAnalytiX; Qiagen/BD, Hombrechtikon, Switzerland), RNA was isolated and purified using PAXgene blood RNA kits (Qiagen, Hilden, Germany). RNA sequencing libraries were prepared using the NEBNext Ultra RNA Library Prep Kit (NEB, Ipswich, MA). Sequencing libraries were multiplexed and analyzed using a 2 × 150 paired-end configuration on the Illumina HiSeq 4000 platform (Illumina, Inc., San Diego, CA). Further details on RNA sequencing methods, including data processing, alignment, and transcript quantification, are in the supplemental materials.11–14
Biomarker selection.
To identify a parsimonious set of biomarkers that may augment mortality prediction, we performed feature selection using random forest and gradient-boosted machine classifier models (mlr, caret, randomForest, and xgboost R packages). Each hyperparameter-tuned classifier, trained to predict 30-day mortality, was applied to log10-transformed concentrations of all 14 biomarkers. For the randomForest classifier, the mtry hyperparameter was selected based on minimization of out-of-bag error estimates, with maximum nodes and trees set to 10 and 1,000, respectively. For the gradient-boosted machine classifier, the eta, max_depth, and nrounds hyperparameters were selected based on maximization of area under the receiver–operating characteristic curve (AUC-ROC) in a grid search approach, with the remaining hyperparameters left at default settings. In random forest and gradient-boosted machine classifiers, the predictive importance of each biomarker was ranked according to Gini impurity and split-gain values, respectively.
Model development and internal validation.
To optimize parsimony, the two biomarkers with the consistently greatest importance across random forest and gradient-boosted machine classifiers were added to physiology-based clinical risk models validated for prediction of severe infection-related outcomes in resource-limited settings Supplemental Table 1). Clinical risk models included the quick Sepsis-Related Organ Failure Assessment (qSOFA) score, the Modified Early Warning Score (MEWS), and the Universal Vital Assessment (UVA) score.5 All clinical and clinical–biomarker (i.e., clinicomolecular) models were adjusted for age, sex, and prehospital illness duration—factors that may affect the host response to severe infection and sepsis mortality risk.
For all adjusted clinical models (qSOFA, MEWS, and UVA), we first determined the incremental predictive accuracy of the corresponding clinicomolecular model using likelihood ratio χ2 tests, followed by computation of 10,000 bootstrap-derived Brier scores and continuous net reclassification improvement (NRI) indices (rms and Hmisc R packages).15–18 Discrimination and calibration of each adjusted clinical and clinicomolecular model were evaluated further using AUC-ROC and calibration curves generated via 100-times–repeated 10-fold cross-validation (caret and MLeval R packages). At this stage, we performed three sensitivity analyses: one in which clinicomolecular models were expanded to include the three most important biomarkers identified across classifier models, another in which all models were additionally adjusted for HIV and malaria status, and one in which patients with unknown 30-day vital status were considered deceased. After selecting the best-performing clinicomolecular model based on optimization of Brier score, NRI indices, AUC-ROC, and calibration curve fit, we applied 100-times–repeated 10-fold cross-validation to generate confusion matrices and related metrics across probability cutoffs of 0.3 to 0.7 (caret R package). Using the probability cutoff that maximized accuracy, we assigned patients to high- versus low-mortality risk groups.
Differential gene expression, biological pathway, and immune cell type deconvolution analyses.
Between patients assigned to high- versus low-mortality risk groups, we performed differential gene expression analysis of whole-blood RNA sequencing data using the DESeq2 R package.19 Genes were considered differentially expressed based on a log2-fold change ≥ |0.26| and a Benjamini–Hochberg-adjusted P value ≤ 0.05. Differentially expressed gene sets were selected for biological pathway analyses (Ingenuity Pathway Analysis, Qiagen), the results of which were examined to infer functional differences between risk groups. To infer the relative abundance of immune cell subsets across risk groups, we performed digital cytometry deconvolution using the CIBERSORTx platform and LM22 hematopoietic gene signature reference matrix.20
Reporting statement.
The Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis guidelines were used to report model development and reporting.21
Statistical analyses.
Analyses were performed using R (v4.2.0, R Foundation for Statistical Computing, Vienna, Austria) via the RStudio environment, with packages specified as above. Biological pathway analysis was performed using Ingenuity Pathway Analysis (Qiagen, Hilden, Germany).
RESULTS
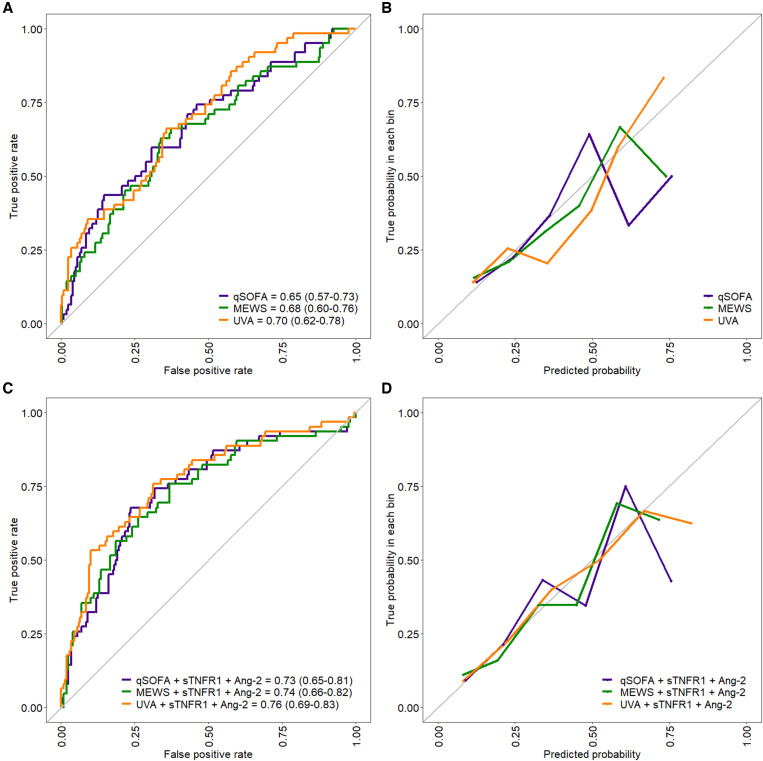
Of the 301 adults enrolled in the RESERVE-U study, 288 (95.6%) had soluble biomarkers quantified in serum samples. Of these 288 patients, 30-day postdischarge vital status was determined for 260 (90.3%), of whom 62 died (23.8%) (Table 1, Supplemental Figure 1). Among adjusted clinical models, UVA demonstrated optimal performance, with the lowest Brier score, highest cross-validated AUC-ROC (0.70; 95% CI 0.62–0.78), and best-fitting calibration curve (Figure 1A and B).
Table 1.
Patient characteristics stratified by UVA–based clinicomolecular risk groups including sTNFR1 and Ang-2
| Variable | All patients (N = 260) | High-risk group (n = 51) | Low-risk group (n = 209) |
|---|---|---|---|
| Female sex, n/N (%) | 154/260 (59.2) | 25/51 (49.0) | 129/209 (61.7) |
| Age, y; median (IQR) | 32 (26–43) | 43 (32–50) | 32 (26–40) |
| Illness duration prior to hospitalization, d; median (IQR)* | 4 (3–7) | 6 (3–7) | 4 (2–6) |
| Received antibacterial or antimalarial agent prior to hospitalization, n/N (%) | 97/260 (37.3) | 21/51 (41.2) | 76/209 (36.4) |
| Temperature ≥ 38°C, n/N (%) | 95/260 (36.5) | 21/51 (41.2) | 74/209 (35.4) |
| Temperature < 36°C, n/N (%) | 73/260 (28.1) | 18/51 (35.3) | 55/209 (26.3) |
| Heart rate, beats/min; median (IQR) | 98 (87–108) | 104 (95–121) | 96 (86–107) |
| Respiratory rate, breaths/min; median (IQR) | 22 (21–26) | 24 (22–28) | 22 (20–26) |
| Systolic blood pressure, mm Hg; median (IQR) | 102 (91–117) | 90 (85–110) | 104 (95–118) |
| Oxygen saturation, %; median (IQR) | 97 (95–98) | 97 (91–98) | 97 (96–98) |
| Encephalopathy, n/N (%)† | 54/260 (20.8) | 27/51 (52.9) | 27/209 (12.9) |
| qSOFA score, points; median (IQR)‡ | 1 (1–2) | 2 (2–3) | 1 (1–2) |
| qSOFA score ≥ 2, n/N (%)‡ | 120/260 (46.2) | 39/51 (76.5) | 81/209 (38.8) |
| qSOFA score ≥ 1, n/N (%)‡ | 229/260 (88.1) | 49/51 (96.1) | 180/209 (86.1) |
| MEWS, points; median (IQR) | 3 (2–5) | 5 (3.50–6.50) | 3 (2–4) |
| UVA score, points; median (IQR) | 3 (1, 4) | 6 (3.50, 8) | 2 (1, 4) |
| Shock, n/N (%)§ | 39/260 (15.0) | 16/51 (31.4) | 23/209 (11.0) |
| Acute respiratory failure, n/N (%)ǁ | 60/260 (23.1) | 22/51 (43.1) | 38/209 (18.2) |
| Severe anemia, n/N (%)¶ | 53/260 (20.4) | 22/51 (43.1) | 31/209 (14.8) |
| Living with HIV, n/N (%) | 139/258 (53.9) | 45/51 (88.2) | 94/207 (45.4) |
| WHO HIV clinical stage 3 or 4, n/N (%) | 111/139 (79.9) | 36/45 (80.0) | 75/94 (79.8) |
| Receiving ART prior to hospitalization, n/N (%) | 82/139 (59.0) | 31/45 (68.9) | 51/94 (54.3) |
| Receiving TMP-SMX prophylaxis prior to hospitalization, n/N (%) | 85/139 (61.2) | 31/45 (68.9) | 54/94 (57.4) |
| Malaria RDT positive, n/N (%) | 57/256 (22.3) | 6/49 (12.2) | 51/207 (24.6) |
| Microbiological TB positive, n/N (%)# | 49/260 (18.8) | 19/51 (37.3) | 30/209 (14.4) |
| Urine TB-LAM positive, n/N (%)** | 38/112 (31.7) | 16/33 (48.5) | 22/79 (27.8) |
| Influenza PCR positive, n/N (%) | 16/235 (6.8) | 2/46 (4.3) | 14/189 (7.4) |
| Death in hospital or transfer, n/N (%) | 38/260 (14.6) | 23/51 (45.1) | 15/209 (7.2) |
| Duration of hospitalization, d; median (IQR)†† | 5 (3–8) | 7 (4–10) | 5 (3–7) |
| KPS ≤ 70 at alive discharge, n/N (%) | 16/220 (7.3) | 6/28 (21.4) | 10/192 (5.2) |
| Death at 30 days post-discharge, n/N (%) | 62/260 (23.8) | 33/51 (64.7) | 29/209 (13.9) |
Ang-2 = angiopoietin 2; ART = antiretroviral therapy; IQR = interquartile range; KPS = Karnofsky Performance Status; LAM = lipoarabinomannan; MEWS = Modified Early Warning Score; PCR = polymerase chain reaction; qSOFA = quick Sepsis-Related Organ Failure Assessment; RDT = rapid diagnostic test; sTNFR1 = soluble tumor necrosis factor receptor 1; TB = tuberculosis; TMP-SMX = trimethoprim–sulfamethoxazole; UVA = Universal Vital Assessment.
Median value of 4 days imputed for one patient with unknown data.
Anything other than “alert” on alert, responsive to voice, responsive to pain, unresponsive (AVPU) mental status assessment.
Systolic blood pressure ≤ 100 mm Hg, respiratory rate ≥ 22 breaths/min, and encephalopathy, with the latter defined using the AVPU scale.
Systolic blood pressure ≤ 90 mm Hg despite administration of ≥ 1 L intravenous fluid.
Oxygen saturation ≤ 90% or respiratory rate ≥ 30 breaths/min.
Hemoglobin < 9 g/dL or administration of blood transfusion.
Positive result by sputum Xpert Ultra or smear or urine TB-LAM.
Performed for patients living with HIV if urine sample obtainable.
Unknown for 10 patients.
Figure 1.
Receiver–operating characteristic (ROC) and calibration curves for (A, B) clinical and (C, D) clinicomolecular models including soluble tumor necrosis factor receptor 1 (sTNFR1) and angiopoietin 2 (Ang-2). All curves were generated using 100-times–repeated 10-fold cross-validation, and reflect models adjusted for age, sex, and illness duration prior to hospitalization. Area under ROC curves are presented with 95% CIs in parentheses. MEWS = Modified Early Warning Score; qSOFA = quick Sepsis-Related Organ Failure Assessment; UVA = Universal Vital Assessment.
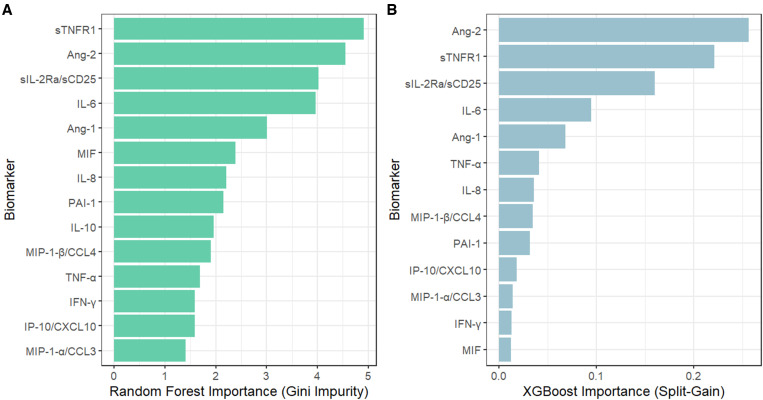
The concentrations of most biomarkers, with the exception of Ang-1 and MIP-1a/CCL3, were greater in patients who died at 30 days compared with those who survived Supplemental Table 2). In random forest and gradient-boosted machine classifier models, sTNFR1 and Ang-2 demonstrated the greatest importance for 30-day mortality prediction (Figure 2). The addition of sTNFR1 and Ang-2 (individually and in combination) to each adjusted clinical model improved 30-day mortality prediction significantly (likelihood ratio χ2 test, P < 0.01 for all biomarker-inclusive models), with lower Brier scores and improved net reclassification (Table 2). The UVA-based clinicomolecular model including sTNFR1 and Ang-2 demonstrated optimal performance, with the lowest Brier score, highest AUC-ROC (0.76; 95% CI, 0.69–0.83) and best-fitting calibration curve (Table 2, Figure 1C and D). The addition of the third most important biomarker (soluble IL-2 receptor alpha/soluble CD25) to clinicomolecular models including sTNFR1 and Ang-2 did not improve performance metrics Supplemental Table 3). Our results were generally consistent when patients with unknown 30-day vital status were considered deceased and when all clinical and biomarker-inclusive models were additionally adjusted for HIV and malaria status Supplemental Tables 4 and 5).
Figure 2.
Importance of immune biomarkers for 30-day mortality prediction in (A) random forest and (B) gradient-boosted machine classifier models ranked according to Gini impurity and split-gain values, respectively. Ang-1 = angiopoietin-1; Ang-2 = angiopoietin-2; CCL3 = chemokine (C-C motif) motif ligand 3; CCL4 = chemokine (C-C motif) ligand 4; IFN = Interferon; IL = Interleukin; IP-10/CXCL10 = IFN-γ-induced protein-10/C-X-C motif chemokine 10; MIF = macrophage migration inhibitory factor; MIP-1α = macrophage inflammatory protein-1-alpha; MIP-1β = macrophage inflammatory protein-1-beta; PAI-1 = plasminogen activator inhibitor-1; sTNFR1 = soluble TNF-receptor type 1; sIL-2RA/sCD25 = soluble IL-2 receptor alpha/soluble CD25; and TNF-α = tumor necrosis factor-alpha.
Table 2.
Performance characteristics of clinical and clinicomolecular models for prediction of 30-day mortality (N = 260)
| Model* | Likelihood ratio χ2 P value† | Optimism-corrected Brier score‡ | NRI (95% CI) | AUC-ROC (95% CI)§ |
|---|---|---|---|---|
| qSOFA | Reference | 0.171 | Reference | 0.65 (0.57–0.73) |
| qSOFA + Ang-2 | < 0.001 | 0.161 | 0.39 (0.11–0.67) | 0.71 (0.63–0.79) |
| qSOFA + sTNFR1 | < 0.001 | 0.160 | 0.50 (0.22–0.77) | 0.73 (0.65–0.81) |
| qSOFA + Ang-2 + sTNFR1 | < 0.001 | 0.157 | 0.69 (0.42–0.96) | 0.73 (0.65–0.81) |
| MEWS | Reference | 0.170 | Reference | 0.68 (0.60–0.76) |
| MEWS + Ang-2 | 0.002 | 0.163 | 0.40 (0.12–0.68) | 0.72 (0.64–0.80) |
| MEWS + sTNFR1 | < 0.001 | 0.161 | 0.38 (0.09–0.66) | 0.74 (0.66–0.82) |
| MEWS + Ang-2 + sTNFR1 | < 0.001 | 0.159 | 0.60 (0.32–0.87) | 0.74 (0.66–0.82) |
| UVA | Reference | 0.163 | Reference | 0.70 (0.62–0.78) |
| UVA + Ang-2 | 0.001 | 0.155 | 0.43 (0.15–0.71) | 0.74 (0.66–0.82) |
| UVA + sTNFR1 | < 0.001 | 0.150 | 0.53 (0.25–0.80) | 0.76 (0.69–0.83) |
| UVA + Ang-2 + sTNFR1 | < 0.001 | 0.149 | 0.68 (0.41–0.95) | 0.76 (0.69–0.83) |
Ang-2 = angiopoietin 2; AUC-ROC = area under the receiver–operating characteristic curve; MEWS = Modified Early Warning Score; NRI = net reclassification improvement; qSOFA = quick Sepsis-Related Organ Failure Assessment; sTNFR1 = soluble tumor necrosis factor receptor 1; UVA = Universal Vital Assessment.
All models adjusted for age, sex, and prehospital illness duration.
P value reflects results of the likelihood ratio χ2 test comparing each biomarker-inclusive (i.e., clinicomolecular) model against the reference clinical model (qSOFA, MEWS, UVA).
Optimism-corrected Brier scores generated using 10,000 bootstraps.
AUC-ROC and 95% CIs generated using 100-times–repeated 10-fold cross-validation.
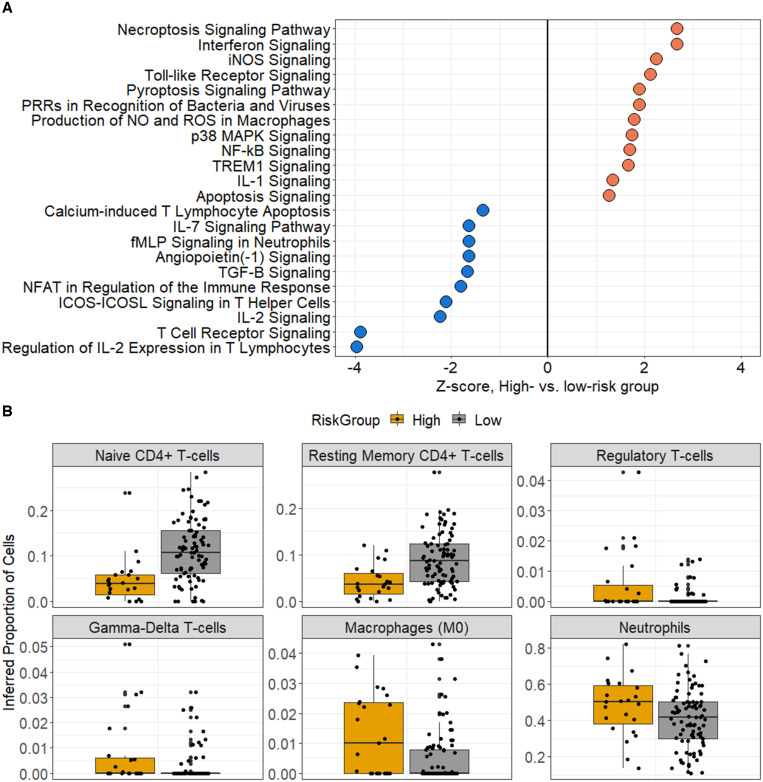
For the UVA-based clinicomolecular model including sTNFR1 and Ang-2, a probability cutoff of 0.40 optimized predictive accuracy (accuracy, 0.80; specificity, 0.90; sensitivity, 0.49; negative predictive value, 0.85; positive predictive value, 0.62), with 51 (19.6%) and 209 (80.4%) patients assigned to high- versus low-mortality risk groups, respectively (Table 1 and Supplemental Tables 6 and 7). Of these 260 patients, whole-blood RNA sequence analysis was performed in 112 (43.1%), including 23 of 51 (45.1%) and 89 of 209 (42.6%) patients in the high- versus low-risk groups, respectively. A total of 2,209 genes were expressed differentially across groups. Functionally, the high-risk subgroup exhibited an immune profile characterized by T-cell exhaustion, including downregulation of T-cell receptor, inducible T-cell co-stimulator, IL-2, and IL-7 signaling (Figure 3A). These findings were observed alongside the upregulation of key proinflammatory innate immune pathways, including Toll-like receptor, nuclear factor-kappa B, triggering receptor expressed on myeloid cells 1, and IL-1 signaling, as well as those related to necroptosis, apoptosis, and production of reactive nitrogen and oxygen species. In contrast, patients in the high-risk subgroup showed downregulation of genes associated with the Ang-1/Tie2 axis, which promotes endothelial stability and mitigates inflammation. In the high-risk group, immune cell-type deconvolution inferred significantly greater quantities of neutrophils, naive (M0) macrophages, regulatory T cells, and gamma-delta T cells, along with lower quantities of CD4 T cells (Figure 3B).
Figure 3.
(A) Ingenuity pathway analysis of canonical signaling gene sets enriched across Universal Vital Assessment–based clinicomolecular risk groups including soluble tumor necrosis factor receptor 1 (sTNFR1) and angiopoietin 2 (Ang-2). Inferred pathway enrichment was based on differentially expressed genes at a log2-fold change ≥ |0.26| and a Benjamini-Hochberg-adjusted P value ≤ 0.05. Key immune pathways with activation Z-score ≥ |1.25| are presented. (B) Inferred abundance of immune cell subsets across the Universal Vital Assessment-based clinicomolecular risk groups including sTNFR1 and Ang-2. Relative immune cell proportions were inferred using the CIBERSORTx platform. Presented cell subsets reflect those with significantly different abundance across groups (P ≤ 0.05, Wilcoxon rank-sum test). IL = interleukin; NOS = inducible nitric oxide synthase; TREM = triggering receptor expressed on myeloid cells.
DISCUSSION
Using immune biomarker and physiological data from a microbiologically diverse cohort of adults with suspected sepsis in Uganda, we developed a novel clinicomolecular risk index for 30-day mortality prediction. Our optimized clinicomolecular model, integrating sTNFR1 and Ang-2 with the UVA risk score, identified a high-risk, HIV-predominant subgroup defined by T-cell exhaustion, innate immune activation, necroptosis, and pyroptosis, and differential expansion of key immune cell subsets. Although hundreds of studies in HICs have evaluated the utility of biomarker-driven patient stratification in adult sepsis, to our knowledge this is the first to apply a clinicomolecular approach to sepsis triage in SSA.22,23
Results from our pilot study suggest that integrated use of immune biomarkers and bedside physiological data could enhance patient stratification and prognostic clinical trial enrichment for sepsis in SSA and other high HIV-burden settings.24 Furthermore, considering that HIC-derived, biologically agnostic treatment strategies have been associated with poor clinical outcomes when implemented in SSA, future sepsis trials in the region must incorporate predictive enrichment strategies rooted in locally relevant immunopathology—an approach that biomarker-driven patient classification may enhance.6–8,25–27 For example, our optimized clinicomolecular model identified a high-risk, HIV-predominant subgroup characterized by T-cell exhaustion and proinflammatory innate immune activation, suggesting a possible role for synergistic host-directed therapeutics targeting T-cell stimulation and innate immune dampening.28,29 In addition, the importance of necroptotic and pyroptotic pathways in the high-risk subgroup suggests that these processes, the upregulation of which have been associated with poor outcomes in animal models of sepsis pathogenesis, may represent important candidates for locally relevant enrichment strategies.30 Operationally, the relatively high specificity and moderately impactful positive likelihood ratio of our optimized clinicomolecular model (in its current iteration) suggest that it would be most applicable to “rule in” high-risk patients with a more dysregulated immune profile, thereby providing a subpopulation target for candidate immunomodulatory agents. As rapid quantification of blood-based biomarkers may be feasible in resource-limited settings with emerging immunoassays, clinicomolecular sepsis triage could conceivably be applied in SSA.
Our study has limitations. First, the single-center nature and limited size of our cohort mandates external validation of our findings. Second, 30-day vital status was unknown for approximately 10% of analyzed patients. However, in a sensitivity analysis considering such patients as deceased, our results were generally consistent. Third, although our optimized clinicomolecular model had high specificity, sensitivity was low and AUC-ROC was modest, reinforcing the limitations of its use in its current form and the need for further refinement. Fourth, although biomarkers were included based on immunopathological relevance, there may be other proteins that better augment mortality prediction in SSA. Fifth, biomarker-driven triage strategies are not immediately implementable in much of SSA. However, accessibility to rapid, point-of-need immunoassays in low-income settings is increasing, and we applied minimally biased feature selection methods to identify the most predictive biomarkers that could be prioritized for such platforms. Next, whole-blood RNA sequencing data was available from approximately 43% of analyzed patients. However, the characteristics of these patients were similar to the larger cohort.10 Last, we relied on computational deconvolution analyses to infer quantities of immune cell subsets because we were unable to isolate peripheral blood mononuclear cells at our study site as a result of resource limitations.
Clinicomolecular stratification of adults with suspected sepsis in Uganda enhanced 30-day mortality prediction and identified a high-risk subgroup with a therapeutically targetable immunological profile. Further studies are needed to advance biologically informed sepsis management strategies in SSA.
Supplemental files
ACKNOWLEDGMENTS
We acknowledge and thank the patients enrolled in this study, clinicians at Entebbe General Referral Hospital, and the staff of the Uganda Virus Research Institute Arbovirology and Emerging and Reemerging Infections Laboratory for their assistance with data and sample collection, and laboratory testing.
Note: Supplemental materials appear at www.ajtmh.org.
REFERENCES
- 1. Rudd KE. et al. , 2020. Global, regional, and national sepsis incidence and mortality, 1990–2017: analysis for the Global Burden of Disease Study. Lancet 395: 200–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singer M. et al. , 2016. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA 315: 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vincent JL. et al. , 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302: 2323–2329. [DOI] [PubMed] [Google Scholar]
- 4. Lewis JM, Feasey NA, Rylance J, 2019. Aetiology and outcomes of sepsis in adults in sub-Saharan Africa: a systematic review and meta-analysis. Crit Care 23: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adegbite BR, Edoa JR, Ndzebe Ndoumba WF, Dimessa Mbadinga LB, Mombo-Ngoma G, Jacob ST, Rylance J, Hänscheid T, Adegnika AA, Grobusch MP, 2021. A comparison of different scores for diagnosis and mortality prediction of adults with sepsis in low-and-middle-income countries: a systematic review and meta-analysis. EClinicalMedicine 42: 101184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maitland K. et al. , 2011. Mortality after fluid bolus in African children with severe infection. N Engl J Med 364: 2483–2495. [DOI] [PubMed] [Google Scholar]
- 7. Andrews B, Muchemwa L, Kelly P, Lakhi S, Heimburger DC, Bernard GR, 2014. Simplified severe sepsis protocol: a randomized controlled trial of modified early goal-directed therapy in Zambia. Crit Care Med 42: 2315–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Andrews B, Semler MW, Muchemwa L, Kelly P, Lakhi S, Heimburger DC, Mabula C, Bwalya M, Bernard GR, 2017. Effect of an early resuscitation protocol on in-hospital mortality among adults with sepsis and hypotension: a randomized clinical trial. JAMA 318: 1233–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cummings MJ. et al. , 2021. Stratifying sepsis in Uganda using rapid pathogen diagnostics and clinical data: a prospective cohort study. Am J Trop Med Hyg 105: 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cummings MJ. et al. , 2022. Multidimensional analysis of the host response reveals prognostic and pathogen-driven immune subtypes among adults with sepsis in Uganda. Crit Care 26: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bolger AM, Lohse M, Usadel B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andrews S, 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed December 14, 2022.
- 13. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liao Y, Smyth GK, Shi W, 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 15. Cook NR, 2018. Quantifying the added value of new biomarkers: how and how not. Diagn Progn Res 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, Vasan RS, 2008. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat Med 27: 157–172. [DOI] [PubMed] [Google Scholar]
- 17. Harrell FE, 2015. Multivariable modeling strategies. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis , 2nd edition. New York, NY: Springer Press, 63–102. [Google Scholar]
- 18. Pencina MJ, D’Agostino RB, Sr, Steyerberg EW, 2011. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 30: 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S, 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Newman AM. et al. , 2019. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol 37: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Collins GS, Reitsma JB, Altman DG, Moons KG, 2015. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): the TRIPOD statement. Ann Intern Med 162: 55–63. [DOI] [PubMed] [Google Scholar]
- 22. Barichello T, Generoso JS, Singer M, Dal-Pizzol F, 2022. Biomarkers for sepsis: more than just fever and leukocytosis: a narrative review. Crit Care 26: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reinhart K, Bauer M, Riedemann NC, Hartog CS, 2012. New approaches to sepsis: molecular diagnostics and biomarkers. Clin Microbiol Rev 25: 609–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stanski NL, Wong HR, 2020. Prognostic and predictive enrichment in sepsis. Nat Rev Nephrol 16: 20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prescott HC, Calfee CS, Thompson BT, Angus DC, Liu VX, 2016. Toward smarter lumping and smarter splitting: rethinking strategies for sepsis and acute respiratory distress syndrome clinical trial design. Am J Respir Crit Care Med 194: 147–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reddy K, Sinha P, O’Kane CM, Gordon AC, Calfee CS, McAuley DF, 2020. Subphenotypes in critical care: translation into clinical practice. Lancet Respir Med 8: 631–643. [DOI] [PubMed] [Google Scholar]
- 27. Maslove DM. et al. , 2022. Redefining critical illness. Nat Med 28: 1141–1148. [DOI] [PubMed] [Google Scholar]
- 28. van der Poll T, van de Veerdonk FL, Scicluna BP, Netea MG, 2017. The immunopathology of sepsis and potential therapeutic targets. Nat Rev Immunol 17: 407–420. [DOI] [PubMed] [Google Scholar]
- 29. Brady J, Horie S, Laffey JG, 2020. Role of the adaptive immune response in sepsis. Intensive Care Med Exp 8 (Suppl 1): 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qu M, Wang Y, Qiu Z, Zhu S, Guo K, Chen W, Miao C, Zhang H, 2022. Necroptosis, pyroptosis, ferroptosis in sepsis and treatment. Shock 57: 161–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.