Abstract
Sterol levels affect the expression of many genes in yeast and humans. We found that the paralogous transcription factors Upc2p and Ecm22p of yeast were sterol regulatory element (SRE) binding proteins (SREBPs) responsible for regulating transcription of the sterol biosynthetic genes ERG2 and ERG3. We defined a 7-bp SRE common to these and other genes, including many genes involved in sterol biosynthesis. Upc2p and Ecm22p activated ERG2 expression by binding directly to this element in the ERG2 promoter. Upc2p and Ecm22p may thereby coordinately regulate genes involved in sterol homeostasis in yeast. Ecm22p and Upc2p are members of the fungus-specific Zn[2]-Cys[6] binuclear cluster family of transcription factors and share no homology to the analogous proteins, SREBPs, that are responsible for transcriptional regulation by sterols in humans. These results suggest that Saccharomyces cerevisiae and human cells regulate sterol synthesis by different mechanisms.
Sterols, essential for life in most, if not all, eukaryotes, are synthesized by a pathway known variously as the mevalonate pathway, the isoprenoid pathway, or the sterol biosynthetic pathway. Although sterols (which include molecules ranging from cholesterol to steroid hormones) are the major products of the sterol biosynthetic pathway, this pathway also produces many important nonsterol compounds that are necessary for cellular processes such as respiration, glycosylation, protein prenylation and photosynthesis. In the heavily studied mammalian sterol biosynthetic pathway, regulation occurs at both the transcriptional and the posttranscriptional levels. At present, the best-understood aspect of this regulation is the transcriptional response to changes in sterol levels. Sterol depletion in mammalian cells causes activation of the transcription factors known as sterol regulatory element (SRE) binding proteins (SREBPs) (8), encoded by the homologous genes SREBP-1 and SREBP-2. When sterols are abundant, the SREBPs are inactive, tethered to the endoplasmic reticulum (ER) membrane by two intrinsic transmembrane helices. When sterol levels drop, regulated proteolysis releases the transcriptional activation domains of the SREBPs from the membrane tether, allowing the activation domains to translocate to the nucleus. Once in the nucleus, these domains activate transcription of genes involved in sterol and fatty acid homeostasis.
The SREBPs bind the same DNA sequences in vitro (21). In vivo, the SREBPs activate an overlapping set of target genes but may have slightly different DNA binding specificities and/or activities (8, 22). Hence, differential activation and synthesis of the SREBPs may make it possible for mammalian cells to respond to a demand for different levels of sterol and nonsterol products.
Many genes in the sterol biosynthetic pathway in Saccharomyces cerevisiae are transcriptionally regulated in response to changes in sterol levels (6, 13). However, less is known about this regulatory mechanism in yeast. Also, yeast lack convincing homologues of the mammalian SREBPs. This work describes the identification of a yeast SRE shared by many genes involved in sterol biosynthesis and the identification of two SREBPs, Upc2p and Ecm22p, which regulate transcription of the sterol biosynthetic genes. Both Upc2p and Ecm22p are members of the Zn[22]-Cys[6] binuclear cluster family of fungal transcription factors (31). UPC2 was originally identified as a semidominant allele that allows sterol uptake under aerobic conditions (11). Upc2p is also involved in the anaerobic activation of expression of the DAN/TIR mannoproteins (1). The ECM22 gene was identified as a mutant sensitive to the cell wall perturbing agent calcofluor white (20). Our data indicated that both Ecm22p and Upc2p were involved in regulation of sterol biosynthesis and identified ERG2 and ERG3 as targets of both proteins.
MATERIALS AND METHODS
Strains and media.
All yeast strains used were isogenic with W303-1a except as noted in Table 1. Gene deletions were made by a PCR-based gene disruption method (2) such that the entire open reading frame (ORF) was replaced by marker sequences from pRS403 (HIS3), pRS404 (TRP1), pRS405 (LEU2), or pRS406 (URA3) (29). The Escherichia coli strain used was ER2508 from New England Biolabs.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303-1a | MATaade2-1 leu2-3,112 his3-1 ura3-52 trp1-100 can1-100 | R. Rothstein |
| JRY7187 | W303-1a erg2Δ::TRP1 | This study |
| JRY7179 | W303-1a upc2Δ::HIS3 | This study |
| JRY7180 | W303-1a ecm22Δ::TRP1 | This study |
| JRY7181 | W303-1a ecm22Δ::TRP1 upc2Δ::HIS3 | This study |
| JRY7182 | W303-1a ERG2::pJR2320 | This study |
| JRY7183 | W303-1a ERG2::pJR2321 | This study |
| JRY7184 | MATa/MATα ade2/ADE2 LYS2/lys2 his3/his3 leu2/leu2 trp1/trp1 ura3/ura3 suc2::HIS3/suc2::HIS3 ERG2::pJR2325/ERG2::pJR2325 LEU2::pJR2326/LEU2::pJR2326 | This study |
| JRY7188 | W303-1a ecm22Δ::TRP1 upc2Δ::HIS3 erg2Δ::TRP1 pJR2331 (ECM22 2μm URA3) | This study |
| JRY7189 | MATα lys2Δ leu2-3,112 his3-1 ura3-52 trp1-100 ecm22Δ::TRP1 erg2Δ::TRP1 | This study |
| JRY7190 | W303-1a URA3::pJR2318 | This study |
| JRY7191 | JRY7179 URA3::pJR2318 | This study |
| JRY7192 | JRY7180 URA3::pJR2318 | This study |
| JRY7193 | JRY7181 URA3::pJR2318 | This study |
Yeast cells were grown in minimal medium (YM; 0.67% Difco yeast nitrogen base with 5% ammonium sulfate without amino acids) containing 2% glucose and required supplements. Solid media contained 2% agar (Bacto). A 25-mg/ml stock solution of lovastatin (a generous gift from J. Bergstrom, Merck) was made as previously described (12). Lovastatin, Geneticin (Gibco), and amphotericin B (Sigma) were used in solid media at final concentrations of 40 μg/ml, 1 mg/ml, and 100 ng/ml, respectively. All drugs were added after autoclaving. Lovastatin plates were incubated at 24°C, Geneticin plates at 30°C and amphotericin B plates at 37°C. Amphotericin B plates were used within 12 h of preparation. Bacteria were grown in LB Amp (1% tryptone, 0.5% NaCl, 0.5% yeast extract, 100 μg of ampicillin/ml).
Plasmids.
The plasmid constructions of this work made extensive use of the pRS vector series (29) as well as the Univector series (19). pJR2325 was an ERG2-lacZ reporter in an integrating TRP1 plasmid based on pRS414. The CEN ARS fragment of pRS414 was removed by cutting with Eco0109I and religated. The plasmid contained the ERG2 promoter fragment from −751 to +12 relative to the ATG initiation codon and an XbaI site fused to the second amino acid of lacZ. The PGK terminator was subcloned 3′ of lacZ.
pJR2326 was an ERG2-KanMX reporter in an integrating LEU2 plasmid based on YIplac128 (14). pJR2326 was made by a three-fragment ligation of the SphI-XbaI fragment (containing the ERG2 promoter) of pJR2325, a KanMX fragment amplified by PCR using primers av100 (5′-TAAACATCTAGAGGTAAGGAAAAGACTCACGTTTCG-3′) and solig191 (5′-GAAAACAAGAATTCTTTTTATTGTCAGTAC-3′) cut with XbaI and EcoRI and the YIplac128 vector backbone cut with SphI and EcoRI.
pJR2307 was a 2μm URA3-marked plasmid based on pLacZi (GenBank accession no. U89671) from Clontech. The 2μm origin was PCR amplified from pRS426 using primers av249 (5′-TTTCCCCGAAAAGTGCCTGCAGAACGAAGCATCTGTGCTTC-3′) and av250 (5′-CATGATAATAATGGTTCTGCAGTATGATCCAATATCAAAGGAAATG-3′) and subcloned as a PstI fragment into the NsiI site of pLacZi to make pJR2307. Plasmids pJR2308-2315 and pJR2327-2329 were made by subcloning a phosphorylated double-stranded oligonucleotide into the EcoRI and XhoI sites of pJR2307. The oligonucleotides were designed to have single-stranded overhangs on either end to facilitate cloning into the EcoRI and XhoI sites. The design of the oligonucleotides was such that the EcoRI site was followed by an EcoRV site (to allow for easy identification of plasmids with inserts), followed by the relevant sequence from the ERG2 promoter and an XhoI site. pJR2316 was made from pJR2307 by subcloning a fragment of the ERG2 promoter PCR-amplified using primers av134 (5′-AAGAGAGTCGACGGTACCCATTTCGGCACTAAAATC-3′) and av67 (5′-TCTTGTCGACCATGGCGCTGCAGATCTATC-3′) into the EcoRI and XhoI sites of pJR2307. pJR2317 was made similarly as pJR2316 but used pJR2320 as a template for the PCR. The ERG2 promoter in this plasmid therefore had a mutated SRE.
pJR2318 was an ERG3-lacZ reporter made by subcloning an ERG3 promoter fragment as an EcoRI-XhoI fragment into the same sites in pLacZi (integrating plasmid carrying URA3) (GenBank accession no. U89671) from Clontech. The ERG3 promoter fragment was generated by PCR amplification from genomic DNA using primers av127 (5′-ATTCCCGAATTCGTCCTGCTTTGAGTCGTTTTC-3′) and av128 (5′-CTCTAACTCGAGCCGCGATGTTTCTTTCGACC-3′).
pJR2320 contained the ERG2 promoter from −751 to +198 as an EcoRI-XhoI fragment in pRS406 (integrating plasmid carrying URA3). The ERG2 SRE was mutated (from 5′-CTCGTATAAG-3′ to 5′-ACGATATCTA-3′) by using a PCR sewing technique (2) using genomic DNA from W303-1a as a template and the primers av134 (5′-GTACTGGAATTCCTGCATTATCATCCCGATTGCT-3′), av194 (5′-AACCACGGCCACGATATCTACCGCAAGGAAAACTACCGGT-3′), av195 (5′-TTCCTTGCGGTAGATATCGTGGCCGTGGTTCGATTCTGCC-3′), and av308 (5′-CCCCGTAACTCGAGTTAAGTGCGTCTCTGACATCCTG-3′). pJR2321 was similar to pJR2320 but had a wild-type SRE. The insert was generated by PCR using the primers av134 and av308 and genomic DNA from W303-1a as a template.
pJR2330 was a 2μm LEU2 plasmid containing a 3.6-kb fragment containing ECM22. The ECM22 fragment was PCR amplified using primers av357 (5′-GACGTTGGATCCGGTAGAGTGGCGCTAGCATAG-3′) and av358 (5′-GCATTTAGGATCCAACGTATACCGGGTGTACCTC-3′), and the resulting fragment was subcloned into the BamHI site of pRS425. pJR2331 contained ECM22 on a 3.6-kb BamHI fragment (same as in pJR2330) subcloned into the BamHI site of pJR2332. pJR2332 was pRS426 (2μm, URA3), cut with EcoRI and SalI and resealed by blunt-end ligation, such that the EcoRI-SalI fragment in the multiple cloning site was missing.
pJR2322 was made from pJR2333 by cre-lox site-specific recombination with pHB2 as previously described (19), resulting in a GST-Ecm22p1–497 fusion protein expressed from the tac promoter. pJR2333 was made by subcloning a PCR-amplified fragment carrying ECM22 (codons 1 to 497 with a STOP codon inserted after codon 497) as an XhoI-SacI fragment into the same sites in pUNI10 (19). The PCR fragment was generated by using DynaZyme EXT (Finnzymes) with primers av352 (5′-CTCCATCTCTCGAGAAATGACATCCGATGATGGGAATG-3′) and av390 (5′-CAAATTCAAGAGCTCTTATCTAGACAGCCCGACTAATTCAG-3′) and plasmid pJR2330 as a template.
pJR2323 was made by subcloning a PCR-amplified fragment containing Upc2p1–395 (amino acids 1 through 395) as an XhoI-NotI fragment into the bacterial expression vector pBAT4 (23). The PCR fragment was generated using primers av570 (5′-ACAGTTCCTCGAGATGAGCGAAGTCGGTATACAGAATC-3′) and av571 (5′-AGTAGTTGCGGCCGCTTATGATTGTAAAGGCCCTTCCTTTACC-3′).
2μm overexpression screen.
The diploid host strain (JRY7184) was transformed with a 2μm URA3 genomic (YEp24-based) library (9) using a lithium acetate-based transformation protocol (17) and allowed to recover for 6 h in liquid minimal medium lacking uracil, selecting only for uracil prototrophy provided by the plasmid. Transformants were subsequently plated on minimal medium lacking uracil and containing Geneticin (1 mg/ml) to select for transformants with elevated expression of ERG2-KanMX. From an estimated 100,000 initial transformants, 70 Geneticin-resistant colonies were selected, and 41 of these isolates also showed increased expression of ERG2-lacZ. Thirteen plasmids were recovered that conferred increased expression of ERG2-lacZ upon retransformation. Eleven of these contained overlapping genomic fragments that included the gene ECM22. A 2μm plasmid with a 3.6-kb PCR fragment containing only the ECM22 gene (pJR2330) was sufficient to increase the expression of ERG2.
RNA blots.
Cultures for RNA preparation were grown in minimal medium containing all necessary supplements. Cultures grown under inducing conditions contained 40 μg of lovastatin/ml. All cultures were inoculated from a fresh stationary-phase culture (grown overnight) and grown for 16 h to an optical density at 600 nm (OD600) of 0.2 to 1.0. Then, 50 ml of these cultures was harvested by centrifugation, and the cell pellet was frozen at −80°C. Total RNA purification and RNA blot hybridization were performed as previously described (7) with 10 μg of total RNA used in all lanes.
β-Galactosidase assays.
β-Galactosidase liquid assays and filter lifts were performed as described previously (2, 33).
Mobility shift assays.
Mobility shift assays were performed essentially as described previously (4). The wild-type probe used was a 60-bp double-stranded DNA made by annealing av401 (5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAACTACCGGTGCTATCGTTCTCGTTTGGA-3′) to av402 (5′-AGGATCCAAACGAGAACGA TAGCACCGG TAG T T T TCC T TGCGGC T TATACGAGGGCCGTG-3′). The 4-bp overhangs (indicated by underscores) were then filled in by using Klenow fragment (New England Biolabs) and radiolabeled dCTP (NEN). The probe was purified on a 3% agarose gel, recovered on a DEAE membrane (NA45; Schleicher & Schuell), eluted in DEAE elution buffer (10 mM Tris-Cl [pH 7.9], 1 mM EDTA [pH 8.0], 1 M NaCl), ethanol precipitated, and resuspended in Tris-EDTA (pH 7.6). The extent of labeling was determined by scintillation counting. A total of 20,000 cpm were used in each reaction, which corresponded to ca. 3 ng of probe. The probe with the 10-bp SRE mutation was made in the same way by using oligonucleotides av403 (5′-CGACCA C GG CCACG ATATC TAC C G CA AG G AAAAC T AC C GG TGC TAT CG T TCTCGTTTGGA-3′) and av404 (5′-AGGATCCAAACGAGAACGATAGCACCGGTAGTTTTCCTTGCGGTAGATATCGTGGCCGTG-3′). Specific competitors were made by annealing two complementary oligonucleotides (without single-strand overhangs). The sequences of the oligonucleotides used were 5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAA-3′ and 5′-TTTTCCTTGCGGCTTATACGAGGGCCGTGGTCG-3′, except for the point mutations shown in the figure. Sheared salmon sperm DNA was used as nonspecific competitor in all experiments.
Bacterial extracts were made as follows. A 1-ml quantity overnight in LB Amp of the bacterial strain ER2508 expressing either glutathione S-transferase (GST) alone (pHB2), the GST-Ecm22p1–497 fusion protein (pJR2322), or Upc2p1–395 (pJR2323) or carrying the plasmid pBAT4 (23) was diluted into 100 ml of LB Amp culture and grown at 37°C to an OD595 of 0.5. Expression of the various fusion proteins was then induced by adding 100 μl of 4 M IPTG (isopropyl-β-d-thiogalactopyranoside) and growing the culture for an additional 4 h. Cells were harvested by centrifugation at 3,000 × g for 5 min, and the cell pellet was frozen at −80°C. A whole-cell bacterial extract was made by adding 1 ml of buffer (2 mM Tris-Cl [pH 7.5], 10% glycerol, 2 mM MgCl2, 10 μM Zn2SO4, 125 μg of sheared salmon sperm DNA/ml) to the frozen pellet. Lysis was achieved by six 15-s sonications, with a 30-s rest on ice between each sonication. The extract was cleared by centrifugation in an Eppendorf centrifuge for 15 min at 2,000 × g. Protein concentrations were determined by Bio-Rad protein assays (Bio-Rad). We used 25 μg of total protein in each reaction. The protein extracts made from strains carrying plasmids pJR2322 and pHB2 differed from those made from strains carrying plasmids pBAT4 and pJR2323 by the addition of 75 mM NaCl.
Transmembrane helix prediction.
Transmembrane helix predictions were obtained using the following programs: PHDhtm (http://www.embl-heidelberg.de /predictprotein/predictprotein.html), TMAP (http://130.237.130.31/tmap/single.html) (24, 25), TMpred (http://www.ch.embnet.org/software/TMPRED_form.html), PSORT (http://psort.nibb.ac.jp/), TMHMM (http://www.cbs.dtu.dk/services /TMHMM-2.0/), and HMMTOP (http://www.enzim.hu/hmmtop/) (32). TMAP and TMpred predicted four transmembrane helices in both proteins. PHDhtm predicted two transmembrane helices in Ecm22p and one in Upc2p, PSORT predicted one transmembrane helix in both proteins, and TMHMM and HMMTOP both predicted no transmembrane helix in either protein.
RESULTS
Overexpression of ECM22 and UPC2 increased expression of ERG2
ERG2, which encodes Δ8-7 sterol isomerase, is one of many genes in the yeast sterol biosynthetic pathway that are transcriptionally regulated in response to changes in sterol levels (6, 13, 30). The proteins responsible for mediating this regulation have not been described. A genetic selection was performed to identify transcription factors responsible for the sterol-mediated regulation of ERG2 expression. Specifically, an overexpression strategy was used to select for genes that, when overexpressed, led to increased expression of ERG2. ERG2 expression was monitored by a pair of integrated reporter genes consisting of ERG2 regulatory sequences fused to the coding regions of KanMX (ERG2-KanMX, pJR2326) and of ERG2 regulatory sequences fused to LacZ (ERG2-lacZ, pJR2325). These reporters allowed detection of increased ERG2 expression by increased resistance to the translational inhibitor Geneticin (conferred by KanMX) and by increased β-galactosidase activity (encoded by lacZ). Yeast transformation is itself mutagenic, and mutations at most steps of ergosterol biosynthesis can lead to elevated reporter expression due to disruption of feedback repression. Because mutations in ergosterol biosynthesis would primarily be recessive, a diploid strain was used to avoid this substantial background.
A diploid strain (JRY7184) homozygous for both ERG2-lacZ and ERG2-KanMX was transformed with a high-copy (2μm, YEp24) genomic library (9) and then plated on minimal medium lacking uracil, selecting for the plasmid-borne URA3 gene. The medium also contained Geneticin to select for transformants with elevated expression of ERG2-KanMX. Eleven plasmids were recovered from this screen that had overlapping genomic fragments containing the gene ECM22. A plasmid with a 3.6-kb fragment containing only the ECM22 gene (pJR2330) was sufficient to increase expression of both ERG2 reporters.
Ecm22p is a member of the Zn[2]-Cys[6] binuclear cluster family of fungal transcription factors (31). The deduced Ecm22p protein sequence is 45% identical to that encoded by UPC2 (Proteome, Inc. [http://www.proteome.com/databases /index.html]), a member of the same family of transcription factors. The sequence similarity between Ecm22p and Upc2p was particularly high in the carboxyl-terminal region (76% identity, 238 of 314), as well as in the amino-terminal region (70% identity, 56 of 80), which contained the presumptive DNA binding domain (77% identity, 30 of 39) (Fig. 1A). No plasmids containing UPC2 were isolated in the initial screen. Nevertheless, overexpression of UPC2 also led to increased expression of an ERG2-lacZ reporter (pJR2316) (data not shown). Thus, overexpression of either ECM22 or UPC2 affected expression of ERG2.
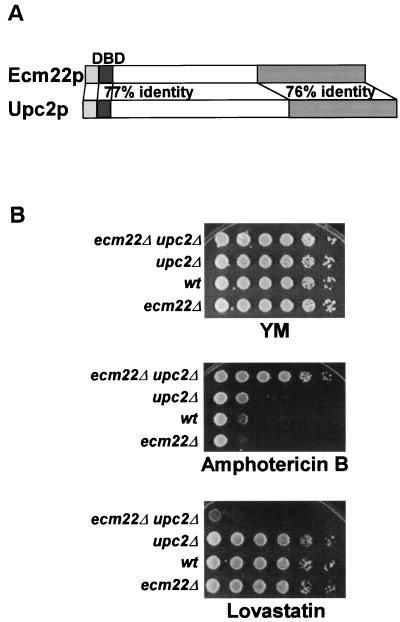
FIG. 1.
(A) Ecm22p and Upc2p show strong sequence similarity. They are 77% identical in the DNA-binding domain (DBD) and 76% identical in the carboxyl-terminal region. (B) ECM22 and UPC2 had overlapping functions. The phenotypes of wild-type (wt; W303-1a), ecm22Δ (JRY7180), upc2Δ (JRY7179), and ecm22Δ upc2Δ (JRY7181) strains were compared by spotting 10-fold serial dilutions of each strain onto minimal medium plates containing 100 ng of amphotericin B or 40 μg of lovastatin/ml.
ECM22 and UPC2 had overlapping functions.
The strong sequence similarity between ECM22 and UPC2 and their shared ability to activate ERG2 expression implied that these genes might have similar or overlapping functions. Evidence of overlapping functions of Ecm22p and Upc2p was also recently reported by Shianna et al. (28). The sensitivity of each of the single null mutants as well as the double null mutant to two sterol-related drugs, lovastatin and amphotericin B, was tested (Fig. 1B). These two inhibitors provide useful measures of the overall expression level of the sterol biosynthetic pathway: reduced flux through the pathway results in lovastatin sensitivity and reduced ergosterol levels in the plasma membrane result in amphotericin B resistance. The ecm22Δ upc2Δ double mutant (JRY7181) was much more resistant to amphotericin B than either of the single mutants (JRY7179 and JRY7180) or the wild-type strain (W303-1a), indicating that the double mutant had reduced levels of ergosterol. The double mutant was also much more sensitive to lovastatin than either of the single mutants or the wild type. Qualitatively, phenotypes such as these might result from a loss of expression of ERG2 alone (as shown below). However, a deletion of the SRE in the ERG2 promoter, to which both Ecm22p and Upc2p bound (described below), did not result in increased resistance to amphotericin B or sensitivity to lovastatin. Furthermore, whereas erg2Δ is viable, a recent report shows that ecm22Δ and upc2Δ are synthetically lethal in some strain backgrounds (28). This result, together with the resistance to amphotericin B and the sensitivity to lovastatin of our ecm22Δ upc2Δ double mutant relative to either single mutant, suggested that the two genes had overlapping functions that extended beyond activation of ERG2, as confirmed below.
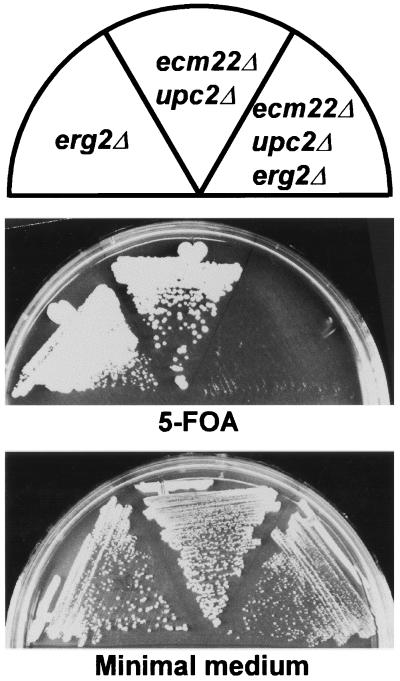
Lovastatin is an inhibitor of sterol biosynthesis and causes a decrease in the production of both sterols and a variety of nonsterol products made by this pathway. The sensitivity of the ecm22Δ upc2Δ double mutant might reflect either the consequences of a lack of sterols or a lack of some other product of the pathway. We distinguished between these two possibilities by inhibiting sterol synthesis after the last branch point of the pathway. A sterol-specific block can be achieved with an erg2Δ null mutation. ERG2 encodes one of several nonessential enzymes in the latter steps of the sterol biosynthetic pathway that modify the basic sterol structure. In the absence of ERG2, cells make a sterol adequate for growth but not the preferred ergosterol. We attempted to generate an erg2Δ ecm22Δ upc2Δ triple mutant by crossing an ecm22Δ upc2Δ mutant (JRY7181) to an erg2Δ ecm22Δ mutant strain (JRY7189). All possible double mutant segregants from this cross (erg2Δ ecm22Δ [2], erg2Δ upc2Δ [2], and ecm22Δ upc2Δ [5]) were viable. However, all five erg2Δ ecm22Δ upc2Δ triple mutant segregants died shortly after spore germination, generating microcolonies of <100 cells (data not shown). The synthetic lethality of the erg2Δ ecm22Δ upc2Δ triple mutant was further evaluated by generating an erg2Δ ecm22Δ upc2Δ triple mutant (JRY7188) kept alive by a URA3-marked plasmid containing a wild-type copy of ECM22 (pJR2331), as evidenced by the inability of the strain to grow on medium containing 5-fluoroorotic acid (5-FOA), which selects against URA3 (Fig. 2). If the lovastatin sensitivity of the ecm22Δ upc2Δ double mutant were due to depletion of a nonsterol product of the pathway, the triple mutant should still be viable but lovastatin sensitive. In contrast, if the lovastatin sensitivity reflected an inability to respond to depletion of late sterol intermediates or ergosterol, the triple mutant should mimic the ecm22Δ upc2Δ double mutant in the presence of lovastatin and be dead. The synthetic lethality of the erg2Δ ecm22Δ upc2Δ triple mutant therefore suggested that the viable ecm22Δ upc2Δ double mutant was deficient in a regulatory response to sterol depletion.
FIG. 2.
erg2Δ was synthetically lethal with ecm22Δ upc2Δ. An erg2Δ (JRY7187) strain, an ecm22Δ upc2Δ (JRY7181) strain, and an ecm22Δ upc2Δ erg2Δ (JRY7188) strain, all carrying the plasmid pJR2331 (ECM22, URA3), were grown on 5-FOA. Only the ecm22Δ upc2Δ erg2Δ strain was unable to grow.
Ecm22p and Upc2p regulated transcription of ERG2 and ERG3 in response to sterol levels.
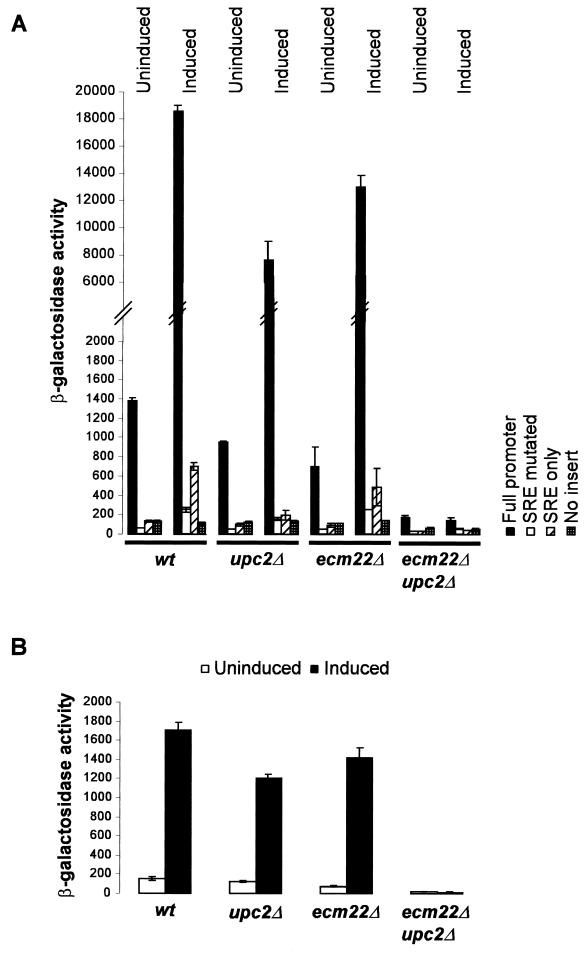
To establish whether or not Ecm22p and Upc2p played a role in sterol-mediated regulation of ERG2 transcription, ERG2 expression levels were measured in an ecm22Δ (JRY7180), an upc2Δ (JRY7179), and an ecm22Δ upc2Δ (JRY7181) mutant grown under inducing and noninducing conditions (Fig. 3A). Inducing conditions differed from noninducing by inclusion of lovastatin in the growth medium. ERG2 expression was measured using a set of ERG2 reporters containing ERG2 regulatory sequences fused to the lacZ coding sequence as described below (see Fig. 5). Either ECM22 or UPC2 was sufficient to confer sterol-mediated regulation to a reporter containing the whole ERG2 promoter. In the absence of both ECM22 and UPC2, no ERG2 induction was detected in response to lovastatin. This regulation was in large part dependent on the presence of an 11-bp SRE (see below) in the ERG2 promoter. Furthermore, Upc2p was capable of activating expression through the 11-bp SRE, indicating that the SRE alone was sufficient for sterol-mediated regulation by Upc2p.
FIG. 3.
(A) UPC2 and ECM22 were necessary for sterol-mediated regulation of ERG2-lacZ. ERG2-lacZ reporters were used to determine the contribution of each of the genes to the regulation of ERG2. “Full promoter” refers to a reporter having ERG2 promoter sequences from −751 to −93 controlling transcription of lacZ. “SRE mutated” has the same promoter except that 10 bp of the SRE have been mutated. “SRE only” has only the 11-bp SRE controlling lacZ. The plasmids were transformed into wild-type (wt; W303-1a), upc2Δ (JRY7179), ecm22Δ (JRY7180), and ecm22Δ upc2Δ (JRY7181) strains. β-Galactosidase assays were performed on the transformants after they were grown for 16 h either in minimal medium (uninduced) or in minimal medium containing 40 μg of lovastatin/ml (induced). The assay values represent the average of two determinations. (B) UPC2 and ECM22 were necessary for sterol-mediated regulation of ERG3-lacZ. An ERG3-lacZ reporter was integrated at the URA3 locus of wild-type (JRY7190), upc2Δ (JRY7191), ecm22Δ (JRY7192), and upc2Δ ecm22Δ (JRY7193) strains. β-Galactosidase assays were performed on the transformants following growth for 16 h in conventional medium (uninduced) or in the presence of 40 μg of lovastatin/ml (induced). The assay values represent the average of two determinations.
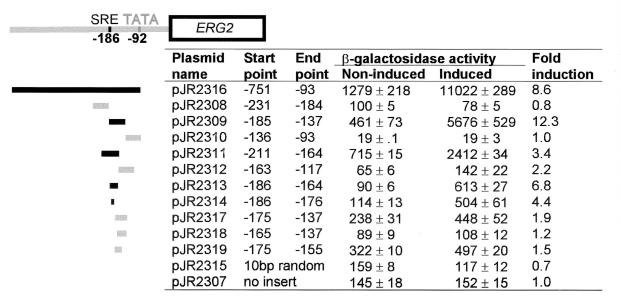
FIG. 5.
An SRE in the ERG2 promoter was sufficient to confer sterol-mediated regulation of expression. Fragments of the ERG2 promoter were subcloned into a CYC1-lacZ reporter. The inserts are represented schematically on the left: black bars indicate inserts that conferred sterol-mediated regulation, and gray bars indicate fragments that did not. pJR2316 had the whole ERG2 promoter from −751 to −93 (numbers refer to base pairs 5′ of the A of the initiation codon). Other sequences inserted were as follows: pJR2308 (5′-GATATCGCACATTCCTGCCCTTACGCTCCAGGGCAGAATCGAACCACGGCCCT-3′), pJR2309 (5′-GATATCGTATAAGCCGCAAGGAAAACTACCGGTGCTATCGTTCTCGTTTGGAT-3′), pJR2310 (5′-GATATCGATTTTCAGTATGGAAGAATTTGGATAGATCTGCAGCGCCATGG-3′), pJR2311 (5′-GATATCTCCAGGGCAGAATCGAACCACGGCCCTCGTATAAGCCGCAAGGAAAA-3′), pJR2312 (5′-GATATCTACCGGTGCTATCGTTCTCGTTTGGATGATTTTCAGTATGGAAGAAT-3′), pJR2313 (5′-GATATCTCGTATAAGCCGCAAGGAAAAC-3′), pJR2314 (5′-GATATCTCGTATAAG-3′), pJR2327 (5′-GATATCGCAAGGAAAACTACCGGTGCTATCGTTCTCGTTTGGAT-3′), pJR2328 (5′-GATATCACTACCGGTGCTATCGTTCTCGTTTGGAT-3′), pJR2329 (5′-GATATCGCAAGGAAAACTACCGGTG-3′), and pJR2315 (5′-GATATCGATACGATT-3′). pJR2307 had no inserted sequences. The resulting plasmids were transformed into the wild-type strain W303-1a. β-Galactosidase assays were performed on the transformants after they were grown for 16 h in conventional minimal medium (uninduced) or in the presence of 40 μg of lovastatin per ml (induced). The assay values represent the average of 2 determinations.
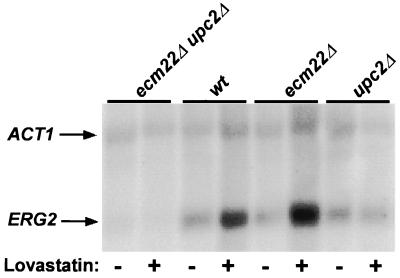
The effect of Ecm22p and Upc2p on sterol-mediated regulation of ERG2 was further investigated by measuring ERG2 mRNA levels in ecm22Δ, upc2Δ, and ecm22Δ upc2Δ mutants (JRY7180, JRY7179, and JRY7181) grown under inducing and noninducing conditions (Fig. 4). Under noninducing conditions, the level of ERG2 mRNA in the upc2Δ mutant was comparable to the level in wild-type cells. Thus, UPC2 was not required for expression of ERG2 per se. In contrast, the upc2Δ strain did not induce ERG2 transcription when grown in medium containing lovastatin, whereas the wild-type strain did. Therefore, UPC2 was necessary for ERG2 induction under low-sterol conditions. The ERG2 mRNA levels in the ecm22Δ mutant were similar to the levels in wild type, under both noninducing and inducing conditions. Thus, ECM22 was not required for uninduced expression of ERG2 and also was not necessary for sterol-mediated regulation of ERG2, at least at the time point assayed. The discrepancy between the mRNA and the β-galactosidase measurements of ERG2 expression in this mutant may simply reflect the higher stability of β-galactosidase (5) compared to ERG2 mRNA. The β-galactosidase measurements integrated most or all of ERG2 induction over the 16-hour induction, whereas the mRNA levels captured a regulatory snapshot at 16 h. Together, these data indicated that either Upc2p or Ecm22p was capable of mediating sterol regulation of ERG2 transcription and suggested that Ecm22p may have a more transient effect on ERG2 transcription in response to sterol limitation. Furthermore, because ERG2 expression under noninducing conditions was reduced in the ecm22Δ upc2Δ double mutant compared to either of the single mutants and wild type, both proteins contributed to the uninduced level of ERG2 transcription.
FIG. 4.
UPC2 was necessary for sterol-mediated regulation of ERG2. ERG2 transcript levels were monitored by RNA blot hybridization. Total RNA was prepared from wild-type (wt; W303-1a), ecm22Δ (JRY7180), upc2Δ (JRY7179), and ecm22Δ upc2Δ (JRY7181) strains grown for 16 h in minimal medium (uninduced) or in the presence of 40 μg of lovastatin/ml (induced). ACT1 served as the loading control.
A similar experiment established that both Ecm22p and Upc2p were involved in the sterol-mediated regulation of the ERG3 gene. As discussed below, ERG3 is one of several sterol biosynthetic genes that had regulatory sequences identical to the ERG2 SRE (Table 2). We used an ERG3 reporter with ERG3 regulatory sequences fused to lacZ (pJR2318) integrated at URA3 (JRY7190, JRY7191, JRY7192, and JRY7193) to study the effect of Ecm22p and Upc2p on ERG3 expression. No induction of ERG3 occurred in response to lovastatin treatment in the ecm22Δ upc2Δ double mutant. Both ECM22 and UPC2 individually were able to confer sterol-mediated regulation of ERG3 transcription (Fig. 3B).
TABLE 2.
Genes with SREs in their promoters
| Gene | Putative SREa | Coordinates
|
|
|---|---|---|---|
| Begin | End | ||
| ERG1 | TTCGTATAGCT | −383 | −373 |
| ERG2 | CTCGTATAAGC | −186 | −176 |
| ERG3 | CTCGTATAAGT | −287 | −277 |
| GTGTATACGAG | −329 | −319 | |
| ERG6 | TTCGTATATGG | −433 | −423 |
| ERG8 | TTCTATACGAG | −155 | −145 |
| ERG11 | TACTATACGAG | −611 | −601 |
| ERG12 | GCCTATACGAA | −675 | −665 |
| ERG13 | TTCGTATATAC | −215 | −205 |
| AATTATACGAG | −101 | −91 | |
| ERG25 | ATCGTATACGG | −464 | −454 |
| TTCGTATACGG | −484 | −474 | |
| GTCGTATAGGA | −716 | −706 | |
| LCB1 | CTTTATACGAG | −444 | −434 |
| LCB2 | TAGTATACGAT | −371 | −361 |
| ATCGTATATCT | −601 | −591 | |
This table shows the location of the putative SREs in the promoters of genes in the sterol biosynthetic pathway, as well as two genes involved in sphingolipid biosynthesis. The seven critical base pairs are shown in boldface. Note that some sequences are of the complementary strand.
An 11-bp SRE in the ERG2 promoter conferred regulation by sterol levels.
Analysis of the ERG2 promoter led to the identification of an SRE. In these experiments, fragments 5′ to the ERG2 coding region were subcloned into a reporter, consisting of the CYC1 promoter directing expression of lacZ (pJR2307). This context allows a regulatory sequence from another gene to be recognized by its ability to activate and/or regulate transcription from the start site provided by the CYC1 promoter. An 11-bp (5′-CTCGTATAAGC-3′) SRE, positioned between bp −186 and −176 relative to the ERG2 ORF, conferred sterol-mediated regulation to the CYC1-lacZ reporter (Fig. 5).
The ERG2 SRE was necessary for induced and uninduced expression but not for basal expression.
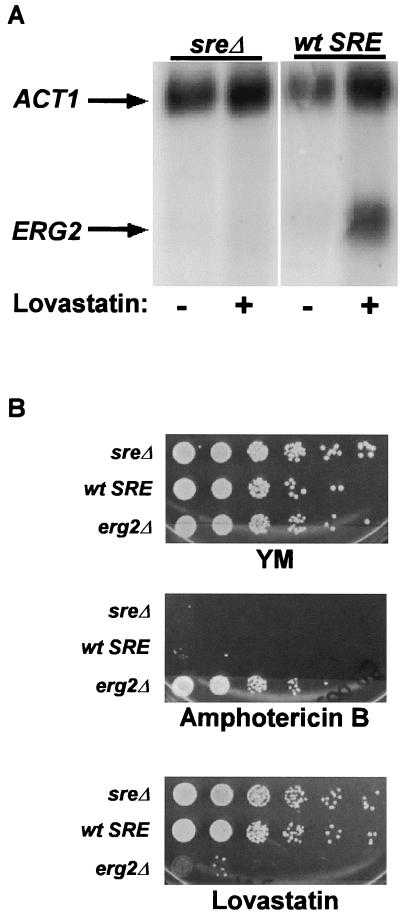
The RNA analysis in Fig. 4 indicated that Ecm22p and Upc2p contributed to the uninduced level of ERG2 expression. To determine whether the SRE was necessary either for basal expression of ERG2 or just for regulation of ERG2 expression, a 10-bp mutation in the SRE in the native ERG2 promoter was constructed. This mutation blocked induction of ERG2 mRNA by lovastatin (Fig. 6A). However, the uninduced level of expression of ERG2 in both mutant and wild-type strains was quite low, raising the possibility that the SRE was required for basal expression, as well as for uninduced and induced expression. Indeed, the lacZ data in Fig. 3A indicated that the SRE contributed to the uninduced level of expression. In the absence of expression, induction would not be reliably measured. To address this possibility, we tested whether the phenotypes of a strain with a deletion of the SRE in the ERG2 promoter (JRY7182) were equivalent to those of a strain with a deletion of the ERG2 gene itself (JRY7187). If the two phenotypes differed, the ERG2 gene must be expressed at some level in the absence of a SRE. The erg2Δ strain was significantly more amphotericin B resistant and lovastatin sensitive than the strain with the SRE deletion (Fig. 6B). Therefore, deletion of the SRE did not eliminate expression of ERG2. Hence, the SRE was a mediator of ERG2 regulation and not of its basal expression. However, the higher levels of expression in the presence of the SRE than in its absence under noninducing conditions (Fig. 3A) implied that a fraction of Ecm22p or Upc2p molecules were active under noninducing conditions.
FIG. 6.
(A) The SRE was necessary for sterol-mediated regulation of ERG2. ERG2 transcript levels were monitored by RNA blot analysis. Total RNA was prepared from strains that had a mutated sreΔ (JRY7182) and wild-type SRE (JRY7183) grown for 16 h in minimal medium (uninduced) or in the presence of 40 μg of lovastatin/ml (induced). ACT1 served as the loading control. (B) The SRE was not necessary for basal expression of ERG2. The phenotypes of strains sreΔ (JRY7182), wild-type (wt) SRE (JRY7183), and erg2Δ (JRY7187) were compared by spotting 10-fold serial dilutions of each strain onto minimal medium plates containing 100 ng of amphotericin B or 40 μg of lovastatin/ml.
Ecm22p and Upc2p bound the ERG2 SRE directly.
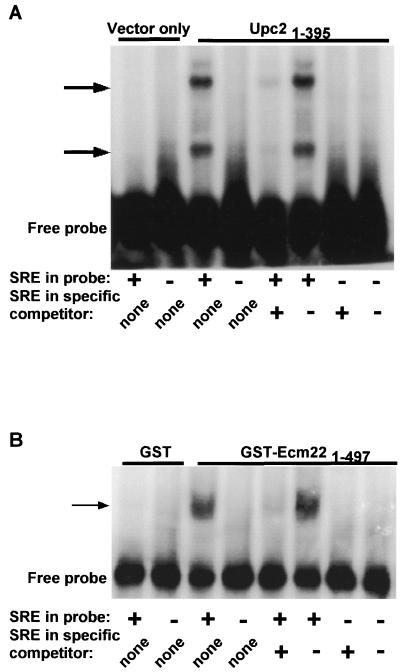
The deduced sequence of Ecm22p and Upc2p indicated that they were both members of the Zn[2]-Cys[6] binuclear cluster family of fungal transcription factors (31), raising the possibility that they were transcriptional activators that bound directly to the SRE. To test this prediction, we developed a mobility shift assay using a radioactively labeled 60-bp probe containing sequences from the ERG2 promoter that included the SRE. A protein extract made from bacteria expressing a truncated version of Upc2p (amino acids 1 through 395) (pJR2323) produced a mobility shift that was dependent on the presence of the SRE (Fig. 7A). The shifted band could be competed away by an excess of an unlabeled double-stranded oligonucleotide that contained the SRE but not by one lacking it. A probe in which the 11 bp in the SRE were replaced by a random sequence was not shifted under the same conditions. Thus, Upc2p bound DNA directly in an SRE-dependent manner, as expected of a transcription factor controlling the expression of ERG2.
FIG. 7.
(A) Upc2p bound directly to the ERG2 promoter in a SRE-dependent manner. Mobility shift assays were performed by using a radiolabeled double-stranded DNA probe containing sequences from the ERG2 promoter overlapping the SRE and 25 μg of bacterial protein extract from a strain expressing a truncated Upc2p1–395 protein (amino acids 1 through 395) (pJR2323) or carrying the vector plasmid pBAT4 (23). Arrows indicate shifted bands. Lanes where the probe contained a wild-type SRE are labeled “+” (5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAAC TACCGG TGC TATCG T TC TCGTTTGGA-3′), and lanes where 10 bp of the SRE were replaced by random sequence (underlined) are labeled “−” (5′-CGACCACGGCCACGATATC TACCGCAAGGAAAAC TACCGGTGC TATCGTTCTCGTTTGGA-3′). “None” indicates no competitor other than salmon sperm DNA was used. Unlabeled double-stranded oligonucleotides (5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAA-3′ is labeled “+”; 5′-CGACCACGGCCACGATATCTACCGCAAGGAAAA-3′ is labeled “−”) were used as specific competitors, where indicated, at a 250-fold molar excess to the radioactively labeled probe. (B) Ecm22p also bound directly to the ERG2 promoter in a SRE-dependent manner. The experiment was performed as for Upc2p, but the protein extract was made from bacteria expressing either GST alone (pHB2) or GST-Ecm22p1–497 (pJR2322).
A similar experiment showed that Ecm22p also bound directly to the ERG2 promoter. A protein extract made from bacteria expressing a truncated version of Ecm22p (amino acids 1 through 497) as a GST fusion protein (pJR2322) produced a mobility shift that was dependent on the presence of the SRE. Hence, Ecm22p, as well as Upc2p, bound DNA directly in a SRE-dependent manner (Fig. 7B).
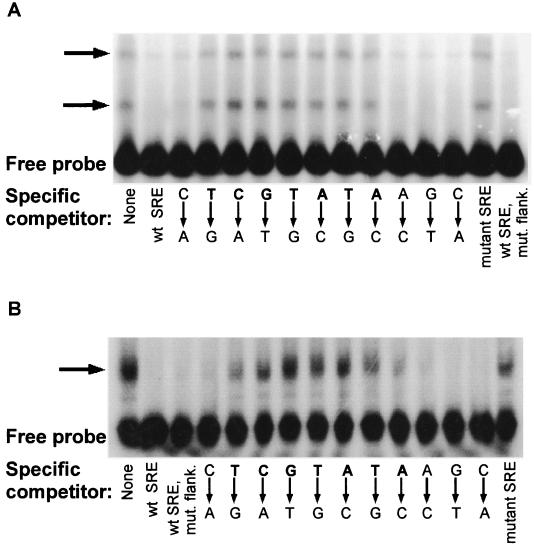
Using this mobility shift assay, 7 of the 11 bp in the SRE were identified to be critical for binding by Upc2p. A series of double-stranded oligonucleotides were constructed with consecutive transversions of each of the 11 bp of the SRE. These unlabeled oligonucleotides were used as specific competitors in mobility shift assays with the radioactively labeled 60-bp probe that contained a wild-type SRE. Only oligonucleotides that could not compete with the labeled probe for Upc2p binding would allow visualization of a shifted band. The critical base pairs in the SRE were the 7-bp sequence TCGTATA (Fig. 8A). In a similar experiment testing the DNA binding specificity of Ecm22p, the same 7-bp sequence was critical for the binding by Ecm22p (Fig. 8B). Therefore, both Upc2 and Ecm22p bound the same 7-bp TCGTATA sequence.
FIG. 8.
(A) The 7-bp sequence TCGTATA was necessary for Upc2p binding to the ERG2 promoter. Mobility shift assays were performed using 25 μg of protein from a bacterial extract expressing a truncated Upc2p1–395 protein (pJR2323) and a radioactively labeled double-stranded DNA probe containing sequences from the ERG2 promoter with a wild-type SRE as described for Fig. 7. Fourteen different specific double-stranded competitors were used. One of the competitors contained a wild-type SRE (wt SRE) and had the sequence 5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAA-3′, one had the entire SRE mutated (5′-CGACCACGGCCACGATATCTACCGCAAGGAAAA-3′, labeled “mutant SRE”), one had a wild-type SRE but all other sequences were mutated (labeled “wt SRE, mut. flank.”), and the rest contained a series of consecutive transversions in each of the base pairs of the SRE. The sequence of these competitors was 5′-CGACCACGGCCCTCGTATAAGCCGCAAGGAAAA-3′ except for the mutations noted in the figure. The competitors were used at a 250-fold excess compared to the probe. (B) Ecm22p bound the same 7-bp sequence (TCGTATA) as did Upc2p. The experiment was performed as for Upc2p except that the protein extract was made from bacteria expressing GST-Ecm22p1–497 (pJR2322).
DISCUSSION
The present study identified an SRE and two SREBPs required for the regulation of the sterol biosynthetic genes ERG2 and ERG3. Upc2p and Ecm22p were necessary for the regulation of ERG2 expression in response to changes in sterol levels, and both bound directly to the ERG2 promoter. Phenotypes of the ecm22Δ upc2Δ double mutant further supported a role for ECM22 and UPC2 in sterol homeostasis.
The SRE in the ERG2 promoter was both necessary and sufficient to confer regulated transcription in response to sterol depletion by lovastatin. Seven base pairs in the ERG2 SRE were necessary for the binding of Upc2p and Ecm22p to DNA. These 7 bp had a perfect match in a 24-bp upstream activating sequence (UAS) found in the ERG3 promoter that is necessary for the regulation of ERG3 in response to sterols (3). ECM22 and UPC2 were also necessary for the sterol regulation of ERG3. Furthermore, the same 7-bp sequence element was found in the promoters of other yeast genes that are regulated by sterols, suggesting that these genes may be coordinately regulated by Upc2p and Ecm22p through their SREs (Table 2).
Sterols and anaerobic growth.
Sterol biosynthesis is dependent on oxygen and can take place only during aerobic growth. Unlike aerobically growing cells, anaerobically growing cells can acquire sterols by taking them up from the growth medium. UPC2 was first identified in a screen for mutants that take up sterols under aerobic conditions (11). The sterol uptake phenotype is due to a semidominant allele containing a point mutation in the carboxyl-terminal part of UPC2. Recently, UPC2 was shown to be necessary for the anaerobic induction of the DAN/TIR genes (1). Due to the oxygen requirement of sterol production, this effect could in principle reflect either a response to oxygen or a response to sterol levels.
Lovastatin inhibits an early step in sterol biosynthesis. Thus, lovastatin treatment will reduce levels of early, nonsterol, products of the pathway in addition to reducing sterol levels. However, the erg2Δ null mutant blocks sterol synthesis specifically, leaving early products of the pathway unaffected. The synthetic lethality of erg2Δ in combination with the ecm22Δ upc2Δ double mutant therefore suggested that a sterol-specific block generated a signal to activate Ecm22p and Upc2p. When this signal failed to upregulate sterol biosynthesis, because the transcription factors responsible for the effect had been deleted, the cells died. This conclusion is supported by the induction of ERG2 expression in an erg24Δ mutant (30). Moreover, ERG3-lacZ expression is induced in an erg2Δ mutant (3). Both of these observations pinpoint a late sterol product as the key product whose level is monitored by Ecm22p and Upc2p. We therefore propose that the principal function of Ecm22p and Upc2p is to regulate genes in response to sterol levels.
Effect of ECM22 and UPC2 on sterol biosynthesis.
Upc2p and Ecm22p activated ERG2 transcription in response to the need for more sterols by binding directly to a 7-bp SRE in the ERG2 promoter. The presence of identical sequence elements in the promoters of several other genes involved in sterol biosynthesis in yeast indicated that Upc2p and Ecm22p may regulate multiple genes in this pathway (Table 2). Sequence elements identical to the ERG2 SRE are within 500 bp 5′ of the coding region of ERG1, ERG2, ERG3, ERG6, ERG8, ERG13, and ERG25, and within the first 1,000 bp 5′ of ERG11 and ERG12. With the exception of ERG13, all of these genes have been shown to be transcriptionally regulated by sterols (3, 6, 13, 18, 30). It therefore seems likely that Upc2p and Ecm22p are involved in the sterol regulation of genes other than ERG2 and ERG3. Hence, Upc2p and Ecm22p may be major transcriptional regulators of sterol-responsive genes in the sterol biosynthetic pathway of yeast.
The SRE motif is found in the promoters of a number of other sterol-regulated genes in yeast. Some of these genes are involved in sphingolipid biosynthesis. Both LCB1 and SUR2 are regulated by sterols (6), and LCB1 contains an SRE within the first 500 bp of the 5′ untranslated region. This raises the possibility that sphingolipid biosynthesis in yeast may be coordinated with sterol biosynthesis, with Upc2p and Ecm22p providing the coordination.
Our data indicated that Upc2p and Ecm22p were transcriptional activators of ERG2 and other genes and that the activities of the proteins were influenced by sterol levels. The glutamine-rich domain in the middle of both Upc2p and Ecm22p was consistent with this hypothesis. However, at least three of the genes with a SRE in the promoter (CYC7, BAP2, and DIP5) are repressed, rather than induced, by the lack of sterols (6). This paradoxical response of SRE-containing genes raises the formal possibility that Upc2p and Ecm22p may also be transcriptional repressors or recruit a repressor under certain conditions. In fact, such a dual effect is seen in the case of the transcriptional repressor Ume6p, another member of the Zn[2]-Cys[6] binuclear cluster family of transcription factors. During mitosis Ume6p recruits the Sin3p-Rpd3p histone deacetylase, resulting in repression of adjacent genes, whereas in meiosis Ume6p recruits the transcriptional activator Ime1p (27), thereby activating transcription. However, if Upc2p and Ecm22p were activators as well as repressors, the nature of their transcriptional effect would have to depend on other DNA-binding proteins in the vicinity of the SRE or upon the precise position of the SRE in the promoter. Alternatively, other proteins in yeast may mediate repression in response to sterol depletion.
Numerous genes shown to be regulated in response to changes in sterol levels (6, 13) lack promoter elements with similarity to the ERG2 SRE. It is possible that Upc2p and Ecm22p could indirectly regulate a broader set of genes than those that have SREs in their promoters by regulating other transcription factors. Alternatively, Upc2p and Ecm22p may bind additional sequence elements different from the SRE identified here. Hap1p, another member of the Zn[2]-Cys[6] binuclear cluster family, can bind two unrelated sequences (15, 26). Moreover, in identifying the 7 bp of the SRE necessary for binding, we tested only one point mutation (a transversion) in each position. In fact, the anaerobic response element (AR1; TCGT TYAG) (10) through which Upc2p induces anaerobically expressed genes (1) differs from the SRE by 2 bp. It is still not known whether or not Upc2p binds directly to AR1. It is therefore possible that further mutations of the binding site may be functional. Hence, it is conceivable that Upc2p and Ecm22p could directly regulate genes lacking a SRE.
Why have two genes for the same function?
Upc2p and Ecm22p had closely overlapping functions, and both proteins were capable of conferring sterol-mediated regulation to ERG2 and ERG3 by binding directly to the same 7-bp regulatory sequence. The major difference between UPC2 and ECM22 in our experiments was that Ecm22p failed to activate transcription from the 11-bp SRE alone, although the effect of Ecm22p on the whole promoter was SRE dependent. Ecm22p binding and/or activation therefore may be dependent on other factors that bound the ERG2 promoter. Based on β-galactosidase assays, the effect of Ecm22p was somewhat weaker than that of Upc2p. Furthermore, Ecm22p seemed to have a more transient effect on ERG2 expression than Upc2p. In a situation where Ecm22p and Upc2p may compete for binding to the same site, different promoters with different relative affinities for Ecm22p and Upc2p may provide a range of sterol-mediated regulatory responses.
Activation of Upc2p and Ecm22p.
The central unanswered question is the mechanism by which sterols influence the activity of Upc2p and Ecm22p. Recent work by Abramova et al. (1) showed that UPC2 transcription, although expressed aerobically, is anaerobically induced, and that the semidominant UPC2-1 mutant, which has a point mutation in the carboxyl-terminal domain, causes constitutive aerobic expression of anaerobically induced genes. Hence, the carboxyl-terminal domain may repress transcriptional activity. Interestingly, the carboxyl-terminal domains of both Ecm22p and Upc2p were predicted to have one or more transmembrane helices, making it a possibility that Ecm22p and Upc2p directly measure sterol levels in membranes. However, the number of transmembrane helices predicted varied considerably depending upon which program was used to make the prediction.
Nevertheless, if Upc2p and Ecm22p are integral membrane proteins, then Upc2p and Ecm22p, much like the mammalian SREBPs, may be released from the membrane in response to the need for more sterols. Regulated proteolysis as a mechanism for activating a membrane-bound transcription factor has been described in yeast (16). Sterols may regulate proteolytic activation by influencing properties of the ER membrane.
Another possibility is that the carboxyl-terminal domain of Ecm22p and Upc2p is a direct sterol sensor, even though it has no similarity at the protein sequence level to any currently known sterol binding domains. Clearly, determining where the Ecm22p and Upc2p proteins reside in the cells under noninducing conditions and whether they are structurally altered upon induction will be decisive in testing these and other models of activation.
The Zn[2]-Cys[6] binuclear cluster family of transcription factors is fungus specific (31) and, as expected, we have not found any proteins with similarity to the amino-terminal DNA binding region of Upc2p and Ecm22p in any nonfungal organism. Perhaps more surprisingly, we have also found no proteins with homology to the carboxyl-terminal domains of either protein outside of fungi. Therefore, proteins homologous to Ecm22p and Upc2p are probably absent from mammals. Consequently, despite seemingly parallel sterol regulatory circuits in yeast and mammals, the transcription factors involved share no sequence similarity and are clearly not homologous proteins. Understanding the mechanism of activation of Ecm22p and Upc2p may therefore reveal a new dimension to how organisms regulate sterol synthesis.
ACKNOWLEDGMENTS
We thank J. Zupicich for assistance with TMH predictions, J. Gin for technical assistance, and S. Okamura and V. Boyartchuk for comments on the manuscript.
A.V. was supported by a doctoral fellowship from the Norwegian Research Council (grant 70429/410). Further research support was provided by a grant from the National Institutes of Health (GM35827), with core support from an NIEHS Mutagenesis Center grant (ESO1896).
REFERENCES
- 1.Abramova N E, Cohen B D, Sertil O, Kapoor R, Davies K J A, Lowry C V. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics. 2001;157:1169–1177. doi: 10.1093/genetics/157.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A, Gottschling D E, Kaiser C A, Stearns T. Methods in yeast genetics. New York, N.Y: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 3.Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 5.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 6.Bammert G F, Fostel J M. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44:1255–1265. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeck R, Lapeyre B, Brown C E, Sachs A B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M S, Goldstein J L. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- 10.Cohen B D, Sertil O, Abramova N E, Davies K J A, Lowry C V. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 2001;29:799–808. doi: 10.1093/nar/29.3.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley J H, Leak F W, Jr, Shianna K V, Tove S, Parks L W. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1998;180:4177–4183. doi: 10.1128/jb.180.16.4177-4183.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimster-Denk D, Thorsness M K, Rine J. Feedback regulation of 3-hydroxy-3-methylglutaryl coenzyme a reductase in Saccharomyces cerevisiae. Mol Biol Cell. 1994;5:655–665. doi: 10.1091/mbc.5.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dimster-Denk D, Rine J, Phillips J, Scherer S, Cundiff P, DeBord K, Gilliland D, Hickman S, Jarvis A, Tong L, Ashby M. Comprehensive evaluation of isoprenoid biosynthesis regulation in Saccharomyces cerevisiae utilizing the genome reporter matrix. J Lipid Res. 1999;40:850–860. [PubMed] [Google Scholar]
- 14.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 15.Hach A, Hon T, Zhang L. A new class of repression modules is critical for heme regulation of the yeast transcriptional activator Hap1. Mol Cell Biol. 1999;19:4324–4333. doi: 10.1128/mcb.19.6.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppe T, Matuschewski K, Rape M, Sclenker S, Ulrich H D, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 17.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leber R, Zenz R, Schrottner K, Fuchsbichler S, Puhringer B, Turnowsky F. A novel sequence element is involved in the transcriptional regulation of expression of the ERG1 (squalene epoxidase) gene Saccharomyces cerevisiae. Eur J Biochem. 2001;268:914–924. doi: 10.1046/j.1432-1327.2001.01940.x. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Mamie Z L, Leibham D, Cortez D, Elledge S J. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr Biol. 1998;8:1300–1309. doi: 10.1016/s0960-9822(07)00560-x. [DOI] [PubMed] [Google Scholar]
- 20.Lussier M, White A M, Sheraton J, di Paolo T, et al. Large-scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics. 1997;147:435–450. doi: 10.1093/genetics/147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magana M M, Osborne T F. Two tandem binding sites for sterol regulatory element binding proteins are required for sterol regulation of fatty-acid synthase promoter. J Biol Chem. 1996;271:32689–32694. doi: 10.1074/jbc.271.51.32689. [DOI] [PubMed] [Google Scholar]
- 22.Pai J T, Guryev O, Brown M S, Goldstein J L. Differential stimulation of cholesterol and unsaturated fatty acid biosynthesis in cells expressing individual nuclear sterol regulatory element-binding proteins. J Biol Chem. 1998;273:26138–26148. doi: 10.1074/jbc.273.40.26138. [DOI] [PubMed] [Google Scholar]
- 23.Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with a modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
- 24.Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- 25.Persson B, Argos P. Topology prediction of membrane proteins. Prot Sci. 1996;5:363–371. doi: 10.1002/pro.5560050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeifer K, Arcangioli B, Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987;49:9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- 27.Rubin-Bejerano I, Mandel S, Robzyk K, Kassir Y. Induction of meiosis in Saccharomyces cerevisiae depends on conversion of the transcriptional represssor Ume6 to a positive regulator by its regulated association with the transcriptional activator Ime1. Mol Cell Biol. 1996;16:2518–2526. doi: 10.1128/mcb.16.5.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shianna K V, Dotson W D, Tove S, Parks L W. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J Bacteriol. 2001;183:830–834. doi: 10.1128/JB.183.3.830-834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soustre I, Dupuy P H, Silve S, Karst F, Loison G. Sterol metabolism and ERG2 gene regulation in the yeast Saccharomyces cerevisiae. FEBS Lett. 2000;470:102–106. doi: 10.1016/s0014-5793(00)01300-4. [DOI] [PubMed] [Google Scholar]
- 31.Todd R B, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 32.Tusnády G E, Simon I. Principles governing amino acid composition of integral membrane proteins: applications to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 33.Xie K, Lambie E J, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]