Abstract
目的
对登革病毒-2(DENV-2)感染人脐静脉内皮细胞(HUVEC)的差异磷酸化蛋白质进行分析以及初步探究DENV-2感染的可能致病机制。
方法
实验设置DENV-2感染HUVEC组和HUVEC空白对照组,每组3个重复。收集细胞沉淀后,利用SDT裂解法获取蛋白质,通过串联质谱标签(TMT)法对磷酸化蛋白质进行定性与定量分析,对鉴定到的差异磷酸化蛋白质进行氨基酸保守基序分析、亚细胞定位分析、蛋白质结构域分析、GO富集分析、KEGG通路分析和蛋白质互作(PPI)等生物信息学分析。利用Western blot检测JUN、MAP2K2和AKT1磷酸化蛋白的表达。
结果
共检测到1385个差异蛋白质上的2918个修饰肽段,其中有1346个修饰肽段显著上调(FC > 1.2,P < 0.05),1572个修饰肽段显著下调(FC < 0.83,P < 0.05)。氨基酸保守基序分析共获得49个磷酸化保守基序。蛋白质结构域分析中富集到的差异磷酸化肽段最多是RNA识别基序、蛋白激酶结构域和PH结构域。通过亚细胞定位分析发现差异的修饰肽段主要定位在细胞核和细胞质中。GO富集和KEGG通路分析结果显示差异肽段主要在刺激反应的调节、含核碱基的小分子生物合成过程、吞噬体和白细胞跨内皮迁移处富集。对上调和下调的差异磷酸化蛋白分别进行蛋白质互作—KEGG联合分析,结果发现上调和下调的差异磷酸化蛋白质共富集到15条通路,且通过Western Blot检测与自噬途径相关的3个差异磷酸化蛋白质即JUN、MAP2K2和AKT1的表达,结果显示DENV-2组与空白组相比,p-JUN表达显著下调(1.48±0.01 vs 1.23±0.01,P < 0.0001);p-AKT1表达显著上调(0.87±0.02 vs 1.01±0.01,P < 0.001);p-MAP2K2表达显著上调(1.10±0.02 vs 1.29±0.05,P < 0.01)。
结论
DENV-2感染HUVEC存在较多差异表达蛋白,p-JUN的下调、p-MAP2K2和p-AKT1的上调提示3个能够调节自噬的磷酸化蛋白可能与DENV-2感染机制有关。
Keywords: 磷酸化组学, 登革病毒-2, 人脐静脉内皮细胞
Abstract
Objective
To analyze the differentially phosphorylated proteins in DENV-2-infected human umbilical venous endothelial cells (HUVECs) and explore the possible pathogenic mechanism of DENV-2 infection.
Methods
The total proteins were extracted from DENV-2-infected HUVECs and blank control HUVEC using SDT lysis method. The phosphorylated proteins were qualitatively and quantitatively analyzed using tandem mass spectrometry (TMT). The identified differentially phosphorylated proteins were analyzed by bioinformatics analyses such as subcellular localization analysis, GO enrichment analysis, KEGG pathway analysis and protein-protein interaction (PPI) analysis. Western blotting was used to detect the expressions of phosphorylated Jun, map2k2 and AKT1 proteins in DENV-2-infected HUVECs.
Results
A total of 2918 modified peptides on 1385 different proteins were detected, and among them 1346 were significantly upregulated (FC > 1.2, P < 0.05) and 1572 were significantly downregulated (FC < 0.83, P < 0.05). A total of 49 phosphorylated conserved motifs were obtained by amino acid conservative motif analysis. The most abundant differentially phosphorylated peptides in protein domain analysis included RNA recognition motif, protein kinase domain and PH domain. Subcellular localization analysis showed that the differentially modified peptides were mainly localized in the nucleus and cytoplasm. GO enrichment and KEGG pathway analysis showed that the differential peptides were mainly enriched in the regulation of stimulation response, biosynthesis of small molecules containing nuclear bases, and migration of phagosomes and leukocytes across the endothelium. PPI and KEGG joint analysis showed that the up-regulated and down-regulated differentially phosphorylated proteins were enriched in 15 pathways. In DENV-2-infected HUVECs, Western blotting detected differential expressions of phosphorylated proteins related with the autophagy pathway, namely JUN, MAP2K2 and AKT1, and among them p-JUN was significantly down-regulated and p-AKT1 and p-MAP2K2 were significantly upregulated (P < 0.01).
Conclusion
DENV-2 infected HUVECs show numerous differentially expressed proteins. The downregulation of p-JUN and upregulation of p-MAP2K2 and p-AKT1 suggest their potential roles in regulating autophagy, which is probably involved in the mechanism of DENV-2 infection.
Keywords: phosphorylation omics, DENV-2, human umbilical venous endothelial cells
登革病毒(DENV)是黄病毒科的RNA病毒,可编码膜蛋白(prM)、包膜蛋白(E)和衣壳蛋白(C)3种结构蛋白和NS1、NS2A、NS2B、NS3、NS4A、NS4B和NS5七种非结构蛋白[1]。DENV可分为4种血清型,即DENV-1、DENV-2、DENV-3和DENV-4[2],其中DENV-2是最主要的血清型[3]。DENV主要在伊蚊居住的地区快速传播,可引起轻度症状的登革热(DF)或严重的登革出血热(DHF)甚至是危及生命的登革休克综合征(DSS)[4-6]。据估计,有36亿人口居住在DF传播风险的区域,其中我国有将近28个省份受到DF的威胁[7],主要区域为西南和东南沿海地区以及海南省。2014年广东地区DENV感染数是2013年感染的10倍,累计报告DENV感染数高达四千多例,死亡病例6例[8]。儿童和青少年受病毒感染病例呈指数增长[9]。据WHO统计,截止2019年DF被认为是威胁全球健康的十大因素之一。
DENV感染是一个极其复杂的过程,目前认为DENV发病机制与抗体依赖增强作用(ADE)、非结构蛋白1(NS1)病毒抗原、登革病毒基因组变异和亚基因组RNA等相关[10]。但目前对DENV的感染机制尚存在争议,由于感染机制的不明,因此仍然没有有效疫苗可用,虽然已有登革热疫苗正在被研发,但目前并没有疫苗或药物被FDA认可[11-14]。
蛋白质组学在多种疾病和病毒的研究中被广泛运用,而磷酸化修饰是研究最广泛的翻译后修饰,在蛋白质的激活和失活、调节信号转导、粘附和细胞通讯中发挥作用[15]。因此,通过磷酸化TMT蛋白质组学探究DENV-2感染HUVEC过程中磷酸化蛋白质可能的调控机制,对DENV-2的发病机制研究具有重要意义。本研究采用TMT定量磷酸化蛋白质组学,利用相关生物信息学分析手段,为DENV-2的致病机制研究提供了新的见解。
1. 材料和方法
1.1. 细胞
HUVEC、C6/36和DENV-2 NGC株均由实验室常规保存。
1.2. 材料与仪器
ECM培养基(Sciencell);ECGS(Sciencell);FBS(Sciencell);RPMI 1640培养基(Gibco);p-AKT1(Abcam),p-JUN(Abcam),p-MEK2(Abclonal),β-actin(Abclonal)。
数显式稳压稳流电泳仪(上海天能);超净工作台(苏州佳宝净化工程设备有限公司);SDS-Acryl/Bis蛋白电泳仪(上海天能)。
1.3. HUVEC细胞和C6/36细胞的培养
取液氮保存的HUVEC和C6/36,置于37 ℃水浴锅中快速融化,再将细胞分别转移至盛有5 mL ECM完全培养基(10% FBS,1% ECGS,1% P/S)的培养瓶和5 mL 1640完全培养基(10% FBS,1% P/S)的培养瓶中,放入培养箱中培养至细胞贴壁。直到细胞生长至90%融合度时,加入适量0.25%胰蛋白酶(含0.02% EDTA)消化,1000 r/min,10 min收集细胞,重悬后在培养箱中继续培养。
1.4. DENV-2的扩增、鉴定与毒力测定
1.4.1. DENV-2的扩增
待C6/36细胞长成90%左右融合。弃去细胞培养液后加入DENV-2毒液500 μL,置于28 ℃ 5% CO2培养箱中培养1 h,摇晃1次/15 min,去掉毒液后用Hanks液洗涤2次,加入5 mL 2%维持液(2% FBS,1%P/S)置于37 ℃、5% CO2培养箱中培养,每天观察现象,若出现明显细胞病变现象即空泡拉丝,即可收集毒液。收毒:将细胞培养瓶置于-80 ℃中,流水冲淋手摇至沙冰状,重复3次。液体收集于离心管中,2000 r/min,离心10 min,收集上清分装于冻存管中,冻于-80 ℃,然后转移到液氮中保存。
1.4.2. DENV-2的鉴定
根据NCBI提供的DENV2 NSl区核苷酸序列设计引物,利用常规Trizol法提取C6/36总RNA,试剂盒将RNA逆转录为cDNA后经PCR特异性扩增,普通琼脂糖凝胶确定扩增部分序列大小为321 bp。
1.4.3. DENV-2的毒力测定
将C6/36细胞接种到96孔板中,28 ℃,5% CO2培养箱中培养过夜。将病毒液用无血清1640培养基进行梯度稀释(10-3~10-10),共8个浓度,向C6/36细胞中加入适量不同浓度的病毒液,每个浓度设置8个复孔,37 ℃,5% CO2培养箱中孵育2 h,弃上清,PBS洗涤2次,每孔添加200 μL 2%维持液继续培养,每天观察并记录细胞病变,5 d后统计各稀释度病变孔数量,设置空白对照;根据Reed-Muench法计算DENV2对C6/36细胞的毒力。
1.5. DENV-2感染HUVEC
HUVEC细胞弃掉培养基,加入适量DENV-2毒液(109TCID50),放入37 ℃ 5% CO2培养箱中,摇晃1次/30 min,重复4次,最后1次将上清倒出,加入足量的2% 维持液并放回培养箱中。
1.6. 样品质谱分析
SDT裂解法提取蛋白质后进行BCA蛋白质定量,胰蛋白酶酶解后用TMT标记试剂盒对定量的肽段进行标记,随后采用High-SelectTM Fe-NTA Phosphopeptides Enrichment Ki试剂盒富集磷酸化肽段,富集后的肽段采用HPLC液相系统Easy nLC进行分离,经色谱分离后用Q-Exactive系列质谱仪进行质谱分析。
1.7. 生物信息学分析
1.7.1. 蛋白质聚类分析和保守基序分析
对目标蛋白质进行归一化处理后采用层次聚类算法对差异表达磷酸化肽段进行分组归类,并以热图的形式展示。提取包含修饰位点及修饰位点上下游(+/-)6个氨基酸共计13个氨基酸长度的序列信息,利用这些序列信息在MeMe网站软件中预测可能存在的保守基序。
1.7.2. 亚细胞定位和蛋白结构域分析
采用CELLO方法进行亚细胞定位预测,利用多重支持向量机的机器学习的方法对公共数据库亚细胞定位信息已知的蛋白质序列数据建模,预测待检索蛋白亚细胞定位信息。蛋白结构域分析使用Pfam数据库,利用InterProScan软件包,以集成的方式从InterPro数据库运行扫描算法对序列进行功能表征,从而获得目标蛋白序列在Pfam数据库中的结构域注释信息。
1.7.3. GO功能注释和KEGG通路注释
利用Blast2GO对目标蛋白质集合进行基因本体注释,GO功能注释主要分为3类:生物过程(BP),分子功能(MF)和细胞组分(CC)。利用KAAS软件,对目标蛋白质集合进行KEGG通路注释。
1.7.4. 蛋白质相互作用网络分析
基于STRING(http://string-db.org/)数据库中的信息查找目标蛋白质之间的直接和间接相互作用关系,并使用CytoScape软件(版本号:3.2.1)生成相互作用网络并对网络进行分析。
1.8. Western blot
从CO2培养箱中取出细胞,弃掉上清,用预冷PBS清洗2~3次,细胞刮刮脱细胞,收集至离心管中离心,弃掉上清,再加入细胞裂解液,用移液枪反复吹打至蛋白析出,离心后分装上清,保存于-80 ℃冰箱。通过BCA检测蛋白浓度,采用SDS-PAGE分离。孵育抗体p-AKT1、p-JUN、p-MEK2和β-actin过夜,再孵育二抗2 h后,进行显色。
1.9. 统计学分析
采用GraphPad Prism9软件进行统计处理,结果均以均数±标准差表示,采用单因素方差分析,以P < 0.05为差异有统计学意义。
2. 结果
2.1. DENV-2感染HUVEC细胞的磷酸化蛋白质组学分析
将采集到的数据进行鉴定与定量结果分析,磷酸化蛋白质组鉴定到2477个修饰蛋白上的5745个可定量的磷酸化肽段和6730个可定量的磷酸化位点(表 1)。在磷酸化的氨基酸中,丝氨酸、苏氨酸和酪氨酸是影响蛋白质磷酸化功能的主要氨基酸。对磷酸化位点进行统计时,发现在丝氨酸处发生磷酸化的比列为89.3%,苏氨酸处的为10.5%,酪氨酸处的为0.21%(图 1A)。在2477个磷酸化蛋白中,有两个磷酸化位点及以上的蛋白有48.53%,其中SMRR2蛋白质上含有高达366个修饰位点(图 1B)。每100个氨基酸修饰位点的平均分布为0.47(图 1C)。
1.
鉴定与定量结果统计表
Statistics of identification and quantitative results
| Phospho sites | Phospho petides | Phospho proteins | |||||
| Identified | Quantified | Identified | Quantified | Identified | Quantified | ||
| 6793 | 6730 | 5794 | 5745 | 2493 | 2477 | ||
1.
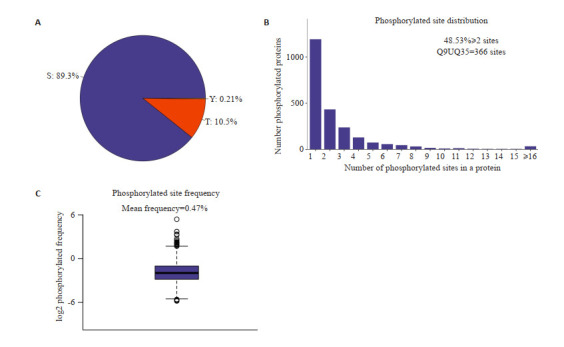
修饰位点分布分析图
Distribution analysis of the modification sites. A: S/T/Y phosphorylation modification site distribution ratio. B: Number distribution of phosphorylation modification sites. C: Distribution frequency map of phosphorylation modification sites. Among all the identified modified proteins, the average number of phosphorylation modification sites per 100 amino acids is 0.47.
利用Corrplot包和Pheatmap包对数据进行相关性分析和聚类分析。结果显示DENV感染组与正常组相比,两组之间的数据模式相似性较低,而组内的数据模式相似性较高,因此可以有效区分组别,且能说明差异表达磷酸化肽段筛选能够代表生物学处理对样本的影响(图 2A、B)。为了分析不同组间具有表达差异的磷酸化蛋白,对鉴定到的2477个磷酸化蛋白质进一步进行差异筛选,1385个显著差异的磷酸化蛋白上检测出2918个差异显著的修饰肽段(P < 0.05),其中有1346显著个上调(FC>1.2且P < 0.05),1572个显著下调(FC < 0.83且P < 0.05)(图 2C)。同时为了直观的比较组间磷酸化修饰肽段的显著性差异,以表达差异倍数和P (t-test)两个因素为标准绘制火山图(图 2D)。
2.
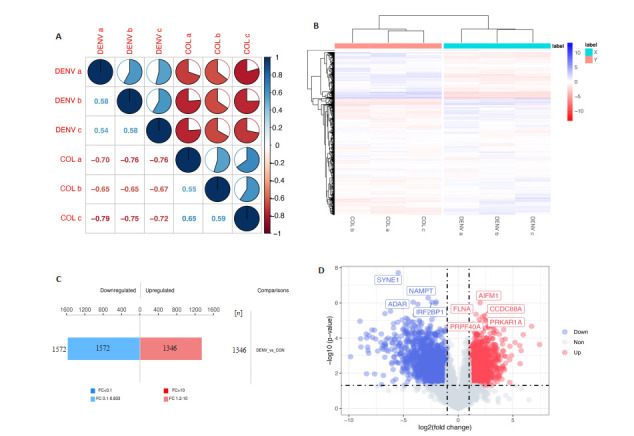
磷酸化组学差异蛋白数量分析
Quantitative analysis of differentially phosphorylated proteins. A: Correlation coefficient diagram. B: Cluster analysis of differentially expressed phosphorylated peptides. C: Histogram of quantitative difference results of phosphorylated peptides. D: Volcano map.
2.2. 氨基酸保守基序分析
利用MEME软件进行Motif分析,结果显示,共得到49个磷酸化保守基序,包括43个pSer基序和6个pThr基序。[sPExxK]、[RxxsPE]和[sPRs]基序在磷酸化肽段富集最为显著(图 4A)。在43个pSer基序中[x_S_Px]、[xG_S_x]和[Rxx_S_Px]基序占磷酸化修饰肽段比列最高,分别为770、240和223个(图 4B)。DENV-2感染组和正常组相比,上调的差异磷酸化蛋白的磷酸化肽段显著富集的Motif是[x_S_PxxxxK]、[Dxxx_S_Px]和[Lxx_S_Px]。而在下调的磷酸化蛋白肽段中显著富集的是[x_T_Px]、[xL_S_Px] 和[xQx_S_Px](图 3C)。
4.
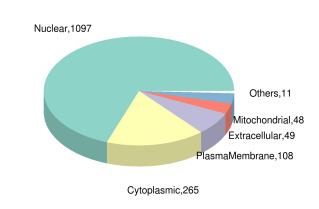
差异表达修饰肽段所属蛋白亚细胞定位饼图
Pie chart of subcellular localization of proteins to which the differentially expressed modified peptides belong.
3.
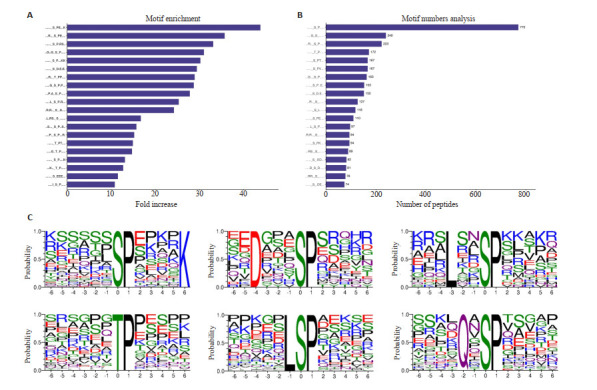
保守基序分析
Conserved motif analysis. A: Predicted conservative motif enrichment statistics (top 20). B: Prediction of the number of phosphorylated modified peptides corresponding to motif (top 20). C: Up-regulated and down-regulated phosphorylated peptides are significantly enriched in the top 3 motifs.
2.3. 亚细胞定位分析
利用亚细胞结构预测软件CELLO对所有差异表达的磷酸化蛋白质进行亚细胞定位分析,并以饼状图形式展示各亚细胞器中的差异表达修饰肽段所属蛋白质数目(图 4)。1097、265、108、49和48个差异表达的磷酸化修饰肽段分别位于细胞核、细胞质、质膜、细胞外和线粒体中,而位于其余部分如内质网、高尔基体、过氧化物酶体的磷酸化肽段有共11个。
2.4. 蛋白质结构域分析
利用结构域预测软件Interproscan对差异表达修饰肽段所属蛋白质进行结构域预测,结果显示,RNA识别基序(又称RRM、RBD或RNP结构域)、蛋白激酶结构域和PH结构域富集到的差异磷酸化肽段最多,分别为48、43、和31(图 5A)。进一步用Fisher精确检验对差异表达修饰所属蛋白质进行结构域富集分析。结果显示,14-3-3蛋白质、PDZ域(也称为DHR或GLGF)、连接组蛋白H1和H5家族、钙调素同源结构域和B盒锌指结构富集度最为显著(图 5B)。
5.
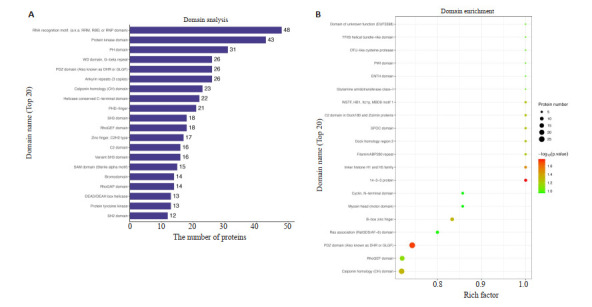
结构域分析
Domain analysis. A: Analysis of protein domains of differentially expressed modified peptides (top 20). B: Domain enrichment analysis diagram.
2.5. 差异表达磷酸化蛋白的GO功能分类
受到调控的差异磷酸化蛋白参与了广泛的细胞生物过程,注释到了13到25个功能组(图 6A)。利用Fisher精确检验对差异表达修饰所属蛋白质进行GO功能富集分析。结果显示在生物过程的范畴中,差异磷酸化蛋白质在刺激反应的调节,含核碱基的小分子生物合成过程,NAD生物合成过程,转运体活性的正调节,烟酰胺核苷酸生物合成过程等重要生物学过程显著富集,在分子功能分类中,蛋白质结合,酶结合,核苷三磷酸酶调节活性,蛋白质结构域特异性结合,钙粘蛋白结合等分子功能显著富集到了差异磷酸化蛋白。在细胞组分分类中,大多数磷酸化蛋白质富集于突触后,不对称突触,突触后密度,突触后特化,神经元间突触等(图 6B、D)。
6.
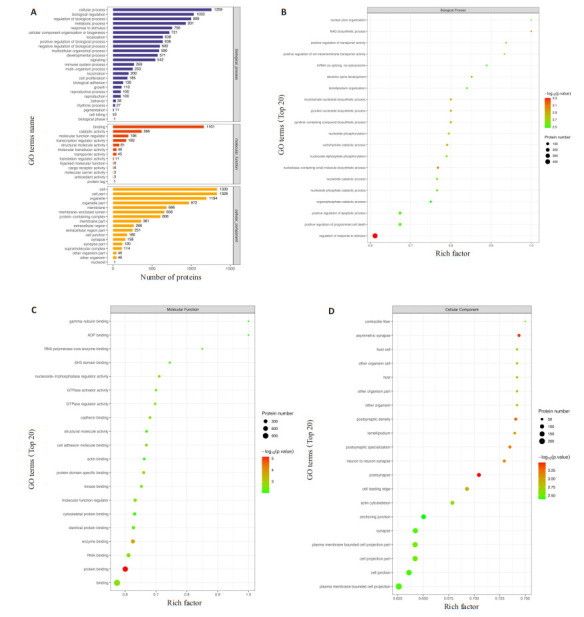
GO功能分析图
GO function analysis diagram. A: GO annotation statistics of proteins to which the differentially expressed modified peptide belongs. B: GO function enrichment bubble diagram (biological process, BP) under biological process classification. C: GO function enrichment bubble diagram (MF) under molecular function classification. D: GO function enrichment bubble diagram (cellular component, CC) under cell component classification.
2.6. 差异表达磷酸化蛋白的KEGG功能分类
结果显示差异磷酸化蛋白质多注释于癌症,RNA转运,内吞,剪接体,肌萎缩侧索硬化等生物合成途径(图 7A)。采用Fisher精确检验对差异表达磷酸化蛋白质进行KEGG通路富集分析。结果表明,差异表达的磷酸化蛋白质在辅助因子生物合成、利什曼病、吞噬体和白细胞跨内皮迁移等重要通路中发生了显著发化(图 7B)。
7.
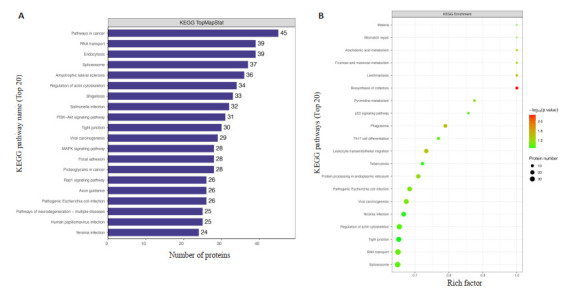
KEGG通路分析图
KEGG pathway analysis. A: KEGG pathway annotation statistics of differentially expressed proteins to which the modified peptides belong (top20). B: KEGG enrichment pathway map of the differentially expressed proteins to which the modified peptide belongs (top20).
2.7. DENV感染后蛋白质相互作用分析
基于STRING数据库,利用Cytoscape_v3.7.2软件,对所有差异表达的磷酸化蛋白构建蛋白质互作网络图(图 8A)。以1346个上调的差异磷酸化蛋白制作PPIKEGG分析网络图,途径富集分析显示10条KEGG通路(P < 0.05,图 8B),分别为剪接体、内吞作用、轴突导向、粘着、肌动蛋白细胞骨架的调节、胰岛素信号通路、细菌侵入上皮细胞、耶尔森菌感染、癌症中的蛋白多糖和肾细胞癌。以1572个下调的磷酸化蛋白制作PPIKEGG分析网络图,结果显示富集到RNA转运、紧密连接、白细胞跨内皮迁移、致病性大肠杆菌感染和志贺菌病五条KEGG通路(P < 0.05,图 8C)。在这些富集通路中,对三个蛋白进行了Western bolt分析,分别是属于活化蛋白1(AP-1)转录因子家族的JUN、MAP激酶家族的MAP2K2和丝氨酸/苏氨酸蛋白激酶之一AKT1。结果显示,在DENV-2感染后,与正常组相比,MAP2K2和AKT1磷酸化水平均显著上调,JUN磷酸化水平显著下调,这与组学结果一致(图 8D)。
8.
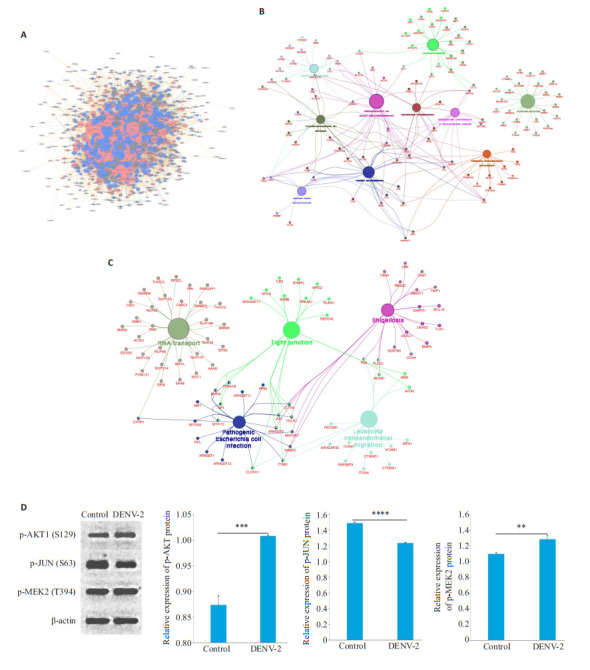
蛋白互作网络分析图及Western blot结果分析统计图
Protein interaction network analysis chart and Western blotting results. A: The interaction network of proteins to which the differentially expressed modified peptides belong. B: Significantly upregulated differentially phosphorylated protein network interaction map. C: Significantly downregulated differentially phosphorylated protein network interaction map. D: Effects of DENV-2 on p-JUN, p-MAP2K2 and p-AKT1 expressions. **P < 0.01, ***P < 0.001, ****P < 0.0001.
3. 讨论
登革病毒属于黄病毒科黄病毒属的单股正链RNA病毒[16]。我国首次经病原学证实的登革热流行发生于1978年的广东省佛山市。我国东南沿海地区、西南地区和海南省为主要流行地区[17, 18]。但由于对其致病机制认识不足,仍然没有公认的药物或疫苗可用[11, 19]。本研究是对DENV-2感染HUVEC后的差异磷酸化蛋白进行分析。磷酸化蛋白质组学共检测到1385个差异显著的磷酸化蛋白质上2918个差异显著的磷酸化修饰肽段,其中1346个肽段显著上调和1572个肽段显著下调。Motif分析结果显示,在上调的磷酸化肽段中,显著富集到[x_S_PxxxxK]、[Dxxx_S_Px]和[Lxx_S_Px]保守基序,下调的磷酸化肽段中[x_T_Px]、[xL_S_Px]和[xQx_S_Px]显著富集,结果提示富集到的Motif在DENV-2感染的过程中可能发挥了重要作用。亚细胞定位分析结果提示大部分的差异磷酸化修饰肽段位于细胞核、细胞质和质膜中,已有研究表明,DENV主要通过网格蛋白介导的内吞作用而进入细胞,病毒膜与囊泡膜融合后允许核衣壳释放入细胞质,从而释放直接用于病毒翻译的RNA[20-22]。病毒的蛋白合成主要发生在内质网中[23],所以我们推测在细胞核、细胞质、内质网等中检测出的差异磷酸化肽段与DENV感染相关。利用Interproscan软件进行结构域分析,结果提示DENV-2感染过程中,可能与14-3-3蛋白质、PDZ域等相关。已有研究表明,14-3-3蛋白与抗病毒相关,14-3-3蛋白家族可以促进MAD5易位到线粒体,从而促进抗病毒[24]。PDZ结构域通常存在于细胞质和膜转接器蛋白中,参与维持细胞间连接、信号转导等多种细胞过程,对病毒感染具有重要意义[25]。在GO富集和KEGG通路分析中发现DENV的感染可能与刺激反应的调节,含核碱基的小分子生物合成过程、辅助因子生物合成、吞噬体和白细胞跨内皮迁移途径密切相关。近年来对DENV感染致病机制的研究处于热门,在Velandia-Romero ML等的研究中表明DENV感染与免疫细胞迁移有关,这与DENV诱发神经系统性疾病密切相关[26]。已有研究证明DENV感染可以调节自噬[27]。自噬能够改变细胞脂质代谢来产生ATP,从而为DENV的复制提供有效的环境,帮助DENV的复制稳固进行。而本课题组前期已经证实DENV-2感染能诱导HUVEC发生自噬[28],但确切感染机制还有待进一步研究。
通过对上调差异蛋白和下调差异磷酸化蛋白进行蛋白质互作网络分析,JUN、MAP2K2和AKT1三个能调控自噬的蛋白引起了我们注意。JUN又称c-JUN,是第1个被称为转录因子的原癌基因[29]。c-JUN是活化蛋白1(AP-1)转录因子家族的关键成员,在细胞增殖、凋亡中发挥作用[30]。MAPK介导细胞内信号,MAP2K2是MAPK信号转导的重要组成部分,能与白介素一起调节细胞成熟、免疫应答[31]。AKT1属于丝氨酸、苏氨酸蛋白激酶,在代谢调节、细胞增值、存活、生长等过程中发挥作用[32-34]。AKT通路在癌症中是最常见的通路之一,癌症中的增值和生存信号主要是通过AKT/MAPK信号传导的。Sun等[35]研究证明,JUN与自噬相关。JUN的下调增加了PI3K/AKT/mTOR信号通路相关蛋白的磷酸化,从而增强自噬的发生。除此之外,JUN还可以激活Beclin-1,上调LC3II,调控自噬[36]。本实验Western bolt结果中,DENV组p-JUN显著下调,提示DENV感染后自噬的发生可能与JUN有关。此外,有研究表明,在饥饿和姜黄素治疗的反应中,RAS-RAFMAP2K/MEK-MAPK/ERK信号通路对自噬起着积极调解作用[37]。Chen等[38]证明,siRNA介导的MAP2K2可以抑制Cory诱导的自噬,并表明MAP2K2在神经元自噬的调节中是必须的。因此,本次实验上调的pMAP2K2调节自噬的发生可能与此途径相关。而AKT是自噬的上游调节因子,AKT通过激活mTORC1来调节自噬。此外,研究表明,ATG3可以通过AKT/mTOR信号通路下调盐霉素诱导的自噬对细胞凋亡的抑制[39]。
本次研究可以说明DENV-2感染HUVEC的机制与调节自噬的JUN、MAP2K2和AKT1具有一定关系,DENV-2感染HUVEC后,能够调节p-MAP2K2、pJUN和p-AKT1的表达,已有研究证明JUN能够调控AKT,而AKT也能通过调控MAPK从而调控癌细胞增值与生长,但JUN、MAP2K2和AKT1三者之间是否存在相互调控作用目前尚不清楚。因此,本课题组将在此研究的基础上,进一步证明JUN、MAP2K2和AKT1三者之间调控自噬的作用以及对DENV致病机制的影响。
Biography
胡盼,硕士研究生,E-mail: hupan1194407713@163.com
Funding Statement
国家自然科学基金(81860289)
Supported by National Natural Science Foundation of China (81860289)
Contributor Information
胡 盼 (Pan HU), Email: hupan1194407713@163.com.
左 丽 (Li ZUO), Email: gzykdxzuoli@163.com.
吴 宁 (Ning WU), Email: wuning@gmc.edu.cn.
References
- 1.Pierson TC, Diamond MS. The continued threat of emerging flaviviruses. Nat Microbiol. 2020;5(6):796–812. doi: 10.1038/s41564-020-0714-0. [Pierson TC, Diamond MS. The continued threat of emerging flaviviruses[J]. Nat Microbiol, 2020, 5(6): 796-812.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waman VP, Kolekar P, Ramtirthkar MR, et al. Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes. PeerJ. 2016;4:e2326. doi: 10.7717/peerj.2326. [Waman VP, Kolekar P, Ramtirthkar MR, et al. Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes[J]. PeerJ, 2016, 4: e2326.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avilés-Vergara PA, Trujillo-Correa A, Gómez-Suárez LA, et al. DENV and ZIKV detection in patients with acute febrile syndrome in Córdoba, Colombia. Int J Infect Dis. 2020;99:458–65. doi: 10.1016/j.ijid.2020.08.008. [Avilés-Vergara PA, Trujillo-Correa A, Gómez-Suárez LA, et al. DENV and ZIKV detection in patients with acute febrile syndrome in Córdoba, Colombia[J]. Int J Infect Dis, 2020, 99: 458-65.] [DOI] [PubMed] [Google Scholar]
- 4.Harapan H, Michie A, et al. Dengue: a minireview. Viruses. 2020;12(8):E829. doi: 10.3390/v12080829. [Harapan H, Michie A, et al. Dengue: a minireview[J]. Viruses, 2020, 12(8): E829.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umakanth M, Suganthan N. Unusual manifestations of dengue fever: a review on expanded dengue syndrome. Cureus. 2020;12(9):e10678. doi: 10.7759/cureus.10678. [Umakanth M, Suganthan N. Unusual manifestations of dengue fever: a review on expanded dengue syndrome[J]. Cureus, 2020, 12(9): e10678.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo HJ, et al. Analyses of clinical and laboratory characteristics of dengue adults at their hospital presentations based on the World Health Organization clinical-phase framework: emphasizing risk of severe dengue in the elderly. J Microbiol Immunol Infect. 2018;51(6):740–8. doi: 10.1016/j.jmii.2016.08.024. [Kuo HJ, et al. Analyses of clinical and laboratory characteristics of dengue adults at their hospital presentations based on the World Health Organization clinical-phase framework: emphasizing risk of severe dengue in the elderly[J]. J Microbiol Immunol Infect, 2018, 51(6): 740-8.] [DOI] [PubMed] [Google Scholar]
- 7.刘 起勇. 我国登革热流行新趋势、防控挑战及策略分析. https://www.cnki.com.cn/Article/CJFDTOTAL-ZMSK202001001.htm. 中国媒介生物学及控制杂志. 2020;31(1):1–6. [刘起勇. 我国登革热流行新趋势、防控挑战及策略分析[J]. 中国媒介生物学及控制杂志, 2020, 31(1): 1-6.] [Google Scholar]
- 8.郭 前方, 崔 国辉, 方 丹云, et al. 2014年广东省登革热大流行的病原体来源及分子进化特点. https://www.cnki.com.cn/Article/CJFDTOTAL-ZSYK201701004.htm. 中山大学学报: 医学科学版. 2017;38(1):21–8. [郭前方, 崔国辉, 方丹云, 等. 2014年广东省登革热大流行的病原体来源及分子进化特点[J]. 中山大学学报: 医学科学版, 2017, 38(1): 21-8.] [Google Scholar]
- 9.Villar L, Rojas D, Besada-Lombana S, et al. Epidemiological trends of dengue disease in Colombia (2000-2011): a systematic review. PLOS. 2015;9(3):e0003499. doi: 10.1371/journal.pntd.0003499. [Villar L, Rojas D, Besada-Lombana S, et al. Epidemiological trends of dengue disease in Colombia (2000-2011): a systematic review[J]. PLOS, 2015, 9(3): e0003499.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatt P, Sabeena SP, Varma M, et al. Current understanding of the pathogenesis of dengue virus infection. Curr Microbiol. 2021;78(1):17–32. doi: 10.1007/s00284-020-02284-w. [Bhatt P, Sabeena SP, Varma M, et al. Current understanding of the pathogenesis of dengue virus infection[J]. Curr Microbiol, 2021, 78 (1): 17-32.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nanaware N, Banerjee A, Mullick Bagchi S, et al. Dengue virus infection: a tale of viral exploitations and host responses. Viruses. 2021;13(10):1967. doi: 10.3390/v13101967. [Nanaware N, Banerjee A, Mullick Bagchi S, et al. Dengue virus infection: a tale of viral exploitations and host responses[J]. Viruses, 2021, 13(10): 1967.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Liu JY, Cheng G. Vaccines and immunization strategies for dengue prevention. Emerg Microbes Infect. 2016;5(7):e77. doi: 10.1038/emi.2016.74. [Liu Y, Liu JY, Cheng G. Vaccines and immunization strategies for dengue prevention[J]. Emerg Microbes Infect, 2016, 5(7): e77.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez DR, Metz SW, Baric RS. Dengue vaccines: the promise and pitfalls of antibody-mediated protection. Cell Host Microbe. 2021;29(1):13–22. doi: 10.1016/j.chom.2020.12.011. [Martinez DR, Metz SW, Baric RS. Dengue vaccines: the promise and pitfalls of antibody-mediated protection[J]. Cell Host Microbe, 2021, 29(1): 13-22.] [DOI] [PubMed] [Google Scholar]
- 14.Nivarthi UK, Swanstrom J, Delacruz MJ, et al. A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans. Nat Commun. 2021;12(1):1102. doi: 10.1038/s41467-021-21384-0. [Nivarthi UK, Swanstrom J, Delacruz MJ, et al. A tetravalent live attenuated dengue virus vaccine stimulates balanced immunity to multiple serotypes in humans[J]. Nat Commun, 2021, 12(1): 1102.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sathe G, Na CH, Renuse S, et al. Phosphotyrosine profiling of human cerebrospinal fluid. Clin Proteomics. 2018;15:29. doi: 10.1186/s12014-018-9205-1. [Sathe G, Na CH, Renuse S, et al. Phosphotyrosine profiling of human cerebrospinal fluid[J]. Clin Proteomics, 2018, 15: 29.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian JN, Yang CC, Chuang CK, et al. A dengue virus type 2 (DENV-2) NS4B-interacting host factor, SERP1, reduces DENV-2 production by suppressing viral RNA replication. Viruses. 2019;11(9):E787. doi: 10.3390/v11090787. [Tian JN, Yang CC, Chuang CK, et al. A dengue virus type 2 (DENV-2) NS4B-interacting host factor, SERP1, reduces DENV-2 production by suppressing viral RNA replication[J]. Viruses, 2019, 11(9): E787.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank C, Höhle M, Stark K, et al. More reasons to dread rain on vacation? Dengue fever in 42 German and United Kingdom Madeira tourists during autumn 2012. Euro Surveill. 2013;18(14):20446. doi: 10.2807/1560-7917.es2013.18.14.20446. [Frank C, Höhle M, Stark K, et al. More reasons to dread rain on vacation? Dengue fever in 42 German and United Kingdom Madeira tourists during autumn 2012[J]. Euro Surveill, 2013, 18(14): 20446.] [DOI] [PubMed] [Google Scholar]
- 18.王 永亮, 钱 成, 左 锋. 中国内陆口岸登革热流行势态及风险评述. 口岸卫生控制. 2018;23(5):17–22. doi: 10.3969/j.issn.1008-5777.2018.05.004. [王永亮, 钱成, 左锋. 中国内陆口岸登革热流行势态及风险评述[J]. 口岸卫生控制, 2018, 23(5): 17-22.] [DOI] [Google Scholar]
- 19.侯 凌欣, 鞠 翰, 展 鹏, et al. 抗登革病毒药物化学研究新进展. https://www.cnki.com.cn/Article/CJFDTOTAL-YXXB202004012.htm. 药学学报. 2020;55(4):669–78. [侯凌欣, 鞠翰, 展鹏, 等. 抗登革病毒药物化学研究新进展[J]. 药学学报, 2020, 55(4): 669-78.] [Google Scholar]
- 20.Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis. J Gen Virol. 2008;89(pt 2):474–84. doi: 10.1099/vir.0.83357-0. [Acosta EG, Castilla V, Damonte EB. Functional entry of dengue virus into Aedes albopictus mosquito cells is dependent on clathrin-mediated endocytosis[J]. J Gen Virol, 2008, 89(pt 2): 474-84.] [DOI] [PubMed] [Google Scholar]
- 21.van der Schaar HM, Rust MJ, Chen C, et al. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells. PLoS Pathog. 2008;4(12):e1000244. doi: 10.1371/journal.ppat.1000244. [van der Schaar HM, Rust MJ, Chen C, et al. Dissecting the cell entry pathway of dengue virus by single-particle tracking in living cells[J]. PLoS Pathog, 2008, 4(12): e1000244.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nayak V, Dessau M, Kucera K, et al. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol. 2009;83(9):4338–44. doi: 10.1128/JVI.02574-08. [Nayak V, Dessau M, Kucera K, et al. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion[J]. J Virol, 2009, 83(9): 4338-44.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie LK, Hoenen A, Morgan G, et al. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J Virol. 2010;84(20):10438–47. doi: 10.1128/JVI.00986-10. [Gillespie LK, Hoenen A, Morgan G, et al. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex[J]. J Virol, 2010, 84(20): 10438-47.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JP, Fan YK, Liu HM. The 14-3-3η chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution. PLoS Pathog. 2019;15(2):e1007582. doi: 10.1371/journal.ppat.1007582. [Lin JP, Fan YK, Liu HM. The 14-3-3η chaperone protein promotes antiviral innate immunity via facilitating MDA5 oligomerization and intracellular redistribution[J]. PLoS Pathog, 2019, 15(2): e1007582.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, et al. Viral PDZ binding motifs influence cell behavior through the interaction with cellular proteins containing PDZ domains. Methods Mol Biol. 2021;2256:217–36. doi: 10.1007/978-1-0716-1166-1_13. [Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, et al. Viral PDZ binding motifs influence cell behavior through the interaction with cellular proteins containing PDZ domains[J]. Methods Mol Biol, 2021, 2256: 217-36.] [DOI] [PubMed] [Google Scholar]
- 26.Velandia-Romero ML, Calderón-Peláez MA, Castellanos JE. In vitro infection with dengue virus induces changes in the structure and function of the mouse brain endothelium. PLoS One. 2016;11(6):e0157786. doi: 10.1371/journal.pone.0157786. [Velandia-Romero ML, Calderón-Peláez MA, Castellanos JE. In vitro infection with dengue virus induces changes in the structure and function of the mouse brain endothelium[J]. PLoS One, 2016, 11(6): e0157786.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YR, Lei HY, Liu MT, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008;374(2):240–8. doi: 10.1016/j.virol.2008.02.016. [Lee YR, Lei HY, Liu MT, et al. Autophagic machinery activated by dengue virus enhances virus replication[J]. Virology, 2008, 374(2): 240-8.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.毛 佳璇, 左 丽, 孔 维莹, et al. DENV-2诱导HUVECs自噬和影响细胞活力的研究. 中国免疫学杂志. 2018;34(8):1131–6. doi: 10.3969/j.issn.1000-484X.2018.08.002. [毛佳璇, 左丽, 孔维莹, 等. DENV-2诱导HUVECs自噬和影响细胞活力的研究[J]. 中国免疫学杂志, 2018, 34(8): 1131-6.] [DOI] [Google Scholar]
- 29.Halazonetis TD, Georgopoulos K, Greenberg ME, et al. C-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55(5):917–24. doi: 10.1016/0092-8674(88)90147-X. [Halazonetis TD, Georgopoulos K, Greenberg ME, et al. C-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities[J]. Cell, 1988, 55(5): 917-24.] [DOI] [PubMed] [Google Scholar]
- 30.Johnson R, Spiegelman B, Hanahan D, et al. Cellular transformation and malignancy induced by ras require c-Jun. Mol Cell Biol. 1996;16(8):4504–11. doi: 10.1128/MCB.16.8.4504. [Johnson R, Spiegelman B, Hanahan D, et al. Cellular transformation and malignancy induced by ras require c-Jun[J]. Mol Cell Biol, 1996, 16(8): 4504-11.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X, Cao X, Zhang X, et al. MEK1/2 inhibitors induce interleukin-5 expression in mouse macrophages and lymphocytes. Biochem Biophys Res Commun. 2016;473(4):939–46. doi: 10.1016/j.bbrc.2016.03.156. [Li X, Cao X, Zhang X, et al. MEK1/2 inhibitors induce interleukin-5 expression in mouse macrophages and lymphocytes[J]. Biochem Biophys Res Commun, 2016, 473(4): 939-46.] [DOI] [PubMed] [Google Scholar]
- 32.Heron-Milhavet L, Khouya N, Fernandez A, et al. Akt1 and Akt2: differentiating the aktion. Histol Histopathol. 2011;26(5):651–62. doi: 10.14670/HH-26.651. [Heron-Milhavet L, Khouya N, Fernandez A, et al. Akt1 and Akt2: differentiating the aktion[J]. Histol Histopathol, 2011, 26(5): 651-62.] [DOI] [PubMed] [Google Scholar]
- 33.Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy. Cell Signal. 2002;14(5):381–95. doi: 10.1016/S0898-6568(01)00271-6. [Nicholson KM, Anderson NG. The protein kinase B/Akt signalling pathway in human malignancy[J]. Cell Signal, 2002, 14(5): 381-95.] [DOI] [PubMed] [Google Scholar]
- 34.Rönnstrand L. Signal transduction via the stem cell factor receptor/c-Kit. Cell Mol Life Sci. 2004;61(19/20):2535–48. doi: 10.1007/s00018-004-4189-6. [Rönnstrand L. Signal transduction via the stem cell factor receptor/c-Kit[J]. Cell Mol Life Sci, 2004, 61(19/20): 2535-48.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y, Chen K, Lin G, et al. Silencing c-Jun inhibits autophagy and abrogates radioresistance in nasopharyngeal carcinoma by activating the PI3K/AKT/mTOR pathway. Ann Transl Med. 2021;9(13):1085. doi: 10.21037/atm-21-2563. [Sun Y, Chen K, Lin G, et al. Silencing c-Jun inhibits autophagy and abrogates radioresistance in nasopharyngeal carcinoma by activating the PI3K/AKT/mTOR pathway[J]. Ann Transl Med, 2021, 9(13): 1085.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klein SR, Piya S, Lu Z, et al. C-Jun N-terminal kinases are required for oncolytic adenovirus-mediated autophagy. Oncogene. 2015;34(41):5295–301. doi: 10.1038/onc.2014.452. [Klein SR, Piya S, Lu Z, et al. C-Jun N-terminal kinases are required for oncolytic adenovirus-mediated autophagy[J]. Oncogene, 2015, 34(41): 5295-301.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinojima N, Yokoyama T, Kondo Y, et al. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy. Autophagy. 2007;3(6):635–7. doi: 10.4161/auto.4916. [Shinojima N, Yokoyama T, Kondo Y, et al. Roles of the Akt/mTOR/p70S6K and ERK1/2 signaling pathways in curcumin-induced autophagy[J]. Autophagy, 2007, 3(6): 635-7.] [DOI] [PubMed] [Google Scholar]
- 38.Chen LL, Wang YB, Song JX, et al. Phosphoproteome-based kinase activity profiling reveals the critical role of MAP2K2 and PLK1 in neuronal autophagy. Autophagy. 2017;13(11):1969–80. doi: 10.1080/15548627.2017.1371393. [Chen LL, Wang YB, Song JX, et al. Phosphoproteome-based kinase activity profiling reveals the critical role of MAP2K2 and PLK1 in neuronal autophagy[J]. Autophagy, 2017, 13(11): 1969-80.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Li F, Liu L, et al. Salinomycin-induced autophagy blocks apoptosis via the ATG3/AKT/mTOR signaling axis in PC-3 cells. Life Sci. 2018;207:451–60. doi: 10.1016/j.lfs.2018.06.034. [Zhang Y, Li F, Liu L, et al. Salinomycin-induced autophagy blocks apoptosis via the ATG3/AKT/mTOR signaling axis in PC-3 cells[J]. Life Sci, 2018, 207: 451-60.] [DOI] [PubMed] [Google Scholar]


