Abstract
G protein-coupled receptors (GPCRs), the largest family of transmembrane proteins, regulate a wide array of physiological processes in response to extracellular signals. Although these receptors have proven to be the most successful class of drug targets, their complicated signal transduction pathways (including different effector G proteins and β-arrestins) and mediation by orthosteric ligands often cause difficulties for drug development, such as on- or off-target effects. Interestingly, identification of ligands that engage allosteric binding sites, which are different from classic orthosteric sites, can promote pathway-specific effects in cooperation with orthosteric ligands. Such pharmacological properties of allosteric modulators offer new strategies to design safer GPCR-targeted therapeutics for various diseases. Here, we explore recent structural studies of GPCRs bound to allosteric modulators. Our inspection of all GPCR families reveals recognition mechanisms of allosteric regulation. More importantly, this review highlights the diversity of allosteric sites and presents how allosteric modulators control specific GPCR pathways to provide opportunities for the development of new valuable agents.
Keywords: G protein-coupled receptors (GPCRs), allosteric modulator, structural investigations, GPCR signaling regulation, allosteric drug discovery
Introduction
G protein-coupled receptors (GPCRs), the most successful class of drug targets, regulate almost all physiological responses by sensing diverse external signals, including light, hormones, ions, and proteins (1–3). GPCRs share a typical architecture with seven transmembrane helices and exhibit conformational dynamics under physiological conditions (4–7). Historically, most marketed pharmaceuticals target orthosteric sites on GPCRs, where endogenous signal molecules are bound, to control conformational changes and regulate signal transduction (8). However, the highly conserved property of orthosteric sites among GPCR subtypes and their complicated signaling pathways cause numerous difficulties for the development of specific and safe therapeutics. Unlike classical orthosteric ligands, allosteric modulators bind to a distinct site on the receptor. Upon binding, allosteric modulators can remotely regulate the conformational transition of GPCRs and specifically regulate their signal transduction pathways, offering new strategies for the development of GPCR-targeted drugs (8–10).
Emerging allosteric modulators of GPCRs are chemically diverse (including proteins, peptides, small molecules, ions, and lipids) but can be divided into three categories according to their pharmacological properties on receptor signaling (11, 12): (i) positive allosteric modulators (PAMs) work synergistically with orthosteric agonists to enhance downstream signals; (ii) negative allosteric modulators (NAMs) modulate the affinity of orthosteric ligands to a receptor, ultimately downregulating or blocking orthosteric agonism; and (iii) neutral allosteric modulators do not have a positive or negative regulatory effect on signal transduction of the receptor after binding to the allosteric site. Some allosteric modulators also exhibit intrinsic agonism (known as ago-PAM) or inverse agonist profiles when used alone (13, 14). Notably, such modulator-mediated allostery depends on orthosteric ligands and receptor signaling pathways, and is therefore deemed to be probe-dependent (14–16). For example, the NTSR1 modulator SBI-55 was found a PAM for β-arrestin recruitment but a NAM-agonist at G protein pathway when cooperating with the orthosteric ligand neurotensin, thus allosterically induced biased signaling (17–19).
According to the Allosteric Database (ASD, http://mdl.shsmu.edu.cn/ASD) (20), four allosteric drugs targeting GPCRs (Cinacalcet, Ticagrelor, Ivermectin, and ATx-201) have been approved by the U.S. Food and Drug Administration (FDA), and another 25 are in clinical trials ( Table 1 ). Among agents currently on the market, Cinacalcet and ATx-201 positively modulate the Gq signaling of extracellular Ca2+-sensing receptor (CaSR) and neuropeptide Y receptor type 4 (NPY4R), respectively (21, 26), while Ticagrelor negatively regulates the Gi pathway of P2Y receptor 12 (26). Avacopan was recently approved for antineutrophil cytoplasmic antibody-associated vasculitis (52) but has not been updated in the ASD database. Avacopan is a NAM of C5a anaphylatoxin chemotactic receptor 1 that inhibits both Gi protein and β-arrestin signals, which may confer signal bias (29). With recent breakthroughs in structural biology, more abundant allosteric sites and regulatory mechanisms of GPCRs have been identified, providing a basis for accelerating the development of allosteric drugs. This mini-review summarizes recent structural investigations of allosteric regulation of Class A, Class B, and Class C GPCRs ( Table 2 ) and exemplars of allosteric modulator-bound GPCR structures to provide insight into the allosteric mechanisms of GPCR transduction signaling.
Table 1.
List of allosteric drugs target GPCR in clinical trials.
| Allosteric drugs | Condition | GPCR Target | Data | Action | Signaling | References |
|---|---|---|---|---|---|---|
| Approved | ||||||
| Cinacalcet | Hyperparathyroidism | CasR | April 2002 | PAM | Ca2+ mobilization | (21–25) |
| Ticagrelor | Stroke; Acute coronary syndrome | P2Y12 | July 2011 | NAM | Gi | (26) |
| Ivermectin | Parasitic roundworm infections | GABAB | 1987 | PAM | Unclear | (27) |
| ATx-201 | viral and bacterial infections; Atopic dermatitis; Cancer; Rheumatoid arthritis; |
NPY4 | 2019 | PAM | Gq | (28) |
| Avacopan | ANCA-Associated Vasculitis | C5aR1 | October 2021 | NAM | Gi/β-arrestin2 | (29) |
| Phase III | ||||||
| Vercirnon | Inflammatory bowel disease | CCR9 | September 2017 (completed) |
NAM | Ca2+ mobilization | (30) |
| BMS-986165 | Plaque psoriasis; Psoriatic arthritis; Crohn’s disease; Systemic lupus erythematosus |
mGluR4 | January 2023 (Recruiting) |
Unclear | Unclear | ASD database |
| mavoglurant | Fragile X syndrome | mGluR5 | March 2016 (Terminated) |
NAM | Gq | (31) |
| ADX-48621 | Parkinson’s disease levodopa- induced dyskinesia |
mGluR5 | April 2022 (Recruiting) |
NAM | Gq | (32) |
| Basimglurant | Fragile X syndrome | mGluR5 | December 2022 (Recruiting) | NAM | Gq | (33) |
| Phase II | ||||||
| ADX-10059 | Gastroesophageal reflux; Migraines | mGluR5 | July 16, 2012 (completed) | NAM | Unclear | (34) |
| T-62 | Neuropathic pain; Postherpetic neuralgia (PHN) |
A1AR | June 8, 2012 (Terminated) |
PAM | Unclear | (35) |
| AZD-8529 | Smoking cessation therapy; Schizophrenia | mGluR2 | November 2017 (completed) |
PAM | Gi | (36) |
| ADX-71149 | Epilepsy; Anxiety disorder; Schizophrenia | mGluR2 | January 2023(Recruiting) | PAM | Gi | (37, 38) |
| MK-7622 | Pain; Schizophrenia; Sleep disorder; Dementia, Alzheimer’s type | M1R | September 2018 (terminated) |
PAM | Gq | (39–41) |
| LY-3154207 | Dementia, Parkinson | DRD1 | July 23, 2021(completed) | PAM | Gs | (42, 43) |
| ASP-4345 | Schizophrenia; Cognitive disorders | DRD1 | May 2022 (completed) | PAM | Unclear | (44) |
| PXT-002331 | Parkinson’s disease | mGluR4 | March 2020 (completed) | PAM | Gi | (45) |
| ASP-8302 | Detrusor underactivity (Underactive bladder) | M3R | July 2022 (completed) |
PAM | Gq | (46) |
| Emraclidine | Schizophrenia | M4R | December 2022 (Recruiting) | PAM | Gi | (47) |
| Phase I | ||||||
| HTL-0014242 | Neurological disorders; Psychiatric disorders | mGluR5 | April 2021 (completed) | NAM | Gq | (48) |
| [11C]JNJ-4229193 | Diagnostics | mGluR2 | Unclear | PAM | Gi | (49) |
| JNJ-2463 | Non-alcoholic steatohepatitis; Nephropathy, diabetic; Non-alcoholic fatty liver disease (NAFLD); Fibrosis; Metabolic Diseases | CB1 | Unclear | NAM | Unclear | https://profiles.biocentury.com/products/namacizumab_(jnj-2463_ryi-018) |
| RG-7342 | Schizophrenia | mGluR5 | Unclear | PAM | Unclear | ASD database |
| JNJ-55375515 | Cognitive disorders; Psychosis | mGluR2 | October 2018 (Completed) | NAM | Unclear | ASD database |
| ODM-106 | Essential tremor | GABAB | December 2016 (Completed) | PAM | Unclear | ASD database |
| MK-6884 | Dementia, Alzheimer’s type | M4R | September 2022 (completed) | PAM | Gq | (50) |
| RGH-618 | Anxiety disorder | mGluR5 | Unclear | NAM | Unclear | ASD database |
| TAK-071 | Lewy body dementia; Neurological Disorders; Dementia, Alzheimer’s type | M1R | December 2022 (Active, not recruiting) | PAM | Gq | (51) |
| VU-319 | Pain; Sleep disorder; Dementia, Alzheimer’s type |
M1R | February 2020 (completed) | PAM | Unclear | ASD database |
Table 2.
Recent structures of GPCRs in complex with allosteric modulators.
| Allosteric modulators | GPCR Target | Action | Binding site | Signaling | PDB code | References |
|---|---|---|---|---|---|---|
| Class A | ||||||
| Cmpd-6FA | β2AR | PAM | Within 7TMD near TM2/TM3/TM4 and ICL2 | Gs | 6N48 | (53) |
| Cmpd-15PA | β2AR | NAM | Intracellular ends of TM1, TM2, TM6, TM7, helix 8, and ICL1 | Gs | 5X7D | (54) |
| AS408 | β2AR | NAM | Outside 7TMD near TM3//TM5 | Gs/β-arrestin | 6OBA | (55) |
| AP8 | GPR40 | ago-PAM | Outside 7TMD near TM3/TM4/TM5 and ICL2 | Gq | 5TZY | (56) |
| MK-8666 | GPR40 | Partial agonist | Outside the 7TMD near TM3/TM4 | Gq | 5TZR | (56) |
| TAK-875 | GPR40 | Partial agonist | Outside the 7TMD near TM3/TM4 | Gq | 4PHU | (57) |
| LY3154207 | DRD1 | PAM | Outside 7TMD near TM3/TM4/TM5 and ICL2 | Gs | 7CKZ | (43, 58) |
| ZCZ011 | CB1 | PAM | Outside 7TMD near TM2/TM3/TM4 | Gi | 7FEE;7WV9 | (59) |
| ORG27569 | CB1 | NAM | Outside 7TMD near TM2/TM3/TM4 | Gi | 6KQI | (60) |
| MIPS521 | A1R | PAM | Outside 7TMD near TM6/TM7 | Gi | 7LD3 | (61) |
| LY2116920 | M2R | PAM | Top of extracellular vestibule | GoA/β-arrestin | 4MQT;7T94;7T96 | (62, 63) |
| 2-PCCA | GPR88 | PAM | Outside 7TMD near the cytoplasmic ends of TM5/TM6 | Gi | 7EJK | (64) |
| ML382 | MRGPRX1 | PAM | Within 7TMD near TM1/TM2/TM3/TM6/TM7 | Gq | 8DWG | (65) |
| BPTU | P2Y1R | antagonist | Outside 7TMD near TM1/TM2/TM3 | unclear | 4XNV | (66) |
| AZ3451 | PAR2 | antagonist | Outside 7TMD near TM2/TM3/TM4 | Gq/β-arrestin | 5NDZ | (67) |
| AZ8838 | PAR2 | antagonist | Extracellular vestibule near TM1/TM2/TM3/TM7 | Gq/β-arrestin | 5NDD | (67) |
| CCX168 | C5aR1 | antagonist | Outside 7TMD near TM3/TM4/TM5 | Gi/β-arrestin2 | 6C1R | (68) |
| NDT9513727 | C5aR1 | antagonist | Outside 7TMD near TM3/TM4/TM5 | Gi/β-arrestin2 | 5O9H | (69) |
| CCR2-RA-[R] | CCR2 | antagonist | Intracelluar surface near TM6/TM7/H8 | Gi | 5T1A | (70) |
| vercirnon | CCR9 | antagonist | Intracellular surface near TM6/TM7/H8 | Gi | 5LWE | (71) |
| Class B | ||||||
| PF-06372222 | GLP-1R | NAM | Outside 7TMD near TM5/TM6/TM7 | Gs | 5VEW | (72) |
| NNC0640 | GLP-1R | NAM | Outside 7TMD near TM5/TM6/TM7 | Gs | 5VEX | (72) |
| LSN3160440 | GLP-1R | PAM | Within 7TMD near TM1/TM2 | Gs | 6VCB | (73) |
| compound 2 | GLP-1R | ago-PAM | Outside 7TMD near TM6 | Gs | 7DUR; 7DUQ;7E14 | (74) |
| NNC0640 | GCGR | NAM | Outside 7TMD near TM5/TM6/TM7 | Gs | 5XEZ;5XF7 | (75) |
| MK-0893 | GCGR | NAM | Outside 7TMD near TM5/TM6/TM7 | Gs | 5EE7 | (76) |
| CP-376395 | CRF1R | NAM | Within 7TMD near TM3/TM5/TM7 | Gs | 4K5Y | (77) |
| Class C | ||||||
| GS39783 | GABAB | PAM | Intracellular tips of TM6-mediated dimerization interface | Gi/o | 6UO8 | (78) |
| rac-BHFF | GABAB | PAM | Intracellular tips of TM5-TM6 of GB1 and TM6 of GB2 | Gi/o | 7CA3;7C7Q;7EB2 | (79–81) |
| Evocalcet | CaSR | PAM | Outside 7TMD near TM2/TM5/TM6/TM7 | Gq | 7DD7 | (82, 83) |
| NPS-2143 | CaSR | NAM | Outside 7TMD near TM3/TM5/TM6/TM7 | Gq | 7DD5;7SIN;7M3J | (82–84) |
| R-568 | CaSR | PAM | Outside 7TMD near TM2/TM5/TM6/TM7 | Gq | 7SIL | (84) |
| Cinacalcet | CaSR | PAM | Outside 7TMD near TM2/TM5/TM6/TM7 | Gq | 7M3F | (83) |
| Etelcalcetide | CaSR | PAM | At the LB2 interface(ECD) | Gq | 7M3G | (83) |
| FITM | mGluR1 | NAM | Outside 7TMD near TM2/TM3/TM4/TM5 and ECL2 | Unclear | 4OR2 | (85) |
| JNJ-40411813 | mGluR2 | PAM | Outside 7TMD near TM3/TM5/TM6/TM7 | Gi | 7E9G | (38) |
| NAM563 | mGluR2 | NAM | Outside 7TMD near TM3/TM5/TM6/TM7 | Gi | 7EPE | (86) |
| NAM597 | mGluR2 | NAM | Outside 7TMD near TM3/TM5/TM6/TM7 | Gi | 7EPF | (86) |
| VU6001966 | mGluR2 | NAM | Unclear | Gi | 7MTQ | (87) |
| ADX55164 | mGluR2 | ago-PAM | Outside 7TMD near TM3/TM5/TM6 | Gi | 7MTR | (87) |
| VU0650786 | mGluR3 | NAM | Outside 7TMD near TM3/TM5/TM6/TM7 | Gi | 7WI6 | (88) |
| Mavoglurant | mGluR5 | NAM | Outside 7TMD near TM2/TM3/TM5/TM6/TM7 | Gq | 4OO9 | (89) |
| Fenobam | mGluR5 | NAM | Outside 7TMD near TM2/TM3/TM5/TM6/TM7 | Gi | 6FFH | (90) |
| M-MPEP | mGluR5 | NAM | Outside 7TMD near TM2/TM3/TM5/TM6/TM7 | Gi | 6FFI | (90) |
| MMPIP | mGluR7 | NAM | Unclear | Gi | 7EPC | (86) |
Allosteric modulation mechanism of Class A GPCRs transduction
Class A GPCRs (also called rhodopsin-like GPCRs), including aminergic receptors, lipid receptors, peptide receptors, and other receptors, are the single most fruitful drug target (91–93). The first structure of rhodopsin was determined two decades ago (94), while the first GPCR signaling complex [β2 adrenergic receptor (β2AR)-Gs bound to an agonist] was reported in 2011 (95). Growing numbers of receptors in complex with orthosteric or allosteric ligands are being published, providing opportunities to understand conformational transitions of receptors upon activation and allosteric modulation.
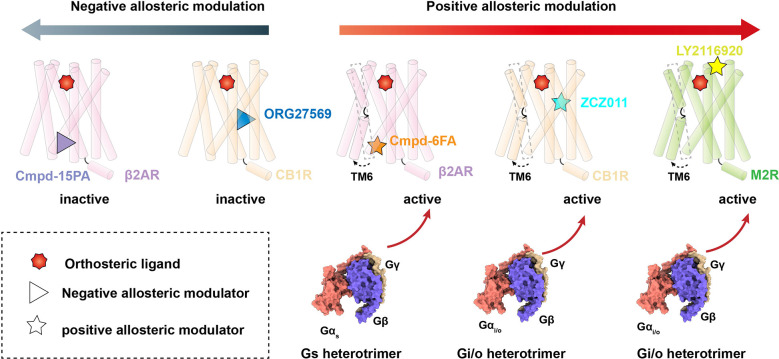
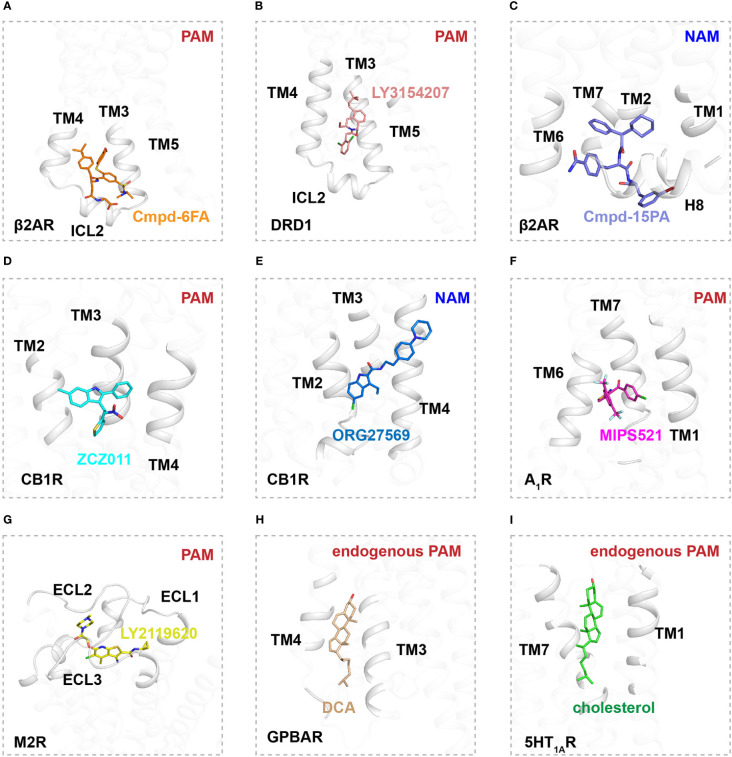
The β2AR is a well-characterized canonical receptor that exhibits dynamic conformational changes in membrane bilayers (96). Ligand Cmpd-6FA, identified as a PAM of β2AR, exhibits robust positive cooperativity with orthosteric agonists to activate Gs signaling. The structure of this receptor complex reveals that the binding pocket of Cmpd-6FA is formed by the intracellular regions of transmembrane helix 2 (TM2), TM3, TM4, and intracellular loop 2 (ICL2) (53)( Figures 1 , 2A ). Upon Cmpd-6FA binding, ICL2 undergoes notable rearrangement from a disordered loop to a helical shape. This stabilization of ICL2 by Cmpd-6FA may explain its allosteric communication with agonists, e.g., enhanced binding affinity of orthosteric agonists ( Figures 1 , 2A ). Similar binding of allosteric modulators to the region above the ICL2 has been reported for other GPCRs, such as GPR40 with ago-PAM AP8(56) and DRD1 with PAM LY3154207 (43, 58) ( Figure 2B ). These findings indicate that certain PAM ligands can stabilize an intermediate receptor state and further activate intracellular effectors. Especially, 2-PCCA is a synthetic ago-allosteric modulator of orphan receptor GPR88, which has two binding sites. One is canonical orthosteric site formed by TM3, TM4, TM5, TM7 and ECL2, another is formed by the cytoplasmic ends of TM5 TM6 and C-terminus of the Gi1 α5 helix. 2-PCCA binding to GPR88 and directly interact with G protein, which stabilizes the active state of the receptor (64). In contrast, Cmpd-15PA, a negative allosteric modulator of β2AR, binds an intracellular allosteric site formed by the intracellular ends of TM1, TM2, TM6, TM7, helix 8, and ICL1 ( Figures 1 , 2C ). The structure of β2AR with Cmpd-15PA reveals that the NAM molecule restricts the inactive conformation of receptor by making direct contacts with residues N692.40, I722.43 and T2746.36, thereby decreasing its binding affinity for the agonist isoproterenol and activation of corresponding signaling (54, 97, 98).
Figure 1.
Allosteric modulation of Class A GPCRs. Negative allosteric modulation of β2AR and CB1R signaling transduction (left) and positive allosteric modulation of β2AR, CB1R and M2R signaling transduction (right).
Figure 2.
Binding sites of allosteric modulators in Class A GPCRs. (A) Interactions of Cmpd-6FA (orange, sticks) with TM2, TM3, TM4, and ICL2 of β2AR (gray, cartoon) (PDB ID: 6N48). (B) Interactions of LY3154207 (pink, sticks) with TM2, TM3, TM4, and ICL2 of DRD1 (gray, cartoon) (PDB ID: 7CKZ). (C) Interactions of Cmpd-15PA (purple, sticks) with TM1, TM2, TM6, TM7, H8 and ICL1 of β2AR (gray, cartoon) (PDB ID: 5X7D). (D) Interactions of ZCZ011 (blue, sticks) with TM2, TM3 and TM4 of CB1R (gray, cartoon) (PDB ID: 7WV9). (E) Interactions of ORG27569 (deep blue, sticks) with TM2, TM3 and TM4 of CB1R (gray, cartoon) (PDB ID: 6KQI). (F) Interactions of MIPS521 (magentas, sticks) with TM6 and TM7 of A1R (gray, cartoon) (PDB ID: 7LD3). (G) Interactions of LY2116920 (yellow, sticks) with extracellular region of M2R (gray, cartoon) (PDB ID: 4MQT). (H) Interactions of DCA (wheat, sticks) with extracellular region of GPBAR (gray, cartoon) (PDB ID: 7CFM). (I) Interactions of cholesterol (green, sticks) with TM1 and TM7 of 5-HT1A (gray, cartoon) (PDB ID: 7E2Y).
Cannabinoid receptor 1 (CB1) is the most abundant GPCR in the central nervous system (CNS), whereby it regulates diverse physiological and pathological processes (99). Plant-derived cannabinoids and synthetic agonists are under clinical trials for treatment of various diseases (100, 101); unfortunately, some have undesirable side effects referred to as “cannabimimetic” effects (102–108). Allosteric modulators undoubtedly release the untapped potential of CB1 by cooperatively or non-cooperatively regulating the efficacy of its signal transduction with orthosteric ligands (109). ZCZ011, a PAM ligand of CB1, was recently reported to bind an extrahelical site in TM2, TM3, and TM4 ( Figures 1 , 2D ). Our molecular dynamics simulations indicate that ZCZ011 could increase the distribution of receptor conformations by promoting rearrangements of TM2 to enhance Gi protein-mediated signaling (59). Distinct from the PAM mechanism of ZCZ011 on CB1, the NAM ORG27569 was found to bind to the lower half of the TM2-TM3-TM4 surface ( Figures 1 , 2E ). Accordingly, the mechanism of allosteric antagonism might involve ORG27569 capturing an intermediate state of CB1 in which toggle-switch residues F3.36–W6.48 at the base of the agonist-binding pocket adopt an inactive conformation, thereby inhibiting Gi-protein activation of CB1 (59).
Allosteric sites in GPCRs are not conserved and exhibit divergent pharmacological properties, providing new therapeutic strategies for a wide array of diseases (13). In particular, MIPS521 was found to act as a PAM of adenosine 1 receptor (A1R) by binding a novel allosteric site formed by TM6 and TM7 ( Figure 2F ). In this interaction, the ligand exerts positive allosteric modulation cooperatively with endogenous adenosine to further stabilize the Gi-protein activation state of A1R (61). In addition to extrahelical and intracellular allosteric sites, allosteric modulators can bind to the extracellular region of receptors. LY2116920, a well-characterized ligand, acts as a PAM of muscarinic acetylcholine receptor M2 ( Figures 1 , 2G ) by binding a new allosteric site formed above the orthosteric pocket to synergistically modulate receptor signal transduction by preventing agonist dissociation from the orthosteric pocket (62, 63). Furthermore, TAK-875 was characterized as an ago-allosteric modulator of GPR40 and is under phase III clinical trials for the treatment of type-2 diabetes, further structural determination reveals that TAK-875 binds to a non-canonical site formed by TM3, TM4, TM5 and ECL2 (57).
Recently, an emerging class of GPCR allosteric modulators, which exert pathway-specific effects on receptor signaling, are defined as biased allosteric modulators (BAMs). For instance, SBI-553 was reported as an arrestin-biased PAM of NTS1R (18, 19). A recent complex structure indicated that SBI-553 is bound to a binding site at the interface between GRK2 and NTSR1, where it can enhance GRK2 binding and phosphorylation of receptor (19). In detail, SBI-553 forms predominately hydrophobic contacts with the intracellular residues from TM2, TM3, TM5, TM6, TM7, and H8 in NTS1R, as well as direct interactions with intracellular effectors (18, 19).
Some endogenous molecules can reportedly behave as allosteric modulators (110, 111). For example, certain bile acid-derivative cholic acids (e.g., deoxycholic acid (DCA), taurocholic acid (TCA), and taurodeoxycholic acid (TDCA) act as PAMs by binding an allosteric pocket formed by TM3, TM4, TM5, and ICL2 in G protein-coupled bile acid receptor (GPBAR) (110) ( Figure 2H ). In addition, certain endogenous lipids in membrane bilayers, such as phospholipid and cholesterol, can regulate signaling transduction by GPCRs (112–116). Cholesterol is an essential component of eukaryotic membranes and plays an important role in GPCR function and pharmacology (112, 117). Approximately 44% of human class A receptors are predicted to have a cholesterol binding site (118, 119). Indeed, high-resolution GPCR structures confirm the presence of lipid-binding sites in GPCRs. For example, a cholesterol molecule was found to bind to the cleft between TM1 and TM7 in the serotonin 1A receptor (5HT1AR), shaping the orthosteric ligand binding pocket by allosteric communication (111, 120) ( Figure 2I ). On the contrary, for β2AR, cholesterol was found to bind the surface of TM1, TM2, TM3 and TM4 and act as a NAM, since the bound cholesterol can increase the affinity for partial inverse agonist timolol and inhibit signaling pathway (118, 119).
Structural basis for Class B GPCR allostery
Class B GPCRs are a small subfamily of 15 receptors, typically with a large N-terminal domain involved in recognition of peptide hormones (77, 121, 122). A prototypical example is glucagon-like peptide-1 receptor (GLP-1R), which predominately couples to the Gs effector and serves as an important drug target for the treatment of type 2 diabetes (123, 124).
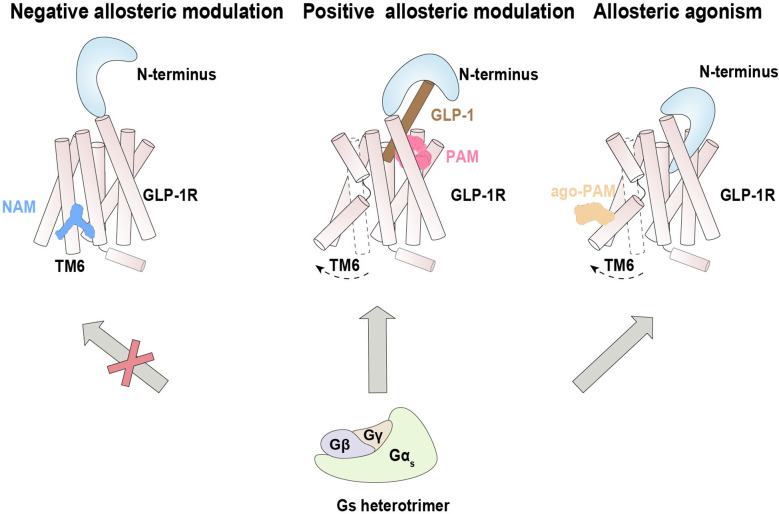
PF-06372222 and NNC0640, two reported NAMs of GLP-1 with distinct scaffolds, bind a common extrahelical binding pocket formed by TM5–TM7 near the intracellular part (72). These NAMs inhibit the Gs protein pathway by restricting the outward movement of TM6 from the inactive state, which is crucial for receptor activation. Coincidentally, previous studies revealed that NNC0640 and MK-0893 (another NAM for the glucagon receptor) also bind in this region, although the allosteric pocket is not fully conserved between these two receptors (75, 76) ( Figure 3 ). These results suggest a common mechanism by which NAMs of Class B GPCRs inhibit Gs protein coupling by preventing the conformational transition of TM6 to an active state, although the allosteric antagonist CP-376395 of CRF1R were identified to bind within the helical bundle of TM3, TM5 and TM7 (77).
Figure 3.
Allosteric modulation of GLP-1R signaling. Negative allosteric modulation of GLP-1R signaling (left), positive allosteric modulation of GLP-1R signaling (middle) and positive allosteric agonism of GLP-1R signaling (right).
LSN3160440 was characterized as a PAM of GLP-1R that enhances both the efficacy and potency of G protein signaling. Structural determination suggests that binding of LSN3160440 within the transmembrane helical bundle near TM1 and TM2 simultaneously interacts with the orthosteric ligand GLP-1(9-36) and GLP-1R (73) ( Figure 3 ). This unique binding mode appears to stabilize the interacting interface between the orthosteric agonist and receptor, thereby enhancing the binding affinity of GLP-1(9-36) and elevating potential receptor activation.
Spatially distinct from LSN3160440, the ago-PAM compound 2 was found to covalently bond to the C3476.36b residue located on the intracellular side of TM6 of GLIP-1R (74). Intriguingly, in a reported structure of compound 2/GLP-1/GLP-1R/Gs, compound 2 seemed to remotely induce insertion of the N-terminal domain into the orthosteric binding pocket, thus triggering activation of G1P-1R underlying its agonistic property ( Figure 3 ). In addition, compound 2 cooperated with diverse orthosteric agonists to positively modulate cAMP signaling (125), enhanced the binding ability of agonists, and strengthened the G protein-receptor interface (74).
Together, the structural discovery of allosteric sites expands our understanding of negative and positive allosteric modulation of downstream signaling of Class B GPCRs.
Structural basis for Class C GPCR allostery
Class C GPCRs mainly include γ-aminobutyric acid B (GABAB) receptors, CaSR, and metabotropic glutamate (mGlu) receptors, which are very important therapeutic targets for the treatment of CNS disorders (126). Class C receptors function in a dimer state (either hetero or homo), each with three domains: Venus flytrap (VFT), a cysteine-rich domain (CRD, except GABAB receptors), and a seven-transmembrane-helices domain (TMD) (127) ( Figure 4 ).
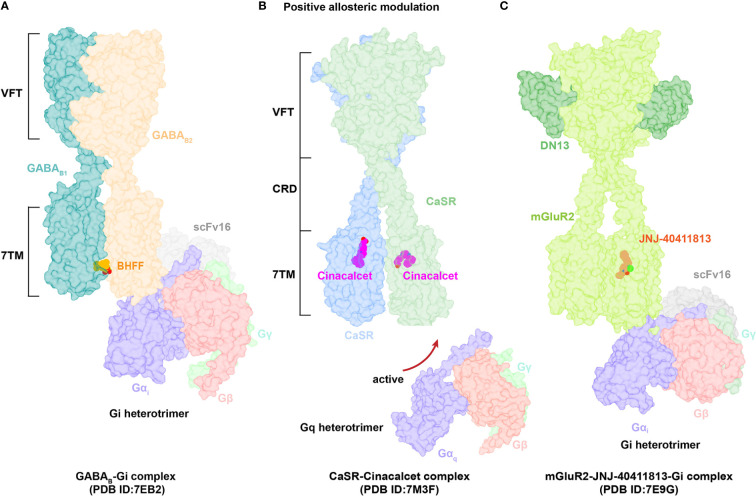
Figure 4.
Structural basis of allosteric regulation in Class C GPCRs. (A) Cryo-EM structural model of active-state GABAB complexed with PAM BHFF (orange). (B) Cryo-EM structural model of active-state CaSR complexed with PAM cinacalcet (magentas). (C) Cryo-EM structural model of active-state mGluR2 complexed with PAM JNJ-40411813 (salmon).
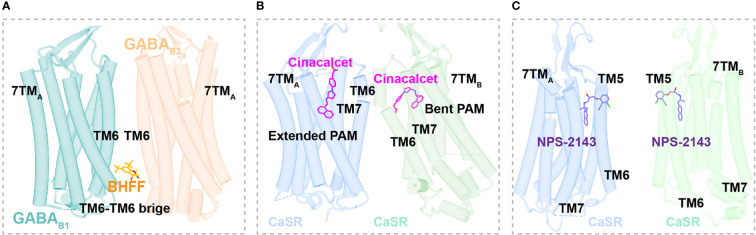
The GABAB heterodimer includes two subunits: GABAB1 (responsible for endogenous ligand binding) and GABAB2 (responsible for Gi/o protein activation) (128). Several compounds reportedly act as PAMs of GABAB receptors, such as CGP7930 (129) (the first characterized PAM of GABAB receptors), R,S-5,7-di-tert-butyl-3-hydroxy-3-trifluoromethyl-3H-benzofuran-2-one (BHFF) (130), and GS39783 (131). Notably, they all exert positive allosteric effects on both ligand binding and the signaling response of GABA (78). A reported structure of the GABAB-Gi signaling complex bound to BHFF and an agonist reveals that the PAM links TM6 from both receptor subunits, forming a TM6-TM6 bridge required for receptor activation (81) ( Figures 4A , 5A ). In contrast, the first identified NAM of the GABAB receptor, CLH304a, can attenuate intracellular signaling (132).
Figure 5.
Binding Sites of allosteric modulators in Class C GPCRs. (A) Close view of binding sites of positive allosteric modulator BHFF (orange, sticks) in GABAB (dark cyan and light salmon, cartoon) (PDB ID: 7EB2). (B) Close view of binding sites of positive allosteric modulator cinacalcet (magentas, sticks) in CaSR (cornflower blue and medium aquamarine, cartoon) (PDB ID: 7M3F). (C) Close view of binding sites of negative allosteric modulator NPS-2143 (medium purple, sticks) in CaSR (cornflower blue and medium aquamarine, cartoon) (PDB ID: 7M3J).
Cinacalcet, the first GPCR allosteric drug approved by the FDA (21, 22), acts as a PAM of CaSR (25). Two other PAMs, evocalcet and etelcalcetide, have been investigated in clinical trials to treat secondary hyperparathyroidism and familial hypocalciuric hypercalcemia type 1 (FHH1) (83). Cinacalcet adopts two different binding conformations that allow it to bend into the seven-transmembrane core of each subunit of CaSR; purportedly, the extended version stabilizes the active state to promote G protein activation ( Figures 4B , 5B ). In addition, L-amino acids can bind to one VFT cleft of CaSR to increase its sensitivity to fast fluctuations of Ca2+ concentrations. Compared with the asymmetric activation of CaSR induced by a PAM, the TMD of CaSR bound to the NAM NPS-2143 is absolutely symmetrical (83) ( Figure 5C ).
mGluRs can be divided into three groups: Group I (mGluR1 and mGluR5) couples to Gq/G11 proteins and activates phospholipase Cβ, resulting in production of inositol 1,4,5-trisphosphate (IP3) and diacylglycerol; Group II (mGluR2 and mGluR3) and Group III (mGluR4, mGluR6–mGluR8) predominantly couple to Gi/o, inhibiting adenylyl cyclase and cAMP production (127, 133). The ligand FTIM was characterized as a NAM of mGluR1; its recognition pocket is constituted by residues from extracellular loop 2, TM2–TM3, and TM5–TM7 (85). Interestingly, VU0424465 (a PAM of mGluR5) could make TMD closer in the absence of an endogenous agonist, further triggering signal transduction (134). Mavoglurant acts as a NAM of mGlu5, which is used to treat fragile X syndrome. In comparison to the position of FITM in mGlu1, mavoglurant is found lower in the mGlu5 allosteric site (89). The ligand JNJ-40411813, a reported PAM of mGluR2, binds to one of the TMD required for Gi protein coupling to potentiate downstream signaling (38) ( Figure 4C ). Upon binding of the NAM VU0650786 to mGluR3, the TMD undergoes structural rearrangements to reduce the distance between TM3 and TM4 helices, subsequently decreasing cAMP inhibition. These findings confirm that VU0650786 stabilizes the inactive state of mGluR3 (88).
Conclusion and perspectives
In this review, we summarized recent structural studies of allosteric regulation of GPCRs. Progress in defining GPCR structures has facilitated understanding of the complex pharmacological features of their allosteric modulation, providing structural clues for ligand optimization and design of novel allosteric therapeutics. Recent studies demonstrate the analgesic efficacy of allosteric modulators of A1R in rats with neuropathic pain (61), whereas SBI-553 (an arrestin-biased allosteric modulator of neurotensin receptor 1) shows efficacy in animal models of psychostimulant abuse (18). Thus, understanding how allosteric modulators bias mechanisms of GPCRs has the potential to improve the precision of treatments for various diseases, while structure-based allosteric agent discovery could accelerate translational studies of GPCR allostery. Nevertheless, some challenges remain to untap the mechanisms of GPCRs, namely: (i) more GPCR structures in complex with allosteric modulators are urgently needed to identify and characterize allosteric sites (e.g., no structure of a GPCR in complex with a biased allosteric modulator has been reported); (ii) divergent cooperative mechanisms of allosteric modulators with orthosteric ligands remain largely elusive; and (iii) recently NMR study of the PAM LY2119620 at the M2R (63) as well as single molecule FRET studies (134–137) that explore conformational changes at the mGlu2 in response to PAMs have begun to provide dynamic structural information for understanding the mechanism of GPCR allostery. Further exploration of the dynamic states of GPCRs in response to different types of ligands using biophysical techniques (e.g., nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR), hydrogen-deuterium exchange (HDX), and time-resolved single molecules) will facilitate additional identification of intermediate receptor states in response to allosteric modulators. In sum, the structural determination of GPCRs in complexes with allosteric modulators will improve our understanding of receptor allostery.
Author contributions
SiS, CZ, CW, and ZS discussed and formulated the focus of the review. SiS, CZ, CW, SuS, and ZL conducted the literature search and drafted the manuscript under the supervision of ZS and WY. ZS and SiS evaluated and revised the manuscript for final submission. All authors contributed to the article and approved the submitted version.
Funding Statement
This work was supported by the National Natural Science Foundation of China (32100988 to WY), Science and Technology Department of Sichuan Province (2021ZYD0080 to WY).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. Park PSH, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: Beyond two-state models and tertiary conformational changes. Annu Rev Pharmacol (2008) 48:107–41. doi: 10.1146/annurev.pharmtox.48.113006.094630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature (2013) 494:185–94. doi: 10.1038/nature11896 [DOI] [PubMed] [Google Scholar]
- 3. Shimada I, Ueda T, Kofuku Y, Eddy MT, Wuthrich K. GPCR drug discovery: integrating solution NMR data with crystal and cryo-EM structures. Nat Rev Drug Discovery (2019) 18:59–82doi: 10.1038/nrd.2018.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leff P. The two-state model of receptor activation. Trends Pharmacol Sci (1995) 16:89–97doi: 10.1016/s0165-6147(00)88989-0 [DOI] [PubMed] [Google Scholar]
- 5. Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, et al. Structural insights into the dynamic process of beta2-adrenergic receptor signaling. Cell (2015) 161:1101–11. doi: 10.1016/j.cell.2015.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weis WI, Kobilka BK. The molecular basis of G protein-coupled receptor activation. Annu Rev Biochem (2018) 87:897–919doi: 10.1146/annurev-biochem-060614-033910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Erlandson SC, McMahon C, Kruse AC. Structural basis for G protein-coupled receptor signaling. Annu Rev Biophys (2018) 47:1–18doi: 10.1146/annurev-biophys-070317-032931 [DOI] [PubMed] [Google Scholar]
- 8. Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discovery (2017) 16:829–42. doi: 10.1038/nrd.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leach K, Sexton PM, Christopoulos A. Allosteric GPCR modulators: taking advantage of permissive receptor pharmacology. Trends Pharmacol Sci (2007) 28:382–9. doi: 10.1016/j.tips.2007.06.004 [DOI] [PubMed] [Google Scholar]
- 10. Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nat Rev Drug Discovery (2013) 12:630–44. doi: 10.1038/nrd4052 [DOI] [PubMed] [Google Scholar]
- 11. Conn PJ, Christopoulos A, Lindsley CW. Allosteric modulators of GPCRs: a novel approach for the treatment of CNS disorders. Nat Rev Drug Discovery (2009) 8:41–54doi: 10.1038/nrd2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Changeux JP, Christopoulos A. Allosteric modulation as a unifying mechanism for receptor function and regulation. Cell (2016) 166:1084–102. doi: 10.1016/j.cell.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 13. Thal DM, Glukhova A, Sexton PM, Christopoulos A. Structural insights into G-protein-coupled receptor allostery. Nature (2018) 559:45–53doi: 10.1038/s41586-018-0259-z [DOI] [PubMed] [Google Scholar]
- 14. Christopoulos A. Advances in G protein-coupled receptor allostery: from function to structure. Mol Pharmacol (2014) 86:463–78. doi: 10.1124/mol.114.094342 [DOI] [PubMed] [Google Scholar]
- 15. Lazareno S, Dolezal V, Popham A, Birdsall NJ. Thiochrome enhances acetylcholine affinity at muscarinic M4 receptors: Receptor subtype selectivity via cooperativity rather than affinity. Mol Pharmacol (2004) 65:257–66. doi: 10.1124/mol.65.1.257 [DOI] [PubMed] [Google Scholar]
- 16. Valant C, Felder CC, Sexton PM, Christopoulos A. Probe dependence in the allosteric modulation of a G protein-coupled receptor: Implications for detection and validation of allosteric ligand effects. Mol Pharmacol (2012) 81:41–52doi: 10.1124/mol.111.074872 [DOI] [PubMed] [Google Scholar]
- 17. Slosky LM, Bai Y, Toth K, Ray C, Rochelle LK, Badea A, et al. β-Arrestin-Biased allosteric modulator of NTSR1 selectively attenuates addictive behaviors. Cell (2020) 181:1364–1379.e1314doi: 10.1016/j.cell.2020.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Krumm BE, DiBerto JF, Olsen RHJ, Kang HJ, Slocum ST, Zhang S, et al. Neurotensin receptor allosterism revealed in complex with a biased allosteric modulator. bioRxiv (2022) 2012:2026.521971. doi: 10.1101/2022.12.26.521971 [DOI] [PubMed] [Google Scholar]
- 19. Duan J, Liu H, Ji Y, Yuan Q, Li X, Wu K, et al. Structure of a G protein-coupled receptor with GRK2 and a biased ligand. bioRxiv (2022) 2010:2019.512855. doi: 10.1101/2022.10.19.512855 [DOI] [Google Scholar]
- 20. Liu X, Lu S, Song K, Shen Q, Ni D, Li Q, et al. Unraveling allosteric landscapes of allosterome with ASD. Nucleic Acids Res (2020) 48:D394–401. doi: 10.1093/nar/gkz958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cook AE, Mistry SN, Gregory KJ, Furness SGB, Sexton PM, Scammells PJ, et al. Biased allosteric modulation at the CaS receptor engendered by structurally diverse calcimimetics. Brit J Pharmacol (2015) 172:185–200doi: 10.1111/bph.12937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davey AE, Leach K, Valant C, Conigrave AD, Sexton PM, Christopoulos A. Positive and negative allosteric modulators promote biased signaling at the calcium-sensing receptor. Endocrinology (2012) 153:1232–41. doi: 10.1210/en.2011-1426 [DOI] [PubMed] [Google Scholar]
- 23. Leach K, Gregory KJ, Kufareva I, Khajehali E, Cook AE, Abagyan R, et al. Towards a structural understanding of allosteric drugs at the human calcium-sensing receptor. Cell Res (2016) 26:574–92. doi: 10.1038/cr.2016.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leach K, Wen A, Cook AE, Sexton PM, Conigrave AD, Christopoulos A. Impact of clinically relevant mutations on the pharmacoregulation and signaling bias of the calcium-sensing receptor by positive and negative allosteric modulators. Endocrinology (2013) 154:1105–16. doi: 10.1210/en.2012-1887 [DOI] [PubMed] [Google Scholar]
- 25. Nemeth EF, Heaton WH, Miller M, Fox J, Balandrin MF, Van Wagenen BC, et al. Pharmacodynamics of the type II calcimimetic compound cinacalcet HCl. J Pharmacol Exp Ther (2004) 308:627–35. doi: 10.1124/jpet.103.057273 [DOI] [PubMed] [Google Scholar]
- 26. Hoffmann K, Lutz DA, Strassburger J, Baqi Y, Muuller CE, Von Kugelgen I. Competitive mode and site of interaction of ticagrelor at the human platelet P2Y(12)-receptor. J Thromb Haemost (2014) 12:1898–905. doi: 10.1111/jth.12719 [DOI] [PubMed] [Google Scholar]
- 27. Krusek J, Zemkova H. Effect of ivermectin on gamma-aminobutyric acid-induced chloride currents in mouse hippocampal embryonic neurones. Eur J Pharmacol (1994) 259:121–8. doi: 10.1016/0014-2999(94)90500-2 [DOI] [PubMed] [Google Scholar]
- 28. Sliwoski G, Schubert M, Stichel J, Weaver D, Beck-Sickinger AG, Meiler J. Discovery of small-molecule modulators of the human Y4 receptor. PloS One (2016) 11:e0157146. doi: 10.1371/journal.pone.0157146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li XX, Lee JD, Massey NL, Guan C, Robertson AAB, Clark RJ, et al. Pharmacological characterisation of small molecule C5aR1 inhibitors in human cells reveals biased activities for signalling and function. Biochem Pharmacol (2020) 180:114156. doi: 10.1016/j.bcp.2020.114156 [DOI] [PubMed] [Google Scholar]
- 30. Walters MJ, Wang Y, Lai N, Baumgart T, Zhao BN, Dairaghi DJ, et al. Characterization of CCX282-b, an orally bioavailable antagonist of the CCR9 chemokine receptor, for treatment of inflammatory bowel disease. J Pharmacol Exp Ther (2010) 335:61–9. doi: 10.1124/jpet.110.169714 [DOI] [PubMed] [Google Scholar]
- 31. Vranesic I, Ofner S, Flor PJ, Bilbe G, Bouhelal R, Enz A, et al. AFQ056/mavoglurant, a novel clinically effective mGluR5 antagonist: Identification, SAR and pharmacological characterization. Bioorg Med Chem (2014) 22:5790–803. doi: 10.1016/j.bmc.2014.09.033 [DOI] [PubMed] [Google Scholar]
- 32. Chae E, Shin YJ, Ryu EJ, Ji MK, Ryune Cho N, Lee KH, et al. Discovery of biological evaluation of pyrazole/imidazole amides as mGlu5 receptor negative allosteric modulators. Bioorg Med Chem Lett (2013) 23:2134–9. doi: 10.1016/j.bmcl.2013.01.116 [DOI] [PubMed] [Google Scholar]
- 33. Jaeschke G, Kolczewski S, Spooren W, Vieira E, Bitter-Stoll N, Boissin P, et al. Metabotropic glutamate receptor 5 negative allosteric modulators: discovery of 2-chloro-4-[1-(4-fluorophenyl)-2,5-dimethyl-1H-imidazol-4-ylethynyl]pyridine (basimglurant, RO4917523), a promising novel medicine for psychiatric diseases. J Med Chem (2015) 58:1358–71. doi: 10.1021/jm501642c [DOI] [PubMed] [Google Scholar]
- 34. Marin JC, Goadsby PJ. Glutamatergic fine tuning with ADX-10059: a novel therapeutic approach for migraine? Expert Opin Investig Drugs (2010) 19:555–61. doi: 10.1517/13543781003691832 [DOI] [PubMed] [Google Scholar]
- 35. Pan HL, Xu Z, Leung E, Eisenach JC. Allosteric adenosine modulation to reduce allodynia. Anesthesiology (2001) 95:416–20. doi: 10.1097/00000542-200108000-00025 [DOI] [PubMed] [Google Scholar]
- 36. Justinova Z, Panlilio LV, Secci ME, Redhi GH, Schindler CW, Cross AJ, et al. The novel metabotropic glutamate receptor 2 positive allosteric modulator, AZD8529, decreases nicotine self-administration and relapse in squirrel monkeys. Biol Psychiatry (2015) 78:452–62. doi: 10.1016/j.biopsych.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lavreysen H, Ahnaou A, Drinkenburg W, Langlois X, Mackie C, Pype S, et al. Pharmacological and pharmacokinetic properties of JNJ-40411813, a positive allosteric modulator of the mGlu2 receptor. Pharmacol Res Perspect (2015) 3:e00096. doi: 10.1002/prp2.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin SL, Han S, Cai XQ, Tan QX, Zhou KX, Wang DJ, et al. Structures of g(i)-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature (2021) 594:583–8. doi: 10.1038/s41586-021-03495-2 [DOI] [PubMed] [Google Scholar]
- 39. Beshore DC, Di Marco CN, Chang RK, Greshock TJ, Ma L, Wittmann M, et al. MK-7622: A first-in-Class M(1) positive allosteric modulator development candidate. ACS Med Chem Lett (2018) 9:652–6. doi: 10.1021/acsmedchemlett.8b00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mandai T, Sako Y, Kurimoto E, Shimizu Y, Nakamura M, Fushimi M, et al. T-495, a novel low cooperative M(1) receptor positive allosteric modulator, improves memory deficits associated with cholinergic dysfunction and is characterized by low gastrointestinal side effect risk. Pharmacol Res Perspect (2020) 8:e00560. doi: 10.1002/prp2.560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Uslaner JM, Kuduk SD, Wittmann M, Lange HS, Fox SV, Min C, et al. Preclinical to human translational pharmacology of the novel M(1) positive allosteric modulator MK-7622. J Pharmacol Exp Ther (2018) 365:556–66. doi: 10.1124/jpet.117.245894 [DOI] [PubMed] [Google Scholar]
- 42. Hao J, Beck JP, Schaus JM, Krushinski JH, Chen Q, Beadle CD, et al. Synthesis and pharmacological characterization of 2-(2,6-Dichlorophenyl)-1-((1S,3R)-5-(3-hydroxy-3-methylbutyl)-3-(hydroxymethyl)-1-methyl-3,4-dihydroisoquinolin-2(1H)-yl)ethan-1-one (LY3154207), a potent, subtype selective, and orally available positive allosteric modulator of the human dopamine D1 receptor. J Med Chem (2019) 62:8711–32. doi: 10.1021/acs.jmedchem.9b01234 [DOI] [PubMed] [Google Scholar]
- 43. Xiao P, Yan W, Gou L, Zhong YN, Kong L, Wu C, et al. Ligand recognition and allosteric regulation of DRD1-gs signaling complexes. Cell (2021) 184:943–956 e918doi: 10.1016/j.cell.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hall A, Provins L, Valade A. Novel strategies to activate the dopamine d-1 receptor: Recent advances in orthosteric agonism and positive allosteric modulation. J Medicinal Chem (2019) 62:128–40. doi: 10.1021/acs.jmedchem.8b01767 [DOI] [PubMed] [Google Scholar]
- 45. Charvin D, Pomel V, Ortiz M, Frauli M, Scheffler S, Steinberg E, et al. Discovery, structure-activity relationship, and antiparkinsonian effect of a potent and brain-penetrant chemical series of positive allosteric modulators of metabotropic glutamate receptor 4. J Med Chem (2017) 60:8515–37. doi: 10.1021/acs.jmedchem.7b00991 [DOI] [PubMed] [Google Scholar]
- 46. Okimoto R, Ino K, Ishizu K, Takamatsu H, Sakamoto K, Yuyama H, et al. Potentiation of muscarinic M(3) receptor activation through a new allosteric site with a novel positive allosteric modulator ASP8302. J Pharmacol Exp Ther (2021) 379:64–73doi: 10.1124/jpet.121.000709 [DOI] [PubMed] [Google Scholar]
- 47. Krystal JH, Kane JM, Correll CU, Walling DP, Leoni M, Duvvuri S, et al. Emraclidine, a novel positive allosteric modulator of cholinergic M4 receptors, for the treatment of schizophrenia: a two-part, randomised, double-blind, placebo-controlled, phase 1b trial. Lancet (2023) 400:2210–20. doi: 10.1016/S0140-6736(22)01990-0 [DOI] [PubMed] [Google Scholar]
- 48. Christopher JA, Aves SJ, Bennett KA, Dore AS, Errey JC, Jazayeri A, et al. Fragment and structure-based drug discovery for a class c GPCR: Discovery of the mGlu5 negative allosteric modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J Med Chem (2015) 58:6653–64. doi: 10.1021/acs.jmedchem.5b00892 [DOI] [PubMed] [Google Scholar]
- 49. Andres JI, Alcazar J, Cid JM, De Angelis M, Iturrino L, Langlois X, et al. Synthesis, evaluation, and radiolabeling of new potent positive allosteric modulators of the metabotropic glutamate receptor 2 as potential tracers for positron emission tomography imaging. J Med Chem (2012) 55:8685–99. doi: 10.1021/jm300912k [DOI] [PubMed] [Google Scholar]
- 50. Tong L, Li W, Lo MM, Gao X, Wai JM, Rudd M, et al. Discovery of [(11)C]MK-6884: A positron emission tomography (PET) imaging agent for the study of M4Muscarinic receptor positive allosteric modulators (PAMs) in neurodegenerative diseases. J Med Chem (2020) 63:2411–25. doi: 10.1021/acs.jmedchem.9b01406 [DOI] [PubMed] [Google Scholar]
- 51. Sako Y, Kurimoto E, Mandai T, Suzuki A, Tanaka M, Suzuki M, et al. TAK-071, a novel M(1) positive allosteric modulator with low cooperativity, improves cognitive function in rodents with few cholinergic side effects. Neuropsychopharmacology (2019) 44:950–60. doi: 10.1038/s41386-018-0168-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lee A. Avacopan: First approval. Drugs (2022) 82:79–85doi: 10.1007/s40265-021-01643-6 [DOI] [PubMed] [Google Scholar]
- 53. Liu X, Masoudi A, Kahsai AW, Huang LY, Pani B, Staus DP, et al. Mechanism of beta2AR regulation by an intracellular positive allosteric modulator. Science (2019) 364:1283–7. doi: 10.1126/science.aaw8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu X, Ahn S, Kahsai AW, Meng KC, Latorraca NR, Pani B, et al. Mechanism of intracellular allosteric beta(2)AR antagonist revealed by X-ray crystal structure. Nature (2017) 548:480–4. doi: 10.1038/nature23652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu X, Kaindl J, Korczynska M, Stossel A, Dengler D, Stanek M, et al. An allosteric modulator binds to a conformational hub in the beta(2) adrenergic receptor. Nat Chem Biol (2020) 16:749–55. doi: 10.1038/s41589-020-0549-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lu J, Byrne N, Wang J, Bricogne G, Brown FK, Chobanian HR, et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat Struct Mol Biol (2017) 24:570–7. doi: 10.1038/nsmb.3417 [DOI] [PubMed] [Google Scholar]
- 57. Srivastava A, Yano J, Hirozane Y, Kefala G, Gruswitz F, Snell G, et al. High-resolution structure of the human GPR40 receptor bound to allosteric agonist TAK-875. Nature (2014) 513:124–7. doi: 10.1038/nature13494 [DOI] [PubMed] [Google Scholar]
- 58. Zhuang Y, Krumm B, Zhang H, Zhou XE, Wang Y, Huang XP, et al. Mechanism of dopamine binding and allosteric modulation of the human D1 dopamine receptor. Cell Res (2021) 31:593–6. doi: 10.1038/s41422-021-00482-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang X, Wang X, Xu Z, Wu C, Zhou Y, Wang Y, et al. Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1. Nat Chem Biol (2022) 18:831–40. doi: 10.1038/s41589-022-01038-y [DOI] [PubMed] [Google Scholar]
- 60. Shao Z, Yan W, Chapman K, Ramesh K, Ferrell AJ, Yin J, et al. Structure of an allosteric modulator bound to the CB1 cannabinoid receptor. Nat Chem Biol (2019) 15:1199–205. doi: 10.1038/s41589-019-0387-2 [DOI] [PubMed] [Google Scholar]
- 61. Draper-Joyce CJ, Bhola R, Wang J, Bhattarai A, Nguyen ATN, Cowie-Kent I, et al. Positive allosteric mechanisms of adenosine A(1) receptor-mediated analgesia. Nature (2021) 597:571–6. doi: 10.1038/s41586-021-03897-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature (2013) 504:101–6. doi: 10.1038/nature12735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Xu J, Wang Q, Hubner H, Hu Y, Niu X, Wang H, et al. Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor. Nat Commun (2023) 14:376. doi: 10.1038/s41467-022-35726-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen G, Xu J, Inoue A, Schmidt MF, Bai C, Lu Q, et al. Activation and allosteric regulation of the orphan GPR88-Gi1 signaling complex. Nat Commun (2022) 13:2375. doi: 10.1038/s41467-022-30081-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu Y, Cao C, Huang XP, Gumpper RH, Rachman MM, Shih SL, et al. Ligand recognition and allosteric modulation of the human MRGPRX1 receptor. Nat Chem Biol (2022) 2022:1–7. doi: 10.1038/s41589-022-01173-6 [DOI] [PubMed] [Google Scholar]
- 66. Zhang D, Gao ZG, Zhang K, Kiselev E, Crane S, Wang J, et al. Two disparate ligand-binding sites in the human P2Y1 receptor. Nature (2015) 520:317–21. doi: 10.1038/nature14287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cheng RKY, Fiez-Vandal C, Schlenker O, Edman K, Aggeler B, Brown DG, et al. Structural insight into allosteric modulation of protease-activated receptor 2. Nature (2017) 545:112–5. doi: 10.1038/nature22309 [DOI] [PubMed] [Google Scholar]
- 68. Liu H, Kim HR, Deepak R, Wang L, Chung KY, Fan H, et al. Orthosteric and allosteric action of the C5a receptor antagonists. Nat Struct Mol Biol (2018) 25:472–81. doi: 10.1038/s41594-018-0067-z [DOI] [PubMed] [Google Scholar]
- 69. Robertson N, Rappas M, Dore AS, Brown J, Bottegoni G, Koglin M, et al. Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature (2018) 553:111–4. doi: 10.1038/nature25025 [DOI] [PubMed] [Google Scholar]
- 70. Zheng Y, Qin L, Zacarias NV, de Vries H, Han GW, Gustavsson M, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature (2016) 540:458–61. doi: 10.1038/nature20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Oswald C, Rappas M, Kean J, Dore AS, Errey JC, Bennett K, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature (2016) 540:462–5. doi: 10.1038/nature20606 [DOI] [PubMed] [Google Scholar]
- 72. Song G, Yang D, Wang Y, de Graaf C, Zhou Q, Jiang S, et al. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature (2017) 546:312–5. doi: 10.1038/nature22378 [DOI] [PubMed] [Google Scholar]
- 73. Bueno AB, Sun B, Willard FS, Feng D, Ho JD, Wainscott DB, et al. Structural insights into probe-dependent positive allosterism of the GLP-1 receptor. Nat Chem Biol (2020) 16:1105–10. doi: 10.1038/s41589-020-0589-7 [DOI] [PubMed] [Google Scholar]
- 74. Cong Z, Chen LN, Ma H, Zhou Q, Zou X, Ye C, et al. Molecular insights into ago-allosteric modulation of the human glucagon-like peptide-1 receptor. Nat Commun (2021) 12:3763. doi: 10.1038/s41467-021-24058-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang H, Qiao A, Yang D, Yang L, Dai A, de Graaf C, et al. Structure of the full-length glucagon class b G-protein-coupled receptor. Nature (2017) 546:259–64. doi: 10.1038/nature22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jazayeri A, Dore AS, Lamb D, Krishnamurthy H, Southall SM, Baig AH, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature (2016) 533:274–7. doi: 10.1038/nature17414 [DOI] [PubMed] [Google Scholar]
- 77. Hollenstein K, Kean J, Bortolato A, Cheng RK, Dore AS, Jazayeri A, et al. Structure of class b GPCR corticotropin-releasing factor receptor 1. Nature (2013) 499:438–43. doi: 10.1038/nature12357 [DOI] [PubMed] [Google Scholar]
- 78. Shaye H, Ishchenko A, Lam JH, Han GW, Xue L, Rondard P, et al. Structural basis of the activation of a metabotropic GABA receptor. Nature (2020) 584:298–303doi: 10.1038/s41586-020-2408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kim Y, Jeong E, Jeong JH, Kim Y, Cho Y. Structural basis for activation of the heterodimeric GABA(B) receptor. J Mol Biol (2020) 432:5966–84. doi: 10.1016/j.jmb.2020.09.023 [DOI] [PubMed] [Google Scholar]
- 80. Mao C, Shen C, Li C, Shen DD, Xu C, Zhang S, et al. Cryo-EM structures of inactive and active GABA(B) receptor. Cell Res (2020) 30:564–73. doi: 10.1038/s41422-020-0350-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shen C, Mao C, Xu C, Jin N, Zhang H, Shen DD, et al. Structural basis of GABA(B) receptor-g(i) protein coupling. Nature (2021) 594:594–8. doi: 10.1038/s41586-021-03507-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wen T, Wang Z, Chen X, Ren Y, Lu X, Xing Y, et al. Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor. Sci Adv (2021) 7(23):eabg1483. doi: 10.1126/sciadv.abg1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gao Y, Robertson MJ, Rahman SN, Seven AB, Zhang C, Meyerowitz JG, et al. Asymmetric activation of the calcium-sensing receptor homodimer. Nature (2021) 595:455–9. doi: 10.1038/s41586-021-03691-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Park J, Zuo H, Frangaj A, Fu Z, Yen LY, Zhang Z, et al. Symmetric activation and modulation of the human calcium-sensing receptor. Proc Natl Acad Sci U.S.A. (2021) 118(51):e2115849118. doi: 10.1073/pnas.2115849118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wu H, Wang C, Gregory KJ, Han GW, Cho HP, Xia Y, et al. Structure of a class c GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science (2014) 344:58–64doi: 10.1126/science.1249489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Du J, Wang D, Fan H, Xu C, Tai L, Lin S, et al. Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature (2021) 594:589–93. doi: 10.1038/s41586-021-03641-w [DOI] [PubMed] [Google Scholar]
- 87. Seven AB, Barros-Alvarez X, de Lapeyriere M, Papasergi-Scott MM, Robertson MJ, Zhang C, et al. G-Protein activation by a metabotropic glutamate receptor. Nature (2021) 595:450–4. doi: 10.1038/s41586-021-03680-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Fang W, Yang F, Xu C, Ling S, Lin L, Zhou Y, et al. Structural basis of the activation of metabotropic glutamate receptor 3. Cell Res (2022) 32:695–8. doi: 10.1038/s41422-022-00623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dore AS, Okrasa K, Patel JC, Serrano-Vega M, Bennett K, Cooke RM, et al. Structure of class c GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature (2014) 511:557–62. doi: 10.1038/nature13396 [DOI] [PubMed] [Google Scholar]
- 90. Christopher JA, Orgovan Z, Congreve M, Dore AS, Errey JC, Marshall FH, et al. Structure-based optimization strategies for G protein-coupled receptor (GPCR) allosteric modulators: A case study from analyses of new metabotropic glutamate receptor 5 (mGlu(5)) X-ray structures. J Med Chem (2019) 62:207–22. doi: 10.1021/acs.jmedchem.7b01722 [DOI] [PubMed] [Google Scholar]
- 91. Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main familiesphylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol (2003) 63:1256–72. doi: 10.1124/mol.63.6.1256 [DOI] [PubMed] [Google Scholar]
- 92. Spehr M, Munger SD. Olfactory receptors: G protein-coupled receptors and beyond. J Neurochem (2009) 109:1570–83. doi: 10.1111/j.1471-4159.2009.06085.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zhou Q, Yang D, Wu M, Guo Y, Guo W, Zhong L, et al. Common activation mechanism of class a GPCRs. Elife (2019) 8:e50279. doi: 10.7554/eLife.50279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science (2000) 289:739–45. doi: 10.1126/science.289.5480.739 [DOI] [PubMed] [Google Scholar]
- 95. Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of the beta2 adrenergic receptor-gs protein complex. Nature (2011) 477:549–55. doi: 10.1038/nature10361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Spijker P, Vaidehi N, Freddolino PL, Hilbers PA. & Goddard, wa., 3rddynamic behavior of fully solvated beta2-adrenergic receptor, embedded in the membrane with bound agonist or antagonist. Proc Natl Acad Sci U.S.A. (2006) 103:4882–7. doi: 10.1073/pnas.0511329103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ahn S, Kahsai AW, Pani B, Wang QT, Zhao S, Wall AL, et al. Allosteric "beta-blocker" isolated from a DNA-encoded small molecule library. Proc Natl Acad Sci U.S.A. (2017) 114:1708–13. doi: 10.1073/pnas.1620645114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Staus DP, Strachan RT, Manglik A, Pani B, Kahsai AW, Kim TH, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature (2016) 535:448–52. doi: 10.1038/nature18636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zou S, Kumar U. Cannabinoid receptors and the endocannabinoid system: Signaling and function in the central nervous system. Int J Mol Sci (2018) 19(3):833. doi: 10.3390/ijms19030833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lemberger L. Potential therapeutic usefulness of marijuana. Annu Rev Pharmacol Toxicol (1980) 20:151–72. doi: 10.1146/annurev.pa.20.040180.001055 [DOI] [PubMed] [Google Scholar]
- 101. Pain S. A potted history. Nature (2015) 525:S10–11. doi: 10.1038/525S10a [DOI] [PubMed] [Google Scholar]
- 102. Nguyen PT, Schmid CL, Raehal KM, Selley DE, Bohn LM, Sim-Selley LJ. Beta-arrestin2 regulates cannabinoid CB1 receptor signaling and adaptation in a central nervous system region-dependent manner. Biol Psychiatry (2012) 71:714–24. doi: 10.1016/j.biopsych.2011.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hampson RE, Deadwyler SA. Cannabinoids reveal the necessity of hippocampal neural encoding for short-term memory in rats. J Neurosci (2000) 20:8932–42. doi: 10.1523/JNEUROSCI.20-23-08932.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Justinova Z, Tanda G, Redhi GH, Goldberg SR. Self-administration of delta9-tetrahydrocannabinol (THC) by drug naive squirrel monkeys. Psychopharmacol (Berl) (2003) 169:135–40. doi: 10.1007/s00213-003-1484-0 [DOI] [PubMed] [Google Scholar]
- 105. Cooper ZD, Haney M. Actions of delta-9-tetrahydrocannabinol in cannabis: relation to use, abuse, dependence. Int Rev Psychiatry (2009) 21:104–12. doi: 10.1080/09540260902782752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hutcheson DM, Tzavara ET, Smadja C, Valjent E, Roques BP, Hanoune J, et al. Behavioural and biochemical evidence for signs of abstinence in mice chronically treated with delta-9-tetrahydrocannabinol. Br J Pharmacol (1998) 125:1567–77. doi: 10.1038/sj.bjp.0702228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Thomas G, Kloner RA, Rezkalla S. Adverse cardiovascular, cerebrovascular, and peripheral vascular effects of marijuana inhalation: what cardiologists need to know. Am J Cardiol (2014) 113:187–90. doi: 10.1016/j.amjcard.2013.09.042 [DOI] [PubMed] [Google Scholar]
- 108. Wei TT, Chandy M, Nishiga M, Zhang A, Kumar KK, Thomas D, et al. Cannabinoid receptor 1 antagonist genistein attenuates marijuana-induced vascular inflammation. Cell (2022) 185:1676–1693 e1623doi: 10.1016/j.cell.2022.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Lu D, Immadi SS, Wu Z, Kendall DA. Translational potential of allosteric modulators targeting the cannabinoid CB(1) receptor. Acta Pharmacol Sin (2019) 40:324–35. doi: 10.1038/s41401-018-0164-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yang F, Mao C, Guo L, Lin J, Ming Q, Xiao P, et al. Structural basis of GPBAR activation and bile acid recognition. Nature (2020) 587:499–504doi: 10.1038/s41586-020-2569-1 [DOI] [PubMed] [Google Scholar]
- 111. Xu P, Huang S, Zhang H, Mao C, Zhou XE, Cheng X, et al. Structural insights into the lipid and ligand regulation of serotonin receptors. Nature (2021) 592:469–73. doi: 10.1038/s41586-021-03376-8 [DOI] [PubMed] [Google Scholar]
- 112. Burger K, Gimpl G, Fahrenholz F. Regulation of receptor function by cholesterol. Cell Mol Life Sci (2000) 57:1577–92. doi: 10.1007/pl00000643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Pucadyil TJ, Chattopadhyay A. Role of cholesterol in the function and organization of G-protein coupled receptors. Prog Lipid Res (2006) 45:295–333doi: 10.1016/j.plipres.2006.02.002 [DOI] [PubMed] [Google Scholar]
- 114. Paila YD, Chattopadhyay A. Membrane cholesterol in the function and organization of G-protein coupled receptors. Subcell Biochem (2010) 51:439–66. doi: 10.1007/978-90-481-8622-8_16 [DOI] [PubMed] [Google Scholar]
- 115. Alemany R, Perona JS, Sanchez-Dominguez JM, Montero E, Canizares J, Bressani R, et al. G Protein-coupled receptor systems and their lipid environment in health disorders during aging. Biochim Biophys Acta (2007) 1768:964–75. doi: 10.1016/j.bbamem.2006.09.024 [DOI] [PubMed] [Google Scholar]
- 116. Damian M, Louet M, Gomes AA.S, M'Kadmi C, Denoyelle S, Cantel S, et al. Allosteric modulation of ghrelin receptor signaling by lipids. Nat Commun (2021) 12:3938. doi: 10.1038/s41467-021-23756-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Simons K, Ikonen E. How cells handle cholesterol. Science (2000) 290:1721–6. doi: 10.1126/science.290.5497.1721 [DOI] [PubMed] [Google Scholar]
- 118. Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, et al. A specific cholesterol binding site is established by the 2.8 a structure of the human beta2-adrenergic receptor. Structure (2008) 16:897–905doi: 10.1016/j.str.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Dehail P, Duclos C, Barat M. Electrical stimulation and muscle strengthening. Ann Readapt Med Phys (2008) 51:441–51. doi: 10.1016/j.annrmp.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 120. Duan J, Xu P, Cheng X, Mao C, Croll T, He X, et al. Structures of full-length glycoprotein hormone receptor signalling complexes. Nature (2021) 598:688–92. doi: 10.1038/s41586-021-03924-2 [DOI] [PubMed] [Google Scholar]
- 121. Hollenstein K, de Graaf C, Bortolato A, Wang MW, Marshall FH, Stevens RC. Insights into the structure of class b GPCRs. Trends Pharmacol Sci (2014) 35:12–22doi: 10.1016/j.tips.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wootten D, Miller LJ, Koole C, Christopoulos A, Sexton PM. Allostery and biased agonism at class b G protein-coupled receptors. Chem Rev (2017) 117:111–38. doi: 10.1021/acs.chemrev.6b00049 [DOI] [PubMed] [Google Scholar]
- 123. Graaf C, Donnelly D, Wootten D, Lau J, Sexton PM, Miller LJ, et al. Glucagon-like peptide-1 and its class b G protein-coupled receptors: A long march to therapeutic successes. Pharmacol Rev (2016) 68:954–1013doi: 10.1124/pr.115.011395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Drucker DJ. Mechanisms of action and therapeutic application of glucagon-like peptide-1. Cell Metab (2018) 27:740–56. doi: 10.1016/j.cmet.2018.03.001 [DOI] [PubMed] [Google Scholar]
- 125. Koole C, Wootten D, Simms J, Valant C, Sridhar R, Woodman OL, et al. Allosteric ligands of the glucagon-like peptide 1 receptor (GLP-1R) differentially modulate endogenous and exogenous peptide responses in a pathway-selective manner: implications for drug screening. Mol Pharmacol (2010) 78:456–65. doi: 10.1124/mol.110.065664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. From the American Association of Neurological Surgeons, ASoNC et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke (2018) 13:612–32. doi: 10.1177/1747493018778713 [DOI] [PubMed] [Google Scholar]
- 127. Chun L, Zhang WH, Liu JF. Structure and ligand recognition of class c GPCRs. Acta Pharmacologica Sin (2012) 33:312–23. doi: 10.1038/aps.2011.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, et al. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature (1998) 396:679–82. doi: 10.1038/25354 [DOI] [PubMed] [Google Scholar]
- 129. Urwyler S, Mosbacher J, Lingenhoehl K, Heid J, Hofstetter K, Froestl W, et al. Positive allosteric modulation of native and recombinant gamma-aminobutyric acid(B) receptors by 2,6-Di-tert-butyl-4-(3-hydroxy-2,2-dimethyl-propyl)-phenol (CGP7930) and its aldehyde analog CGP13501. Mol Pharmacol (2001) 60:963–71. doi: 10.1124/mol.60.5.963 [DOI] [PubMed] [Google Scholar]
- 130. Malherbe P, Masciadri R, Norcross RD, Knoflach F, Kratzeisen C, Zenner MT, et al. Characterization of (R,S)-5,7-di-tert-butyl-3-hydroxy-3-trifluoromethyl-3H-benzofuran-2-one as a positive allosteric modulator of GABAB receptors. Br J Pharmacol (2008) 154:797–811doi: 10.1038/bjp.2008.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Urwyler S, Pozza MF, Lingenhoehl K, Mosbacher J, Lampert C, Froestl W, et al. N,N'-Dicyclopentyl-2-methylsulfanyl-5-nitro-pyrimidine-4,6-diamine (GS39783) and structurally related compounds: novel allosteric enhancers of gamma-aminobutyric acidB receptor function. J Pharmacol Exp Ther (2003) 307:322–30. doi: 10.1124/jpet.103.053074 [DOI] [PubMed] [Google Scholar]
- 132. Chen LH, Sun B, Zhang Y, Xu TJ, Xia ZX, Liu JF, et al. Discovery of a negative allosteric modulator of GABAB receptors. ACS Med Chem Lett (2014) 5:742–7. doi: 10.1021/ml500162z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Pin JP, Acher F. The metabotropic glutamate receptors: structure, activation mechanism and pharmacology. Curr Drug Targets CNS Neurol Disord (2002) 1:297–317doi: 10.2174/1568007023339328 [DOI] [PubMed] [Google Scholar]
- 134. Nasrallah C, Cannone G, Briot J, Rottier K, Berizzi AE, Huang CY, et al. Agonists and allosteric modulators promote signaling from different metabotropic glutamate receptor 5 conformations. Cell Rep (2021) 36:109648. doi: 10.1016/j.celrep.2021.109648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Lecat-Guillet N, Quast RB, Liu H, Møller TC, Rovira X, Soldevila S, et al. Dissecting conformational rearrangements and allosteric modulation in metabotropic glutamate receptor activation. bioRxiv (2022) 2001:2007.474531. doi: 10.1101/2022.01.07.474531 [DOI] [Google Scholar]
- 136. Liauw BW, Foroutan A, Schamber MR, Lu W, Samareh Afsari H, Vafabakhsh R. Conformational fingerprinting of allosteric modulators in metabotropic glutamate receptor 2. Elife (2022) 11(2022):e78982. doi: 10.7554/eLife.78982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Cao AM, Quast RB, Fatemi F, Rondard P, Pin JP, Margeat E. Allosteric modulators enhance agonist efficacy by increasing the residence time of a GPCR in the active state. Nat Commun (2021) 12:5426. doi: 10.1038/s41467-021-25620-5 [DOI] [PMC free article] [PubMed] [Google Scholar]