Abstract
The active 17S U2 small nuclear ribonucleoprotein particle (snRNP), which binds to the intron branch site during the formation of the prespliceosome, is assembled in vitro by sequential interactions of the essential splicing factors SF3b and SF3a with the 12S U2 snRNP. We have analyzed the function of individual subunits of human SF3a (SF3a60, SF3a66, and SF3a120) by testing recombinant proteins, expressed in insect cells, in various in vitro assays. The recombinant subunits readily form the SF3a heterotrimer, where SF3a60 and SF3a66 interact with SF3a120, but not with each other. All SF3a subunits are essential for the formation of the mature 17S U2 snRNP and the prespliceosome. Single subunits engage in interactions with the 15S U2 snRNP (consisting of the 12S U2 snRNP and SF3b), and SF3a60 appears to play a major role in recruiting SF3a120 into the U2 particle. Analysis of functional domains in SF3a60 and SF3a66 identified interaction sites for SF3a120 in their N-terminal portions. C2H2-type zinc finger domains mediate the integration of SF3a60 and SF3a66 into the U2 snRNP, and we propose a model in which protein-protein interactions between the zinc finger domains and the Sm proteins, common to all spliceosomal snRNPs, contribute to the assembly of the 17S U2 snRNP. Finally, we demonstrate that all domains required for interactions within the SF3a heterotrimer and the formation of the 17S U2 snRNP are also necessary to assemble the prespliceosome.
The small nuclear ribonucleoprotein particles (snRNPs) U1, U2, U4/U6, and U5 are essential for pre-mRNA splicing. They play major roles in the accuracy and specificity of intron removal by recognizing conserved sequence elements in the pre-mRNA substrate (for review, see references 9, 32, 43, and 53). In concert with a large number of non-snRNP protein factors, they assemble on the pre-mRNA in a highly ordered and dynamic fashion. A multitude of interactions between the pre-mRNA, snRNPs, and protein factors promotes the formation of a large catalytically active complex, the spliceosome, which is the site of two transesterification reactions that result in intron removal.
The formation of the early splicing complex (complex E) involves base pairing between the RNA moiety of the U1 snRNP with the 5′ splice site and the recognition of elements at the 3′ splice site by protein factors (for review, see references 43 and 54). These elements include the intron branch site, a polypyrimidine tract, and the 3′ splice site AG dinucleotide. Recent data suggest that the U2 snRNP also represents a component of complex E and is required for the formation of this complex (16). During the ATP-dependent transition of complex E to presplicing complex A, a short helix is formed between U2 snRNA and the branch site, defining the adenosine that functions as the nucleophile in the first catalytic step. The U4/U6 and U5 snRNPs, in the form of a tri-snRNP, associate with complex A to generate complex B, and the active splicing complex C is formed by a conformational rearrangement.
The snRNPs consist of one or two small nuclear RNAs and a number of common and particle-specific proteins (53). Seven common proteins (the Sm or core proteins B/B′, D1, D2, D3, E, F, and G) bind to the Sm binding site present in U1, U2, U4, and U5 snRNAs. A set of related proteins (the Lsm proteins) bind to U6 snRNA (24). All snRNPs also contain particle-specific proteins, which are more or less tightly associated with the core snRNPs.
The U2 snRNP was initially isolated as a 12S particle containing the Sm proteins and two U2-specific proteins, A′ and B" (53). Milder purification conditions led to the identification of the 17S U2 snRNP containing nine additional polypeptides (4), seven of which correspond to the subunits of splicing factors SF3a and SF3b (7, 33). Consistent with an essential role of SF3a and SF3b in spliceosome assembly (8), the 12S U2 snRNP is inactive in splicing, whereas the 17S particle represents the active U2 snRNP (3). In HeLa cell nuclear extracts, and probably also in vivo, most of the U2 snRNP is present in the 17S form (4, 7). The dissociation into the 12S particle, SF3a, and SF3b during chromatographic fractionation is most likely a consequence of the salt-sensitive association of SF3a and SF3b with the U2 snRNP (4, 7, 33). In vitro, the 17S U2 snRNP is formed in two distinct steps. SF3b binds to the 12S U2 snRNP, generating an intermediate particle of 15S. SF3a cannot bind to either SF3b or the 12S U2 snRNP alone and interacts only with the 15S U2 snRNP to form the 17S particle (7, 33). Biochemical and electron microscopic studies initially suggested that most of the 17S U2-specific proteins associated with the 5′ portion of the U2 snRNP (4). Additional experiments indicated that SF3b interacts with the 5′ half of U2 snRNA, whereas SF3a associates with the 3′ portion of the U2 snRNP and possibly also interacts with SF3b (33).
SF3a purified from HeLa cells contains three polypeptides of 60, 66, and 120 kDa (SF3a60, SF3a66, and SF3a120) in a 1:1:1 ratio (8, 16). SF3b consists of four subunits (SF3b49, SF3b130, SF3b145, and SF3b155) (33). All polypeptides have also been identified as spliceosome-associated proteins, which are detected in complexes E and A and remain associated with the spliceosome through both catalytic steps (5, 16, 21, 48). cDNAs for all subunits have been isolated, and homologues in Saccharomyces cerevisiae are essential splicing factors that function in the formation of the prespliceosome (10, 15, 22, 26, 27, 32, 40, 46, 49, 51).
In addition to a phylogenetic conservation of structural features of the SF3a and SF3b subunits (see below), interactions between individual polypeptides are conserved. For example, SF3a60 and SF3a66 interact with SF3a120, but not with one another, thus generating the SF3a heterotrimer (6, 13, 31). Similarly, Prp9p, Prp11p, and Prp21p (the yeast homologues of SF3a60, SF3a66, and SF3a120, respectively) form a heteromeric complex in which Prp9p and Prp11p physically interact with Prp21p (36, 37, 52). Contacts between SF3b subunits have also been established and are conserved in yeast (11, 15, 19, 27). To date, only one direct interaction between subunits of SF3a and SF3b, namely that of Prp9p and Rse1p (the yeast homologue of SF3b130) (10, 16, 40), has been detected (19). Direct contacts of SF3a or SF3b with Sm proteins or the U2-specific proteins A′ and B" have not been reported. However, Prp11p and Hsh49p (yeast SF3b49) bind to the U6-associated proteins Lsm4p and Lsm8p, respectively (19, 20).
SF3a and SF3b are essential for prespliceosome assembly (8) and are thought to recruit the U2 snRNP to the branch site (22, 23). Except for SF3b130, all subunits UV cross-link to a 20-nucleotide region in the intron located upstream of the branch site (termed “anchor site”), and SF3b155 also cross-links to sequences immediately downstream of the branch site (22, 23). In agreement with the variability of the anchor site in different pre-mRNAs, RNA binding of the proteins is sequence independent (22). Moreover, the SF3a and SF3b subunits only cross-link to RNA in the context of the prespliceosome; direct interactions of individual subunits with RNA in the absence of other spliceosomal components have not been observed (11).
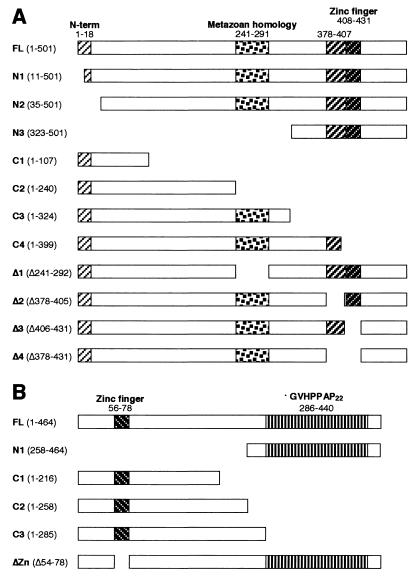
Given the multisubunit nature of SF3a and SF3b, we have begun to analyze the roles that individual SF3a subunits play in the formation of the active 17S U2 snRNP and the prespliceosome. In addition, we have studied domains in SF3a60 and SF3a66 that are essential for function. Metazoan, plant, and yeast homologues of SF3a60 share the highest similarity in the C-terminal ∼120 amino acids, which contain a zinc finger domain of the C2H2 type (13, 30, 35, 39; data not shown) (see Fig. 1). With the exception of S. cerevisiae Prp9p, several blocks of homology are apparent in the N-terminal half of all homologues. Within this region, Prp9p shares highest homology with other SF3a60 counterparts in the N-terminal ∼25 amino acids. A central portion of 50 amino acids is almost identical in the human and Drosophila proteins (39) and highly conserved in Caenorhabditis elegans (accession no. Q22469) and Arabidopsis thaliana (accession no. AP002544). In contrast, Prp9p and SF3a60 proteins from Schizosaccharomyces pombe (accession no. AI023589) and Neurospora crassa (accession no. AL353819) contain a second C2H2 zinc finger domain in the central region.
FIG. 1.
Schematic representation of the structure of recombinant SF3a60 and SF3a66 proteins. Recombinant SF3a60 (A) and SF3a66 (B) proteins carried N-terminal His6 tags derived from the expression vector. Amino acids present in or deleted from the recombinant proteins are given in parentheses behind the names of the proteins. Conserved regions are indicated above the diagrams and numbered according to the amino acids in full-length SF3a60 and SF3a66.
SF3a66 proteins from human (6), mouse (18), Drosophila (1), C. elegans (accession no. Q19335), A. thaliana (accession no. AP002544), and S. pombe (accession no. AL157918) exhibit high sequence identity in the entire N-terminal half (data not shown). S. cerevisiae Prp11p (12) is clearly related in sequence, but more divergent. A C2H2 zinc finger domain with sequence homology to the zinc finger domain of SF3a60 is located near the N terminus (35). In addition, sequences immediately N terminal to the zinc finger domains of SF3a60 and SF3a66 are related in all species (30; data not shown). The C-terminal halves of human and mouse SF3a66 comprise 22 and 25 heptad repeats, respectively (6, 18). Two related repeats are found in A. thaliana and Drosophila melanogaster SF3a66. Conserved repeats are absent from the C-terminal portion of the C. elegans counterpart, and SF3a66 proteins from S. cerevisiae and S. pombe lack an equivalent of the C-terminal domain.
We show here that all subunits of SF3a are essential for the formation of the active 17S U2 snRNP and the prespliceosome. Domains in SF3a60 and SF3a66 that are required for the interaction with SF3a120 are confined to the N-terminal portions of both subunits, whereas the zinc finger domains are necessary for the association with the 15S U2 snRNP. Both regions have to be intact for prespliceosome assembly to occur.
MATERIALS AND METHODS
Cloning procedures.
For the expression of proteins with N-terminal His6 tags, cDNAs encoding full-length and truncated versions of SF3a60 and SF3a66 were cloned into the pFastBac HT series of baculovirus expression vectors (Gibco BRL). Sequences encoding 3a60-FL, -N1, -N2, -C1, and -C2 (Fig. 1A) were amplified with the Expand High Fidelity PCR System (Roche Molecular Biochemicals) and appropriate primers from SF3a60 cDNA (30). PCR products were cloned into pFastBac HTb. Internal deletions (Δ1 to Δ4) were generated by PCR of sequences upstream and downstream of the desired deletion, and amplified fragments were cloned into the BamHI site of pFastBac HTb by three-molecular ligation. Deleted amino acids in the recombinant proteins were replaced by glycine and threonine encoded by a KpnI site joining the two amplified fragments. 3a60-C3 and -C4 were generated by subcloning appropriate restriction fragments into the BamHI-XbaI and BamHI-HindIII sites, respectively, of pFastBac HTb. Sequences encoding 3a60-N3 were subcloned into the XbaI site of pFastBac HTa.
A cDNA encoding SF3a66 was isolated from a 5′ stretch HeLa cDNA library in λgt11 (Clontech). The correct sequence (6) was verified by sequencing both strands. Restriction fragments containing sequences of 3a66-FL and -C2 (Fig. 1B) were subcloned into the BamHI and BamHI-StuI sites, respectively, of pFastBac HTb. A restriction fragment encoding sequences of 3a66-N1 was cloned into the StuI-PstI sites of pFastBac HTa. Sequences of 3a66-C1 and -C3 were amplified by PCR and ligated into the BamHI-KpnI sites of pFastBac HTb. 3a66ΔZn was generated by PCR as described above for internal deletions in SF3a60. A glutathione S-transferase (GST)-tagged version of SF3a66 was obtained by cloning the entire coding sequence into the BamHI site of the baculovirus expression vector pACG2T (Pharmingen).
The full-length SF3a120 coding sequence (31) was cloned into the EcoRI site of pFastBac HTa, generating plasmid 3a120-FL.
Expression of recombinant SF3a proteins in insect cells.
Recombinant baculoviruses were generated in Spodoptera frugiperda (Sf21) cells with the Bac-to-Bac Baculovirus Expression System (Gibco BRL) following the supplier's instructions. After verification of protein expression by Western blotting with antipolyhistidine antibodies (Sigma), virus isolates were amplified and Trichoplusia ni (Tn5) cells were infected at a multiplicity of infection of >3. Cells were harvested 48 h postinfection, and recombinant His6-tagged proteins were purified with Ni-nitrilotriacetic acid-agarose (Qiagen) in the Tris buffer system essentially as described by the supplier, except that the concentration of Nonidet P-40 in the lysis buffer was decreased to 0.1%. For the purification of insoluble proteins (3a120-FL and 3a66-C2), the KCl concentration in the lysis buffer was increased to 300 mM. All proteins were dialyzed against buffer D (17) and stored in aliquots at −80°C.
Far-Western blotting.
Recombinant proteins (1.5 μg) were separated in a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel and transferred to nitrocellulose (34). The membrane was probed with [35S]methionine-labeled in vitro-translated SF3a120 as described previously (44). Input proteins separated in a parallel gel were visualized by Western blotting with monoclonal antipolyhistidine antibodies (Sigma) and the ECL (enhanced chemiluminescence) detection system (Amersham).
Immunoprecipitation.
Mouse anti-SF3a66 (mAb66) (8) was coupled to 20 μl of packed protein A-Sepharose in 0.4 ml of NET-2 (50 mM Tris-HCl [pH 7.9], 150 mM NaCl, 0.05% Nonidet P-40, 0.5 mM dithiothreitol) overnight at 4°C. The beads were washed three times with NET-2 to remove unbound material. Recombinant proteins (1.0 μg) were incubated with protein A-Sepharose-coupled immunoglobulin G (IgG) in 0.4 ml of NET-2 for 90 min at 4°C with slow rotation. Beads were collected by centrifugation and washed three times with NET-2. Bound proteins were eluted with SDS sample solution, separated by 12% SDS–polyacrylamide gel electrophoresis (PAGE), and visualized by silver staining.
For immunoprecipitation with affinity-purified guinea pig anti-SF3b155 antibodies (46), recombinant proteins (1.2 μg) were incubated for 30 min at 30°C with Mono Q-purified 15S U2 snRNP (33), added to protein G-Sepharose-coupled IgG in buffer D (17) containing 3 mM MgCl2, and further processed as described above.
GST pull-down assay.
Interactions with GST-tagged SF3a66 were analyzed as described by Rain et al. (44). In some reactions (see Fig. 3B), the glutathione agarose was washed with buffer containing 400 mM NaCl before elution of bound proteins.
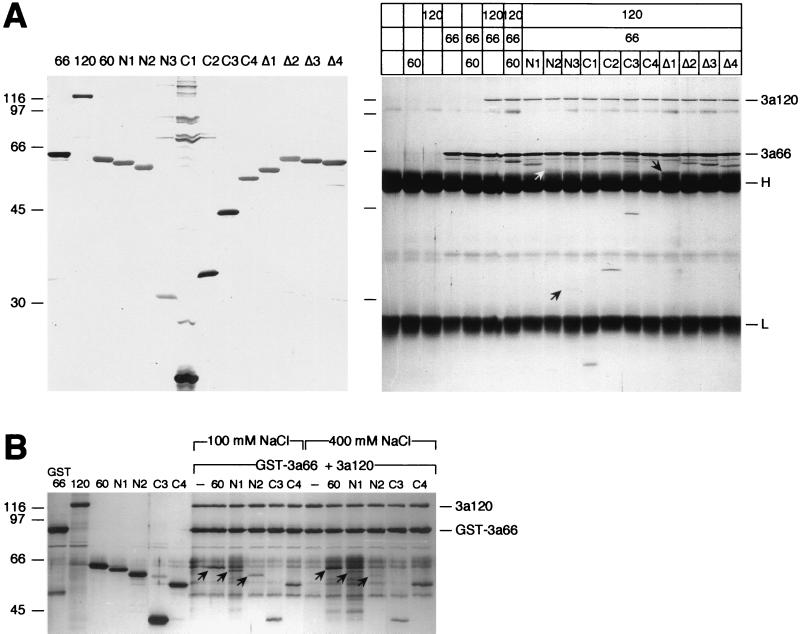
FIG. 3.
Amino acids 35 to 107 of SF3a60 are sufficient for its incorporation into SF3a. (A) Coimmunoprecipitation assay. Recombinant SF3a60 proteins were separated by SDS-PAGE and stained with silver (left panel). Identical amounts of the proteins were precipitated with mAb66 bound to protein A-Sepharose in the presence or absence of SF3a66 and/or SF3a120 as indicated above the figure. Bound proteins were separated by SDS-PAGE and visualized by silver staining (right panel). The positions of SF3a120, SF3a66, and the IgG heavy and light chains are indicated. Arrows point to 3a60-N2 and -N3, which bound inefficiently to SF3a120, and to 3a60Δ1 migrating just above the IgG heavy chain. The migration of molecular mass standards (in kilodaltons) is indicated to the left of each panel. (B) GST pull-down. Recombinant SF3a proteins (as indicated above the figure) were either loaded directly onto a SDS-polyacrylamide gel or incubated with GST-SF3a66 and SF3a120. Before elution of bound proteins, the gluthathione agarose was washed with buffer containing 100 or 400 mM NaCl as indicated. Arrows indicate full-length SF3a60, 3a60-N1, and 3a60-N2. The positions of SF3a120 and GST-SF3a66 are shown on the right, and the migration of molecular mass standards (in kilodaltons) is indicated on the left.
Mobility shift assays.
Recombinant proteins (0.5 μg each) were incubated with Mono Q-purified 15S U2 snRNP (33). The reaction conditions used were those described by Brosi et al. (7). Mono S-purified SF3a (8) was used as a control. Reaction products were resolved in native 4% polyacrylamide gels and visualized by autoradiography (28). The interaction of SF3a60 with the 15S U2 snRNP was more easily detected when gel electrophoresis was performed in the cold room. Mobility shifts in the presence of SF3a66 were more pronounced upon gel electrophoresis at room temperature, and binding of SF3a120 was unaffected by temperature (data not shown). Therefore, gel electrophoresis of complexes formed with SF3a60 mutant proteins was performed in the cold room.
Presplicing complex assembly.
Presplicing complexes were assembled as described (29). Recombinant proteins (0.25 μg each) were preincubated for 30 min at 30°C before addition of SF1, U2AF, SR proteins, U1 snRNP, the 12S U2 snRNP, and SF3b partially purified from HeLa cells (8). Reactions performed with HeLa cell nuclear extract (17) or with partially purified SF3a (8) served as controls.
RNA-binding assays.
Recombinant proteins (0.8 μg) were UV cross-linked to [α-32P]UTP-labeled synthetic pre-mRNA derived from the adenovirus major late (AdML) transcription unit in a total volume of 10 μl under standard splicing conditions (2). After UV light irradiation, unprotected RNA was digested with RNase A (0.3 mg/ml) and RNase T1 (200 U/ml) for 30 min at 37°C. Proteins were separated in a 10% SDS–polyacrylamide gel and visualized by autoradiography.
RESULTS
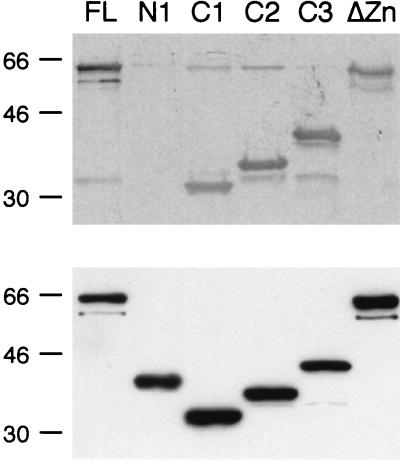
To study the function of individual subunits of splicing factor SF3a, His6-tagged, full-length proteins and derivatives with N-terminal, C-terminal, or internal deletions (Fig. 1) were produced in insect cells via baculovirus expression plasmids and purified by Ni2+ chelate chromatography. As a prerequisite for the present study, the recombinant full-length subunits were active in prespliceosome assembly when combined with other splicing components in an in vitro reconstitution assay (see Fig. 6).
FIG. 6.
The N-terminal regions and the zinc finger domains of SF3a60 and SF3a66 are essential for prespliceosome formation. The assembly of presplicing complex A was tested in the presence of buffer (−), HeLa cell nuclear extract (NE), or fractions containing partially purified SF1, U1 snRNP, the 12S U2 snRNP, U2AF, and SF3b. HeLa cell SF3a or recombinant proteins were added as indicated. Splicing complexes were separated in a native 4% polyacrylamide gel. The positions of pre-mRNA, complex H, presplicing complex A, and splicing complexes B and C are indicated on the left.
The SF3a120 binding site of SF3a60 is located between amino acids 35 and 107.
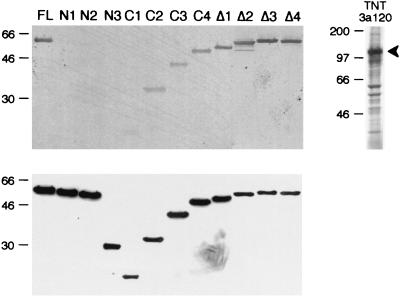
In the SF3a heterotrimer, SF3a60 physically interacts with SF3a120 (13, 30). Sequences in SF3a60 required for this interaction were initially studied by far-Western blotting. Recombinant proteins were separated by SDS-PAGE and transferred to nitrocellulose, which was incubated with in vitro-translated [35S]methionine-labeled SF3a120 (Fig. 2). SF3a120 bound efficiently to full-length SF3a60, but deletion of as few as 10 amino acids from the N terminus (mutants N1 to N3) abolished the interaction. SF3a120 bound to SF3a60 proteins with C-terminal truncations (C1 to C4); however, the interaction became weaker with decreasing size of the recombinant proteins. Deletion of the highly conserved internal domain of SF3a60 (Δ1), the zinc finger domain (Δ2), 30 amino acids N terminal to the zinc finger (Δ3), or the entire zinc finger region (Δ4) did not interfere with the interaction between SF3a60 and SF3a120. These results identify the N terminus of SF3a60 as essential for the interaction with SF3a120.
FIG. 2.
The N terminus of SF3a60 is required for binding to SF3a120. Recombinant SF3a60 proteins, as indicated above the figure, were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was incubated with in vitro-translated [35S]methionine-labeled SF3a120 (TNT 3a120), and bound SF3a120 was visualized by autoradiography (top). Input proteins separated in a parallel gel were visualized by Western blotting with antipolyhistidine antibodies (bottom). The migration of molecular mass standards (in kilodaltons) is indicated to the left of each panel.
Taking advantage of the interaction between SF3a66 and SF3a120, the SF3a120 interaction domain of SF3a60 was further delineated by coimmunoprecipitation (6, 31). In control reactions, the SF3a subunits (either alone or in combination) were incubated with a monoclonal anti-SF3a66 antibody (mAb66) coupled to protein A-Sepharose. SF3a66 was precipitated efficiently, but, as expected, neither SF3a60 nor SF3a120 bound to mAb66 (Fig. 3A). Precipitation of SF3a120 occurred in the presence of SF3a66, whereas SF3a60 was only precipitated in the presence of both SF3a66 and SF3a120. In agreement with previous data, these results demonstrate that SF3a60 and SF3a66 do not directly interact with each other, but form the SF3a heterotrimer by association with SF3a120 (6, 13, 30, 31).
Next, mutant SF3a60 proteins were precipitated in the presence of SF3a66 and SF3a120. In contrast to the results obtained by far-Western blotting, 3a60-N1 was found in the precipitate (Fig. 3A). In addition, small amounts of 3a60-N2 and a faint band of 3a60-N3 were detected. SF3a60 proteins with C-terminal (C2 to C4) or internal deletions (Δ1 to Δ4) were efficiently precipitated. 3a60-C4 comigrated with the IgG heavy chain and could not be detected in this experiment. However, it formed a complex with SF3a120 and GST-tagged SF3a66 (Fig. 3B). 3a60-C1, which was barely recognized by 35S-labeled SF3a120 in the far-Western assay (Fig. 2), was clearly coprecipitated with SF3a120 (Fig. 3A), although, compared to the other C-terminal deletions, the efficiency of binding appeared slightly reduced. The discrepancy between the results obtained by the different methods used could be explained by a failure to detect weak protein-protein interactions by far-Western blotting or a stabilization of weak interactions due to the presence of SF3a66 in the coimmunoprecipitation assay. Several observations argue in favor of the former possibility. First, 3a60-N1 was efficiently coimmunoprecipitated with SF3a120 by anti-SF3a120 in the absence of SF3a66 (data not shown), indicating that SF3a66 does not stabilize this interaction. Second, both 3a60-N1 and -N2, but not -N3, bound to SF3a120 in the presence of GST-SF3a66 in a GST pull-down experiment (Fig. 3B). Notably, when the glutathione agarose was washed with 400 mM instead of 100 mM NaCl before elution, binding of 3a60-N1 and -N2, but not of full-length SF3a60 and 3a60-C3 and -C4, was reduced. Comparable results were obtained by coimmunoprecipitation with mAb66 (data not shown). Third, although it is possible that proteins with N-terminal deletions do not refold properly during the blotting procedure, we believe this is unlikely, because the corresponding in vitro-translated proteins failed to bind to SF3a120, when HeLa nuclear proteins were immobilized on nitrocellulose (data not shown). Therefore, the most likely explanation for our results is that the conditions for far-Western blotting are too stringent to detect weaker interactions between SF3a120 and SF3a60 proteins lacking the N-terminal 34 amino acids.
Together, these results demonstrate that the major site for contact with SF3a120 is confined to amino acids 35 to 107 of SF3a60. The N-terminal 34 amino acids stabilize this interaction, and sequences located between amino acids 108 and 240 may also contribute to a tight association. Sequences further C terminal, including the internal conserved region or the zinc finger domain, are not involved in binding of SF3a60 to SF3a120. Moreover, we have found no evidence that SF3a66 affects the SF3a60-SF3a120 interaction.
The N-terminal half of SF3a66, excluding the zinc finger domain, mediates binding to SF3a120.
Domains in SF3a66 required for binding to SF3a120 were examined by the same strategy as above. SF3a120 bound to full-length SF3a66 in a far-Western blot (Fig. 4). No interaction was observed with the C-terminal portion of SF3a66 (N1), consisting almost entirely of 22 heptad repeats (6). The N-terminal 216 amino acids were sufficient for binding to SF3a120 (C1). Moreover, the zinc finger domain is not involved in these interactions, since SF3a120 efficiently bound to 3a66ΔZn. Identical results were obtained by coimmunoprecipitation with anti-SF3a60 (data not shown). We therefore conclude that the N-terminal 216 amino acids of SF3a66, excluding the zinc finger domain, are necessary and sufficient for binding to SF3a120.
FIG. 4.
The N-terminal half of SF3a66 is required and sufficient for binding to SF3a120. The recombinant SF3a66 proteins indicated above the figure were separated by SDS-PAGE and transferred to nitrocellulose. The membrane was incubated with in vitro-translated [35S]methionine-labeled SF3a120 (see Fig. 2) and bound SF3a120 was visualized by autoradiography (top). Input proteins separated in a parallel gel were visualized by Western blotting with antipolyhistidine antibodies (bottom). The migration of molecular mass standards (in kilodaltons) is indicated to the left of each panel.
Individual SF3a subunits interact with the 15S U2 snRNP.
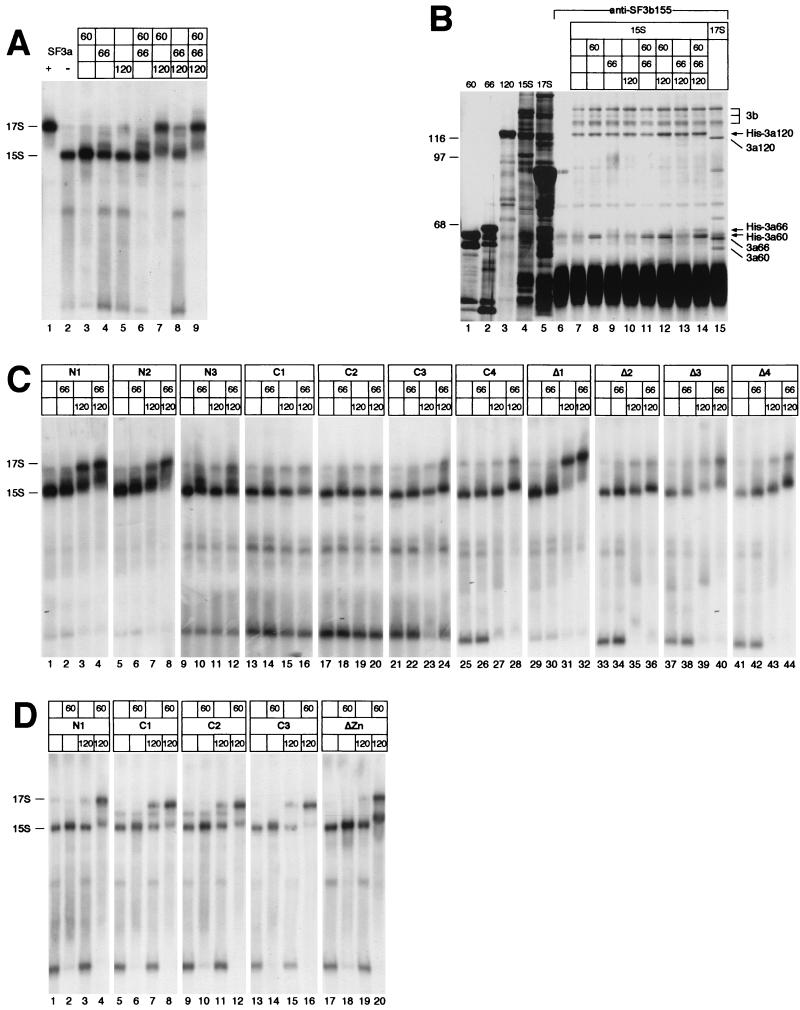
The 17S U2 snRNP forms in vitro by binding of SF3b to the 12S U2 snRNP, followed by interaction of SF3a with the intermediate 15S particle (7). Due to the salt-sensitive nature of the 17S U2 snRNP, the particle dissociates into SF3a, SF3b, and the 12S and 15S U2 snRNPs during chromatographic fractionation (8, 33). To determine the role of individual SF3a subunits in the formation of the mature 17S U2 snRNP, full-length proteins were incubated with Mono Q-purified 15S U2 snRNP. An end-labeled oligoribonucleotide complementary to the 5′ end of U2 snRNA was added to the reactions to detect snRNP complexes after electrophoresis in native polyacrylamide gels (Fig. 5A). A control experiment showed that the 15S U2 snRNP (lane 2) was quantitatively converted into the lower-mobility 17S particle in the presence of SF3a purified from HeLa cells (lane 1) or the three recombinant SF3a subunits (lane 9), demonstrating that the recombinant proteins substitute for HeLa SF3a.
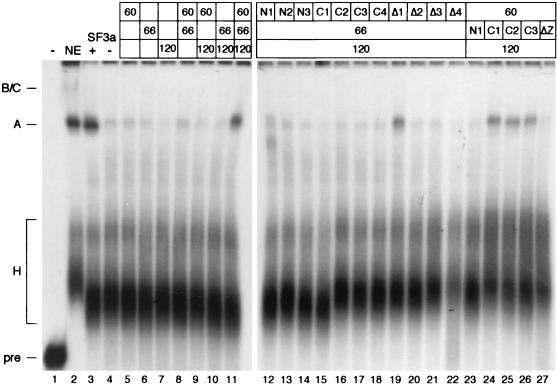
FIG. 5.
The zinc finger domains in SF3a60 and SF3a66 are required for their integration into the active 17S U2 snRNP. (A) SF3a purified from HeLa cells (+), buffer (−), or recombinant SF3a subunits, as indicated above the figure, were incubated under splicing conditions with partially purified 15S U2 snRNP in the presence of a 5′-end-labeled oligoribonucleotide complementary to the 5′ end of U2 snRNA. RNP complexes were separated in a native 4% polyacrylamide gel at room temperature and exposed to X-ray film. The positions of the 15S and 17S U2 snRNPs are indicated to the left. (B) Recombinant SF3a subunits were incubated with the 15S U2 snRNP in the combinations shown above the figure and subjected to immunoprecipitation with anti-SF3b155 antibodies coupled to protein G-Sepharose (lanes 6 to 14). Precipitation of the 17S U2 snRNP served as a control (lane 15). Input (lanes 1 to 5) and bound proteins were separated by SDS-PAGE and stained with silver. The positions of SF3b155, SF3b145, and SF3b130 are indicated by a bracket labeled 3b. Recombinant SF3a proteins and SF3a subunits associated with the 17S U2 snRNP are indicated by arrows and lines, respectively. The migration of molecular mass standards (in kilodaltons) is indicated to the left. (C) Derivatives of SF3a60 were incubated with the 15S U2 snRNP and recombinant full-length SF3a66 and SF3a120 as indicated above the figure. The analysis of RNP complexes was performed as in panel A, except that the complexes were resolved at 4°C. (D) Derivatives of SF3a66 were incubated with the 15S U2 snRNP and recombinant full-length SF3a60 and SF3a120 as indicated above the figure. The analysis of RNP complexes was performed as in panel A.
When individual SF3a subunits were incubated with the 15S U2 snRNP, subunit-specific mobility shifts were observed. SF3a60 increased the intensity of the band migrating at the position of the 15S U2 snRNP, in addition to reducing the abundance of faster-migrating complexes of unknown composition (Fig. 5A, lane 3). In the presence of SF3a66, one or two bands appeared that migrated more slowly than the 15S U2 snRNP (lane 4). Addition of SF3a120 generated a weak band migrating at the position of the 17S U2 snRNP (lane 5). In the experiment shown, this band is barely visible; however, it was readily detected in other experiments. Combining SF3a60 and SF3a66 with the 15S U2 snRNP had additive effects, suggesting that these proteins associate with the 15S particle independently (lane 6). Incubation of SF3a60 and SF3a120 with the 15S U2 snRNP resulted in a complex migrating at the position of the 17S U2 snRNP in addition to a band migrating slightly slower than the 15S particle (lane 7). Formation of these complexes was highly efficient, suggesting that SF3a60 and SF3a120 stabilize each other's binding to the 15S U2 snRNP. A combination of SF3a66 and SF3a120 appeared mostly additive in nature, except for a more efficient formation of a complex comigrating with the 17S U2 snRNP (lane 8).
In summary, each SF3a subunit appears to bind independently to the 15S U2 snRNP, resulting in subunit-specific mobility shifts in native polyacrylamide gels. Addition of SF3a66 in combination with either SF3a60 or SF3a120 had mostly additive effects on the mobility of the 15S U2 snRNP, whereas SF3a60 and SF3a120 may interact with the 15S U2 snRNP in a cooperative fashion.
Because the mobility shifts with certain combinations of SF3a subunits were rather weak, the incorporation of the proteins into the 15S U2 snRNP was tested by coimmunoprecipitation with anti-SF3b155 (Fig. 5B). The antibody precipitated four high-molecular-mass proteins from the 15S U2 snRNP fraction, the three largest of which represent SF3b155, SF3b145, and SF3b130 (lane 7). The fourth protein (of ∼120 kDa) has previously been shown to cosediment with the 15S U2 snRNP in glycerol gradients, but its identity is unknown (33). The small SF3b subunit of 49 kDa was obscured by the IgG heavy chain. When recombinant SF3a subunits were preincubated with the 15S U2 snRNP, SF3a60 was efficiently bound in all combinations tested (lanes 8, 11, 12, and 14). Binding of SF3a66 was barely detectable; however, it was clearly precipitated in the presence of SF3a60 and SF3a120 (lane 14). Although SF3a120 comigrated with the 120-kDa protein present in the 15S U2 snRNP fraction, the intensity of the 120-kDa band was increased in immunoprecipitations performed in the presence of SF3a60 and/or SF3a66 (as confirmed by quantification of the bands; data not shown), suggesting that SF3a120 is incorporated into the 15S U2 snRNP in the presence of the other subunits (lanes 12 to 14). These results are in agreement with the data presented in Fig. 5A, in that a low degree of coprecipitation of SF3a66 and SF3a120 by anti-SF3b155 correlates with weak changes in the mobility of the 15S U2 snRNP. Only SF3a60 is efficiently bound to the 15S U2 snRNP in the absence of the other subunits, suggesting that it plays a major role in the recruitment of at least SF3a120 into the U2 snRNP.
The C terminus of SF3a60 including the zinc finger domain is essential for the formation of the 17S U2 snRNP.
To determine sequences in SF3a60 required for integration into the 17S U2 snRNP, SF3a60 deletion mutants were incubated with the 15S U2 snRNP, either alone or in combination with SF3a66 and/or SF3a120, and tested in mobility shift assays. SF3a60 proteins with N-terminal deletions caused mobility shifts similar to full-length SF3a60 when incubated with the 15S U2 snRNP alone (Fig. 5C, lanes 1, 5, and 9) or in the presence of SF3a66 (lanes 2, 6, and 10), indicating that amino acids 1 to 322 of SF3a60 are dispensable for binding to the 15S particle. However, deletion of more than 10 N-terminal amino acids negatively affected the ability of the truncated proteins to shift the 15S U2 snRNP in the presence of SF3a120 or SF3a66 and SF3a120. Whereas 3a60-N1 behaved as full-length SF3a60 (cf. lanes 3 and 4 in Fig. 5C to lanes 7 and 9 in Fig. 5A), the conversion of the 15S U2 snRNP into complexes of slower mobility was reduced upon incubation with 3a60-N2 (cf. lanes 7 and 8 in Fig. 5C with lanes 7 and 9 in Fig. 5A) and highly impaired in the presence of 3a60-N3 (cf. lanes 11 and 12 in Fig. 5C to lanes 7 and 9 in Fig. 5A). These data show that the N terminus of SF3a60 is not necessary for interaction with the 15S U2 snRNP per se. However, because this region is required for binding of SF3a60 to SF3a120 (Fig. 3), we conclude that the interaction between the two proteins is important for the formation of the 17S U2 snRNP.
Incubation of the 15S U2 snRNP with SF3a60 proteins deleted for C-terminal sequences (C1 to C4) did not promote mobility changes in any combination tested (Fig. 5C, lanes 13 to 28), suggesting that the C-terminal 100 amino acids of SF3a60 are essential for its association with the 15S U2 snRNP. (Please note that in control reactions, similar to those presented in Fig. 5A, shifts of the 15S U2 snRNP in the presence of full-length SF3a60 and SF3a66 or SF3a60 and SF3a120 were less pronounced than those seen in Fig. 5A.) To test whether the zinc finger domain in the C-terminal region is involved in binding of SF3a60 to the 15S U2 snRNP, proteins with internal deletions covering the zinc finger domain and neighboring sequences were examined. Incubation of the 15S U2 snRNP in the presence of SF3a60 lacking only the zinc finger domain (Δ2, lanes 33 to 36), the region N terminal of this motif (Δ3, lanes 37 to 40), or the entire zinc finger region (Δ4, lanes 41 to 44) did not significantly change the mobility of the 15S particle regardless of whether SF3a66 or SF3a120 were present. These data show that the zinc finger and 30 amino acids N terminal to this domain are essential for binding of SF3a60 to the U2 snRNP. Moreover, although all SF3a60 proteins with C-terminal truncations or internal deletions of the zinc finger region bound SF3a120 (Fig. 3), this interaction apparently does not suffice to stably tether SF3a60 to the U2 snRNP.
Finally, we tested whether the region of high homology in metazoan SF3a60 (Δ1) was necessary for recruitment into the U2 snRNP. Deletion of these sequences did not impair binding of SF3a60 to the 15S particle in any combination tested (Fig. 5C, lanes 29 to 32), indicating that this region is dispensable for the formation of the 17S U2 snRNP.
The zinc finger domain of SF3a66 is required for 17S U2 snRNP formation.
Analysis of truncated derivatives of SF3a66 demonstrated that the region encompassing the C-terminal heptad repeats (N1) did not interact with the 15S U2 snRNP nor promote the formation of the 17S U2 snRNP in the presence of the other subunits (Fig. 5D, lanes 1 to 4), because all mobility shifts resembled those generated in the absence of SF3a66 in the control reactions (Fig. 5A). In contrast, SF3a66 proteins containing the N-terminal 216 or 258 amino acids (C1 and C2) bound to the 15S U2 snRNP either alone or in the presence of SF3a60 or SF3a120 and converted it into a complex of slower mobility in the presence of both subunits (lanes 5 to 12). Surprisingly, this complex migrated faster than the 17S U2 snRNP assembled in control reactions with full-length SF3a66. Because complexes formed in the absence of SF3a66 migrated at the same position as the mature 17S particle (cf. lanes 7 and 9 in Fig. 5A), it is unlikely that the smaller sizes of 3a66-C1 and -C2 are responsible for the faster migration. Possibly, the conformation of these truncated proteins is such that their incorporation into the U2 snRNP results in an aberrant mobility of the particle. As shown below, both proteins were active in prespliceosome assembly, suggesting that the U2 snRNP formed in their presence is functional. Incubation of 3a66-C3 with the 15S U2 snRNP in the absence or presence of the other subunits resulted in barely detectable mobility shifts (lanes 13 to 16). However, the complex formed together with SF3a60 and SF3a120 migrated similarly to the complex assembled in the corresponding reactions with 3a66-C1 and -C2 (cf. lane 16 with lanes 8 and 12). Moreover, 3a66-C3 contains all sequences present in 3a66-C1 and -C2 and is active in prespliceosome assembly, suggesting that 3a66-C3 is incorporated into the U2 snRNP. Similar to what we observed with SF3a60, deletion of the zinc finger domain of SF3a66 (ΔZn) abolished the interaction with the 15S U2 snRNP regardless of the presence of the other subunits (lanes 17 to 20), since all characteristic mobility shifts generated in the presence of SF3a66 in the control reactions (Fig. 5A) were absent. Therefore, the zinc finger domain of SF3a66 represents at least one site of interaction with the 15S U2 snRNP and is required for the formation of the 17S U2 snRNP.
Assembly of presplicing complex A requires all SF3a subunits.
Presplicing complex A forms upon binding of the 17S U2 snRNP to the pre-mRNA (3). This reaction can be reproduced in vitro by incubation of the 12S U2 snRNP, SF3b, and SF3a with other partially purified splicing factors (U1 snRNP, U2AF, SF1, and SR proteins) (8, 33). Complex A and a small amount of splicing complexes B and C were assembled in a control reaction with HeLa cell nuclear extract (Fig. 6, lane 2). In the reconstituted system, complex A was efficiently formed in the presence of HeLa SF3a (lane 3), but not in its absence (lane 4). When HeLa SF3a was exchanged for recombinant full-length subunits, complex formation occurred only in the presence of all three proteins (lane 11). These results clearly demonstrate that the recombinant SF3a subunits can assemble into a functional SF3a heterotrimer and all subunits are essential for spliceosome assembly.
Sequences in SF3a60 and SF3a66 necessary for binding to SF3a120 and assembly of the 17S U2 snRNP are essential for prespliceosome formation.
To analyze sequences in SF3a60 and SF3a66 required for A complex formation, truncated proteins or proteins with internal deletions were added to the reconstitution reaction together with the remaining two subunits. In the presence of 3a60-N1, two minor complexes formed, one migrating more slowly, the other faster than complex A (Fig. 6, lane 12). We have not investigated the nature of these complexes; however, since 3a60-N1 interacted weakly with SF3a120 and promoted the formation of the 17S U2 snRNP (Fig. 3 and 5), an inefficient assembly of complex A (or related complexes) may be expected. Further deletion of N-terminal sequences (N2 and N3) completely abolished the ability of the proteins to participate in A complex formation (Fig. 6, lanes 13 and 14). The same result was obtained with all C-terminal deletions (C1 to C4, lanes 15 to 18), and proteins carrying deletions in the zinc finger region (Δ2 to Δ4, lanes 20 to 22). Only the deletion of the conserved internal region (Δ1, lane 19) was without effect on A complex assembly. Together, these results demonstrate that the N-terminal sequences required for stable interaction of SF3a60 with SF3a120 and the zinc finger region, which is necessary for incorporation of SF3a into the 17S U2 snRNP, are essential for SF3a60 function in spliceosome assembly.
Sequences in SF3a66 necessary for A complex assembly are confined to the N-terminal 216 amino acids (N1 and C1 to C3; Fig. 6, lanes 23 to 26), and the zinc finger domain is also required for this reaction (ΔZn, lane 27). Thus, the N-terminal half of SF3a66 is essential and sufficient for its function at early steps of splicing and again, no role for the C-terminal heptad repeats could be established. In conclusion, all domains in SF3a60 and SF3a66 that participate in the formation of the SF3a heterotrimer and the incorporation of SF3a into the mature U2 snRNP are essential for the assembly of the prespliceosome.
Recombinant SF3a66 and SF3a120 bind nonspecifically to RNA.
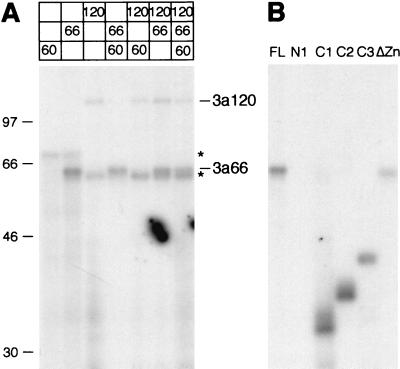
The SF3a subunits contact a region upstream of the intron branch site in isolated prespliceosomes in a sequence-independent manner (22). When SF3a purified from HeLa cells was used in UV cross-linking assays in the absence of other splicing factors, none of the subunits bound to RNA (8). This raised the question of whether the SF3a subunits bound pre-mRNA only in the presence of other splicing components or whether the concentration of purified SF3a was insufficient for detection of protein-RNA interactions. To address this issue, recombinant SF3a subunits were incubated with a radiolabeled pre-mRNA derived from the AdML transcription unit in the absence of other components. Proteins were UV cross-linked to RNA and separated by SDS-PAGE after digestion of unprotected RNA. As shown in Fig. 7A, SF3a66 and SF3a120 cross-linked to the pre-mRNA, whereas SF3a60 did not, even when tested in the presence of SF3a66 and/or SF3a120. Thus, SF3a60 appears to bind RNA only in the context of the prespliceosome (22), suggesting that components other than SF3a66 and SF3a120 are necessary for its binding to pre-mRNA. Binding of SF3a66 and SF3a120 was not sequence specific, since both proteins also cross-linked to a control RNA derived from vector sequences (data not shown). This result is consistent with the sequence variability of the anchor site (22).
FIG. 7.
Recombinant SF3a66 and SF3a120 bind RNA. Full-length SF3a60, SF3a66, and SF3a120 (A) or full-length and mutant SF3a66 proteins (B) were UV cross-linked to synthetic [α-32P]UTP-labeled RNA, treated with RNases A and T1, and separated by SDS-PAGE. The gel was dried and exposed to X-ray film. The positions of SF3a66 and SF3a120 are shown. Asterisks indicate binding of unrelated proteins. Molecular mass standards (in kilodaltons) are shown on the left.
Analysis of mutant proteins by UV cross-linking showed that deletion of the N-terminal 257 amino acids of SF3a66 (N1) completely abolished RNA binding, whereas SF3a66 deleted for C-terminal sequences (C1 to C3) bound to RNA (Fig. 7B). SF3a66 lacking the zinc finger domain (ΔZn) cross-linked to RNA; however, it did so consistently less efficiently than proteins containing this domain. Thus, similar to other functions tested, sequences essential for RNA binding of SF3a66 are restricted to the N-terminal half of the protein. The zinc finger domain is dispensable for RNA binding, but may contribute to binding efficiency.
DISCUSSION
As a first step toward understanding the roles of individual SF3a subunits, recombinant SF3a60, SF3a66, and SF3a120 have been expressed in insect cells. Purified recombinant proteins readily formed the SF3a heterotrimer, which behaved identically to SF3a purified from HeLa cells. All subunits are essential for the assembly of the 17S U2 snRNP and for the function of SF3a in prespliceosome formation.
Interactions between SF3a subunits and with the 15S U2 snRNP.
Recombinant SF3a subunits interacted with each other as predicted from previous studies (6, 13, 31): SF3a60 and SF3a66 bound to SF3a120, but did not associate with one another (Fig. 3 and 4). Moreover, the binding of SF3a60 or SF3a66 to SF3a120 was not influenced by the presence of the third subunit (data not shown), suggesting that SF3a60 and SF3a66 probably do not contact each other within the SF3a heterotrimer.
Earlier analyses showed that SF3a does not associate with either SF3b or the 12S U2 snRNP alone, but only binds to the 15S U2 snRNP (7, 33). In the present study, single SF3a subunits were shown to interact with the 15S U2 snRNP independently, resulting in subunit-specific mobility changes of the particle (Fig. 5A), which suggests the existence of different binding sites for each subunit in the 15S U2 snRNP. The combined results of the mobility shift assay and coimmunoprecipitation of the SF3a subunits with the 15S U2 snRNP by anti-SF3b155 suggest that SF3a60 binds efficiently to the 15S U2 snRNP, but interactions of SF3a66 and SF3a120 with the particle are weak (Fig. 5A and B). Binding was not considerably strengthened when the 15S U2 snRNP was incubated with combinations of SF3a60 and SF3a66 or SF3a66 and SF3a120, indicating that these subunits do not, or only weakly, influence each other's interaction with the 15S U2 snRNP. In contrast, the 15S U2 snRNP was quantitatively converted into more slowly migrating complexes in the presence of SF3a60 and SF3a120, and both subunits were precipitated with the 15S U2 snRNP by anti-SF3b155. These results suggest that the binding of SF3a120 to the U2 particle is stabilized by its interaction with SF3a60. In support of this, an intact SF3a120 binding site of SF3a60 is required for efficient formation of the 17S U2 snRNP. Although a large proportion of the particles formed in the presence of SF3a60 and SF3a120 migrated at the position of the 17S U2 snRNP, these particles did not support prespliceosome assembly (Fig. 6). SF3a66 was required to convert the U2 snRNP into its active form, most likely through stabilization of existing interactions and/or a conformational change within the particle to promote the interaction of the U2 snRNP with the intron branch site.
In summary, our results indicate that SF3a60 plays a major role in recruiting SF3a120 into the U2 snRNP. Weak interactions of SF3a66 and SF3a120 with specific binding sites in the 15S U2 snRNP appear to be stabilized through protein-protein contacts within the SF3a heterotrimer, and all three subunits fulfill essential functions in the assembly of the active U2 snRNP.
Functional domains in SF3a60.
Analysis of SF3a60 proteins with N- or C-terminal or internal deletions revealed a modular structure of functional domains. The N-terminal portion interacts with SF3a120, whereas the C-terminal region (including the zinc finger and sequences immediately N terminal to this domain) establishes direct contacts with the 15S U2 snRNP. Both regions are required for the assembly of the prespliceosome.
SF3a60 binds SF3a120 through its N-terminal 107 amino acids (Fig. 2 and 3), extending previous results that deletion of the N-terminal 92 amino acids abolished the interaction with SF3a120 (13). An essential binding site is located between amino acids 35 and 107 of SF3a60, whereas the N-terminal 34 amino acids appear to stabilize the interaction with SF3a120. At present, we cannot predict specific amino acids in SF3a60 that mediate binding to SF3a120. The N-terminal region of the protein does not contain any protein motifs indicative of a role in protein-protein interactions, and only a few, relatively short stretches of high sequence conservation are apparent in multiple sequence alignments (see the introduction). The N-terminal 25 amino acids of human SF3a60 are well conserved in all homologues, making this region a good candidate for a site that stabilizes the SF3a60-SF3a120 interaction. Mutational analysis of S. cerevisiae Prp9p has shown that amino acids 78 and 177 are required for binding to Prp21p (37). Amino acid 78 of Prp9p is conserved in metazoan and yeast SF3a60 counterparts and is present in a block of relatively high sequence similarity corresponding to amino acids 77 to 97 of human SF3a60, suggesting that this region may represent (at least) one binding site. In contrast, amino acid 177 of Prp9p lies outside of the region that is sufficient for the SF3a60-SF3a120 interaction and is not conserved.
N-terminal sequences are not essential for direct physical contact between SF3a60 and the 15S U2 snRNP, but appear to stabilize the incorporation of both SF3a60 and SF3a120 into the mature U2 particle (Fig. 5C). Binding of SF3a60 to the 15S U2 snRNP is established through sequences located at the C terminus encompassing the C2H2 zinc finger domain. SF3a60 with an internal deletion of the zinc finger domain failed to bind to the U2 snRNP, whether or not the remaining subunits were present. This result is consistent with findings with S. cerevisiae, in which deletion of the second zinc finger domain of Prp9p (which corresponds to the single zinc finger domain in metazoan and plant SF3a60) resulted in a dominant lethal phenotype (37). Based on this and other results, Legrain et al. (37) proposed that the Prp9p zinc finger binds to a factor X. Given our finding that the zinc finger domain of human SF3a60 is required for binding to the U2 particle, factor X most likely represents a component of the yeast equivalent of the 15S U2 snRNP.
In addition to the zinc finger domain, 30 amino acids N terminal to this domain appear to participate in the binding of SF3a60 to the 15S U2 snRNP. These sequences are highly conserved among all SF3a60 homologues. Moreover, the region encompassing the SF3a60 zinc finger domain and the N-terminal conserved sequences is exchangeable for the corresponding sequences in Prp9p (30), suggesting that this entire region carries out an evolutionarily conserved and crucial function. How the sequences N terminal to the zinc finger domain contribute to 15S U2 snRNP binding is not known. Since these sequences are necessary for binding of SF3a60 to the 15S U2 snRNP in the absence of the other subunits, they could either be directly involved in contacts with 15S U2 snRNP components or stabilize interactions mediated by the zinc finger domain. As noted previously (30, 35; data not shown), ∼13 amino acids N terminal to the SF3a60 zinc finger domain are well conserved with the corresponding sequences in SF3a66. Although we have not tested whether these sequences are necessary for the binding of SF3a66 to the 15S U2 snRNP, we believe this is highly likely, given that the zinc finger domains of both proteins are essential for incorporation into the mature U2 snRNP. Whether additional sequences N or C terminal to the SF3a60 zinc finger region contribute to U2 snRNP binding remains to be tested.
No function could be attributed to an internal sequence that is highly conserved in metazoans and A. thaliana SF3a60 (39; see the introduction). Homologues from S. cerevisiae, S. pombe, and N. crassa contain a second C2H2 zinc finger domain in the corresponding region (35; see the introduction). When the cysteine and histidine residues within this domain of S. cerevisiae Prp9p were mutated, only a substitution of the first histidine destroyed the function of the protein (35), but the role of this domain is unknown. The divergence of the internal sequences could indicate specialized functions specific to lower eukaryotes and metazoans. Nevertheless, the internal region of human SF3a60 is dispensable for prespliceosome assembly, and whether it plays a role in later steps in the splicing reaction or for the regulation of SF3a60 activity remains to be tested.
Functional domains in SF3a66.
The N-terminal 216 amino acids of SF3a66 are necessary and sufficient for the interaction with SF3a120 and the 15S U2 snRNP, prespliceosome assembly, and RNA binding (Fig. 4, 5D, 6, and 7). Interestingly, in multiple sequence alignments, the entire sequences of S. pombe and S. cerevisiae SF3a66 extend only two and six amino acids, respectively, beyond amino acid 216 of human SF3a66. These data support our definition of the N-terminal region of SF3a66 as containing all domains required for function in the basic splicing reaction, which is conserved from yeast to humans.
Furthermore, our results clearly show that the zinc finger domain of SF3a66, similar to that of SF3a60, plays a crucial role in direct binding to the 15S U2 snRNP. This feature appears to be conserved in S. cerevisiae, since Prp11p with a deletion of its zinc finger domain exhibited a dominant lethal phenotype (36). Because deletion of the SF3a66 zinc finger domain resulted in a reduction in general RNA binding (Fig. 7), this domain could also participate in the binding of SF3a66 to the pre-mRNA (22). The C-terminal half of human SF3a66, comprising 22 heptad repeats (6), is dispensable for all functions tested. Given the divergence of the C-terminal sequences between mammals, other metazoans, and plants and their absence in S. cerevisiae and S. pombe, it is possible that these regions function in regulatory events that differ from one organism to another.
Potential targets in the 15S U2 snRNP for interaction with the SF3a60 and SF3a66 zinc finger domains.
Zinc finger domains were initially identified as nucleic acid-binding domains; however, a number of zinc fingers (including those of the C2H2 type) mediate protein-protein interactions (for review, see reference 38). We favor the hypothesis that the zinc finger domains of SF3a60 and SF3a66 contact protein components of the 15S U2 snRNP for the following reason. The zinc finger domains of the two proteins are related to the zinc finger domain in the U1 snRNP-specific protein C (30, 35). U1 C does not bind to naked U1 snRNA, but requires the U1 70,000-molecular-weight (70K) protein for association with the core U1 snRNP, which consists of the seven Sm proteins bound to U1 snRNA (41, 42). Incorporation of U1 C into the U1 snRNP is mediated by its zinc finger domain, and chemical cross-linking revealed interactions between U1 C and the Sm proteins B and B′ (41, 42). In analogy to these results, we propose that the incorporation of SF3a into the U2 snRNP occurs through protein-protein interactions of the zinc finger domains of SF3a60 and SF3a66 with one or more of the Sm proteins. Our results suggest that SF3a60 and SF3a66 have distinct binding sites within the 15S U2 snRNP. As the Sm proteins share a common evolutionarily conserved domain (14, 25, 47), contacts of the zinc finger domains of SF3a60 and SF3a66 with different Sm proteins can be envisioned. The fact that direct interactions between the SF3a subunits and Sm proteins have not been reported does not contradict our hypothesis. As previously suggested (33), binding of SF3b to the 12S U2 snRNP may induce a conformational change in the structure of the Sm domain of the U2 snRNP, thus generating a favorable platform for binding of SF3a. Interactions of one or more SF3a subunits with the Sm domain of the U2 snRNP are also compatible with biochemical and electron microscopic studies indicating that SF3a primarily binds to the 3′ portion of the U2 snRNP (33). The Sm-related protein Lsm4p has recently been found as a partner of Prp11p in a two-hybrid screen (20). Although domains that mediate the Prp11p-Lsm4p interaction have not been reported, this finding may be taken as further evidence that an interaction between SF3a66 (or SF3a60) and Sm or Sm-like proteins would in principal be possible.
Interactions of SF3a60 and SF3a66 with Sm proteins would not exclude contacts with other proteins, for example, the subunits of SF3b or the U2 snRNP-specific A′ and B" proteins. To date, a physical interaction between S. cerevisiae Prp9p and Rse1p (the homologue of SF3b130) (10, 16, 40) has been demonstrated in a two-hybrid screen (19), and Prp11p interacts genetically with Cus1p (the homologue of SF3b145) (51).
The zinc finger domains of SF3a60 and SF3a66 could also bind directly to U2 snRNA; however, we consider this possibility less likely. Although all SF3a subunits bind to the pre-mRNA in the context of the spliceosome (22), and recombinant SF3a66 and SF3a120 cross-link to RNA in the absence of other components (Fig. 7), our finding that the zinc finger domain of SF3a66 is not essential for RNA binding argues against an interaction of the zinc finger domain with the U2 snRNA. The yeast counterparts of the SF3a subunits interact genetically with U2 snRNA (45, 50, 55); however, these interactions could be indirect and, for example, be mediated via SF3b subunits (51, 55). Also, we never observed cross-linking of any SF3a subunit to synthetic U2 snRNA under conditions in which cross-links with SF3b subunits were readily detected (K. Gröning and A. Krämer, unpublished observation).
In the present study, we have analyzed functions and functional domains of single polypeptides of a multisubunit splicing factor by testing recombinant proteins in in vitro reconstitution assays. With the system described, it should also be possible to dissect functional domains in SF3a120 and address the roles of individual subunits of SF3b. In addition, our analysis could be extended to reveal functions of individual SF3a or SF3b subunits at later steps of the splicing reaction.
ACKNOWLEDGMENTS
We thank A. Lamond for the U2a oligoribonucleotide, M. Schmidt-Zachmann for affinity-purified anti-SF3b155 antibodies, F. Mulhauser for cloning of SF3a66, A.-M. Tourmel for technical assistance, and G. Bilbe, G. Moreau, and J. Steitz for comments on the manuscript. D.N. thanks P. Grüter for helpful discussions throughout this work.
D.N. was a recipient of an EMBO long-term fellowship. This work was supported by grants from the Schweizerischer Nationalfonds and the Canton of Geneva to A.K.
REFERENCES
- 1.Adams M D, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 2.Arning S, Grüter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens S-E, Galisson F, Legrain P, Lührmann R. Evidence that the 60-kDa protein of 17S U2 small nuclear ribonucleoprotein is immunologically and functionally related to the yeast PRP9 splicing factor and is required for the efficient formation of prespliceosomes. Proc Natl Acad Sci USA. 1993;90:8229–8233. doi: 10.1073/pnas.90.17.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 6.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1993;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 7.Brosi R, Gröning K, Behrens S-E, Lührmann R, Krämer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 8.Brosi R, Hauri H P, Krämer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 9.Burge C, Tuschl T, Sharp P. Splicing of precursors to mRNA by the spliceosome. In: Gesteland R, Cech T, Atkins J, editors. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 10.Caspary F, Shevchenko A, Wilm M, Séraphin B. Partial purification of the yeast U2 snRNP reveals novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 12.Chang T-H, Clark M W, Lustig A J, Cusick M E, Abelson J. RNA11 protein is associated with the yeast spliceosome and is localized in the periphery of the cell nucleus. Mol Cell Biol. 1988;8:2379–2393. doi: 10.1128/mcb.8.6.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiara M D, Champion-Arnaud P, Buvoli M, Nadal-Ginard B, Reed R. Specific protein-protein interactions between the essential mammalian spliceosome-associated proteins SAP 61 and SAP 114. Proc Natl Acad Sci USA. 1994;91:6403–6407. doi: 10.1073/pnas.91.14.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper M, Parkes V, Johnston L H, Beggs J D. Identification and characterization of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das B K, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dresser D W, Hacker A, Lovell-Badge R, Guerrier D. The genes for a spliceosome protein (SAP62) and the anti-Mullerian hormone (AMH) are contiguous. Hum Mol Genet. 1995;4:1613–1618. doi: 10.1093/hmg/4.9.1613. [DOI] [PubMed] [Google Scholar]
- 19.Fromont-Racine M, Rain J-C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 20.Fromont-Racine M, Mayes A E, Brunet-Simon A, Rain J-C, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs J D, Legrain P. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast. 2000;17:95–110. doi: 10.1002/1097-0061(20000630)17:2<95::AID-YEA16>3.0.CO;2-H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins (SAPs) and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 23.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18:4752–4760. doi: 10.1128/mcb.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol. 2000;12:346–350. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 25.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodges P E, Beggs J D. RNA splicing. U2 fulfils a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 27.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 28.Krämer A. Pre-splicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988;2:1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- 29.Krämer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krämer A, Legrain P, Mulhauser F, Gröning K, Brosi R, Bilbe G. Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 1994;22:5223–5228. doi: 10.1093/nar/22.24.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krämer A, Mulhauser F, Wersig C, Gröning K, Bilbe G. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PRP21p of S. cerevisiae. RNA. 1995;1:260–272. [PMC free article] [PubMed] [Google Scholar]
- 32.Krämer A. The structure and function of proteins involved in nuclear pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 33.Krämer A, Grüter P, Gröning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyhse-Anderson J. Electroblotting of multiple gels. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- 35.Legrain P, Choulika A. The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J. 1990;9:2775–2781. doi: 10.1002/j.1460-2075.1990.tb07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legrain P, Chapon C. Interaction between PRP11 and SPP91 yeast splicing factors and characterization of a PRP9-PRP11-SPP91 complex. Science. 1993;262:108–110. doi: 10.1126/science.8211114. [DOI] [PubMed] [Google Scholar]
- 37.Legrain P, Chapon C, Galisson F. Interactions between PRP9 and SPP91 splicing factors identify a protein complex required in prespliceosome assembly. Genes Dev. 1993;7:1390–1399. doi: 10.1101/gad.7.7b.1390. [DOI] [PubMed] [Google Scholar]
- 38.Mackay J P, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1–4. doi: 10.1016/s0968-0004(97)01168-7. [DOI] [PubMed] [Google Scholar]
- 39.Meyer V, Oliver B, Pauli D. Multiple developmental requirements of Noisette, the Drosophila homolog of the U2 snRNP-associated polypeptide SF3a60. Mol Cell Biol. 1998;18:1835–1843. doi: 10.1128/mcb.18.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mintz P J, Patterson S D, Neuwald A F, Spahr C S, Spector D L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelissen R L, Heinrichs V, Habets W J, Simons F, Lührmann R, van Venrooij W J. Zinc finger-like structure in U1-specific protein C is essential for specific binding to U1 snRNP. Nucleic Acids Res. 1991;19:449–454. doi: 10.1093/nar/19.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelissen R L, Will C L, van Venrooij W J, Lührmann R. The association of the U1-specific 70K and C proteins with U1 snRNPs is mediated in part by common U snRNP proteins. EMBO J. 1994;13:4113–4125. doi: 10.1002/j.1460-2075.1994.tb06729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nilsen T. RNA-RNA interactions in nuclear pre-mRNA splicing. In: Simons R, Grunberg-Manago M, editors. RNA structure and function. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. pp. 279–307. [Google Scholar]
- 44.Rain J-C, Rafi Z, Rhani Z, Legrain P, Krämer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4:551–565. doi: 10.1017/s1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruby S W, Chang T-H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt-Zachmann M, Knecht S, Krämer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wells S E, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr Cus1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 52.Wiest D K, O'Day C L, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- 53.Will C, Lührmann R. SnRNP structure and function. In: Krainer A, editor. Eukaryotic mRNA processing. IRL Press, Oxford, United Kingdom. 1997. pp. 130–173. [Google Scholar]
- 54.Will C L, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 55.Yan D, Ares M., Jr Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol. 1996;16:818–828. doi: 10.1128/mcb.16.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]