A comprehensive analysis of gene expression and DNA regulation in ccRCC using scATAC-seq and scRNA-seq reveals the DNA regulatory programs of ccRCC at the single-cell level.
Abstract
Clear cell renal cell carcinoma (ccRCC) frequently features a high level of tumor heterogeneity. Elucidating the chromatin landscape of ccRCC at the single-cell level could provide a deeper understanding of the functional states and regulatory dynamics underlying the disease. Here, we performed single-cell RNA sequencing (scRNA-seq) and single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) on 19 ccRCC samples, and whole-exome sequencing was used to understand the heterogeneity between individuals. Single-cell transcriptome and chromatin accessibility maps of ccRCC were constructed to reveal the regulatory characteristics of different tumor cell subtypes in ccRCC. Two long noncoding RNAs (RP11-661C8.2 and CTB-164N12.1) were identified that promoted the invasion and migration of ccRCC, which was validated with in vitro experiments. Taken together, this study comprehensively characterized the gene expression and DNA regulation landscape of ccRCC, which could provide new insights into the biology and treatment of ccRCC.
Significance:
A comprehensive analysis of gene expression and DNA regulation in ccRCC using scATAC-seq and scRNA-seq reveals the DNA regulatory programs of ccRCC at the single-cell level.
Introduction
Kidney cancer is one of the most common cancers in the urinary system, accounting for 179,368 deaths worldwide in 2020 (1). In addition to the large number of people dying from this disease, the incidence of kidney cancer is increasing annually (2, 3). Clear cell renal cell carcinoma (ccRCC) is the most common type of kidney cancer, accounting for approximately 60% to 80% of all primary cases (1, 3). In recent years, due to the extensive and in-depth research on the molecular biology and genetics of ccRCC, as well as the improved understanding of the mechanism of its occurrence and development, ccRCC targeted therapy and immune checkpoint inhibitor therapy have achieved a number of breakthroughs and great progress (4, 5). However, the overall improvement of disease-free survival rate is still very limited; advanced renal cell carcinoma (RCC) still confers a high mortality (4, 5).
ccRCC is a disease with a potentially high level of tumor heterogeneity and inherited predisposition (6, 7). ccRCC arises from genes involved in regulating cellular metabolism, also known as the “Warburg effect” (6). VHL gene mutation is the most frequent mutation of ccRCC, which is closely associated with tumor pathogenesis (8). In addition, genes such as PBRM1, BAP1, and SETD2 were found to be altered in ccRCC (9–11). Here, The Cancer Genome Atlas (TCGA) project on ccRCC, which studied a large cohort of patients with ccRCC at the DNA, RNA, and protein levels and important molecular pathways, has illustrated well the genetic characteristics of ccRCC (12). Although TCGA project on ccRCC showed significant findings at that time, it still appears to have limitations due to using the bulk tissue samples, which caused difficulty in the replacement of individual tumor cells. Considering the intratumor heterogeneity of ccRCC, tumors evolve to become masses of cells with distinctive genomic profiles (7). Therefore, in recent years, many single-cell RNA sequencing (scRNA-seq) studies of ccRCC have revealed the characteristics of tumor cells, tumor-specific markers, and tumor immune microenvironment at the single-cell level (13–17). However, previous studies mostly focused on the single-cell RNA level, neglecting the epigenetic regulatory of ccRCC.
Variation between tumor cells or individuals is a common feature of life, also known as tumor heterogeneity (18). Single-cell assay for transposase-accessible chromatin using sequencing (scATAC-seq) is a robust method to understand the fundamental mechanisms of variability from identical DNA sequences (19). In a previous study, the human single-cell atlas of chromatin accessibility was established by scATAC-seq, which provides a foundation for the analysis of gene regulatory programs in human cell types across tissues (20). In addition, the single-cell chromatin landscape of immune cells in ccRCC has been mapped to provide a rich resource for understanding the functional states and regulatory dynamics of immune cells (21). However, few scATAC-seq studies have focused on tumor cells, especially on ccRCC. The DNA regulatory programs of ccRCC remain elusive at the single-cell level. To address these challenges, we performed single-cell multiomics studies on 19 ccRCC samples. In this study, we revealed the epigenetic regulation characteristics of ccRCC by integrating scATAC-seq and scRNA-seq of 19 ccRCC samples (∼164,000 cells in total), combined with whole-exome sequencing (WES).
Materials and Methods
Ethics approval of ccRCC samples and cell lines
This study was approved by the Institutional Review Board of the First Affiliated Hospital of Guangxi Medical University (Nanning, P.R. China), and the authors obtained written informed consent from the patients. The authors confirmed that the study was conducted in accordance with Declaration of Helsinki.
The samples were obtained from 19 patients (Supplementary Table S1) undergoing laparoscopic partial nephrectomy or radical nephrectomy at the First Affiliated Hospital of Guangxi Medical University in China. Postoperative pathologic results revealed that all tumor samples were ccRCC. Considering tumor heterogeneity, we adopted the multisampling method, and at least four tumor tissues from different sites of each sample were selected. Human ccRCC cell lines 786-O and Caki-2 were purchased from Procell. The 786-O cells were cultured at 37°C and 5% CO2 atmosphere with RPMI1640 (WISENT, 350-006-CL) plus 10% FBS (Gibco, 10099141). Caki-2 cells were cultured at 37°C and 5% CO2 atmosphere with McCoy's 5A (Procell, PM150710A) plus 10% FBS (Gibco, 10099141). All ccRCC cell lines were confirmed within 6 months before use by using a short tandem repeat profiling and confirmed negative for Mycoplasma contamination by MycoBlue Mycoplasma Detector (Vazyme, D101-01).
Isolation of single-cell suspension
This step has been described in our previous study (22). Briefly, fresh tumor samples were obtained from the hospital and transferred to the laboratory in cold transfer buffer Hanks’ Balanced Salt Solution (HBSS; Gibco, 24020117; 5% FBS, Gibco, 10099141 and 1% penicillin/streptomycin, Gibco, 15240062) within 30 minutes. Each sample included at least four different tumor sites and avoided areas of tumor necrosis.
After being washed with 4°C Dulbecco's PBS (WISENT, 311-425-CL), the tissues were cut into 2–4 mm pieces with sterile scissors, washed and resuspended twice. The tissue specimens were digested for 30 minutes at 37°C in a digestion solution [1 mg/mL collagenase I (Gibco, 5401020001) and 1 mg/mL DNaseI (Roche, 10104159001) in HBSS]. The whole digestion process was terminated by DMEM (WISENT, 319-006-CL) with 10% FBS (Gibco, 10099141). Then, we used a 70 μm cell strainer (Falcon) to filter out large tissue fragments. Red blood cells (RBC) were removed by RBC lysis buffer (10× diluted to 1×; BioLegend, 420301). This process lasted for 5 minutes on ice. The samples were filtered through a 40 μm cell strainer (Falcon). Viable cells were counted after trypan blue (Gibco, 15250-061) staining. We ensured the cell viability of more than 80% in each sample. Here, some of the living cells were used for scRNA-seq directly, whereas some were subsequently isolated from single-cell nuclei. The rest were cryopreserved.
Isolation of single-cell nuclei
We added single cells into a 1.5 mL microcentrifuge tube, which were centrifuged at 300 × g for 5 minutes at 4°C. Then, we removed the supernatant without disrupting the cell pellet. Lysis buffer [10 mmol/L Tris-HCl (pH 7.4), 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.1% Tween-20, 0.1% Nonidet P40 Substitute, 0.01% digitonin, and 1% BSA] and wash buffer [10 mmol/L Tris-HCl (pH 7.4), 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.1% Tween-20, and 1% BSA] should be prepared on ice. About 100 μL of fresh lysis buffer was added to the pellet and pipette mixed 10 times. Then, we incubated the fresh lysis buffer with single cells on ice for 3–4.5 minutes. Here, we strongly recommend designing four different time gradients (3, 3.5, 4, and 4.5 minutes) because we found that the quality of nuclei was best in this range. We could select the optimal group for downstream sequencing according to the quality of the cell nucleus under the microscope. After lysis, we added 1 mL of wash buffer to the lysed cells and fully blended the samples. Next, the samples were centrifuged at 500 × g for 5 minutes at 4°C, and the supernatant was removed without disrupting the nuclei pellet. Finally, the single-cell nuclei were resuspended in chilled 1× Nuclei Buffer (10x Genomics, 2000153) at approximately 5,000–7,000 nuclei/μL.
cDNA library construction and sequencing for scRNA-seq
scRNA-seq was performed on the above single-cell suspensions in accordance with the standard protocol of 10x Genomics “Chromium Single Cell 3′ Reagent Kits User Guide” (https://support.10xgenomics.com/single-cell-gene-expression/index/doc/user-guide-chromium-single-cell-3-reagent-kits-user-guide-v3-chemistry). Briefly, the concentration of the single-cell suspensions was adjusted to 2,000 cells/μL by a hemocytometer. The number of target cells captured in each sample was 10,000. Then, we followed the steps of “Chromium Single Cell 3′ Reagent Kits User Guide” to construct the cDNA library.
The samples were sequenced by NovaSeq 6000 (Illumina). We used the following sequence parameters: read 1 for 150 cycles, read 2 for 150 cycles, and index for 14 cycles. Preliminary sequencing files (.bcl) were converted to FASTQ files by CellRanger (version 3.1.0, https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/what-is-cell-ranger) with the cellranger mkfastq function. FASTQ files were compared with the human genome reference sequence GRCh38 by the cellranger count function. After using the cellranger count function, the “filtered_feature_bc_matrix” file will be generated for secondary analysis.
DNA library construction and sequencing for scATAC-seq
Suitable nuclei concentrations were adjusted from the above experiment. As mentioned in the previous study (avoiding potential cell duplets; ref. 23), the number of target nucleus captured in each sample was 6,000. We adopted 10x Genomics “Chromium Single Cell ATAC Reagent Kits User Guide” for library preparation (https://support.10xgenomics.com/single-cell-atac/library-prep/doc/ technical-note-chromium-next-gem-single-cell-atac-v11-reagent-workflow-and-software-updates). Briefly, the ATAC Buffer (10x Genomics, 200122) and ATAC Enzyme (10x Genomics, 200123) were added to the 5 μL nuclei, which was incubated for 60 minutes at 37°C. We prepared “Master Mix,” including 56.5 μL of Barcoding Reagent B (10x Genomics, 2000194), 1.5 μL of Reducing Agent B (10x Genomics, 2000087) and 2 μL of Barcoding Enzyme (10x Genomics, 2000125). After mixing the nuclei with the Master Mix, we added it to the Chromium Chip E (10x Genomics, 2000121). At the same time, we added Partitioning Oil (10x Genomics, 220088) and Chromium Single Cell ATAC Gel Beads (10x Genomics, 2000132) to the Chromium Chip E. The Chromium Chip E was placed into a Chromium Single Cell Controller instrument (10x Genomics) before attaching a 10x Gasket (10x Genomics, 3000072). Then, the Chromium Single Cell Controller Instrument could be run.
The gel bead in emulsions (GEM) was generated after the above steps. Subsequently, GEMs were incubated and cleaned by using Dynabeads (10x Genomics, 2000048) and SPRIselect (Beckman Coulter, B23318). scATAC-seq library construction eliminates the process of reverse transcription compared with scRNA-seq. PCR was performed after “Sample Index PCR Mix” and Single Index Kit N Set A (10x Genomics, 1000212) were added into the samples. The details of PCR parameters could be referred to Chromium Single Cell ATAC Reagent Kits User Guide. DNA libraries were sequenced by NovaSeq 6000 (Illumina). Consistent with previous studies, we followed the 2 × 50 paired-end sequencing scheme: 50 bp read 1N, 8 bp i7 index, 16 bp i5 index, and 50 bp read 2N. Preliminary sequencing files (.bcl) were converted to FASTQ files by CellRanger ATAC (version 1.2.0, https://support.10xgenomics.com/single-cell-atac/software/pipelines/latest/ what-is-cell-ranger-atac) with the cellranger-atac mkfastq function. Then, FASTQ files were compared with the human genome reference sequence GRCh38 by the cellranger-atac count function. Here, 10x CellRanger ATAC also performed peak calling step, where a CellRanger peak merged distinct MACS2 (24) peaks into a single region. Finally, data were generated for downstream analysis.
scRNA-seq data analysis
We used R (version 4.0.2, https://www.r-project.org/) for secondary analysis of scRNA data. First, we used Seurat (25) R package for data quality control (QC) and downstream analysis. In general, one cell should be at least 500 genes, and the number of genes is less than twice of the median number of detected genes (potential cell duplets). The proportion of mitochondrial genes was recommended to be less than 10% (Supplementary Fig. S1). According to the above conditions, we filtered the cells that were obtained from the primary analysis and finally obtained high-quality cells for downstream analysis (Supplementary Table S2).
Nineteen samples were merged together by using the merge command. After data normalization, the highly variable genes were identified, and the top 2,000 genes were used for downstream analysis. Considering the cell-cycle genes in tumor cells that interfere with cell clustering, we need to remove this effect based on a previous study (26). Before cell clustering, we should eliminate the batch effect between 19 samples. Here, we used the Harmony (27) package to eliminate batch effect and generate uniform manifold approximation and projection (UMAP). We calculated the significant principal components from each sample by Jackstraw function. Thirty principal components were chosen as the appropriate cell clustering parameter, which affected the distribution of UMAP. At a resolution of 0.5, all cells were clustered by the FindClusters function from Seurat. We then used FindAllMarkers to calculate differentially expressed genes (DEG) between each cell type (Supplementary Table S3).
Reconstructing cell differentiation trajectories by RNA velocity and monocle3
Initially, RNA velocity (http://velocyto.org, version 0.6; ref. 28) was used to estimate the direction and rate of differentiation of tumor cells. Using the Bam file generated by CellRanger processing, we applied the velocyto function to generate the .loom file. Then, the SeuratWrappers package made it easy to integrate Seurat objects with .loom files. Finally, the rate and direction of each cell were calculated by RunVelocity function. We chose the following parameters: deltaT to 1, kCells to 25 and fit.quantile to 0.02.
The Monocle3 (29) R package (version 1.0.0) was used to reconstruct the cell fate decisions and pseudotime trajectories of tumor cells. After the tumor cell, subtypes (ccRCC 1, ccRCC 2, ccRCC 3, and ccRCC 4) were classified by Seurat, the differentiation trajectory of different cell subtypes was reconstructed. SeuratWrappers can convert Seurat objects into Monocle3 objects using the as.cell_data_set function. On the basis of the root of tumor cells, which was found by RNA velocity, we could reconstruct the trajectories of tumor cells using orderCells function.
Copy-number variation analysis of single-cell data
The inferCNV (https://github.com/broadinstitute/inferCNV) was used to perform copy-number variation (CNV) analysis on scRNA-seq data. The inferCNV is a R package that can estimate the chromosome copy-number characteristics from scRNA-seq data. In this study, we estimated CNV from four tumor cell types, whereas monocytes served as the control group. In general, the cut-off value of 0.1 was chosen for 10x Genomics scRNA-seq data.
Comparing present scRNA-seq data with those of previous work
The scRNA-seq data of tumor adjacent normal kidney tissues came from our previous study (GSE131685; ref. 30). Renal epithelial cells were selected and compared with the tumor cells in this study.
The relationship between tumor cell subtypes and prognosis
First, we selected the top 100 DEGs of four tumor cell subpopulations from Supplementary Table S3. TCGA database on ccRCC was used to detect the association of these genes with prognosis. We obtained three results: positive prognosis, negative prognosis, and no correlation with prognosis. The genes associated with positive and negative prognoses were counted. We compared the prognosis of tumor cell subtypes by χ2 test.
Gene ontology enrichment analysis on tumor cell subpopulations
In accordance with the DEGs (Supplementary Table S3) calculated using the Seurat FindAllMarkers function, the top 50 DEGs in each tumor cell type (ccRCC 1, ccRCC 2, ccRCC 3, and ccRCC 4) were selected for gene ontology (GO) enrichment analysis by DAVID (https://david.ncifcrf.gov/). Finally, each tumor cell type underwent enrichment analysis of biological process (BP), and the five most significant BPs were shown.
Ligand–receptor interactions
The ligand–receptor interactions were calculated by CellPhoneDB (https://www.cellphonedb.org/; ref. 31). In brief, we created the Seurat objects of endothelial, tumor, and immune cells. First, we calculated the number of ligand–receptor interactions between tumor cells and endothelial cells by CellPhoneDB. Then, we calculated the number of ligand–receptor interactions between different immune cell types and the significant ligand–receptor pairs. According to a previous study (32), the ligand–receptor interaction score between tumor-associated macrophages (TAM) and tumor cells was calculated. We showed the ligand–receptor pairs with a score greater than 1.
scATAC-seq data analysis
Here, Signac (https://github.com/timoast/signac; https://satijalab.org/signac/; ref. 33) was applied for secondary analysis of scATAC-seq data. Signac is an R toolkit for analyzing and visualizing single-cell chromatin data in a way that allows interoperability with the Seurat R package, and is designed for analyzing multimodal single-cell data (33). First, we need to merge 19 samples, which we cannot use the merge command directly, compared with scRNA-seq data. If peak calling was performed on each dataset independently, the peaks are unlikely to be exactly the same. Thus, we created a common set of peaks across all the samples to be merged. After loading the cell metadata for each sample, we created Fragment objects using the CreateFragmentObject function. Subsequently, we could create a matrix of peaks × cell for each sample using the FeatureMatrix function and use the quantified matrices to create a Seurat object for each sample, storing the Fragment object for each sample in the assay. Finally, we merged 19 scATAC-seq samples into one object for downstream analysis. We examined the chromatin accessibility region of each sample and demonstrated this by randomly selecting three different chromosomal regions. We found that the overall accessibility of each sample was similar (Supplementary Fig. S2A–S2C). Meanwhile, compared with the two different results of bulk ATAC-seq, the same area of open accessibility existed, but the accessible area of bulk ATAC was wider (Supplementary Fig. S2A–S2C).
Then, we need to perform QC of the single-cell nuclei information and remove the low-quality nuclei. Here, we need to calculate the peak region fragments, ratio reads in genomic blacklist regions (blacklist ratio), nucleosome signal, and transcription start site (TSS) enrichment score of each nucleus (Supplementary Fig. S3A). According to the filtration strategy of a previous study (34, 35), we followed the parameters (1,000 < peak region fragments < 20,000, blacklist ratio < 0.05, nucleosome signal < 4 and TSS enrichment score > 1) to filter the nuclei, resulting in high-quality nuclei (Supplementary Fig. S3B). We analyzed the counts of different fragment lengths and found that most fragments were less than 400 bp (Supplementary Fig. S3C), which was consistent with a previous study (33). As a result, we obtained 61,693 high-quality single-cell nuclei for downstream analysis (Supplementary Table S4). We created objects that contained genomic coordinates for each feature by GRanges function and gene annotations by “hg19” from University of California Santa Cruz (Santa Cruz, CA). Next, we performed data normalization, feature selection, and dimension reduction (for details, refer to https://satijalab.org/signac/). In brief, Signac performs term frequency–inverse document frequency (IDF) normalization and runs singular value decomposition (SVD) on the term frequency-inverse document frequency (TD-IDF) matrix using the features (peaks) selected above. The combined steps of TD-IDF followed by SVD are also called latent semantic indexing (36). We selected the second to 40th dimensions and created a Seurat object, which identified crude clusters using Seurat's shared nearest neighbor graph clustering with FindClusters function with a resolution of 0.4.
We considered the cell definition of scATAC-seq to be elusive and uncertain compared with scRNA-seq. To further define cell subtypes, we combined the following three methods: (i) creating a gene activity matrix, in which gene coordinates are extracted and extended to include the 2 kb upstream region; (ii) analysis of cell-type-specific peaks, wherein the FindAllMarkers function was used to determine the cell type–specific peaks and the gene location corresponding to those peaks (Supplementary Table S5); (iii) transcription factor (TF) analysis.
Integration of genome-wide association study and scATAC-seq analysis
All the genome-wide association study (GWAS) loci associated with RCC were downloaded from the GWAS catalog (downloaded February 20, 2022; ref. 37). Using RCC as the keyword, we searched in the GWAS catalog and downloaded the data for downstream analysis. The genes with P value greater than 5 × 10−8 were filtered out (Supplementary Table S6). Then, we analyzed whether the chromatin locations of these susceptible loci overlapped with the accessibility of single-cell chromatin.
Motif and TF footprinting analysis
Motif analysis was mainly based on the chromVAR (38) R package. In brief, we run the AddMotifs function to add the DNA sequence motif information required for motif analyses. Then, we could calculate a per-cell motif activity score by running chromVAR and identify differential activity scores between cell types. Motif activity scores were normalized by z-scores, and the differential activity scores between cell types were replaced with “avg_diff.” TF footprinting was gathered by Footprint function and plotted by PlotFootprint function.
Integrating scATAC-seq and scRNA-seq analysis
Here, we mainly used Seurat and Signac to process scRNA-seq and scATAC-seq data, respectively. Cells from scRNA-seq have been previously annotated on the basis of transcriptomic state. We predicted annotations for the scATAC-seq cells. First, to identify anchors between scRNA-seq and scATAC-seq experiments, we generated a rough estimate of the transcriptional activity of each gene by quantifying ATAC-seq counts in the 2 kb upstream region and gene body using the GeneActivity function in the Signac package. The ensuing gene activity scores from the scATAC-seq data were used as input for canonical correlation analysis, along with gene expression quantifications from scRNA-seq. Then, we identified anchors between scRNA-seq and scATAC-seq datasets using the FindTransferAnchors function. We predicted annotations for the scATAC-seq cells. To perform coembedding in UMAP plot, we imputed the RNA expression into the scATAC-seq cells based on the previously computed anchors and merged the datasets.
WES
WES was performed by Novogene, Ltd. These 19 samples corresponded to the previous single-cell sequencing samples, among which, three were tumor cell samples (RCC101, RCC106, and RCC112) and the rest were tissue samples. These tissue samples were restricted to those that contained at least 65% tumor nuclei (median 80%) by pathologic review. In brief, after we extracted the DNA samples, the following methods were used for detecting DNA samples: (i) The degree of DNA degradation and RNA contamination were analyzed by agarose gel electrophoresis; (ii) DNA purity (OD260/280 ratio) was detected by using Nanodrop; (iii) DNA concentration was accurately quantified using Qubit, in which DNA samples had optical density values between 1.8 and 2.0. The content above 1.5 μg was used for library construction.
The SureSelect Human All Exon kit (Agilent, 5191-7411) was used for library construction and capture process. We performed the experimental operation procedure according to the manufacturer's protocol. DNA fragments with length of 180–280 bp were randomly interrupted by Covaris platform. DNA libraries were prepared by connecting splicing at both ends of fragments after terminal repair and a-tail addition. We used liquid chip capture system (Agilent) to efficiently enrich human DNA in the whole-exon region and then performed high-throughput and high-depth sequencing on Illumina NovaSeq 6000. The sequencing depth of each sample was set at 10 Gb. For the data obtained from sequencing, we carried out QC, requiring the average Q30 ratio to be above 80% and the average error rate to be below 0.1% (Supplementary Table S7). Subsequently, we used samtools (version, 1.0) for SNP and insertion/deletion (INDEL) analysis. For the detected variants, we used ANNOVAR (39) software to annotate the structure and function. Then, we used MuTect (40) software to find the somatic single-nucleotide variant (SNV) locus, and Strelka (41) software to detect the information of somatic INDEL for each sample (Supplementary Tables S8 and S9). According to the results of SNV and INDEL, we could detect the mutations of the common mutated genes in these 19 samples (Supplementary Table S10).
Bulk ATAC-seq
Here, two single-cell samples of ccRCC (ID: ccRCC 61 and ccRCC 76) were selected for bulk ATAC-seq. Briefly, we prepared these two single-cell suspensions (ccRCC 61 and ccRCC 76) as described above. A total of 10,000 cells were lysed in lysis buffer [10 mmol/L Tris-HCl (pH 7.4), 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.1% Tween-20, 0.1% Nonidet P40 Substitute, 0.01% digitonin, and 1% BSA] for 4 minutes on ice to isolate the nuclei. To perform Tn5 transposition, the nuclei collected by centrifugation for 500 × g for 5 minutes at 4°C were incubated with the reaction mix (10 μL of 5×TTBL, 5 μL of TTE Mix V50, 35 μL of ddH2O; TruePrep DNA Library Prep Kit V2 for Illumina, Vazyme, TD501-01) at 37°C for 30 minutes. After using 2× VAHTS DNA Clean Beads (Vazyme, N411) for purification, the DNA was added in the TruePrep Index Kit V2 for Illumina (Vazyme, TD202) and then amplified by PCR using the following PCR conditions (72°C for 3 minutes; 98°C for 30 seconds; and 15 cycles at 98°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; 72°C 5 minutes; 4°C hold). Finally, DNA libraries were purified with the 1.2× VAHTS DNA Clean beads and subjected to sequencing (Illumina NovaSeq 6000). The authors used cutadapt (version, 3.1) tool to filter raw data (.fastq files). Quality control included the removal of sequencing primers and adaptors, low-quality bases at both ends of reads, reads with extremely high number of N bases and reads with truncated single-end reads less than 75 bp in length. Then, samtools (version, 1.10) software was used to calculate the sequencing depth, coverage, alignment rate, repetition rate, etc. This process generated the .bam files. Then, we converted the .bam files to .bed files by using bedtools (version, 2.27.1) software. The bamCoverage (version, 3.1.1) software was used to convert .bam files into .bw files, which could be visualised by IGV (version, 2.11.9) software.
Separate nuclear/cytoplasm and qRT-PCR
To determine the subcellular localization of long noncoding RNA (lncRNA; RP11-661C8.2, CTB-164N12.1, RP11-267A15.1, and CTB-32H22.1), we isolated nuclear and cytoplasm fractions and then performed qRT-PCR. The Ambion PARIS kit (Invitrogen, AM1921) was used to isolate nuclear and cytoplasmic RNA according to the manufacturer's protocol. Nuclear/cytoplasmic RNA was separately reverse transcribed by HiScript II Q RT SuperMix for qPCR (+gDNA wiper; Vazyme, R223). Then, diluted cDNA was used for qRT-PCR using FastStart Essential DNA Green Master (Roche, 6924204001) on a Light Cycler 96 (Roche) in line with the guidelines. The relative expression was calculated using the 2−ΔΔCt method and normalized by GAPDH.
Design and construction of antisense oligonucleotide
According to the results of scATAC-seq and subcellular localization of lncRNA, we discovered four tumor cell–specific lncRNAs, which were expressed in the nucleus. Therefore, antisense oligonucleotide (ASO) was designed to interfere with two lncRNAs (RP11-661C8.2 and CTB-164N12) in ccRCC cell lines (Caki-2 and 786-O). Here, we used ASO-5717 (RiBoBio, ASO-h-ENST00000518605_001, target sequence: CACAGGCATTATCGGGACTA) and ASO-5608 (RiBoBio, ASO-h-ENST00000507989_001, target sequence: GTCCCAGAAAGAACGGCAGC) to interfere with the expression of CTB-164N12.1 and RP11-661C8.2, respectively. We verified that ASO-5717 hit the CTB-164N12.1 specifically, while ASO-5608 hit the RP11-661C8.2 by qRT-PCR.
Cell counting kit-8 and 5-ethynyl-2′-deoxyuridine assay of RCC lines after ASO treatment
The viability of cells was assessed by cell counting kit-8 (CCK-8) assay (Vazyme, A311-01). The Caki-2 (Procell, CL-0326, RRID: CVCL_0235) and 786-O (Procell, CL-0010, RRID: CVCL_1051) cells were incubated in a 96-well plate and then treated with ASO Negative Control (ASO-control; RiBoBio, lnc6N0000001-1-10), RP11-661C8.2 ASO (ASO-5608), and CTB-164N12.1 ASO (ASO-5717) for 48 hours. Subsequently, the medium was replaced, and CCK-8 solution was added to each well. After incubation for 2 hours at 37°C, the absorbance of cells was measured at 450 nm using a microplate reader (BioTek Inc.).
After 48 hours ASO treatment, the proliferation of transfected cells was determined by 5-ethynyl-2′-deoxyuridine (EdU) assay. EdU assay was performed using Cell Light EdU Apollo 488 In Vitro Kit (RiBoBio, C10338-1) according to the manufacturer's instructions. The proliferation rate was calculated as the ratio of the number of EdU-positive cells to the number of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells. EdU assay was used to determine the proliferation of cells. The transfected cells were incubated with EdU work solution for 2 hours at 37°C. Then, the cells were fixed with 4% paraformaldehyde for 30 minutes and treated with 0.1% Triton X-100 for 10 minutes. After rinsing with PBS three times, the cells were exposed to staining reaction solution for 30 minutes and then incubated with DAPI or Hoechst 33342 to stain the cell nuclei for 30 minutes. Images were captured using a microscope (Olympus). The ratio of EdU-positive cells (Apollo488+cell) was defined as the proliferation rate.
Annexin V-FITC/PI and cell-cycle analysis (propidium iodide staining) detection by flow cytometry
An Annexin V-FITC/PI Apoptosis Detection Kit (BD, 556547) was used to examine cell apoptosis by flow cytometry (FC; BD, C6 Plus). The Caki-2 and 786-O cells after ASO treatment were collected by centrifugation at 300 × g for 5 minutes at 4°C and then stained with 5 μL of Annexin V-FITC and 5 μL of propidium iodide (PI; 100 μL/1 × 10e5 cells) for 15 min at room temperature in the dark. The samples were analyzed by FC within 1 hour.
After the ASO treatments, cell-cycle distribution was detected by Cell Cycle Staining Kit (MultiSciences, 70-CCS012) according to the manufacturer's protocol. Briefly, the transfected cells were harvested and fixed with 70% cold ethanol. After fixation, the PI staining solution with RNase A was added and incubated in the dark at 37°C for 30 minutes. The stained samples were tested by FC (BD C6 Plus).
Wound healing of cell migration and transwell assay
Here, migration and transwell assays were performed on two different ccRCC cell lines, namely, Caki-2 (Procell, CL-0326) and 786-O (Procell, CL-0010). Each cell line was divided into three observation groups: negative control (NC) group, ASO-5717 treatment group and ASO-5608 treatment group. A total of 2 × 10e4 cells were seeded in 6-well plates and reached 90% confluence after 48 hours of ASO transfection. A scratch of uniform width was made with a sterile pipette tip. The same portion of the scratch was photographed at 0, 24, and 48 hours to assess the scratch width, and the scratch area was measured. These differences were statistically analyzed by t test.
Approximately 6 × 10e4 786-O or Caki-2 cells were incubated in 0.1% FBS medium in a transwell chamber (Corning, 3422) for 24 hours. Invasion measurements were made according to the manufacturer's instructions. Cells were incubated in 150 μL of 0.1% FBS medium in the upper compartment, and 500 μL of complete culture medium was added to the lower compartment. After 24 hours invasion, the cells were washed with PBS, and under 100% methanol for 20 minutes. Subsequently, the cells were washed with PBS and stained at room temperature with Crystal Violet Stain solution (0.1%, Solarbio, G1063) for 20 minutes. The cells on the upper surface of the membrane were washed with PBS, and the cells on the lower surface of the membrane were erased. Five locations were randomly selected using an optical microscope (Mingmei, China) to count the cells stained on the submembrane.
RNA pulldown and protein mass spectrometry
An RNA pulldown kit (BersinBio, Bes5102) was used for efficient enrichment and identification of RNA-binding proteins by desthiobiotin end-labeled RNA and streptavidin-labeled magnetic beads. Desthiobiotin-labeled specific RNA probes were designed for the two lncRNA sequences (RP11-661C8.2 and CTB-164N12.1) and incubated with 786-O cell protein extracts. The target RNA probe–protein complexes were obtained by coupling magnetic beads and elution. Then, we identified these two proteins (RP11-661C8.2 and CTB-164N12.1) by silver staining.
Protein mass spectrometry was performed by IEMed Guangzhou Biomedical Technology Co., Ltd. In brief, reductive alkylation and enzymatic hydrolysis were performed on the proteins obtained by the above process. Subsequently, polypeptide desalting and pristine detection were performed. The offline data of mass spectrometry need to be searched in the protein database. The selection of the database is a key step in the whole information analysis process, and the final protein sequences are identified from the selected database. Here, the raw files were directly imported to Proteome Discoverer (version 2.2) for database retrieval. We only retained the trusted peptides and proteins and performed FDR validation to remove the peptides and proteins with FDR greater than 1% (Supplementary Tables S11 and S12). According to the results, we could enrich proteins that closely interacted with RP11-661C8.2 and CTB-164N12.1. Here, peptide coverage in protein amino acid sequence (Coverage) and protein score (Score) were key parameters to measure the interaction between proteins (Supplementary Tables S13 and S14). The proteins identified above were annotated by GO (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) to understand the functional properties of different proteins (Supplementary Tables S15–S18).
Immunoblotting
Briefly, the ccRCC cell lines of Caki-2 (ASO-5717 treatment, 3 × 10e6 cells), Caki-2 (ASO-5608 treatment, 3 × 10e6 cells), Caki-2 (NC, 3×10e6 cells), 786-O (ASO-5717 treatment, 3 × 10e6 cells), 786-O (ASO-5608 treatment, 3 × 10e6 cells), and 786-O (NC, 3 × 10e6 cells) were lysed with RIPA lysis buffer (Beyotime, P0013B) containing both protease inhibitor (Beyotime, P1045-1) and phosphatase inhibitor (Beyotime, P1045-2) on ice. After centrifugation at 12,000 rpm for 5 minutes, the supernatants were harvested, and the protein concentrations were determined using a BSA Quantification Kit (Beyotime, P0010S). Protein samples (100 μL) from supernatants were separated, and then SDS-PAGE was performed using an SDS-PAGE Kit (Sangon Biotech, C631100). After washing thrice with transferred buffer (Sangon Biotech, C520039), samples were transferred onto polyvinylidene difluoride membrane (Biosharp, BS-PVDF-22). The membrane was blocked for 30 minutes with blocking buffer (Beyotime, P0252). After washing thrice with Tris-buffered saline/Tween 20 (TBST, Sangon Biotech, C520009), the membranes were incubated at 4°C overnight with primary antibodies. The primary antibodies were anti-E-cadherin (rabbit anti-human, 1:5,000, Thermo Fisher Scientific, 701134, RRID: AB_2532405) and anti-GAPDH (mouse anti-human/mouse/rat, 1:5000, Abcam, ab8245, RRID: AB_2107448). The membranes were washed thrice with TBST and incubated with horseradish peroxidase–conjugated secondary antibody (goat anti-rabbit, 1:1,000, Cell Signaling Technology, 7074, RRID: AB_2099233; goat anti-mouse, 1:1,000, Solarbio, SE131, RRID: AB_2797595) at room temperature for 1 hour. Then, they were washed thrice again. Immunoreactivity using the ECL Kit for Western blot analysis (Thermo Fisher Scientific, 34577) was visualized by an imager (GE Healthcare, ImageQuant LAS 500). GAPDH was used as a loading control. The presented results are from at least three repetitions of Western blot analysis.
Data availability
The raw data of protein mass spectrometry can be accessed in Figshare (http://www.doi.org/10.6084/m9.figshare.19603750). The results of Flow Cytometry experiments can be accessed in FlowRepository (http://flowrepository.org/id/FR-FCM-Z5JS). The scRNA-seq and scATAC-seq raw data after processing in this article can be accessed in Gene Expression Omnibus datasets (GSE207493). Bulk ATAC-seq data are available through NCBI BioProject, accession number PRJNA866658. WES data have been deposited in the NCBI BioProject under project PRJNA861705.
Code availability
All the code used to perform the analysis is available at Figshare (http://www.doi.org/10.6084/m9.figshare.19603750) and github (https://github.com/lessonskit/Single-cell-multi-omics-profiling-of-ccRCC.git).
Results
Single-cell multiomics studies mapping the human ccRCC transcriptome landscape
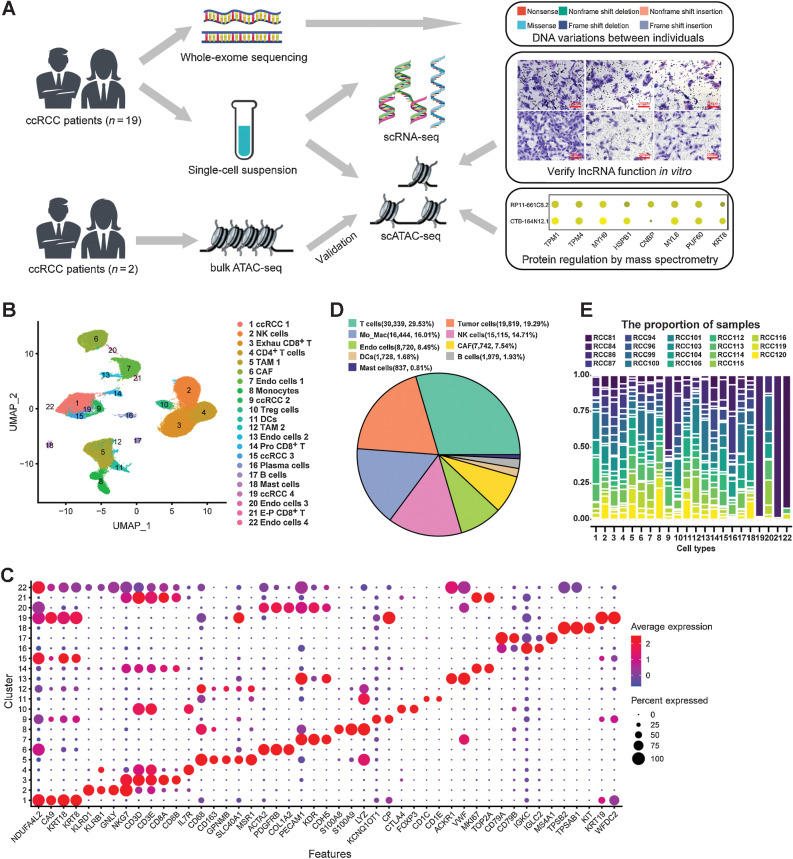
Considering the number of previous single-cell transcriptome studies in ccRCC (13–16), we hope to further discover the epigenetic regulatory mechanisms through single-cell multiomics scheme (Fig. 1A). In this study, we collected fresh surgical resections from 19 patients who suffered from localized ccRCC (Supplementary Table S1). Single-cell suspensions were prepared by enzyme lysis (Materials and Methods). Part of the single-cell suspension was used for scRNA-seq, while the rest was used for scATAC-seq by 10x Genomics platform (Fig. 1A). Additional tissue or single-cell suspensions were used for bulk ATAC-seq or WES, where available (Fig. 1A).
Figure 1.
Overview of single-cell transcriptomic atlas of ccRCC sample. A, Schematic of the experimental design for this study. The same images from Fig. 6G and J were used for the right panel in A. B, UMAP plot representation of ccRCC with 22 distinct cell types. The exhausted (Exhau) CD8+ T cells, TAMs, CAFs, DCs, proliferative (Pro) CD8+ T cells, and exhausted-proliferative (E-P) CD8+ T cells. C, Bubble chart showing the marker genes of each cluster. Dot size represents the proportion of cells, and the color represents gene expression with high or low. D, Pie graph showing the fraction of main cell types. E, Proportion of 19 ccRCC samples in each cell type.
After QC by Seurat (Supplementary Fig. S1, Materials and Methods; ref. 25), we captured 102,723 high-quality single-cell transcriptome information from ccRCC (Supplementary Table S2). To mitigate possible batch effects, we used Harmony (27) to aggregate cells across samples and jointly cluster them in an unsupervised manner. On the basis of the expression of marker genes and cell annotation in our previous studies (13, 22), we classified ccRCC into 22 independent cell types, including four tumor cell subtypes (ccRCC 1, ccRCC 2, ccRCC 3, and ccRCC 4), four endothelial cell subtypes (Endo cells 1, Endo cells 2, Endo cells 3, and Endo cells 4), etc. (Fig. 1B and C). In ccRCC, we found that T cells (30,339, 29.53%) were the most numerous followed by tumor cells (19,819, 19.29%; Fig. 1D). Compared with previous single-cell studies of advanced ccRCC (15, 42), this study captured more tumor cells and classified tumor cells into subpopulations (Fig. 1B). In addition, endothelial cells were also more diverse and abundant than that in previous studies (14, 15, 42). Because of the tumor heterogeneity of ccRCC (7), we performed scRNA-seq on tumor samples from 19 different individuals. We found that most cell types were derived from each sample, whereas individual differences between tumor cells and endothelial cells were more pronounced (Fig. 1E). For example, tumor cells (ccRCC 4) were only derived from patient RCC84, and Endo cells 4 were derived from only three individuals (Fig. 1E). Consistent with previous studies (14, 15, 42), extensive infiltration of immune cells, especially T cells and TAMs, characterized the immune microenvironment of ccRCC (Fig. 1B and D). In addition, CD8+ T cells infiltration was more remarkable in the ccRCC tissue, as reported in previous studies (15, 42, 43).
Characterization of the tumor cell molecular subtypes by scRNA-seq
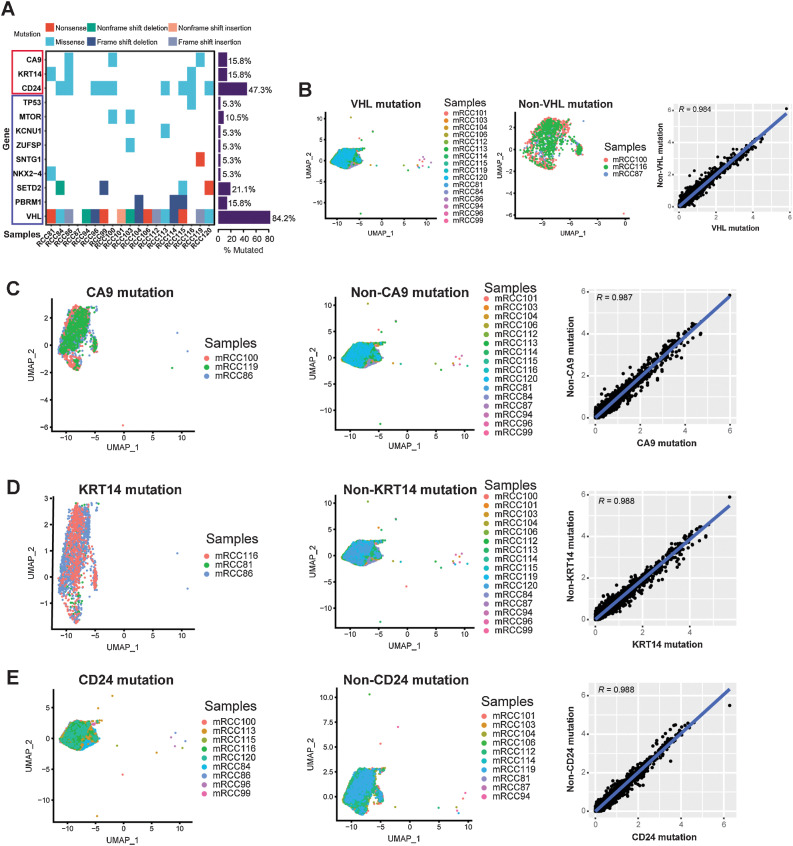
Considering the inherited predisposition of ccRCC (6), we performed WES on all ccRCC samples to understand the DNA variations between individuals (Supplementary Tables S8 and S9). We found that variations in these common mutated genes were similar to those found in previous studies (Fig. 2A; refs. 12, 15). VHL was the most common mutated gene, whose mutation rate was 84.2% in this study (Fig. 2A). Interestingly, we discovered that CA9 (15.8%), KRT14 (15.8%), and CD24 (47.3%) also showed a high mutation rate, which were missense (Fig. 2A). We classified tumor cells into VHL, CA9, KRT14, and CD24 mutated or nonmutated groups and compared their gene expression differences (Fig. 2B–E). We found that gene expressions in the mutated and nonmutated tumor cells were very similar, with a Pearson correlation coefficient greater than 0.98 (Fig. 2B–E). Thus, this also indicated that a single mutated gene in ccRCC did not cause great changes in all gene expression of tumor cells.
Figure 2.
scRNA-seq revealed the relationship between gene mutation and gene expression in ccRCC. A, WES for ccRCC samples. Each row represents a gene, and the frequency of mutations is indicated on the right side of the bars. B–E, Left, cluster of tumor cells in VHL (B), CA9 (C), KRT14 (D), and CD24 (E) mutated samples are shown. Middle, cluster of tumor cells in VHL (B), CA9 (C), KRT14 (D), and CD24 (E) nonmutated samples are shown. Right, scatterplot showing the log1p of the average expression per gene of VHL (B), CA9 (C), KRT14 (D), and CD24 (E) mutated/nonmutated samples.
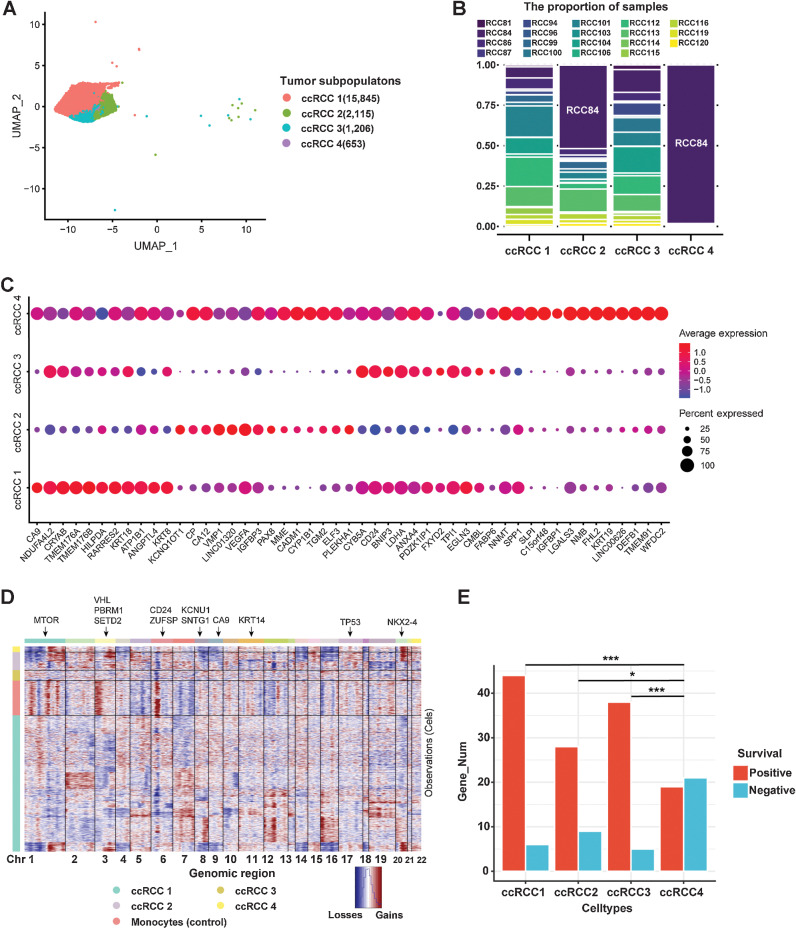
According to the gene expression characteristics, tumor cells were unbiased clustering into four cell subtypes (ccRCC 1, ccRCC 2, ccRCC 3, and ccRCC 4) by scRNA-seq (Fig. 3A). Except for ccRCC 4, the remaining three tumor cell subtypes were derived from each patient (Fig. 3B). Although individual differences still existed, there was a good representation of cell subpopulation classification in tumor cells. Initially, to objectively reveal the specific gene expression of tumor cells, we compared that with adjacent normal epithelial cells, which were obtained from our previous study (30). We classified these adjacent normal epithelial cells into proximal tubular cells, proximal straight tubule cells, proximal convoluted tubule cells, glomerular parietal epithelial cells, distal tubule cells, collecting duct principal cells and collecting duct intercalated cells, which integrated with four tumor cell subtypes in this study (Supplementary Fig. S4A). We discovered that some genes, such as CRYAB, CYB5A, CD24, SPP1, KRT18, KRT8, and GPX3, were conserved, which expressed in both normal epithelial cells and tumor cells (Supplementary Fig. S4B). Distinct differences in gene expression were found in different tumor cell subtypes. We found that ccRCC 1 highly expressed CA9 and NDUF4AL2, whereas ccRCC 2 highly expressed KCNQ1OT1 and CP (Fig. 3C). ccRCC 3 highly expressed NDUF4AL2 and CYB5A, whereas ccRCC 4 expressed most of the highly expressed genes in tumor cells (Fig. 3C). CA9 and NDUF4AL2 have been identified as marker genes for ccRCC in previous studies (12). CP was confirmed as a specific marker of ccRCC in our previous single-cell study (13), which was confirmed from a larger sample of ccRCC in this study. KCNQ1OT1 is an oncogenic lncRNA, which has been reported in a previous study (44). However, this is the first time that KCNQ1OT1 was found to be highly expressed in a specific cell subtype of ccRCC by scRNA-seq.
Figure 3.
Heterogeneity of tumor cells in human ccRCC. A, UMAP plot of four subtypes of ccRCC tumor cells (ccRCC1, ccRCC2, ccRCC3, and ccRCC4). B, Proportion of samples in each tumor cell type. C, Bubble chart showing DEGs in each tumor cluster (color represents the expression level, and dot sizes represent the relative abundance). D, CNV landscape of tumor cells. Monocytes were used as the reference cells. Red, gains of copy number; blue, losses of copy number. The annotated gene represents its chromosomal location (arrows). E, The number of genes associated with prognosis is found by integrating differentially expressed genes from tumor cells into TCGA database on ccRCC. *, P < 0.05; ***, P < 0.001.
According to the scRNA-seq data, we conducted CNV analysis and found that there were different CNV characteristics in these tumor cell subtypes (Fig. 3D). We found that most tumor cells were characterized by losses on chromosome 3 (chr3) and gains of chromosome 5 (chr5), consistent with TCGA project on ccRCC (12). This result was more concentrated in ccRCC 1, but not in ccRCC 4 (Fig. 3D). This may be caused by cloning or subcloning of tumor cells. Therefore, we can use scRNA-seq results to predict the tumor cells in the root state and the direction of tumor cell evolution. Here, we combined RNA velocity (28) and Monocle3 (29) to reconstruct the evolution trajectory of tumor cells. First, we identified the root of tumor cells and the direction of differentiation by RNA velocity (Supplementary Fig. S4C). Then, we reconstructed the evolution trajectory of tumor cells by Monocle3 (Supplementary Fig. S4D). We found that ccRCC 4 was located in the root, which suggests that it might be stem-like cells and could differentiate into ccRCC 1, ccRCC 2, and ccRCC 3 (Supplementary Fig. S4C and S4D).
To further evaluate the biological functions of each tumor cell type, we performed GO enrichment analysis based on their DEGs by DAVID (https://david.ncifcrf.gov/). The BPs of ccRCC 1 were concentrated in “response to hypoxia,” “cellular response to hypoxia,” and “glycolytic process” (Supplementary Fig. S5A). A previous study had reported that the pathogenesis of ccRCC is related to the hypoxia caused by VHL mutation (45). CNV analysis showed that ccRCC 1 is characterized by losses on chr3, where VHL is located. Therefore, ccRCC1 may be associated with these processes. The BPs of ccRCC 3 were similar to those of ccRCC 1, namely, “cytoplasmic translation,” “translation,” and “response to hypoxia” (Supplementary Fig. S5B). The ccRCC 2 was mainly concentrated in “positive regulation of angiogenesis” and “negative regulation of endopeptidase activity” (Supplementary Fig. S5C), whereas ccRCC 4 was concentrated in “epithelial cell differentiation” and “innate immune response” (Supplementary Fig. S5D).
With the subtypes of ccRCC in our hands, we hope to predict the prognosis of these four tumor cell types. On the basis of the results of scRNA-seq, we can calculate the DEGs of each tumor cell type (Supplementary Table S3). We selected the top 100 DEGs in each cell subtype and determined the prognosis based on the survival analysis results in TCGA database on ccRCC. We found that most of the DEGs in ccRCC 1, ccRCC 2, and ccRCC 3 suggested positive survival, whereas more DEGs in ccRCC 4 indicated poor prognosis, which was significant compared with the three previous cell subtypes (Fig. 3E).
scRNA-seq identified endothelial cell subtypes
Similar to tumor cells, endothelial cells also included four cell subtypes in ccRCC, which were identified by the characteristics of gene expression (Supplementary Fig. S6A and S6B). We found that some marker genes of endothelial cells were highly expressed in them, such as PECAM1, VWF, and CDH5 (Supplementary Fig. S6B). Interestingly, we identified a subtype of endothelial cells (Endo cells 3) with cancer-associated fibroblast (CAF) markers (TAGLN, PDGFRB, COL1A2, and ACTA2), consistent with our previous study (Supplementary Fig. S6B; ref. 13). Considering that a previous study has reported that endothelial cells and fibroblasts can transition into each other in the tumor microenvironment (46), our results supported the possibility that this transition process existed in ccRCC. On the other hand, as the number of ccRCC samples increased, there were more subtypes of endothelial cells than in our previous studies (13, 22), indicating the heterogeneity of endothelial cells. Ligand–receptor interactions between endothelial and tumor cells were determined by using Cellphone DB (31). We found that most endothelial and tumor cell subtypes interacted with a large number of ligand–receptor pairs, especially in Endo cells 3 (Supplementary Fig. S6C and S6D). All endothelial and tumor cell subtypes interacted with some ligand–receptor pairs, such as “ITGB1-VEGFA” and “ITGB1-SPP1” (Supplementary Fig. S6E–S6H). ITGB1 and VEGFA played an important role in the interaction between endothelial and tumor cells (Supplementary Fig. S6E–S6H). As reported previously, drugs that inhibit VEGF and its receptor VEGFR are effective therapeutics for metastatic ccRCC (47).
scRNA-seq revealed the characteristics of the immune microenvironment in ccRCC
Previous studies have reported comprehensive immune microenvironment characteristics of ccRCC, which provided more insights for the treatment of ccRCC (14, 15, 42). In this study, immune cells accounted for a high proportion in ccRCC tissues, which were about 64.7% of all cells (Fig. 1D), consistent with previous studies (14, 15, 42). Not only the number of immune cells but cell types were also abundant, which could be classified into 13 cell types (Supplementary Fig. S7A). Moreover, in the tumor microenvironment, the interaction of immune cells is very close, especially TAM 1, dendritic cells (DC), and proliferative CD8+ T cells (Supplementary Fig. S7B and S7C). Consistent with previous studies (15, 42), we found that these ligand–receptor pairs, such as CD74-MIF, CD74-COPA, and CD74-APP, played a significant role in TAM and other immune cells (Supplementary Fig. S7D). CD74 is expressed on TAM, which promotes the secretion of growth factor by TAM.
T cells were the most abundant immune cells in ccRCC, accounting for 29.53% of all cells (Fig. 1D). According to their corresponding markers, T cells can be clustered into five cell subtypes, namely, exhausted CD8+ T cells, CD4+ T cells, regulatory T cells, proliferative CD8+ T cells, and exhausted proliferative CD8+ T cells (Supplementary Fig. S7E). PD-1 (PDCD1 encoding) is a key receptor of immune checkpoint, which may be important for the effect of immunotherapy (48). Because of scRNA-seq technology, we were able to calculate the expression of PDCD1 in each T cell. Here, T cells with an average expression of PDCD1 greater than 1 were considered as PD-1 positive. We discovered that the positive rate of PD-1 in ccRCC was about 21.49% (Supplementary Fig. S7F). In ccRCC, macrophages mainly exist in the form of TAM, which were characterized by the expression of GPNMB, SLC40A1, and MSR1 in addition to the classical macrophage marker CD68 (Supplementary Fig. S7G; refs. 49, 50). Similar to a previous study (13), TAM can be divided into two cell subtypes (TAM 1 and TAM 2) in ccRCC, one of which had high expression of CD163, CSF1R, and CD86, and the other had low expression of these genes (Supplementary Fig. S7G). Next, we analyzed cell interactions between TAM 1 and TAM 2 with all tumor cell subtypes. We found that the cellular interactions between TAM and tumor cells were very close, especially TAM 1 (Supplementary Fig. S7H -S7I). In addition to the common ligand–receptor interactions of CD74-APP and CD63-TIMP1, we also discovered an interesting ligand–receptor pair, SLC40A1-CP (Supplementary Fig. S7H and S7I). SLC40A1 is a marker for TAM (49), whereas CP is a marker for ccRCC tumor cells (13). Thus, the biological effects of SLC40A1-CP binding are worthy of further study.
Construction of single-cell chromatin accessibility landscape in ccRCC
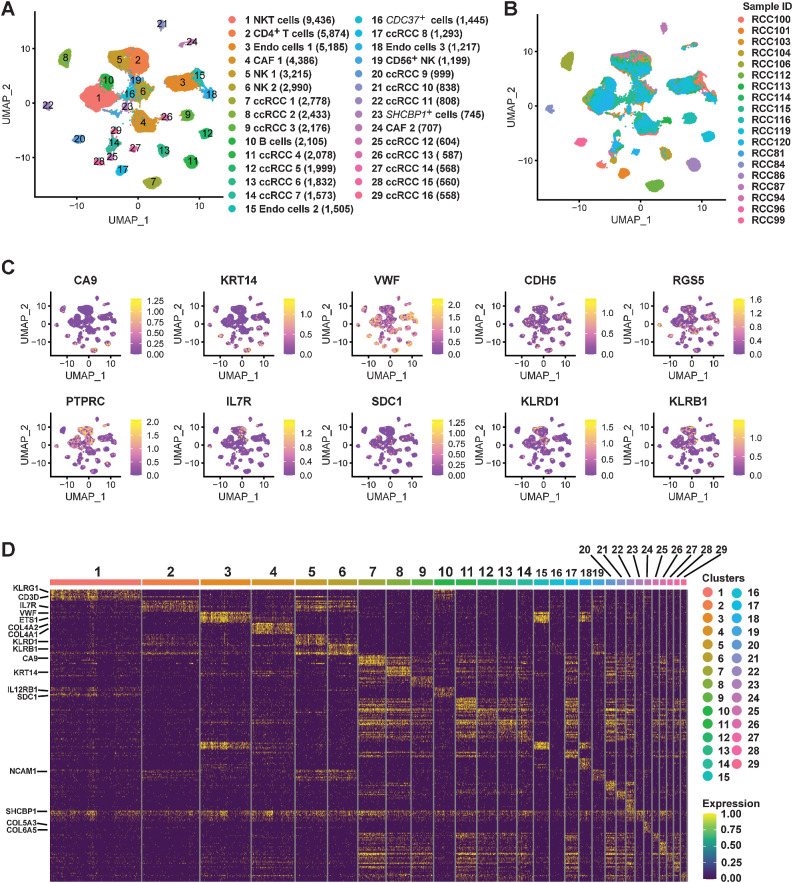
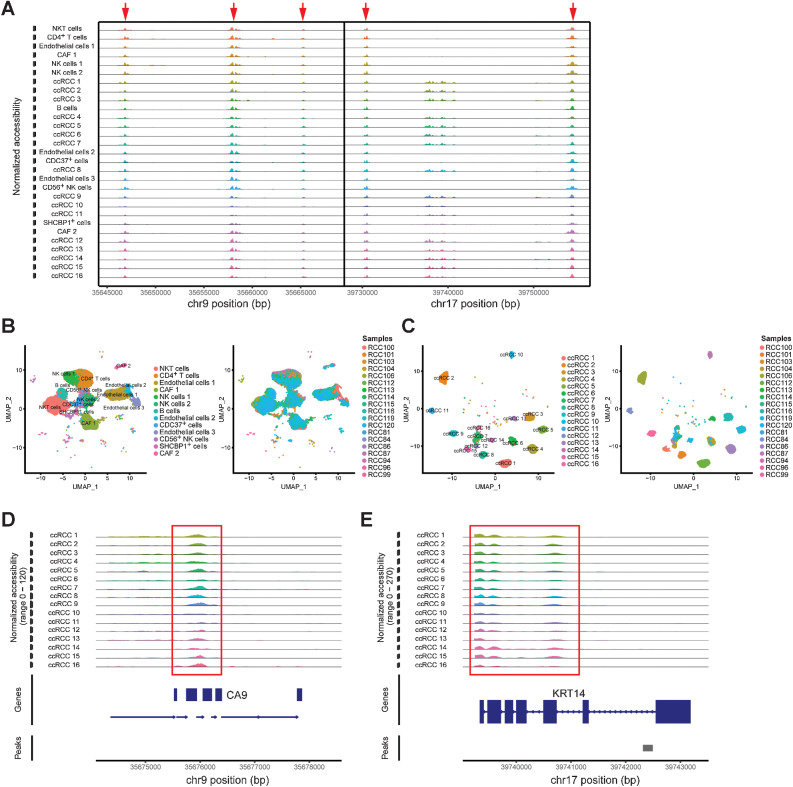
A previous study has constructed a single-cell map of dynamic chromatin landscapes of immune cells in RCC, which provided a rich resource for understanding the functional states and regulatory dynamics of immune cells in ccRCC (21). However, a more comprehensive single-cell chromatin accessibility landscape of ccRCC with large samples, including tumor cells, needs to be constructed, which may reveal the regulatory characteristics of tumor cells. In this study, we performed scATAC-seq on 19 ccRCC samples (Supplementary Table S1) by 10x Genomics platform. After QC by Signac (33), a total of 61,693 high-quality nuclei and 190,916 unique peaks were captured from 19 ccRCC samples (Supplementary Table S4). On the basis of the LSI algorithm (Materials and Methods), we classified these cells into 29 different cell types and identified the sample source of each cell type (Fig. 4A and B; Supplementary Fig. S8). For cell annotation of scATAC-seq, we combined three strategies to identify: gene activity scores, cell type–specific peaks and TF analysis. Here, we calculated the marker gene activity scores of tumor cells (CA9 and KRT14), endothelial cells (VWF and CDH5), CAF (RGS5), immune cells (PTPRC), CD4+ T cells (IL7R), B cells (SDC1), and NK cells (KLRD1 and KLRB1; Fig. 4C). Subsequently, cell type–specific peaks were analyzed, and then the corresponding gene regions of these peaks were identified (Fig. 4D). On the basis of the above, 29 different cell types in ccRCC were defined relatively reliably (Fig. 4A).
Figure 4.
Single-cell chromatin accessibility landscape in ccRCC. A, UMAP plot shows the cell landscape of ccRCC identified by scATAC-seq. B, Proportion of 19 ccRCC samples in each cell type. C, UMAP plots shows the gene activity scores of marker genes identified from each cell cluster. The color represents the grade. D, Heatmap represents the top 10 peaks for each cell cluster identified by scATAC-seq.
We identified the coaccessible chromatin regions of all cell types, such as in chr9 and chr17, by using scATAC-seq (Fig. 5A). These coaccessible chromatin regions also indicated that their regulatory processes were not cell type specific. Therefore, cell type–specific regulation processes could be discovered by identifying the cell type–specific coaccessible chromatin regions. In addition, our scATAC-seq results showed that the chromatin accessibility of nontumor cells, such as immune cells, endothelial cells and CAF, was consistent across individuals (Fig. 5B). Each cell type had good sample universality and derived from almost all samples (Fig. 5B). However, our results showed that tumor cells could be classified into 16 cell subtypes based on chromatin accessibility (Fig. 5C). The majority of tumor cell subtypes were derived from a single ccRCC sample (Fig. 5C). Similar to previous study on basal cell carcinoma (23), there was significant interindividual heterogeneity in chromatin accessibility of tumor cells in ccRCC.
Figure 5.
Single-cell chromatin accessibility characteristics in ccRCC. A, Chromatin accessibility profiles of each cluster in chr 9 and chr 17. B, UMAP of nontumor cells identifies cell clusters (left) and sample origin (right). C, UMAP of tumor cells identifies cell clusters (left) and sample origin (right). D and E, Specific chromatin accessibility region of tumor cells identified by scATAC-seq.
scATAC-seq revealed the characteristics of chromatin regulation in ccRCC tumor cells
Although a previous study revealed the regulatory dynamics of immune cells in ccRCC (21), the regulatory characteristics of tumor cells remain unknown at single-cell level. To investigate the characteristics of chromatin regulation in tumor cells, we identified 16 tumor cell subtypes and the chromatin accessibility regions of each cell subtype by using scATAC-seq (Fig. 5C; Supplementary Table S5). The advantage of scATAC-seq was that these chromatin accessibility regions could be pinpointed to cell types (Supplementary Table S5). Here, we found that CA9 and KRT14 served as markers of accessible regions in ccRCC cells, which maintained chromatin accessibility in each tumor cell subtype (Fig. 5D and E). To better discover the heterogeneity amongst tumor cell subtypes, we enriched the top 10 chromatin accessibility regions in 16 cell subtypes and mapped these regions to genes (Supplementary Fig. S9). We found that these chromatin accessibility regions were both overlapping and different, presenting an irregular state. This may also indicate inherited predisposition at the DNA level in ccRCC.
In addition, these chromatin accessible regions included not only protein encoding genes, lncRNA and rRNA in coding regions, but also accessibility to noncoding regions, which cannot be revealed by scRNA-seq. Tumor cell–specific chromatin accessibility regions were discovered by scATAC-seq, such as IGSF21, ATRNL1, and HPSE2, which were protein-coding regions (Supplementary Fig. S10A). Some tumor cell–specific rRNAs were identified by scATAC-seq (Supplementary Fig. S10B). Although previous GWAS of RCC identified many susceptibility loci (51, 52), many of them were not in the coding region, making it difficult to study their biological functions. We used GWAS Catalog (52) to find the susceptibility loci of RCC (Materials and Methods) and then integrated them into scATAC-seq data for analysis. We found that the chromatin regions of most susceptible loci were not open, except for rs4903064, which is located at Chr14:72812712. Moreover, the chromatin accessibility of this susceptibility locus was also cell-type specific, which was only open in tumor cell subtypes (Supplementary Fig. S10C).
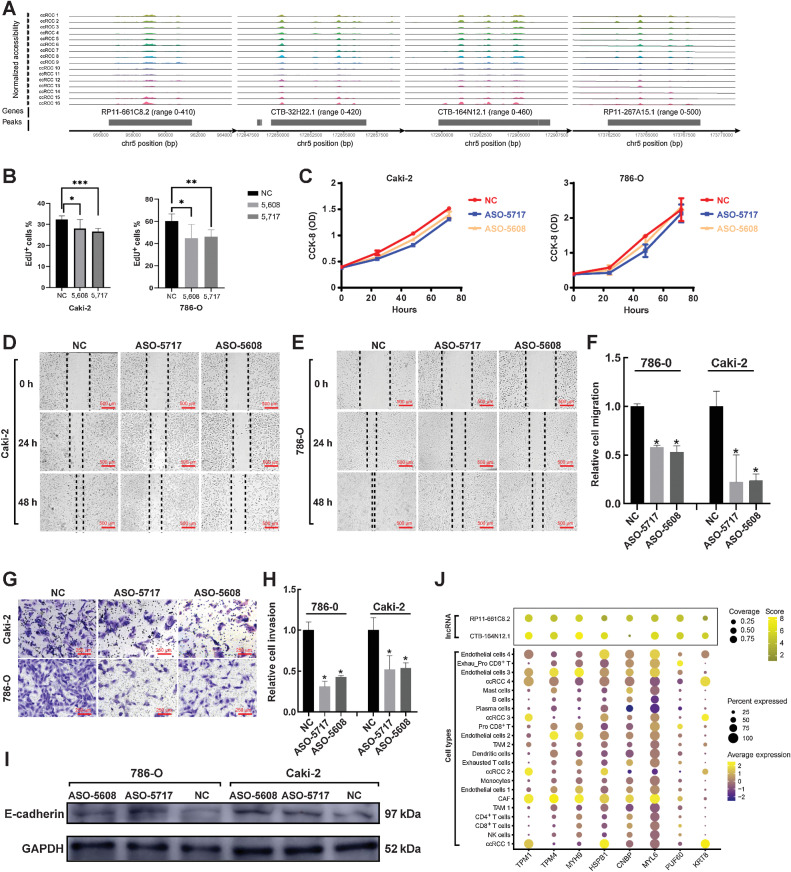
Interestingly, we discovered four lncRNAs (RP11-661C8.2, CTB-32H22.1, CTB-164N12.1, and RP11-267A15.1) that were specifically accessible to tumor cell subtypes by scATAC-seq (Fig. 6A). All of these lncRNAs had a common feature that was located on chromosome 5 (Fig. 6A). Through various mechanisms, cis-acting lncRNAs have been demonstrated to activate, repress or otherwise modulate the expression of target genes (53). Considering that TCGA results on ccRCC (12) and this study have confirmed that ccRCC has the feature with gains of copy number on chromosome 5, we hypothesized that these lncRNAs may play a specific regulatory role in ccRCC. Thus, we selected two lncRNAs (RP11-661C8.2 and CTB-164N12.1) for the following experiments, which verified the biological functions of these lncRNAs.
Figure 6.
Discovery and validation of specific lncRNAs in ccRCC tumor cells. A, Specific lncRNAs in ccRCC tumor cells were identified by scATAC-seq. B, ASO-5717 hit the CTB-164N12.1 specifically, while ASO-5608 hit the RP11-661C8.2. Cell proliferation was assessed by immunofluorescence using the EdU incorporation assay. C, Absorbance of 786-O and Caki-2 cell lines was measured at 450 nm after ASO treatment. D and E, Wound-healing assay reflected the migration of ccRCC after ASO treatment. Scale bar, 500 μm. F, Results of wound-healing assay were statistically analyzed. G, Transwell assay reflected the invasion of ccRCC after ASO treatment. Scale bar, 250 μm. H, Results of transwell assay were statistically analyzed. I, Western blotting analysis after ASO treatment in 786-O and Caki-2 cells. J, Integrating protein mass spectrometry (top) and scRNA-seq results (bottom), mapping the closely binding proteins found by protein mass spectrometry to gene expression in cell types identified by scRNA-seq. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
scATAC-seq revealed the biological function of lncRNA in promoting ccRCC invasion and migration in vitro
First, we had to identify whether these four lncRNAs were mainly expressed in the nucleus or cytoplasm, so that we could correctly select interference mediators later. Through cytoplasmic/nuclear separation experiments, we confirmed the expression of these four lncRNAs in the nucleus (Supplementary Fig. S11A; Supplementary Table S19). Here, we constructed ASO-5608 to interfere with the expression of RP11-661C8.2 specifically, while ASO-5717 to CTB-164N12.1, which verify their biological functions in reverse. Two cell lines of ccRCC, 786-O and Caki-2, were used as cell models. After 48 hours of treatment by ASO-5608 and ASO-5717, we found that the expression of RP11-661C8.2 and CTB-164N12.1 in 786-O and Caki-2 decreased significantly compared with the NC group (Supplementary Fig. S11B; Supplementary Table S20). This result also indicated that the inhibitory effect of ASO-5608 and ASO-5717 was credible.
Next, EdU detections were performed on ccRCC cell lines. We found that the proliferation activity of ccRCC cell lines after ASO treatment was significantly decreased, including 786-O group and Caki-2 group (Fig. 6B; Supplementary Fig. S11C and S11D). The CCK-8 experiment also supported the result that the proliferation of ccRCC cell lines after ASO treatment was inhibited in 24, 48, and 72 hours compared with the NC group (Fig. 6C). Subsequently, we carried out cell scratch experiment to observe the wound healing of ccRCC cell lines after being scratched in 0, 24, and 48 hours. We discovered that the wound healing of the NC group was better than that of the ASO treatment group, both in 786-O and Caki-2 (Fig. 6D–F). In addition, transwell assay was performed to indicate changes in migration function of ccRCC cell lines after ASO treatment. These results indicated that the migrations of 786-O and Caki-2 were significantly decreased after ASO treatment (Fig. 6G and H). The epithelial cell–cell adhesion molecule cadherin 1 (E-cadherin) is a well-known growth and invasion suppressor, which is known to suppress tumorigenicity and tumor dissemination via complex mechanisms (54). We discovered that after ASO treatment, the expressions of E-cadherin in ccRCC cell lines were significantly increased compared with those in the NC group by Western blot analysis (Fig. 6I). Although there were no significant changes in apoptosis and cell cycle of ccRCC cell lines after ASO treatment by FC (Supplementary Fig. S12A–S12D), this may be the characteristic of these lncRNAs. Thus, these results confirmed the function of RP11-661C8.2 and CTB-164N12.1, which promoted ccRCC invasion and migration in vitro.
Regulation of RP11-661C8.2 and CTB-164N12.1 was identified as cell-type specific by protein mass spectrometry
On the basis of the above results, we have confirmed some functions of RP11-661C8.2 and CTB-164N12.1. However, the regulatory mechanisms of these lncRNAs still remain unknown. We hope to explore them in future studies. Here, proteins interacting with RP11-661C8.2 and CTB-164N12.1 were identified by protein mass spectrometry. First, the proteins interacting with RP11-661C8.2 and CTB-164N12.1 were obtained by RNA pulldown (Materials and Methods). We confirmed these two kinds of proteins by silver staining (Supplementary Fig. S13A). Then, we performed protein mass spectrometry for the two kinds of proteins. By using Proteome Discoverer (version 2.2) software to analyze the results of mass spectrometry, we only retained the trusted peptides and proteins and performed FDR validation to remove the peptides and proteins with FDR greater than 1% (Supplementary Tables S11 and S12). According to the trusted peptides and proteins, we enriched proteins that closely interact with RP11-661C8.2 and CTB-164N12.1 (Supplementary Tables S13 and S14). Therefore, we discovered many proteins closely bound to the two lncRNAs and showed the most significant proteins, such as TPM1, TPM4, MYH9, HSPB1, CNBP, MYL6, PUF60, and KRT8 (Fig. 6J). We found that the proteins bound by the two lncRNAs were very similar, except for CNBP, which was more closely bound by RP11-661C8.2 (Fig. 6J). Subsequently, the genes encoding these proteins were placed into scRNA-seq data to estimate their expression in each cell subtype. Interestingly, these genes were almost highly expressed in CAF and Endo cells 3 (Fig. 6J). Some genes, such as TPM1, HSPB1, and KRT8, were highly expressed in tumor cells (Fig. 6J). Thus, we considered that the regulatory function of RP11-661C8.2 and CTB-164N12.1 was cell-type specific, mainly regulating CAF, endothelial cells, and tumor cells.
Next, to further understand the biological processes of the proteins, GO analysis was performed on the genes encoding the proteins closely bound to RP11-661C8.2 and CTB-164N12.1. We found that the biological processes of the genes encoding the proteins bound to these two lncRNAs were very similar, mainly reflected in “RNA splicing,” “ribonucleoprotein complex,” “biogenesis,” etc. (Supplementary Fig. S13B and S13C; Supplementary Tables S15 and S16). In addition, to understand the pathways, we performed KEGG analysis on the two gene clusters. We found that it was enriched in the pathways of “Spliceosome,” “Ribosome,” “Salmonella infection,” etc. (Supplementary Fig. S13D and S13E; Supplementary Tables S17 and S18).
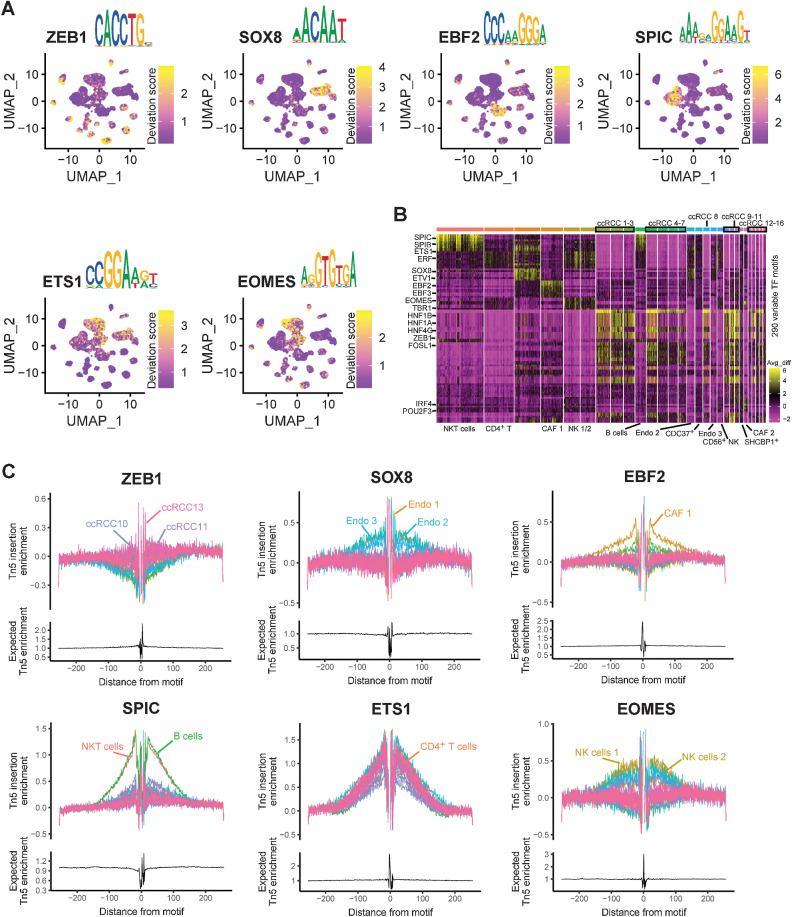
scATAC-seq revealed transcriptional regulation factors in ccRCC
Another advantage of scATAC-seq was the discovery of specific regulatory processes, especially TFs. Because of the single-cell study, it was possible to pinpoint these TFs specifically to cell types. Fortunately, in this study, we discovered many TFs with cell-type specificity and TF binding motifs (Supplementary Table S21). Consistent with a previous study (23), we found that SPIC and EOMES were specific TFs in B cells and natural killer (NK) cells, respectively (Fig. 7A). The DNA base sequence of motifs was also very similar compared with a previous study (Fig. 7A; ref. 23). Interestingly, we discovered that ZEB1 acts as a TF that binds tumor cells universally, whereas ETV4 is universally bound by nontumor cells (Fig. 7A; Supplementary Fig. S14A). In addition, we identified the specific TFs and motifs of endothelial cells (SOX8), CAF (EBF2), and T cells (ETS1) by scATAC-seq (Fig. 7A). By enriching the top 10 most significant TFs in each cell type, we found that these TFs had cell type specificity, especially between tumor cells and immune cells (Fig. 7B). Therefore, we also mentioned earlier that TFs need to be considered when defining cell types in scATAC-seq.
Figure 7.
Characteristics of TFs in ccRCC were identified by scATAC-seq. A, UMAP plot highlighting TF motif scores for ZEB1, SOX8, EBF2, SPIC, ETS1, and EOMES. B, Heatmap represents the 290 variable TF motifs from each cluster by scATAC-seq. C, TF footprints of ZEB1, SOX8, EBF2, SPIC, ETS1, and EOMES, with motifs in each cluster by scATAC-seq. The Tn5 insertion bias track is shown below.
To predict the exact binding location of these cell type–specific TFs, TF footprint analysis was performed (Fig. 7C). In addition, we identified many tumor cell–specific binding TFs in ccRCC, such as the HNF family, NFIB and TEAD3 (Supplementary Fig. S14B–S14G; Supplementary Fig. S15A–S15G). The TF hepatocyte nuclear factor 1β (HNF1B) represents the most commonly known monogenic cause of developmental kidney disease (55). HNF1B has also been reported to be associated with a rare type of RCC, whose mechanism is unclear (56). Thus, our results should provide many new ideas for studying the pathogenesis of ccRCC.
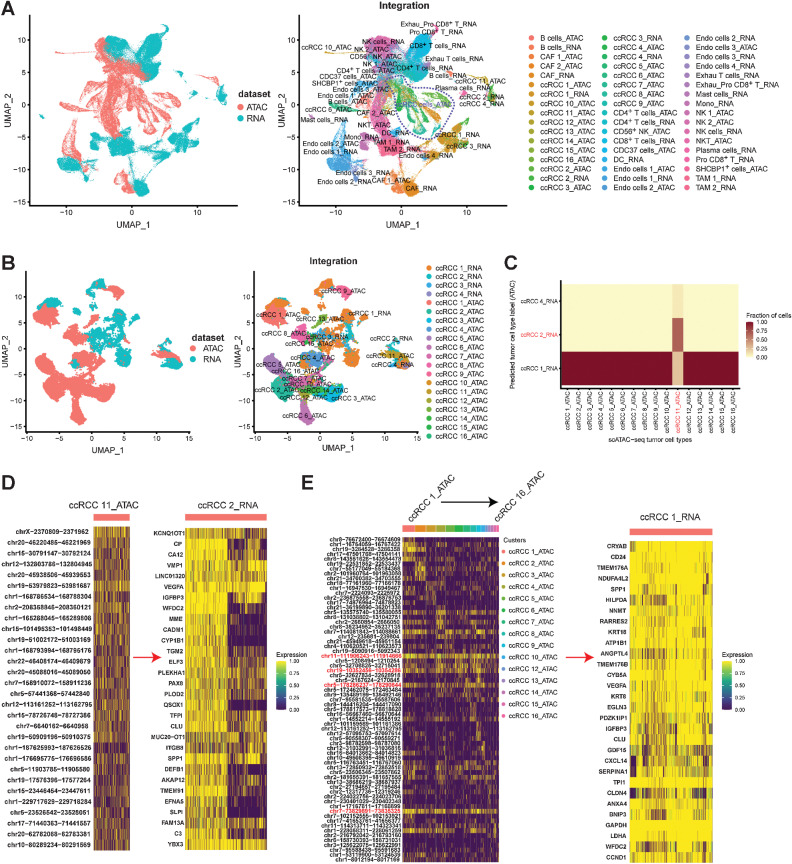
Integrating scATAC-seq and scRNA-seq analysis revealed the regulatory characteristics of tumor cells
The regulation of gene expression is a very complex process, and it is influenced by epigenetic regulation, lncRNAs, enhancers, promoters, and other factors (53). In this study, we aimed to discover the regulation rules of ccRCC by integrating the results of scATAC-seq and scRNA-seq. A map including the correspondence for all cell types between scATAC-seq and scRNA-seq was established (Supplementary Fig. S16A). The major cell types in scATAC-seq, such as tumor, T, NK, and endothelial cells and CAF, were predicted correctly based on the annotation of scRNA-seq (Supplementary Fig. S16B). Subsequently, we constructed the single-cell chromatin accessibility and single-cell transcriptome coembedding map (Fig. 8A). Although differences were recognized in the data of single-cell transcriptome and single-cell chromatin accessibility, as reflected by spatial projections, the distributions between the same cell types were very similar (Fig. 8A).
Figure 8.
Integrating scRNA-seq and scATAC-seq analysis. A, Integrating all cell types from scRNA-seq and scATAC-seq by UMAP plot. B, Integrating tumor cell types from scRNA-seq and scATAC-seq by UMAP plot. C, Heatmap showing the proportions of tumor cells from each scATAC-seq cluster (x-axis) that were annotated with cluster labels transferred from scRNA-seq clusters (y-axis). D, ccRCC 11 (scATAC-seq cluster)–specific chromatin accessibility regulated the gene expression characteristics of ccRCC 2 (scRNA-seq cluster). E, ccRCC 1–10, 12–16 (scATAC-seq cluster)–specific chromatin accessibility regulated the gene expression characteristics of ccRCC 1 (scRNA-seq cluster).
Then, by integrating all tumor cells in scATAC-seq (16 subtypes) and scRNA-seq (four subtypes), we found a correlation between chromatin accessibility and gene expression in the tumor cell subtypes (Fig. 8B; Supplementary Fig. S16C). The gene expression of ccRCC 1 (scRNA-seq subtype) was associated with the chromatin accessibility of 15 tumor cell types (scATAC-seq subtypes; Fig. 8C), which indicated the complexity of tumor cell regulation. We also discovered a pair of tumor cell types, namely, ccRCC 2 (scRNA-seq subtype) and ccRCC 11 (scATAC-seq subtypes), which implied a correspondence between chromatin accessibility and gene expression (Fig. 8C). In previous results, ccRCC 2 and ccRCC 11 were mainly derived from sample RCC84 (Fig. 2C; Supplementary Fig. S8). Therefore, revelation of the gene expression characteristics of ccRCC 2 regulated by the accessible chromatin region of ccRCC 11 was accurate (Fig. 8D). However, the regulatory characteristics of ccRCC 1 (scRNA-seq subtype) were complex and included the regulatory features of 15 scATAC-seq tumor cell subtypes (Fig. 8E). Regardless, we discovered several common accessible chromatin regions in all 15 tumor cell subtypes: “chr11-111906243-111914666,” “chr19-10352456-10354286,” “chr5-178286237-178290844,” and “chr7-73829891-73835325” (Fig. 8E). “chr11-111906243-111914666” and “chr5-178286237-178290844” were located on DLAT and ZNF354B, respectively (Supplementary Fig. S16D). DLAT encodes component E2 of the multienzyme pyruvate dehydrogenase complex. Notably, the low expression of DLAT signature was associated with reduced overall patient survival in TCGA ccRCC cohort (P = 3.6e−7; Supplementary Fig. S16E). We also observed that “chr19-10352456-10354286” and “chr7-73829891-73835325” were located in the DNA noncoding regions, which are close to the MRPL4 and CLIP2, respectively (Supplementary Fig. S16D). These findings suggest that individual differences existed in the accessible chromatin region of the studied ccRCC tumor cells, and common regulatory regions were present.
Discussion
Because of a complex interplay between genetic and nongenetic determinants of somatic evolution, the study of cancer requires the integration of multiple heritable dimensions at the single-cell resolution (57). Although researchers have previously done a great deal of work at the single-cell transcriptome level (13–17, 22, 42) and exomes (58) in ccRCC, this is just one dimension of information for cancer research (57). For a tumor cell, it should contain more information, such as TFs, histone modifications, chromatin accessibility, DNA methylation, copy-number alterations, protein expression, SNVs, and INDELs. With the development of single-cell multiomics technology in recent years (59, 60), many unknown fields have been explored. However, there are a few single-cell multiomics studies in oncology, especially on ccRCC. Thus, in this study, we hope to integrate scRNA-seq, scATAC-seq, WES, and protein mass spectrometry techniques to comprehensively understand ccRCC.
We captured 102,723 high-quality single-cell transcriptome information from 19 ccRCC samples by scRNA-seq (Fig. 1B). More cell numbers and larger sample sizes will better reveal the tumor heterogeneity of ccRCC. Here, we performed molecular classification of tumor cells based on the gene expression characteristics, which is a supplement to the prognostic classification of ccRCC in TCGA project (12). In addition, by integrating the results of single-cell transcriptome and WES, the authors found that gene expressions in the mutated and nonmutated tumor cells were very similar, with a Pearson correlation coefficient greater than 0.98 (Fig. 3A–D). This also indicated that a single mutated gene in ccRCC did not cause great changes in all gene expression of tumor cells. Interestingly, we discovered a specific ligand–receptor pair (SLC40A1-CP) in the tumor immune microenvironment, mainly TAM interacting with tumor cells (Fig. 4H and I). Previous studies have reported that SLC40A1 (49) is a marker of TAM, which is a cell membrane protein that may be involved in iron export from duodenal epithelial cells. CP (13) is a marker of ccRCC tumor cells, which is a metalloprotein that binds most of the copper in plasma and is involved in the peroxidation of Fe(II) transferrin to Fe(III) transferrin. Therefore, the biological effects of SLC40A1-CP binding are worthy of further study.
scATAC is a robust method for studying DNA epigenetic regulation, which reveals epigenetic regulatory features down to cell types and even individual cells (19, 20). In this study, we discovered some chromatin accessible regions on lncRNAs with tumor cell specificity (Fig. 6A). In addition, our experiments verified that these lncRNAs could promote ccRCC invasion and migration in vitro. This is going to be an interesting discovery because previous studies have shown that it is difficult to find each lncRNA that contributes a lot to the adaptability of organisms, whereas large numbers of lncRNAs contribute substantially when they are considered in aggregate (61). On the basis of these lncRNAs found by scATAC-seq, we can further search for the binding proteins through protein mass spectrometry, so as to understand their regulatory mechanism. Moreover, scATAC-seq is efficient and accurate in identifying tumor cell–specific TFs. Thus, scATAC-seq may be a reliable method to study the regulatory networks of DNA in tumor cells.
Although our study obtained a large amount of information about transcriptome and chromatin accessible regions, it is still very difficult to fully understand the regulatory relationship between them. The regulation of gene expression by cis-acting lncRNAs is a very complex process that involves promoters and enhancers (53). More explorations and research are needed to fully elucidate the regulatory network of ccRCC. On the other hand, we found it difficult to define cell types based on scATAC-seq results compared with scRNA-seq. In this study, the two cell types (CDC37+ cells and SHCBP1+ cells) identified by scATAC-seq were not matched with scRNA-seq results. More and more powerful analysis methods for scATAC-seq data may need to be developed in the future. Although scRNA-seq and scATAC-seq were performed simultaneously on the same tube single-cell from the same RCC sample in this study, which did not capture both mRNA and nucleus in the same cell with labeling. Collectively, our study demonstrates the comprehensive gene expression and DNA regulation information of ccRCC, discovering and validating lncRNAs that promote tumor cell invasion and migration, which will provide new insights into the treatment of ccRCC.
Supplementary Material
Details of each sample
Table S3, S5-S7, S10-S18, S21
SNV was detected by whole-exome sequencing of 19 ccRCC samples.
INDEL was detected by whole-exome sequencing of 19 ccRCC samples.
qPCR results of the expression of 4 deferent lncRNA in cytoplasm and nucleus. Three PCR by-wells were prepared for each sample.
qPCR results after ASO (5717 and 5608) treatment of 2 lncRNAs in caki-2 and 786-O cell lines.Three or six PCR by-wells were prepared for each sample. NC, negative control.
Figure S1-16
Acknowledgments
The authors wish to thank the laboratory members for their helpful advice and technical assistance. This work was supported by the grants from the National Natural Science Foundation of China (81770759), the National Key R&D Program of China (2017YFC0908000), Major Project of Guangxi Innovation Driven (AA18118016), Guangxi key Laboratory for Genomic and Personalized Medicine (grant no. 16-380-54, 17-259-45, 19-050-22, 19-185-33, 20-065-33), Guangxi Key Research and Development Project (Guike AB21196022), Guangxi Science and Technology Major Project (Guike AA22096032), and Guangxi Science and Technology Major Project (GuikeAA22096030). The funder of above financial support is Z. Mo.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
Z. Yu: Data curation, software, formal analysis, visualization, writing–original draft. Y. Lv: Validation, methodology, writing–original draft. C. Su: Validation, methodology. W. Lu: Validation, methodology. R. Zhang: Validation, methodology. J. Li: Software. B. Guo: Validation, methodology. H. Yan: Resources. D. Liu: Resources. Z. Yang: Resources. H. Mi: Resources. L. Mo: Resources. Y. Guo: Validation. W. Feng: Software. H. Xu: Validation. W. Peng: Validation. J. Cheng: Resources, supervision. A. Nan: Conceptualization, supervision. Z. Mo: Conceptualization, supervision, funding acquisition, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Capitanio U, Bensalah K, Bex A, Boorjian SA, Bray F, Coleman J, et al. Epidemiology of renal cell carcinoma. Eur Urol 2019;75:74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xu W, Atkins MB, McDermott DF. Checkpoint inhibitor immunotherapy in kidney cancer. Nat Rev Urol 2020;17:137–50. [DOI] [PubMed] [Google Scholar]
- 4. Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 2007;356:115–24. [DOI] [PubMed] [Google Scholar]
- 5. Wan X, Zhang Y, Tan C, Zeng X, Peng L. First-line nivolumab plus ipilimumab vs sunitinib for metastatic renal cell carcinoma: a cost-effectiveness analysis. JAMA Oncol 2019;5:491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: a metabolic disease. Nat Rev Urol 2010;7:277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gnarra JR, Tory K, Weng Y, Schmidt L, Wei MH, Li H, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85–90. [DOI] [PubMed] [Google Scholar]
- 9. Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010;463:360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guo G, Gui Y, Gao S, Tang A, Hu X, Huang Y, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet 2011;44:17–9. [DOI] [PubMed] [Google Scholar]
- 11. Varela I, Tarpey P, Raine K, Huang D, Ong CK, Stephens P, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 2011;469:539–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Network TCGAR. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Su C, Lv Y, Lu W, Yu Z, Ye Y, Guo B, et al. Single-cell RNA sequencing in multiple pathologic types of renal cell carcinoma revealed novel potential tumor-specific markers. Front Oncol 2021;11:719564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Krishna C, DiNatale RG, Kuo F, Srivastava RM, Vuong L, Chowell D, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell 2021;39:662–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bi K, He MX, Bakouny Z, Kanodia A, Napolitano S, Wu J, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 2021;39:649–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science 2018;361:594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Narayanan SP, Mannan R, Raskind G, Wang X, Vats P, et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc Natl Acad Sci U S A 2021;118:e2103240118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 2015;523:486–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang K, Hocker JD, Miller M, Hou X, Chiou J, Poirion OB, et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 2021;184:5985–6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kourtis N, Wang Q, Wang B, Oswald E, Adler C, Cherravuru S, et al. A single-cell map of dynamic chromatin landscapes of immune cells in renal cell carcinoma. Nat Cancer 2022;3:885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu Z, Lu W, Su C, Lv Y, Ye Y, Guo B, et al. Single-cell RNA-seq identification of the cellular molecular characteristics of sporadic bilateral clear cell renal cell carcinoma. Front Oncol 2021;11:659251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, et al. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 2019;37:925–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016;352:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 2019;16:1289–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liao J, Yu Z, Chen Y, Bao M, Zou C, Zhang H, et al. Single-cell RNA sequencing of human kidney. Sci Data 2020;7:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vento-Tormo R, Efremova M, Botting RA, Turco MY, Vento-Tormo M, Meyer KB, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018;563:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, et al. Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep 2018;25:1458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stuart T, Srivastava A, Madad S, Lareau CA, Satija R. Single-cell chromatin state analysis with Signac. Nat Methods 2021;18:1333–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mei Y, Xiao W, Hu H, Lu G, Chen L, Sun Z, et al. Single-cell analyses reveal suppressive tumor microenvironment of human colorectal cancer. Clin Transl Med 2021;11:e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang H, Mei Y, Luo C, Huang Q, Wang Z, Lu GM, et al. Single-cell analyses reveal mechanisms of cancer stem cell maintenance and epithelial-mesenchymal transition in recurrent bladder cancer. Clin Cancer Res 2021;27:6265–78. [DOI] [PubMed] [Google Scholar]
- 36. Cusanovich DA, Daza R, Adey A, Pliner HA, Christiansen L, Gunderson KL, et al. Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 2015;348:910–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 2017;45:D896–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schep AN, Wu B, Buenrostro JD, Greenleaf WJ. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat Methods 2017;14:975–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 42. Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell 2021;39:632–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Obradovic A, Chowdhury N, Haake SM, Ager C, Wang V, Vlahos L, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell 2021;184:2988–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cagle P, Qi Q, Niture S, Kumar D. KCNQ1OT1: an oncogenic long noncoding RNA. Biomolecules 2021;11:1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baldewijns MM, van Vlodrop IJ, Vermeulen PB, Soetekouw PM, van Engeland M, de Bruïne AP. VHL and HIF signalling in renal cell carcinogenesis. J Pathol 2010;221:125–38. [DOI] [PubMed] [Google Scholar]
- 46. LeBleu VS, Kalluri R. A peek into cancer-associated fibroblasts: origins, functions and translational impact. Dis Model Mech 2018;11:dmm029447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Escudier B, Porta C, Schmidinger M, Rioux-Leclercq N, Bex A, Khoo V, et al. Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v58–68. [DOI] [PubMed] [Google Scholar]
- 48. Dizman N, Philip EJ, Pal SK. Genomic profiling in renal cell carcinoma. Nat Rev Nephrol 2020;16:435–51. [DOI] [PubMed] [Google Scholar]
- 49. Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell 2019;179:829–45. [DOI] [PubMed] [Google Scholar]
- 50. Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An immune atlas of clear cell renal cell carcinoma. Cell 2017;169:736–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schödel J, Bardella C, Sciesielski LK, Brown JM, Pugh CW, Buckle V, et al. Common genetic variants at the 11q13.3 renal cancer susceptibility locus influence binding of HIF to an enhancer of cyclin D1 expression. Nat Genet 2012;44:420–5, S1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scelo G, Purdue MP, Brown KM, Johansson M, Wang Z, Eckel-Passow JE, et al. Genome-wide association study identifies multiple risk loci for renal cell carcinoma. Nat Commun 2017;8:15724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet 2020;21:102–17. [DOI] [PubMed] [Google Scholar]
- 54. van Roy F. Beyond E-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer 2014;14:121–34. [DOI] [PubMed] [Google Scholar]
- 55. Clissold RL, Hamilton AJ, Hattersley AT, Ellard S, Bingham C. HNF1B-associated renal and extra-renal disease-an expanding clinical spectrum. Nat Rev Nephrol 2015;11:102–12. [DOI] [PubMed] [Google Scholar]
- 56. Bockenhauer D, Jaureguiberry G. HNF1B-associated clinical phenotypes: the kidney and beyond. Pediatr Nephrol 2016;31:707–14. [DOI] [PubMed] [Google Scholar]
- 57. Nam AS, Chaligne R, Landau DA. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet 2021;22:3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 2012;148:886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Argelaguet R, Clark SJ, Mohammed H, Stapel LC, Krueger C, Kapourani CA, et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019;576:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li L, Guo F, Gao Y, Ren Y, Yuan P, Yan L, et al. Single-cell multi-omics sequencing of human early embryos. Nat Cell Biol 2018;20:847–58. [DOI] [PubMed] [Google Scholar]
- 61. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136:629–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of each sample
Table S3, S5-S7, S10-S18, S21
SNV was detected by whole-exome sequencing of 19 ccRCC samples.
INDEL was detected by whole-exome sequencing of 19 ccRCC samples.
qPCR results of the expression of 4 deferent lncRNA in cytoplasm and nucleus. Three PCR by-wells were prepared for each sample.
qPCR results after ASO (5717 and 5608) treatment of 2 lncRNAs in caki-2 and 786-O cell lines.Three or six PCR by-wells were prepared for each sample. NC, negative control.
Figure S1-16
Data Availability Statement
The raw data of protein mass spectrometry can be accessed in Figshare (http://www.doi.org/10.6084/m9.figshare.19603750). The results of Flow Cytometry experiments can be accessed in FlowRepository (http://flowrepository.org/id/FR-FCM-Z5JS). The scRNA-seq and scATAC-seq raw data after processing in this article can be accessed in Gene Expression Omnibus datasets (GSE207493). Bulk ATAC-seq data are available through NCBI BioProject, accession number PRJNA866658. WES data have been deposited in the NCBI BioProject under project PRJNA861705.