Abstract
Background and objective
The development of epigenetics holds great promise for diagnosis and treatment of lung adenocarcinoma (LUAD). The purpose of this work was to analyze the correlation between Ras Homolog Gene Family Member H (RHOH) expression and methylation in patients with LUAD and its association with survival.
Methods
Data related to gene expression, DNA methylation, and clinical features of LUAD from The Cancer Genome Atlas (TCGA) database were analyzed. A total of 50 patients were included in verification group. The methylation level of RHOH in verification group was detected by bisulfite amplicon sequencing.
Results
The RHOH methylation level in TCGA cohort was significantly and negatively correlated with its expression level (Cor = −0.5, p = 2.687e−33). Patients with hypermethylation and low expression of RHOH had significantly worse prognosis than patients with hypomethylation and low expression of RHOH (TCGA: p = 0.004; validation cohort: p = 0.006, HR: 4.740, 95% CI: 1.567–14.340).
Conclusion
Our research revealed that RHOH may prove to be a new potential prognostic predictor for LUAD patients.
Keywords: lung adenocarcinoma, methylation, prognosis, RHOH, TCGA
RHOH methylation and gene expression were both related to the prognosis of LUAD patients, and the combined analysis of RHOH methylation and expression could better predict the prognosis of LUAD patients.
1. INTRODUCTION
Lung cancer is one of the most prevalent malignant tumors with high mortality worldwide, 1 with lung adenocarcinoma (LUAD) as the most common pathological type. In recent years, epigenetic alterations have been described to be closely related to the occurrence and development of LUAD, among which DNA methylation has become the most extensively studied epigenetic form. 2 Aberrant methylation of genes can occur in the early stage of tumorigenesis, and the degree of aberrant methylation increases with tumor progression. Since the discovery of genome instability caused by genome‐wide hypomethylation, inactivation of tumor suppressor genes due to CpG islands hypermethylation, 3 , 4 and inactivation of miRNA due to DNA methylation, DNA methylation has been considered pivotal in LUAD development and progression. Therefore, DNA methylation analysis may provide us with promising targets for the diagnosis and treatment of LUAD.
As a fusion transcript with transcriptional repressor LAZ3/CL6, Ras Homolog Gene Family Member H (RHOH) can negatively modulate IL3 signaling by regulating the JAK‐STAT pathway. 5 Recent studies have demonstrated that RHOH plays an important role in tumor immune response. 6 , 7 Low expression of RHOH was found to be an independent adverse prognostic factor for overall survival (OS) and disease‐free survival in patients with acute myeloid leukemia. 8 In addition, RHOH has been revealed to regulate the tumor microenvironment. 9 However, the clinical significance of RHOH in solid tumors has rarely been elucidated, although it was reported as a valuable factor for tumorigenesis. 10 The aim of the present study was to gain more insights into RHOH and explore the association between RHOH methylation and its expression in LUAD. The results showed that RHOH methylation and gene expression were both related to the prognosis of LUAD patients, and the combined analysis of RHOH methylation and expression could better predict the prognosis of LUAD patients.
2. MATERIALS AND METHODS
2.1. Cohort
Data related to DNA methylation, mRNA expression, and prognosis of LUAD patients were downloaded from The Cancer Genome Atlas (TCGA) project (https://tcga-data.nci.nih.gov/tcga/). The mRNA expression data included 594 samples (normal: 59 cases and tumor: 535 cases), and the sample size of the methylation profile was 616 cases (normal: 53 cases and tumor: 563 cases). Samples from 50 LUAD patients who underwent surgical resection at our hospital were used as the validation group upon the approval from the ethics committee of the hospital. The final pathological type was determined by postoperative paraffin pathology. Tissues were obtained upon written informed consent from each subject.
2.2. Gene ontology (GO) analysis
GO analysis for differentially methylated genes was performed with DAVID 6.8 (https://david.ncifcrf.gov/), knowing that GO can provide a comprehensive resource of computable knowledge about the function of genes and gene products, including the biological process, molecular function, and cellular component. The p value < 0.05 was considered statistically significant. GOplot package in R was used to visualize the results.
2.3. DNA isolation and methylation analysis
Genomic DNA was extracted using E.Z.N.A.TM Tissue DNA Kit (Omega Bio‐Tek), and its DNA quality and quantity were determined by NanoDrop. Bisulfite amplicon sequencing was performed to detect DNA methylation. A total of 500 ng of genomic DNA were treated with bisulfite using the EZ Methylation‐Gold Kit. In the present study, we used the following Polymerase Chain Reaction (PCR) primers for RHOH methylation sequencing: left primer: TGTTTAATAAAAGTAGGTGAAAATAAAAG and right primer: AAAAAATCATTTAAAACTCTTCTCAATC. After purification of the PCR product, the DNA library was constructed by using the VAHTSTM Turbo DNA library preparation kit for Illumina. All procedures were carried out in accordance with the manufacturers' standard protocol. Deep sequencing was carried out by Illumina Miseq.
2.4. Statistical analysis
All TCGA data were standardized before analysis. |Cor| > 0.3 and p < 0.05 were considered statistically significant in the correlation analysis of mRNA expression and DNA methylation. Gene expression levels and gene methylation levels were divided into high and low groups by median. Kaplan–Meier survival analysis and Cox regression hazards model were used to estimate the correlation between gene expression, DNA methylation, and the prognosis of LUAD patients. The correlation between RHOH methylation level and clinical characteristics of the LUAD patients was analyzed by chi‐square test (Bonferroni correction was set, and the p value for significance was at p < 0.006). The correlation between the clinical features and RHOH in TCGA was analyzed by MEXPRESS. 11 The p < 0.05 was considered statistically significant. The correlation between RHOH expression and the prognosis of LUAD patients was analyzed by PREdiction of Clinical Outcomes from Genomic profiles (https://precog.stanford.edu), and the selected Gene Expression Omnibus chips and public database accession numbers were the following: ca00153, ca00182, ca00191, GSE19188, GSE31547, GSE3141, GSE4716, GSE8894, GSE29013, GSE31210, GSE10245, GSE11969, and GSE13213. The software used in this study was Perl, R. The R packages used were "limma", "lush", and "survival".
3. RESULTS
3.1. Sources of clinical data
A total of 50 LUAD patients were enrolled as the verification cohort of the present study. The clinicopathological data including age, sex, Tumor Node Metastasis (TNM) stage, pathological subtype, and smoking status were collected. The baseline clinical characteristics of the LUAD patients are shown in Table 1.
TABLE 1.
Correlation between clinicopathological features and RHOH methylation in lung adenocarcinoma
| Features | n | RHOH hypomethylation | RHOH hypermethylation | χ2 | p |
|---|---|---|---|---|---|
| Age | |||||
| <61 | 24 | 14 (58.3%) | 10 (41.7%) | 1.974 | 0.160 |
| ≥61 | 26 | 10 (38.5%) | 16 (61.5%) | ||
| Gender | |||||
| Male | 22 | 9 (40.9%) | 13 (59.1%) | 0.791 | 0.374 |
| Female | 28 | 15 (53.6%) | 13 (46.4%) | ||
| pT | |||||
| 1 | 45 | 24 (53.3%) | 21 (46.7%) | 5.128 | 0.077 |
| 2 | 3 | 0 (0.0%) | 3 (100.0%) | ||
| 3 | 2 | 0 (0.0%) | 2 (100.0%) | ||
| 4 | — | — | — | ||
| pN | |||||
| 0 | 46 | 23 (50.0%) | 23 (50.0%) | 1.923 | 0.382 |
| 1 | 2 | 0 (0.0%) | 2 (100.0%) | ||
| 2 | 2 | 1 (50.0%) | 1 (50.0%) | ||
| 3 | — | — | — | — | — |
| pM | |||||
| 0 | 47 | 24 (51.1%) | 23 (48.9%) | 2.946 | 0.086 |
| 1 | 3 | 0 (0.0%) | 3 (100.0%) | ||
| Stage | |||||
| 1 | 44 | 23 (52.3%) | 21 (47.7%) | 4.017 | 0.260 |
| 2 | 2 | 1 (50.0%) | 1 (50.0%) | ||
| 3 | 1 | 0 (0.0%) | 1 (100.0%) | ||
| 4 | 3 | 0 (0.0%) | 3 (100.0%) | ||
| Tumor size | |||||
| Small (≤15 mm) | 25 | 16 (64.0%) | 9 (36.0%) | 5.128 | 0.024 |
| Large (>15 mm) | 25 | 8 (32.0%) | 17 (68.0%) | ||
| Pathology | |||||
| Acinar | 34 | 21 (52.9%) | 13 (47.1%) | 10.319 | 0.035 |
| Solid | 8 | 3 (50.0%) | 5 (50.0%) | ||
| Papillary | 4 | 0 (33.3%) | 4 (66.7%) | ||
| Lepidic | 3 | 0 (0.0%) | 3 (100.0%) | ||
| Micropapillary | 1 | 0 (0.0%) | 1 (100.0%) | ||
| Smoking | |||||
| Nonsmoker | 38 | 20 (52.6%) | 18 (47.4%) | 1.361 | 0.243 |
| Current or ex‐smoker | 12 | 4 (33.3%) | 8 (66.7%) | ||
Note: Median age = 61 years.
Abbreviation: RHOH, Ras Homolog Gene Family Member H.
3.2. GO analysis
The expression and methylation of genes related to OS of LUAD patients were analyzed, and the result showed that the expression level of 22 genes was significantly correlated with the methylation status (|Cor| > 0.3 and p < 0.05). Further GO analysis of these genes showed four major GOs: structural molecule activity, negative regulation of GTPase activity, cornified envelope, and peptide cross‐linking (Figure 1A). The methylation level of the related genes was relatively low in LUAD (Figure 1B). The molecular function of CLDN23, DSP, and SPRR1B was characterized by structural molecule activity, and the biological processes involved in RGN and RHOH were associated with the negative regulation of GTPase activity (Figure 1C). In the present study, we selected RHOH for further research. The predicted functional partners of RHOH are shown in Figure S1.
FIGURE 1.
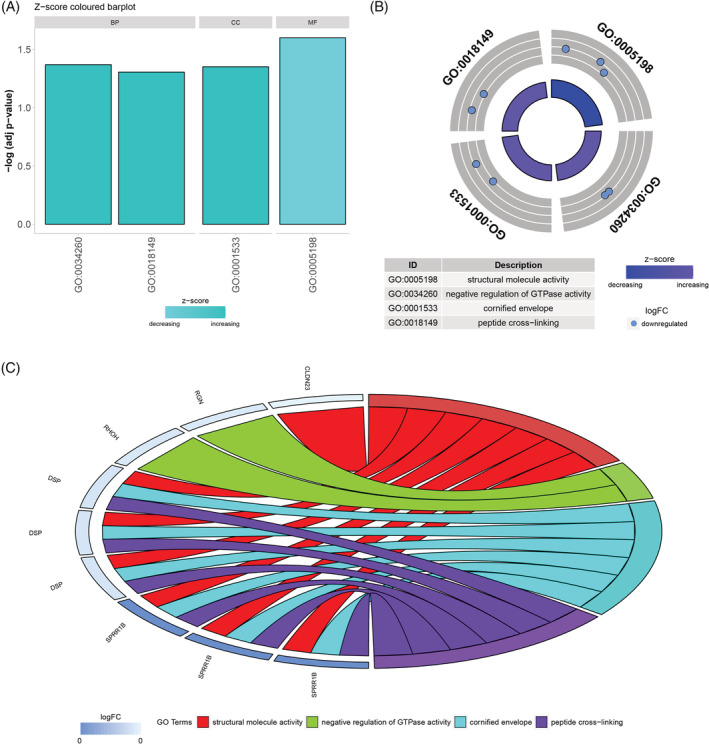
GO analysis of 22 genes related to the prognosis of LUAD patients whose expression level was related to DNA methylation. (A) Structural molecule activity, negative regulation of GTPase activity, cornified envelope, and peptide cross‐linking were found significant different based on those genes by DAVID 6.8. (B) Four GO were downregulated in LUAD. (C) Genes included in the four GO. BP, biological process; CC, cellular component; LUAD, lung adenocarcinoma; MF, molecular function; RHOH, Ras Homolog Gene Family Member H
3.3. RHOH methylation is negatively correlated with RHOH expression
In the current study, we focused on the correlation between DNA methylation and mRNA expression of RHOH and found that the DNA methylation level of RHOH was significantly and negatively correlated with its mRNA expression (Cor = −0.5, p value = 2.687e−33) (Figure 2A). TCGA methylation database showed that each methylation site of RHOH was negatively correlated with its gene expression (Figure 2B–K). Among them, four methylation sites were significantly correlated with the expression of RHOH: cg00804392 (Cor = −0.676, p value = 1.097e−68), cg11903057 (Cor = −0.602, p value = 6.412e−51), cg05658107 (Cor = −0.449, p value = 2.441e−26), and cg26296101 (Cor = −0.43, p value = 4.074e−24).
FIGURE 2.
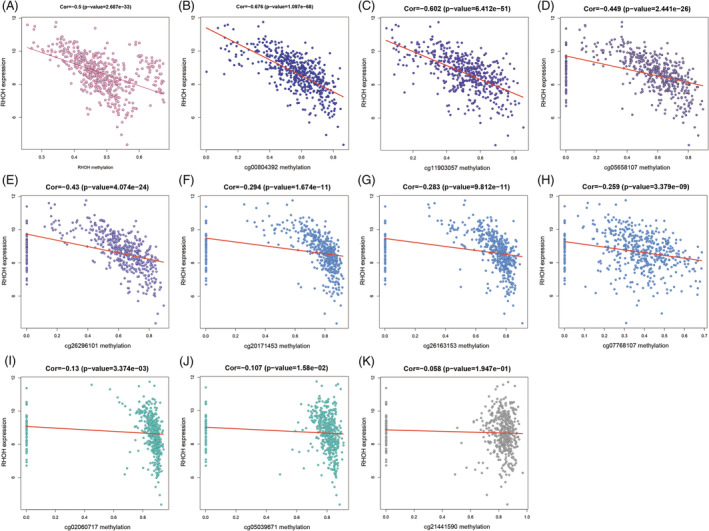
Relationship between Ras Homolog Gene Family Member H (RHOH) expression and DNA methylation. (A) Total. (B–K) Each methylation site
3.4. Correlation between RHOH methylation, expression, and prognosis
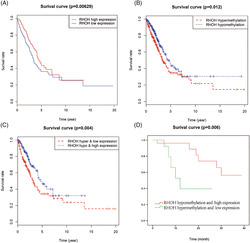
We identified a significant correlation between RHOH expression and the prognosis of LUAD patients (p = 0.00629) (Figure 3A) and found that patients with high RHOH expression had a higher OS rate than those with low RHOH expression. To investigate whether RHOH was associated with OS of LUAD patients, we further collected data of RHOH expression and patient prognosis in PREdiction of Clinical Outcomes from Genomic profiles. It is worth noting that in most datasets, patients with high RHOH expression levels were more likely to survive longer (Figure S2). We next analyzed the association between RHOH methylation and OS in LUAD patients and found that patients with RHOH hypermethylation had a significantly worse prognosis than those with RHOH hypomethylation (p = 0.012) (Figure 3B). Integrative analysis of DNA methylation and mRNA expression of RHOH revealed that patients with hypermethylation and low expression of RHOH had a significantly worse prognosis than those with hypomethylation and low expression of RHOH in both TCGA (p = 0.004) (Figure 3C) and validation cohort (p = 0.006, HR: 4.740, 95% CI: 1.567–14.340) (Figure 3D).
FIGURE 3.
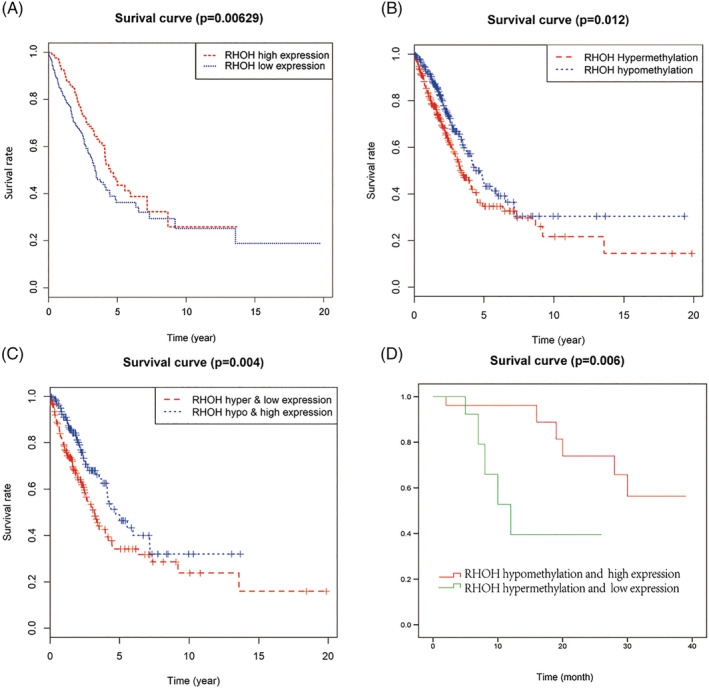
Survival curves of lung adenocarcinoma patients in The Cancer Genome Atlas and validation cohorts. (A) The blue line indicates RHOH high expression patients, and the red line indicates RHOH low expression patients, p = 0.00629. (B) The blue line indicates RHOH hypomethylation patients, and the red line indicates RHOH hypermethylation patients, p = 0.012. (C) Combined survival analysis of RHOH expression and methylation, p = 0.004. (D) Survival curves of lung adenocarcinoma patients in the validation cohort, p = 0.006. RHOH, Ras Homolog Gene Family Member H
3.5. Correlation between RHOH methylation and clinicopathological features
The correlation between RHOH expression and the clinical pathologic characteristics of LUAD patients was assessed by MEXPRESS. As shown in Figure 4, the mRNA expression level of RHOH was related to age (p = 0.023) and the tumor stage (p = 0.027) of LUAD patients. By measuring the methylation of RHOH in LUAD and normal lung tissues from the patients, we investigated the correlation between the clinicopathological features and RHOH methylation level (Table 1).
FIGURE 4.
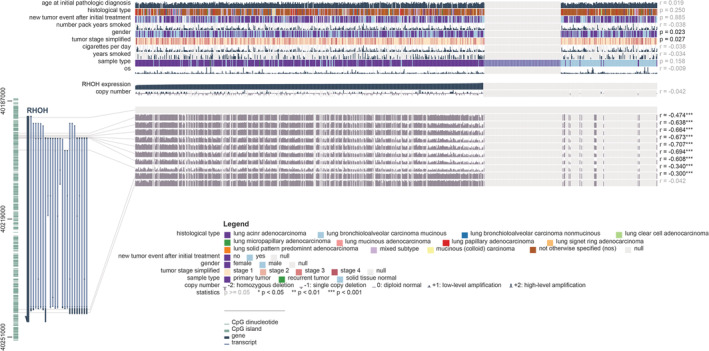
Correlation analysis of RHOH expression and methylation and relationship between RHOH expression and clinical features in The Cancer Genome Atlas cohort by MEXPRESS. RHOH, Ras Homolog Gene Family Member H
4. DISCUSSION
Studies have revealed the significance of DNA methylation as a promising biomarker for the diagnosis and prognosis of various malignant tumors including lung cancer. 12 , 13 DNA methylation and gene expression profiles hold great promise for the diagnosis and treatment of LUAD. Gene methylation alterations of PTEN, RASSF1A, DAPK, 14 APC, 15 and TGFBI 16 were found to be related to the prognosis of LUAD patients. In the present study, we further investigated RHOH by integrative analysis of DNA methylation, mRNA expression, and clinical prognosis of LUAD in TCGA.
Aberrant expression of Rho GTPase has been found in a variety of tumors. The Rho subfamily of GTPase was found to play a vital role in carcinogenesis and tumor progression 17 and act as a “molecular switch” in many signaling pathways. 18 High expression of RhoB in testicular cancer and breast cancer was found to be related to tumor progression, but whether it plays a tumor‐promoting or tumor‐suppressing role in colon cancer remains controversial. 19 , 20 Overexpression of RhoB in breast cancer was related to tumor progression, 21 whereas the loss of RhoB expression in advanced lung cancer suggested that it may play a tumor‐suppressing role. 22 RhoC was involved in the invasion and metastasis of breast cancer, lung cancer, pancreatic cancer, gastric cancer, and other malignant tumors. 23 , 24 , 25 , 26 RHOH was considered as proto‐oncogene in diffuse large B‐cell lymphoma, suggesting that overexpression of RHOH was associated with poor prognosis. 27 As RHOH is expressed in hematopoietic cells, it can be used as a liquid biopsy marker for differential diagnosis of tumors. 28 Abnormal expression of RHOH usually occurs in hematological disease, but it is rarely reported in solid tumors. It was reported that RHOH could promote migration of prostate cancer cells, and the prognosis of prostate cancer patients with high expression of RHOH was relatively poor. 29 It was found in our study that RHOH was hypermethylated and low‐expressed in LUAD, and the LUAD patients with high expression of RHOH had a favorable prognosis. In addition, the expression of RHOH was related to age and tumor stage; larger‐sample studies are required to verify our findings and conclusions.
RHOH is a membrane‐bound adaptor protein involved in proximal T cell receptor signaling, playing a key role in T cell differentiation. In a study on the screening of markers for early diagnosis of metastatic melanoma, RHOH was identified as central gene involved in immune response and tumor cell progression. In intestinal flora infection, RHOH was involved in the process of immunity and metabolism. 30 But whether RHOH is involved in the immune response and tumorigenesis of LUAD needs to be further studied.
The important role of Rho GTPase in hematological tumors has been confirmed in recent years, 31 , 32 but its role in LUAD has not been reported. We revealed the difference between tumor and normal lung tissue in RHOH methylation based on TCGA database and verified it with Asian population samples, indicating that RHOH may be a potential target for lung cancer treatment. This study has potential limitations that should be taken into account when interpreting the results. In view of the relatively small number of patients included in the validation cohort, it is necessary to conduct a larger study and have sufficient statistical capacity to verify the conclusions. The biological function of RHOH methylation in LUAD needs further study.
In summary, our study demonstrated a correlation between RHOH expression and methylation, which could be used as a biomarker for predicting the prognosis of LUAD patients. In addition, the correlation between RHOH expression and methylation in LUAD was correlated with the clinicopathological features of LUAD patients. In our ongoing study, we will further explore this correlation with immune metabolism in LUAD with the expectation that RHOH could become a new target for the treatment of LUAD.
AUTHOR CONTRIBUTIONS
Muyu Kuang performed data analysis and wrote the manuscript, and Zhenhua Zhou and Zhongyuan Lu carried out DNA extraction of tissues. Weina Shen, Haiyan Ge, and Xiaoting Tao helped to perform bioinformatics analysis. Yue Zhao and Lingdun Zhuge collected samples and information of clinical cases. Yihua Sun, Dongmei Ji, and Huibiao Zhang conceived of the study and participated in its designation and helped to draft the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST
The authors have stated that they have no conflicts of interest.
ETHICS STATEMENT
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of Huadong Hospital affiliated to Fudan University.
Supporting information
Figure S1. STRING protein interaction analysis of RHOH.
Figure S2. Survival curves concerning RHOH expression level of LUAD patients in PRECOG. LUAD, lung adenocarcinoma.
ACKNOWLEDGMENTS
This work was supported by the Research Foundation of Huadong Hospital (No. 2019jc006).
Kuang M, Zhou Z, Lu Z, et al. Prognostic prediction of lung adenocarcinoma by integrative analysis of RHOH expression and methylation. Clin Respir J. 2023;17(3):148‐156. doi: 10.1111/crj.13574
Muyu Kuang, Zhenhua Zhou, and Zhongyuan Lu contributed equally to this work.
Funding information Research Foundation of Huadong Hospital, Grant/Award Number: 2019jc006
Contributor Information
Yihua Sun, Email: sun_yihua76@hotmail.com.
Dongmei Ji, Email: jid09@fudan.edu.cn.
Huibiao Zhang, Email: huibiao_zhang@163.com.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2. Rizzi G, Lee JR, Dahl C, et al. Simultaneous profiling of DNA mutation and methylation by melting analysis using magnetoresistive biosensor array. ACS Nano. 2017;11(9):8864‐8870. doi: 10.1021/acsnano.7b03053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang P, Qiu W, Dudgeon C, et al. PUMA is directly activated by NF‐κB and contributes to TNF‐α‐induced apoptosis. Cell Death Differ. 2009;16(9):1192‐1202. doi: 10.1038/cdd.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sakai T, Toguchida J, Ohtani N, Yandell DW, Rapaport JM, Dryja TP. Allele‐specific hypermethylation of the retinoblastoma tumor‐suppressor gene. Am J Hum Genet. 1991;48(5):880‐888. [PMC free article] [PubMed] [Google Scholar]
- 5. Gündogdu MS, Liu H, Metzdorf D, et al. The haematopoietic GTPase RhoH modulates IL3 signalling through regulation of STAT activity and IL3 receptor expression. Mol Cancer. 2010;9(1):225. doi: 10.1186/1476-4598-9-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muro R, Nitta T, Nakano K, Okamura T, Takayanagi H, Suzuki H. γδTCR recruits the Syk/PI3K axis to drive proinflammatory differentiation program. J Clin Invest. 2018;128(1):415‐426. doi: 10.1172/jci95837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pan YR, Chen CC, Chan YT, et al. STAT3‐coordinated migration facilitates the dissemination of diffuse large B‐cell lymphomas. Nat Commun. 2018;9(1):3696. doi: 10.1038/s41467-018-06134-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Troeger A, Johnson AJ, Wood J, et al. RhoH is critical for cell‐microenvironment interactions in chronic lymphocytic leukemia in mice and humans. Blood. 2012;119(20):4708‐4718. doi: 10.1182/blood-2011-12-395939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwasaki T, Katsumi A, Kiyoi H, et al. Prognostic implication and biological roles of RhoH in acute myeloid leukaemia. Eur J Haematol. 2008;81(6):454‐460. doi: 10.1111/j.1600-0609.2008.01132.x [DOI] [PubMed] [Google Scholar]
- 10. Wang LX, Li Y, Chen GZ. Network‐based co‐expression analysis for exploring the potential diagnostic biomarkers of metastatic melanoma. PLoS ONE. 2018;13(1):e0190447. doi: 10.1371/journal.pone.0190447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koch A, De Meyer T, Jeschke J, Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16(1):636. doi: 10.1186/s12864-015-1847-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baglietto L, Ponzi E, Haycock P, et al. DNA methylation changes measured in pre‐diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer. 2017;140(1):50‐61. doi: 10.1002/ijc.30431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A. 2017;114(28):7414‐7419. doi: 10.1073/pnas.1703577114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site‐specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126(7):1630‐1639. doi: 10.1002/ijc.24896 [DOI] [PubMed] [Google Scholar]
- 15. Feng H, Zhang Z, Qing X, Wang X, Liang C, Liu D. Promoter methylation of APC and RAR‐beta genes as prognostic markers in non‐small cell lung cancer (NSCLC). Exp Mol Pathol. 2016;100(1):109‐113. doi: 10.1016/j.yexmp.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 16. Seok Y, Lee WK, Park JY, Kim DS. TGFBI promoter methylation is associated with poor prognosis in lung adenocarcinoma patients. Mol Cells. 2019;42(2):161‐165. doi: 10.14348/molcells.2018.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haga RB, Ridley AJ. Rho GTPases: regulation and roles in cancer cell biology. Small GTPases. 2016;7(4):207‐221. doi: 10.1080/21541248.2016.1232583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mitin N, Rossman KL, Der CJ. Signaling interplay in Ras superfamily function. Curr Biol. 2005;15(14):R563‐R574. doi: 10.1016/j.cub.2005.07.010 [DOI] [PubMed] [Google Scholar]
- 19. Dopeso H, Rodrigues P, Bilic J, et al. Mechanisms of inactivation of the tumour suppressor gene RHOA in colorectal cancer. Br J Cancer. 2018;118(1):106‐116. doi: 10.1038/bjc.2017.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jeong D, Park S, Kim H, et al. RhoA is associated with invasion and poor prognosis in colorectal cancer. Int J Oncol. 2016;48(2):714‐722. doi: 10.3892/ijo.2015.3281 [DOI] [PubMed] [Google Scholar]
- 21. Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87(6):635‐644. doi: 10.1038/sj.bjc.6600510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mazieres J, Antonia T, Daste G, et al. Loss of RhoB expression in human lung cancer progression. Clin Cancer Res. 2004;10(8):2742‐2750. doi: 10.1158/1078-0432.ccr-03-0149 [DOI] [PubMed] [Google Scholar]
- 23. Griner EM, Dancik GM, Costello JC, et al. RhoC is an unexpected target of RhoGDI2 in prevention of lung colonization of bladder cancer. Mol Cancer Res. 2015;13(3):483‐492. doi: 10.1158/1541-7786.mcr-14-0420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wynn ML, Yates JA, Evans CR. RhoC GTPase is a potent regulator of glutamine metabolism and N‐acetylaspartate production in inflammatory breast cancer cells. J Biol Chem. 2016;291(26):13715‐13729. doi: 10.1074/jbc.M115.703959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin M, DiVito MM, Merajver SD, Boyanapalli M, van Golen KL. Regulation of pancreatic cancer cell migration and invasion by RhoC GTPase and caveolin‐1. Mol Cancer. 2005;4(1):21. doi: 10.1186/1476-4598-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu Y, Chen YC, Sang JR, Xu WR. RhoC protein stimulates migration of gastric cancer cells through interaction with scaffold protein IQGAP1. Mol Med Rep. 2011;4(4):697‐703. doi: 10.3892/mmr.2011.482 [DOI] [PubMed] [Google Scholar]
- 27. Duquette ML, Huber MD, Maizels N. G‐rich proto‐oncogenes are targeted for genomic instability in B‐cell lymphomas. Cancer Res. 2007;67(6):2586‐2594. doi: 10.1158/0008-5472.can-06-2419 [DOI] [PubMed] [Google Scholar]
- 28. Zhang YH, Huang T, Chen L, et al. Identifying and analyzing different cancer subtypes using RNA‐seq data of blood platelets. Oncotarget. 2017;8(50):87494‐87511. doi: 10.18632/oncotarget.20903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tajadura‐Ortega V, Garg R, Allen R, et al. An RNAi screen of Rho signalling networks identifies RhoH as a regulator of Rac1 in prostate cancer cell migration. BMC Biol. 2018;16(1):29. doi: 10.1186/s12915-018-0489-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun Y, Zhuang Z, Wang X, Huang H, Fu Q, Yan Q. Dual RNA‐seq reveals the effect of the flgM gene of Pseudomonas plecoglossicida on the immune response of Epinephelus coioides. Fish Shellfish Immunol. 2019;87:515‐523. doi: 10.1016/j.fsi.2019.01.041 [DOI] [PubMed] [Google Scholar]
- 31. Voena C, Chiarle R. RHO family GTPases in the biology of lymphoma. Cell. 2019;8(7):646. doi: 10.3390/cells8070646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Que F, Zhang L, Wang T, Xu M, Li W, Zang S. RHOA G17V induces T follicular helper cell specification and involves angioimmunoblastic T‐cell lymphoma via upregulating the expression of PON2 through an NF‐κB‐dependent mechanism. Onco Targets Ther. 2022;11(1):2134536. doi: 10.1080/2162402x.2022.2134536 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. STRING protein interaction analysis of RHOH.
Figure S2. Survival curves concerning RHOH expression level of LUAD patients in PRECOG. LUAD, lung adenocarcinoma.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


