Abstract
The Saccharomyces cerevisiae U6 RNA gene, SNR6, possesses upstream sequences that allow productive binding in vitro of the RNA polymerase III (Pol III) transcription initiation factor IIIB (TFIIIB) in the absence of TFIIIC or other assembly factors. TFIIIC-independent transcription of SNR6 in vitro is highly sensitive to point mutations in a consensus TATA box at position −30. In contrast, the TATA box is dispensable for SNR6 transcription in vivo, apparently because TFIIIC bound to the intragenic A block and downstream B block can recruit TFIIIB via protein-protein interactions. A mutant allele of SNR6 with decreased spacing between the A and B blocks, snr6-Δ42, exhibits increased dependence on the upstream sequences in vivo. Unexpectedly, we find that in vivo expression of snr6-Δ42 is much more sensitive to mutations in a (dT-dA)7 tract between the TATA box and transcription start site than to mutations in the TATA box itself. Inversion of single base pairs in the center of the dT-dA tract nearly abolishes transcription of snr6-Δ42, yet inversion of all 7 base pairs has little effect on expression, indicating that the dA-dT tract is relatively orientation independent. Although it is within the TFIIIB footprint, point mutations in the dT-dA tract do not inhibit TFIIIB binding or TFIIIC-independent transcription of SNR6 in vitro. In the absence of the chromatin architectural protein Nhp6, dT-dA tract mutations are lethal even when A-to-B block spacing is wild type. We conclude that the (dT-dA)7 tract and Nhp6 cooperate to direct productive transcription complex assembly on SNR6 in vivo.
Initiation of transcription by an RNA polymerase at the start site of a gene involves multiple protein-DNA interactions, but one protein-DNA interaction often has a dominant role in specifying the site and efficiency of transcription complex assembly. For example, a central step in initiation by eukaryotic RNA polymerase II (Pol II) on many protein-coding genes is binding of transcription factor (TF) IID, via its TATA-binding protein (TBP) subunit, to an upstream TATA box promoter element. A key step in initiation by Pol III on tRNA genes is binding of TFIIIC to two intragenic promoter elements, the A and B blocks. TFIIIC then places TFIIIB upstream of the transcription start site (reviewed in reference 9). These distinct promoter recognition steps in Pol II and Pol III transcription imply divergent mechanisms for initiation complex assembly. Yet further characterization of Pol II and Pol III transcription initiation complexes has revealed striking similarities. For example, like TFIID, TFIIIB contains the TATA-binding protein as a subunit. Although most Pol III transcription units lack a consensus TATA box in the TFIIIB-binding region, A/T-rich sequences 15 to 30 base pairs upstream of the start site are known to contribute to the efficiency of tRNA and 5S rRNA gene transcription in vitro (29, 38) and in vivo (23, 40). In addition, an increasing number of Pol II transcription units have been found to contain TFIID-binding promoter elements 20 to 30 base pairs downstream of the transcription start site in a position analogous to that of the Pol III A block element (3, 33). These findings suggest that Pol II and Pol III transcription initiation complexes may in fact have similar architectures but vary in the relative contributions of different protein-DNA and protein-protein interactions to the assembly pathway.
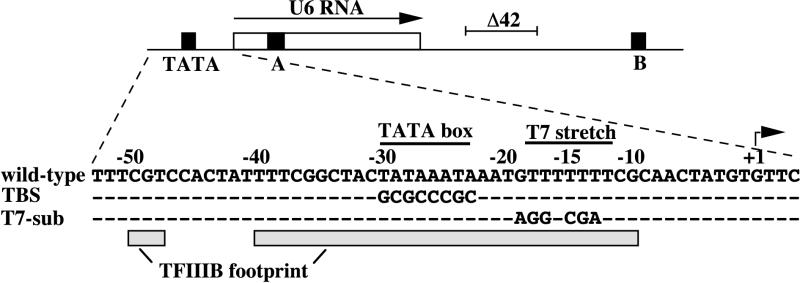
In this study we investigate a noncanonical Pol III transcription unit that contains a consensus upstream TATA box element, the Saccharomyces cerevisiae U6 RNA gene SNR6 (Fig. 1). SNR6 also contains the canonical A and B block promoter elements found in tRNA genes, but the B block is in a unique location 100 base pairs downstream of the transcribed region and 200 base pairs downstream of the A block (8). TFIIIB binding to SNR6 is more sequence specific than on most tRNA genes, perhaps because the broad separation of the A and B block elements reduces the ability of TFIIIC to precisely position TFIIIB. The presence of a relatively weak A block and an overlapping, cryptic A block in SNR6 may further increase the dependence of TFIIIB positioning on upstream sequences (10). The SNR6 upstream sequence is so highly adapted to binding TFIIIB that the yeast U6 gene can be transcribed in vitro with only purified TFIIIB and Pol III, in the complete absence of the assembly factor TFIIIC (14, 28). In contrast, only a few yeast tRNA genes are transcribed in a TFIIIC-independent manner in vitro, and these genes also contain a consensus TATA box (7). In vivo the SNR6 A and B blocks are essential promoter elements (8, 15), while the SNR6 TATA box is dispensable (8). The essential function of TFIIIC in vivo may reflect constraints on TFIIIB access to DNA packaged in chromatin (4, 10) or competition of TFIIIB with other soluble DNA-binding proteins. Nevertheless, the TATA box does contribute to TFIIIB placement in vivo, as its complete substitution (TATAbox-sub: TATAAATA→GCGCCCGC), while viable, results in degenerate start site selection (8).
FIG. 1.
At the top is a linear portrayal of SNR6, showing previously identified promoter elements as black boxes. The open box designates the segment of DNA that codes for the U6 RNA, and an arrow indicates the direction of SNR6 transcription. The line labeled Δ42 shows the portion of SNR6 that is deleted to create the Δ42 allele. The SNR6 sequence is shown for the wild-type, TATA box-sub (TBS) and T7-sub alleles. Dashes indicate no change from the wild-type sequence. The start site of transcription is indicated by an arrow at +1. The region protected by TFIIIB from DNase I digestion is indicated by the shaded boxes.
Deletion of 42 base pairs of noncoding sequence between the SNR6 terminator and downstream B block, creating the snr6-Δ42 allele (Fig. 1), decreases transcription in vivo and in cell extracts three- to fourfold (8) and disrupts the native chromatin structure over the SNR6 TATA box (10). This and other evidence suggests that the DNA between the SNR6 A and B blocks is packaged in a chromatin structure that brings the distant elements into the proper relative position for binding of TFIIIC (10, 27). The snr6-Δ42 allele appears to exhibit an increased dependence of TFIIIB binding on sequence-specific DNA contacts, since the TATAbox-sub mutation is lethal in the snr6-Δ42 allele (10).
Here we have utilized the snr6-Δ42 allele to identify upstream sequences important for productive transcription complex assembly in vivo. Surprisingly, even in the snr6-Δ42 allele the TATA box is relatively resistant to mutation of up to 4 of the 8 positions, perhaps because an overlapping nonconsensus TATA box 4 base pairs downstream can also direct TFIIIB placement. In contrast, a stretch of 7 dT-dA base pairs centered between the TATA box and the transcription start site is strikingly sensitive to mutation in the snr6-Δ42 allele. Inversion of 1, 3, or 5 base pairs in the T7 stretch is synthetically lethal with snr6-Δ42, as is repositioning the T7 stretch 4 base pairs downstream. In contrast, inversion of all 7 dT-dA base pairs has little effect on transcription of the snr6-Δ42 allele. Electrophoretic mobility shift assays (EMSAs) show that recombinant TBP and TFIIIB bind to a DNA probe bearing mutations in the T7 stretch with similar or higher affinity than to the wild-type SNR6 upstream region. Furthermore, transcription of T7 stretch mutant alleles with TFIIIB and purified Pol III is at least as efficient as transcription of wild-type SNR6. Thus, the T7 stretch may facilitate transcription complex assembly only on a chromatin template. Consistent with this idea, we find that the absence of the chromatin architectural protein Nhp6, recently implicated in SNR6 transcription (18, 25), is synthetically lethal with mutations in the T7 stretch. In addition, purified Nhp6 stimulates transcription of the snr6-Δ42 allele in yeast subcellular extract more than 20-fold. These results indicate that Nhp6 and the T7 stretch act cooperatively to facilitate transcription complex assembly on SNR6 in a chromatin environment.
MATERIALS AND METHODS
Plasmid construction.
Plasmids used in these experiments are derivatives of either p-539H6 (2) or pΔ139-178 (hereafter pΔ42) (8). Briefly, p-539H6 contains S. cerevisiae sequence from −539 to +630 relative to the start site of SNR6 transcription cloned into pUC118. pΔ42 was generated from the plasmid p-539H6 and contains a 42-base-pair deletion downstream of the transcription terminator. Plasmids used for in vivo study were generated by digestion of p-539H6 or pΔ42 with EcoRI-PstI, gel purification, and subsequent ligation into EcoRI-PstI-cut yeast shuttle vector pRS314 (TRP1, CEN6, ARSH4) (37). Plasmids containing the TATA-sub, AATA-sub, and TATAbox-sub mutations were described previously (8).
Oligonucleotide-directed mutagenesis was employed essentially as described by Kunkel et al. (21) to create mutant SNR6 alleles. TATA box mutagenesis was done on pRS314-Δ42 and used the oligonucleotide TATA box-sat. T7 stretch mutagenesis was done on pRS314-Δ42 using the following oligonucleotides: BFP-down-sat, U6T-15B, U6T-14B, and U6T-13B. Oligonucleotides T7-flip, T5-flip, T3-flip, T7-slide2, and T7-slide4 were used to mutagenize either pRS314-539H6 or pRS314-Δ42 to create alleles of the same name. The T6-flipU/Δ42 and T6-flipD/Δ42 alleles were made using the plasmid pRS314-T5-flip/Δ42 and the oligonucleotide T-12A/T-18A mut. This strategy also afforded new T7-flip/Δ42 and T5-flip/Δ42 alleles to confirm previous results. Following mutagenesis, the plasmids were sequenced from the TATA box to the B block to confirm that the only mutations present were those intentionally introduced. Plasmids used for in vitro transcription were generated by digestion of pRS314-T3-flip or pRS314-T3-flip/Δ42 with EcoRI and PstI, gel purification, and subsequent ligation into EcoRI-PstI-cut pUC118 (41). These plasmids were purified by CsCl gradient centrifugation prior to use in transcription reactions.
Oligonucleotides.
The following oligonucleotides were used in mutagenesis, where N is A, C, G, or T in a ratio of 70%:10%:10%:10% wild-type nucleotide to other three nucleotides, B is G, T, or C in equal amounts, and W is A or T in equal amounts: TATA box-sat, 5′-GTTGCGAAAAAAACATTNNNNNNNNGTAGCCGAAAATAGTGG; BFP-down-sat, 5′-GAACACATAGTTGCGAAANNNNNNNNTATTTATAGTAGCCG; U6T-15B, 5′-CATAGTTGCGAAABAAACATTTATTTATAGTAGC; U6T-14B, 5′-CATAGTTGCGAABAAAACATTTATTTATAGTAGC; U6T-13B, 5′-CATAGTTGCGABAAAAACATTTATTTATAGTAGC; T7-flip, 5′-CGCGAACACATAGTTGCGTTTTTTTCATTTATTTATAGTAGCC; T5-flip, 5′-CGCGAACACATAGTTGCGATTTTTACATTTATTTATAGTAGCC; T3-flip, 5′-CGCGAACACATAGTTGCGAATTTAACATTTATTTATAGTAGCC; T-12A/T-18A mut, 5′-CGCGAACACATAGTTGCGWTTTTTWCATTTATTTATAGTAGCC; T7-slide 2, 5′-CGCGAACACATAGTTGAAAAAAACACATTTATATATAGTAGCC; T7-slide 4, 5′-CGCGAACACATAGTAAAAAAACATTCATTTATATATAGTAGCC.
The following oligonucleotides were used in primer extension analysis: 6D, 5′-AAAACGAAATAAATCTCTTTG; 14C, 5′-ACAATCTCGGACGAATCCTC; 5B, 5′-AAGTTCCAAAAAATATGGCAAGC.
The following oligonucleotides were used to create T7 RNA polymerase templates for RNase protection probes:
5′ U6 PCR/XhoI, 5′-TCCGCTCGAGGTTCGCGAAGTAACCCTT; T7-U6R, 5′-TAATACGACTCACTATAGGGAAAACGAAATAAATCTC; SNR7-5′, 5′-TAGTATTCTCATCACGATTAACG; U5R-T7, 5′-TAATACGACTCACTATAGGGAAGTTCCAAAAAATATGGCAAGCCC.
In vivo analysis of gene function.
To test if an SNR6 allele can provide viable levels of U6 RNA, a plasmid shuffle was performed. Mutant alleles cloned into pRS314 plasmid were tested in the strain MWK027 (15), which has a chromosomal SNR6 deletion and carries pseudo-wild-type U6 (2) on a YCp50 (URA3 CEN4 ARS1) (35) plasmid. Trp+ Ura+ transformants of MWK027 were grown in yeast extract-peptone-dextrose (YEPD) overnight followed by plating to −Trp media containing 0.75 mg of 5-fluoroorotic acid (5-FOA)/ml. 5-FOA selects against cells that harbor a functional URA3 gene, thus assuring that the sole copy of SNR6 is the pRS314 plasmid-borne allele. Cells that produce a sufficient amount of U6 RNA from the mutant allele survive; those that do not die.
Total cellular RNA was isolated using the guanidinium thiocyanate method, including a 65°C phenol extraction (43). Cells were grown under conditions that selected for maintenance of both the pseudo-wild-type U6 plasmid and SNR6 plasmid. 32P-labeled oligonucleotides 6D and 14C were used in primer extensions of 0.25 to 0.75 μg of RNA as described by Eschenlauer et al. (8), except that the annealing temperature was held at 45°C. RNase protection assays were done according to the procedure outlined by Gilman (11), with a 4-h incubation of probe and RNA at 37°C. Reverse transcription and RNase protection reaction products were run on 6% polyacrylamide–8.3 M urea gels and visualized with a PhosphorImager (Molecular Dynamics). Data were quantitated with Molecular Dynamics ImageQuant software.
Strain construction.
Strain MM082 (snr6-Δ::LEU2 nhp6A-Δ2::ura3 nhp6B-Δ1::HIS3, pRS316-pseudo-wild-type U6) was used to test the effect of deletion of the NHP6A and NHP6B genes (nhp6-Δ) on SNR6 transcription. To construct this strain, PCR-generated DNA fragments containing the NHP6A and NHP6B loci of DKY625 (6) were transformed sequentially to knock out the corresponding loci in strain MM032 (MWK027 with pseudo-wild-type U6 in pRS317 rather than YCp50), and knockouts were confirmed by PCR and phenotype. Knockout strains were grown on plates containing 5-FOA to select for a mutation in the URA3 gene at the NHP6A locus. 5-FOA-resistant strains were transformed with pRS316-pseudo-wild-type U6 and selected on −Ura plates. Transformants were next grown on −Ura plates containing 2 mg of α-aminoadipate/ml to select for loss of the pRS317-pseudo-wild-type plasmid, thus creating MM082. Candidate MM082 strains were screened by 5-FOA sensitivity to confirm their identity.
EMSAs.
Complementary oligonucleotides corresponding to positions −45 to −8 of SNR6 or snr6-T3-flip were gel-purified and 5′-end labeled with polynucleotide kinase and [γ-32P]ATP. Oligonucleotides were annealed according to the protocol given by the manufacturer (Gibco). Double-stranded DNA was separated from unannealed oligonucleotides on a 12% native gel prior to passive elution at 37°C overnight. Protein-DNA complexes were formed in a volume of 20 μl at 20°C under the following conditions: 5 mM Tris-Cl (pH 8.0), 70 mM NaCl, 5 mM MgCl2, 2.5 mM KOAc, 25 μM EDTA, 0.1 mg of acetylated bovine serum albumin/ml, 4% glycerol, 5 ng of poly(dG-dC)/μl, and 0.5 fmol of probe. Reactions were run on 6% (29:1 acrylamide:bisacrylamide) native gels that included 50 mM Tris-borate, 1 mM EDTA, 1 mM MgCl2, 0.5 mM dithiothreitol, and 3% glycerol. Running buffer lacked dithiothreitol and glycerol. Gels (10 cm by 10 cm by 1 mm) were run at room temperature for 15 to 20 min at 15 V/cm before exposure to a PhosphorImager screen overnight. Recombinant yeast TFIIIB subunits were purified as described previously (16) and were a generous gift from George Kassavetis and Peter Geiduschek (University of California at San Diego). Nhp6A was purified as described previously (46) and was kindly provided by Reid Johnson (University of California at Los Angeles).
In vitro transcription.
TFIIIB-DNA complexes for TFIIIC-independent in vitro transcription were formed in 19.5-μl volumes containing 40 mM Tris-Cl (pH 8), 7 mM MgCl2, 3 mM dithiothreitol, 0.1 mg of acetylated bovine serum albumin/ml, 70 mM NaCl, and 200 ng of supercoiled plasmid DNA. Reactions contained 1 pmol of TBP, 360 fmol of Brf, and 300 fmol of B" (recombinant TFIIIB subunits; see above) and were incubated at 20°C for 60 min. Five microliters of nucleoside triphosphate (NTP) mix was then added to give final concentrations of 200 μM ATP, 100 μM CTP, 100 μM UTP, and 25 μM [α-32P]GTP (∼6 μCi/reaction). Purified yeast Pol III (a kind gift from G. Kassavetis and E. P. Geiduschek) was added 2 min after the NTP mix and was incubated for 60 min at 20°C. Transcription was stopped by addition of 25 μl of stop mix containing 10 mM Tris-Cl (pH 8), 20 mM EDTA, 0.2% sodium dodecyl sulfate (SDS), 200 μg of sheared salmon sperm DNA/ml, and a radiolabeled recovery marker. Following phenol-chloroform extraction and ethanol precipitation, samples were run on a 10% acrylamide (19:1 acrylamide:bisacrylamide)–8.3 M urea denaturing gel that included 50 mM Tris-borate and 1 mM EDTA. Gels (39 cm by 15.5 cm by 0.5 mm) were run for 85 min at 0.9 W/cm and exposed to a PhosphorImager screen overnight.
Transcription reactions using a subcellular extract were performed as previously described (8). Briefly, 19.5-μl reaction mixtures containing 40 mM HEPES (pH 7.9), 65 mM (NH4)2SO4, 7 mM MgCl2, 3 mM dithiothreitol, and 100 ng of plasmid DNA were assembled at room temperature. Three microliters of purified Nhp6A or protein dilution buffer (20 mM HEPES [pH 7.5], 100 mM potassium acetate, 1 mM EDTA, and 50% glycerol) was added, and reaction mixtures were placed at 25°C. After 5 min of incubation, 3 μl of subcellular extract (55.2 μg of protein) was added, and incubation was continued for 15 min. An additional 30 min of incubation followed addition of 2.5 μl of NTP mix containing 6 mM concentrations each of ATP, CTP, and UTP and 250 μM [α-32P]GTP (∼8 μCi/reaction). Reactions were brought to 0.1% SDS and 1 mg of proteinase K/ml and incubated for 5 min at 25°C then stopped and run on a discontinuous gel (23 cm by 16.5 cm by 0.75 mm) at 35 mA as described previously (15). The fixed and dried gel was exposed to a PhosphorImager screen overnight.
RESULTS
Sequence specificity of SNR6 TATA box function.
To determine the sequence specificity of TATA box function in vivo in the context of SNR6, we examined the effects on gene expression of a range of TATA box mutations in the snr6-Δ42 allele. From a combination of random and site-directed mutagenesis of the 8-base-pair consensus sequence (Fig. 1), 38 mutant alleles were obtained (Table 1). The in vivo function of each mutant allele as the sole copy of SNR6 was tested by the plasmid shuffle technique (see Materials and Methods). The growth phenotype at 30°C was scored by quantitative serial dilution. None of the 11 single base pair substitutions resulted in a detectable growth phenotype. However, 2 of the 17 double substitutions, 2 of the 4 triple substitutions, and 2 of the 5 quadruple substitutions resulted in a detectable decrease in growth rate (Table 1). The range of growth phenotypes obtained is illustrated in Fig. 2A, which shows the growth on solid medium of fivefold serial dilutions of cultures of several mutant strains.
TABLE 1.
Results of random mutagenesis of the SNR6 TATA box
| Allele no. | TATAAATAa | Growthb | Fxn. +5c | Total U6d |
|---|---|---|---|---|
| Δ42 | -------- | +++ | 0 | 0.27 |
| 101 | C------- | +++ | ||
| 102 | G------- | +++ | ||
| 103 | -C------ | +++ | ||
| 104 | -G------ | +++ | ||
| 105 | --A----- | +++ | 0 | |
| 106 | --C----- | +++ | ||
| 107 | --G----- | +++ | ||
| 108 | ---G---- | +++ | ||
| 109 | ----T--- | +++ | 0.05 | |
| 110 | -----G-- | +++ | ||
| 111 | -----T-- | +++ | ||
| 112 | A-----A- | +++ | ||
| 113 | CC------ | +++ | ||
| 114 | CT------ | +++ | ||
| 115 | G---T--- | +++ | 0.06 | |
| 116 | G------C | +++ | ||
| 117 | -G----C- | +++ | ||
| 118 | --AC---- | +++ | ||
| 119 | --AG---- | +++ | ||
| 120 | --A---A- | +++ | 0.04 | 0.20 |
| 121 | --G-G--- | +++ | 0.19 | 0.28 |
| 122 | ---C--C- | ++ | 0.08 | 0.21 |
| 123 | ---G--C- | +++ | ||
| 124 | ---T-C-- | +++ | 0.2 | |
| 125 | ---CG--- | ++ | 0.44 | 0.14 |
| 126 | ----T-A- | +++ | ||
| 127 | -----C-T | +++ | ||
| 128 | -----TG- | +++ | ||
| 129 | CC---G-- | +++ | ||
| 130 | -C-G--C- | ++ | 0.29 | 0.21 |
| 131 | -T-CG--- | + | 0.54 | 0.11 |
| 132 | ---CTC-- | +++ | 0 | |
| 133 | AG-TT--- | + | 0.38 | 0.16 |
| 134 | AC---G-C | ++ | 0 | 0.19 |
| 135 | -C--TTA- | +++ | ||
| 136 | GCGC---- | +++ | 0.4e | |
| 137 | ----CCGC | +++ | ||
| 138 | GCGCCCGC | − | 0.01 |
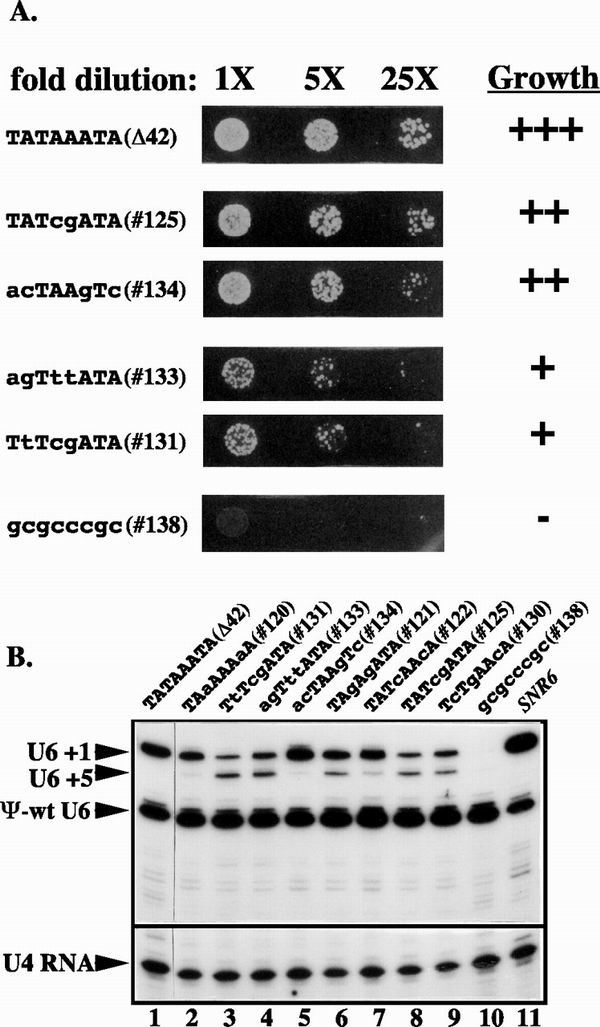
FIG. 2.
TATA box mutagenesis in the Δ42 allele results in growth defects and aberrant start site selection. Lower-case sequences indicate mutations in the wild-type TATA box. (A) Fivefold dilutions of representative mutants to assign growth values. 1X equals an optical density at 600 nm of 0.25. (B) Primer extension of transcripts produced by TATA box mutant alleles using primers complementary to U6 and U4 RNAs. Cells were grown under selection for the plasmids containing the mutant SNR6 allele and the pseudo-wild-type U6 allele prior to isolation of whole-cell RNA. U6 +1 indicates the U6 RNA that has initiated at the normal start site. U6 +5 indicates an aberrant start site 4 nucleotides downstream. ψ-wt U6 is pseudo-wild-type U6 RNA and is 13 nucleotides shorter than wild-type U6. U4 RNA is used as a normalization standard in this experiment.
To directly examine in vivo RNA synthesis from selected mutant alleles, transcripts were detected by primer extension using total cellular RNA from the mutant strains as template. Because the copy number of centromere-containing plasmids can increase severalfold under selective pressure, strains bearing the mutant SNR6 alleles also contained a pseudo-wild-type SNR6 allele, which provides functional U6 RNA but generates shorter primer extension products than the alleles to be analyzed (26). In the presence of the pseudo-wild-type SNR6 allele, the copy number of the plasmids bearing the mutant alleles is not expected to vary significantly between strains.
Figure 2B shows a representative primer extension experiment. Two effects on RNA synthesis are seen. First, most mutations result in some amount of misdirection of initiation to position +5. The percentage of transcripts that initiate at +5 varies from 0 to 54% (Table 1). Second, the total amount of U6 RNA that is synthesized varies. When RNA loading is normalized using the U4 RNA level as a control, the snr6-Δ42 allele produces 27% of the wild-type level of U6 RNA (Fig. 2B, compare lanes 1 and 11). Substitution of all eight positions of the TATA box (allele 138 is TATAbox-sub of reference 8) reduces the transcript level to 1% or less of wild-type (Fig. 2B, compare lanes 10 and 11). The tested 2-, 3-, and 4-base-pair substitution mutations result in 11 to 28% of the wild-type level of RNA synthesis (Table 1).
From these data we can draw two conclusions regarding the sequence specificity of SNR6 TATA box function in vivo. First, mutations that alter both halves of the TATA box result in the strongest growth phenotypes. Substitution of positions 1 to 4, as in allele 136 (TATA-sub of reference 8), has no effect on growth. In contrast, mutation of positions 4 and 5, as in allele 125, significantly impairs growth. The apparent requirement for mutations in both halves of the consensus TATA box to produce a growth defect is likely due to the fact that SNR6 has an overlapping TATA box sequence, −26/aATAAATg/-19. The nonconsensus TATA-like sequence is 4 base pairs downstream of the consensus TATA box and appears to be responsible for initiation at position +5. Second, mutations A4C and A5G in the consensus TATA box seem to be particularly deleterious to its function. These mutations appear in combination in alleles 125 and 131, which exhibit the greatest shift to initiation at position +5.
The T7 stretch: a novel promoter element between the SNR6 TATA box and transcription start site.
Given that the in vitro and in vivo footprints of TFIIIB extend well beyond the SNR6 TATA box (Fig. 1), it seemed possible that additional sequence-specific interactions might be detectable in the snr6-Δ42 background. A 6-base-pair substitution in the T-rich region immediately downstream of the TATA box (called T7-sub; Fig. 1) decreases SNR6 transcription approximately fivefold in vitro, although it has no effect in vivo (10). However, when introduced in the snr6-Δ42 allele, the T7-sub mutation is lethal. Analysis of RNA produced in vivo from the snr6-T7-sub/Δ42 allele documents a synergistic effect of the two mutations on transcript levels, resulting in approximately 1% of wild-type transcription (data not shown). Thus, mutations downstream of the TATA box act similarly to mutations in the TATA box when present in combination with the Δ42 mutation.
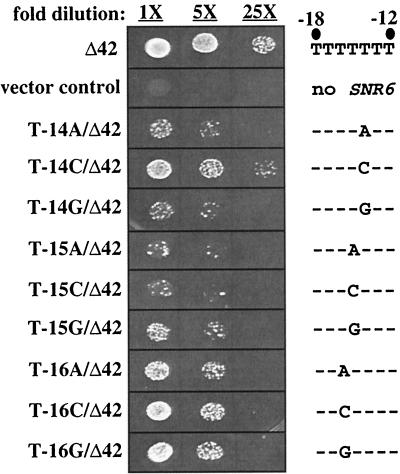
To determine which residues downstream of the TATA box are important for transcription of the Δ42 allele, we randomly mutagenized residues −22 to −15 of snr6-Δ42. Thirty-three mutant alleles were obtained (11 single, 16 double, 5 triple, and 1 quadruple substitution), and the function of each as the sole SNR6 allele was tested as described for the TATA box mutants. The only alleles that resulted in growth defects were those that have substitutions in positions −17, −16, or −15, which correspond to positions 2, 3, and 4 of the T7 stretch (data not shown). To systematically examine the sequence requirement for the T7 stretch, we made every possible single substitution at the middle five positions. Remarkably, the phenotype resulting from some of these point mutations is more severe than that of the most severe quadruple substitution in the TATA box. Figure 3 shows the growth phenotype of point mutations in the middle three positions of the T7 stretch in a Δ42 allele. The T7 stretch positions that are most sensitive to mutation are T-14 and T-15. In general, T-to-A transversions in the middle of the T7 stretch resulted in the strongest growth phenotype. The more severe T7 stretch mutations exhibit a papillate growth phenotype similar to that of a B block mutation that was shown to grow weakly when present on a centromeric plasmid but was lethal when integrated (15).
FIG. 3.
Point mutations at positions T-14 and T-15 in the Δ42 allele have the greatest effect on growth. Cells were grown overnight in YEPD followed by plating fivefold dilutions on 5-FOA, −Trp. 1X equals an optical density at 600 nm of 0.13. Plates were incubated at 30°C for 2 days. The sequence of the T7 stretch of each SNR6 allele is shown to the right of the figure.
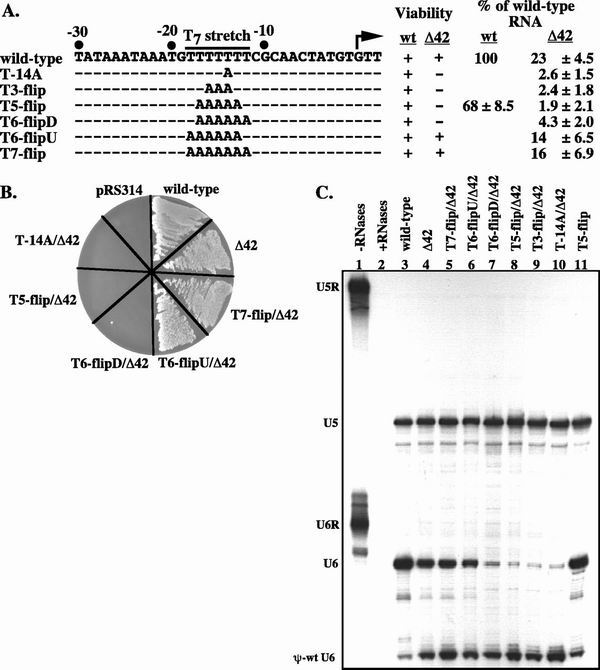
The above results imply that dT residues in the nontemplate strand and/or dA residues in the template strand at positions −14 and −15 are important to facilitate transcription of SNR6 by Pol III. Point mutations in the T7 stretch could alter TFIIIB base-specific interactions or disrupt the structure provided by a contiguous stretch of dT-dA base pairs. To distinguish between these two possibilities, additional mutations were made in the T7 stretch. Since poly(dT-dA) elements that perform a structural role in transcription have previously been shown to function in an orientation-independent manner (13, 47), we inverted the entire T7 stretch sequence to place the dT residues in the template DNA strand of the snr6-Δ42 allele (called T7-flip; Fig. 4A). Plasmid shuffle demonstrated that snr6-T7-flip/Δ42 produces enough U6 RNA for normal growth (Fig. 4B). Using RNase protection assays to analyze RNA steady-state levels, U6 RNA levels from snr6-T7-flip/Δ42 are only slightly lower than the levels produced by the Δ42 mutation alone (Fig. 4C, compare lanes 4 and 5). This result suggests that the T7 stretch is acting as an orientation-independent structural element that is crucial for transcription of snr6-Δ42, although there is slight preference for dT residues in the nontemplate strand.
FIG. 4.
Full and partial inversions of the T7 stretch suggest that it acts as a structural element in promoting transcription. (A) Sequence upstream of the transcription start site in wild-type and T7 stretch mutant alleles in the nontemplate strand; the TATA box is located at position −30 to −23. Dashed lines indicate no change from the wild-type sequence. Plus and minus signs indicate viability and inviability, respectively. The amount of U6 RNA present in each strain is designated as the percent of the wild-type (wt) value. Values listed are the averages of at least four experiments and are shown ± standard deviation. (B) Viability test to determine if any of the T7 stretch mutants are synthetically lethal with Δ42. Cells were grown in YEPD followed by plating to 5-FOA, −Trp, to select against the plasmid harboring the pseudo-wild-type U6 allele. The plates were incubated at 30°C for 2 days. pRS314 contains no SNR6 allele and should therefore die on 5-FOA, −Trp. (C) Total cellular RNA from strains containing the indicated SNR6 allele and the pseudo-wild-type U6 allele on separate plasmids was analyzed by RNase protection assay. Samples were run on an 8.3 M urea–6% polyacrylamide gel and exposed to a PhosphorImager screen overnight. U5 RNA is a normalization standard in this experiment. The undigested probes and probes digested in the absence of total cellular RNA are shown in lanes 1 and 2, respectively.
If the structure of the T7 stretch is necessary for efficient transcription, there should be a minimal length that is sufficient to adopt the structure found in (dA-dT) elements. Any element that is shorter than the minimal length will no longer facilitate transcription. Two new mutants were constructed that invert the middle five T residues (T5-flip) or the middle three T residues (T3-flip) of the T7 stretch. In contrast to T7-flip, either T5-flip or T3-flip is lethal in snr6-Δ42 (Fig. 4B; see Fig. 6). RNase protection assays show that T5-flip/Δ42 produces about 2% of wild-type U6 RNA steady-state levels, a 10-fold reduction in the amount of U6 RNA found in Δ42 strains (Fig. 4C, compare lanes 4 and 8). T3-flip/Δ42 also exhibits a severe transcription defect. Thus, dA-dT tracts of less than 6 base pairs are insufficient to facilitate transcription of the Δ42 allele.
FIG. 6.
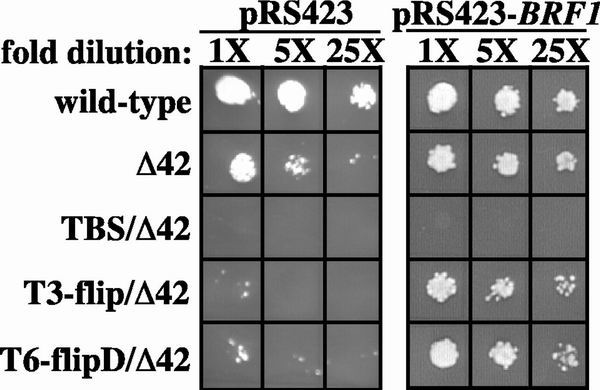
Overexpression of BRF1 suppresses the synthetic lethality of T7 stretch/Δ42 mutations. Cells were grown overnight in YEPD prior to plating on 5-FOA (−Trp, −His). Fivefold dilutions were grown at 30°C for 2 days. 1X equals an optical density at 600 nm of 0.08. The SNR6 alleles are listed to the left of the panels. pRS423 is a HIS3-marked high-copy vector, and cells were tested with either the empty vector or a vector containing BRF1 (pRS423-BRF1).
Two different 6-base-pair dA-dT stretches were constructed to determine whether a 6-base-pair tract could promote transcription of SNR6. The upstream and downstream 6 base pairs of the T7 stretch were inverted in the Δ42 allele to create T6-flipU/Δ42 and T6-flipD/Δ42 (Fig. 4A). When present as the sole copy of SNR6 in a cell, snr6-T6-flipU/Δ42 generates sufficient U6 RNA to provide viability (Fig. 4B), an amount nearly equivalent to that produced by T7-flip/Δ42 (Fig. 4C, compare lanes 5 and 6). However, the T6-flipD/Δ42 mutation does not promote transcription to the same degree. In fact, T6-flipD is synthetically lethal with Δ42 (Fig. 4B) and is transcribed only slightly better than T5-flip/Δ42 (Fig. 4C, compare lanes 7 and 8). These results suggest that a 6-base-pair dA-dT tract is marginally sufficient to adopt the structure necessary for snr6-Δ42 transcription. These two 6-base-pair tracts are not functionally equivalent in nature, possibly resulting from an altered position relative to the TATA box (see below) or differences in the dinucleotide steps present at the 5′ and 3′ tract junctions.
The T7 stretch cannot reposition the transcription start site.
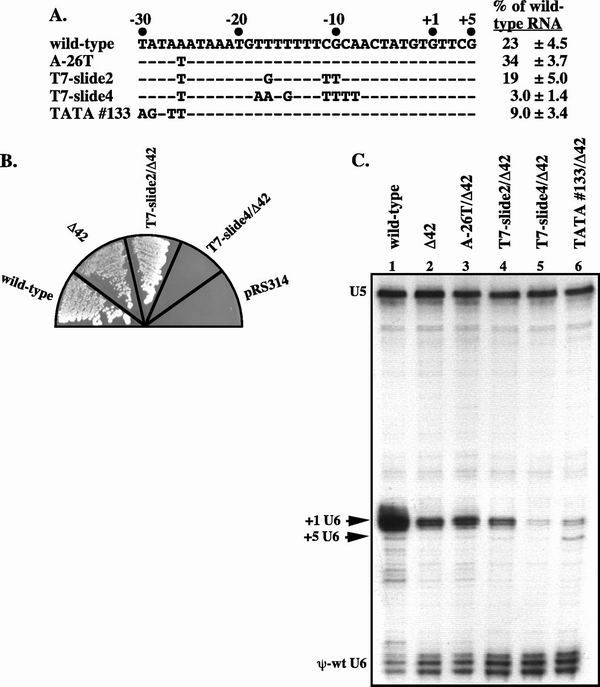
To determine if the location of the T7 stretch can influence start site selection, as expected if it is a strong determinant of TFIIIB positioning, we moved it 2 or 4 base pairs downstream of its normal position. To facilitate a switch of TFIIIB from the −30 TATA box to the nonconsensus −26 TATA box, the two elements were made identical in sequence at positions 1 to 4, 6, and 7 by replacing −26A with T in the Δ42 allele (A-26T, Fig. 5A). The A-26T/Δ42 allele produces a U6 RNA population that contains 5% of transcripts initiating at +5. If the T7 stretch affects the positioning of TFIIIB, transcription initiation should shift from +1 to +5 as the T7 stretch is moved downstream in the A-26T/Δ42 allele. We hypothesized that the T7-slide4/Δ42 allele (Fig. 5A) would produce the most +5 U6 RNA, since this mutation places the −26 TATA box and T7 stretch at the same spacing as is present in the wild-type allele. Surprisingly, we found that T7-slide4 is synthetically lethal with Δ42, while the T7-slide2/Δ42 strain is viable (Fig. 5B) and has only a minor transcription defect (Fig. 5C, lane 4). Neither strain exhibits an increase in the amount of +5 U6 RNA that is synthesized (Fig. 5C, lane 5). We conclude that the T7 stretch does not play a major role in positioning TFIIIB, but the location of the T7 stretch appears to be critical for the efficiency of SNR6 transcription.
FIG. 5.
Positioning the T7 stretch further downstream from the TATA box does not affect transcription start site selection but does decrease overall transcription. (A) Sequence of mutant alleles used to test the effect of moving the T7 stretch. Dashes represent no change from the wild-type sequence. Both the normal (+1) and aberrant (+5) start sites are shown. Values for the in vivo transcription activity of these mutant alleles are compiled from the data shown in panel C of this figure. These values are the averages of three experiments and are shown ± standard deviations. (B) Growth phenotype when the mutant allele is the sole copy of SNR6. Cells were grown overnight in YEPD before streaking onto 5-FOA, −Trp. Plates were incubated at 30°C for 2 days. The plasmid-borne allele of SNR6 is indicated in each sector, except for pRS314, which has no SNR6 allele and serves as a negative control. (C) RNase protection of whole-cell RNA from strains expressing the indicated mutant SNR6 alleles and the pseudo-wild-type U6 RNA. Products specific to both +1 and +5 start sites are shown. U5 RNA is used as a control.
T7 stretch mutations do not disrupt TFIIIB binding or TFIIIC-independent transcription of SNR6.
Mutations in the T7 stretch may affect SNR6 transcription by lowering the affinity of TFIIIB for SNR6 upstream sequences. This hypothesis was tested in vivo by increasing the effective amount of TFIIIB present in the cell. It has previously been shown that overexpression of the Brf1 subunit of TFIIIB increases transcription from mutant but not wild-type Pol III promoters (36). We transformed cells containing various SNR6 alleles with a high-copy plasmid harboring either BRF1 or no insert. Cells were subsequently grown on 5-FOA (−Trp, His) to determine the ability of BRF1 overexpression to suppress the synthetic lethalities of TBS/Δ42, T3-flip/Δ42, and T6-flipD/Δ42 (Fig. 6). BRF1 overexpression partially suppresses the synthetic lethalities of both T3-flip/Δ42 and T6-flipD/Δ42. Although it is possible that BRF1 overexpression suppresses Δ42 and not the upstream mutations, this seems unlikely since the TBS/Δ42 mutation was not rescued. The presence of an empty vector had no effect on cell growth in any strain. These results suggest that the T7 stretch mutations inhibit the assembly of TFIIIB on SNR6 in vivo. However, we cannot exclude indirect effects due to BRF1 overexpression.
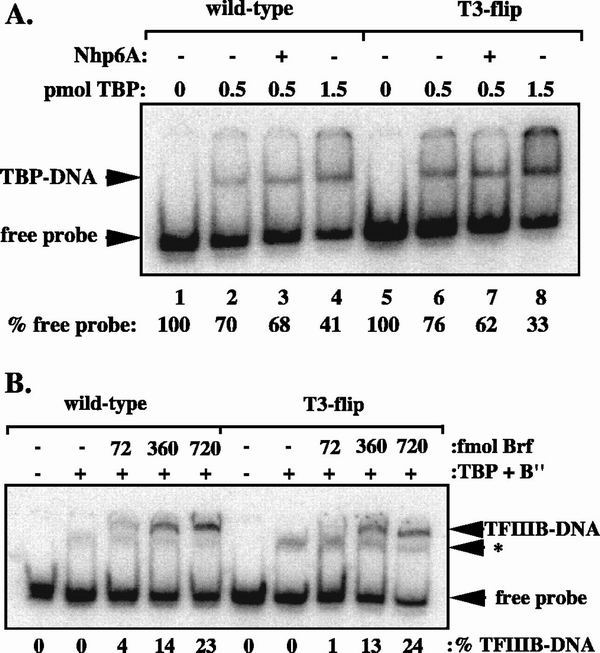
To examine directly whether T7 stretch mutations result in decreased affinity for TFIIIB and/or its individual subunits (TBP, Brf, B"), we performed EMSAs using recombinant TFIIIB components. All EMSA experiments were done using wild-type and T3-flip probes that contained SNR6 sequence from −45 to −8, which was found to be the minimal sequence required to assemble a TFIIIB-DNA complex (5). Binding of TBP might be affected by the T3-flip mutation, since in vitro analysis of TBP binding demonstrates a preference for specific sequences flanking the TATA box (24, 45). However, TBP exhibited no marked preference for binding wild-type or mutant probes (Fig. 7A, compare lanes 2 and 4 with lanes 6 and 8). The kinetics of TBP binding were also examined; mutant and wild-type SNR6 probes exhibit no difference in rate of association with TBP (data not shown).
FIG. 7.
The T3-flip mutation does not inhibit TFIIIB binding to the SNR6 promoter in vitro. (A) In vitro binding of TBP to SNR6 promoters. Free probe is expressed as a percentage of the amount in the protein-free lane. Wild-type and T3-flip probes were incubated with the indicated amount of TBP for 45 min, followed by loading on a 6% polyacrylamide (30:1 acrylamide:bisacrylamide) gel. The presence of 13 pmol of Nhp6 is indicated by a plus sign. (B) Brf binds with similar affinity to wild-type and T3-flip probes. The amounts of added TBP and B" are 1 pmol and 300 fmol, respectively. Reaction conditions were identical to those described for panel A, except heparin was added to 180 μg/ml prior to loading to remove nonspecific complexes from the DNA. An asterisk indicates a heparin-resistant complex that is present when TBP and B" are added to the probe. The percentage of probe in TFIIIB-DNA complex is listed below each lane.
Given our Brf overexpression results and the fact that Brf is the only TFIIIB subunit that cross-links efficiently to the region corresponding to the SNR6 T7 stretch (1, 31), we also tested whether Brf incorporation is affected by the T3-flip mutation. TFIIIB complex formation was assayed at a fixed TBP concentration, with saturating levels of B" and various amounts of Brf. Any TBP-Brf-DNA complexes should be chased into heparin-resistant TFIIIB-DNA complexes under these conditions. We found that Brf incorporation is not impaired on the T3-flip probe (Fig. 7B). For both wild-type and T3-flip DNAs, 23 to 24% of the probe was shifted into a TFIIIB-DNA band with the highest amount of Brf. (Interestingly, the T3-flip DNA yielded ∼3-fold higher amounts of a novel heparin-resistant complex formed in reactions containing only TBP and B". The significance of this result is unclear.) Furthermore, when the cloned T3-flip/Δ42 and T-14A/Δ42 alleles were incubated with recombinant TFIIIB subunits, purified Pol III and NTPs, transcription was at least as efficient as for wild-type SNR6 (data not shown). Thus, T7 stretch mutations do not inhibit TFIIIC-independent transcription in vitro, and so may hinder SNR6 transcription in vivo by interfering with TFIIIC function or altering chromatin structure.
Nhp6 cooperates with the T7 stretch in SNR6 transcription complex assembly.
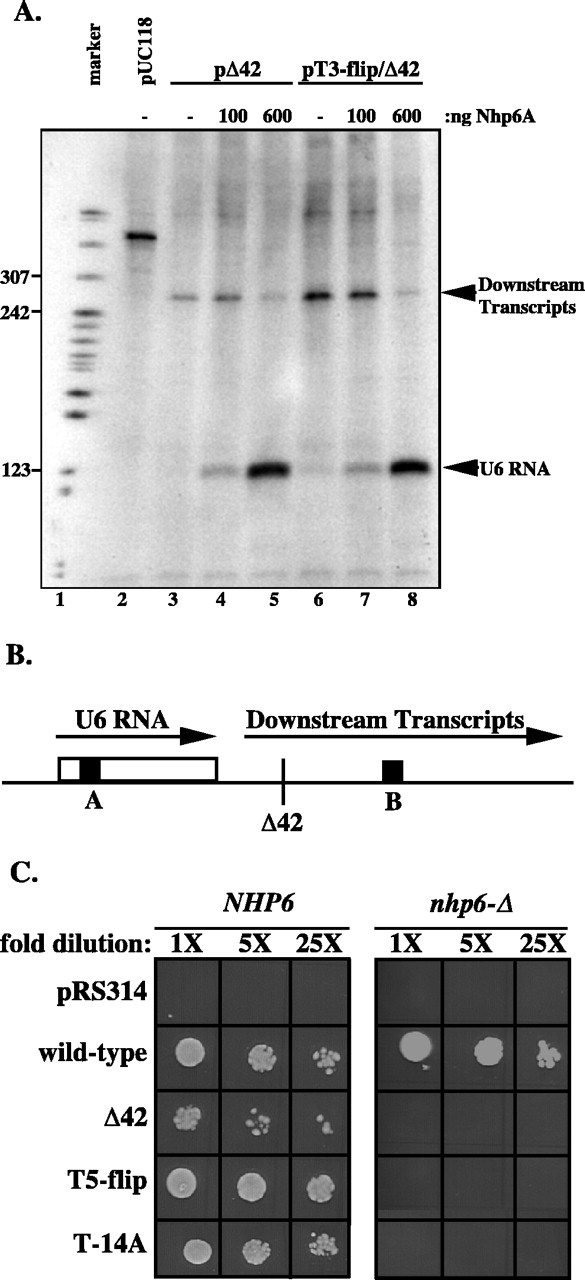
Recently published studies by Kruppa et al. (18) and Lopez et al. (25) implicate the yeast nonhistone chromatin protein Nhp6 in stimulation of SNR6 transcription. Deletion of both of the redundant and nonessential NHP6A and NHP6B genes decreases U6 RNA accumulation, while overexpression of Nhp6A or Nhp6B suppresses mutations in the SNR6 TATA box or B block. Two models have been proposed for the function of Nhp6 at the SNR6 promoter: (i) it may stimulate binding of TBP (and thus TFIIIB) to the SNR6 TATA box, as it has been shown to do for a Pol II promoter (30a), or (ii) it may package DNA between the SNR6 A and B blocks, thus stimulating binding of TFIIIC to these distantly spaced promoter elements. We find that purified Nhp6A does not appear to stimulate binding of TBP or TFIIIB to wild-type or T3-flip SNR6 upstream DNA in vitro (Fig. 7A, lanes 3 and 7 and data not shown). However, purified Nhp6A greatly stimulates transcription of the snr6-Δ42 and snr6-T3-flip/Δ42 alleles in yeast subcellular extract (Fig. 8A). At the highest Nhp6 concentration, U6 RNA yield was increased >20-fold, whereas transcription from a wild-type SNR6 allele increased only about 4-fold under these conditions (18, 25, and data not shown). Importantly, stimulation of U6 RNA synthesis is coincident with inhibition of downstream transcript synthesis. The downstream transcripts arise from misinitiation downstream of the SNR6 terminator (Fig. 8B) (8) when B block-bound TFIIIC fails to recognize the SNR6 A block. Packaging of DNA between the SNR6 A and B blocks into a protein complex would be expected to suppress such misinitiation and stimulate proper initiation. Thus, our results favor the latter model for Nhp6 function on SNR6.
FIG. 8.
Nhp6 stimulates transcription of SNR6. (A) In vitro transcription of SNR6 is stimulated by Nhp6A while downstream transcripts are suppressed. Plasmids pUC118 (vector control), pΔ42, and pT3-flip/Δ42 were incubated with 0, 100, or 600 ng of purified Nhp6A for 5 min before addition of subcellular extract. After a 15-min incubation nucleoside triphosphates were added, and incubation continued for 30 min. Samples were treated with SDS and proteinase K prior to loading on a denaturing gel. Marker consists of pBR322 cut with MspI. (B) Schematic of U6 RNA and downstream transcripts from the Δ42 allele. Endpoints of transcription are indicated with an arrowhead. Downstream transcripts terminate in an oligo(dT) stretch at +378 of the Δ42 allele. The location of the Δ42 deletion is indicated, and the open box represents the U6 RNA coding region. (C) Promoter mutations in SNR6 are lethal in the absence of Nhp6. Strains that are isogenic except for deletions of NHP6A and NHP6B were grown over- night in YEPD before dilutions were plated to 5-FOA, −trp. 1X equals an optical density at 600 nm of 0.1. Strains with wild-type NHP6 were grown at 30°C for 2 days, while nhp6-Δ strains were grown at 30°C for 3 days. SNR6 alleles are listed to the left of the panels, with pRS314 being a vector control.
To determine if Nhp6 similarly influences transcription of mutant SNR6 alleles in vivo, we tested whether nhp6-Δ enhances the effect of our SNR6 mutations. A nhp6-Δ snr6-Δ strain was transformed with various SNR6 alleles on centromeric plasmids. Plasmid shuffle experiments showed that snr6-Δ42 is synthetically lethal with nhp6-Δ, while wild-type SNR6 exhibits a slow growth phenotype characteristic of nhp6-Δ (Fig. 8C). The T-14A and T5-flip mutations, which have no effect on growth and little effect on transcription in an otherwise wild-type SNR6 allele, are lethal in the absence of Nhp6 (Fig. 8C). The marked enhancement of T7 stretch and Δ42 mutations in the absence of Nhp6 strongly implies a cooperative function of Nhp6 and the T7 stretch in assembly of a functional transcription complex on SNR6 in vivo.
DISCUSSION
In this study we mutated the SNR6 TATA box and T7 stretch upstream sequences and studied their function in vivo, making use of an SNR6 allele with an increased dependence on upstream contacts for Pol III transcription. Although the SNR6 TATA box is relatively refractory to mutation in vivo, we could nevertheless discern a sequence preference for promoter activity. In particular, mutations at positions 4 and 5 of the snr6-Δ42 TATA box produced the greatest decrease in transcription and misdirection of initiation to position +5. These results imply similar sequence requirements for binding of the TBP-containing initiation factors TFIID and TFIIIB to Pol II and Pol III TATA boxes, respectively, since substitution of either position 4 or 5 of a consensus TATA box with dC or dG results in a severe decrease in Pol II transcription (44).
Surprisingly, the SNR6 T7 stretch is much more sensitive to mutation in vivo than the TATA box. The orientation-independent function of the T7 stretch suggests that a particular DNA structure is critical to facilitate SNR6 transcription. There is evidence suggesting that oligo(dA-dT) tracts induce an overall DNA curvature or bend in solution and that this altered DNA structure is more likely to form as the length of a dA-dT tract increases (17). Consistent with natural curvature induced by the T7 stretch, Grove and coworkers (12) found that an increase in DNA flexure due to insertion of single-stranded loops or missing nucleosides in a region centered at position −15 of a yeast tRNA gene stimulates TFIIIC-independent formation of the TFIIIB-DNA complex. This result suggests that TFIIIB binding induces an extended DNA deformation between the TATA box and transcription start site. Conceivably, the SNR6 T7 stretch is naturally curved in a way that promotes TFIIIB function. However, we were unable to detect any inhibition by T7 stretch mutations of TFIIIB binding or TFIIIC-independent transcription in vitro.
The TATA box and the T7 stretch appear to have different functions in directing the transcription of SNR6 by Pol III. The TATA box is responsible for the appropriate placement of TFIIIB, via binding of its TBP subunit, in order to initiate transcription at +1. Mutations in the TATA box generally alter start site selection. The presence of a consensus TATA box in SNR6 may have evolved in order to direct precise initiation of the U6 RNA at +1. Precise initiation may be more important for U6 RNA, which does not undergo trimming of its 5′ end, than for other Pol III transcripts, such as tRNA and RNase P RNA, that are 5′-end trimmed during maturation. In contrast to the TATA box, our results indicate that the T7 stretch does not play a major role in start site selection. Placement of the T7 stretch downstream of its normal location results in decreased transcription, but no aberrant start site selection is associated with these mutations. We conclude that the T7 stretch participates in forming active Pol III complexes on SNR6 in vivo but cannot position TFIIIB. This is consistent with the absence of a marked effect of the T3-flip mutation on TFIIIB binding in vitro, demonstrating that the T7 stretch is not the major determinant of TFIIIB binding to SNR6.
Two possible functions of the T7 stretch are interaction with TFIIIC and opening of the chromatin structure at the TFIIIB binding site. Both of these functions are expected to be important in vivo but unnecessary in the TFIIIC-independent in vitro transcription assay. TFIIIC directs the selective recruitment of TFIIIB rather than TFIID to the SNR6 promoter in vivo (34), as well as the proper orientation of TFIIIB on the near-symmetric SNR6 TATA box in vitro (42). The second-largest subunit of TFIIIC, Tfc4 (Pcf1), reaches far upstream of the start site in the transcription initiation complex and cross-links efficiently to the region corresponding to the SNR6 T7 stretch (1). Tfc4 is the TFIIIC subunit that directs recruitment of TFIIIB to the promoter, primarily via interaction with Brf (9). It seems possible that the T7 stretch may mediate the interaction between Tfc4 and Brf, given that both proteins cross-link to the corresponding region of Pol III promoters. Such a function would, of course, not be required in TFIIIC-independent transcription but could be particularly crucial in vivo if TFIIIC binding to the A block is inhibited by the Δ42 mutation or by the absence of Nhp6.
The T7 stretch could also aid transcription in vivo by precluding nucleosome assembly over the upstream portion of SNR6. Poly(dA-dT) elements have been shown to inhibit assembly of nucleosomes (20, 32) and to increase accessibility of DNA within a nucleosome (47). Also, in vitro experiments show that nucleosomes are passed upstream during Pol III transcription (39). If this phenomenon occurs in vivo, an element antagonistic to nucleosome assembly might prevent TFIIIB displacement by nucleosomes, resulting in faster recycling of Pol III. TFIIIC-independent transcription of SNR6 by purified TFIIIB and Pol III becomes TFIIIC dependent when SNR6 is first assembled into chromatin (4). Thus, TFIIIC appears to function in derepression of chromatin. Indeed, human TFIIIC exhibits histone acetyltransferase activity (19). It is therefore reasonable that a nucleosome-disrupting activity of the T7 stretch would become essential when TFIIIC interaction with the SNR6 upstream region is disrupted by the Δ42 mutation or by the absence of Nhp6.
If the T7 stretch facilitates TFIIIB recruitment in vivo by either of the proposed mechanisms, one might expect to find it in other Pol III promoters, perhaps in conjunction with a consensus TATA box. Indeed, three yeast tRNA genes possess a consensus TATA box and a T-rich sequence in a position analogous to that of SNR6 (7). These tRNA genes are similar to SNR6 in possessing abnormally large A-to-B block spacing due to the presence of introns (7). Interestingly, RPR1, which encodes the RNase P RNA and is transcribed by Pol III in yeast, possesses an upstream T7 stretch interrupted by a single C residue but lacks a good match to the SNR6 TATA box or the consensus B block (22). Furthermore, the RPR1 and SNR6 T-rich sequences are flanked by the same dinucleotides (underlined)—TGTTTcTTTTCG (lowercasing indicates a nucleotide present in RPR1 but not SNR6)—and are present in the same position relative to the transcription start site (22). Finally, a T-rich sequence 14 to 21 base pairs upstream of the start site of a silkworm tRNA gene contributes to the efficiency of transcription in crude nuclear extracts (30). Thus, a T7 stretch centered between 10 and 20 base pairs upstream of a Pol III transcription start site may be a common adaptation to suboptimal position or sequence of the A and B block promoter elements.
ACKNOWLEDGMENTS
We are very grateful to George Kassavetis for generously providing purified proteins and instruction in the TFIIIB-binding assays. We also thank Reid Johnson, Ian Willis, and David Kolodrubetz for providing purified proteins, plasmids, and yeast strains, respectively. We are grateful to George Kassavetis for critical reading of the manuscript and to members of the Brow and Dahlberg labs for discussions and suggestions. We thank David Kolodrubetz and Mike Kruppa for sharing unpublished results.
This work was supported by National Institutes of Health grant GM44665 to D.A.B. V.L.G. was a trainee of National Institutes of Health training grant GM07215.
REFERENCES
- 1.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brow D A, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. doi: 10.1101/gad.4.8.1345. [DOI] [PubMed] [Google Scholar]
- 3.Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- 4.Burnol A F, Margottin F, Huet J, Almouzni G, Prioleau M N, Mechali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- 5.Colbert T, Lee S, Schimmack G, Hahn S. Architecture of protein and DNA contacts within the TFIIIB-DNA complex. Mol Cell Biol. 1998;18:1682–1691. doi: 10.1128/mcb.18.3.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costigan C, Kolodrubetz D, Snyder M. NHP6A and NHP6B, which encode HMG1-like proteins, are candidates for downstream components of the yeast SLT2 mitogen-activated protein kinase pathway. Mol Cell Biol. 1994;14:2391–2403. doi: 10.1128/mcb.14.4.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dieci G, Percudani R, Giuliodori S, Bottarelli L, Ottonello S. TFIIIC-independent in vitro transcription of yeast tRNA genes. J Mol Biol. 2000;299:601–613. doi: 10.1006/jmbi.2000.3783. [DOI] [PubMed] [Google Scholar]
- 8.Eschenlauer J B, Kaiser M W, Gerlach V L, Brow D A. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol. 1993;13:3015–3026. doi: 10.1128/mcb.13.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geiduschek E P, Kassavetis G A. The RNA polymerase III transcription apparatus. J Mol Biol. 2001;310:1–26. doi: 10.1006/jmbi.2001.4732. [DOI] [PubMed] [Google Scholar]
- 10.Gerlach V L, Whitehall S K, Geiduschek E P, Brow D A. TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol Cell Biol. 1995;15:1455–1466. doi: 10.1128/mcb.15.3.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilman M. Ribonuclease protection assay. In: Ausubel R B F M, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1993. pp. 4.7.1–4.7.8. [Google Scholar]
- 12.Grove A, Kassavetis G A, Johnson T E, Geiduschek E P. The RNA polymerase III-recruiting factor TFIIIB induces a DNA bend between the TATA box and the transcriptional start site. J Mol Biol. 1999;285:1429–1440. doi: 10.1006/jmbi.1998.2347. [DOI] [PubMed] [Google Scholar]
- 13.Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joazeiro C A, Kassavetis G A, Geiduschek E P. Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes. Mol Cell Biol. 1994;14:2798–2808. doi: 10.1128/mcb.14.4.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser M W, Brow D A. Lethal mutations in a yeast U6 RNA gene B block promoter element identify essential contacts with transcription factor-IIIC. J Biol Chem. 1995;270:11398–11405. doi: 10.1074/jbc.270.19.11398. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis G A, Letts G A, Geiduschek E P. The RNA polymerase III transcription initiation factor TFIIIB participates in two steps of promoter opening. EMBO J. 2001;20:2823–2834. doi: 10.1093/emboj/20.11.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koo H S, Wu H M, Crothers D M. DNA bending at adenine and thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 18.Kruppa M, Moir R D, Kolodrubetz D, Willis I M. Nhp6: an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol Cell. 2001;7:309–318. doi: 10.1016/s1097-2765(01)00179-4. [DOI] [PubMed] [Google Scholar]
- 19.Kundu T K, Wang Z, Roeder R G. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunkel G R, Martinson H G. Nucleosomes will not form on double-stranded RNA or over poly(dA)-poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981;9:6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 22.Lee J Y, Evans C F, Engelke D R. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci USA. 1991;88:6986–6990. doi: 10.1073/pnas.88.16.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Linning R M, Kondo K, Honda B M. Differential expression of individual suppressor tRNA(Trp) gene gene family members in vitro and in vivo in the nematode Caenorhabditis elegans. Mol Cell Biol. 1998;18:703–709. doi: 10.1128/mcb.18.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Librizzi M D, Brenowitz M, Willis I M. The TATA element and its context affect the cooperative interaction of TATA-binding protein with the TFIIB-related factor, TFIIIB70. J Biol Chem. 1998;273:4563–4568. doi: 10.1074/jbc.273.8.4563. [DOI] [PubMed] [Google Scholar]
- 25.Lopez S, Livingstone-Zatchej M, Jourdain S, Thoma F, Sentenac A, Marsolier M C. High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene. Mol Cell Biol. 2001;21:3096–3104. doi: 10.1128/MCB.21.9.3096-3104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madhani H D, Bordonne R, Guthrie C. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev. 1990;4:2264–2277. doi: 10.1101/gad.4.12b.2264. [DOI] [PubMed] [Google Scholar]
- 27.Marsolier M C, Tanaka S, Livingstone-Zatchej M, Grunstein M, Thoma F, Sentenac A. Reciprocal interferences between nucleosomal organization and transcriptional activity of the yeast SNR6 gene. Genes Dev. 1995;9:410–422. doi: 10.1101/gad.9.4.410. [DOI] [PubMed] [Google Scholar]
- 28.Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J. 1990;9:271–277. doi: 10.1002/j.1460-2075.1990.tb08105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton D G, Sprague K U. In vitro transcription of a silkworm 5S RNA gene requires an upstream signal. Proc Natl Acad Sci USA. 1984;81:5519–5522. doi: 10.1073/pnas.81.17.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palida F A, Hale C, Sprague K U. Transcription of a silkworm tRNA(cAla) gene is directed by two AT-rich upstream sequence elements. Nucleic Acids Res. 1993;21:5875–5881. doi: 10.1093/nar/21.25.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Paull T T, Carey M, Johnson R C. Yeast HMG proteins NHP6A/B potentiate promoter-specific transcriptional activation in vivo and assembly of pre-initiation complexes in vitro. Genes Dev. 1996;10:2769–2781. doi: 10.1101/gad.10.21.2769. [DOI] [PubMed] [Google Scholar]
- 31.Persinger J, Sengupta S M, Bartholomew B. Spatial organization of the core region of yeast TFIIIB-DNA complexes. Mol Cell Biol. 1999;19:5218–5234. doi: 10.1128/mcb.19.7.5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA)-poly(dT) EMBO J. 1982;1:173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purnell B A, Emanuel P A, Gilmour D S. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- 34.Roberts S, Colbert T, Hahn S. TFIIIC determines RNA polymerase III specificity at the TATA-containing yeast U6 promoter. Genes Dev. 1995;9:832–842. doi: 10.1101/gad.9.7.832. [DOI] [PubMed] [Google Scholar]
- 35.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 36.Sethy-Coraci I, Moir R D, Lopez-de-Leon A, Willis I M. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 1998;26:2344–2352. doi: 10.1093/nar/26.10.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sprague K U, Larson D, Morton D. 5′ Flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980;22:171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- 39.Studitsky V M, Kassavetis G A, Geiduschek E P, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 40.Trivedi A, Young L S, Ouyang C, Johnson D L, Sprague K U. A TATA element is required for tRNA promoter activity and confers TATA-binding protein responsiveness in Drosophila Schneider-2 cells. J Biol Chem. 1999;274:11369–11375. doi: 10.1074/jbc.274.16.11369. [DOI] [PubMed] [Google Scholar]
- 41.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 42.Whitehall S K, Kassavetis G A, Geiduschek E P. The symmetry of the yeast U6 RNA gene's TATA box and the orientation of the TATA-binding protein in yeast TFIIIB. Genes Dev. 1995;9:2974–2985. doi: 10.1101/gad.9.23.2974. [DOI] [PubMed] [Google Scholar]
- 43.Wise J A, Tollervey D, Maloney D, Swerdlow H, Dunn E J, Guthrie C. Yeast contains small nuclear RNAs encoded by single copy genes. Cell. 1983;35:743–751. doi: 10.1016/0092-8674(83)90107-1. [DOI] [PubMed] [Google Scholar]
- 44.Wobbe C R, Struhl K. Yeast and human TATA-binding proteins have nearly identical DNA sequence requirements for transcription in vitro. Mol Cell Biol. 1990;10:3859–3867. doi: 10.1128/mcb.10.8.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong J M, Bateman E. TBP-DNA interactions in the minor groove discriminate between A:T and T:A base pairs. Nucleic Acids Res. 1994;22:1890–1896. doi: 10.1093/nar/22.10.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yen Y M, Wong B, Johnson R C. Determinants of DNA binding and bending by the Saccharomyces cerevisiae high mobility group protein NHP6A that are important for its biological activities. Role of the unique N terminus and putative intercalating methionine. J Biol Chem. 1998;273:4424–4435. doi: 10.1074/jbc.273.8.4424. [DOI] [PubMed] [Google Scholar]
- 47.Zhu Z, Thiele D J. A specialized nucleosome modulates transcription factor access to a C. glabrata metal responsive promoter. Cell. 1996;87:459–470. doi: 10.1016/s0092-8674(00)81366-5. [DOI] [PubMed] [Google Scholar]