Abstract
Environmental sustainability is an increasingly important issue in industry. As an environmentally friendly and sustainable way, constructing microbial cell factories to produce all kinds of valuable products has attracted more and more attention. In the process of constructing microbial cell factories, systems biology plays a crucial role. This review summarizes the recent applications of systems biology in the design and construction of microbial cell factories from four perspectives, including functional genes/enzymes discovery, bottleneck pathways identification, strains tolerance improvement and design and construction of synthetic microbial consortia. Systems biology tools can be employed to identify functional genes/enzymes involved in the biosynthetic pathways of products. These discovered genes are introduced into appropriate chassis strains to build engineering microorganisms capable of producing products. Subsequently, systems biology tools are used to identify bottleneck pathways, improve strains tolerance and guide design and construction of synthetic microbial consortia, resulting in increasing the yield of engineered strains and constructing microbial cell factories successfully.
Keywords: Microbial cell factories, Systems biology, Functional genes/enzymes discovery, Bottleneck pathways, Strains tolerance, Synthetic microbial consortia
1. Introduction
Environmental sustainability is an increasingly important issue in industry. So, it is critical to find a sustainable, green and clean way to produce enzymes, fuels, bulk chemicals and natural products for maintaining a sustainable social economy. As an environmentally friendly and sustainable way, biotechnological industry has attracted more and more attention. With the increased size of the bio-based production market, microbial cell factories offer a promising approach to manufacturing valuable products from renewable resources [1]. More and more high-value chemicals have been successfully industrialized through microbial cell factories, such as raspberry ketonel [2], salidroside [3], gastrodin [4], salvianic acid A [5] and artemisinin [6]. One noteworthy example is the development of a process for the production of 1,4-butanediol directly in Escherichia coli by the company Genomatica (San Diego, CA), which reached the production scale of 30,000 tons/year [7]. Thus, we can see the great potential of using microbial cell factories to produce all kinds of valuable products.
However, from the design to the construction of microbial cell factories, there are some crucial problems to resolve. Firstly, the biosynthetic pathways of most desired products are not fully elucidated and enzymes that catalyze proposed pathways remain unknown. Therefore, finding crucially functional genes/enzymes involved in the biosynthetic pathways is the first key step to construct microbial cell factories. After the biosynthetic pathways are constructed in suitable hosts, it's key to find bottleneck pathways that limit the improvement in the yield of desired products, and then alleviate these bottleneck pathways to facilitate the construction of microbial cell factories. In the process of constructing microbial cell factories, substrates, toxic intermediates, products or other environmental conditions may limit the growth ability of strains and the yield of products. Therefore, it's also crucial to improve the tolerance of strains to various stresses. In addition, during the process of constructing microbial cell factories, using single engineered strain to produce chemicals often faces a large metabolic burden due to the long and complex biosynthetic pathways, resulting in low production efficiency. Design and construction of synthetic microbial consortia is an effective strategy to relive metabolic burden and improve production efficiency.
Systems biology, as an important tool, plays an important role in solving these mentioned problems to facilitate the design and construction of microbial cell factories. The scope of systems biology is to investigate biological systems in a holistic manner to elucidate the mechanisms underlying the cellular behavior [8]. With the continuous development of systems biology, it has been possible to comprehensively understand the metabolic network of strains from the genomic scale, including the structural genes that constitute metabolic pathways, the complex regulatory mechanisms of cell metabolism, and the effects of genetic and environmental disturbances on cell global metabolism, so as to establish metabolic models to evaluate and predict the possible effects of genetic engineering operations [9,10]. And omics technologies (e.g. genomics, transcriptomics, proteomics, metabolomics, and fluxomics) are the major analytical tools of systems biology. By analyzing the metabolic network of strains obtained by genetic engineering, we can better guide the metabolic engineering and improve the physiological function and production efficiency of strains.
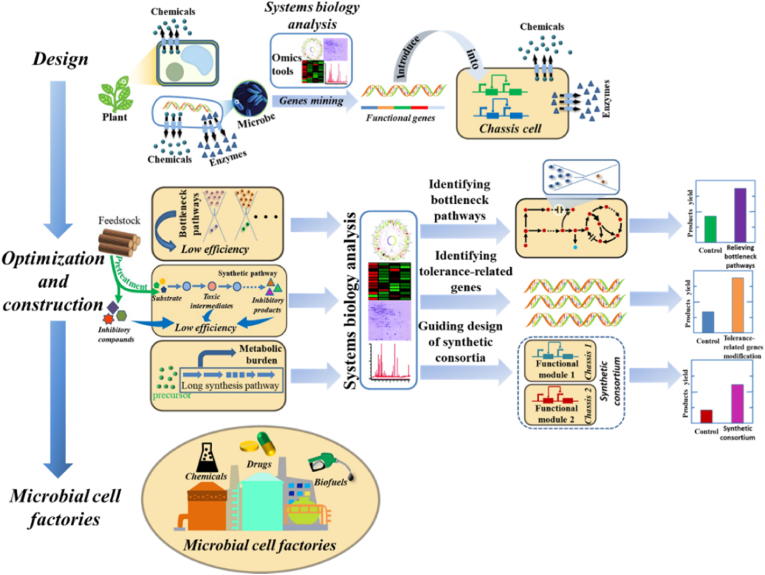
Genomics, transcriptomics and proteomics are the most common tools to discover functional genes/enzymes involved in the biosynthetic pathways of desired products. To identify bottlenecks in metabolic pathways, metabolomics is one of the most effective tools. Through the detection of relevant metabolic perturbations, metabolomics can identify potential bottleneck pathways for strains improvement [11]. For improving the tolerance of strains, transcriptomics, proteomics and metabolomics can be used to identify potential tolerance-related genes, proteins and metabolites to guide strains tolerance engineering. For designing and constructing microbial consortia, systems biology analysis can be employed to systematically analyze the genetic and metabolic pathways in microbial consortia, contributing to elucidating comprehensive molecular mechanisms of interactions in microbial consortia. In conclusion, from design to construction of microbial cell factories, systems biology plays an important role [12]. The general process of design and construction of microbial cell factories based on systems biology is shown in Fig. 1. First, systems biology tools are employed to identify functional genes/enzymes involved in the biosynthetic pathways of products. These discovered genes will be introduced into appropriate chassis strains to build engineering strains capable of producing products. Then, in order to improve the yield of engineered strains for facilitating the construction of microbial cell factories, systems biology tools are used to identify bottleneck pathways, improve strains tolerance and guide design and construction of synthetic microbial consortia. After these optimization steps, microbial cell factories capable of high-yielding products are successfully constructed.
Fig. 1.
Design and construction of microbial cell factories based on systems biology analysis.
This review summarizes the recent applications of systems biology in the design and construction of microbial cell factories. And it is summarized from four perspectives, including functional genes/enzymes discovery, bottleneck pathways identification, strains tolerance improvement and design and construction of synthetic microbial consortia.
2. Strains design based on systems biology
For design and construction of microbial cell factories, the first step is to introduce appropriate biosynthetic pathways of products into chassis strains to design engineered strains capable of producing desired products. However, the biosynthetic pathways of most desired product are not fully elucidated and enzymes that catalyze proposed pathways remain unknown. Therefore, to design strains capable of producing products, it's essential to mine functional genes/enzymes involved in synthetic pathways of products. The numerous enzymes and pathways present from plants and microorganisms represent a large resource for constructing synthetic pathways to produce chemicals or degrade pollutants. With the development of systems biology tools, more and more potentially valuable enzymes and pathways are identified, and some of them have been successfully applied to microbial cell factories [13].
Among all omics tools, genomics, transcriptomics and proteomics are most commonly used to identify functional genes. Many potential genes/enzymes, involved in producing variously valuable chemicals, such as ibogaine, ferruginol, olivetolic acid and so on, have been found by systems biology tools (Table 1). And these genes/enzymes are mainly derived from various of plants in nature. In addition to discovering genes/enzymes for biosynthesis of high-value chemicals, systems biology tools are used to identify functional genes/enzymes to degrade polymer pollutants, such as plastics, petroleum and so on, which is helpful to construct microbial cell factories. For example, the drastically increasing amount of plastic waste is causing an environmental crisis. Therefore, some novel enzymes discovered by systems biology tools are summarized, which can degrade various plastic waste, such as polyurethane (PU), poly (ethylene terephthalate) (PET) and so on (Table 1). These enzymes are mainly derived from variously environmental microbial community, which is different from the genes/enzymes involved in synthetic pathways of high-value products. The complex structures of mentioned compounds in Table 1 are shown in Figure S1.
Table 1.
Discovered genes/enzymes for producing desired chemicals/degrading targeted pollutants.
| Desired chemicals/Targeted pollutants | Pathway Genes/Enzymes discovered | Origin | Omics tools | Reference | |
|---|---|---|---|---|---|
| Desired chemicals | ibogaine | ibogamine 10-hydroxylase, oribogaine-10-O-methyltransferase | Tabernanthe iboga | genomics/transcriptomics | [14] |
| ferruginol | CYP76AH1 | Salvia miltiorrhiza | transcriptomics | [15] | |
| O-glc- echinocystic acid | CYP716Y1 | Bupleurum falcatum | transcriptomics | [16] | |
| cucurbitacin B and E | Cm180, CmBi, CmACT, Cm890, CmBt, Cl180, ClBi, ClACT, Cl890A, Cl890B, ClBt | Cucumis melo L, Citrullus lanatus | genomics/transcriptomics | [17] | |
| wogonin | O-methyltransferase (PFOMT5) | Scutellaria baicalensis Georgi | genomics | [18] | |
| saponins | oxidosqualene cyclases, CYP450s, UDP-glucuronosyltransferases | Maesa lanceolate, Chenopodium quinoa | transcriptomics | [19] | |
| olivetolic acid | olivetolic acid cyclase | Cannabis flowers | transcriptomics | [20] | |
| Targeted pollutants | PET | a PET hydrolytic enzyme | Actinobacteria | metagenomics | [21] |
| PU | gene sequences similar to PU hydrolytic enzyme | Landfill microbial community | metagenomics | [22] | |
| synthetic polyesters: PLA, PCL, PBSA | synthetic polyesters hydrolytic enzyme belonging to new esterase families | Environmental microbial community | metagenomics | [23] | |
| PCL, PET | a novel thermostable cutinase homologue, leaf and branch compost cutinase | Microbial community of a leaf-branch compost with copious natural plant-derived polymers | metagenomics | [24] | |
| poly DEGA | esterases capable of hydrolyzing poly DEGA | Moss-associated microorganisms. | metagenomics | [25] | |
| PBAT | esterases capable of hydrolyzing synthetic copolyester PBAT | Soil compost microorganisms | metagenomics | [25] | |
| PBAT | a novel PBAT degrading polyesterase- PpEst | Pseudomonas pseudoalcaligenes and Knufia chersoneso | proteomics | [26] | |
| PHB | a PHB depolymerase ALC24_4107 | Alcanivorax sp. 24 | proteomics | [27] | |
2.1. Functional genes/enzymes discovery for producing high-value chemicals
To discover functional genes/enzymes for producing high-value chemicals, genomics and transcriptomics are the most commonly used tools. Next-generation sequencing, an important genomic tool, is extensively used to discover potentially valuable genes/enzymes, contributing to construct microbial cell factories. Farrow et al. [14] used next-generation sequencing to generate the first iboga transcriptome and leveraged homology-guided gene discovery to identify the penultimate hydroxylase ibogamine 10-hydroxylase and final O-methyltransferase steps noribogaine-10-O-methyltransferase in ibogaine biosynthesis, which is greatly significant for the construction of microbial cell factory to produce ibogaine. There is another example, which used a next-generation sequencing approach combined with comparative transcriptomics analysis to identify six candidate cytochrome P450 CYP genes responsible for converting miltiradiene to tanshinones, bioactive compounds from Chinese medicinal herb. One of these cytochromes, CYP76AH1, was demonstrated to catalyze a unique four-electron oxidation cascade on miltiradiene to produce ferruginol both in vitro and in vivo. Expressing the CYP76AH1 and phyto-CYP genes in Saccharomyces cerevisiae resulted in heterologous production of 10.5 mg/L ferruginol, the precursor of tanshinones [15].
Comparative analysis of the genomes is also a common genomic tool for discovering potentially functional genes. Cucurbitacins, a group of bitter and highly oxygenated tetracyclic triterpenes, are mainly produced by the plant family Cucurbitaceae. These compounds have similar structures but differ in their antitumour activities and ecophysiological roles. Zhou et al. [17] identified conserved syntenic loci encoding metabolic genes for distinct cucurbitacins by comparative analysis of the genomes of cucumber, melon and watermelon. Characterization of the cytochrome P450s (CYPs) identified from these loci led to find a novel multi-oxidation CYP for the tailoring of the cucurbitacin core skeleton as well as two other CYPs responsible for the key structural variations among cucurbitacins C, B and E. They also discovered a syntenic gene cluster of transcription factors that regulates the tissue-specific biosynthesis of cucurbitacins and may confer the loss of bitterness phenotypes associated with convergent domestication of wild cucurbits. Baicalein, wogonin, and their glycosides baicalin and wogonoside, are the major flavone compounds with explicit pharmacological activities in extracts of the roots of Scutellaria baicalensis. Comparative genomic analysis was used to analyze the published genomes of Salvia miltiorrhiza, Salvia splendens, and Salvia indicum, for elucidating the final step for biosynthesis of wogonin from norwogonin [18]. The O-methyltransferases were finally identified to be responsible for wogonin biosynthesis and confirmed by in vivo assays in yeast as well as by RNAi experiments in hairy roots of S. baicalensis.
Differential expression gene analysis based on the RNA-seq data (comparative transcriptomics) is a common strategy for the gene function prediction, which makes full use of the different gene expression between the reference and test groups. For instance, saponin accumulation was dramatically activated upon MeJA treatment in many plant tissues, which caused the elevated expression of related genes in biosynthetic pathways [28]. By comparison of MeJA-treated samples and untreated control, numerous oxidosqualene cyclases, CYP450s, and UDP-glucuronosyltransferases were screened out and verified involved in the biosynthesis of saponins in many plants such as Maesa lanceolate and Chenopodium quinoa [19]. Likewise, some key genes were identified to produce olivetolic acid, which was proposed to be the first intermediate to form the polyketide nucleus of the cannabinoids in Cannabis sativa. Gagne et al. [20] identify an olivetolic acid cyclase that catalyzes a C2-C7 intramolecular aldol condensation with carboxylate retention to form olivetolic acid by analyzing the transcriptome of cannabis flowers. Combined expression of a tetraketide synthase and this olivetolic acid cyclase in yeast achieved a 0.48 mg/L yield of olivetolic acid when cultures were supplied with exogenous hexanoate.
In a different example, to identify the genes involved in methanol metabolism in methylotrophic yeast Pichia pastoris, a comparative proteomic analysis was employed to analyze Pichia pastoris cultivated in minimal media containing methanol and glucose, respectively. The result demonstrated the transaldolase isoenzyme was highly up-regulated in methanol medium cultivation, which plays an important role in methanol utilization [29].
2.2. Functional genes/enzymes discovery for degrading pollutants
In addition to discovering genes/enzymes involved in synthetic pathways of high-value chemicals, systems biology tools are used to identify functional genes/enzymes able to degrade polymer pollutants such as plastics, petroleum and so on, which is helpful to construct microbial cell factories to produce enzymes, capable of degrading polymer substance. During these pollutants, the drastically increasing amount of plastic waste is causing an environmental crisis. Therefore, in this section we take novel plastic-degrading enzymes discovery as example to explain how to discover functional genes/enzymes for degrading pollutants via omics-based approaches.
Metagenomics has shown great potential to facilitate the discovery of novel enzymes from environment. Specifically, there are two different methods to mine enzymes via metagenomics: sequence-based screening and function-based screening [30]. Sequence-based screening takes advantage of sequence similarity comparison and functional gene annotation by searching bioinformatic databases [31]. For example, a PET hydrolytic enzyme was uncovered through in silico sequence based screening from metagenome databases [21]. In the past few years, many gene sequences similar to those encoding known enzymes capable of degrading PU plastics were identified from landfill-derived metagenomes [22]. Alternatively, function-based screening uses activity assays to search for the desired phenotypes from metagenomic libraries. Compared with sequence-based screening, function-based screening is particularly advantageous in mining completely novel groups of enzymes for which the sequences are more divergent from existing homologous ones. For example, multiple enzymes, capable of hydrolyzing different polyesters including poly(lactic acid) (PLA), poly(ε-caprolactone) (PCL), and poly(butylene succinate-co-adipate) (PBSA), were found from environmental metagenomes by function-based screening [23]. And the plastic-degrading enzymes are mostly discovered from the environments abundant with biopolymeric substances. For example, by function-based screening, a novel thermostable cutinase homologue, leaf and branch compost cutinase (LCC), which exhibited hydrolytic activity towards PCL and PET, was identified from the metagenome of a leaf-branch compost with copious natural plant-derived polymers [24]. Likewise, esterases with hydrolytic activity towards poly(diethylene glycol adipate) (poly DEGA) and synthetic copolyester poly(butylene adipate-co-terephthalate) (PBAT) were identified in metagenomic libraries constructed from Sphagnum moss and soil compost, respectively [25].
The proteomics-based approach directly detects and quantifies protein expression and has been proven as a huge potential tool to identify new enzymes from a broadly microbial source. Since pollutants such as plastics are polymer substance, unable to enter the microbial cell, discovery of degrading enzymes from exocrine proteins is the principal target when screening for potential plastic-degrading enzymes [32]. Comparative proteomics is most commonly used to identify plastic-degrading enzymes. For example, several novel putative polyesterases involved in PBAT degradation were identified by comparative analysis of the exoproteomes of the bacteria Pseudomonas pseudoalcaligenes and the fungus Knufia chersonesos, demonstrating the effectiveness of this method in identifying plastic-degrading enzymes [26]. In another study, a polyhydroxybutyrate (PHB) depolymerase ALC24_4107, capable of hydrolyzing many kinds of polyesters, was discovered from Alcanivorax sp. 24 via the comparative proteomic approach [27]. Although there are some research of discovering plastic-degrading enzymes via the proteomics-based approach. Due to the difficulty in extracting high-quality protein and limited databases for downstream bioinformatic analysis, directly discovering enzymes degrading plastics from complex environmental samples via metaproteomic is still challenging [33].
2.3. Modification of microbial chassis
When the synthetic pathways of desired products are fully elucidated, and genes involved in proposed pathways are discovered, the following step is to introduce the discovered genes involved in synthetic pathways of desired products into microbial chassis. Then engineered strains capable of producing desired products can be obtained.
However, in some emerging microbial chassis, less basic research results in a lack of available biological parts such as promoters, functional for the expression of heterologous genes. The lack of available biological parts limits the expression of heterologous genes. Hence, it is necessary to identify more biological parts for accurate heterologous genes expression. Omics tools of systems biology are often employed to predict and identify regulatory biological parts.
For instance, transcriptomics was used to screen strong endogenous promoter in Pichia pastoris. Based on transcriptomics analysis of Pichia pastoris cultured in three frequently-used media, ten potential strong endogenous promoters contributing to higher transcriptional expression levels were screened out [34]. In another example, due to limited constitutive promoters in Streptomycetes, transcriptomics was employed to screen more available constitutive promoters. A total of 941 qualified genes were selected based on systematic analysis of five sets of time-series transcriptome microarray data of Streptomyces coelicolor M145 cultivated under different conditions. Then, 166 putative constitutive promoters were selected by following a rational selection workflow [35]. In a different example, transcriptomics was performed to help identify 5′ Untranslated Regions(5′UTRs) in Zymomonas mobilis as regulatory biological parts. 101 potential 5′UTRS in Z. mobilis were identified by transcriptomic data. Then, an in vivo fluorescence-based screening system was developed to confirm the responsiveness of 36 5′UTR candidates to ethanol, acetate, and xylose stresses [36].
Besides, during the process of further modifying microbial chassis, the clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) system can be used to edit a genome through gene knockouts or homology-mediated knockins to control transcription of exogenous or endogenous genes and has been one of the most efficient genome-editing technologies to engineer microorganisms for improving production [37]. Therefore, identifying the CRISPR-Cas systems within the candidate chassis microorganisms is important for constructing microbial cell factories. And the CRISPR-Cas systems can be identified by mining omics data.
David et al. [38] used genome-resolved metagenomics to identify a number of CRISPR–Cas systems, including the first reported Cas9 in the archaeal domain of life. Two previously unknown systems, CRISPR–CasX and CRISPR–CasY, which are among the most compact systems yet discovered were also been identified. In another example, Yan et al. [39] systematically discovered additional subtypes of type V CRISPR-Cas systems by mining the metagenomic database.
3. Construction of microbial cell factories based on systems biology
After the biosynthetic pathways of desired products are introduced into chassis strains, engineered strains capable of producing desired products can be acquired. However, the products yield of these engineered strains may be low. To further construct microbial cell factories, it's necessary to optimize these engineered strains to improve the yield of products. There are mainly three strategies to improve the products yield for constructing microbial cell factories, including bottleneck pathways identification, strains tolerance improvement and design and construction of synthetic microbial consortia. Systems biology, as an important tool, plays an important role in achieving these strategies to facilitate the construction of microbial cell factories.
3.1. Bottleneck pathways identification
In the process of constructing microbial cell factories, some metabolic pathways of desired products are introduced into suitable chassis stains to obtain engineered bacteria. However, the metabolic pathways of these products are generally composed of multi-step enzymatic reactions, and it is often difficult to completely coordinate and match the catalytic efficiency of these enzymes. Among them, the reactions with low enzymatic catalytic efficiency become the bottleneck restricting the metabolic pathway. These bottleneck pathways greatly limit the production yield and efficiency of the product. Therefore, identification of bottleneck pathways is an important step to improve cell production performance and thus facilitate the construction of microbial cell factories.
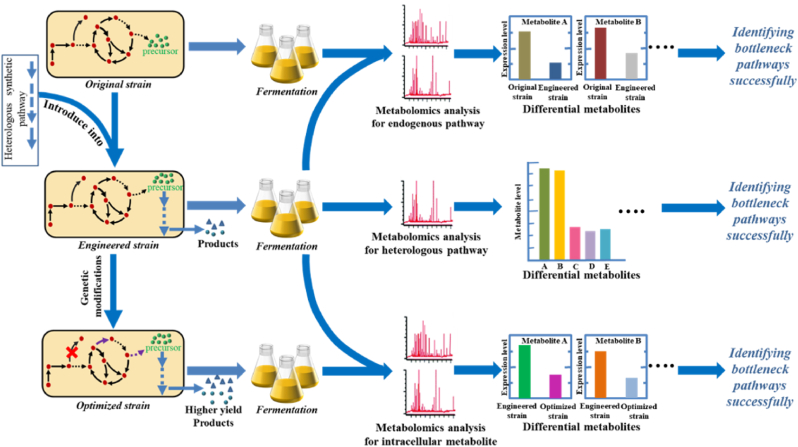
In the process of constructing microbial cell factories, the introduction of heterologous synthetic pathways may result in bottleneck pathways. First, bottleneck pathways may occur in the introduced heterologous synthetic pathways. In addition, the introduction of heterologous synthetic pathways may lead to changes in the gene expression levels of original strain, resulting in new bottlenecks. After constructing engineered strains, genetic modifications are often performed to improve the products yield. But these genetic modifications may disrupt the gene expression levels of microorganism, resulting in new bottleneck pathways. And metabolomics is one of the most common and effective tools, which can identify the bottlenecks of metabolic pathways by quantitatively analyzing intracellular metabolite concentrations and finding metabolites with significantly different concentrations [40]. And the specific process of identifying the above-mentioned bottlenecks via metabolomics is shown in Fig. 2.
Fig. 2.
Identifying bottleneck pathways via metabolomics.
3.1.1. Identifying bottleneck pathways of heterologous synthetic pathways
When an engineered strain capable of producing products is constructed by introducing heterologous biosynthetic pathway, if the production yield is relatively low, the bottleneck pathways may be considered to exist in the heterologous synthetic pathways. Metabolomics is employed to quantitatively analyze the concentration of intermediates in the heterologous synthetic pathways and find metabolites with significantly different concentrations, contributing to identify the potential bottleneck pathways (Fig. 2). For example, a heterologous MVA pathway was constructed in E. coli for the high-yield production of C5 alcohols. To identify bottleneck pathways for optimizing the pathway, targeted metabolomics was used to quantify MVA pathway intermediates in this engineered strain [41]. Isopentenyl diphosphate (IPP) concentrations were found to be much higher than any other intermediates involved in the MVA pathway, revealing that NudB, the protein capable of conversing IPP into 3-methyl-3-buten-1-ol, was the bottleneck pathway. To alleviate the bottleneck and increase the yield, the Shine-Dalgarno sequence of nudB was engineered, finally achieving a decrease in IPP accumulation by 4-fold and a 60% increase in 3-methyl-3-buten-1-ol yield. Likewise, quantitative metabolomics analysis of heterologous IPP-bypass pathway intermediates in engineered S. cerevisiae allowed for the identification of the bottleneck pathway. The last step of the pathway to produce isoprenol (a hydrolysis of IPP to isoprenol) was identified as the bottleneck. Subsequently, a promiscuous phosphatase that is more efficient in IPP hydrolysis was screened, relieving this bottleneck and improving isoprenol titer significantly [42]. In another research of engineering cyanobacteria to produce 1-butanol by introducing into the heterologous CoA pathway, the quantitative metabolomics analysis of intermediate involved the CoA pathway indicated that the reduction reaction of butyl-CoA to butanol may be the rate-limiting step in the heterologous pathway [43].
3.1.2. Identifying bottleneck pathways of endogenous pathways
In addition to those bottleneck pathways from heterologous synthetic pathways, the introduction of heterologous synthetic pathways may affect the gene expression levels of endogenous pathways in original strain, leading to some new bottleneck pathways. And these new bottleneck pathways mainly derive from the central metabolic pathway and the upstream pathway of heterologous synthetic pathways in endogenous pathways. Metabolomics can be employed to quantitatively analyze the involved metabolite concentrations between origin strains and engineered strains and find metabolites with significantly different concentrations, contributing to identifying the bottleneck pathways (Fig. 2). For example, the heterologous 2-keto acid pathway for isobutanol biosynthesis was firstly constructed in Methylobacterium extorquens AM1. And the KDC enzyme, essential for catalyzing 2-ketoisovalerate to isobutanol, had been demonstrated as a bottleneck for isobutanol production [44]. To further improve the yield, metabolomics was used to analyze the central metabolic pathway, including EMC pathway, serine cycle, and TCA cycle, and the upstream pathway of heterologous synthetic pathways between engineered strain and the control strain. The result showed that all interest intermediates in the central metabolic pathway had similar pool levels, while 2-ketoisovalerate, a direct precursor for isobutanol biosynthesis, appeared a five-fold reduction in isobutanol producer strain. This revealed that the introduction of the heterologous isobutanol synthetic pathway did not disrupt the central carbon metabolism and insufficient carbon flux through 2-ketoisovalerate may be one bottleneck, resulting in the production inhibition [45]. In a different example, to construct an engineered Corynebacterium glutamicum simultaneously utilizing both d-glucose and l-arabinose, LC-MS/MS based metabolomics was used to quantify central carbon metabolism intermediates in C. glutamicum, allowing for the identification of pyruvate kinase as the bottleneck in the co-utilization of l-arabinose with d-glucose [46].
3.1.3. Identifying bottleneck pathways resulting from genetic modifications
During the process of constructing microbial cell factories, genetic modifications are often performed to improve the yield of engineered microorganism, such as deletion of genes involved in producing byproducts and overexpression of some key genes involved in the synthetic pathway. But these genetic modifications may disrupt the gene expression levels of microorganism, resulting in bottleneck pathways. And metabolomics can be employed to determine and compare intracellular metabolite profiles of microorganisms before and after genetic modifications to find metabolites with significantly different concentrations, contributing to identifying new bottleneck pathways (Fig. 2). For example, to increase shikimate yield in engineered C. glutamicum, sugar phosphotransferase system (PTS) was inactivated to improve the availability of phosphoenolpyruvate (PEP), a key upstream precursor for shikimate biosynthesis. Subsequently, intracellular concentrations of glycolytic metabolites were analyzed by LC-MS/MS based metabolomics, indicating that the non-PTS strain accumulated 1,3-dihydroxyacetone (DHA) and glycerol and those glyceraldehyde-3-phosphate dehydrogenase (GAPDH) upstream reaction in non-PTS strain were much higher than the original strain. This metabolomics analysis identified GAPDH as a new metabolic bottleneck for producing shikimate [47].
The same metabolomics method was used to reveal a CoA imbalance resulting from deletion of phosphate acetyltransferase, blocking the production of 1-butanol in E. coli. And the reduction of butanol-CoA to butanal catalyzed by alcohol dehydrogenase AdhE2 was determined as a rate-limiting step. To alleviate the bottleneck, AdhE2 was engineered through a ribosome binding site library, resulting in an improvement of the 1-butanol titer from 15 to 18.3 g/L [48]. In another study, to further improve 1-butanol yield, metabolomics was employed to quantitatively analyze intracellular metabolite levels in strains before and after alleviating the above-mentioned bottleneck. The results demonstrated an increase in glyoxylate accumulation, indicating that glyoxylate accumulation may be a new bottleneck [49].
3.2. Strains tolerance improvement
In the process of constructing microbial cell factories to produce products, strains are usually exposed to variously different stresses. These stresses can be derived from feedstocks, toxic intermediates and products, limiting production capacity and growth ability of strains [50,51]. For example, in the process of constructing microbial cell factories to produce bioenergy, such as ethanol and butanol, these low-carbon alcohol products have a certain toxic effect on strains, so it's vital to improve product tolerance, which can improve the growth ability of strains and the yield of target products [52]. Therefore, strains tolerance improvement plays an important role in constructing microbial cell factories [53]. To improve strains tolerance, tolerance-related genes identification is crucial. These discovered tolerance-related genes can be rationally modified to improve strains tolerance.
There are mainly two strategies to discover tolerance-related genes via systems biology tools. Transcriptomics, proteomics and metabolomics can be respectively employed to measure the gene expression levels of strains from different perspectives, contributing to finding tolerance-related genes. The first strategy is to discover tolerance-related genes from strains under different stresses. Systems biology tools can be employed to measure the gene expression levels of strains under different stress, contributing to finding genes with significantly different expression levels. By further analysis, tolerance-related genes can be screened from genes with significantly different expression levels. The second strategy is to discover the tolerance-related genes from mutant strains with tolerance improvement, which can be acquired by random mutation strategy or other semi-rational and rational strategies. Systems biology tools can measure the gene expression levels of mutant strains and parental strains. Genes with significantly different expression levels can be identified, and tolerance-related genes can be screened from these genes by further analysis. As shown in Table 2, many tolerance-related genes have been successfully identified by the above-mentioned strategies.
Table 2.
Applications of systems biology tools on improving strains tolerance.
| Strategy | Stress | Strains | Omics tools | Tolerance-related genes/enzymes | Reference |
|---|---|---|---|---|---|
| Identifying tolerance-related genes from strains under different stresses | furfural | Clostridium beijerinckii | transcriptomics | two enzymes encoded by Cbei_3974 and Cbei_3904 belonging to aldo/keto reductase (AKR) and short-chain dehydrogenase/reductase (SDR families) | [54] |
| phenolic aldehydes | Z. mobilis | transcriptomics | genes encoding key reductases: ZMO1696, ZMO1116, and ZMO1885 | [55] | |
| p-benzoq-uinone | Z. mobilis | transcriptomics | Zinc-binding alcohol dehydrogenase ZMO1696, NAD(P)H dehydrogenase ZMO1949, short-chain dehydrogenase/reductase ZMO1576, aldo-keto reductase ZMO1984, fatty acid hydroxylase ZMO1399 | [56] | |
| formate, acetate | S. cerevisiae | transcriptomics | transcriptional/translational machinery-related genes: RTC3 and ANB1 | [57] | |
| butanol | Cyanobacteria | transcriptomics | the small heat shock protein HspA | [58] | |
| isopen-tanol | E. coli | transcriptomics | genes related to oxidative stress response (fpr), general stress response (metR, yqhD, and gidB), heat shock-related response (ibpA), and transport (mdlB) | [59] | |
| butanol | Chlamydomonas reinhardti | proteomics | Cre.770 peroxidase | [60] | |
| limonene | Y. lipolytica | transcriptomics | YALI0F19492g gene encoding a 169 amino acid protein YALI0F19492p | [61] | |
| acetic acid | S. cerevisiae | metabolomics | key genes of pentose phosphate pathway: transketolase gene TKL1 and transaldolase gene TAL1 | [62] | |
| Identifying tolerance-related genes from mutant strains | ethanol | Cyanobacterium synechocystis | metabolomics | two transcriptional regulators (TR) and one eukaryotic-like protein phosphatases (PP) | [63] |
| 1-butanol | S. cerevisiae | metabolomics | genes related to accumulation of threonine and reduction of citric acid | [64] | |
| ethanol | S. cerevisiae | metabolomics | LEU4 and LEU9 genes (related to accumulation of valine) or INM1 and INM2 genes (related to reduction of inositol) | [65] | |
| butanol | C. acetobutylicum | proteomics | chaperones and solvent formation related genes | [66] | |
| sabinene | E. coli | transcriptomics | ybcK gene of the DLP12 family, inner membrane protein gene ygiZ, methylmalonyl-CoA mutase gene scpA | [67] | |
| ferulic acid | Y. lipolytica | transcriptomics | YALI0_E25201g, YALI0_B18854g, YALI0_F16731g | [68] | |
| salt | Z. mobilis | metabolomics | gene ZZ6_1149 encoding carboxyl-terminal protease | [69] | |
| methanol | O. polymorpha | genomics | gene LPL1 (encoding a putative lipase) and gene IZH3 (encoding a membrane protein related to zinc metabolism) | [70] |
3.2.1. Identifying tolerance-related genes from strains under different stresses
To improve strains tolerance, discovering tolerance-related genes is crucial. And these discovered tolerance-related genes can be rationally modified to improve strains tolerance. The tolerance-related genes can be identified from strains under different stress. Systems biology tools can be employed to measure the gene expression levels of strains under different stresses, contributing to finding genes with significantly different expression levels. By further analysis, tolerance-related genes can be screened from genes with significantly different expression levels.
For example, to improve the tolerance to phenolic aldehydes of strain Z. mobilis, transcriptomics analysis by microarray was employed to analyze the transcript levels of Z. mobilis ZM4 exposed to phenolic aldehyde inhibitors. Compared with the parental strain, 272 genes including 36 gene clusters were found to up-regulate. Furthermore, ZMO1696, ZMO1116, and ZMO1885 were found to encode key reductases, playing important roles in reducing phenolic aldehydes into the corresponding phenolic alcohols. To improve the cellulosic ethanol production, three key genes were overexpressed in Z. mobilis ZM4, finally achieving increased phenolic aldehydes tolerance and ethanol productivity [55]. The same strategy was performed in another study. Metabolomics was used to analyze the metabolite levels between engineered S. cerevisiae exposed to acetic acid. The result revealed that metabolites involved in the non-oxidative pentose phosphate (PPP) pathway were significantly accumulated when exposed to acetic acid, demonstrating that acetic acid slowed down the downstream metabolic flux of the PPP pathway. Based on this discovery, the key genes of the pentose phosphate pathway (the transketolase gene TKL1 and the transaldolase gene TAL1) were overexpressed, which significantly improved tolerance of the strain to acetic acid [62]. And many other tolerance-related genes have been identified by the same strategy, as shown in Table 2.
3.2.2. Identifying tolerance-related genes from mutant strains
In addition to identifying tolerance-related genes from strains under different stresses, the tolerance-related genes can be discovered from mutant strains with tolerance improvement, which can be acquired by random mutation strategy or other semi-rational and rational strategies. Systems biology tools can be performed to measure the gene expression levels of mutant strains and parental strains. Genes with significantly different expression levels can be identified, and tolerance-related genes can be screened from these genes by further analysis.
For instance, in one study, mutant Clostridium acetobutylicum with higher butanol tolerance and higher butanol yield was successfully constructed. To reveal the mechanism of butanol tolerance, comparative proteomics was employed to analyze the differentially expressed proteins between wild type C. acetobutylicum and mutant with higher butanol tolerance and butanol yield. The result showed that chaperones and solvent formation related proteins were upregulated in both stages, while both proteins related to amino acid metabolism and protein synthesis were downregulated, providing potential target proteins for butanol tolerance modification to enhanced butanol tolerance and butanol yield [66]. In another example, to improve tolerance to 1-butanol in S. cerevisiae, 19 single-gene knockout strains with different growth rates under 1-butanol were constructed. GC-MS based metabolomics was used to measure the metabolite profiles of the 19 strains under 1-butanol stress and developed a regression model between metabolite abundance and stress growth rate. This model revealed that metabolites positively correlated with growth rate was identified as threonine, while metabolites negatively correlated with growth rate was identified as citric acid. This result demonstrated that genes related to accumulation of threonine and reduction of citric acid were key genes related to 1-butanol tolerance [64].
In a different example, adaptive laboratory evolution was employed to improve tolerance to methanol in methylotrophic yeast Ogataea polymorpha. Whole-genome sequencing of the adapted strains reveals that inactivation of LPL1 (encoding a putative lipase) and IZH3 (encoding a membrane protein related to zinc metabolism) preserve cell survival in methanol by restoring phospholipid metabolism [70]. And some other tolerance-related genes identified by this strategy were also shown in Table 2.
3.3. Design and construction of synthetic microbial consortia
During the process of constructing microbial cell factories, using single engineered strain to produce chemicals often faces a large metabolic burden due to the long and complex biosynthetic pathways, resulting in low production efficiency. Reversely, constructing synthetic microbial consortia became more attractive, as they could not only perform more complicated tasks, but also endure changeable environments [71]. And synthetic microbial consortia have been employed to produce various products [[72], [73], [74], [75], [76]]. To design and construct target microbial consortia, it's necessary to understand the molecular mechanisms of cell–cell interactions (including exchange of metabolites and cell-cell communication). Systems biology analysis could offer a global view of all members in the synthetic microbial consortia, which can systematically analyze the genetic and metabolic pathways in microbial consortia, contributing to elucidating comprehensive molecular mechanisms of interactions in microbial consortia [71]. In conclusion, systems biology analysis plays a crucial role in the design and construction of synthetic microbial consortia.
Taking the vitamin C fermentation process as an example, synthetic microbial consortia consisting of Ketogulonicigenium vulgare and Bacillus megaterium or Bacillus cereus was constructed to produce 2-KGA, and systems biology approaches were used to analyze the molecular mechanisms of cell–cell interactions for optimizing the yield of vitamin C. Zhou et al. [77] employed metabolomics to analyze the interaction mechanism between the two microorganisms, revealing that the interactions between K. vulgare and B. megaterium were a synergistic combination of mutualism and antagonism. To further reveal the interaction mechanism of the ecosystem, Ma et al. [78] used integrated time-series proteomic and metabolomic to analyze the microbial consortia, discovering that proteins involved in pentose phosphate pathway, tricarboxylic acid cycle, amino acids metabolism, l-sorbose pathway and proteins defending against intracellular reactive oxygen stress (ROS) were up-regulated when B. megaterium lysed. The result demonstrated that the cell lysis of B. megaterium might promote the growth and metabolism of K. vulgare by supplying key elements necessary for K. vulgare.
In addition to the conventional two-step fermentation method for producing 2-KGA, a synthetic microbial consortium consisted of Gluconobacter oxydans, K. vulgare and B. endophyticus was conducted for one-step vitamin C fermentation [79]. Furthermore, an integrated proteomic and metabolomic analysis was conducted to investigate the cell–cell interaction. The results revealed that the existence of G. oxydans and B. endophyticus together promoted the growth of K. vulgare and the 2-KGA production by supplying more nutrients and substrate, respectively. Meanwhile, the growth of G. oxydans and B. endophyticus was impaired by K. vulgare competition for nutrients, promoting efficient 2-KGA production. Wang et al. [80] also reorganized a synthetic consortium of G. oxydans and K. vulgare for one-step vitamin C fermentation. And the competition between the two microbes was alleviated and their mutualism was enhanced by deleting genes involved in sorbose metabolism of G. oxydans. Furthermore, metabolomics was used to investigate metabolic interaction between the two strains, which verified the enhancement of the symbiotic relationship. These examples suggest that systems biology analysis can give a better understanding of synthetic consortia and provide potential strategies for optimizing synthetic consortia and promoting the production of desired products.
Systems biology tools are also used to reveal the mechanisms of cell–cell interactions and guide the optimization of many other microbial consortia, such as yeast and cyanobacteria consortium for lipid production [81], Synechococcus elongates and E. coli consortium for isoprene production [82], Chlorella sacchrarophila and xylanolytic bacterium consortium for algal lipid production [83].
4. Conclusions
Constructing microbial cell factories to produce various products is an environmentally friendly and sustainable way, which has attracted more and more attention. This review summarizes the recent applications of systems biology in the design and construction of microbial cell factories. And it is summarized from four perspectives, including functional genes/enzymes discovery, bottleneck pathways identification, strains tolerance improvement and design and construction of synthetic microbial consortia. In conclusion, systems biology plays a crucial role in the design and construction of microbial cell factories. And this review is expected to provide reference for the construction of microbial cell factories for more valuable products in the future from the perspective of systems biology.
Author contributions
W.Y.: conceptualization, writing—original draft; Z.C.: writing—review and editing; M.D.: conceptualization, writing—review and editing, supervision; Y·Y.: supervision. All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgement
This work was funded by the National Key Research and Development Program of China (2019YFA0706900) and National Natural Science Foundation of China (22278310).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.synbio.2022.11.001.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Wang Y., et al. Microbial cell factories for green production of vitamins. Front Bioeng Biotechnol. 2021;9:473. doi: 10.3389/fbioe.2021.661562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee D., et al. Heterologous production of raspberry ketone in the wine yeast Saccharomyces cerevisiae via pathway engineering and synthetic enzyme fusion. Microb Cell Factories. 2016;15:1–7. doi: 10.1186/s12934-016-0446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai Y., et al. Production of salidroside in metabolically engineered Escherichia coli. Sci Rep. 2014;4:1–8. doi: 10.1038/srep06640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bai Y., et al. De novo biosynthesis of Gastrodin in Escherichia coli. Metab Eng. 2016;35:138–147. doi: 10.1016/j.ymben.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Luo Y., et al. Engineered biosynthesis of natural products in heterologous hosts. Chem Soc Rev. 2015;44(15):5265–5290. doi: 10.1039/c5cs00025d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J.-Q., et al. Side Products of recombinant amorpha-4,11-diene synthase and their effect on microbial artemisinin production. J Agric Food Chem. 2021;69(7):2168–2178. doi: 10.1021/acs.jafc.0c07462. [DOI] [PubMed] [Google Scholar]
- 7.Liu H., Lu T. Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab Eng. 2015;29:135–141. doi: 10.1016/j.ymben.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Asl H., et al. Systems biology solutions for biochemical production challenges. Curr Opin Biotechnol. 2017;45:85–91. doi: 10.1016/j.copbio.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 9.Liu D., Hoynes-O'Connor A., Zhang F.Z. Bridging the gap between systems biology and synthetic biology. Front Microbiol. 2013;4:211. doi: 10.3389/fmicb.2013.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brul S., et al. Microbial systems biology: new frontiers open to predictive microbiology. Int J Food Microbiol. 2008;128(1):16–21. doi: 10.1016/j.ijfoodmicro.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 11.Noguchi S., et al. Quantitative target analysis and kinetic profiling of acyl-CoAs reveal the rate-limiting step in cyanobacterial 1-butanol production. Metabolomics. 2016 doi: 10.1007/s11306-015-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song H., et al. Synthetic microbial consortia: from systematic analysis to construction and applications. Chem Soc Rev. 2014;43(20):6954–6981. doi: 10.1039/c4cs00114a. [DOI] [PubMed] [Google Scholar]
- 13.Hansen A.S.L., et al. Systems biology solutions for biochemical production challenges. Curr Opin Biotechnol. 2017;45:85–91. doi: 10.1016/j.copbio.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 14.Farrow S.C., et al. Cytochrome P450 and O-methyltransferase catalyze the final steps in the biosynthesis of the anti-addictive alkaloid ibogaine from Tabernanthe iboga. J Biol Chem. 2018;293(36):13821–13833. doi: 10.1074/jbc.RA118.004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J., et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci USA. 2013;110(29):12108–12113. doi: 10.1073/pnas.1218061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moses T., et al. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16 alpha hydroxylase from Bupleurum falcatum. Proc Natl Acad Sci USA. 2014;111(4):1634–1639. doi: 10.1073/pnas.1323369111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y., et al. Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nature Plants. 2016;2(12):1–8. doi: 10.1038/nplants.2016.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Q., et al. The reference genome sequence of Scutellaria baicalensis provides insights into the evolution of wogonin biosynthesis. Mol Plant. 2019;12(7):935–950. doi: 10.1016/j.molp.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Fiallos-Jurado J., et al. Saponin determination, expression analysis and functional characterization of saponin biosynthetic genes in Chenopodium quinoa leaves. Plant Sci. 2016;250:188–197. doi: 10.1016/j.plantsci.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 20.Gagne S.J., et al. Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc Natl Acad Sci USA. 2012;109(31):12811–12816. doi: 10.1073/pnas.1200330109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danso D., et al. New insights into the function and global distribution of Polyethylene Terephthalate (PET)-degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl Environ Microbiol. 2018;84(8) doi: 10.1128/aem.02773-17. e02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaytan I., et al. Degradation of recalcitrant polyurethane and xenobiotic additives by a selected landfill microbial community and its biodegradative potential revealed by proximity ligation-based metagenomic analysis. Front Microbiol. 2020;10:2986. doi: 10.3389/fmicb.2019.02986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hajighasemi M., et al. Screening and characterization of novel polyesterases from environmental metagenomes with high hydrolytic activity against synthetic polyesters. Environ Sci Technol. 2018;52(21):12388–12401. doi: 10.1021/acs.est.8b04252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sulaiman S., et al. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol. 2012;78(5):1556–1562. doi: 10.1128/aem.06725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller C.A., et al. Discovery of polyesterases from moss-associated microorganisms. Appl Environ Microbiol. 2017;83(4) doi: 10.1128/aem.02641-16. e02641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wallace P.W., et al. PpEst is a novel PBAT degrading polyesterase identified by proteomic screening of Pseudomonas pseudoalcaligenes. Appl Microbiol Biotechnol. 2017;101(6):2291–2303. doi: 10.1007/s00253-016-7992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zadjelovic V., et al. Beyond oil degradation: enzymatic potential of Alcanivorax to degrade natural and synthetic polyesters. Environ Microbiol. 2020;22(4):1356–1369. doi: 10.1111/1462-2920.14947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., et al. Effect of methyl jasmonate on the ginsenoside content of Panax ginseng adventitious root cultures and on the genes involved in triterpene biosynthesis. Res Chem Intermed. 2013;39(5):1973–1980. doi: 10.1007/s11164-012-0730-7. [DOI] [Google Scholar]
- 29.Hou R., et al. Comparative proteomics analysis of Pichia pastoris cultivating in glucose and methanol. Synthetic and Systems Biotechnology. 2022;7(3):862–868. doi: 10.1016/j.synbio.2022.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ufarte L., et al. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol Adv. 2015;33(8):1845–1854. doi: 10.1016/j.biotechadv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 31.Subramanian S.H.S., et al. RemeDB: tool for rapid prediction of enzymes involved in bioremediation from high-throughput metagenome data sets. J Comput Biol. 2020;27(7):1020–1029. doi: 10.1089/cmb.2019.0345. [DOI] [PubMed] [Google Scholar]
- 32.Urbanek A.K., et al. Biochemical properties and biotechnological applications of microbial enzymes involved in the degradation of polyester-type plastics. Biochim Biophys Acta, Proteins Proteomics. 2020;1868(2) doi: 10.1016/j.bbapap.2019.140315. [DOI] [PubMed] [Google Scholar]
- 33.Biswas R., Sarkar A. In: Advances in soil microbiology: recent trends and future prospects. Adhya T.K., et al., editors. Vol. 1. 2018. Omics' tools in soil microbiology: the state of the art; pp. 35–64. (Soil-microbe interaction). [Google Scholar]
- 34.Dou W., et al. Screening and evaluation of the strong endogenous promoters in Pichia pastoris. Microb Cell Factories. 2021;20(1):1–12. doi: 10.1186/s12934-021-01648-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S., et al. Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microb Cell Factories. 2015;14(1):1–11. doi: 10.1186/s12934-015-0351-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho S.H., et al. Identification and characterization of 5' Untranslated Regions (5'UTRs) in Zymomonas mobilis as regulatory biological parts. Front Microbiol. 2018;8:2432. doi: 10.3389/fmicb.2017.02432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donohoue P.D., Barrangou R., May A.P. Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol. 2017;36(2):134–146. doi: 10.1016/j.tibtech.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Burstein D., et al. New CRISPR–Cas systems from uncultivated microbes. Nature. 2016;542(7640):237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan W.X., et al. Functionally diverse type V CRISPR-Cas systems. Science. 2018;363(6422):88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vavricka C.J., Hasunuma T., Kondo A. Dynamic metabolomics for engineering biology: accelerating learning cycles for bioproduction. Trends Biotechnol. 2020;38(1):68–82. doi: 10.1016/j.tibtech.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 41.George K.W., et al. Metabolic engineering for the high-yield production of isoprenoid-based C-5 alcohols in E-coli. Sci Rep. 2015;5:1–12. doi: 10.1038/srep11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim J., et al. Engineering Saccharomyces cerevisiae for isoprenol production. Metab Eng. 2021;64:154–166. doi: 10.1016/j.ymben.2021.02.002. [DOI] [PubMed] [Google Scholar]
- 43.Noguchi S., et al. Quantitative target analysis and kinetic profiling of acyl-CoAs reveal the rate-limiting step in cyanobacterial 1-butanol production. Metabolomics. 2016;12(2):1–10. doi: 10.1007/s11306-015-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Higashide W., et al. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl Environ Microbiol. 2011;77(8):2727–2733. doi: 10.1128/aem.02454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Z.-x., et al. Metabolomic analysis improves bioconversion of methanol to isobutanol in Methylorubrum extorquens AM1. Biotechnol J. 2021;16(6) doi: 10.1002/biot.202000413. [DOI] [PubMed] [Google Scholar]
- 46.Kawaguchi H., et al. Metabolome analysis-based design and engineering of a metabolic pathway in Corynebacterium glutamicum to match rates of simultaneous utilization of D-glucose and L-arabinose. Microb Cell Factories. 2018;17:1–16. doi: 10.1186/s12934-018-0927-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kogure T., et al. Metabolic engineering of Corynebacterium glutamicum for shikimate overproduction by growth-arrested cell reaction. Metab Eng. 2016;38:204–216. doi: 10.1016/j.ymben.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 48.Ohtake T., et al. Metabolomics-driven approach to solving a CoA imbalance for improved 1-butanol production in Escherichia coli. Metab Eng. 2017;41:135–143. doi: 10.1016/j.ymben.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Nitta K., et al. Metabolome analysis revealed the knockout of glyoxylate shunt as an effective strategy for improvement of 1-butanol production in transgenic Escherichia coli. J Biosci Bioeng. 2019;127(3):301–308. doi: 10.1016/j.jbiosc.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 50.Ran H., et al. Analysis of biodegradation performance of furfural and 5-hydroxymethylfurfural by Amorphotheca resinae ZN1. Biotechnol Biofuels. 2014;7:1–12. doi: 10.1186/1754-6834-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu Z., et al. A short-chain dehydrogenase plays a key role in cellulosic D-lactic acid fermentability of Pediococcus acidilactici. Bioresour Technol. 2020;297 doi: 10.1016/j.biortech.2019.122473. [DOI] [PubMed] [Google Scholar]
- 52.Gong Z., Nielsen J., Zhou Y.J. Engineering robustness of microbial cell factories. Biotechnol J. 2017;12(10) doi: 10.1002/biot.201700014. [DOI] [PubMed] [Google Scholar]
- 53.Alper H., et al. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314(5805):1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 54.Zhang Y., Ezeji T.C. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 to elucidate role of furfural stress during acetone butanol ethanol fermentation. Biotechnol Biofuels. 2013;6:1–17. doi: 10.1186/1754-6834-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi X., et al. Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels. 2015;8:1–15. doi: 10.1186/s13068-015-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan Z., et al. Mechanism of tolerance to the lignin-derived inhibitor p-benzoquinone and metabolic modification of biorefinery fermentation strains. Appl Environ Microbiol. 2019;85(22) doi: 10.1128/aem.01443-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hasunuma T., Sakamoto T., Kondo A. Inverse metabolic engineering based on transient acclimation of yeast improves acid-containing xylose fermentation and tolerance to formic and acetic acids. Appl Microbiol Biotechnol. 2016;100(2):1027–1038. doi: 10.1007/s00253-015-7094-z. [DOI] [PubMed] [Google Scholar]
- 58.Anfelt J., et al. Using transcriptomics to improve butanol tolerance of Synechocystis sp strain PCC 6803. Appl Environ Microbiol. 2013;79(23):7419–7427. doi: 10.1128/aem.02694-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Foo J.L., et al. Improving microbial biogasoline production in Escherichia coli using tolerance engineering. mBio. 2014;5(6) doi: 10.1128/mBio.01932-14. e01932-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y., et al. Metabolic responses to ethanol and butanol in Chlamydomonas reinhardtii. Biotechnol Biofuels. 2017;10:1–16. doi: 10.1186/s13068-017-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li J., et al. Simultaneous improvement of limonene production and tolerance in Yarrowia lipolytica through tolerance engineering and evolutionary engineering. ACS Synth Biol. 2021;10(4):884–896. doi: 10.1021/acssynbio.1c00052. [DOI] [PubMed] [Google Scholar]
- 62.Hasunuma T., et al. Metabolic pathway engineering based on metabolomics confers acetic and formic acid tolerance to a recombinant xylose-fermenting strain of Saccharomyces cerevisiae. Microb Cell Factories. 2011;10:1–13. doi: 10.1186/1475-2859-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu Y., et al. Metabolomic analysis reveals functional overlapping of three signal transduction proteins in regulating ethanol tolerance in cyanobacterium Synechocystis sp PCC 6803. Mol Biosyst. 2015;11(3):770–782. doi: 10.1039/c4mb00651h. [DOI] [PubMed] [Google Scholar]
- 64.Teoh S.T., et al. A metabolomics-based strategy for identification of gene targets for phenotype improvement and its application to 1-butanol tolerance in Saccharomyces cerevisiae. Biotechnol Biofuels. 2015;8:1–14. doi: 10.1186/s13068-015-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohta E., et al. Metabolomic approach for improving ethanol stress tolerance in Saccharomyces cerevisiae. J Biosci Bioeng. 2016;121(4):399–405. doi: 10.1016/j.jbiosc.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Mao S., et al. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res. 2010;9(6):3046–3061. doi: 10.1021/pr9012078. [DOI] [PubMed] [Google Scholar]
- 67.Wu T., et al. Improvement of sabinene tolerance of Escherichia coli using adaptive laboratory evolution and omics technologies. Biotechnol Biofuels. 2020;13(1):1–15. doi: 10.1186/s13068-020-01715-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z.D., et al. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl Microbiol Biotechnol. 2021;105(4):1745–1758. doi: 10.1007/s00253-021-11130-3. [DOI] [PubMed] [Google Scholar]
- 69.Fuchino K., Bruheim P. Increased salt tolerance in Zymomonas mobilis strain generated by adaptative evolution. Microb Cell Factories. 2020;19(1):1–11. doi: 10.1186/s12934-020-01406-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gao J.Q., et al. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nature Metabolism. 2022;4(7):932. doi: 10.1038/s42255-022-00601-0. [DOI] [PubMed] [Google Scholar]
- 71.Ding M.Z., et al. Design and construction of synthetic microbial consortia in China. Synthetic and Systems Biotechnology. 2016;1(4):230–235. doi: 10.1016/j.synbio.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang Y.J., et al. Designing synthetic microbial consortia for biofuel production. Trends Biotechnol. 2020;38(8):828–831. doi: 10.1016/j.tibtech.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 73.Sgobba E., et al. Synthetic Escherichia coli-Corynebacterium glutamicum consortia for L-lysine production from starch and sucrose. Bioresour Technol. 2018;260:302–310. doi: 10.1016/j.biortech.2018.03.113. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y., et al. Construction of synthetic microbial consortia for 2-keto-L-gulonic acid biosynthesis. Synthetic and Systems Biotechnology. 2022;7(1):481–489. doi: 10.1016/j.synbio.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sgobba E., Wendisch V.F. Synthetic microbial consortia for small molecule production. Curr Opin Biotechnol. 2020;62:72–79. doi: 10.1016/j.copbio.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y., et al. A three-species microbial consortium for power generation. Energy Environ Sci. 2017;10(7):1600–1609. doi: 10.1039/c6ee03705d. [DOI] [Google Scholar]
- 77.Zhou J., et al. Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility. Appl Environ Microbiol. 2011;77(19):7023–7030. doi: 10.1128/aem.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma Q., et al. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PLoS One. 2011;6(10) doi: 10.1371/journal.pone.0026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma Q., et al. Integrated proteomic and metabolomic analysis of a reconstructed three-species microbial consortium for one-step fermentation of 2-keto-l-gulonic acid, the precursor of vitamin C. J Ind Microbiol Biotechnol. 2019;46(1):21–31. doi: 10.1007/s10295-018-2096-3. [DOI] [PubMed] [Google Scholar]
- 80.Wang E.-X., et al. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation. Microb Cell Factories. 2016;15:1–11. doi: 10.1186/s12934-016-0418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li T., et al. Mimicking lichens: incorporation of yeast strains together with sucrose-secreting cyanobacteria improves survival, growth, ROS removal, and lipid production in a stable mutualistic co-culture production platform. Biotechnol Biofuels. 2017;10(1):1–11. doi: 10.1186/s13068-017-0736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu H., et al. Study on the isoprene-producing co-culture system of Synechococcus elongates–Escherichia coli through omics analysis. Microb Cell Factories. 2021;20(1):1–18. doi: 10.1186/s12934-020-01498-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xie Z., Lin W., Luo J. Co-cultivation of microalga and xylanolytic bacterium by a continuous two-step strategy to enhance algal lipid production. Bioresour Technol. 2021;330 doi: 10.1016/j.biortech.2021.124953. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.