Abstract
The yeast Isw2 chromatin remodeling complex functions in parallel with the Sin3-Rpd3 histone deacetylase complex to repress early meiotic genes upon recruitment by Ume6p. For many of these genes, the effect of an isw2 mutation is partially masked by a functional Sin3-Rpd3 complex. To identify the full range of genes repressed or activated by these factors and uncover hidden targets of Isw2-dependent regulation, we performed full genome expression analyses using cDNA microarrays. We find that the Isw2 complex functions mainly in repression of transcription in a parallel pathway with the Sin3-Rpd3 complex. In addition to Ume6 target genes, we find that many Ume6-independent genes are derepressed in mutants lacking functional Isw2 and Sin3-Rpd3 complexes. Conversely, we find that ume6 mutants, but not isw2 sin3 or isw2 rpd3 double mutants, have reduced fidelity of mitotic chromosome segregation, suggesting that one or more functions of Ume6p are independent of Sin3-Rpd3 and Isw2 complexes. Chromatin structure analyses of two nonmeiotic genes reveals increased DNase I sensitivity within their regulatory regions in an isw2 mutant, as seen previously for one meiotic locus. These data suggest that the Isw2 complex functions at Ume6-dependent and -independent loci to create DNase I-inaccessible chromatin structure by regulating the positioning or placement of nucleosomes.
The regulation of RNA synthesis is a complex process affected by many factors that influence the initiation, elongation, and termination of transcription. These factors include transcriptional activators and repressors, as well as general transcription factors. In eukaryotes, the compaction of DNA into chromatin provides an additional level of complexity, due to the fact that chromatin structure is inhibitory to many protein-DNA interactions required for transcription (43). Consequently, chromatin structure must be dynamically regulated for many genes whose transcription is repressed or induced at different times of the cell cycle, at different phases of development, or by changes in growth conditions.
A large number of proteins from several organisms have been found to modify chromatin structure. These factors can be grouped into two classes based on their biochemical activities: histone-modifying enzymes and ATP-dependent chromatin remodeling complexes (for reviews, see references 29, 30, 36, 42, 45, 54, and 55). Histone-modifying enzymes use several different enzymatic mechanisms to covalently modify histone proteins. These modifications include acetylation, phosphorylation, ubiquitination, and methylation. The second class of factors uses the energy of ATP hydrolysis to alter histone-DNA contacts, often leading to repositioning of histone octamers on DNA (56).
The role of histone acetylation in the regulation of transcription has been studied extensively (11, 42, 57). With some exceptions, high levels of histone acetylation correlate with transcriptionally active regions of chromatin, while lower levels of acetylation are found in transcriptionally inactive regions. Histone acetylation is regulated by the activities of histone acetyltransferase and histone deacetylase (HDAC) complexes, many of which function as transcriptional activators or repressors, respectively (42, 44, 45).
Three major families of HDACs have been conserved in eukaryotes, represented by the Rpd3, Hda1, and Sir2 proteins (12, 32). The RPD3 gene was first identified genetically in yeast as a transcriptional repressor of several genes (52, 53). It was later found that Rpd3 proteins in several species associate with Sin3p and other Sin3-associated proteins to form a multiprotein complex (1, 24, 59). Recruitment of this complex by transcriptional repressors leads to local deacetylation and repression of transcription (14, 15, 24, 26, 33, 37, 59).
A variety of ATP-dependent chromatin remodeling factors have also been identified from several organisms and can be grouped into three classes based on the ATPase subunit present in each complex: SWI/SNF, ISWI, and CHD1 (6, 22, 29, 38). Members of the ISWI class of chromatin remodeling factors were first identified biochemically in Drosophila, following their ATP-dependent activity in disrupting nucleosomes or imparting regular spacing on nucleosome arrays in vitro. These studies led to the identification of three ISWI-containing complexes, NURF (48, 50), CHRAC (51), and ACF (20). Subsequently, several other ISWI-containing complexes were identified in humans (3, 34, 35, 39), yeasts (49), and Xenopus (13, 28), based on sequence homology to Drosophila ISWI.
Although the biochemical activities of several ISWI complexes have been studied in detail, the in vivo functions of this class of factors are only beginning to be identified. The Drosophila ISWI gene is essential for development and cell viability (9). Drosophila ISWI mutants also have reduced expression of the Ubx and engrailed genes, suggesting the requirement of one or more ISWI-containing complexes for the expression of these genes. In addition, ISWI mutants are compromised for male X chromosome integrity (9).
It was recently found that one of the two yeast ISWI complexes, the Isw2 complex, functions during vegetative growth to repress genes induced early in meiosis (10). Many early meiotic genes are repressed under these conditions by the Sin3-Rpd3 HDAC complex upon recruitment by the sequence-specific DNA-binding protein, Ume6p (24, 25, 40). Like the Sin3-Rpd3 complex, the Isw2 complex appears to be recruited to the promoters of early meiotic genes by Ume6p; however, repression by the Isw2 complex occurs independently of the Sin3-Rpd3 complex. Analysis of chromatin structure of one Ume6 target gene in wild-type and various mutant cells reveals that the Isw2 complex functions to create DNase I-inaccessible chromatin structure by altering the positions of nucleosomes upstream of the Ume6p-binding site. These data reveal that the Isw2 complex functions in parallel with the Sin3-Rpd3 complex to repress the transcription of common target genes.
While it is clear that Isw2 and Sin3-Rpd3 complexes collaborate to repress Ume6 target genes, it is not clear whether this collaboration extends to any Ume6-independent genes. To investigate this possibility, we compared the full genome transcriptional profiles of mutants defective in one or more of these factors. We also competed single mutants of sin3 and rpd3 with isw2 sin3 and isw2 rpd3 double mutants directly on microarray slides to identify genetic interactions between Isw2 and Sin3-Rpd3 complexes that are not always detectable in traditional mutant versus wild-type competitions. We find that while Ume6 target genes represent a major group of genes repressed by these complexes, a large number of Ume6-independent genes are derepressed in mutants defective in Sin3-Rpd3 and Isw2 complex functions. A comparison of isw2 and rpd3 deletion and catalytically inactive mutants reveals differences in both phenotype and transcriptional profiles which may result from functions of these proteins independent of their known catalytic activities, inhibition of related factors by catalytically inactive proteins, or a combination of the two. We also find that ume6 mutants have a reduced fidelity of chromosome segregation which results in higher rates of chromosome gains and losses relative to those in wild-type cells, suggesting a role for Ume6p in chromosome segregation that is independent of Isw2 and Sin3-Rpd3 complexes. Chromatin structure analyses of two Ume6-independent genes that require Sin3-Rpd3 and Isw2 complexes for proper regulation reveal that ISW2 function is required in both instances for the formation of DNase I-inaccessible chromatin structure, as previously observed for the Ume6 target gene REC104 (10). These data suggest that Sin3-Rpd3 and Isw2 complexes collaborate to repress the transcription of Ume6-dependent and some Ume6-independent genes and that the Isw2 complex functions to create DNase I-inaccessible chromatin structure at the promoters of many of these genes.
MATERIALS AND METHODS
Strains.
Unless indicated, all yeast strains were derived from W1588-4C. This strain is congenic to W303-1A, except that a weak rad5 mutation in the original strain W303 is repaired (60). Deletion and catalytically inactive mutations of the ISW2, SIN3, RPD3, and UME6 genes were described previously (10). The CFIII minichromosome (41) was introduced into isogenic wild-type and mutant strains by crossing an isw2 ume6 double mutant (YTT1065) with SBY475 (courtesy of Sue Biggins, Fred Hutchinson Cancer Research Center). The colony sectoring assay for measuring rates of minichromosome loss was done as previously described (16, 58).
RNA isolation.
RNA samples were prepared from mutant and wild-type cells grown at 30°C in YEPD medium (2% Bacto Peptone, 1% yeast extract, 2% glucose) to early log phase (optical density at 660 nm, 0.7) using acid phenol extraction. To ensure identical growth conditions, strains were grown in aliquots of media from a common preparation. mRNA was prepared using Oligotex beads (Qiagen) as described by the manufacturer.
Production of spotted microarrays.
Microarray construction and hybridization protocols were modified from those described elsewhere (8). Yeast microarrays were constructed using a set of ∼6,200 open reading frame (ORF)-specific PCR primer pairs (Research Genetics, Huntsville, Ala.), which were used to amplify each ORF of the yeast genome. Individual PCR products were verified as unique via gel electrophoresis and purified using ArrayIt 96-well PCR purification kits (TeleChem International, Sunnyvale, Calif.). Purified PCR products in 3× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate [pH 7.0]) were mechanically spotted onto polylysine-coated microscope slides using an OmniGrid high-precision robotic gridder (GeneMachines, San Carlo, Calif.).
Microarray hybridizations and data analysis.
The protocol used for cDNA labeling was a modification of a protocol described elsewhere (http://cmgm.stanford.edu/pbrown/protocols/aadUTPCouplingProcedure.htm). Briefly, labeled cDNA targets were prepared by reverse transcription of 2 μg of mRNA using oligo(dT)18 primer in the presence of 0.2 mM 5-(3-aminoallyl)-2′-deoxyuridine 5′-triphosphate (Sigma-Aldrich Company, St. Louis, Mo.), 0.3 mM dTTP, and 0.5 mM each dATP, dCTP, and dGTP. Following cDNA synthesis, either Cy3 or Cy5 monoreactive fluor (Amersham Life Sciences, Arlington Heights, Ill.) was covalently coupled to the cDNA-incorporated aminoallyl linker in the presence of 50 mM sodium bicarbonate (pH 9.0). Two color expression profiles were generated using microarrays in which reference and experimental cDNA targets were labeled with different fluors. Following cohybridization to the chip, a fluorescent image of a microarray was collected at both emission wavelengths using a GenePix 4000 fluorescence scanner (Axon Instruments, Inc., Foster City, Calif.), and image analysis was performed using GenePix Pro microarray acquisition and analysis software.
Four microarray hybridizations were carried out for each comparison of mutant versus wild type or mutant versus mutant (two sets of two reverse fluor combinations). We carried out a Bayesian background correction, described in detail by Kooperberg et al. (31). Briefly, we assume that both the foreground and the background intensities of both channels for each spot come from a normal distribution with an unknown mean and variance. After estimating the variance parameters from the data, we can estimate for each channel the posterior distribution of the difference between the foreground and the background means as well as their ratio, which is the quantity of interest. To do this, we assume an uninformative uniform prior on the intensities. The effect of this procedure is that for genes where the intensities are high, the estimated ratio is very similar to the traditional estimate, but for spots where the foreground intensity is close to the background intensity, the estimate for the ratio is shrunk slightly toward one, yielding a substantial reduction in variance for these spots.
All calculations involving expression ratios were carried out on the (natural) log scale by computing averages and standard errors (SEs). Averages and SEs were then converted to ratios by assuming that the average of the log ratios follows the normal distribution (which seems reasonable given both the error distribution of log ratios and the central-limit theorem). The average ratio then has a log-normal distribution. This average and its SE are related, as described previously (21), to the average and the SE of the log ratios by the following equations:
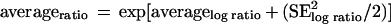 |
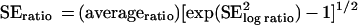 |
For analysis of the genomic locations of misregulated genes and cis element searches of promoter sequences, we used the GENESPRING software package (Silicon Genetics).
Chromatin structure analysis.
Digestion of chromatin was performed with crude preparations of nuclei (17) as described previously (10). After digestion with micrococcal nuclease or DNase I, DNA was purified, digested with EcoRI and PstI (for POT1) or EcoRI and XbaI (for SUC2), and subjected to indirect end labeling. The probe used for indirect end labeling of the POT1 locus was a PCR product extending from +406 to +623 with respect to the initiation codon. The probe used for indirect end labeling of the SUC2 locus was a PCR product extending from −885 to −661 with respect to the initiation codon.
RESULTS
Overlapping functions of Isw2 and Sin3-Rpd3 complexes.
Recently, it was found that the Isw2 complex functions during mitotic growth in Ume6p-dependent repression of early meiotic genes in a pathway parallel to that of the Sin3-Rpd3 HDAC complex (10). While it is clear that Sin3-Rpd3 and Isw2 complexes function in the repression of Ume6 target genes, it is not known whether this collaboration extends to genes not regulated by Ume6p. To determine the full extent to which Sin3-Rpd3 and Isw2 complexes function in parallel pathways of transcriptional repression, we used cDNA spotted microarrays representing >96% of all yeast genes to analyze the genome-wide expression profiles for mutants defective in one or both of these complexes. For each mutant, four independent mutant versus wild-type microarray hybridizations were carried out, and the average expression ratio and standard error for each spot were determined (see Materials and Methods for details; the full data set can be obtained at ftp://milano.fhcrc.org/ArrayLab/Fazzio/). Initially, we focused on isw2, rpd3, and sin3 single null mutants, as well as isw2 rpd3 and isw2 sin3 double null mutants. In addition, we analyzed expression profiles for ume6 and isw2 ume6 null mutants; these data are discussed below. We observed relatively few changes in transcript levels for the isw2 single mutant, as previously described for an isw2/isw2 homozygous mutant diploid strain (18). Because it was previously found that Isw2 and Sin3-Rpd3 complexes can each partially compensate for the lack of the other in repression of common target genes, we focused on genes that require defects in both complexes for moderate levels of derepression. As shown in Table 1, larger numbers of genes are moderately derepressed (>3-fold) in isw2 rpd3 and isw2 sin3 double mutants than in single mutants. Similarly, more genes are derepressed to higher levels (>5-fold, >10-fold) in double mutants than in single mutants, revealing that Sin3-Rpd3 and Isw2 complexes function in parallel pathways to repress the transcription of a considerable group of genes.
TABLE 1.
Genes with increased expression in various deletion mutantsa
| Deletion mutant | No. of genes with the following fold increases in expressionb
|
||
|---|---|---|---|
| >3 | >5 | >10 | |
| isw2 | 3 (0) | 0 | 0 |
| rpd3 | 35 (8) | 9 (3) | 1 (1) |
| isw2 rpd3 | 114 (21) | 24 (13) | 6 (2) |
| sin3 | 42 (10) | 8 (3) | 2 (1) |
| isw2 sin3 | 93 (22) | 31 (12) | 9 (5) |
| ume6 | 113 (40) | 45 (20) | 17 (10) |
| isw2 ume6 | 320 (52) | 80 (24) | 22 (13) |
A total of 720 genes were more than 3-fold derepressed in one or more of the seven mutants listed above; of these, the expression ratios for 630 were statistically significantly higher than 2 at the 0.05 level. The ratios of 178 of the 197 genes that were more than 5-fold derepressed were statistically significantly higher than 3 at the 0.05 level; and the expression ratios of 53 of the 57 genes that were more than 10-fold derepressed were statistically significantly higher than 5 at the 0.05 level. b Values in parentheses are the numbers of genes that contain the URS1 core sequence (GGCGGC) within 500 bp upstream of their initiation codons.
As expected, many genes found to be repressed by Sin3-Rpd3 and Isw2 complexes include known targets of Ume6p and genes predicted to be Ume6 targets by virtue of an upstream Ume6p-binding site (URS1) (Table 1). We also searched for a second Ume6-binding sequence previously identified in the promoter of the PHR1 gene (46); however, this sequence was not overrepresented in the promoters of genes derepressed in any of our mutants. In addition to genes repressed by Ume6p, we observed many genes that require Sin3-Rpd3 and Isw2 complexes for repression that lack URS1 sequences. The majority (82%) of these genes are not substantially derepressed in the ume6 mutant, suggesting that derepression in isw2 sin3 and isw2 rpd3 mutants is not a secondary consequence of the upregulation of early meiotic genes (see the full data set for details). Together, these data reveal that a substantial group of genes requires Sin3-Rpd3 and Isw2 complexes for repression, independent of Ume6p.
In addition to the role of the Isw2 complex in repression, we also found that the transcription of a small number of genes is reduced in the isw2 mutant (see the full data set for details). For these genes, the relationship between the Isw2 complex and Sin3-Rpd3 complex in the activation or maintenance of basal transcription is less clear. Few genes have significantly reduced expression in the isw2 deletion mutant (zero genes reduced >3-fold and one gene reduced >2-fold). In addition, many genes with reduced expression in the sin3 or rpd3 single mutant are unaffected or oppositely affected by the addition of an isw2 mutation in these backgrounds. These data suggest a relatively minor role of the Isw2 complex in the transcriptional activation or maintenance of basal transcription under the conditions used (logarithmic growth in rich glucose medium).
Hidden functions of the Isw2 complex revealed in sin3 and rpd3 mutant backgrounds.
The data in Table 1 suggest that many targets of Isw2-mediated repression may be masked in isw2 mutant cells by the presence of a functional Sin3-Rpd3 complex. To identify ISW2 target genes affected in this way, we performed two experiments. First, we divided isw2 rpd3/wild-type expression ratios by rpd3/wild-type ratios for all genes (Table 2); similarly, isw2 sin3/wild-type expression ratios were divided by sin3/wild-type ratios (Table 2). In theory, the ratios obtained through these calculations should reveal defects in transcriptional regulation resulting from the loss of Isw2 function in an rpd3 or sin3 mutant background. However, when we compared these data to those obtained by Northern blotting (Table 2), we noticed several inconsistencies. For several meiotic genes previously found to be targets of Isw2-dependent repression, the ratios derived from one or both calculations described above do not reflect known derepression due to the isw2 mutation (cf. HOP1, SPO11, REC104, and SPO1). Under the growth conditions used for this experiment (logarithmic growth of haploid cells in rich glucose medium), many genes are tightly repressed in wild-type cells. In microarray experiments, expression ratios for genes with a low signal in the wild-type channel are often underestimated relative to ratios measured by Northern blotting (31). As a result, isw2 rpd3/ wild-type expression ratios for these genes measured using microarrays can be similar to rpd3/wild-type ratios; likewise, isw2 sin3/wild-type expression ratios can be similar to sin3/wild-type ratios. Consequently, when double mutants are divided by single mutants to compare their expression profiles, some differences observed by Northern blotting are not revealed.
TABLE 2.
Hidden functions of the Isw2 complex revealed by direct competition of double mutants with single mutants
| Gene | Ratioa for:
|
|||||
|---|---|---|---|---|---|---|
| isw2 rpd3/rpd3b | isw2 rpd3 vs wt/rpd3 vs wtc | isw2 rpd3/rpd3d | isw2 sin3/sin3b | isw2 sin3 vs wt/sin3 vs wtc | isw2 sin3/sin3d | |
| SPO11 | 2.0 | 1.1 (0.3) | 1.6 (0.2) | 1.6 | 1.4 (0.3) | 1.6 (0.3) |
| SPO1 | 3.3 | 1.3 (0.2) | 1.7 (0.2) | 2.5 | 1.6 (0.1) | 1.8 (0.2) |
| HOP1 | 4.1 | 1.5 (0.1) | 1.9 (0.1) | 2.4 | 1.8 (0.1) | 2.2 (0.3) |
| REC104 | 5.5 | 2.5 (0.2) | 2.9 (0.4) | 4.0 | 1.4 (0.4) | 3.3 (0.3) |
| IME2 | 2.8 | 2.2 (0.5) | 1.8 (0.1) | 1.6 | 1.8 (0.2) | 1.9 (0.2) |
| SGA1 | 1.7 | 1.5 (0.2) | 1.8 (0.1) | 1.4 | 1.9 (0.2) | 1.8 (0.2) |
| SIP4 | 2.7 | 1.9 (0.3) | 2.9 (0.4) | 2.0 | 1.6 (0.3) | 1.9 (0.4) |
| POT1 | 3.7 | 3.1 (0.5) | 6.1 (3.2)e | 3.2 | 3.3 (0.7) | 4.7 (0.8) |
| SPO13 | 4.7 | 4.2 (1.1) | 3.4 (0.3) | 6.8 | 2.9 (0.4) | 3.0 (0.4) |
| INO1 | 7.3 | 6.4 (0.8) | 8.3 (0.4) | 2.9 | 3.6 (0.7) | 5.0 (0.6) |
Means (SEs) of ratios were computed as means (SEs) of log ratios and backtransformed to the ratio scale assuming that the log ratios have a normal distribution, so that the ratios have a log-normal distribution (21). Bold type indicates genes for which derepression due to the isw2 mutation is more evident in direct hybridization experiments than in array-array comparisons. wt, wild type.
Ratios of Northern quantitations of genes. mRNA levels measured in isw2 rpd3 and isw2 sin3 double mutants were divided by mRNA levels measured in sin3 and rpd3 single mutants, respectively.
Ratios were obtained by dividing the results from one mutant-wild type microarray experiment by those from another (array-array comparisons). isw2 sin3/wild-type ratios were divided by sin3/wild-type ratios for each gene. Similarly, isw2 rpd3/wild-type ratios were divided by rpd3/wild-type ratios for each gene.
isw2 sin3/sin3 expression ratios were measured directly on microarray slides (direct hybridization). Similarly, isw2 rpd3/rpd3 expression ratios were measured directly on microarray slides.
For one of the replicates for POT1, the background level was very high, and as a result the posterior variance of the estimate of the expression log ratio was very high. If this one realization is excluded, the estimate of the expression ratio changes to 3.4, with an SE of 0.8.
To circumvent this problem, we measured isw2 rpd3/rpd3 ratios and isw2 sin3/sin3 ratios directly by labeling single- and double-mutant RNA samples with opposing dyes and hybridizing them to the same microarray slide. In contrast to the typical method of measuring mutant/wild-type ratios, this approach uses rpd3 or sin3 single mutants as “references” against which double-mutant expression levels are compared. As a result, the problem of underestimated expression ratios resulting from minimal wild-type expression is significantly reduced for genes derepressed in rpd3 or sin3 mutants. For both the isw2 rpd3-rpd3 and the isw2 sin3-sin3 comparisons, the expression ratio obtained from the direct (mutant-versus-mutant) hybridization is higher for most genes examined than that obtained by dividing two different mutant/wild-type expression ratios (Table 2). In addition, when expression ratios obtained by direct hybridization are compared to those calculated from mutant–versus–wild-type experiments, we find that direct hybridization more accurately detects derepression observed by Northern blotting. While the values of expression ratios measured for mutant-versus-mutant experiments were not always identical to those calculated from Northern data, this method qualitatively identified derepression due to the isw2 mutation for all 10 genes analyzed in both rpd3 and sin3 mutant backgrounds.
In an attempt to categorize the targets of Isw2-dependent repression, we analyzed more closely the set of genes identified above that are derepressed in isw2 rpd3 or isw2 sin3 double mutants relative to sin3 or rpd3 single mutants. This class of genes requires the Isw2 complex for repression in the absence of a functional Sin3-Rpd3 complex. We focused on the set of genes derepressed at least 1.7-fold in one or both of the direct hybridization experiments, as this cutoff level allowed for inclusion of ∼90% of genes known to be repressed by the Isw2 complex (as measured by Northern blotting) (Table 2) but did not include any genes found by Northern blotting to be unaffected in isw2 mutant cells (data not shown). By these criteria, 315 genes (∼5% of all yeast genes) belonging to many different functional categories were found to be derepressed when an isw2 mutation was present in rpd3 and/or sin3 mutant backgrounds; representatives of this group of genes are listed in Table 3. For comparison, only 112 genes met this 1.7-fold threshold in the isw2 single mutant. As expected, a significant portion (∼20%) of the 315 genes in this group contained the core Ume6p-binding site, 5′-GGCGGC-3′, in the 500 bp upstream of their initiation codons; the majority (∼72%) of these genes were also found to be derepressed in ume6 and isw2 ume6 mutant backgrounds (see the full data set). No other sequences were significantly overrepresented in the promoters of these genes. We also observed 80 genes whose expression is decreased more than 1.7-fold in one or both double mutants relative to rpd3 or sin3 single mutants, consistent with a lesser role for the Isw2 complex in the activation of transcription. The majority of these genes are uncharacterized ORFs, and no functional category of genes is highly represented in this group.
TABLE 3.
Subset of genes that require ISW2 function for repression in sin3 or rpd3 mutant backgrounda
| Category | Gene |
|---|---|
| ATP synthesis | STF1 |
| ATP10 | |
| ATP20 | |
| Secretion | BFR2 |
| SED5 | |
| NCE103 | |
| Transcriptional regulation | SMP1 |
| CAT8 | |
| SIP4 | |
| MTH1 | |
| KAR4 | |
| SPO1 | |
| RTG1 | |
| PHD1 | |
| YAP6 | |
| CIN5 | |
| CYC8 | |
| SUB1 | |
| NRD1 | |
| FUR4 | |
| Cu2+ ion homeostasis | SLF1 |
| CUP1-1 | |
| CUP1-2 | |
| Cell cycle | PCL5 |
| PCL1 | |
| SYF2 | |
| CDC26 | |
| DNA repair | MSH5 |
| MAG1 | |
| RNR3 | |
| DIN7 | |
| ALK1 | |
| MGT1 | |
| RAD51 | |
| Ethanol utilization | ALD3 |
| ALD2 | |
| ALD4 | |
| Glucose metabolism | HSP12 |
| GIP2 | |
| PIG2 | |
| GPH1 | |
| GPM2 | |
| Amino acid synthesis | GCV1 |
| LYS1 | |
| MET3 | |
| Protein degradation or modification | SMT3 |
| RPN4 | |
| SRT1 | |
| Cytoskeleton | SRO9 |
| TPM2 | |
| BNR1 | |
| HSP42 | |
| Phosphate metabolism | PHO11 |
| PHO89 | |
| PHO12 | |
| Meiosis or sporulation | SPO13 |
| MEK1 | |
| SPO1 | |
| HOP2 | |
| NDJ1 | |
| MSH4 | |
| REC104 | |
| HOP1 | |
| MEI5 | |
| SHC1 | |
| SPS100 | |
| SGA1 | |
| SPS19 | |
| NOS1 | |
| Tricarboxylic acid cycle | IDP1 |
| IDP2 | |
| CIT3 | |
| Transport | FUR4 |
| AGP2 | |
| BPH1 | |
| SIT1 | |
| FTR1 | |
| JEN1 | |
| TPO1 | |
| SUL2 | |
| THI7 | |
| FET3 | |
| ALP1 | |
| YOR071C | |
| CTP1 | |
| AQY2 | |
| AQY1 | |
| Unknown | BOP2 |
| BTN2 | |
| COS1 | |
| COS4 | |
| COS7 | |
| COS8 | |
| CTL1 | |
| EBP2 | |
| FKS3 | |
| FRE7 | |
| GIT1 | |
| KIN2 | |
| PGU1 | |
| PIR3 | |
| SIP18 | |
| SMT4 | |
| SOL4 | |
| SPI1 | |
| YCL035C | |
| YDR374C | |
| YER037W | |
| YET1 | |
| YFR026C | |
| YGL117W | |
| YGR079W | |
| YIL037C | |
| YIR043C | |
| YJL045W | |
| YKL071W | |
| YLR179C | |
| YMR269W |
For a full list of genes that require ISW2 function for repression in sin3 and rpd3 mutant backgrounds, see ftp://milano.fhcrc.org/ArrayLab/Fazzio/.
Catalytically inactive isw2 and rpd3 mutants differ from deletion mutants in phenotype and transcriptional profiles.
Isw2p has nucleosome-stimulated ATPase activity that is required for both chromatin remodeling in vitro (49) and repression of early meiotic genes in vivo (10). Similarly, HDAC activity of Rpd3p is required for normal levels of repression of target genes (7, 10, 23). However, several deacetylase-defective mutants of rpd3 were previously found to be only partially defective in the repression of a LexA reporter construct (23), suggesting one or more functions of Rpd3p that are independent of its deacetylase activity. In addition, overexpression of catalytically inactive Rpd3 protein in wild-type cells results in a partial dominant-negative phenotype (23). It is therefore possible that catalytically inactive mutants of isw2 and rpd3 exhibit defects in transcriptional regulation not seen in deletion mutants. In deletion mutants, related chromatin remodeling factors may partially compensate for the deleted proteins, whereas catalytically inactive proteins may prevent these factors from accessing chromatin. In addition, accessory proteins shared by multiple chromatin remodeling factors may be titrated by catalytically inactive proteins. Alternatively, deletion mutants may have defects in transcriptional regulation not observed in catalytically inactive mutants due to functions of these proteins that are independent of their known catalytic activities.
To determine the differences between catalytically inactive and deletion mutants of isw2 and rpd3, we analyzed the transcriptional profiles of isw2 and rpd3 single catalytically inactive mutants as well as the isw2 rpd3 double catalytically inactive mutant. For this purpose, we analyzed an rpd3 mutation by which a conserved histidine residue at position 151 is changed to alanine; this mutation was previously found to eliminate the deacetylase activity of Rpd3p (23). Similarly, we analyzed a previously characterized isw2 substitution mutation (which changes lysine 214 to arginine), known to eliminate the ATPase activity of Isw2p.
While many of the same genes were misregulated in deletion and catalytically inactive mutants of isw2 and rpd3, in each instance there were considerable differences in transcriptional profiles between the two types of mutants (Fig. 1A). While relatively few genes are derepressed at least twofold in either isw2 mutant, a substantial amount of nonoverlap is apparent for the two transcriptional profiles. A moderate number of genes are derepressed at least twofold in the rpd3 deletion mutant but not in the catalytically inactive mutant. More strikingly, a large number of genes that are derepressed at least twofold in the rpd3 catalytically inactive mutant do not meet this threshold in the deletion mutant (Fig. 1A, right panel). Most of these genes are not significantly derepressed in sin3 mutant cells, suggesting that Rpd3p is required for the repression of this group of genes independent of Sin3p. In contrast, most genes derepressed in the sin3 mutant are also derepressed in one or both rpd3 mutants, with the largest single group of sin3-responsive genes being those derepressed in all three mutants. These data suggest that Sin3p acts primarily in conjunction with Rpd3p to repress the transcription of common target genes, whereas Rpd3p appears to be required for the repression of some genes independent of Sin3p.
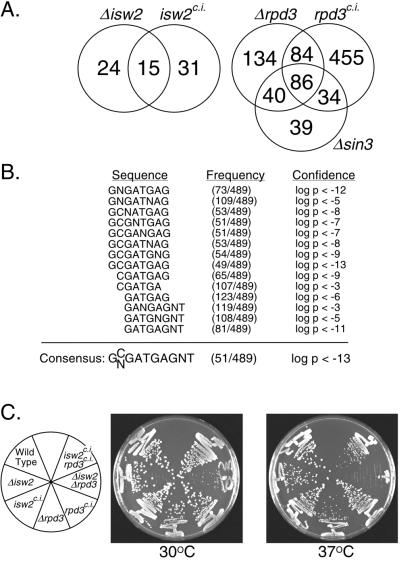
FIG. 1.
Catalytically inactive and deletion mutations of isw2 and rpd3 have different effects on transcription and growth. (A) The numbers of genes derepressed at least twofold in catalytically inactive (c.i.) and deletion (Δ) mutants of isw2 and rpd3 are compared in Venn diagrams. The number of genes derepressed at least twofold in the Δsin3 mutant is included for comparison. It should be noted that the Venn diagrams tended to overestimate the differences between two mutants, since genes derepressed in both mutants can be derepressed slightly more than twofold in one mutant and slightly less than twofold in the other mutant. Nevertheless, real differences in transcriptional profiles between deletion and catalytically inactive mutants are evident for both isw2 and rpd3. (B) Summary of a regulatory element search of the 489 genes derepressed at least twofold in the rpd3 catalytically inactive mutant but not in the rpd3 deletion mutant. The search parameters allowed for oligonucleotides of eight bases or fewer, permitting degeneracies. Highly significant elements belonging to the same consensus were aligned, and their frequencies and confidence estimates (calculated with the GENESPRING software package) are indicated. (C) Wild-type, deletion, and catalytically inactive mutant yeast cells were streaked on rich (YEPD) plates and incubated at 30 or 37°C. The genotypes of the yeast cells are shown to the left.
To further investigate the differences in transcriptional profiles between deletion and catalytically inactive mutants of rpd3, we searched the 489 genes derepressed in the catalytically inactive mutant but not in the deletion mutant for common sequences within their presumed regulatory regions (within 500 bp upstream of their initiation codons). While we did not find any known regulatory elements, several elements with overlapping sequences were overrepresented in this group to a high level of significance (Fig. 1B). The consensus for these sequences, 5′-GNGATGAGNT-3′, is present in the upstream 500 bp of 174 yeast genes. Of these, 56 (32%) are derepressed at least 2-fold and 131 (75%) are derepressed at least 1.5-fold in the rpd3 catalytically inactive mutant. This sequence was previously identified by two independent computational approaches and found to be overrepresented in the regulatory regions of a group of genes with a common expression pattern during the cell cycle (5, 47). These data cannot distinguish whether this sequence functions directly in Rpd3-mediated repression or whether the rpd3 catalytically inactive mutation indirectly leads to derepression of these genes. However, the fact that this sequence is overrepresented in the regulatory regions of genes derepressed in the same mutant as well as coexpressed during the cell cycle suggests that it may function in transcriptional regulation. Among the genes that contain this sequence in their upstream regulatory regions are those encoding components of the RNA processing and degradation machinery, as well as subunits of RNA polymerases I and III (47) (data not shown). This element may therefore coordinate the regulation of genes involved in several different aspects of RNA metabolism.
Consistent with the differences in transcriptional profiles, we also observed differences in synthetic growth phenotypes between deletion and catalytically inactive mutants of isw2 and rpd3. It was previously found that the isw2 rpd3 double deletion mutant exhibits a slow-growth phenotype at 30°C and fails to form colonies at 37°C (10). In contrast, the double catalytically inactive mutant grows at a rate similar to that of the wild type at 30°C and grows slightly more slowly than the wild type at 37°C (Fig. 1C). We also find that the rpd3 single deletion mutant grows slightly more slowly than the wild type at 37°C, while growth of the single catalytically inactive mutant is unaffected. These results suggest that the isw2 rpd3 double deletion mutant is impaired in one or more functions required for cell growth or division and that this defect is enhanced at higher temperatures. The phenotype of the double catalytically inactive mutant suggests that it is less severely impaired in these functions. The partial growth defect of the double catalytically inactive mutant at 37°C is very similar to that observed previously for an isw2 sin3 double mutant (10). These data, combined with the transcriptional data discussed above, suggest one or more functions of Rpd3p that require neither deacetylase activity nor Sin3p. However, the fact that many more genes are derepressed in the rpd3 catalytically inactive mutant than in the deletion mutant suggests that the catalytically inactive protein may inhibit other transcription or chromatin remodeling factors. The differences in growth phenotypes and transcriptional profiles between deletion and catalytically inactive mutants may therefore result from a combination of these two effects.
Genomic instability of ume6 mutants.
The Isw2 complex is recruited by Ume6p and functions in repression of genes that contain the Ume6p-binding site, URS1 (10). Since repression of early meiotic genes by Isw2 and Sin3-Rpd3 complexes is fully dependent on Ume6p, we wished to determine whether UME6 function is required for repression of nonmeiotic genes by these complexes. To determine the extent to which Isw2 and Sin3-Rpd3 complexes depend on Ume6p, as well as the extent to which Ume6p functions through these complexes, we analyzed the expression profiles for ume6 and isw2 ume6 mutants.
Surprisingly, more genes were derepressed to high levels in ume6 and isw2 ume6 mutants than in isw2 rpd3 or isw2 sin3 mutants (Table 1), suggesting the presence of a number of Ume6 targets that are not regulated by Isw2 and Sin3-Rpd3 complexes. In addition, we found that many genes with no nearby URS1 sequence were derepressed in ume6 and isw2 ume6 mutants (Table 1). Upon closer inspection, a much higher-than-average density of derepressed genes was located on chromosome 16 (for the ume6 mutant) or chromosomes 9 and 16 (for the isw2 ume6 double mutant) (data not shown). Recently, Hughes et al. observed similar chromosomal expression biases in ∼8% of a large number of expression studies (19). This group examined the nature of the expression biases for each of these mutants and found, for nearly every one examined, that the biases resulted from duplications or deletions of chromosomal segments or whole chromosomes.
To determine whether ume6 and isw2 ume6 mutants contained chromosomal duplications, we isolated DNA from mutant and wild-type cells, labeled each, and performed competitive hybridizations on microarray slides as described previously (19). We analyzed the DNA contents of two independent ume6 mutants and three independent isw2 ume6 mutants constructed in two different strain backgrounds in our laboratory, as well as one ume6 mutant obtained from another laboratory (Table 4). For all of the mutants, we found evidence that at least one and sometimes two or more chromosomes were duplicated. During the course of this work, independent expression data for a ume6 mutant were published (2); analysis of these data reveals chromosomal expression biases that strongly suggest the presence of chromosomal duplications (data not shown), supporting our findings. Independent DNA isolates from the same ume6 mutant revealed variability in which chromosomes were duplicated, suggesting that duplicated chromosomes were not always maintained during growth (Table 4). While the most common duplications were of chromosomes 9 and 16, we also observed, less frequently, duplications of chromosomes 1, 2, 3, and 8. Hughes et al. (19) found that for several mutants, the duplicated chromosomes or chromosomal segments contained genes homologous to the mutated gene, suggesting that selection for extra copies of homologous genes might partially compensate for the lack of the mutated gene. However, for UME6, we have found no close homolog within the yeast genome. In addition, the high degree of variability in chromosomes duplicated in ume6 mutants argues against the possibility that these mutants are selected for extra copies of particular genes.
TABLE 4.
Chromosomal duplications in ume6 mutants
| Straina | Background | Genotype | Chromosomal duplication(s)
|
|
|---|---|---|---|---|
| Partialb | Fullc | |||
| YTT570 | W303 | ume6 | 1, 6 | 9, 16 |
| 1, 3, 6 | 9, 16 | |||
| 1, 3, 6, 9 | 16 | |||
| 1, 3, 6 | 16 | |||
| 1, 3, 5, 6, 9 | 16 | |||
| RSY431 | W303d | ume6 | 1, 3, 6 | 2, 9 |
| YTT622 | S288C | ume6 | 1, 6 | 9 |
| YTT572 | W303 | isw2 ume6 | 3, 6 | 1 |
| 1, 3, 6 | 9, 16 | |||
| 1, 3, 6 | 9, 16 | |||
| 1, 3, 6 | 9, 16 | |||
| 1, 3, 6 | 9, 16 | |||
| YTT624 | S288C | isw2 ume6 | 1, 6 | 9 |
| YTT625 | S288C | isw2 ume6 | 1, 6 | 3, 8, 9 |
Five independent DNA isolates were analyzed for YTT570 (ume6) and YTT572 (isw2 ume6).
Chromosomes with DNA contents that were moderately higher than normal were considered partial duplications. In every instance, the DNA contents of genes along the entire length of the partially duplicated chromosome were moderately increased, suggesting that the entire chromosome was duplicated in a portion of cells from which DNA was prepared, rather than that a portion of the chromosome was duplicated in all cells.
Chromosomes with DNA contents that were substantially higher than normal were considered full duplications (duplicated in all or nearly all cells).
RSY431 is a ume6 mutant in the W303 yeast background (obtained from Randy Strich).
Alternatively, chromosomal duplications in ume6 mutants may result from a general defect in chromosome segregation. In this scenario, chromosome loss would be predicted to occur in addition to chromosomal duplications. However, in our expression studies, we would only have observed chromosomal duplications, since chromosome loss is lethal to haploid cells. To test this possibility, we constructed strains containing a minichromosome harboring the SUP11 gene, which suppresses the ade2-101 mutation in our yeast strains, rendering colonies white. Because this chromosome is dispensable for cell viability, chromosome loss rates can easily be measured by monitoring differences in colony color after nonselective growth (16, 41). With this assay, we found that ume6 and isw2 ume6 mutant cells lose the minichromosome roughly 26- to 62-fold more often than wild-type cells (Table 5). These results suggest that chromosomal duplications observed in ume6 mutants result from chromosome missegregation events.
TABLE 5.
Minichromosome loss in ume6 mutants
| Genotype | No. of colonies:
|
Rate of loss/generationa | ||
|---|---|---|---|---|
| Red | Half-sector | Total | ||
| Wild type | 19 | 4 | 6,944 | 5.8 × 10−4 |
| isw2 | 47 | 5 | 5,248 | 9.6 × 10−4 |
| ume6 | 183 | 36 | 1,225 | 3.6 × 10−2 |
| isw2 ume6 | 154 | 26 | 1,924 | 1.5 × 10−2 |
Half-sector colonies/(total colonies − red colonies) (58).
The Isw2 complex is required for the formation of DNase I-inaccessible chromatin structure at two Ume6-independent loci.
Chromatin structure analysis of one gene (REC104) targeted for repression by Isw2 and Sin3-Rpd3 complexes via Ume6p revealed that the Isw2 complex forms DNase I-inaccessible chromatin structure upstream of the Ume6p-binding site (URS1) (10). In isw2 mutants, increased accessibility of chromatin near the URS1 sequence appeared to be due to a shift in the positions of two or three nucleosomes directly upstream of this site. The formation of an inaccessible chromatin structure at this site by the Isw2 complex requires a functional UME6 gene but is unaffected by mutation of the RPD3 gene.
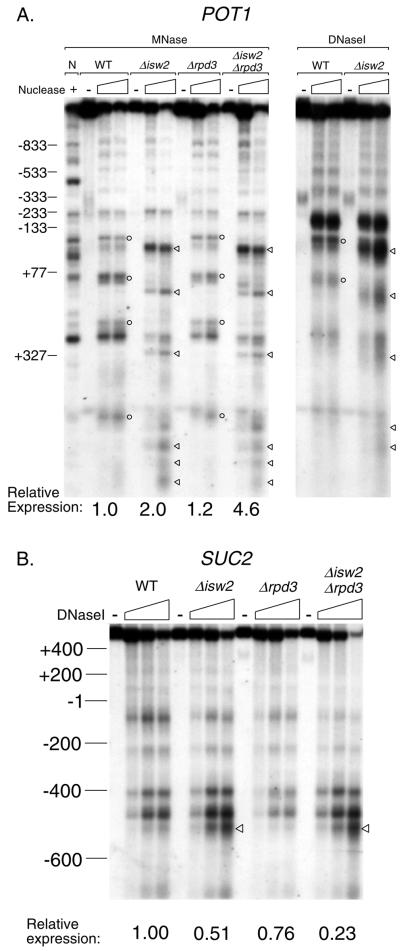
Because the formation of DNase I-inaccessible chromatin structure by the Isw2 complex occurs directly adjacent to the Ume6p binding site, it is possible that Ume6p modifies or regulates the activity of the Isw2 complex upon recruitment, in a manner unique to Ume6 target genes. If this theory is correct, the Isw2 complex may modify chromatin structure differently at loci where it functions independently of Ume6p. Alternatively, the Isw2 complex may function similarly to create inaccessible chromatin structure at all genes for which it regulates chromatin structure, regardless of the mechanism of recruitment. To distinguish between these possibilities, we analyzed the chromatin structure of the POT1 gene using micrococcal nuclease and DNase I digestions of chromatin, followed by indirect end labeling (Fig. 2A). The POT1 gene was selected because it is repressed in parallel pathways by Isw2 and Sin3-Rpd3 complexes, yet data from both microarray and Northern blotting experiments confirmed little or no change in POT1 expression in a ume6 mutant (Fig. 2A) (see the full microarray data set). In addition, no Ume6p-binding site (URS1) is present in the POT1 upstream regulatory region. As previously observed for the REC104 gene, the positions of three nucleosomes at the POT1 locus appear to change in the isw2 mutant, while no change is observed in the rpd3 mutant. As with the REC104 gene, these changes are accompanied by an increase in DNase I hypersensitivity near the promoter. In contrast to the situation for the REC104 locus, chromatin structure changes at the POT1 locus extend well into the coding region in isw2 mutant cells. Nevertheless, DNase I-inaccessible chromatin structure established at the POT1 locus in the presence of the Isw2 complex is very similar to chromatin structure at the REC104 locus. These data suggest that the Isw2 complex may function similarly to remodel chromatin at these two loci, despite the fact that repression of REC104 by the Isw2 complex requires UME6 function, while repression of POT1 transcription is UME6 independent.
FIG. 2.
TheIsw2 complex is required for the formation of DNase I-inaccessible chromatin structure at two Ume6-independent loci. (A) Increased DNase I accessibility and improperly positioned nucleosomes at the POT1 locus in isw2 mutant cells. Chromatin structure analysis was performed using micrococcal nuclease (MNase) or DNase I digestion followed by indirect end labeling. MNase and DNase I cleavage sites enhanced in wild-type (WT) or rpd3 mutant cells are marked with circles; those enhanced in isw2 mutant cells are marked with triangles. N, naked DNA control. Relative expression levels indi- cated below each mutant were measured by Northern blotting. For reference, POT1 expression measured by Northern blotting in a ume6 mutant is 1.2-fold that in the wild type. (B) Decreased expression of the SUC2 gene in isw2 mutants despite increased DNase I accessibility of the upstream regulatory region. Chromatin structure analysis was performed using DNase I digestion followed by indirect end labeling. The DNase I cleavage site enhanced in isw2 mutant cells is marked with triangles. Relative expression levels indicated below each mutant were measured by Northern blotting.
Although the Isw2 complex appears to have only a small role in transcriptional activation or maintenance of basal transcription, we found that transcription of one gene (SUC2) was significantly reduced in isw2, sin3, rpd3, isw2 sin3, and isw2 rpd3 mutants (see the full data set). The level of expression of SUC2 was lower in isw2 sin3 and isw2 rpd3 double mutants than in any single mutant, indicating that Isw2 and Sin3-Rpd3 complexes affect SUC2 transcription independently. These data were confirmed by Northern blotting (Fig. 2B, Relative expression). Under the conditions of RNA isolation used for these studies (2% glucose media), the SUC2 gene is actively repressed. Therefore, our results suggest that Isw2 and Sin3-Rpd3 complexes are required for low levels of basal SUC2 transcription. The chromatin structure of the SUC2 promoter has been studied extensively, revealing a role for the Swi-Snf complex in the creation of nuclease-accessible chromatin during transcriptional activation (17). Since increased transcription is often associated with a more accessible chromatin structure, it is possible that the Isw2 complex also functions to make chromatin more accessible at the SUC2 locus, in contrast to its functions to create DNase I-inaccessible chromatin structure at the REC104 and POT1 loci.
To test this possibility, we probed the chromatin structure of the SUC2 locus using DNase I digestions of chromatin, followed by indirect end labeling (Fig. 2B). The DNase I digestion patterns of wild-type chromatin and isw2 mutant chromatin are very similar for the SUC2 ORF and much of the upstream regulatory region. However, a notable increase in DNase I cleavage is observed approximately 500 bp upstream of the initiation codon in the isw2 mutant. As previously found for the REC104 and POT1 loci, the DNase I digestion pattern of rpd3 mutant chromatin was very similar to that of wild-type chromatin. Thus, at the SUC2 locus, as at the REC104 and POT1 loci, the Isw2 complex, but not the Sin3-Rpd3 complex, is required for the creation of DNase I-inaccessible chromatin structure. However, unlike the situation for the REC104 and POT1 genes, the increase in DNase I accessibility is associated with a decrease in SUC2 transcription in the isw2 mutant. These data suggest that the Isw2 complex functions by a common mechanism at Ume6-dependent and -independent loci to create DNase I-inaccessible chromatin structure and that ISW2-dependent inaccessible chromatin structure can affect transcription both positively and negatively.
DISCUSSION
Because it was previously found that the functional Sin3-Rpd3 complex could often compensate for the loss of Isw2 function in repression of common target genes, we compared the transcriptional profiles of mutants defective in one or both complexes to determine the full extent of their overlap. In addition, we wished to determine the extent to which the repressive functions of Isw2 and Sin3-Rpd3 complexes depend on Ume6p. Our data indicate four main conclusions. First, the Isw2 complex functions in a pathway parallel to that of the Sin3-Rpd3 complex to repress transcription of a substantial number of genes. We find that synthetic phenotypes (those observed only when an isw2 mutation is combined with a sin3 or rpd3 mutation) are more consistently revealed using double mutant versus single mutant hybridizations, especially for genes that are tightly repressed in wild-type cells. While the largest single group of genes repressed by these complexes consists of Ume6 target genes, the majority of genes that require Isw2 and Sin3-Rpd3 complexes for repression are Ume6 independent.
Second, chromatin structure analyses suggest that the Isw2 complex is required to create DNase I-inaccessible chromatin structure in the upstream regulatory regions of two Ume6-independent genes. At this time, we cannot exclude the possibility that the observed changes in chromatin structure and transcription are due to indirect effects of isw2 mutation, since we have been unable to localize the Isw2 complex to specific chromosomal loci. The rpd3 mutant is defective in regulation of the POT1 and SUC2 genes to a similar extent as the isw2 mutant. However, only cells with the isw2 mutation show the mutant nuclease digestion patterns at these loci, arguing against the possibility that chromatin structure defects in isw2 mutant cells result from misregulation of transcription. Because SUC2 transcription is decreased in isw2 mutant cells, it appears that Isw2-dependent DNase I-inaccessible chromatin structure can have different effects on transcription at different loci. While it is not clear why inaccessible chromatin structure results in increased transcription of the SUC2 gene, one possibility is that it partially inhibits the binding of a transcriptional repressor of SUC2. Consistent with this possibility, a binding site for the Mig1 repressor is located very close (at −499 with respect to the initiation codon) (4) to the DNase I-hypersensitive site present only in isw2 mutant cells.
Recently, Kent et al. showed that the Isw2 complex is required for the formation of wild-type chromatin structure upstream of three randomly selected Ume6-independent genes, FIG1, MET17, and PHO3, as revealed by differences in their MNase cleavage patterns in isw2 mutant cells relative to wild-type cells (27). However, transcription of the FIG1 and PHO3 genes is unaffected by the isw2 mutation, and MET17 transcription is only slightly increased in isw2 mutant cells (see the full data set). It is therefore possible that regulation of chromatin structure at these loci by the Isw2 complex serves a function other than transcriptional regulation. Alternatively, additional chromatin remodeling factors may function in parallel to regulate the transcription of these genes. In this case, loss of Isw2 function may be masked by these parallel factors, analogous to the relationship between Isw2 and Sin3-Rpd3 complexes in the repression of some early meiotic genes. Taken together, the data suggest that the Isw2 complex functions by a common mechanism at Ume6-dependent and -independent loci to create nuclease-inaccessible chromatin structure and that the ISW2-dependent inaccessible chromatin structure can affect transcription positively, negatively, or not at all, depending on the specific context.
Third, there are considerable differences in transcriptional profiles between deletion and catalytically inactive mutants of isw2 and rpd3. These differences are most evident for rpd3 mutants, suggesting the presence of deacetylase-independent and Sin3-independent functions of Rpd3p, as well as the possibility that catalytically inactive Rpd3p may inhibit other repressors of transcription. Consistent with these differences, rpd3 deletion mutations show a synthetic growth defect in an isw2 mutant background, whereas catalytically inactive mutations of rpd3 and deletion mutations of sin3 show less severe synthetic growth defects in this background. In contrast, a recently published comparison of the transcriptional profiles of rpd3 and sin3 deletion mutants suggests that virtually all Rpd3p functions require Sin3p (2). The major difference between this report and our data is our finding that a large number of genes require RPD3 but not SIN3 for repression. These conflicting conclusions may be explained by differences in strain background or conditions used for the growth of yeast cells. However, we previously observed phenotypic differences between rpd3 and sin3 deletion mutants in two different yeast strain backgrounds with regard to their synthetic growth defect in an isw2 mutant background (10). These data support the conclusion that Rpd3 has some Sin3-independent functions in vivo.
Fourth, Ume6p has one or more roles in chromosome segregation during haploid mitotic growth that are independent of Isw2 and Sin3-Rpd3 complexes. As a result, haploid ume6 mutants tend to accumulate chromosomal duplications. For this reason, expression data for the ume6 and isw2 ume6 mutants should be interpreted with caution, since chromosomal duplications result in artificial inflation of genes on the duplicated chromosomes and may indirectly affect the expression of additional genes. Despite this fact, we find that a large group of genes appears to be regulated by Isw2 and Sin3-Rpd3 complexes independently of Ume6p. It is noteworthy that the expression profiles for sin3, rpd3, isw2 sin3, and isw2 rpd3 mutants show no signs of chromosomal duplications in these mutants. In contrast, Hughes et al. previously found chromosomal duplications associated with rpd3/rpd3 and sin3/sin3 homozygous mutant diploids (19). These differences may result from differences in strain background or growth media or possibly from different effects of sin3 and rpd3 mutations in haploid and diploid cells. Nevertheless, our data suggest that Ume6p has one or more functions in chromosome segregation during haploid mitotic growth that are independent of Sin3-Rpd3 and Isw2 complexes under the conditions tested. It is possible that Ume6p regulates the transcription of specific genes independently of Sin3-Rpd3 and Isw2 complexes. Alternatively, Ume6p may function more directly in some aspect of chromosome segregation. The bias observed for duplications of chromosomes 9 and 16 may reflect a growth advantage associated with extra copies of these chromosomes in ume6 mutant cells. However, it is also possible that these chromosomes are more susceptible than others to missegregation in ume6 mutants.
ACKNOWLEDGMENTS
We thank Sue Biggins, Marnie Gelbart, Mark Groudine, Cedar McKay, and Jay Vary for helpful discussions and critical reading of the manuscript. We also thank Sue Biggins and Randy Strich for yeast strains and Christine Jasoni for help with data processing.
This work was supported by a Pew Charitable Trust Biomedical Scholars fellowship and NIH grant GM58465 to T.T. The FHCRC DNA Array Facility is funded in part by NCI Cancer Center support grant 5P30 CA15704-28. T.G.F. is supported by a predoctoral fellowship from HHMI. C.K. is supported by NIH grant CA74841.
REFERENCES
- 1.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho R A. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein B E, Tong J K, Schreiber S L. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bochar D A, Savard J, Wang W, Lafleur D W, Moore P, t. J C, Shiekhattar R. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci USA. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bu Y, Schmidt M C. Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 1998;26:1002–1009. doi: 10.1093/nar/26.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussemaker H J, Li H, Siggia E D. Regulatory element detection using correlation with expression. Nat Genet. 2001;27:167–171. doi: 10.1038/84792. [DOI] [PubMed] [Google Scholar]
- 6.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Fernandez J, Mische S, Courey A J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Deuring R, Fanti L, Armstrong J A, Sarte M, Papoulas O, Prestel M, Daubresse G, Verardo M, Moseley S L, Berloco M, Tsukiyama T, Wu C, Pimpinelli S, Tamkun J W. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 10.Goldmark J P, Fazzio T G, Estep P W, Church G M, Tsukiyama T. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 11.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 12.Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- 13.Guschin D, Geiman T M, Kikyo N, Tremethick D J, Wolffe A P, Wade P A. Multiple ISWI ATPase complexes from xenopus laevis. Functional conservation of an ACF/CHRAC homolog. J Biol Chem. 2000;275:35248–35255. doi: 10.1074/jbc.M006041200. [DOI] [PubMed] [Google Scholar]
- 14.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 15.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 16.Hieter P, Mann C, Snyder M, Davis R W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- 17.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 18.Hughes T R, Marton M J, Jones A R, Roberts C J, Stoughton R, Armour C D, Bennett H A, Coffey E, Dai H, He Y D, Kidd M J, King A M, Meyer M R, Slade D, Lum P Y, Stepaniants S B, Shoemaker D D, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend S H. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 19.Hughes T R, Roberts C J, Dai H, Jones A R, Meyer M R, Slade D, Burchard J, Dow S, Ward T R, Kidd M J, Friend S H, Marton M J. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- 20.Ito T, Bulger M, Pazin M J, Kobayashi R, Kadonaga J T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 21.Johnson N L, Kotz S. Continuous univariate distributions. Vol. 1. 1970. Houghton-Mifflin, Boston, Mass. [Google Scholar]
- 22.Kadonaga J T. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 23.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998;12:797–805. doi: 10.1101/gad.12.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 25.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deacetylase complex generates a highly localized domain of repressed chromatin in vivo. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasten M M, Dorland S, Stillman D J. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997;17:4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent N A, Karabetsou N, Politis P K, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kikyo N, Wade P A, Guschin D, Ge H, Wolffe A P. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science. 2000;289:2360–2362. doi: 10.1126/science.289.5488.2360. [DOI] [PubMed] [Google Scholar]
- 29.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 30.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 31.Kooperberg, C., T. G. Fazzio, J. J. Delrow, and T. Tsukiyama. Improved background correction for spotted DNA microarrays. J. Comp. Biol., in press. [DOI] [PubMed]
- 32.Kuo M H, Allis C D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 33.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 34.LeRoy G, Loyola A, Lane W S, Reinberg D. Purification and Characterization of a Human Factor That Assembles and Remodels Chromatin. J Biol Chem. 2000;275:14787–14790. doi: 10.1074/jbc.C000093200. [DOI] [PubMed] [Google Scholar]
- 35.LeRoy G, Orphanides G, Lane W S, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 36.Maldonado E, Hampsey M, Reinberg D. Repression: targeting the heart of the matter. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 37.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 38.Peterson C L. SWI/SNF complex: dissection of a chromatin remodeling cycle. Cold Spring Harbor Symp Quant Biol. 1998;63:545–552. doi: 10.1101/sqb.1998.63.545. [DOI] [PubMed] [Google Scholar]
- 39.Poot R A, Dellaire G, Hulsmann B B, Grimaldi M A, Corona D F, Becker P B, Bickmore W A, Varga-Weisz P D. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. EMBO J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 41.Spencer F, Gerring S L, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 43.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 44.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 45.Suka N, Carmen A A, Rundlett S E, Grunstein M. The regulation of gene activity by histones and the histone deacetylase RPD3. Cold Spring Harbor Symp Quant Biol. 1998;63:391–399. doi: 10.1101/sqb.1998.63.391. [DOI] [PubMed] [Google Scholar]
- 46.Sweet D H, Jang Y K, Sancar G B. Role of UME6 in transcriptional regulation of a DNA repair gene in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6223–6235. doi: 10.1128/mcb.17.11.6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tavazoie S, Hughes J D, Campbell M J, Cho R J, Church G M. Systematic determination of genetic network architecture. Nat Genet. 1999;22:281–285. doi: 10.1038/10343. [DOI] [PubMed] [Google Scholar]
- 48.Tsukiyama T, Daniel C, Tamkun J, Wu C. ISWI, a member of the SWI2/SNF2 ATPase family, encodes the 140 kDa subunit of the nucleosome remodeling factor. Cell. 1995;83:1021–1026. doi: 10.1016/0092-8674(95)90217-1. [DOI] [PubMed] [Google Scholar]
- 49.Tsukiyama T, Palmer J, Landel C C, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 51.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 52.Vidal M, Gaber R F. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal M, Strich R, Esposito R E, Gaber R F. RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991;11:6306–6316. doi: 10.1128/mcb.11.12.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wade P A, Wolffe A P. Transcriptional regulation: SWItching circuitry. Curr Biol. 1999;9:R221–R224. doi: 10.1016/s0960-9822(99)80134-1. [DOI] [PubMed] [Google Scholar]
- 55.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 56.Wu C, Becker P B, Tsukiyama T. ATP-dependent chromatin remodeling by the ISWI complexes. In: Elgin S C R, Workman J L, editors. Chromatin structure and gene expression. New York, N.Y: Oxford University Press; 2000. pp. 114–134. [Google Scholar]
- 57.Wu J, Grunstein M. 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 58.Yoon H J, Carbon J. Participation of Bir1p, a member of the inhibitor of apoptosis family, in yeast chromosome segregation events. Proc Natl Acad Sci USA. 1999;96:13208–13213. doi: 10.1073/pnas.96.23.13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell. 1997;89:357–364. doi: 10.1016/s0092-8674(00)80216-0. [DOI] [PubMed] [Google Scholar]
- 60.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]