Abstract
Signal transduction pathways regulate gene expression in part by modulating the stability of specific mRNAs. For example, the mitogen-activated protein kinase (MAPK) p38 pathway mediates stabilization of tumor necrosis factor alpha (TNF-α) mRNA in myeloid cells stimulated with bacterial lipopolysaccharide (LPS). The zinc finger protein tristetraprolin (TTP) is expressed in response to LPS and regulates the stability of TNF-α mRNA. We show that stimulation of RAW264.7 mouse macrophages with LPS induces the binding of TTP to the TNF-α 3′ untranslated region. The p38 pathway is required for the induction of TNF-α RNA-binding activity and for the expression of TTP protein and mRNA. Following stimulation with LPS, TTP is expressed in multiple, differentially phosphorylated forms. We present evidence that phosphorylation of TTP is mediated by the p38-regulated kinase MAPKAPK2 (MAPK-activated protein kinase 2). Our findings demonstrate a direct link between a specific signal transduction pathway and a specific RNA-binding protein, both of which are known to regulate TNF-α gene expression at a posttranscriptional level.
The cytoplasmic concentration of a given mRNA is a function of its rates of synthesis and degradation. The regulation of mRNA stability is therefore an important means of modulating gene expresssion (9, 16, 20, 36). For example, the rapid and transient induction of several genes is mediated by transient transcriptional activation and transient stabilization of intrinsically unstable mRNAs. Control of mRNA stability is mediated by cis-acting sequences within 5′ or 3′ untranslated regions (UTRs) or, in some cases, within the coding region. The best-characterized regulatory elements are the adenosine/uridine-rich elements (AREs) within 3′ UTRs of cytokine, growth factor, and proto-oncogene mRNAs, often containing several copies of the motif AUUUA (4, 8, 46). Overlapping AUUUA motifs or single copies within a uridine-rich context are able to confer instability upon otherwise stable reporter mRNAs (8, 28, 46, 57). It is assumed that the regulation of mRNA stability is mediated by trans-acting RNA-binding factors which interact with these cis-acting elements. ARE-binding regulators of mRNA stability include AUF1 (3, 12, 32, 45, 47, 56), HuR (1, 11, 15, 38) and tristetraprolin (TTP) (6, 7, 29, 48).
There is growing evidence that AU-rich elements do not simply confer mRNA instability; rather, they also participate in the dynamic regulation of gene expression in response to external stimuli. For example, AREs from the 3′ UTRs of interleukin-8 (IL-8) or cyclooxygenase-2 mRNA mediate regulation of mRNA stability by the mitogen-activated protein kinase (MAPK) p38 pathway (33, 34, 55). This pathway is activated by proinflammatory stimuli such as bacterial lipopolysaccharide (LPS), IL-1, tumor necrosis factor alpha (TNF-α), and UV light (21, 27, 35, 43). MAPK p38 is phosphorylated and activated by MAPK kinase MKK6 or MKK3 (14, 22, 37, 39) and in turn phosphorylates its own substrates, including the kinase MAPK-activated protein kinase 2 (MAPKAPK2) (17, 41). The effects of p38 upon mRNA stability are mediated by MAPKAPK2 (34, 55), although the relevant substrate(s) of MAPKAPK2 remain(s) to be identified conclusively. The stability of TNF-α mRNA is regulated by the p38 pathway in myeloid cell lines (2, 42, 53). Its 3′ UTR contains an ARE with five (mouse) or six (human) repeats of the AUUUA motif, which is able to mediate the stabilization of a reporter mRNA by the p38 pathway in HeLa cells (2). The deletion of this region from the mouse genomic TNF-α locus results in elevated basal and LPS-induced TNF-α expression, chronic inflammatory arthritis, and inflammatory bowel disease (25). The stability of the modified TNF-α transcript appears to be increased, and the negative regulation of TNF-α expression by a p38 inhibitor is ablated.
A similar syndrome of inflammatory arthritis and bowel disease is observed in mice deficient in TTP (otherwise known as Nup475, TIS11, or Zfp36) (49). This is a member of a novel class of RNA-binding proteins containing two CCCH zinc fingers and three tetraproline (PPPP) motifs and was initially described as being encoded by an immediate-early mitogen-induced gene (13, 19, 31, 52). In macrophages, its expression is also induced by LPS or by TNF-α itself (7). The inflammatory syndrome of TTP-null mice is caused by increased stability of TNF-α mRNA and consequent overexpression of the cytokine (5, 7, 49). In transfected HEK293 cells, TTP destabilizes a coexpressed TNF-α reporter mRNA (29). In extracts from transfected HEK293 cells, binding of overexpressed TTP to the TNF-α ARE can be detected by electrophoretic mobility shift assays (EMSAs) or by UV cross-linking (7, 29, 30). The binding of endogenous TTP protein to this regulatory element has not previously been described.
In summary, TTP is believed to destabilize TNF-α mRNA in an ARE-dependent manner, forming a negative-feedback loop which functions to restrain TNF-α biosynthesis. TTP is known to exist as a phosphoprotein in vivo and to be phosphorylated by MAPK p42 in vitro (51). However the mechanisms of regulation of TTP expression and function have not been characterized. Here, we demonstrate that an LPS-inducible TNF-α ARE-binding factor in mouse RAW264.7 macrophage cells contains TTP. Following stimulation of RAW264.7 cells with LPS, TTP protein is expressed in several, differentially phosphorylated forms. We present evidence that the p38 signal transduction pathway regulates both the expression and the posttranslational modification of TTP.
MATERIALS AND METHODS
Materials.
Salmonella enterica serovar Typhimurium LPS was from Sigma-Aldrich Company, Ltd., and was used at a concentration of 10 ng ml−1. Sheep anti-rabbit MAPKAPK2 antibody was from Upstate Biotechnology, Inc. The rabbit antiserum to the C-terminal peptide of MAPK p38 has been described previously (44). Rabbit antisera were raised against the peptides SAIYESLQSMSHDLSC and CPRRLPIFNRISVSE (Babraham Institute, Cambridge, United Kingdom), corresponding to, respectively, the amino-terminal and the carboxy-terminal 16 amino acids of murine TTP. Recombinant MAPKAPK2 has been described previously (40). Recombinant human hsp27 was from Bioquote, Ltd. Recombinant protein phosphatase 2A (PP2A) was prepared by S. Sarsfield at the Kennedy Institute of Rheumatology. SB203580 was from Calbiochem-Novabiochem, Ltd.
A baculovirus glutathione S-transferase (GST)-MKK6b expression vector was the gift of A. Finch (UCSF Cancer Center), and a bacterial GST-p38α expression vector was the gift of S. Lumb (Celltech, Slough, United Kingdom). The mouse TNF-α 3′ UTR was amplified by PCR from a plasmid containing the entire TNF-α locus (gift from A. Shakov). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TNF-α 3′ UTR fragments were amplified by PCR from genomic DNA. PCR products were subcloned into pBluescript KS(+) (Stratagene, La Jolla, Calif.), and sequenced. A full-length murine TTP cDNA (I.M.A.G.E. clone 3823873) was obtained from the American Type Culture Collection (Manassas, Va.). The coding region was amplified by PCR, cloned in frame into pGEX2T (Amersham Pharmacia, Little Chalfont, United Kingdom), and sequenced. Sequences of oligonucleotides used in PCR are available on request from the corresponding author.
Preparation of cell extracts.
Human monocytes were prepared by elutriation from peripheral blood (10) and treated with macrophage colony-stimulating factor (M-CSF) (100 ng/ml) overnight before stimulation with LPS. RAW264.7 cells (ATCC TIB-71) were maintained as described previously (2). Cell extracts were prepared as described previously (24). All operations were performed at 0 to 4°C. The cells were cooled on ice for 5 min, rinsed once with ice-cold phosphate-buffered saline, and harvested by scraping. Then, the cells were pelleted by centrifugation at 600 × g for 10 min and washed once more in phosphate-buffered saline. The cells were lysed by the addition of buffer A (20 × 106 cells/100 μl), containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 40 mM KCl, 2 mM dithiothreitol (DTT), 5% glycerol, 0.5% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 10 μM E64, 4 μg of aprotinin per ml, 4 μg of pepstatin per ml, 100 μM sodium vanadate, and 1 μM microcystin. After gentle pipetting, the cells were left on ice for 15 min to swell and burst. Nuclei were removed by centrifugation at 600 × g for 10 min. The supernatant, designated the cytoplasmic extract, was aliquoted and snap-frozen at −70°C. Protein concentrations were determined by Bradford protein assay.
In vitro transcription.
RNA probes were prepared as follows. Template DNA was linearized to completion by appropriate restriction digestion and then purified by phenol-chloroform extraction and ethanol precipitation. Labeled transcripts were synthesized by in vitro transcription of the linearized templates (1 μg of DNA) with 20 U of T7 polymerase in the presence of transcription buffer (Promega), 10 mM DTT, 20 U of recombinant RNasin RNase inhibitor (Sigma), 125 μM (each) ATP, GTP, and CTP, 12 μM UTP, and 20 μCi of [α-32P]UTP (800 Ci/mmol; Amersham Pharmacia) in a final volume of 20 μl. Transcription was carried out for 1 h at 37°C and was stopped by the addition of 10 U of RNase-free DNase (Life Technologies). Reaction mixtures were brought to a volume of 100 μl with distilled water and extracted once with phenol-chloroform. The riboprobe was then purified on a S200 spin column (Amersham Pharmacia) and stored at −20°C.
EMSA.
RNA band shift assays were performed essentially as described previously (24). The protein extracts were incubated with the indicated RNA probes in RNA-binding buffer containing 20 mM HEPES (pH 7.6), 3 mM MgCl2, 40 mM KCl, 2 mM DTT, and 5% glycerol in a total volume of 20 μl. Typically, 10 μg of protein was incubated with 400,000 to 500,000 cpm of α-32P-labeled RNA probe, corresponding to approximately 20 fmol of RNA. The reaction mixture was incubated on ice for 20 min. RNase T1 and heparin sulfate were added to final concentrations of 50 U/ml and 5 mg/ml, respectively, and the reaction was allowed to continue for a further 20 min on ice. Quantities (3 μl) of loading buffer (90% glycerol, 0.025% bromophenol blue) were added to the samples, which were then resolved by electrophoresis on a 0.5× Tris-borate-EDTA nondenaturing 4% polyacrylamide gel at 150 V for 6 h at 4°C. Gels were dried, autoradiographed, and analyzed using a phosphorimager (FLA2000; Fuji).
Western blotting.
RAW264.7 mouse macrophage extracts were prepared as described above, separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and electrophoretically transferred to polyvinylidene fluoride microporous membrane (NEN Life Sciences). The membrane was probed with a rabbit polyclonal antiserum raised against the C terminus of mouse TTP and then with a peroxidase-coupled secondary antibody (Dako). Proteins were detected using an enhanced chemiluminescence system (Amersham Pharmacia).
Northern blotting.
RNA was purified from RAW264.7 cells using QIAamp RNA blood kits (Qiagen, Crawley, United Kingdom). At each experimental time point, 20 μg of RNA was subjected to Northern blotting as described previously (2). The probe was a 1-kb full-length murine TTP cDNA fragment, labeled using ReadyToGo reagents (Amersham Pharmacia). Prior to transfer and blotting of the gel, 18S and 28S ribosomal RNAs were visualized by staining with Sybr green (Molecular Probes) and quantified using a phosphorimager (FLA2000; Fuji).
p38 MAPK and MAPKAPK2 kinase assays.
GST-MKK6b was expressed in baculovirus-infected Sf9 cells. GST-TTP, GST-p38α, and His6-tagged MAPKAPK2 were expressed in Escherichia coli DH5α. Purification of GST fusion proteins was by glutathione Sepharose affinity chromatography with Amersham Pharmacia reagents by the corresponding protocols. Purification of His6-tagged MAPKAPK2 was by Ni-nitrilotriacetic acid (NTA) chromatography with Qiagen reagents, using the corresponding protocols.
Assays using recombinant proteins were performed by mixing the purified kinases with 4 μCi of [γ-32P]ATP in 30 μl of kinase buffer (20 mM HEPES [pH 7.6], 20 mM sodium β-glycerophosphate, 200 mM NaCl, 10 mM MgCl2, 10 mM NaF, 2 mM DTT, 0.5 mM EDTA, 0.5 mM EGTA, and 0.1 mM sodium orthovanadate). The quantities of proteins were as follows: for GST-MKK6b, 1 μg; for GST-p38α, 1 μg; for His6-tagged MAPKAPK2, 2 μg; for GST-TTP, 2 μg; and for hsp27, 2 μg.
For the immune-complex kinase assays, RAW264.7 cells were stimulated with LPS for the time periods indicated in the legends to the figures and the lysates were prepared as described above. Lysate (300 μg) was used in kinase assays that were carried out as described previously (2, 10).
RESULTS
One LPS-inducible and five constitutive factors bind to the TNFα 3′-UTR.
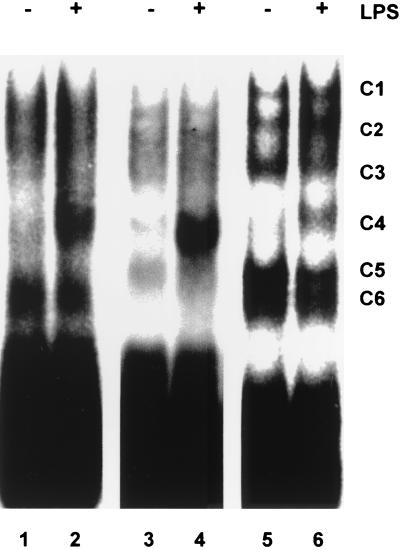
To detect RNA-protein interactions, cytoplasmic protein extracts were prepared from M-CSF-treated, unstimulated, or LPS-stimulated human peripheral blood monocytes. These extracts were used in EMSAs with a radiolabeled RNA probe corresponding to the full-length 3′ UTR of human TNF-α. An LPS-inducible complex of intermediate mobility was readily detected (Fig. 1, lanes 1 and 2). The experiment was repeated using extracts of untreated or LPS-stimulated RAW264.7 mouse macrophage cells, which produce large quantities of TNF-α in response to LPS. Five complexes, designated, respectively, C1, C2, C3, C5, and C6, were detected in control extracts (Fig. 1, lanes 5 and 6). Complexes C5 and C6 migrated very similarly but could be distinguished on the basis of competition or mapping experiments (data not shown). Stimulation of RAW264.7 cells for 2 h with LPS (10 ng/ml) led to the formation of an additional complex, C4, of similar mobility to the monocyte LPS-inducible complex. The mouse TNF-α 3′ UTR is 68% identical to the human sequence but 91% identical within the central AU-rich region. Similar patterns of protein-RNA complexes were observed using a mouse TNF-α 3′ UTR probe (Fig. 1, lanes 3 and 4), with the exception that C6 was not detected. The apparent induction of C4 varied between experiments (with levels in the range of 4- to 10-fold). C4 was not detected if the phosphatase inhibitors sodium orthovanadate and microcystin were omitted from the lysis buffer (data not shown). The cytoplasmic extract of RAW264.7 cells was further fractionated by high-speed centrifugation at 100,000 × g. C4 was detected in both the soluble supernatant (S100) and the solubilized pellet (P100) generated by this step (data not shown). The P100 fraction contains high-molecular-weight complexes, including ribosomes, and is the location of most of the TNF-α mRNA in LPS-stimulated RAW264.7 cells (11). Unfractionated cytoplasmic extracts were used in all subsequent experiments. Murine and human full-length TNF-α 3′ UTR probes were used interchangeably, since C4 was detected with both probes.
FIG. 1.
LPS-inducible binding of a factor to human and mouse TNF-α 3′ UTRs. M-CSF-treated human peripheral blood monocytes or RAW264.7 cells were stimulated with 10 ng of LPS per ml for 2 h, and then cytoplasmic extracts were prepared. EMSAs were performed using 10 μg of cytoplasmic extract and 20 fmol of 32P-labeled human or mouse full-length TNF-α 3′ UTR probes. Lanes 1 and 2, cytoplasmic extract of unstimulated or LPS-stimulated, M-CSF-treated human peripheral blood monocytes probed with 32P-labeled human full-length TNF-α 3′ UTR probes; lanes 3 and 4, cytoplasmic extract of unstimulated or LPS-stimulated RAW264.7 cells probed with mouse full-length TNF-α 3′ UTR probes; lanes 5 and 6, cytoplasmic extract of unstimulated or LPS-stimulated RAW264.7 cells probed with 32P-labeled human full-length TNF-α 3′ UTR probes. The well-resolved complexes evident in lanes 5 and 6 are labeled C1 to C6.
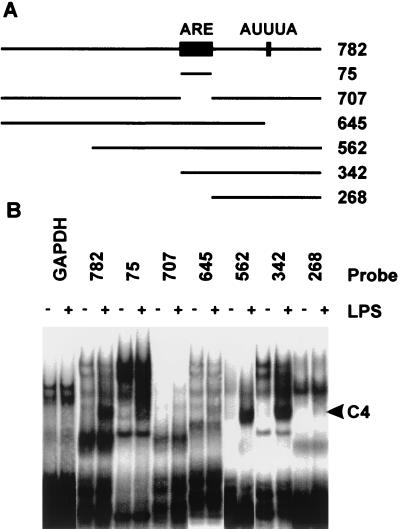
Formation of C4 is dependent upon the TNF-α ARE.
In order to map TNF-α mRNA sequences involved in the formation of C4, EMSAs were performed using cytoplasmic extracts of unstimulated or LPS-stimulated RAW264.7 cells, as well as a series of truncated 3′ UTR probes, shown schematically in Fig. 2A. A control GAPDH 3′ UTR probe formed no LPS-inducible complexes (Fig. 2B); therefore, stimulation of RAW264.7 cells with LPS does not simply induce nonspecific RNA-binding activity. A TNF-α 3′ UTR probe lacking the ARE (probe 707) failed to generate C4 under these conditions. A probe containing only the ARE (probe 75) generated C4, although relatively weakly. The presence of additional 3′ sequences (in probes 562 and 342) enhanced the formation of C4, yet a 3′ UTR probe lacking the ARE (probe 268) failed to generate C4. A probe containing all but the last 137 nucleotides of the TNF-α 3′ UTR (probe 645) generated a relatively weak C4. Therefore the central AU-rich region of the TNF-α 3′ UTR is necessary but not sufficient for optimal formation of an LPS-inducible protein–RNA complex, and additional sequences lying 3′ to the ARE contribute to complex formation.
FIG. 2.
Mapping of protein interactions with the TNF-α 3′ UTR. (A) Schematic of truncated TNF-α 3′UTR probes. (B) Results for EMSAs in which 20 fmol of each probe was used with 10 μg of cytoplasmic extract prepared from untreated RAW264.7 cells (−) or from cells stimulated for 2 h with 10 ng of LPS per ml (+).
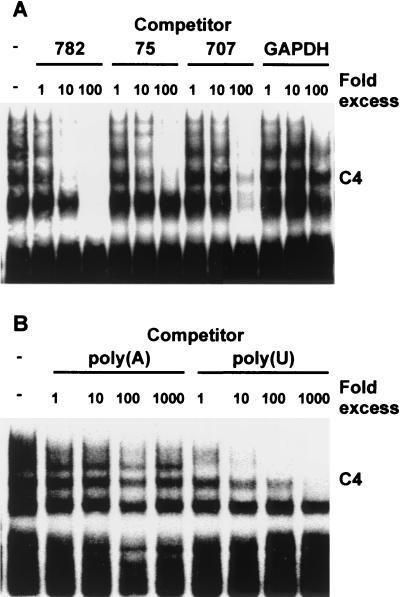
The specificity of protein interactions with the TNF-α 3′ UTR was further analyzed by competition assays (Fig. 3). C4 was not competed by a control GAPDH RNA that was devoid of AUUUA motifs, but it was competed efficiently by an excess of unlabeled full-length (782-nucleotide [nt]) TNF-α 3′ UTR. The 75-nt TNF-α ARE was a less efficient competitor for the formation of C4, and a 707-nt competitor lacking the ARE was less efficient still, with residual complex remaining detectable in the presence of a 100-fold molar excess of competitor. This experiment suggests that competitor probes 75 and 707 bind C4 protein(s) with affinities roughly 1 and 2 orders of magnitude lower, respectively, than that of the full-length TNF-α 3′ UTR. Again, this indicates that the ARE plays a critical role in the formation of C4 and that sequences lying outside the ARE may provide additional sites of protein-RNA interaction. The higher-mobility complexes C5 and/or C6 (not resolved in these experiments) were competed similarly by 782- and 707-nt RNAs but not by the 75-nt ARE, and they therefore presumably involve sequences lying outside the ARE. Finally, as indicated in Fig. 3B, C4 was efficiently competed by poly(U) but not by poly(A), -(C), or -(G) RNA [data from poly(C) and poly(G) competitions not shown]. Therefore, in common with several RNA-binding factors thought to be involved in the regulation of mRNA stability, the proteins present in C4 bind specifically to AUUUA repeats or to U-rich RNA.
FIG. 3.
Specificity of protein interactions with the TNF-α 3′ UTR. EMSAs were performed as described for Fig. 1 and 2, using a full-length human TNF-α 3′ UTR probe and 10 μg of cytoplasmic extract from LPS-stimulated RAW264.7 cells. Competitor RNA was added to the binding reaction mixtures 20 min prior to the addition of labeled probe. (A) Protein interactions with specific RNA competitors present, as indicated, in 1- to 100-fold molar excess over the probe. (B) Protein interactions with poly(U) or poly(A) RNA present, as indicated, in 1- to 1,000-fold excess (by mass) over the probe.
Induction of C4 is relatively slow and requires de novo gene expression.
The kinetics of the induction of C4 by LPS were examined following LPS stimulation of RAW264.7 cells (Fig. 4A). C4 was not detectable until at least 1 h after the administration of the stimulus but thereafter was sustained for at least 16 h. The relative slowness of this induction suggested a requirement for de novo gene expression, which was assessed using inhibitors of transcription and translation (Fig. 4B). In order to minimize exposure of cells to the toxins actinomycin D and cycloheximide, LPS stimulation was performed for only 1 h in these experiments; hence, the induction of C4 was submaximal. The inhibition of either transcription or translation blocked the induction of C4, confirming a requirement for de novo gene expression. In contrast, C3, which contains the RNA-stabilizing protein HuR (11), was upregulated in the cytoplasm following treatment with actinomycin D. Actinomycin D has been reported to stimulate the translocation of HuR from the nucleus to the cytoplasm (38).
FIG. 4.
LPS-induction of TNF-α 3′ UTR binding activity requires de novo gene expression. (A) Cytoplasmic extracts prepared from RAW264.7 cells at the indicated times after stimulation with 10 ng of LPS per ml. Ten micrograms of each extract was used in an EMSA with 20 fmol of full-length human TNF-α 3′ UTR probe. The first lane (marked with a minus sign) contains RNA probe but no protein. (B) RAW264.7 cells preincubated with vehicle, with cycloheximide (CHX [10 μg/ml]) or with actinomycin D (AmD [1 μg/ml]) for 15 min and then stimulated with LPS for 1 h prior to the preparation of cytoplasmic extracts. Ten micrograms of each extract was used in an EMSA with 20 fmol of full-length human TNF-α 3′ UTR probe.
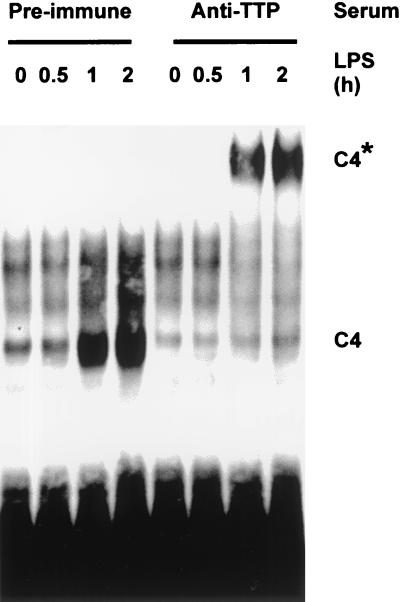
LPS-induced complex contains TTP.
Expression of TTP mRNA is induced following stimulation of mouse macrophages with LPS, peaking roughly 1 h after the administration of the stimulus and remaining elevated for at least 4 h (7). Overexpressed TTP binds to the TNF-α 3′ UTR (29, 30), although the regulated binding of endogenous protein to this RNA has not been described. To test the hypothesis that C4 contains TTP, antibodies were raised against the amino and carboxy termini of the mouse protein. In EMSAs, C4 was induced over a 2-h time course as previously described (Fig. 5). No supershift of C4 was observed in the presence of a preimmune serum. In the presence of an immune serum raised against the amino terminus of TTP, a low-mobility, supershifted complex was generated, with a corresponding total depletion of the LPS-inducible complex. Identical results were obtained using an antiserum raised against the carboxy terminus of TTP (data not shown). The LPS-inducible complex therefore contains TTP.
FIG. 5.
LPS-induced complex contains TTP. Cytoplasmic extracts of LPS-stimulated RAW264.7 cells were prepared, and EMSAs were performed as described for Fig. 4, except that the probe was murine, and 1 μl of preimmune serum or anti-TTP serum was present in each 20-μl binding reaction mixture. The supershifted C4 is indicated with an asterisk.
Induction of TTP gene expression by LPS is dependent upon MAPK p38.
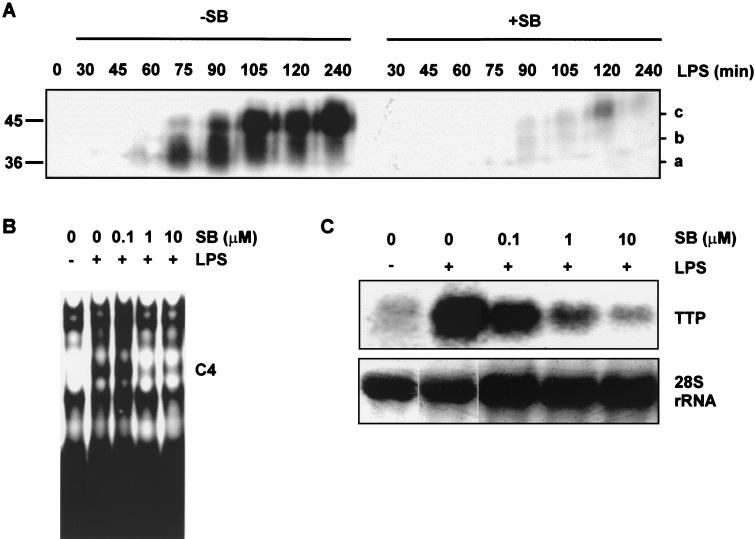
The induction of TTP protein following LPS stimulation of RAW264.7 cells was assessed by Western blotting using the amino-terminal TTP antibody described above (Fig. 6A). Unlike other investigators (50), we were unable to detect TTP protein in unstimulated cells. Antigen was first detected 45 min after stimulation, was maximally induced by 2 h after stimulation, and remained elevated for at least 4 h. The expression of TTP was accompanied by an apparent change in molecular size from a high-mobility form of approximately 36 kDa (Fig. 6A, band a) to an intermediate form (Fig. 6A, band b), and finally to a low-mobility form of approximately 45 kDa (Fig. 6A, band c [also, see Fig. 7]). This alteration of mobility suggests a posttranslational modification such as phosphorylation. As the MAPK p38 pathway is known to regulate TNF-α mRNA stability, a p38 inhibitor was used to investigate the involvement of this pathway in the expression and modification of TTP. At a concentration of 1 μM, SB203580 inhibits p38 activity by approximately 90% in LPS-stimulated RAW264.7 cells and has minimal effects upon other MAPK pathways (2). The preincubation of RAW264.7 cells with 1 μM SB203580 almost completely blocked the induction of TTP protein, as assessed by Western blotting (Fig. 6A, right). Correspondingly, 1 μM SB203580 inhibited the formation of C4 (Fig. 6B). LPS stimulation strongly induced the expression of TTP mRNA, and this induction was dose-dependently inhibited by SB203580 (Fig. 6C). Therefore, the MAPK p38 pathway is required for the expression of TTP mRNA following stimulation with LPS.
FIG. 6.
MAPK p38 activity is required for the expression of TTP in response to LPS. (A) RAW264.7 cells preincubated for 15 min with vehicle or 1 μM SB203580 and then stimulated with LPS for the times indicated. Cytoplasmic extracts were prepared and separated by SDS-polyacrylamide gel electrophoresis on a 10% gel and then analyzed by Western blotting using an anti-N-terminal TTP antiserum. The letters at the right (a, b, and c) indicate TTP bands of differing mobility. The positions of molecular size markers are indicated at the left of the gel. (B) RAW264.7 cells preincubated for 15 min with vehicle or 0.1 to 10 μM SB203580, as indicated, and then stimulated with 10 ng of LPS per ml for 2 h. Cytoplasmic extracts were prepared and used in EMSAs with 20 fmol of a full-length human TNF-α 3′ UTR probe. (C) RAW264.7 cells treated as described for panel B, with RNA then extracted and subjected to Northern blotting using a 1-kb TTP cDNA probe. As a loading control, 28S rRNA was visualized by staining of the gel with Sybr green prior to transfer and Northern blotting.
FIG. 7.
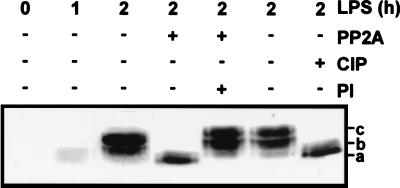
TTP is differentially phosphorylated following an stimulation with LPS. RAW264.7 cells were stimulated with 10 ng of LPS per ml for 0 to 2 h, as indicated, and then cytoplasmic extracts were prepared. A 200-μg quantity of extract was treated with recombinant PP2A (72 mU) or calf intestinal phosphatase (CIP [50 U]) in the presence or absence of the phosphatase inhibitor microcystin (PI [10 μM]). The extracts were then subjected to Western blotting as described for Fig. 6A. The letters to the right (a, b, and c) indicate TTP bands of differing mobility.
LPS-induced TTP is expressed in several differentially phosphorylated forms.
Changes in the electrophoretic mobility of TTP in LPS-stimulated RAW264.7 cells were not observed in a previous study (50). However, in Western blotting, stimulation of NIH 3T3 cells with serum caused a similar shift in the mobility of TTP, which shift was ascribed to the phosphorylation of the protein (50, 51). To test whether the LPS-induced shift of TTP was caused by phosphorylation, cytoplasmic extracts of RAW264.7 cells were treated with phosphatases in the absence or presence of phosphatase inhibitors, and then subjected to Western blotting (Fig. 7). Treatment of extracts with the serine/threonine phosphatase PP2A resulted in the disappearance of TTP bands b and c, leaving only the highest-mobility form, and this alteration in mobility was completely blocked by the PP2A inhibitor microcystin. Calf intestinal phosphatase caused an identical shift from bands b and c to a. LPS treatment of RAW264.7 cells therefore causes a strong induction of TTP protein, which is accompanied by its phosphorylation at serine and/or threonine residues.
TTP is a substrate of MAPKAPK2.
It is difficult to assess the role of the p38 pathway in the phosphorylation of TTP in vivo, as little TTP protein is synthesized in the presence of a p38 inhibitor. The TTP protein detected under these conditions appears to be a lower-mobility form, suggesting that it is phosphorylated. This phosphorylation could be mediated by a signal transduction pathway other than the p38 cascade or by residual activity of the p38 pathway under these experimental conditions. Therefore, two approaches were used to investigate the phosphorylation of TTP via the p38 pathway in vitro.
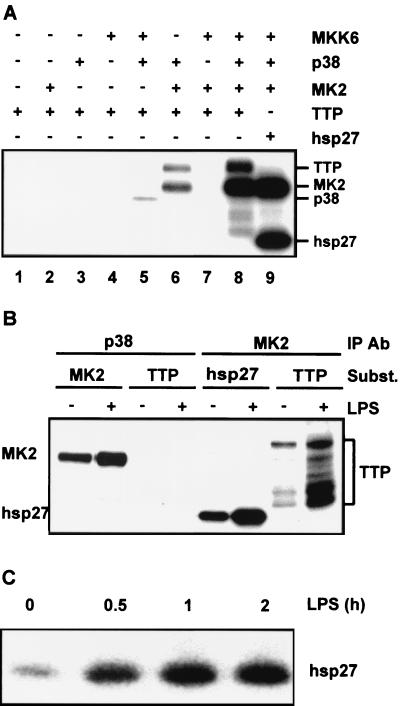
Firstly, the p38 signal transduction pathway was reconstituted using purified recombinant kinases (Fig. 8A). GST-MKK6 was expressed in Sf9 cells (using a baculovirus system) and purified by glutathione Sepharose affinity chromatography. GST-p38, GST-TTP, and His6-tagged MAPKAPK2 were expressed in E. coli and purified by glutathione Sepharose chromatography or Ni-NTA chromatography. Purified GST-MKK6 was active, GST-p38 was weakly active, and His6-tagged MAPKAPK2 was inactive (data not shown). No contaminants of the different preparations were detectable by SDS-PAGE or Coomassie blue staining, but the recombinant GST-TTP was extensively degraded in spite of the presence of protease inhibitors during purification. Reconstitution of the cascade was demonstrated by the strong phosphorylation of hsp27 in the presence of MKK6, p38, and MAPKAPK2 (Fig. 8A, lane 9). Activated p38 did not phosphorylate GST-TTP (Fig. 8A, lane 5); however, strong phosphorylation of GST-TTP was observed in the presence of MKK6, p38, and MAPKAPK2 (Fig. 8A, lane 8), suggesting that TTP is a substrate of MAPKAPK2 in vitro. The only other combination of kinases which generated detectable phosphorylation of GST-TTP was that of p38 and MAPKAPK2 (Fig. 8A, lane 6), presumably because of the basal activity of the unphosphorylated p38 and the consequent activation of MAPKAPK2 (G. Sully and J. Saklatvala, unpublished observations).
FIG. 8.
TTP is a substrate of MAPKAPK2. (A) Purified recombinant kinases MKK6, p38, and MAPKAPK2 (MK2) were mixed with the substrate GST-TTP (TTP) or hsp27 in the combinations indicated. Phosphorylation reactions were allowed to proceed for 30 min, and then the phosphorylated products were separated by SDS-PAGE and visualized by phosphorimaging. Lane 1, TTP alone; lane 2, MK2 and TTP; lane 3, p38 and TTP; lane 4, MKK6 and TTP; lane 5, MKK, p38, and TTP; lane 6, p38, MK2, and TTP; lane 7, MKK6, MK2, and TTP; lane 8, MKK6, p38, MK2, and TTP; lane 9, MKK6, p38, MK2, and hsp27. (B) RAW264.7 cells were left untreated (−) or stimulated with 10 ng of LPS per ml for 2 h (+), and then lysates were prepared, p38 or MAPKAPK2 was immunoprecipitated, and immune-complex kinase assays were performed using hsp27, His6-tagged MAPKAPK2 or GST-TTP as substrate (Subst). Phosphorylated products were separated by SDS-PAGE and visualized by phosphorimaging. (C) RAW264.7 cells were stimulated with 10 ng of LPS per ml for the times indicated, and then immune-complex assays of MAPKAPK2 activity were performed as described for panel B, using recombinant hsp27 as the substrate. Phosphorylated products were separated by SDS-PAGE and visualized by phosphorimaging.
The ability of mammalian p38 and MAPKAPK2 to phosphorylate TTP was also assessed by means of immune-complex kinase assays. Active kinases were immunoprecipitated from LPS-stimulated RAW264.7 cells (Fig. 8B). Activated p38 phosphorylated MAPKAPK2 but not TTP. Activated MAPKAPK2 phosphorylated both hsp27 and TTP, confirming that TTP is a substrate for MAPKAPK2 in vitro. Phosphorylation of GST by immunoprecipitated, active kinases was not observed, nor was GST-TTP phosphorylated by control (immunoglobulin class-matched irrelevant antibody) immunoprecipitates (data not shown). Identical results were obtained using activated kinases immunoprecipitated from IL-1-stimulated HeLa cells (data not shown). In both recombinant and immune-complex kinase assay systems, degradation products of GST-TTP were phosphorylated by MAPKAPK2. Taking these phosphorylations into account, TTP appeared to be a MAPKAPK2 substrate almost as effective as hsp27. Immune-complex kinase assays were performed to determine whether MAPKAPK2 activity overlaps with TTP protein expression in LPS-stimulated RAW264.7 cells. MAPKAPK2 activition was sustained for at least 2 h after stimulation with LPS (Fig. 8C); therefore, active kinase is present during the period in which endogenous TTP becomes phosphorylated.
DISCUSSION
In order to better understand the regulation of mRNA stability by signal transduction pathways we sought to identify RNA-binding proteins whose behavior is modulated by the p38 cascade. It was previously shown that TTP gene expression is induced by LPS stimulation of myeloid cells and that overexpressed TTP is capable of binding to TNF-α mRNA. Here, we demonstrated that endogenous TTP was present in a TNF-α ARE-binding complex induced following the stimulation of RAW264.7 cells with LPS. The induction of TTP mRNA, protein expression and RNA-binding activity was blocked by inhibition of p38. This establishes a direct connection between an RNA-binding protein and a signal transduction pathway, both of which are known to regulate the expression of TNF-α at a posttranscriptional level.
Following stimulation with LPS, TTP is expressed in a number of differentially phosphorylated forms. It is difficult to establish whether the p38 pathway is involved in this phosphorylation in vivo, as the phosphorylation status of TTP cannot be assessed in the absence of p38 activity. We demonstrated that recombinant TTP is phosphorylated in vitro by MAPKAPK2 immunoprecipitated from LPS-stimulated RAW264.7 cells and that the kinetics of induction of MAPKAPK2 activity and TTP expression overlap in LPS-stimulated RAW264.7 cells. It is therefore likely that TTP is a substrate of MAPKAPK2 in vivo, although the functional significance of this phosphorylation remains unclear. In mouse spleen cells lacking functional MAPKAPK2, steady-state TNF-α mRNA levels and mRNA stability, measured at a single time point after LPS stimulation, do not appear to be altered (26). The kinetics of TNF-α mRNA expression are altered by deletion of the AU-rich element in mice (25) or by inhibition of the p38 pathway in human monocytes (53). A role for MAPKAPK2 in the regulation of TNF-α mRNA stability cannot be ruled out without determining both the expression of TTP and the kinetics of TNF-α mRNA induction in mouse cells lacking MAPKAPK2 activity.
The accumulation of the hyperphosphorylated form of TTP is relatively slow, peaking approximately 4 h after stimulation with LPS. The dynamic nature of TNF-α mRNA stability regulation is illustrated by the decrease in half-life of the mRNA at late time points following stimulation with LPS (23). It is an interesting possibility that phosphorylation of TTP may be required for TNF-α mRNA destabilizing activity. In longer exposures of Western blots, small quantities of phosphorylated TTP can be detected as early as 1 h after stimulation with LPS, coinciding with the appearance of binding activity in the EMSA. The EMSA is exquisitely sensitive and may detect extremely low concentrations of RNA-binding factors. These observations leave open the question of whether phosphorylation is necessary for the interaction of TTP with the TNF-α 3′ UTR. Our failure to detect TTP-binding activity in extracts prepared without phosphatase inhibitors suggests that this may be the case. It is also possible that phosphorylation by MAPKAPK2 influences other aspects of TTP biology, such as subcellular localization, stability, or interaction with other proteins. TTP may also be phosphorylated by MAPK p42 (51), which is stimulated by LPS stimulation of RAW264.7 cells (18). The principal site of phosphorylation of TTP by p42 has been mapped; however, the functional significance of this phosphorylation remains to be demonstrated (29, 51). It is possible that phosphorylation of TTP via different MAPK pathways modulates TNF-α biosynthesis in response to different environmental cues. Clearly, the role of phosphorylation in the regulation of TTP function merits further investigation. We intend to map the site or sites of phosphoryation by MAPKAPK2 and to develop an appropriate myeloid cell system in which to test the effects of phosphorylation site mutations.
TTP regulates the stability of TNF-α, granulocyte-macrophage colony-stimulating factor and IL-3 mRNAs (6, 7, 29, 48), yet the specificity of binding of TTP to AU-rich RNA is not known. In our hands, clustered AUUUA pentamers appeared to be insufficient for high-affinity binding of TTP, which requires additional sequences from the TNF-α 3′ UTR. These distal sequences are well conserved between eight mammalian species, from human to bottle-nosed dolphin, and include an absolutely conserved AUUUA motif (Fig. 2A). It is possible that distal 3′ sequences provide additional RNA-binding contacts for TTP or contribute to secondary structures of the TNF-α 3′ UTR which favor TTP binding. It has been shown that the TNF-α 3′ UTR forms higher-order structures which influence its interaction with RNA-binding proteins (54). A more detailed analysis of the RNA-binding specificity of TTP may provide insights into its cellular function.
The pattern of expression of TTP has not been studied in detail; however, the pathology of the TTP knockout mouse appears to be restricted to myeloid and lymphoid cells (5, 6, 49). We did not detect the protein in HeLa cells (data not shown), which are able to stabilize ARE-containing transcripts in response to activation of the p38 pathway (2, 33, 34, 55). Stimulation of RAW264.7 cells with LPS leads to a rapid and p38-dependent stabilization of TNF-α mRNA, at a time preceding the appearance of detectable TTP antigen (2; M. Brook and A. R. Clark, unpublished observations). It is thus unlikely that TTP is involved in the active stabilization of labile mRNAs via the p38 pathway. The properties described here and elsewhere are consistent with a cell type-specific role in the off-phase of expression of TNF-α and other genes. It is intriguing that the p38 pathway stabilizes TNF-α mRNA but is also required for the expression of TTP, a TNF-α mRNA destabilizing protein. Such a dual function of the p38 pathway could represent a mechanism for the restraint of TNF-α biosynthesis, in which an LPS-induced signal transduction cascade is intimately involved in both the on phase and the off phase of TNF-α gene expression. According to this model, the stabilization phase is controlled by rapid phosphorylation of a preexisting factor of unknown identity and the destabilization phase is controlled by the more gradual synthesis and perhaps phosphorylation of a second factor, TTP.
ACKNOWLEDGMENTS
M.B. and J.L.E.D. contributed equally to this work.
K.R.M. was supported by a Ph.D. studentship from the Charing Cross Hospital Trustees.
We are grateful to Cathleen Ciesielski for assistance with purification and culture of human monocytes, to Simon Sarsfield for preparation of recombinant PP2A, and to Andrew Finch and Simon Lumb for provision of reagents.
REFERENCES
- 1.Brennan C M, Steitz J A. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook M, Sully G, Clark A R, Saklatvala J. Regulation of tumour necrosis factor alpha mRNA stability by the mitogen-activated protein kinase p38 signalling cascade. FEBS Lett. 2000;483:57–61. doi: 10.1016/s0014-5793(00)02084-6. [DOI] [PubMed] [Google Scholar]
- 3.Buzby J S, Brewer G, Nugent D J. Developmental regulation of RNA transcript destabilization by A+U-rich elements is AUF1-dependent. J Biol Chem. 1999;274:33973–33978. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- 4.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carballo E, Gilkeson G S, Blackshear P J. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 (−/−) mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFα overproduction. J Clin Investig. 1997;100:986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carballo E, Lai W S, Blackshear P J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 7.Carballo E, Lai W S, Blackshear P J. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 8.Chen C Y, Shyu A B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 9.Day D A, Tuite M F. Post-transcriptional gene regulatory mechanisms in eukaryotes: an overview. J Endocrinol. 1998;157:361–371. doi: 10.1677/joe.0.1570361. [DOI] [PubMed] [Google Scholar]
- 10.Dean J L, Brook M, Clark A R, Saklatvala J. p38 mitogen-activated protein kinase regulates cyclooxygenase-2 mRNA stability and transcription in lipopolysaccharide-treated human monocytes. J Biol Chem. 1999;274:264–269. doi: 10.1074/jbc.274.1.264. [DOI] [PubMed] [Google Scholar]
- 11.Dean J L, Wait R, Mahtani K R, Sully G, Clark A R, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMaria C T, Brewer G. AUF1 binding affinity to A+U-rich elements correlates with rapid mRNA degradation. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 13.DuBois R N, McLane M W, Ryder K, Lau L F, Nathans D. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J Biol Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- 14.Enslen H, Raingeaud J, Davis R J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 15.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontes A M, Ito J, Jacobs-Lorena M. Control of messenger RNA stability during development. Curr Top Dev Biol. 1999;44:171–202. doi: 10.1016/s0070-2153(08)60470-6. [DOI] [PubMed] [Google Scholar]
- 17.Freshney N W, Rawlinson L, Guesdon F, Jones E, Cowley S, Hsuan J, Saklatvala J. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell. 1994;78:1039–1049. doi: 10.1016/0092-8674(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 18.Geppert T D, Whitehurst C E, Thompson P, Beutler B. Lipopolysaccharide signals activation of tumor necrosis factor biosynthesis through the ras/raf-1/MEK/MAPK pathway. Mol Med. 1994;1:93–103. [PMC free article] [PubMed] [Google Scholar]
- 19.Gomperts M, Pascall J C, Brown K D. The nucleotide sequence of a cDNA encoding an EGF-inducible gene indicates the existence of a new family of mitogen-induced genes. Oncogene. 1990;5:1081–1083. [PubMed] [Google Scholar]
- 20.Guhaniyogi J, Brewer G. Regulation of mRNA stability in mammalian cells. Gene. 2001;265:11–23. doi: 10.1016/s0378-1119(01)00350-x. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Lee J D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 22.Han J, Lee J D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 23.Han J H, Beutler B, Huez G. Complex regulation of tumor necrosis factor mRNA turnover in lipopolysaccharide-activated macrophages. Biochim Biophys Acta. 1991;1090:22–28. doi: 10.1016/0167-4781(91)90032-h. [DOI] [PubMed] [Google Scholar]
- 24.Hel Z, Skamene E, Radzioch D. Two distinct regions in the 3′ untranslated region of tumor necrosis factor alpha mRNA form complexes with macrophage proteins. Mol Cell Biol. 1996;16:5579–5590. doi: 10.1128/mcb.16.10.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontoyiannis D, Pasparakis M, Pizarro T T, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- 26.Kotlyarov A, Neininger A, Schubert C, Eckert R, Birchmeier C, Volk H D, Gaestel M. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat Cell Biol. 1999;1:94–97. doi: 10.1038/10061. [DOI] [PubMed] [Google Scholar]
- 27.Kyriakis J M, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 28.Lagnado C A, Brown C Y, Goodall G J. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A) Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai W S, Carballo E, Thorn J M, Kennington E A, Blackshear P J. Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J Biol Chem. 2000;275:17827–17837. doi: 10.1074/jbc.M001696200. [DOI] [PubMed] [Google Scholar]
- 31.Lai W S, Stumpo D J, Blackshear P J. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 32.Laroia G, Cuesta R, Brewer G, Schneider R J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 33.Lasa M, Brook M, Saklatvala J, Clark A R. Dexamethasone destabilizes cyclooxygenase 2 mRNA by inhibiting mitogen-activated protein kinase p38. Mol Cell Biol. 2001;21:771–780. doi: 10.1128/MCB.21.3.771-780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lasa M, Mahtani K R, Finch A, Brewer G, Saklatvala J, Clark A R. Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol. 2000;20:4265–4274. doi: 10.1128/mcb.20.12.4265-4274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee J C, Young P R. Role of CSB/p38/RK stress response kinase in LPS and cytokine signaling mechanisms. J Leukoc Biol. 1996;59:152–157. doi: 10.1002/jlb.59.2.152. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 37.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, Nishida E, Hagiwara M. A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J Biol Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 38.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raingeaud J, Whitmarsh A J, Barrett T, Derijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridley S H, Dean J L, Sarsfield S J, Brook M, Clark A R, Saklatvala J. A p38 MAP kinase inhibitor regulates stability of interleukin-1-induced cyclooxygenase-2 mRNA. FEBS Lett. 1998;439:75–80. doi: 10.1016/s0014-5793(98)01342-8. [DOI] [PubMed] [Google Scholar]
- 41.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 42.Rutault K, Hazzalin C A, Mahadevan L C. Combinations of ERK and p38 MAPK inhibitors ablate tumor necrosis factor-alpha (TNF-alpha) mRNA induction. Evidence for selective destabilization of TNF-alpha transcripts. J Biol Chem. 2001;276:6666–6674. doi: 10.1074/jbc.M005486200. [DOI] [PubMed] [Google Scholar]
- 43.Saklatvala J, Davis W, Guesdon F. Interleukin 1 (IL1) and tumour necrosis factor (TNF) signal transduction. Philos Trans R Soc Lond B. 1996;351:151–157. doi: 10.1098/rstb.1996.0011. [DOI] [PubMed] [Google Scholar]
- 44.Saklatvala J, Rawlinson L, Waller R J, Sarsfield S, Lee J C, Morton L F, Barnes M J, Farndale R W. Role for p38 mitogen-activated protein kinase in platelet aggregation caused by collagen or a thromboxane analogue. J Biol Chem. 1996;271:6586–6589. doi: 10.1074/jbc.271.12.6586. [DOI] [PubMed] [Google Scholar]
- 45.Sela-Brown A, Silver J, Brewer G, Naveh-Many T. Identification of AUF1 as a parathyroid hormone mRNA 3′-untranslated region-binding protein that determines parathyroid hormone mRNA stability. J Biol Chem. 2000;275:7424–7429. doi: 10.1074/jbc.275.10.7424. [DOI] [PubMed] [Google Scholar]
- 46.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 47.Sirenko O I, Lofquist A K, DeMaria C T, Morris J S, Brewer G, Haskill J S. Adhesion-dependent regulation of an A+U-rich element-binding activity associated with AUF1. Mol Cell Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoecklin G, Ming X F, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol Cell Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor G A, Carballo E, Lee D M, Lai W S, Thompson M J, Patel D D, Schenkman D I, Gilkeson G S, Broxmeyer H E, Haynes B F, Blackshear P J. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 50.Taylor G A, Thompson M J, Lai W S, Blackshear P J. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol Endocrinol. 1996;10:140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- 51.Taylor G A, Thompson M J, Lai W S, Blackshear P J. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 52.Varnum B C, Lim R W, Sukhatme V P, Herschman H R. Nucleotide sequence of a cDNA encoding TIS11: a message induced in Swiss 3T3 cells by the tumor promoter tetradecanoyl phorbol acetate. Oncogene. 1989;4:119–120. [PubMed] [Google Scholar]
- 53.Wang S W, Pawlowski J, Wathen S T, Kinney S D, Lichenstein H S, Manthey C L. Cytokine mRNA decay is accelerated by an inhibitor of p38-mitogen-activated protein kinase. Inflamm Res. 1999;48:533–538. doi: 10.1007/s000110050499. [DOI] [PubMed] [Google Scholar]
- 54.Wilson G M, Sutphen K, Chuang K, Brewer G. Folding of A+U-rich RNA elements modulates AUF1 binding. Potential roles in regulation of mRNA turnover. J Biol Chem. 2001;276:8695–8704. doi: 10.1074/jbc.M009848200. [DOI] [PubMed] [Google Scholar]
- 55.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen C Y, Shyu A B, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zubiaga A M, Belasco J G, Greenberg M E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]