Abstract
t(8;21) and t(16;21) create two fusion proteins, AML-1–ETO and AML-1–MTG16, respectively, which fuse the AML-1 DNA binding domain to putative transcriptional corepressors, ETO and MTG16. Here, we show that distinct domains of ETO contact the mSin3A and N-CoR corepressors and define two binding sites within ETO for each of these corepressors. In addition, of eight histone deacetylases (HDACs) tested, only the class I HDACs HDAC-1, HDAC-2, and HDAC-3 bind ETO. However, these HDACs bind ETO through different domains. We also show that the murine homologue of MTG16, ETO-2, is also a transcriptional corepressor that works through a similar but distinct mechanism. Like ETO, ETO-2 interacts with N-CoR, but ETO-2 fails to bind mSin3A. Furthermore, ETO-2 binds HDAC-1, HDAC-2, and HDAC-3 but also interacts with HDAC-6 and HDAC-8. In addition, we show that expression of AML-1–ETO causes disruption of the cell cycle in the G1 phase. Disruption of the cell cycle required the ability of AML-1–ETO to repress transcription because a mutant of AML-1–ETO, Δ469, which removes the majority of the corepressor binding sites, had no phenotype. Moreover, treatment of AML-1–ETO-expressing cells with trichostatin A, an HDAC inhibitor, restored cell cycle control. Thus, AML-1–ETO makes distinct contacts with multiple HDACs and an HDAC inhibitor biologically inactivates this fusion protein.
The acute myeloid leukemia 1 (AML-1) gene is one of the most frequently mutated genes in human leukemia and is disrupted by multiple chromosomal translocations in AML, including t(8;21) and t(16;21) (9, 35, 38). t(8;21) is the most frequent of these translocations, and it contains the AML-1 DNA binding domain fused to a transcriptional corepressor, ETO (also known as MTG8) (4, 5, 34). t(16;21), although rarer, fuses the AML-1 DNA binding domain to an ETO-related protein, MTG16 (9). AML-1 is also indirectly affected by inv(16), which fuses CBFβ, an allosteric regulator of AML-1, to a smooth muscle myosin heavy chain (25).
ETO is highly related to MTG16 and a third family member, MTGR1, in mammalian cells and Nervy in Drosophila (6). The mammalian family members are highly conserved throughout the proteins, with four domains conserved in Nervy. These regions are an N-terminal domain that is also homologous to the transcriptional coactivator TAF110 (17), a hydrophobic heptad repeat (HHR) that mediates dimerization (3, 21), a domain of unknown function termed the Nervy domain, and a domain containing two zinc finger motifs that are required for contacting the central domain of N-CoR (29). The murine homologue of MTG16 was identified by low-stringency screening of a cDNA library by using an ETO cDNA as a probe (3). It shares 77% overall identity with human ETO, but within three of four conserved domains, these proteins are 92 to 96% identical, implying that they function similarly. The Nervy domain is the least conserved domain among family members and is 86% identical between these two proteins.
ETO is a component of a high-molecular-weight complex containing histone deacetylases (HDACs) (29), and ETO associates with N-CoR or SMRT and mSin3A independently (10, 29). Incapable of binding DNA directly, ETO and the associated corepressors can be recruited to chromatin through interactions with DNA binding proteins. For example, ETO binds the promyelocytic zinc finger protein PLZF, which is a DNA binding transcriptional repressor that is disrupted by t(11;17) in acute promyelocytic leukemia (31). The binding of ETO to this transcription factor potentiated the repression of a PLZF responsive promoter (32), suggesting that ETO acts as a corepressor.
While ETO associates with mSin3 (29), nuclear hormone corepressors (10, 29, 41), and HDACs (29), the nature and extent of these associations have not been fully addressed. In addition, little is known about the mechanism of action of ETO-2 (MTG16). Therefore, we have further characterized the mechanisms of action of ETO and ETO-2. While it was anticipated that ETO-2 would act as a repressor when fused to a DNA binding domain, we unexpectedly found that ETO-2 did not interact with mSin3A. Based on this observation, we demonstrated that the ETO dimerization motif mediated one contact with mSin3A. In addition, we found that HDAC-1, HDAC-2, and HDAC-3 all bound ETO, but with different characteristics, suggesting that ETO may serve as a platform for multiple HDACs and corepressors. Moreover, constitutive repressors of AML-1-regulated genes, such as the inv (16) fusion protein or engineered AML-1–repression domain fusion proteins, slow cell cycle progression (2, 26, 27). Similarly, exogenous expression of AML-1–ETO inhibited cell cycle progression in the G1 phase of the cell cycle. However, addition of the HDAC inhibitor trichostatin A (TSA) to the cell culture media restored normal cell cycle progression, indicating that AML-1–ETO physically and functionally interacts with HDACs to disrupt biological functions.
MATERIALS AND METHODS
Plasmids.
The GAL4-thymidine kinase (TK)-luciferase construct used in the transcription assays was described previously (7). Several plasmids used in our assays were kindly provided to us by the following investigators: FLAG-tagged HDAC-1 to -6 constructs were given to us by E. Seto (12), and Myc–HDAC-8 was provided by E. Hu (18). Hemagglutinin (HA)–HDAC-7 and full-length N-CoR tagged with the FLAG epitope were kindly provided by R. Evans (20). CMV5–Myc–mETO-2 was created in two steps in order to preserve the Myc tag present in pJM mETO-2 (3). A HindIII-XbaI fragment was generated using an internal HindIII site and XbaI site in the vector 3′ of the cDNA insert placed into HindIII-XbaI-cut pCMV5 and was called pCMV5 mETO-2 C-term. A second fragment generated by the HindIII digest using a HindIII site in the vector 5′ of the Myc tag was isolated and placed into the HindIII site of pCMV5 mETO-2 C-term, and orientation was determined. CMV5–HA–mETO-2 and GAL mETO-2 were made by cutting mETO-2 out of pJ3omega mETO-2 with XbaI and placing it into the XbaI sites of pCMV5-HA2 and pCMV5 M2 (pCMV5 M2 was described previously [7]). The pCMV5-HA series of vectors was made by subcloning oligonucleotides encoding a start methionine and the HA epitope into the EcoRI site of the pCMV5 vector. GAL4-ETO was constructed by releasing ETO from pBS-ETO with XhoI and placing it into the SalI site of pCMV5 M2. Regions of ETO were made by PCR and subcloned into the EcoRI and SalI sites of pCMV5 M2. PCR primers pairs used were as follows: ETO amino acids (aa) 217 to 385, 5′-CGGAATTCGCTGCTCTGGATGCCAGCAC-3′ and 5′-CCGCTCGAGGTCTGCTTCTTGACACCGCCT-3′; ETO aa 379 to 499, 5′-CCGCAATTGGCGGTGTCAAGAAGCAGAC-3′ and 5′-CGCTCGAGCTCCGCCGCCTGCCGTTTGGC-3′; ETO aa 493 to 559, 5′-GGAATTCCAAACGGCAGGCGGCGGAG-3′ and 5′-CGCTCGAGCTGCTGGGCCTGCAGGGTCTG-3′; ETO aa 553 to 604, 5′-CGGAATTCCCAGACCCTGCAGGCCCAGCAG-3′ and 5′-CCGCTCGAGCTAGCGAGGGGTTGTCTCTAT-3′; ETO aa 217 to 301, the same 5′ primer as aa 217 to 385 and 5′-CCGCTCGAGACGGTAACGCTGAGGTGGAGG-3′; ETO aa 259 to 343, 5′-CGGAATTCGCACTCAGAACATCCAAGCAAG-3′ and 5′-CCGCTCGAGGTGACTAATCATTTCTTCTTGACG; ETO aa 300 to 385, 5′-CGGAATTCCTACCGTTTGGATGATATGGCC-3′ and the same 3′ primer as the aa 217 to 385 fragment. To create the pCMV5 hETO–mETO-2 chimeric protein, a BamHI site conserved between the two DNAs and one in the vector on the 3′ side of the inserted cDNA were used to release a C-terminal ETO-2 fragment. This fragment was placed into a BamHI-cut pCMV5 hETO construct. A similar strategy was used to create pCMV5 Myc-mETO-2/hETO. pCMV5 Myc-mETO-2/hETO Δ394-446 was made by taking advantage of a BglII site conserved between hETO and mETO-2 located at nucleotides 1416 and 1483, respectively. A BglII-BamHI fragment was released from ETO and placed into a BamHI-cut pCMV5 Myc-ETO-2 maintaining the open reading frame. For the pCMV5 Myc-mETO2/hETO constructs which delete the Nervy domain, mETO-2/hETO Δ447-488 and mETO/hETO Δ394-488, the same strategy was used with a construct having a deletion of this region, described previously (28). The site-directed mutations DH358,359NS and D347E were generated using overlap extension PCR. The primers that were used to generate the DH-to-NS mutation were 5′-AGTGGAAACATCTTAACGATCTGTTAAACTGCAT-3′ and 5′-ATGCAGTTTAACAGACTGTTAAGATGTTTCCACT-3′ (the changed codons are italicized). The D347E mutant was made using the following oligonucleotides: 5′-CACAGACTAACAGAAAGAGAATGGGCA-3′ and 5′-TGCCCATTCTCTTTCTGTTAGTCTGTG. All of the constructs generated by PCR were sequenced through the Vanderbilt University sequencing core facility to verify that no mutations were introduced.
Transcription assays.
NIH 3T3 cells were transfected using the Superfect reagent (Qiagen) with 1 μg of GAL4-TK-luciferase, 100 ng of the appropriate GAL4-ETO, GAL4–mETO-2, or GAL4-ETO region expression plasmids, and 200 ng of pCMV5-secreted alkaline phosphatase (SEAP) plasmid as an internal control. Firefly luciferase activities were measured using the Luciferase Assay System (Promega) and normalized to SEAP activity. Cells were harvested approximately 48 to 52 h posttransfection.
Cell lines and cell cycle analysis.
C33A cells and Cos-7 cells were maintained in Dulbecco modified Eagle medium (DMEM; BioWhittaker Inc.,Walkersville, Md.) containing 10% fetal bovine serum, 50 U of penicillin/ml, 50 μg of streptomycin/ml, and 2 mM l-glutamine (all from BioWhittaker). NIH 3T3 cells were maintained in DMEM containing 10% bovine serum. The culture of parental 32D.3 myeloid progenitor cells and establishment of stable cell lines was performed as previously described (15, 37). Murine erythroleukemia (MEL) cells were maintained as suspension cultures in DMEM supplemented with 10% fetal bovine serum (BioWhittaker), 2 mM l-glutamine (BioWhittaker), 1% penicillin G-streptomycin (Gibco-BRL, Life Technologies, Inc.) at 37°C in a 5% CO2 humidified incubator. MEL cells were electroporated with the pTET.TAK.HYG vector (kindly provided by Brian Van Ness, University of Minnesota). MEL cells expressing the tet repressor DNA-binding domain fused to the VP16 transcriptional activation domain were electroporated with pTET.AML1-ETO.Neo and selected for G418 resistance. Single-cell clones expressing AML-1–ETO protein were identified by immunoblot analysis. 32D.3 cells stably transfected with AML-1B, A/E, or A/E Δ469 were constructed as previously described (15, 37) by using the sheep metallothionine promoter plasmid pMT-CB6. G418-resistant cells were cloned in methylcellulose or by limiting dilution.
Cell cycle analysis was performed by flow cytometry as described previously (37). Ten thousand total events were analyzed per sample. The ModFit computer program (Verity Software House, Topsham, Maine) was used to generate the histograms and determine the percentage of cells within the G1 phase of the cell cycle.
Coimmunoprecipitations and immunoblotting.
Cos-7 cells (3 × 106 cells in 100-mm-diameter dishes) were transfected using Lipofectamine (BRL) with 1.0 μg of expression plasmids and an appropriate amount of pCMV5 vector to maintain the Lipofectamine-to-DNA ratio. For coexpression experiments, 1 to 2.5 μg of pCMV5-GAL4-ETO and 2.5 μg of Flag-tagged N-CoR or HDACs were cotransfected. Approximately 48 to 52 h posttransfection, cells were harvested and extracted with phosphate-buffered saline supplemented with 1 μg of leupeptin/ml, 1 μg of pepstatin/ml, 0.2 mM phenylmethylsulfonyl fluoride, and 0.1 trypsin inhibitor unit of aprotinin/ml and containing 0.5% Triton X-100, 0.1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate (SDS) unless otherwise noted. For immunoprecipitations, cell lysates were sonicated and then incubated with 50 μl of formalin-fixed Staphylococcus aureus cells (Pansorbin; CalBiochem) for 30 min to remove proteins nonspecifically binding to protein A. After centrifugation for 5 min at 4°C, the supernatants were collected, and a portion was removed for immunoblot analysis and then incubated for 1 h with affinity-purified primary antibody (K 20 anti-mSin3A [Santa Cruz Biotechnology], anti-Myc(9E10) [Babco], and anti-HA [Babco] antibodies). A 20-μl volume of a 50% slurry of protein A-Sepharose (Pharmacia Biotech, Uppsala, Sweden) or protein G-Sepharose (Sigma) was then added for 30 min to collect the immune complexes, and these complexes were washed three times at 4°C with lysis buffer. For FLAG coimmunoprecipitations, 20 μl of a 50% slurry of anti-FLAG M2 beads (Sigma) was added to the lysates, incubated for 90 min at 4°C, and washed three times at 4°C with lysis buffer. Protein A-Sepharose, protein G-Sepharose, and FLAG beads were blocked in PBS containing 1% bovine serum albumin prior to addition to the lysate.
For immunoblot analysis, the cell lysate was electrophoresed through 10% polyacrylamide gels or Criterion 4 to 20% gradient gels (Bio-Rad), transferred onto Immobilon-P (Millipore) or nitrocellulose (Schleicher and Schuell), and probed with the indicated antibodies. Immune complexes were detected using the Super Signal chemiluminescence detection system (Pierce) and a horseradish peroxidase-labeled goat anti-rabbit or rabbit anti-mouse secondary antibody (Sigma). Proteins were detected using purified rabbit polyclonal antibodies specific to the N- or C-terminal domains of ETO, rabbit polyclonal antibody to the AML RHD, and mouse monoclonal antibodies to the GAL4 DNA binding domain (Santa Cruz), FLAG (Sigma), HA (Babco), or Myc (Babco) epitopes. For immunoblot analysis, one-tenth of the total lysate from the immunoprecipitations or 100 μg of protein (quantitated with the Bio-Rad DC protein assay) or immune complexes were boiled in Laemmli buffer for 3 min, fractionated by SDS-polyacrylamide gel electrophoresis (PAGE), and transferred to nitrocellulose. These membranes were blocked for 1 h with 5% milk, and incubated with the indicated primary antibody overnight at 4°C. Proteins were visualized by chemiluminesence (Super Signal; Pierce).
RESULTS
ETO-2, like ETO, represses transcription.
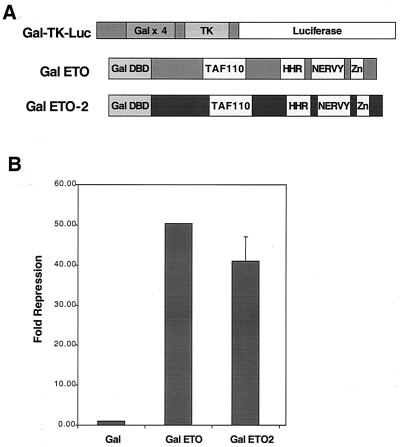
ETO is a transcriptional repressor when fused to the GAL4 DNA binding domain and its activity is measured using a promoter consisting of four GAL4 binding sites upstream of a minimal TK promoter (39). To test the ability of ETO-2 to repress transcription, chimeric proteins were made which fused the GAL4 DNA binding domain to the N terminus of ETO or ETO-2 (Fig. 1A). These plasmids were cotransfected with GAL-TK-luciferase reporter plasmids, and the luciferase activity was normalized to an internal control for transfection efficiency (Fig. 1B). Both ETO and ETO-2 were potent repressors in this assay (Fig. 1B).
FIG. 1.
ETO-2 represses transcription when tethered to a promoter. (A) Schematic diagram of the Gal4-TK-luciferase reporter plasmid and the GAL4-ETO and GAL4–ETO-2 fusion proteins. (B) ETO and ETO-2 repress transcription. GAL4-ETO and GAL4–ETO-2 were transfected with the GAL4 reporter plasmid into NIH 3T3 cells, and relative light units were measured. The data are graphed as the relative fold repression after correction for transfection efficiency.
ETO-2 does not associate with mSin3A but binds N-CoR.
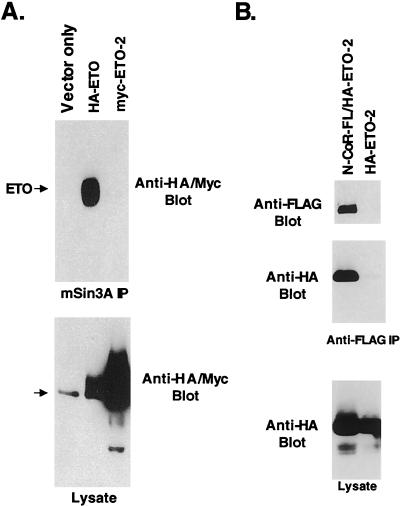
ETO represses transcription by associating with several corepressor molecules, including mSin3A, N-CoR, SMRT, HDAC-1, and HDAC-2 (10, 29, 41), though the nature of these contacts has yet to be defined. To determine if ETO-2 represses transcription through a similar mechanism, epitope-tagged forms of ETO and ETO-2 were transiently expressed, cell lysates were immunoprecipitated with anti-mSin3A antibody, and ETO and ETO-2 were detected by immunoblot analysis. As shown previously (29), ETO copurified with mSin3A (Fig. 2A). Unexpectedly, ETO-2 failed to associate with mSin3A, suggesting a mechanism of repression distinct from ETO. In addition, we coexpressed ETO-2 with FLAG-tagged N-CoR to detect a possible association. HA–ETO-2 was found in the anti-FLAG immune complexes only when FLAG–N-CoR was coexpressed (Fig. 2B). Similar interactions were seen with the N-CoR-related SMRT corepressor (data not shown). Therefore, ETO-2 contacts N-CoR but not mSin3A.
FIG. 2.
ETO and ETO-2 associate with corepressors. Cos-7 cells were transfected with HA-ETO or Myc–ETO-2, and cell lysates were immunoprecipitated (IP) with anti-mSin3A antibody (A) or were transfected with FLAG-tagged N-CoR and HA–ETO-2 or HA–ETO-2 alone and immunoprecipitated with anti-FLAG antibody (B). Proteins were detected by immunoblot analysis using anti-mSin3A, anti-HA, and anti-Myc antibodies. The bottom panels show the level of expression of ETO and ETO-2. The arrow indicates the presence of a background band.
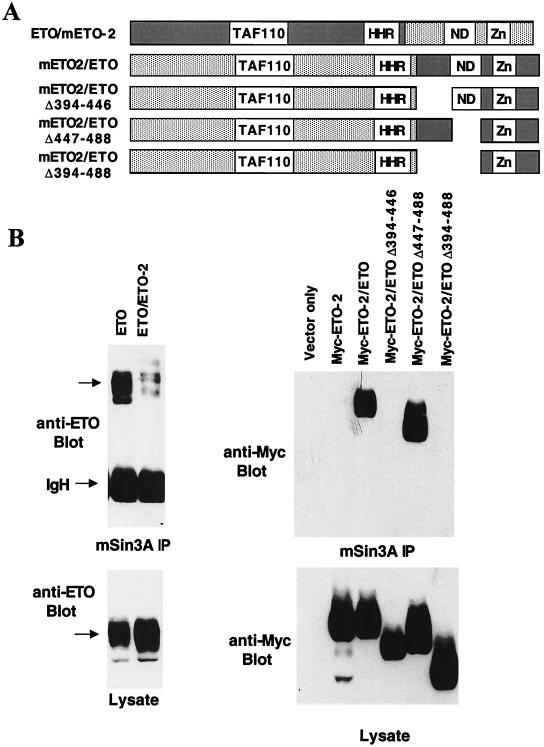
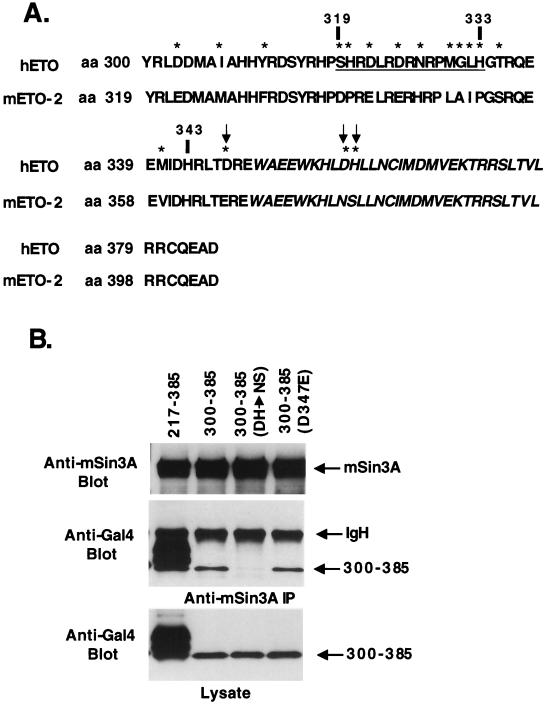
The observation that ETO, but not ETO-2, bound mSin3A suggested that the mSin3A interaction occurred through residues that are not conserved between ETO and ETO-2. The fact that deletion of several conserved regions of ETO, including the HHR, Nervy, and ZnF motifs, failed to eliminate mSin3A binding lends further support to this conclusion (29). We took advantage of this distinction to begin to define the mSin3A binding motifs in ETO by creating chimeric ETO/ETO-2 and ETO-2/ETO proteins (Fig. 3A). The chimeric proteins were constructed using unique BamHI and BglII restriction sites that are conserved in both cDNAs and maintain the open reading frame. The chimeric proteins were expressed in Cos-7 cells and tested for their ability to bind mSin3A in immunoprecipitation assays. The ETO/ETO-2 protein copurified with mSin3A, but the amount of associating protein was reduced compared to wild-type ETO (Fig. 3B, left). By contrast, the ETO-2/ETO chimeric protein associated with mSin3A to a degree similar to ETO (Fig. 3B, right panel). Therefore, we were able to create deletions within the ETO-2/ETO chimeric protein to further define the sequences required for mSin3A binding (Fig. 3A). Deletion of the conserved nervy domain did not affect mSin3A binding, but deletion of the region between the HHR and the nervy domain eliminated the mSin3A interaction (Fig. 3B, right).
FIG. 3.
Mapping the C-terminal mSin3A-binding domain. (A) Schematic diagram of ETO/ETO-2 and ETO-2/ETO chimeric proteins. (B) Lysates were made from cells expressing the indicated chimeric proteins and immunoprecipitated with anti-mSin3A antibody. The chimeric proteins were detected using anti-ETO or anti-Myc antibody. Arrows indicate the position of ETO.
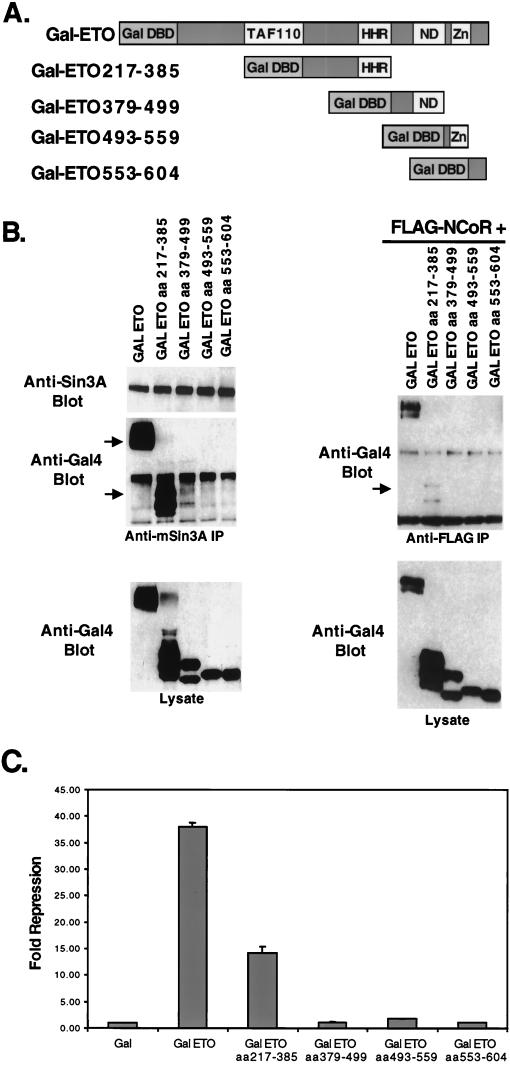
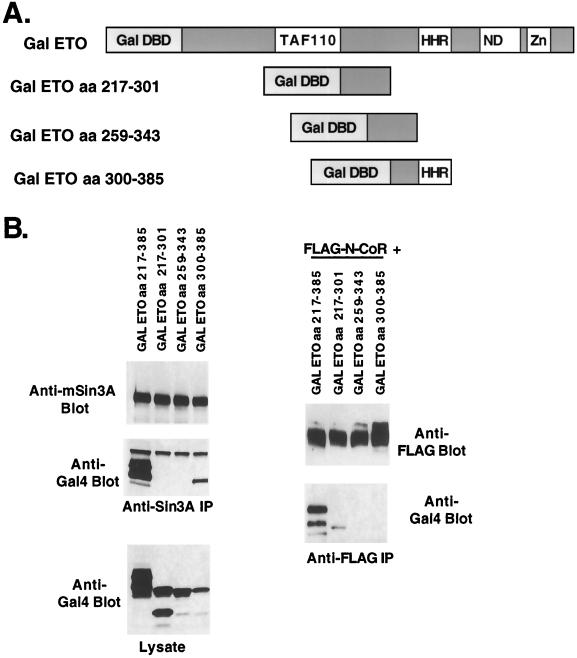
To further define the mSin3 interaction domain observed in the ETO/ETO-2 chimeric protein, we created a series of GAL4 DNA binding domain-ETO fusion proteins to determine what sequences were sufficient for mSin3A binding (Fig. 4A). Unfortunately, a GAL4-ETO protein encompassing only the N-terminal 217 aa of ETO was not expressed to levels high enough to make a clear determination of corepressor binding (data not shown). However, consistent with the analysis of ETO/ETO-2 chimeric proteins, two domains of ETO were sufficient for mSin3A copurification encompassing sequences 217 to 385 and 379 to 499 (Fig. 4B, left), although in this assay, residues 217 to 385 appeared to bind mSin3A better than the 379-to-499 domain.
FIG. 4.
Mapping the N-terminal mSin3A and N-CoR binding sites. (A) Schematic diagram of the GAL4-ETO chimeric proteins. (B) Lysates were made from cells expressing the indicated chimeric proteins and immunoprecipitated with anti-mSin3A antibody (left). Plasmids expressing FLAG-tagged N-CoR were cotransfected into cells with the GAL4-ETO chimeric proteins (right) and immunoprecipitated with anti-FLAG antibody. Chimeric proteins were detected using anti-GAL4 antibody. Arrows indicate the position of GAL4-ETO or the deletion mutants. (C) GAL4-ETO and GAL4-ETO deletion mutants were transfected with the GAL4 reporter plasmid into NIH 3T3 cells and relative light units were measured. The data are graphed as relative fold repression after correction for transfection efficiency.
Next, we used the GAL4-ETO proteins to define the domains of ETO that are sufficient for N-CoR binding. Although the zinc finger motifs are sufficient to bind the central region of N-CoR in yeast two-hybrid assays and mammalian two-hybrid assays (29, 45), these sequences are dispensable for the interaction of AML-1–ETO with full-length N-CoR, suggesting the presence of a second N-CoR binding domain (29). Indeed, only the first domain tested (residues 217 to 385) copurified with N-CoR immune complexes (Fig. 4B, right), suggesting that this domain contains the previously predicted second N-CoR binding motif.
Some of the advantages of using GAL4-ETO fusion proteins are that the GAL4 motif provides both a nuclear localization signal and the ability to measure transcriptional activity of the fusion proteins using a GAL4-dependent reporter plasmid (Fig. 1A). Therefore, the ability of the ETO domains to repress transcription was defined. Relative to wild-type ETO, each individual domain was impaired for repression, consistent with the presence of multiple corepressor binding sites in full-length ETO (Fig. 4C). However, GAL4-ETO(217-385), which appears to bind mSin3A and N-CoR better than the C-terminal fragments, was the only domain that was capable of significant repression. This result agrees with the observation that deletion of the zinc finger motif in the context of AML-1–ETO did not dramatically affect repression by the fusion protein (23).
ETO interacts with class I HDACs independent of mSin3A or N-CoR.
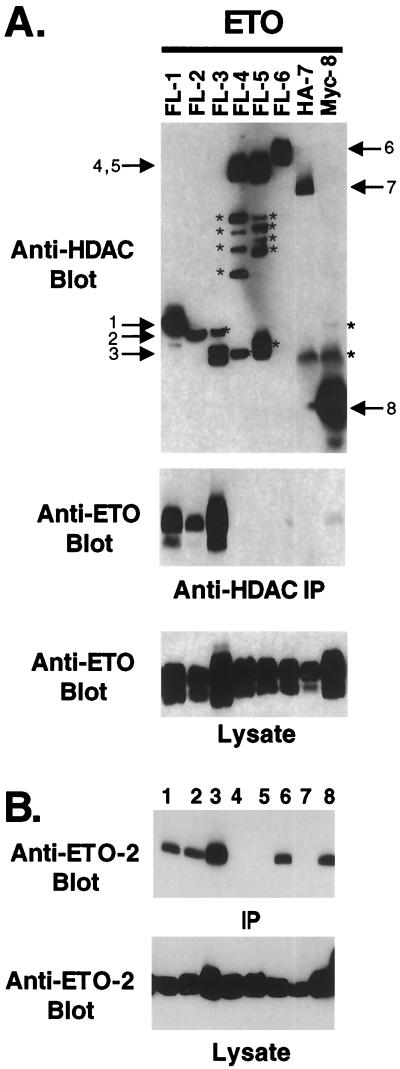
Previously, we had demonstrated that ETO associated with HDAC-1 and HDAC-2 (29). To extend this analysis, we determined whether ETO could contact any of the known class I or class II HDACs. Epitope-tagged forms of HDACs 1 to 8 were coexpressed with ETO, and the levels of ETO that copurified with specific HDACs were measured by immunoblot analysis (Fig. 5A). The class I HDACs HDAC-1, HDAC-2, and HDAC-3 all bound ETO whereas HDAC-8 only weakly associated with ETO (Fig. 5A). However, none of the class II HDACs showed a significant interaction with ETO. Therefore, ETO interacts with a subset of the class I HDACs.
FIG. 5.
ETO and ETO-2 bind multiple HDACs. (A) ETO binds HDAC-1, HDAC-2, and HDAC-3. The numbers above the lanes indicate the epitope-tagged form of HDAC-1 to -8 that was coexpressed with ETO. After immunoprecipitation (IP) with anti-FLAG (FL), anti-HA (HA), or anti-Myc (Myc) antibody, copurifying ETO was detected by immunoblot analysis (middle). The bottom panel is an immunoblot of the cell lysates prior to immunoprecipitation. ∗, nonspecific bands and proteolytic fragments. (B) ETO-2 binds HDACs 1, 2, 3, 6, and 8. The experiment was performed exactly as in panel A, but using epitope-tagged mETO-2. The numbers above the lanes indicate the epitope-tagged HDAC that was coexpressed with ETO-2 as described for panel A. Note that anti-HDAC antibody connotes that the experiment was performed with antibodies to the epitope tag for that HDAC.
Because ETO-2 interacted with N-CoR, but not mSin3A, the HDACs that ETO-2 can bind were determined by copurification (Fig. 5B). Like ETO, mETO-2 associated with HDAC-1, HDAC-2, and HDAC-3. However, mETO-2 also interacted with HDAC-6, which has a duplicated catalytic domain and the fourth class I family member, HDAC-8. Therefore, ETO and mETO-2 are capable of interacting with different subgroups of HDACs to repress transcription.
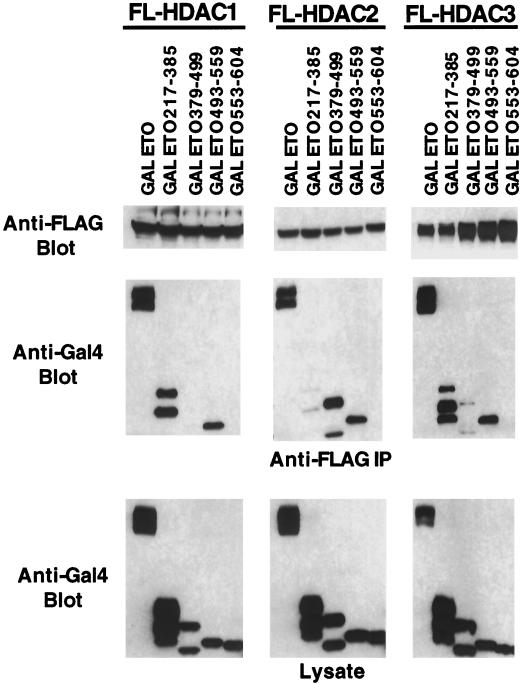
N-CoR interacts with the class II HDACs 4, 5, and 7 (19, 20) and with the class I HDAC HDAC-3 (13, 24, 43), whereas mSin3A binds HDAC-1, HDAC-2, and HDAC-7 (14, 20, 22, 36). Therefore, it is possible that HDAC binding by ETO was via N-CoR or mSin3A. To test this possibility, we coexpressed FLAG-tagged HDAC-1, HDAC-2, and HDAC-3 with the GAL4-ETO chimeric proteins to determine whether mSin3A and HDAC binding cosegregated. All three HDACs tested interacted with ETO residues 493 to 559 (Fig. 6), which were not sufficient for the copurification of either mSin3A or N-CoR (Fig. 4B). Thus, the HDACs bind ETO independent of mSin3 and N-CoR. In addition, HDAC-1 and HDAC-3 bound ETO residues 217 to 385, which also contact mSin3A and N-CoR. HDAC-3 also weakly associated with ETO residues 379 to 499 (Fig. 6). By contrast, HDAC-2 poorly bound ETO residues 217 to 385 but copurified with ETO residues 379 to 499, which also contain a second mSin3A-binding site (compare Fig. 6 and 4B). Therefore, even the homologous class I HDACs bind ETO via separable domains.
FIG. 6.
Mapping of the HDAC-binding sites on ETO. FLAG-tagged HDAC-1 to -3 were coexpressed with the GAL4-ETO chimeric proteins from Fig. 4. Copurifying GAL4-ETO proteins were detected using anti-GAL4 immunoglobulin G for immunoblot analysis. Shown are the control anti-FLAG antibody blot showing that each HDAC was expressed and immunoprecipitated (top), copurifying ETO fragments (middle), and the levels of expression of the GAL4-ETO chimeric proteins (bottom).
HHR-oligomerization domain of ETO is required for mSin3A, but not N-CoR interactions.
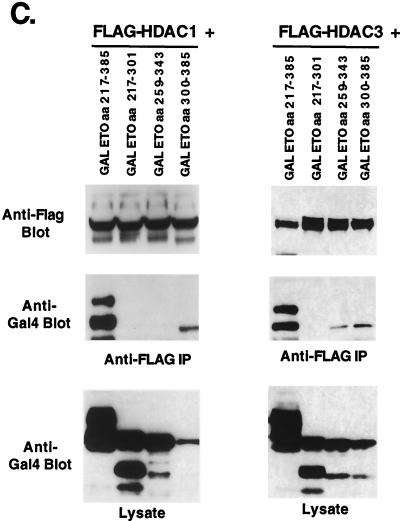
Because mSin3A, N-CoR, HDAC-1, and HDAC-3 all interacted with ETO residues 217 to 385, we further subdivided this domain to define the interacting motifs (Fig. 7A). Three overlapping fragments were fused to the GAL4 DNA binding domain and tested for binding each corepressor in coimmunoprecipitation assays (Fig. 7B). Only ETO residues 300 to 385, containing the HHR-dimerizaton domain, were sufficient to mediate mSin3A binding. By contrast, the first ETO fragment containing residues 217 to 301 weakly bound N-CoR (Fig. 7B). The second fragment (residues 259 to 343) also bound N-CoR in some assays, suggesting a very weak association (data not shown). HDAC-3 associated with fragments containing either ETO residues 259 to 343 or 300 to 385, whereas HDAC-1 interacted only with GAL4-ETO(300-385) (Fig. 7C). Therefore, HDAC-3 either binds ETO twice within this domain or this interaction is mediated by the residues that overlap between these fragments (aa 300 to 343) and it binds differently than HDAC-1. Thus, N-CoR binds a domain distinct from mSin3A, and HDAC-1 and HDAC-3 have different binding patterns that are separable from N-CoR.
FIG. 7.
Fine mapping of the N-terminal repression domain of ETO. (A) Schematic diagram of the GAL4-ETO chimeric proteins used. (B) mSin3A and N-CoR bind to distinct ETO domains. (Left) Lysates were made from cells expressing the indicated chimeric proteins and immunoprecipitated (IP) with anti-mSin3A antibody. Immune complexes were analyzed for the presence of mSin3A (top) and GAL4-ETO proteins (anti-GAL4 immunoblot; middle). Shown also are the levels of expression of the GAL4-ETO chimeric proteins (bottom). (Right) GAL4-ETO chimeric proteins were coexpressed with FLAG–N-CoR, and cell lysates containing FLAG-tagged N-CoR were immunoprecipitated with anti-FLAG antibody. Chimeric proteins were detected using anti-GAL4 antibody (below). (C) HDACs bind different domains in ETO. GAL4-ETO chimeric proteins were coexpressed with FLAG–HDAC-1 or FLAG–HDAC-3, and cell lysates containing expressed FLAG-tagged HDAC-1 or HDAC-3 were immunoprecipitated with anti-FLAG antibody. Chimeric proteins copurifying were detected using anti-GAL4 antibody (middle). Shown are the levels of expression of the GAL4-ETO chimeric proteins (bottom) and immunoprecipitated HDAC (top).
Given that ETO residues 300 to 385 are sufficient to contact mSin3A and that ETO-2 failed to bind mSin3A, we compared the primary sequence of ETO and ETO-2 in this region (Fig. 8A). Because GAL4-ETO(259-343) overlaps with GAL4-ETO(300-385) but does not bind mSin3A, it is likely that residues 343 to 385 (including the HHR-dimerization motif) are required for this interaction. In addition, the region with the least homology within this sequence (residues 319 to 333; Fig. 8A), could be deleted in the context of GAL4-ETO(217-385) without the loss of mSin3A binding (data not shown). Within the sequence from aa 343 to 385, there are only three differences between ETO and ETO-2 that could alter the association with mSin3A. Therefore, we used site-specific mutagenesis to change these residues of ETO to match those found in ETO-2 within the GAL4-ETO(300-385) fusion protein. The alteration of the two amino acids within the HHR-dimerization domain (D358N, H359S) significantly impaired mSin3A binding (Fig. 8B), whereas the conservative D347E change just upstream of the HHR had no effect (Fig. 8B). Thus, one of the mSin3A binding domains of ETO overlaps with the HHR-dimerization motif.
FIG. 8.
The HHR-dimerization motif of ETO contacts mSin3A. (A) Comparison of the sequences of ETO and ETO-2 within the HHR-dimerization domain. Stars highlight the differences between ETO and ETO-2. The HHR is shown in italics. Arrows indicate the amino acids that were changed and analyzed in panel B. The underlined sequence indicates a region of low homology. (B) The HHR is required for binding mSin3A. Lysates were made from cells expressing the indicated GAL4-ETO chimeric proteins and were immunoprecipitated with anti-mSin3A antibody. Shown are immunoprecipitated mSin3A (top), associated GAL4-ETO proteins (middle), and the expression of the GAL4-ETO fusion protiens (bottom). The chimeric proteins were detected using anti-GAL4 antibody.
HDAC inhibitor biologically inactivates t(8;21) fusion protein.
The strong association of ETO with HDACs implied that the t(8;21) fusion protein uses HDACs to alter biological phenotypes. Because of the large number of contacts (some overlapping; see Fig. 11) between ETO and corepressors, we could not readily determine whether AML-1–ETO function cosegregated with mutants that eliminated specific contacts. Therefore, we used the HDAC inhibitor TSA to probe the role of HDACs in AML-1–ETO function. While AML-1–ETO impairs myeloid cell differentiation (1, 44), HDAC inhibitors promote hematopoietic cell differentiation irrespective of AML-1–ETO action. Therefore, we attempted to establish an alternate assay. Previously, we found that AML-1 (RUNX-1) overexpression in 32D.3 myeloid progenitor cells led to an acceleration of S-phase entry and accumulation of S-phase cells when compared to 32D.3 cells expressing control vector alone (40). In addition, expression of the inv(16) or an artificial AML-1-repression domain fusion protein slowed cell cycle progression in the G1 phase (2, 26). Because AML-1–ETO and inv(16) have similar biological actions, we tested whether AML-1–ETO could also impair cell cycle progression.
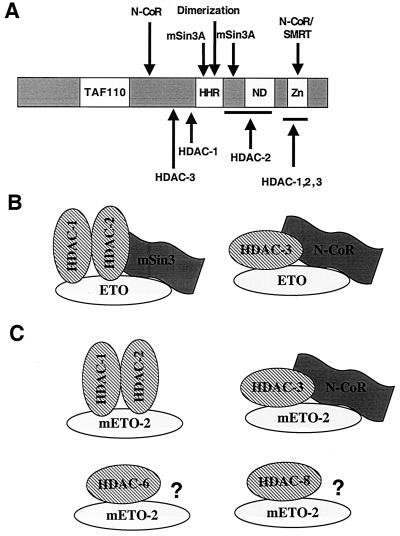
FIG. 11.
Hypothetical model of ETO and ETO-2 corepression complexes. (A) Schematic diagram of the domains of ETO that bind corepressors. Arrows indicate where the stated proteins contact ETO based on the analysis in this work and work previously described (10, 29, 41). TAF110, a region of ETO with homology to Drosophila TAF110; HHR, hydrophobic heptad repeat; ND, nervy homology domain; Zn, the predicted dual zinc finger motif. (B) Model of ETO-containing complexes. (C) Model of ETO-2-containing complexes. ?, potential corepressor interactions yet to be identified.
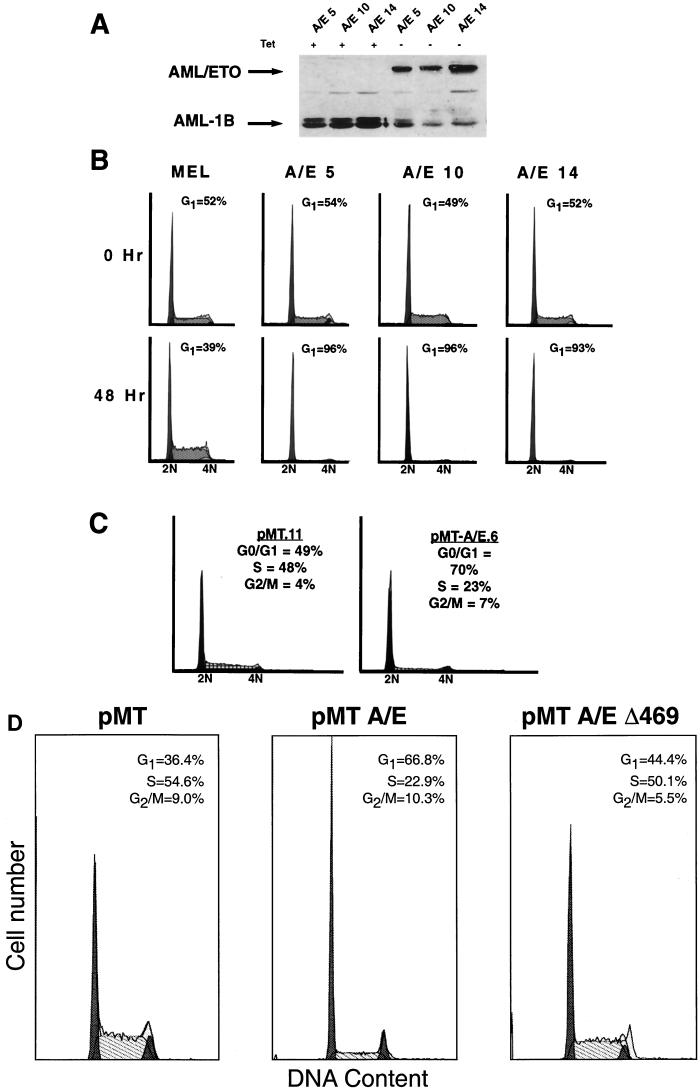
To avoid potential problems in establishing cell lines expressing high levels of the fusion protein, we created tetracycline-inducible AML-1–ETO-expressing MEL cells (11). AML-1–ETO protein was not detected in the presence of tetracycline, but it was strongly induced in the absence of tetracycline (Fig. 9A). In the presence of tetracycline, the AML-1–ETO-expressing cells grew normally; however, when AML-1–ETO expression was induced, these cells grew slowly. The numbers of cells in each cell cycle phase were determined using flow cytometry to measure DNA content after staining with propidium iodide. AML-1–ETO-expressing cells accumulated in G1 with a commensurate decrease in the number of cells in S phase (Fig. 9B).
FIG. 9.
AML-1–ETO-expressing cells accumulate in the G1 phase. (A) Immunoblot analysis of AML-1–ETO in the presence and absence of tetracycline. Cells were cultured in the presence (+) or absence (−) of tetracycline for 48 h prior to the preparation of whole-cell extracts for separation by SDS-PAGE. Proteins were detected using antibodies directed to residues 50 to 177 of AML-1 (the RHD). Arrows indicate the AML-1–ETO and AML-1B bands. Note that the endogenous AML-1B is repressed by AML-1–ETO expression. The numbers following the AML-1–ETO designations indicate individual clonal cell lines. (B) Cell cycle analysis of AML-1–ETO-expressing MEL cells. Cells were cultured in the absence of tetracycline for 48 h before DNA content analysis was determined by flow cytometry after staining the DNA with propidium iodide. (C) Cell cycle analysis of AML-1–ETO-expressing 32D.3 myeloid progenitor cells. These cell lines expressing AML-1–ETO were characterized previously (45). DNA content analysis was determined by flow cytometry after propidium iodide staining. Flow cytometric histograms for the indicated cell lines are shown and the number of cells in each cell cycle phase were determined using the ModFit algorithm. (D) A transcriptionally inactive deletion mutant of AML-1–ETO (Δ469) does not disrupt cell cycle function. Cell cycle analysis of 32D.3 cells expressing AML-1–ETO, the deletion mutant Δ469, or pMT control vector alone were induced with 75 μM ZnSO4 for 16 h, and aliquots were processed for flow cytometric analysis. The names of the 32D.3-derived cell lines are labeled at the top of each panel and the percentages of cells in each phase of the cell cycle, as determined by the ModFit analysis program, are shown within each panel. The apoptotic cells and debris were gated and are not shown in the histograms.
To confirm that this effect was not cell type specific, we used our previously characterized 32D.3 cell lines expressing the fusion protein from the zinc-inducible metallothionine promoter (44). We measured the number of cells in each cell cycle phase by using propidium iodide staining of DNA and flow cytometry to measure DNA content 48 h after zinc induction of AML-1–ETO expression (Fig. 9C). AML-1–ETO-expressing cells, but not control cells, accumulated with a 2N DNA content indicative of a slowing of G1 phase transit.
As a further control for this phenotype, we used a C-terminal deletion mutant of AML-1–ETO (AML-1–ETO Δ469), which removes ETO sequences C terminal to the TAF110 domain (including both mSin3A binding sites, one N-CoR binding site, and the majority of HDAC-binding sites; Fig. 2 to 6) and no longer represses transcription (23). This mutant failed to alter the cell cycle profile of 32D.3 cells (Fig. 9D, right). By contrast, cells expressing AML-1–ETO showed a significant G1 arrest, with a concomitant decrease in S phase (Fig. 9D, middle) when compared to control 32D.3 cells containing the empty vector (Fig. 9D, left). The results with the Δ469 mutant suggest that transcriptional repression mediated by the AML-1–ETO fusion protein may be critical for induction of this aberrant cell cycle phenotype.
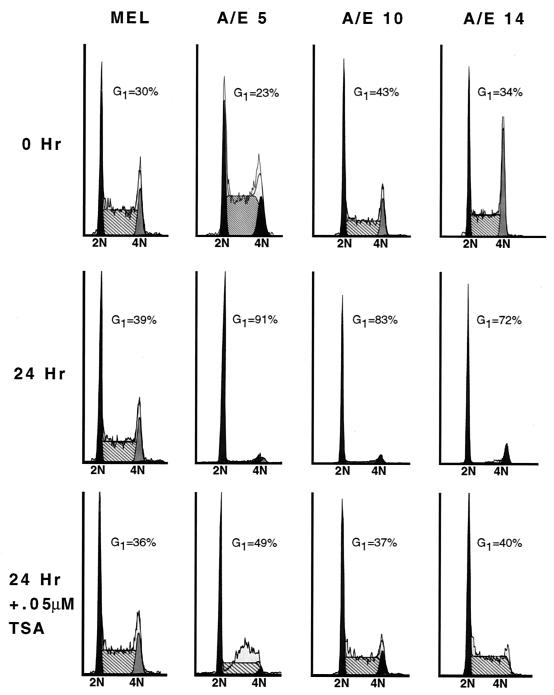
Next, we used the AML-1–ETO-mediated inhibition of cell cycle progression to biologically test the role of HDACs in AML-1–ETO function. MEL cells expressing AML-1–ETO were cultured in the absence of tetracycline to induce protein expression in the presence or absence of 0.05 μM TSA. Consistent with previous experiments, AML-1–ETO overexpression caused cells to arrest in the G1 phase. TSA effectively abrogated the G1 phase accumulation induced by AML-1–ETO but had no effect on control cell lines (Fig. 10). Therefore, AML-1–ETO likely inhibits cell cycle progression by repressing AML-1 target gene expression and HDAC inhibitors specifically inactivate AML-1–ETO.
FIG. 10.
TSA ablates the AML-1–ETO-induced G1 phase accumulation of cells. Cell cycle analysis of AML-1–ETO-expressing (A/E) and control (MEL) cell lines by using propidium iodide staining and flow cytometry were used to measure DNA content. The indicated cell lines (lines 5, 10, and 14) were cultured in media containing or lacking tetracycline. After washing the cells to remove tetracycline, half of the cells were cultured in the presence of 0.05 μM TSA. Cell cycle histograms were compiled using the ModFit algorithm.
DISCUSSION
The molecular contacts between the ETO zinc finger motif and the nuclear hormone corepressors N-CoR and SMRT have been defined by yeast two-hybrid assays, mammalian two-hybrid assays, and purification studies (10, 29, 41, 45). However, our approach of defining domains of ETO that are sufficient for corepressor binding by using chimeric ETO/ETO-2 and GAL4-ETO proteins has defined multiple contacts between ETO, N-CoR, mSin3A, HDAC-1, HDAC-2, and HDAC-3 (the known molecular contacts for ETO are summarized in Fig. 11A). By using overlapping fragments of ETO as GAL4-ETO fusion proteins, we were able to separate closely adjoining binding sites for corepressors. For example, HDAC-3 bound both GAL4-ETO(259-343) and GAL4-ETO(300-385), indicating that the binding site likely resides in the region of overlap between these fragments (residues 300 to 343). By contrast, mSin3A bound to only GAL4-ETO(300-385), placing this binding site C terminal to HDAC-3 (Fig. 11A). These results are consistent with the recent identification of an mSin3A binding site that overlapped the HHR domain (16). However, our results using point mutations indicated that the HHR-dimerization domain of ETO contacts mSin3A (Fig. 8). Although we were able to define a second mSin3A binding site between the HHR and nervy domain by using the ETO/ETO-2 fusion proteins, we were not able to use this strategy to map the second HDAC-2 binding site because HDAC-2 also bound ETO-2 (Fig. 5B).
ETO is the prototype of a family of highly homologous evolutionarily conserved proteins. Like ETO, ETO-2 is a potent repressor when tethered to a promoter by fusing it to the GAL4 DNA binding domain (Fig. 1). Therefore, it was unexpected that ETO, but not ETO-2, interacted with mSin3A. In fact, while the human family members ETO and MTG16a are 67% identical, the regions that specify mSin3A binding include one of the least conserved regions of ETO and a subtle alteration in the HHR-oligomerization motif. Because HDAC-1 and HDAC-2 heterodimerize and copurify with mSin3, whereas HDAC-3 copurified with N-CoR (24, 43), we propose a model in which ETO can recruit at least two different corepression complexes (Fig. 11B). This model is consistent with the sucrose gradient sedimentation pattern of ETO because ETO cosedimentated with both N-CoR and mSin3A but was found in fractions that contained mSin3A and not N-CoR (29). However, as ETO can bind HDACs independent of either mSin3A or N-CoR, it remains possible that ETO is a component of multiple corepression complexes.
The lack of mSin3A binding by ETO-2 and its ability to associate with five HDACs indicates that it is a component of a corepressor complex(es) that is distinct from ETO-containing complexes (Fig. 11C). If ETO and ETO-2 have distinct mechanisms of action or distinct targets for repression, this would explain why multiple family members are expressed in the same cell type. Alternatively, the fact that ETO family members can homo- and heterodimerize through the HHR domain adds a further level of complexity to transcriptional regulation by this family and may be a way of fine tuning transcriptional control in a particular tissue (21, 28).
It has been postulated that oligomerization of ETO is critical for interactions with N-CoR or SMRT and for AML-1–ETO biological actions (33, 45). However, in light of our definition of corepressor binding sites within and adjacent to the HHR-dimerization domain of ETO, it is more likely that it was the deletion of these corepressor binding sites that affected AML-1–ETO function (46). In fact, the AML-1–ETO deletion that was used to suggest a biological role for dimerization of AML-1–ETO (deletion of ETO residues 340 to 440 as numbered here) impinged upon one HDAC-1 binding motif, both mSin3A binding domains, and possibly binding sites for HDAC-2 and HDAC-3 (46) (Fig. 11A). By contrast, a more specific deletion of the HHR-dimerization motif that affected only one mSin3A-binding site had modest effects on GAL4-ETO (45) and AML-1–ETO-mediated transcriptional repression (28). This mutation reduced, but did not eliminate, binding to mSin3A, N-CoR, or HDACs (references 28 and 29 and this work). Thus, it is possible that dimerization contributes to repression, but dimerization is not required for corepressor binding. If indeed ETO family proteins heterodimerize, our results indicate that association between ETO and ETO-2 could lead to the recruitment of a distinct set of corepressors and HDACs given that ETO binds mSin3A, but ETO-2 does not, and ETO-2 binds HDAC-6 and HDAC-8. This might broaden the biological action of ETO and, by extension, AML-1–ETO.
Though counterintuitive, inhibition of cell cycle progression by AML-1–ETO has a precedent in that the inv (16) fusion protein, which also represses AML-1 target genes, caused a slowing of cell cycle progression in the G1 phase (2). In fact, t(8;21)-containing leukemia cell lines (e.g., Kasumi-1) grow very slowly. Although it is difficult to ascertain whether this phenotype is related to leukemogenesis, it does afford us a highly reproducible biological assay with which to probe the mechanism of action of AML-1–ETO. TSA induces cell differentiation, cell death, and, in some cases, cell cycle inhibition, perhaps due to induction of p21waf1/cip1 (30). Indeed, upon longer exposure of these MEL cells to TSA, we observed partial differentiation and cell cycle arrest in both control and AML-1–ETO-expressing cells (data not shown). However, in the short term, we observed that TSA biologically inactivated the t(8;21) fusion protein.
The t(15;17) and t(11;17) fusion proteins also used HDACs to repress transcription and preliminary work using inhibitors of these enzymes in therapy was successful (42). In addition, t(8;21)-containing blasts are sensitive to a combination of retinoic acid and TSA (8). Our results begin to provide a mechanistic basis for the sensitivity to TSA and further emphasize that HDAC inhibitors can biologically inactivate AML-1–ETO. This work also demonstrates the necessity of the development of HDAC inhibitors that target specific classes of HDACs or specific HDACs. Although N-CoR and mSin3A bind class I and class II HDACs, the independent association of HDAC-1, HDAC-2, and HDAC-3 with ETO suggests that treatment of patients harboring t(8;21) may require only inhibition of the class I deacetylases, thereby decreasing any potential toxicity associated with a broad-spectrum HDAC inhibitor such as TSA. Moreover, our work demonstrates the interactions that can take place, but the constellation of corepressors and HDACs that are expressed in t(8;21)-containing leukemic blasts will ultimately define the critical therapeutic targets.
ACKNOWLEDGMENTS
We thank Yue Hou and Dana King for expert technical assistance and the Vanderbilt Ingram Cancer Center sequencing facility for support.
This work was supported by National Institutes of Health grants RO1-CA76186 (SM), RO1-AG13726, RO1-CA64140, and RO1-CA77274, American Cancer Society grants JFRA-591 (S.W.H.) and PO1 CA71907 (J.R.D.), and a Center grant from the National Cancer Institute (CA68485), the Vanderbilt-Ingram Cancer Center, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. J.N. is a Leukemia Society of America Special Fellow (grant 3827-99).
REFERENCES
- 1.Ahn M Y, Huang G, Bae S C, Wee H J, Kim W Y, Ito Y. Negative regulation of granulocytic differentiation in the myeloid precursor cell line 32Dcl3 by ear-2, a mammalian homolog of Drosophila seven-up, and a chimeric leukemogenic gene, AML1/ETO. Proc Natl Acad Sci USA. 1998;95:1812–1817. doi: 10.1073/pnas.95.4.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao W, Britos-Bray M, Claxton D F, Kelley C A, Speck N A, Liu P P, Friedman A D. CBF beta-SMMHC, expressed in M4Eo AML, reduced CBF DNA-binding and inhibited the G1 to S cell cycle transition at the restriction point in myeloid and lymphoid cells. Oncogene. 1997;15:1315–1327. doi: 10.1038/sj.onc.1201305. [DOI] [PubMed] [Google Scholar]
- 3.Davis J N, Williams B J, Herron J T, Galiano F J, Meyers S. ETO-2: a new member of the ETO-family of nuclear proteins. Oncogene. 1999;18:1375–1383. doi: 10.1038/sj.onc.1202412. [DOI] [PubMed] [Google Scholar]
- 4.Erickson P, Gao J, Chang K S, Look T, Whisenant E, Raimondi S, Lasher R, Trujillo J, Rowley J, Drabkin H. Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood. 1992;80:1825–1831. [PubMed] [Google Scholar]
- 5.Erickson P F, Robinson M, Owens G, Drabkin H A. The ETO portion of acute myeloid leukemia t(8;21) fusion transcript encodes a highly evolutionarily conserved, putative transcription factor. Cancer Res. 1994;54:1782–1786. [PubMed] [Google Scholar]
- 6.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Identification of homeotic target genes in Drosophila melanogaster including nervy, a proto-oncogene homologue. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenrick R, Amann J M, Lutterbach B, Wang L, Westendorf J J, Downing J R, Hiebert S W. Both TEL and AML-1 contribute repression domains to the t(12;21) fusion protein. Mol Cell Biol. 1999;19:6566–6574. doi: 10.1128/mcb.19.10.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrara F F, Fazi F, Bianchini A, Padula F, Gelmitti V, Minucci S, Mancini M, Pelicci P G, Lo Coco F, Nervi C. Histone deacetylase-targeted treatment restores retinoic acid signaling and differentiation in acute myeloid leukemia. Cancer Res. 2001;61:2–7. [PubMed] [Google Scholar]
- 9.Gamou T, Kitamura E, Hosoda F, Shimizu K, Shinohara K, Hayashi Y, Nagase T, Yokoyama Y, Ohki M. The partner gene of AML1 in t(16;21) myeloid malignancies is a novel member of the MTG8(ETO) family. Blood. 1998;91:4028–4037. [PubMed] [Google Scholar]
- 10.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grozinger C M, Hassig C A, Schreiber S L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guenther M G, Lane W S, Fischle W, Verdin E, Lazar M A, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1: a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 14.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 15.Hiebert S W, Packham G, Strom D K, Haffner R, Oren M, Zambetti G, Cleveland J L. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hildebrand D, Tiefenbach J, Heinzel T, Grez M, Maurer A B. Multiple regions of ETO cooperate in transcriptional repression. J Biol Chem. 2001;276:9889–9895. doi: 10.1074/jbc.M010582200. [DOI] [PubMed] [Google Scholar]
- 17.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 18.Hu E, Chen Z, Fredrickson T, Zhu Y, Kirkpatrick R, Zhang G F, Johanson K, Sung C M, Liu R, Winkler J. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 19.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 20.Kao H Y, Downes M, Ordentlich P, Evans R M. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 21.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 23.Lenny N, Meyers S, Hiebert S W. Functional domains of the t(8;21) fusion protein, AML-1/ETO. Oncogene. 1995;11:1761–1769. [PubMed] [Google Scholar]
- 24.Li J, Wang J, Nawaz Z, Liu J M, Qin J, Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J. 2000;19:4342–4350. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu P, Tarle S A, Hajra A, Claxton D F, Marlton P, Freedman M, Siciliano M J, Collins F S. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science. 1993;261:1041–1044. doi: 10.1126/science.8351518. [DOI] [PubMed] [Google Scholar]
- 26.Lou J, Cao W, Bernardin F, Ayyanathan K, Rauscher I F, Friedman A D. Exogenous cdk4 overcomes reduced cdk4 RNA and inhibition of G1 progression in hematopoietic cells expressing a dominant-negative CBF—a model for overcoming inhibition of proliferation by CBF oncoproteins. Oncogene. 2000;19:2695–2703. doi: 10.1038/sj.onc.1203588. [DOI] [PubMed] [Google Scholar]
- 27.Lutterbach B, Hou Y, Durst K L, Hiebert S W. The inv(16) encodes an acute myeloid leukemia 1 transcriptional corepressor. Proc Natl Acad Sci USA. 1999;96:12822–12827. doi: 10.1073/pnas.96.22.12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutterbach B, Sun D, Scheutz J, Hiebert S W. The MYND motif is required for repression of basal transcription from the multidrug resistance-1 promoter by the t(8;21) fusion protein. Mol Cell Biol. 1998;18:3604–3611. doi: 10.1128/mcb.18.6.3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, Glass C, Seto E, Hiebert S W. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marks P A, Richon V M, Rifkind R A. Cell cycle regulatory proteins are targets for induced differentiation of transformed cells: molecular and clinical studies employing hybrid polar compounds. Int J Hematol. 1996;63:1–17. doi: 10.1016/0925-5710(95)00428-9. [DOI] [PubMed] [Google Scholar]
- 31.Melnick A, Westendorf J, Arai S, Lutterbach B, Ball H, Pllinger A, Licht J, Hiebert S. The PLZF leukemia protein physically and functionally interacts with the ETO leukemia protein suggesting a common link of transcriptional repression in AML. Blood. 1998;92:479a. [Google Scholar]
- 32.Melnick A M, Westendorf J J, Polinger A, Carlile G W, Arai S, Ball H J, Lutterbach B, Hiebert S W, Licht J D. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol Cell Biol. 2000;20:2075–2086. doi: 10.1128/mcb.20.6.2075-2086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, Di Croce L, Giavara S, Matteucci C, Gobbi A, Bianchini A, Colombo E, Schiavoni I, Badaracco G, Hu X, Lazar M A, Landsberger N, Nervi C, Pelicci P G. Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell. 2000;5:811–820. doi: 10.1016/s1097-2765(00)80321-4. [DOI] [PubMed] [Google Scholar]
- 34.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy L, Kao H Y, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 37.Nip J, Strom D K, Fee B E, Zambetti G, Cleveland J L, Hiebert S W. E2F-1 cooperates with topoisomerase II inhibition and DNA damage to selectively augment p53-independent apoptosis. Mol Cell Biol. 1997;17:1049–1056. doi: 10.1128/mcb.17.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nucifora G, Rowley J D. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995;86:1–14. [PubMed] [Google Scholar]
- 39.Shibata H, Nawaz Z, Tsai S Y, O'Malley B W, Tsai M J. Gene silencing by chicken ovalbumin upstream promoter-transcription factor I (COUP-TFI) is mediated by transcriptional corepressors, nuclear receptor-corepressor (N-CoR) and silencing mediator for retinoic acid receptor and thyroid hormone receptor (SMRT) Mol Endocrinol. 1997;11:714–724. doi: 10.1210/mend.11.6.0002. [DOI] [PubMed] [Google Scholar]
- 40.Strom D K, Nip J, Westendorf J J, Linggi B, Lutterbach B, Downing J R, Lenny N, Hiebert S W. Expression of the AML-1 oncogene shortens the G(1) phase of the cell cycle. J Biol Chem. 2000;275:3438–3445. doi: 10.1074/jbc.275.5.3438. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Hoshino T, Redner R L, Kajigaya S, Liu J M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warrell R P, Jr, He L Z, Richon V, Calleja E, Pandolfi P P. Therapeutic targeting of transcription in acute promyelocytic leukemia by use of an inhibitor of histone deacetylase. J Natl Cancer Inst. 1998;90:1621–1625. doi: 10.1093/jnci/90.21.1621. [DOI] [PubMed] [Google Scholar]
- 43.Wen Y D, Perissi V, Staszewski L M, Yang W M, Krones A, Glass C K, Rosenfeld M G, Seto E. The histone deacetylase-3 complex contains nuclear receptor corepressors. Proc Natl Acad Sci USA. 2000;97:7202–7207. doi: 10.1073/pnas.97.13.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Westendorf J J, Yamamoto C M, Lenny N, Downing J R, Selsted M E, Hiebert S W. The t(8;21) fusion product, AML-1-ETO, associates with C/EBP-alpha, inhibits C/EBP-alpha-dependent transcription, and blocks granulocytic differentiation. Mol Cell Biol. 1998;18:322–333. doi: 10.1128/mcb.18.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Hug B A, Huang E Y, Chen C W, Gelmetti V, Maccarana M, Minucci S, Pelicci P G, Lazar M A. Oligomerization of ETO is obligatory for corepressor interaction. Mol Cell Biol. 2001;21:156–163. doi: 10.1128/MCB.21.1.156-163.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]