Abstract
Introduction
NRAS mutations are common in melanoma and confer a worse prognosis. Although most patients with metastatic melanoma receive immune checkpoint inhibitors (ICIs), the impact of NRAS mutational status on their efficacy remains under debate.
Methods
We performed a comprehensive literature search across several large databases. Inclusion criteria were trials, cohorts, and large case series that analyzed the primary outcome of objective response rate by NRAS mutational status in patients with melanoma treated with any line of ICI. At least two reviewers independently screened studies using Covidence software, extracted data, and assessed risk of bias. Standard meta-analysis was performed in R with sensitivity analysis and tests for bias.
Results
Data on 1770 patients from ten articles were pooled for meta-analysis, and the objective response rate to ICIs was calculated to compare NRAS-mutant and NRAS-wildtype melanoma. The objective response rate was 1.28 (95% confidence interval: 1.01–1.64). Sensitivity analysis identified the study by Dupuis et al. with influential impact on the pooled effect size and heterogeneity, favoring NRAS-mutant melanoma.
Discussion
In this meta-analysis evaluating the impact of NRAS mutational status on objective response to ICIs in metastatic melanoma, NRAS-mutant cutaneous melanoma demonstrated an increased likelihood of partial or complete tumor response, relative to NRAS-wildtype cutaneous melanoma. Genomic screening for NRAS mutations in patients with metastatic melanoma may improve predictive ability when initiating ICIs.
Keywords: metastatic melanoma, cutaneous melanoma, immunotherapy, checkpoint inhibitor, NRAS, objective response rate, disease control rate, NRAS-mutant melanoma
1. Introduction
Melanoma is the sixth-leading cause of cancer diagnosis in the United States and is responsible for the most deaths from skin cancer (1). The Surveillance, Epidemiology, and End Results database indicates that incidence of cutaneous melanoma has increased over the past two decades (2). Whereas FDA approval of multiple immune checkpoint inhibitors (ICIs) in the past decade has revolutionized therapy, five-year survival still hovers around 30% for metastatic melanoma (2–6). While new strategies are being investigated, we are interested in understanding whether and why molecular subtypes of melanoma may respond differently to ICIs (7).
The contemporaneous emergence of next-generation sequencing has improved our abilities to characterize individual patients’ melanomas and to research potential genetic targets. NRAS mutations are present in approximately 20% of all melanomas and portend a worse prognosis than does NRAS-wildtype status (8, 9). Although the lack of effective targeted therapy makes NRAS-mutant melanoma a more controversial subtype than BRAF-mutant melanoma, studies suggest that NRAS mutational status is an independent prognostic factor for metastatic melanoma and may be associated with immunotherapy response (9–15). Immunotherapy is considered the standard of care for most patients with locally advanced or metastatic melanoma without a contraindication, but their comparative efficacy in NRAS-mutant versus NRAS-wildtype melanoma remains unclear (9, 10). Some studies have suggested that in advanced melanoma treated with ICIs, NRAS-mutation-positive status is associated with higher rates of tumor objective response and/or prolonged survival (16, 17), while others do not support this association (18, 19). To our knowledge, a comprehensive synthesis of the available evidence of the outcomes of ICIs for melanoma based on NRAS mutational status has not been undertaken.
Therefore, we conducted a systematic review and meta-analysis to critically appraise and synthesize the published data comparing objective response rate (ORR) to ICIs by NRAS mutational status in patients with melanoma.
2. Manuscript
2.1. Methods
This systematic review was registered in the PROSPERO database (ID: CRD42021273588) and reported according to PRISMA and MOOSE guidelines. The Washington University Institutional Review Board determined this project to be non-human subjects research.
2.1.1. Search strategy
A clinical librarian (AH) executed a comprehensive search in PubMed, Embase, Scopus, Web of Science, Cochrane Library (including CENTRAL), and Clinicaltrials.gov databases from database origin to 14 June 2022. The full search strategy is available in the Supplementary Materials. Results were limited to English and human studies.
Two reviewers independently screened titles and abstracts for eligibility (ZJJ, NSR) and, subsequently, relevant full texts (ZJJ, NSR) using the Covidence systematic review management program (20). One reviewer performed forward and reverse citation searches (ZJJ), i.e., screenings of articles citing and cited by included studies, respectively. Two or three reviewers independently extracted data (ZJJ, NSR, NKAM). Disagreements were resolved by consensus.
2.1.2. Eligibility criteria
We included randomized, controlled trials and cohort studies that analyzed ORR by NRAS mutational status in patients with melanoma treated with any line of ICI. We excluded unpublished work, gray literature, duplicate studies, animal studies, in vitro studies, case reports, expert opinions, and reviews.
The primary outcome was ORR for NRAS-mutant and NRAS-wildtype melanoma, defined as the percentage of patients with complete (CR) or partial response (PR) by Response Evaluation Criteria in Solid Tumors v1.1 or immune-related response criteria at the time of last follow-up (21, 22). We defined the comparison group as either confirmed to be triple-negative for NRAS, BRAF, and NF1 mutations or at least confirmed NRAS-wildtype; if comparison groups included melanoma with other mutations, previous or current targeted therapy merited exclusion. If the genotype data were not explicitly stated, we contacted authors for clarification. Patients with stable (SD) or progressive disease (PD) were considered unresponsive to treatment. A secondary outcome was the disease control rate (DCR), denoted by the percentage of patients with CR, PR, or SD. Other extracted data included patient sex and age, melanoma subtype, stage at ICI initiation, ICI class and line, and tumor mutational burden (TMB).
2.1.3. Quality assessment
Three reviewers (ZJJ, NSR, and NKAM) independently assessed the overall quality of evidence of each study via the Oxford Centre for Evidence-Based Medicine (OCEBM) Levels of Evidence and modified Newcastle-Ottawa scale (mNOS) (23, 24). We designated OCEBM study quality per the “Therapy/Prevention, Aetiology/Harm” category from 1a (best) to 5 (poorest), with mNOS scored from 0 (poorest) to 9 (best). To test for potential publication bias, we visually inspected a contour-enhanced funnel plot, graphically evaluated using the trim-and-fill method (25), and statistically evaluated with the Egger regression test (26).
2.1.4. Data synthesis and statistical analysis
We used R version 4.1.2 (packages “metafor” and “metaviz”) for all data analysis. We performed a meta-analysis of the pooled relative risk ratios of ORR of cutaneous melanoma using a random-effects model. The Yates correction was employed for cells with zero to estimate the 95% confidence intervals (CI). The results were plotted on a forest plot with 95% CI of the relative risk ratios of ORR. We evaluated heterogeneity between studies with the I2 statistic (greater than 50% suggesting moderate heterogeneity) and Q statistic (greater than the degrees of freedom suggesting significant heterogeneity). We repeated these methods for DCR. A sensitivity analysis was performed to identify influential studies in the ORR meta-analysis.
2.2. Results
2.2.1. Literature search
After removal of duplicates, 473 records were screened, 157 full-text articles were assessed, and 16 studies met inclusion criteria for the systematic review, with 10 of those being amenable to meta-analysis (Figure 1) (16, 18, 19, 27–39). Data on 1770 patients from two prospective studies and eight retrospective cohort studies were pooled for analysis.
Figure 1.

PRISMA flow diagram.
2.2.2. Study characteristics
Most included studies were retrospective cohorts and took place in the 2010s in North America, Europe, and Asia (Table 1). Three of four prospective trials were carried out in Asia, with the other trial by Ascierto et al. occurring in Italy (27, 33, 35, 38). Participants were generally middle-aged adults with an approximate pooled median of 55–65 years, with a slight male predominance (54%). Cutaneous melanoma was the predominant subtype in 11 studies, with varying prevalence of non-cutaneous subtypes (mucosal, acral, uveal, and unknown primary) comprising the remainder. Most of the studies of patients with non-cutaneous melanoma occurred in Asia; these studies were included for the systematic review but excluded from meta-analysis (33, 35, 36, 38). All studies involved patients with metastatic melanoma, many with predominantly Stage IV (78% among those reporting stage). ICI regimens varied, ranging from anti-CTLA-4 (8/16, 50%) or anti-PD-1/PD-L1 monoclonal antibody (14/16, 88%) monotherapy to combinations thereof (5/16, 31%), some combined with chemotherapy (3/16, 19%). Only five studies included patients on combination ICI therapy, ranging from 1 to 40% of the study population. Data were unavailable for individual participants by mutational status, precluding subgroup analysis by ICI line and class.
Table 1.
Summary data and quality assessment.
| Author | Year | Country | Study type | Female, n (%) | Age (in years) | Predominant melanoma type | Class of ICIa | OCEBM/mNOS Qualityb |
|---|---|---|---|---|---|---|---|---|
| Ascierto et al. | 2014 | Italy | Prospective cohort | 395 (46) | Median 61, Min–maxc 16–88 | Cutaneous | Anti-CTLA-4 monotherapy | 2b/7 |
| Lipson et al. | 2014 | United States | Case series | 4 (50) | Median 58, Min–max 43–79 | Cutaneous | Anti-CTLA-4 or anti-PD-L1 monotherapy | 4/4 |
| Johnson et al. | 2015 | United States | Retrospective cohort | 50 (28) | 61% 60+ | Cutaneous | Anti-PD-1, anti-PD-L1, anti-CTLA-4, or IL-2 monotherapy | 2b/8 |
| Johnson et al. | 2016 | United States | Retrospective cohort | 27 (42) | Initial: Median 55, Min-max 33–80; Validation: Median 62, Min-max 32–85 |
Cutaneous | Anti-PD-1 or anti-PD-L1 monotherapy | 2b/8 |
| Jamal et al. | 2017 | Canada | Prospective cohort | 8 (27) | Median 55, Min–max 26–74 | Cutaneous | Anti-CTLA-4 monotherapy | 2b/7 |
| Afzal et al. | 2018 | United States | Retrospective cohort | 21 (38) | Mean 63, SDd 15 | Cutaneous | Anti-PD-1 monotherapy or anti-CTLA-4/anti-PD-1 combination therapy | 2b/9 |
| Dupuis et al. | 2018 | France | Retrospective cohort | 29 (41) | 61% 60+ | Cutaneous | Anti-PD-1 monotherapy | 2b/8 |
| Kirchberger et al. | 2018 | Germany | Retrospective cohort | 154 (42) | Median 64, Min–max 20–87 | Cutaneous | Anti-CTLA-4 or anti-PD-1 monotherapy or combination therapy | 2b/8 |
| Sheng et al. | 2019 | China | Phase 1B clinical trial | 20 (61) | Median 54, Min–max 27–70 | Mucosal | Anti-PD-1 monotherapy | 2b/7 |
| Loo et al. | 2020 | United States | Retrospective cohort | 22 (33) | Median 59, Min–max 27–93 | Cutaneous | Anti-CTLA-4 or anti-PD-1 monotherapy or combination therapy | 2b/7 |
| Tang et al. | 2020 | China | Phase 2 clinical trial | 70 (55) | Mean 52, SD 11 | Acral, mucosal | Anti-PD-1 monotherapy | 2b/7 |
| Byeon et al. | 2021 | Korea | Retrospective cohort | 90 (51) | Median 61, Min–max 23–84 | Acral, cutaneous, mucosal | Anti-PD-1 monotherapy | 2b/8 |
| Guida et al. | 2021 | Italy | Retrospective cohort | 127 (38) |
NRAS+: Median 63, IQe min–max 53–74; NRAS-: Median 65, IQ min–max 54–73 |
Cutaneous | Anti-PD-1 or anti-CLTA-4 monotherapy or combination therapy | 2b/9 |
| Li et al. | 2021 | China | Retrospective cohort | 32 (50) | Median 54, Min–max 25–72 | Cutaneous | Anti-PD-1 monotherapy | 2b/8 |
| Zhou et al. | 2021 | China | Prospective trials | 119 (58) | 15% 65+ | Non-cutaneous, cutaneous | Anti-PD-1 monotherapy | 2b/9 |
| Zhang et al. | 2022 | International | Retrospective cohort | 199 (36) | 51% 61+ | Cutaneous | Anti-CTLA-4, anti-PD-1, or anti-PD-L1 monotherapy or combination therapy | 2b/8 |
aICI: immune checkpoint inhibitor. bOCEBM Quality Assessment: Oxford Centre for Evidence-based Medicine Levels of Evidencem; NOS: modified Newcastle-Ottawa scale. cMin–max: minimum to maximum. dSD: standard deviation. eIQ: interquartile.
2.2.3. Meta-analysis
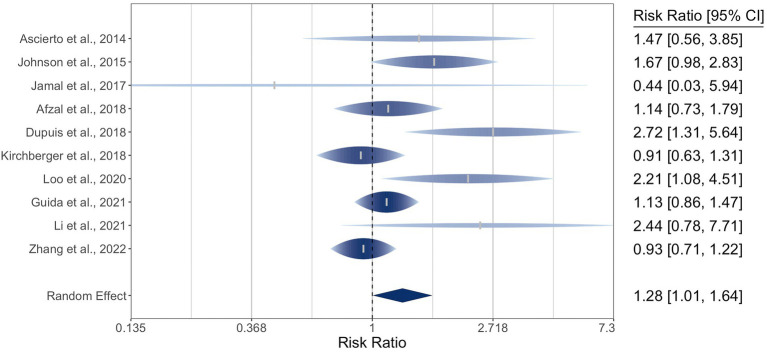
For the ten included studies encompassing 1770 participants, the pooled relative risk ratio (RR) of ORR by NRAS mutational status was 1.28 (95% CI: 1.01–1.64). The I2 statistic was 53%, indicating moderate-to-high heterogeneity, and Q (df = 9) was 17 (p = 0.05). The meta-analysis of ORR is graphically represented in a rainforest plot in Figure 2.
Figure 2.
Forest plot of the random-effects meta-analysis for relative risk ratios of objective response rates (ORR) of cutaneous melanoma by NRAS mutational status. NRAS-mut: NRAS-mutant melanoma; NRAS-wt: NRAS-wildtype melanoma; OR: Objective response; NR: No objective response.
There were eight studies reporting DCR for 1,650 patients with metastatic cutaneous melanoma. The pooled RR by NRAS mutational status was 1.11 (95% CI: 0.87–1.41). There was moderate-to-high heterogeneity, with I2 at 79% (95% CI: 44–97) and Q (df = 7) at 28 (p = 0.0002). The three studies in the ORR meta-analysis that did not provide DCR data reported RRs of 1.70, 2.21, and 2.72, favoring NRAS-mutant melanoma (29, 32, 34). The meta-analysis of DCR is graphically represented in a rainforest plot in Figure 3.
Figure 3.
Forest plot of the random-effects meta-analysis for relative risk ratios of disease control rates (DCR) of cutaneous melanoma by NRAS mutational status. NRAS-mut: NRAS-mutant melanoma; NRAS-wt: NRAS-wildtype melanoma; DC: Disease control; PD: Progressive disease.
2.2.4. Sensitivity analysis
Leave-one-out sensitivity analysis of ORR produced overall effect sizes ranging from 1.16 to 1.38 (lower bound of 95% CI: 0.96–1.05; upper bound: 1.40–1.83). The I2 statistic ranged from 27 to 61%, and Q ranged from 11.3 to 16.9. Omission of the study by Zhang et al. maximized the lower bound at 1.05, greater than the null RR of 1 (39). Individual omission of the study by Dupuis et al. minimized I2 to 27% and the 95% CI bounds to 0.96 and 1.40 (26); it also minimized the pooled estimate to 1.16(), while the remainder ranged from 1.20() to 1.38 (32, 34, 39).
2.2.5. Quality assessment and risk of publication bias
Oxford Centre for Evidence-Based Medicine quality ratings (Table 1) ranged from 2b to 4, with all earning a grade of 2b except the case series (28). mNOS ranged from 7 to 9 indicating generally low risk of bias, except for the case series (4). Inspection of the contour-enhanced funnel plot of the ORR meta-analysis revealed some asymmetry, with larger studies tending to report positive effect sizes and one small study reporting a non-significant negative result. The trim-and-fill method imputed two moderate-to-large studies, one with a non-significant negative effect size, the other on the borderline of significance, and the Egger regression test was statisticallysignificant for asymmetry (p = 0.02). The Baujat plot identified the study by Dupuis et al. (32, 40) and as particularly influential on the overall results. Graphical representations of statistical tests for bias are included in the Supplementary material.
2.2.6. Tumor mutational burden
Six studies reported outcome data on TMB (29, 33, 35–37, 39). Four of these reported increased ORR in higher-TMB groups ranging from 30 to 85%, with only Johnson et al.’s (29, 33, 35–37) paper achieving statistical significance (85%, p < 0.001). Byeon et al. (36) reported better one-year progression-free survival for the higher-TMB group on ICIs at approximately 45 vs. 30%, though it was not quite statistically significant (p = 0.051). Zhang et al. (39) found higher TMB associated with prolonged overall survival (p < 0.001).
2.3. Discussion
The seminal paper by Johnson et al. (16) in 2015 supported positive NRAS mutational status as a predictor of good response to ICIs. In 2018, Dupuis et al. (32) also found an increased ORR for NRAS-mutant melanoma. That same year, however, Kirchberger et al. (18) demonstrated equivocal predictive value of NRAS mutational status. In 2021, Guida et al. (19) explored this hypothesis further and discovered a similar questionable effect with non-significant confidence intervals. In contribution to the debate surrounding the effect of the NRAS mutational status of melanoma on ICI response, this meta-analysis demonstrated a benefit for NRAS-mutant cutaneous melanoma (RR 1.28), i.e., a higher ICI response rate was seen for NRAS-mutant melanoma relative to wildtype. Implications of this conclusion could include, for instance, increased pertinence of NRAS mutational status ordering for patients with newly diagnosed metastatic cutaneous melanoma, or increased likelihood of initiating ICIs in metastatic cutaneous melanoma found to harbor an NRAS mutation. Further research into the potential immunological mechanism should continue to be explored. Interestingly, the results for DCR were lower and centered around the null; this could be partially explained by the exclusion of three studies with positive RR for ORR that did not report data for DCR.
Quality ratings of included studies were consistent with observational data. The funnel plot and trim-and-fill method indicated moderate concern for significant publication bias, and the Egger regression test supported the presence of asymmetry. Sources of bias such as heterogeneity between subgroups, selective outcome reporting, or chance, could be contributing to asymmetry of the funnel plot (41). In further exploration of heterogeneity, the Baujat plot indicated that the study by Dupuis et al. exerted disproportionate influence on the meta-analysis, and the sensitivity analysis was consistent with its impact on both the pooled effect size and heterogeneity measures (32). Taken together, the overall certainty of the evidence for ORR was rated as moderate (42).
The included studies largely assessed cutaneous melanoma patients based in Europe and the United States and excluded non-cutaneous melanoma studies due to their relative paucity and to avoid unlike comparisons. The few non-cutaneous-predominant studies by Sheng et al., Tang et al., Byeon et al., and Zhou et al. (33, 35, 36, 39) generally reported low ORR for NRAS-mutant melanoma. It is well known that non-cutaneous melanoma subtypes carry unique mutation patterns distinct from ultraviolet radiation-related signatures and often respond poorly to current therapies (43–45). Further investigation of this diverse population is warranted to better guide therapy and improve outcomes (46, 47).
The collected literature offers several potential explanations for the observed effects of ICIs on NRAS-mutant melanoma. Johnson et al. conducted a separate analysis which found higher PD-L1 expression in NRAS-mutant melanoma, though this failed to reach significance and other larger cohorts have not found PD-L1 expression to necessarily be consequential to ICI response (16, 48). In contrast, Byeon et al. (36) speculated NRAS-mutant melanoma could be associated with ICI resistance, perhaps due to alterations in cell surface proteins necessary for T-cell response similar to TP53-mutant tumors. Still others have hypothesized a synergistic effect of MAP kinase and ICI due to greater melanoma antigen production, though these preclinical findings need further real-world confirmation (49). Interestingly, in a subset of NRAS-mutant melanoma patients receiving immunotherapy, a frequently co-occurring PBAF complex mutation was associated with greater progression-free and overall survival (50). Most frequently, however, authors posit that TMB may be central to the ICI-NRAS interaction.
Quantifying TMB has become more critical in understanding cancer therapeutics and may help predict ICI response. In theory, higher TMB produces a greater degree of passenger mutations, increasing tumor neoantigenicity, T-lymphocyte infiltration, and antitumor immune response (7, 48, 51). A large-scale retrospective analysis of several primary tumor types found that TMB status may explain 55% of ORR variation to ICIs (52). Similarly, CheckMate 067 trial data revealed ICI responders were more likely to have higher TMB and higher inflammatory signatures than non-responders (53). Six studies included in this meta-analysis reported TMB data associating higher TMB with higher ORR, though an accurate pooled effect cannot be calculated here (29, 33, 35–37, 39). A meta-analysis that focuses on the role of TMB on melanoma outcomes in patients initiating ICIs is needed.
NRAS-mutant melanoma has proven challenging to treat, especially for those unresponsive to immunotherapy. The NRAS protein itself is difficult to target directly, and upstream and downstream effectors have demonstrated equivocal efficacy, with a few showing promise (9). Although testing is available from commercial entities, it may not be routinely performed due to a lack of effective target therapy. MEK inhibitors, alone or in combination with pan-RAF, CDK4/6, or focal adhesion kinase inhibitors, are under investigation in clinical trials for metastatic NRAS-mutant melanoma (14). Others are studying the predictive value of biomarkers and genomic profiles that might shed light on subpopulations more or less responsive to certain immunotherapy regimens (54, 55). One study identified NF1 mutational status as a predictor of poor response (56), while another found good tumor response in NF1-mutant melanoma (57); clearly, there is a need for further investigation. Indeed, the field of melanoma continues to head toward a “multiomics”-driven approach to mutational and molecular-level stratification that could reveal predictors of therapeutic response or novel targets and treatments (58–60).
2.3.1. Limitations
The review presented several challenges. Despite a sensitive literature search, the dearth of eligible articles, combined with the novelty of ICIs, resulted in a relatively small number of included studies. Some heterogeneity among variables and outcomes of interest was present, leading to possible reporting bias. This was partially mitigated by some authors responding to share supplementary data, but with more administrative resources, a meta-analysis with individual participant data and meta-regression would better characterize subgroups and adjust for potentially confounding variables.
2.3.2. Conclusion
Melanoma is a leading cause of cancer diagnosis in the United States and a diagnosis that dermatologists frequently encounter in both inpatient and outpatient settings. At high stages, prognosis is poor, and even more so when NRAS mutational status is positive. In this meta-analysis comparing objective response to ICI therapy between NRAS-mutant and NRAS-wildtype metastatic melanoma, there was an increased likelihood of response for NRAS-mutant cutaneous melanoma. Genomic screening for patients diagnosed with metastatic melanoma may improve predictive ability for those harboring NRAS mutations. In the era of precision medicine with checkpoint inhibition and combination therapies, more prospective research on all types of melanoma is warranted, especially randomized, controlled trials on NRAS mutational status reporting complete demographics, molecular data, tumor response, and survival outcomes.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at: https://pubmed.ncbi.nlm.nih.gov/24885479/; https://pubmed.ncbi.nlm.nih.gov/33399091/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6048096/; https://pubmed.ncbi.nlm.nih.gov/33530579/; https://pubmed.ncbi.nlm.nih.gov/29157311/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4351797/; https://pubmed.ncbi.nlm.nih.gov/27671167/; https://pubmed.ncbi.nlm.nih.gov/29843107/; https://pubmed.ncbi.nlm.nih.gov/34181096/; https://pubmed.ncbi.nlm.nih.gov/25516806/; https://pubmed.ncbi.nlm.nih.gov/32564504/; https://pubmed.ncbi.nlm.nih.gov/31403867/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8787219/; https://pubmed.ncbi.nlm.nih.gov/29966520/; https://pubmed.ncbi.nlm.nih.gov/34290710/; https://pubmed.ncbi.nlm.nih.gov/32321714/.
Author contributions
ZJ, NR, DC, GA, and LC: study conception and design. ZJ, NR, NM, AH, and LC: data collection. ZJ, NR, NM, DC, GA, and LC: analysis and interpretation of results and draft manuscript preparation. All authors meet ICMJE criteria for authorship, and all those who meet ICMJE criteria are listed as authors. All authors contributed to the article and approved the submitted version.
Funding
Discretionary research funds received from the Division of Dermatology, John T. Milliken Department of Medicine, Washington University School of Medicine in St. Louis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank Graham Colditz and Carrie Stoll for their helpful advice and instructional guidance in the initial design of the review, as well as Sierra Jaeger for assistance with R is a statistical analysis software: https://www.r-project.org/about.html.
Glossary
Abbreviations
- ICI
immune checkpoint inhibitor
- ORR
objective response rate
- CR
complete response
- PR
partial response
- SD
stable disease
- PD
progressive disease
- DCR
disease control rate
- TMB
tumor mutational burden
- CI
confidence interval
- RR
relative risk
- OCEBM
Oxford Centre for Evidence-based Medicine
- mNOS
modified Newcastle-Ottawa scale
- ALM
acral lentiginous melanoma
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmed.2023.1090737/full#supplementary-material
References
- 1.U.S. Cancer Statistics Working Group . U.S. Cancer statistics data visualizations tool, based on 2021 submission data (1999-2019): U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. (2022) Available at: www.cdc.gov/cancer/dataviz, released in June 2022. Accessed June 20, 2022.
- 2.Surveillance, Epidemiology, and End Results Program . Cancer stat facts: melanoma of the skin. U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute. (2022) Available at: https://seer.cancer.gov/statfacts/html/melan.html. Accessed June 20, 2022.
- 3.Hodi FS, O'day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. (2010) 363:711–23. doi: 10.1056/NEJMoa1003466, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, Chiarion-Sileni V, Gonzalez R, Rutkowski P, Grob JJ, Cowey CL, et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. (2017) 377:1345–56. doi: 10.1056/NEJMoa1709684, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuryk L, Bertinato L, Staniszewska M, Pancer K, Wieczorek M, Salmaso S, et al. From conventional therapies to immunotherapy: melanoma treatment in review. Cancers (Basel). (2020) 12:3057. doi: 10.3390/cancers12103057, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meric-Bernstam F, Larkin J, Tabernero J, Bonini C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet. (2021) 397:1010–22. doi: 10.1016/S0140-6736(20)32598-8 [DOI] [PubMed] [Google Scholar]
- 7.Maleki VS. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J Immunother Cancer. (2018) 6:157. doi: 10.1186/s40425-018-0479-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akbani R, Akdemir KC, Aksoy BA, Albert M, Ally A, Amin SB, et al. Genomic classification of cutaneous melanoma. Cells. (2015) 161:1681–96. doi: 10.1016/j.cell.2015.05.044, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaddour K, Maahs L, Avila-Rodriguez AM, Maamar Y, Samaan S, Ansstas G. Melanoma targeted therapies beyond BRAF-mutant melanoma: potential druggable mutations and novel treatment approaches. Cancers (Basel). (2021) 13:5847. doi: 10.3390/cancers13225847, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boespflug A, Caramel J, Dalle S, Thomas L. Treatment of NRAS-mutated advanced or metastatic melanoma: rationale, current trials and evidence to date. Ther Adv Med Oncol. (2017) 9:481–92. doi: 10.1177/1758834017708160, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakob JA, Bassett RL, Jr, Ng CS, Curry JL, Joseph RW, Alvarado GC, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. (2012) 118:4014–23. doi: 10.1002/cncr.26724, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devitt B, Liu W, Salemi R, Wolfe R, Kelly J, Tzen CY, et al. Clinical outcome and pathological features associated with NRAS mutation in cutaneous melanoma. Pigment Cell Melanoma Res. (2011) 24:666–72. doi: 10.1111/j.1755-148X.2011.00873.x, PMID: [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Alvarez A, Ortiz C, Munoz-Couselo E. Current perspectives and novel strategies of NRAS-mutant melanoma. Onco Targets Ther. (2021) 14:3709–19. doi: 10.2147/OTT.S278095, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randic T, Kozar I, Margue C, Utikal J, Kreis S. NRAS mutant melanoma: towards better therapies. Cancer Treat Rev. (2021) 99:102238. doi: 10.1016/j.ctrv.2021.102238, PMID: [DOI] [PubMed] [Google Scholar]
- 15.Zhao J, Galvez C, Beckermann KE, Johnson DB, Sosman JA. Novel insights into the pathogenesis and treatment of NRAS mutant melanoma. Expert Rev Precis Med Drug Dev. (2021) 6:281–94. doi: 10.1080/23808993.2021.1938545, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DB, Lovly CM, Flavin M, Panageas KS, Ayers GD, Zhao Z, et al. Impact of NRAS mutations for patients with advanced melanoma treated with immune therapies. Cancer Immunol Res. (2015) 3:288–95. doi: 10.1158/2326-6066.CIR-14-0207, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadetsky N, Lambert P, Julian C, Chen J, Yan Y. Comprehensive genomic profiling and outcomes among metastatic melanoma patients (pts) treated with first-line cancer immunotherapy (CIT) in a real-world setting. Ann Oncol. (2019) 30:vii2. doi: 10.1093/annonc/mdz413.011 [DOI] [Google Scholar]
- 18.Kirchberger MC, Ugurel S, Mangana J, Heppt MV, Eigentler TK, Berking C, et al. MEK inhibition may increase survival of NRAS-mutated melanoma patients treated with checkpoint blockade: results of a retrospective multicentre analysis of 364 patients. Eur J Cancer. (2018) 98:10–6. doi: 10.1016/j.ejca.2018.04.010, PMID: [DOI] [PubMed] [Google Scholar]
- 19.Guida M, Bartolomeo N, Quaglino P, Madonna G, Pigozzo J, di Giacomo A, et al. No impact of NRAS mutation on features of primary and metastatic melanoma or on outcomes of checkpoint inhibitor immunotherapy: an italian melanoma intergroup (IMI) study. Cancers. (2021) 13:1–15. doi: 10.3390/cancers13030475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Covidence Systematic Review Software . Veritas health innovation (2017), Melbourne, Australia. Available at: www.covidence.org. Accessed June 20, 2022.
- 21.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. (2009) 45:228–47. doi: 10.1016/j.ejca.2008.10.026, PMID: [DOI] [PubMed] [Google Scholar]
- 22.Wolchok JD, Hoos A, O'Day S, Weber JS, Hamid O, Lebbé C, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. (2009) 15:7412–20. doi: 10.1158/1078-0432.CCR-09-1624, PMID: [DOI] [PubMed] [Google Scholar]
- 23.Oxford Centre for Evidence-Based Medicine . Levels of evidence (2009). Available at: https://www.cebm.ox.ac.uk/resources/levels-of-evidence/oxford-centre-for-evidence-based-medicine-levels-of-evidence-march-2009. Accessed June 20, 2022.
- 24.Wells G, Shea B, O'Connell D, Peterson JE, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Non-Randomized Studies in Meta-analyses. (2000). Ottawa Hospital Research Institute. Available at: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp
- 25.Duval S, Tweedie R. Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. (2000) 56:455–63. doi: 10.1111/j.0006-341X.2000.00455.x, PMID: [DOI] [PubMed] [Google Scholar]
- 26.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ascierto PA, Simeone E, Sileni VC, Pigozzo J, Maio M, Altomonte M, et al. Clinical experience with ipilimumab 3 mg/kg: real-world efficacy and safety data from an expanded access programme cohort. J Transl Med. (2014) 12:116. doi: 10.1186/1479-5876-12-116, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipson EJ, Velculescu VE, Pritchard TS, Sausen M, Pardoll DM, Topalian SL, et al. Circulating tumor DNA analysis as a real-time method for monitoring tumor burden in melanoma patients undergoing treatment with immune checkpoint blockade. J Immunother Cancer. (2014) 2:42. doi: 10.1186/s40425-014-0042-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DB, Frampton GM, Rioth MJ, Yusko E, Xu Y, Guo X, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. (2016) 4:959–67. doi: 10.1158/2326-6066.CIR-16-0143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamal R, Lapointe R, Cocolakis E, Thébault P, Kazemi S, Friedmann JE, et al. Peripheral and local predictive immune signatures identified in a phase II trial of ipilimumab with carboplatin/paclitaxel in unresectable stage III or stage IV melanoma. J Immunother Cancer. (2017) 5:83. doi: 10.1186/s40425-017-0290-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Afzal MZ, Mercado RR, Shirai K. Efficacy of metformin in combination with immune checkpoint inhibitors (anti-PD-1/anti-CTLA-4) in metastatic malignant melanoma. J Immunother Cancer. (2018) 6:64. doi: 10.1186/s40425-018-0375-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupuis F, Lamant L, Gerard E, Torossian N, Chaltiel L, Filleron T, et al. Clinical, histological and molecular predictors of metastatic melanoma responses to anti-PD-1 immunotherapy article. Br J Cancer. (2018) 119:193–9. doi: 10.1038/s41416-018-0168-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheng X, Yan X, Chi Z, Si L, Cui C, Tang B, et al. Axitinib in combination with toripalimab, a humanized immunoglobulin G 4 monoclonal antibody against programmed cell death-1, in patients with metastatic mucosal melanoma: an open-label phase Ib trial. J Clin Oncol. (2019) 37:2987–99. doi: 10.1200/JCO.19.00210, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loo K, Gauvin G, Soliman I, Renzetti M, Deng M, Ross E, et al. Primary tumor characteristics and next-generation sequencing mutations as biomarkers for melanoma immunotherapy response. Pigment Cell Melanoma Res. (2020) 33:878–88. doi: 10.1111/pcmr.12909, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang B, Chi Z, Chen Y, Liu X, Wu D, Chen J, et al. Safety, efficacy, and biomarker analysis of toripalimab in previously treated advanced melanoma: results of the POLARIS-01 multicenter phase II trial. Clin Cancer Res. (2020) 26:4250–9. doi: 10.1158/1078-0432.CCR-19-3922, PMID: [DOI] [PubMed] [Google Scholar]
- 36.Byeon S, Cho HJ, Jang KT, Kwon M, Lee J, Kim ST. Molecular profiling of Asian patients with advanced melanoma receiving check-point inhibitor treatment. ESMO Open. (2021) 6:100002. doi: 10.1016/j.esmoop.2020.100002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li JJ, Wang JH, Dingv Y, Li DD, Wen XZ, Zhao JJ, et al. Efficacy and safety of anti-PD-1 inhibitor combined with nab-paclitaxel in Chinese patients with refractory melanoma. J Cancer Res Clin Oncol. (2021) 148:1159–69. doi: 10.1007/s00432-021-03700-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou L, Wang X, Chi Z, Sheng X, Kong Y, Mao L, et al. Association of NRAS mutation with clinical outcomes of anti-PD-1 monotherapy in advanced melanoma: a pooled analysis of four Asian clinical trials. Front Immunol. (2021) 12:691032. doi: 10.3389/fimmu.2021.691032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang WJ, Kong YJ, Li YT, Shi F, Lyu J, Sheng C, et al. Novel molecular determinants of response or resistance to immune checkpoint inhibitor therapies in melanoma. Front Immunol. (2022) 12:12. doi: 10.3389/fimmu.2021.798474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baujat B, Mahe C, Pignon JP, Hill C. A graphical method for exploring heterogeneity in meta-analyses: application to a meta-analysis of 65 trials. Stat Med. (2002) 21:2641–52. doi: 10.1002/sim.1221, PMID: [DOI] [PubMed] [Google Scholar]
- 41.Sterne JA, Sutton AJ, Ioannidis JP, Terrin N, Jones DR, Lau J, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. (2011) 343:d4002. doi: 10.1136/bmj.d4002, PMID: [DOI] [PubMed] [Google Scholar]
- 42.Schünemann H, Vist G, Higgins J, Santesso N, Deeks JJ, Glasziou P. Chapter 15: interpreting results and drawing conclusions In: Higgins J, Thomas J, Chandler J, et al., editors. Cochrane Handbook for Systematic Reviews of Interventions Version 61. Cochrane: Wiley-Blackwell. (2020) [Google Scholar]
- 43.Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, et al. Whole-genome landscapes of major melanoma subtypes. Nature. (2017) 545:175–80. doi: 10.1038/nature22071, PMID: [DOI] [PubMed] [Google Scholar]
- 44.Jamerson T, Rebecca VW, Aguh C. Genetic characteristics and response to systemic therapies of acral lentiginous melanoma at a tertiary care center—a retrospective review. J Natl Med Assoc. (2022) 114:7–11. doi: 10.1016/j.jnma.2021.08.034, PMID: [DOI] [PubMed] [Google Scholar]
- 45.Raval NS, Hodges WT, Ugwu-Dike PO, Godoy F, Ansstas G, Cornelius LA, et al. Racial and socioeconomic differences in acral lentiginous melanoma outcomes: a Surveillance, epidemiology, and end results analysis. J Am Acad Dermatol. (2021) 87:866–7. doi: 10.1016/j.jaad.2021.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kailas A, Dawkins M, Taylor SC. Suggestions for increasing diversity in clinical trials. JAMA Dermatol. (2017) 153:727. doi: 10.1001/jamadermatol.2017.0850 [DOI] [PubMed] [Google Scholar]
- 47.Kamal K, Imadojemu S, Charrow A. Why diversity in dermatology clinical trials should no longer be optional: dismantling structural racism in dermatology. JAMA Dermatol. (2022) 158:353–4. doi: 10.1001/jamadermatol.2021.5190, PMID: [DOI] [PubMed] [Google Scholar]
- 48.Klempner SJ, Fabrizio D, Bane S, Reinhart M, Peoples T, Ali SM, et al. Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncologist. (2020) 25:e147–59. doi: 10.1634/theoncologist.2019-0244, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilmott JS, Hersey P, Long GV, Scolyer RA. Synergistic effects of MAPK and immune checkpoint inhibitors in melanoma: what is the best combination strategy? Melanoma Manag. (2015) 2:15–9. doi: 10.2217/mmt.14.26, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway JR, Kofman E, Mo SS, Elmarakeby H, Van Allen E. Genomics of response to immune checkpoint therapies for cancer: implications for precision medicine. Genome Med. (2018) 10:93. doi: 10.1186/s13073-018-0605-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rose AA, Armstrong SM, Hogg D, Butler MO, Saibil SD, Arteaga DP, et al. Biologic subtypes of melanoma predict survival benefit of combination anti-PD1+anti-CTLA4 immune checkpoint inhibitors versus anti-PD1 monotherapy. J Immunother Cancer. (2021) 9:e001642. doi: 10.1136/jitc-2020-001642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. (2019) 4:e126908. doi: 10.1172/jci.insight.126908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hodi FS, Wolchok JD, Schadendorf D, Larkin J, Long GV, Qian X, et al. TMB and inflammatory gene expression associated with clinical outcomes following immunotherapy in advanced melanoma. Cancer Immunol Res. (2021) 9:1202–13. doi: 10.1158/2326-6066.CIR-20-0983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jessurun CAC, Vos JAM, Limpens J, Luiten RM. Biomarkers for response of melanoma patients to immune checkpoint inhibitors: a systematic review. Front Oncol. (2017) 7:233. doi: 10.3389/fonc.2017.00233, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jiang J, Ding Y, Wu M, Chen Y, Lyu X, Lu J, et al. Integrated genomic analysis identifies a genetic mutation model predicting response to immune checkpoint inhibitors in melanoma. Cancer Med. (2020) 9:8498–518. doi: 10.1002/cam4.3481, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cirenajwis H, Lauss M, Ekedahl H, Törngren T, Kvist A, Saal LH, et al. NF1-mutated melanoma tumors harbor distinct clinical and biological characteristics. Mol Oncol. (2017) 11:438–51. doi: 10.1002/1878-0261.12050, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thielmann CM, Chorti E, Matull J, Murali R, Zaremba A, Lodde G, et al. NF1-mutated melanomas reveal distinct clinical characteristics depending on tumour origin and respond favourably to immune checkpoint inhibitors. Eur J Cancer. (2021) 159:113–24. doi: 10.1016/j.ejca.2021.09.035, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Conway JR, Dietlein F, Taylor-Weiner A, AlDubayan S, Vokes N, Keenan T, et al. Integrated molecular drivers coordinate biological and clinical states in melanoma. Nat Genet. (2020) 52:1373–83. doi: 10.1038/s41588-020-00739-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shoushtari AN, Chatila WK, Arora A, Sanchez-Vega F, Kantheti HS, Rojas Zamalloa JA, et al. Therapeutic implications of detecting MAPK-activating alterations in cutaneous and unknown primary melanomas. Clin Cancer Res. (2021) 27:2226–35. doi: 10.1158/1078-0432.CCR-20-4189, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Newell F, Pires da Silva I, Johansson PA, Menzies AM, Wilmott JS, Addala V, et al. Multiomic profiling of checkpoint inhibitor-treated melanoma: identifying predictors of response and resistance, and markers of biological discordance. Cancer Cell. (2022) 40:88–102.e7. doi: 10.1016/j.ccell.2021.11.012, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found at: https://pubmed.ncbi.nlm.nih.gov/24885479/; https://pubmed.ncbi.nlm.nih.gov/33399091/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6048096/; https://pubmed.ncbi.nlm.nih.gov/33530579/; https://pubmed.ncbi.nlm.nih.gov/29157311/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4351797/; https://pubmed.ncbi.nlm.nih.gov/27671167/; https://pubmed.ncbi.nlm.nih.gov/29843107/; https://pubmed.ncbi.nlm.nih.gov/34181096/; https://pubmed.ncbi.nlm.nih.gov/25516806/; https://pubmed.ncbi.nlm.nih.gov/32564504/; https://pubmed.ncbi.nlm.nih.gov/31403867/; https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8787219/; https://pubmed.ncbi.nlm.nih.gov/29966520/; https://pubmed.ncbi.nlm.nih.gov/34290710/; https://pubmed.ncbi.nlm.nih.gov/32321714/.