Abstract
Introduction
To investigate whether biochemical and haematological changes due to the patient’s host response (CoLab algorithm) in combination with a SARS-CoV-2 viability PCR (v-PCR) can be used to determine when a patient with COVID-19 is no longer infectious.
We hypothesise that the CoLab algorithm in combination with v-PCR can be used to determine whether or not a patient with COVID-19 is infectious to facilitate the safe release of patients with COVID-19 from isolation.
Methods and analysis
This study consists of three parts using three different cohorts of patients. All three cohorts contain clinical, vital and laboratory parameters, as well as logistic data related to isolated patients with COVID-19, with a focus on intensive care unit (ICU) stay. The first cohort will be used to develop an algorithm for the course of the biochemical and haematological changes of the host response of the COVID-19 patient. Simultaneously, a second prospective cohort will be used to investigate the algorithm derived in the first cohort, with daily measured laboratory parameters, next to conventional SARS-CoV-2 reverse transcriptase PCRs, as well as v-PCR, to confirm the presence of intact SARS-CoV-2 particles in the patient. Finally, a third multicentre cohort, consisting of retrospectively collected data from patients with COVID-19 admitted to the ICU, will be used to validate the algorithm.
Ethics and dissemination
This study was approved by the Medical Ethics Committee from Maastricht University Medical Centre+ (cohort I: 2020-1565/300523) and Zuyderland MC (cohorts II and III: METCZ20200057). All patients will be required to provide informed consent. Results from this study will be disseminated via peer-reviewed journals and congress/consortium presentations.
Keywords: COVID-19, INTENSIVE & CRITICAL CARE, Diagnostic microbiology, Clinical chemistry, HAEMATOLOGY, Epidemiology
STRENGTHS AND LIMITATIONS OF THIS STUDY.
The algorithm/model is based on routinely tested blood parameters and standardised laboratory tests.
Multicentre approach with a good distribution of hospitals covering various regions of the Netherlands.
Large temporal range of the retrospective cohort III enables model validation for SARS-CoV-2 virus variants of concern.
Viability PCR is not performed in cohorts I and III.
The focus is limited to (de)isolation of patients with COVID-19 in the intensive care unit.
Introduction
The COVID-19 pandemic is globally disruptive regarding the continuation of regular healthcare. Hospitalised patients with COVID-19 need to be isolated and separated from patients without COVID-19. This aspect paired with the large influx of patients with COVID-19 and limited availability of hospital and isolation beds exerts enormous pressure on regular non-COVID-19 healthcare, but also on healthcare professionals. In addition, the need for treatment and support in an intensive care unit (ICU) for a substantial subset of patients with COVID-19 and the limited availability in the number of ICU beds contribute to these effects. Deisolation as early as possible could improve the quality of life for the affected patients, as well as decrease the pressure on the healthcare system and its professionals.
Several study protocols described methods to determine if COVID-19-infected patients can be deisolated: based on clinical signs,1 using reverse transcriptase PCR (RT-PCR)2 or with rapid antigen tests.3 RT-PCR testing is currently the gold standard to determine whether a patient is SARS-CoV-2 positive.4 To deisolate an ICU patient with COVID-19 in the Netherlands two consecutive negative PCR tests are required. However, it can be hypothesised that SARS-CoV-2 RT-PCR positivity does not relate per se with the actual presence of intact, infectious viruses.5 6 Because RT-PCR detects nucleic acids and does not make a distinction between an intact infectious virus and non-intact non-infectious viral particles, this may result in persistently positive RT-PCR test results, which hampers timely deisolation.4
An alternative RT-PCR-based method to detect intact viral particles is to eliminate incomplete viral particles and RNA remnants before the actual RT-PCR is performed. Propidium monoazide (PMA) is a dye that binds irreversibly to DNA/RNA and cannot penetrate cell membranes.7 Pretreatment of a sample with PMA results in the amplification of only intact particles. This so-called viability PCR (v-PCR) has been shown to successfully measure the number of viable micro-organisms, such as Chlamydia trachomatis, in a sample.8 In the present study, we want to adapt and validate this concept for the detection of intact viable RNA-containing SARS-CoV-2 virus. Preliminary data have confirmed its applicability for SARS-CoV-2 diagnostics.9 The adapted v-PCR will be used in the study herein presented to confirm the state of viability and thus potential infectivity of SARS-CoV-2 in patients.
An alternative approach is to assess the host response of the suspected patient to the virus. One of the methods to assess the host response to SARS-CoV-2 is the CoLab score. This score has been developed using an adaptive least absolute shrinkage and selection operator (LASSO) regression technique and requires the input of the numerical results of 10 blood parameters and the age of the patient.10 The required parameters are blood tests that are requested frequently and routinely for emergency room (ER) as well as ICU patients. This score has previously been developed and validated and has been implemented in the ER departments of two large Dutch teaching hospitals, with very high negative predictive value (99.5%) and sensitivity (96.9%).10 The score is also used to exclude COVID-19 in a screening setting for healthcare workers with COVID-19 suspected complaints.11
Preliminary analysis of serially collected data in a pilot set of ICU patients showed a decrease in the CoLab score resulting in normalisation before a patient is discharged (unpublished data). For that reason, we hypothesise that the biochemical and haematological changes in blood parameters necessary to calculate the CoLab score rapidly return to normal values after the host clears the SARS-CoV-2 infection.
This study aims to investigate whether biochemical and haematological changes due to the patient’s host response (CoLab algorithm) and/or the v-PCR can be reliably and validly used to determine, at an earlier stage in comparison with a conventional SARS-CoV-2 RT-PCR, when a patient with COVID-19 is no longer infectious.
Methods and analysis
Cohorts
This study is composed of three cohorts, two prospective cohorts (local and regional) and one retrospective cohort (national), which all consist of serially (ie, daily) collected clinical and laboratory variables of patients with COVID-19 in isolation at an ICU. We intend to include all patients admitted to one of our COVID-19 ICU isolation rooms.
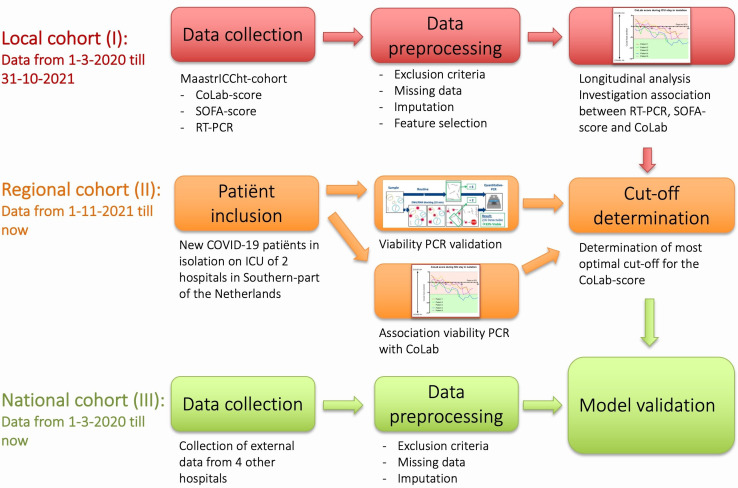
More specifically, the three different cohorts will be used to study the CoLab score over time (local cohort I), to determine a cut-off point related to the intact infectious viral load (regional cohort II) and to validate the CoLab algorithm (national cohort III) on a national level with an external data set (figure 1). While not developed specifically for models using machine learning,12 the study will follow the guidelines of the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis.13
Figure 1.
Overview of the study. ICU, intensive care unit; RT-PCR, reverse transcriptase PCR; SOFA, Sequential Organ Failure Assessment.
Local single-centre prospective cohort (I)
The first single-centre local cohort is the prospective Maastricht Intensive Care COVID (MaastrICCht) cohort, previously described by Tas et al.14 The CoLab score is calculated for each time point using this comprehensively characterised cohort.14–20 In addition, the daily Sequential Organ Failure Assessment (SOFA)15 19 scores are available as well as all conventional SARS-CoV-2 RT-PCRs that are measured within this cohort. The aim is to investigate the development of the CoLab score over time. To possibly deisolate patients, the CoLab score should at least decrease over time in a way that is independent of disease severity and similar for survivors and non-survivors. Therefore, we hypothesise that the CoLab score decreases over time in both survivors and non-survivors in a way that is independent of disease severity over time measured by serial SOFA scores. To have an additional value above conventional RT-PCR-based deisolation, the decrease in CoLab score should occur before deisolation by RT-PCR is done. We hypothesise that a CoLab score decrease is present before RT-PCR-based deisolation. We will explore the association between CoLab score over time and the moment of RT-PCR-driven deisolation. If the CoLab score behaves over time in the ICU as hypothesised above, the next step is to quantify what decrease in CoLab score over time (or what cut-off CoLab score per day) precedes the transition from RT-PCR positive to negative. This decrease in CoLab score over time can be used to develop a diagnostic prediction model for deisolation. Whether this prediction model can be used as the gold standard for deisolation (CoLab prediction model alone, or in combination with conventional SARS-CoV-2 RT-PCR and/or v-PCR) is part of this study protocol.
Regional dual-centre prospective cohort (II)
In the second part, we hypothesise that excluding infectiousness, contributing to deisolation can be done more accurately by using v-PCR instead of RT-PCR. A second prospectively collected dual-centre regional cohort of patients with COVID-19 from the ICU department of both Zuyderland Medical Centre and Maastricht University Medical Centre+ (MUMC+) will be used to evaluate the usability of the v-PCR for the above-mentioned hypothesis. Inclusion of all consecutive ICU patients with COVID-19 will be pragmatic based on the development of the pandemic and related incidence of ICU admission, starting from 1 November 2021. We aim to include a minimum of 90 patients. In this cohort, serial data related to the CoLab algorithm will be collected daily. In addition, both conventional (RT-PCR) and v-PCR testing for the detection of SARS-CoV-2 will be performed three times a week. The aim of this regional cohort (II) is to determine a cut-off point or a certain decrease in CoLab score over time that precedes the transition from positive to negative RT-PCR and v-PCR results.
National multicentre retrospective cohort (III)
For the third part of the study, a retrospectively collected multicentre national cohort will be used. This retrospective cohort will consist of ICU data derived from four other hospitals located in the Netherlands. This data set will contain serially collected data necessary for determining the CoLab score (10 blood parameters and age, see below) next to conventional SARS-CoV-2 RT-PCR results. This cohort will be used to determine whether the CoLab algorithm developed and validated in cohorts I and II in specific contexts is generalisable to and valid in other contexts (cohort III). An additional aim is to test the CoLab algorithm for different variants of concern (VOCs) of SARS-CoV-2 (see also below). For this purpose, we will use data from all ICU patients positive for COVID-19 between March 2020 and September 2022 (estimated at least 250 patients per participating centre).
Context and setting
Data from six hospitals will be used to create the different cohorts of this study. An overview of the number of hospital and ICU beds per participating hospital and per cohort is shown in online supplemental table 1.
bmjopen-2022-069455supp001.pdf (49.9KB, pdf)
The local single-centre cohort I aims to use data obtained at MUMC+ (27 ICU and six high/medium care beds in the prepandemic era), a university medical centre located in the southern part of the Netherlands. During the COVID-19 pandemic, a maximum of 52 ICU beds were available for patients with COVID-19, and 12 for patients without COVID-19. Using this local cohort, the CoLab score will be observed over time.
The regional dual-centre cohort II consists of data from ICU patients from both Zuyderland MC (36 ICU beds) and MUMC+. These two hospitals are both located in Limburg in the Netherlands with an existing close cooperation for clinical purposes. Both hospitals are large teaching hospitals. This regional cohort will be used to assess whether the CoLab score can be used to determine whether patients are SARS-CoV-2 free according to the results of the v-PCR.
The national multicentre cohort III consists of retrospectively collected data from four additional hospitals: Leiden UMC, Radboud UMC, Medical Centre Leeuwarden and Catharina Hospital. The hospitals in this cohort are located in separate provinces leading to a good geographical representation of the national spread of the Dutch patient population with COVID-19. Since Leiden UMC and Radboud UMC are university medical centres and Medical Centre Leeuwarden and Catharina Hospital are large teaching hospitals, both hospital types are represented equally. This national cohort will serve to further validate the model created using cohorts I and II in broader contexts (see online supplemental table 1 for details of the different hospitals contributing to the consortium).
Patient and public involvement
The national patient organisation for lung diseases (Longfonds) has a panel of patients who have experienced isolation process due to COVID-19 in the ICU. These patients have read the study protocol and gave advice that has been implemented in the protocol. The patient panel will also be involved during the study to provide feedback regarding the execution of this study and to provide input for the implementation of the results.
Inclusion and exclusion criteria
For the three cohorts, the same inclusion and exclusion criteria are applicable. All patients with a proven primary and/or secondary SARS-CoV-2 infection are eligible to participate in the study. Exclusion criteria include only patients with extreme laboratory values (more than 10 times the SD).
Parameters
Blood parameters
Blood samples are used to determine a variety of biochemical and haematological parameters in routine diagnostics and disease monitoring, from hospitalisation until discharge of a patient with COVID-19. This has led to a large accumulation of blood-related biomarker data. Previous studies found biochemical and haematological changes measured in peripheral blood samples that characterised SARS-CoV-2 infection.21–23 For instance, in the early stage of COVID-19 disease, haematological changes in immunocompetent leucocytes are associated with a more severe disease progression.23
CoLab score
The CoLab score10 uses the erythrocytes (×1012/L), leucocytes (×109/L), eosinophils (×109/L), basophils (×109/L), log10 of bilirubin (µmol/L), log10 of lactate dehydrogenase (U/L), log10 of alkaline phosphatase (IU/L), log10 of γ-glutamyltransferase (U/L), albumin (g/L), C-reactive protein (mg/L) and age (years accurate to two decimals). These parameters are routinely measured in ICU patients and can be automatically extracted from the laboratory information system. The CoLab algorithm yields a score in the range of −20 to 5 (the so-called CoLab linear predictor10), with a lower score associated with the exclusion of a SARS-CoV-2 infection and a higher score reflecting an increased risk of SARS-CoV-2 infection.
In an emergency department study population, a cut-off of the CoLab linear predictor was determined to classify patients as being COVID-19 negative. This cut-off was originally set to −5.83 to minimise the amount of false negative results, with a score below −5.83 being negative for COVID-19.10 How the CoLab algorithm corresponds with a negligible intact infectious viral load (see the section below) is part of the present study: a cut-off or a certain decrease in CoLab score over time. The CoLab score will be calculated daily for all participating patients, either prospectively or retrospectively.
Clinical parameters
In addition to the blood parameters, the clinical variables of patients are collected in the different cohorts. These include comorbidities and clinical scores as well as ventilation, biometric and physical parameters.15–20 One clinical score of interest is the SOFA score. This score has previously been associated with the survival chance of mechanically ventilated patients with COVID-19.15 A decrease in SOFA score is associated with survival. This sequentially determined SOFA score is measured over time and will be used to investigate whether the association between the CoLab score over time and infectiousness is independent of the SOFA score. This will provide evidence whether the CoLab score operationalises a different dimension of the host response, beyond multiorgan failure and in an independent way with regard to survival. This will generate evidence whether the CoLab score generates new information, beyond existing scores and has potential for diagnosis of deisolation.
Viability PCR
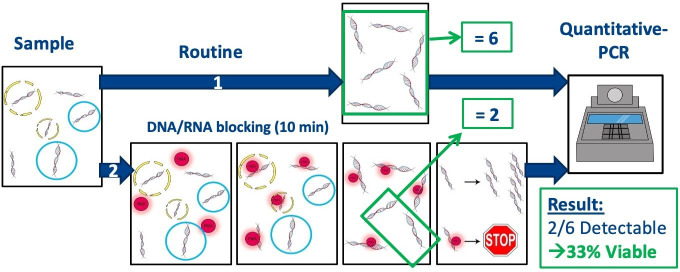
A v-PCR9 is performed to assess the presence of intact viruses and will be compared with the conventional SARS-CoV-2 RT-PCR test.24 Briefly, nasopharyngeal samples are collected in viral transport medium (VTM). The VTM sample is divided into two parts. One part is directly used for a conventional RT-PCR for SARS-CoV-2. For the v-PCR, PMA is added to the other half of the VTM sample.25 After pretreating this sample it is used for the v-PCR (see also figure 2). The difference in cycle time (Ct) values between these two PCR tests will be reported as ΔCt, which is a reliable indication of the amount of intact virus in the sample.
Figure 2.
Schematic representation of the principles of the conventional SARS-CoV-2 reverse transcriptase PCR (RT-PCR) (route 1) in comparison to the viability PCR (route 2). Route 1: all RNA is isolated from the sample and amplified using RT-PCR. Route 2: propidium monoazide (PMA) irreversibly binds to free RNA and RNA from non-intact virus particles. Only RNA from intact virus particles is isolated and amplified by RT-PCR.
The implementation of the v-PCR in the routine diagnostics would add some processing time to the existing SARS-CoV-2 PCR protocols. The v-PCR method is currently not (yet) automated and might as such not fit in every COVID-19 diagnostic workflow. However, the added value of the v-PCR would be the determination of complete virus particles.
Variants of concern
Due to the rapid mutation potential observed in viruses, it is necessary to ensure the robustness of the CoLab algorithm to VOCs of this SARS-CoV-2 virus. This study will address VOCs retrospectively as well as prospectively. Cohort III, spanning from March 2020 until the present, contains data on the Wuhan original SARS-CoV-2 and data from at least three VOCs. Demographic studies showed that during this period three VOCs of the SARS-CoV-2 were present next to the original SARS-CoV-2 virus (March 2020 to January 2021): the B1.1.7 Alpha variant (February 2021 to June 2021), the B1.617.2 Delta variant (July 2021 to December 2021) and the B1.1.529 Omicron variant (January 2022 to present).26 We use time periods to characterise VOCs in cohort III. In contrast, in cohort II VOCs will be measured with variant-specific next generation sequencing.27
Statistical analysis
Analyses will be performed with R V.4.2.0 and with RStudio V.4.2.0,28 combined with the packages Tidyverse,29 lme4,30 multiple imputations by chained equations (MICE),31 MissForest32 and Caret.33 Missing values for numerical variables will be imputed using MICE.31 Mixed-effects regression model analysis will be used to observe the CoLab score over time (cohort I), to determine whether the CoLab score is independent of survival and SOFA score (cohort I) and to determine the association between the CoLab score and the v-PCR (potentially cohort I and particularly cohort II). The reason for this is to determine the maximal cut-off value for the CoLab score to predict negligible viral load. If necessary, the CoLab model can be adjusted using LASSO regression to determine the optimal parameters used in this score. Finally, the CoLab model will be validated using receiver operating characteristic curves, confusion matrices and calibration curves in the analysis of cohort III.
For local cohort I, a prospective serially collected data set of 390 patients positive for COVID-19 admitted to the ICU of MUMC+ is available. This also includes a subset of immunocompromised patients (n=60). Adding interaction terms with immunocompromised groups to the mixed models will test whether the development of the CoLab score over time differs for these patients compared with non-immunocompromised patients. A similar approach will be taken to investigate whether results for sex differ.
For regional cohort II, a negative v-PCR will be considered as the moment when a patient is not infectious anymore. To assess whether a normalised CoLab score can pinpoint this moment, we expect that 95% of the patients will have a normalised CoLab score within a time frame of 2 days before and after the negative v-PCR. Using this proportion of 95% with a total width of the CI of 10%, and an alpha of 5%, we need to include at least 88 new patients with COVID-19 admitted to the ICU for mechanical ventilation.
For national cohort III, we aim to include serially collected data from all patients positive for COVID-19 admitted to the ICU of the other participating hospitals for validation.
Sample size calculation
For the local cohort, as stated above, a prospectively serially collected data set of 390 patients positive for COVID-19 admitted to the ICU of MUMC+ is already available. This also includes a subset of immunocompromised patients (n=60). If hypothesised that the course of the CoLab score does not differ between immunocompromised and non-immunocompromised patients with COVID-19 (the mean difference between these two groups=0), and using a power of 80%, an alpha of 0.05, an SD in CoLab linear predictor of 1.5 and a margin of ±1, then we need to analyse at least 39 patients per group (in our data set we have data available of 60 immunocompromised patients).
For the regional cohort (prospective), a negative v-PCR will be considered as the moment when a patient is not infectious anymore. To assess whether a normalised CoLab score can pinpoint this moment, we expect that 95% of the patients will have a normalised CoLab score within a time frame of 2 days before and after the negative v-PCR. Using this proportion of 95% with a total width of the CI of 10%, and an alpha of 5%, we need to include at least 88 new patients with COVID-19 admitted to the ICU for mechanical ventilation.
For the national cohort (retrospective), we want to include serially collected data sets of at least 250 COVID-19 patients admitted to the ICU of the other participating hospitals for the purpose of validation. These data are already available in the different laboratory information systems of the different hospitals, but needed to be extracted, collected and cleaned.
Ethics and dissemination
Ethical approval for study part I (METC number: 2020-1565/300523) was granted by the Medical Ethical Committee from MUMC+ (Maastricht, the Netherlands). During the pandemic, the board of directors of MUMC+ adopted a policy to inform patients and ask for their consent to use the collected data and to store blood samples for COVID-19 research purposes. The Medical Ethical Committee from Zuyderland Medical Centre (Heerlen/Sittard-Geleen, the Netherlands) approved study parts II (METCZ20210091—CoLaIC study) and III (METCZ20200057). The study is conducted in accordance with the Declaration of Helsinki. Patients will be informed about the purpose and procedures of the study via verbal and written information and informed consent will be obtained. If the patient is not able to communicate himself/herself, for example, due to ICU treatment, the next of kin will be approached. Patients will be asked for consent later when they have recovered. Results from this study will be disseminated via peer-reviewed journals, congress presentations and consortium presentations. The data generated will also be available on request in a public, open-access repository.
Supplementary Material
Footnotes
Collaborators: Dutch CoLaIC Consortium: Stephanie MC Ament (MUMC+, Maastricht); M Sesmu Arbous (LUMC, Leiden); Otto Bekers (MUMC+, Maastricht); Miranda van Berkel (Radboud UMC, Nijmegen); Arjen-Kars Boer (Catharina Hospital, Eindhoven); Dirck W van Dam (Zuyderland MC, Sittard-Geleen/Heerlen); Ruben Deneer (Catharina Hospital, Eindhoven); William PTM van Doorn (MUMC+, Maastricht); Tom P Dormans (Zuyderland MC, Sittard-Geleen/Heerlen); Silvia MAA Evers (Maastricht University, Maastricht); Tim Frenzel (Radboud UMC, Nijmegen); Judith Gillis (LUMC, Leiden); Iwan CC van der Horst (MUMC+, Maastricht); W Nadia H Koek (Medical Centre Leeuwarden, Leeuwarden); Kitty CFM Linssen (Zuyderland MC, Sittard-Geleen); Steven JR Meex (MUMC+, Maastricht); Guy JM Mostard (Zuyderland MC, Sittard-Geleen); Remy LM Mostard (Zuyderland MC, Sittard-Geleen); Luuk C Otterspoor (Catharina Hospital, Eindhoven); Natal AW van Riel (Technical University, Eindhoven); Frans Stals (Zuyderland MC, Sittard-Geleen); Albert Wolthuis (Medical Centre Leeuwarden, Leeuwarden).
Contributors: Study design: BCTvB, IHMvL, PFGW, WNKAvM and MPGL. Development of the study protocol: BCTvB, IHMvL, PFGW, WNKAvM, SHMG, WPHGV-vdV and MPGL. Patient recruitment: BCTvB and WNKAvM. Data collection: TS and FvR. Manuscript preparation: TS, BCTvB, PFGW, IHMvL, WPHGV-vdV, WNKAvM and MPGL. The members of the CoLaIC Consortium codesigned the study protocol, selected potential participants, assisted in their recruitment, collected the data, and set up, prepared and hosted the COVID-19 databases. All authors read and approved the final manuscript.
Funding: This publication is part of the CoLaIC project (project number: 10430102110002) of the COVID-19 research programme which is (partly) financed by the Netherlands Organisation for Health Research and Development (ZonMw).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
CoLaICconsortium:
Stephanie MC Ament, M Sesmu Arbous, Otto Bekers, Miranda van Berkel, Arjen-Kars Boer, Dirck W van Dam, Ruben Deneer, William PTM van Doorn, Tom P Dormans, Silvia MAA Evers, Tim Frenzel, Judith Gillis, Iwan CC van der Horst, W Nadia H Koek, Kitty CFM Linssen, Steven JR Meex, Guy JM Mostard, Remy LM Mostard, Luuk C Otterspoor, Natal AW van Riel, Frans Stals, and Albert Wolthuis
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Kang SW, Park H, Kim JY, et al. Clinical scoring system to predict viable viral shedding in patients with COVID-19. J Clin Virol 2022;157:105319. 10.1016/j.jcv.2022.105319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Syue L-S, Hung Y-P, Li C-W, et al. De-isolation criterion of real-time PCR test in patients with COVID-19: two or three consecutive negative nasopharyngeal swabs? J Microbiol Immunol Infect 2021;54:136–8. 10.1016/j.jmii.2020.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alshukairi AN, Al-Omari A, Al Hroub MK, et al. De-isolation of vaccinated COVID-19 health care workers using rapid antigen detection test. J Infect Public Health 2022;15:902–5. 10.1016/j.jiph.2022.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moghadas SM, Fitzpatrick MC, Sah P, et al. The implications of silent transmission for the control of COVID-19 outbreaks. Proc Natl Acad Sci U S A 2020;117:17513–5. 10.1073/pnas.2008373117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marik PE, Iglesias J, Varon J, et al. A scoping review of the pathophysiology of COVID-19. Int J Immunopathol Pharmacol 2021;35:20587384211048026. 10.1177/20587384211048026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Kampen JJA, van de Vijver D, Fraaij PLA, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun 2021;12:267. 10.1038/s41467-020-20568-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods 2006;67:310–20. 10.1016/j.mimet.2006.04.015 [DOI] [PubMed] [Google Scholar]
- 8. Janssen KJH, Hoebe CJPA, Dukers-Muijrers NHTM, et al. Viability-PCR shows that NAAT detects a high proportion of DNA from non-viable Chlamydia trachomatis. PLoS One 2016;11:e0165920. 10.1371/journal.pone.0165920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hong W, Xiong J, Nyaruaba R, et al. Rapid determination of infectious SARS-cov-2 in PCR-positive samples by SDS-PMA assisted RT-qpcr. Sci Total Environ 2021;797:149085. 10.1016/j.scitotenv.2021.149085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boer A-K, Deneer R, Maas M, et al. Development and validation of an early warning score to identify COVID-19 in the emergency department based on routine laboratory tests: a multicentre case-control study. BMJ Open 2022;12:e059111. 10.1136/bmjopen-2021-059111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leers MPG, Deneer R, Mostard GJM, et al. Use of an algorithm based on routine blood laboratory tests to exclude COVID-19 in a screening-setting of healthcare workers. PLoS One 2022;17:e0270548. 10.1371/journal.pone.0270548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet 2019;393:1577–9. 10.1016/S0140-6736(19)30037-6 [DOI] [PubMed] [Google Scholar]
- 13. Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. The TRIPOD group. Circulation 2015;131:211–9. 10.1161/CIRCULATIONAHA.114.014508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tas J, van Gassel RJJ, Heines SJH, et al. Serial measurements in COVID-19-induced acute respiratory disease to unravel heterogeneity of the disease course: design of the Maastricht intensive care COVID cohort (maastriccht). BMJ Open 2020;10:e040175. 10.1136/bmjopen-2020-040175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bels JLM, van Kuijk SMJ, Ghossein-Doha C, et al. Decreased serial scores of severe organ failure assessments are associated with survival in mechanically ventilated patients; the prospective maastricht intensive care COVID cohort. J Crit Care 2021;62:38–45. 10.1016/j.jcrc.2020.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghossein MA, Driessen RGH, van Rosmalen F, et al. Serial assessment of myocardial injury markers in mechanically ventilated patients with SARS-cov-2 (from the prospective maastriccht cohort). Am J Cardiol 2022;170:118–27. 10.1016/j.amjcard.2022.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heines SJH, van Bussel BCT, Jong MJA, et al. Pulmonary pathophysiology development of COVID-19 assessed by serial electrical impedance tomography in the maastriccht cohort. Sci Rep 2022;12:14517. 10.1038/s41598-022-18843-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hulshof A-M, Brüggemann RAG, Mulder MMG, et al. Serial extem, FIBTEM, and TPA rotational thromboelastometry observations in the maastricht intensive care COVID cohort-persistence of hypercoagulability and hypofibrinolysis despite anticoagulation. Front Cardiovasc Med 2021;8:654174. 10.3389/fcvm.2021.654174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martens B, Driessen RGH, Brandts L, et al. Coronary artery calcifications are associated with more severe multiorgan failure in patients with severe coronavirus disease 2019 infection: longitudinal results of the Maastricht intensive care COVID cohort. J Thorac Imaging 2022;37:217–24. 10.1097/RTI.0000000000000648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mulder MMG, Brandts L, Brüggemann RAG, et al. Serial markers of coagulation and inflammation and the occurrence of clinical pulmonary thromboembolism in mechanically ventilated patients with SARS-cov-2 infection; the prospective maastricht intensive care COVID cohort. Thromb J 2021;19:35. 10.1186/s12959-021-00286-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Linssen J, Ermens A, Berrevoets M, et al. A novel haemocytometric COVID-19 prognostic score developed and validated in an observational multicentre European hospital-based study. Elife 2020;9:e63195. 10.7554/eLife.63195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lippi G, Plebani M. The critical role of laboratory medicine during coronavirus disease 2019 (COVID-19) and other viral outbreaks. Clin Chem Lab Med 2020;58:1063–9. 10.1515/cclm-2020-0240 [DOI] [PubMed] [Google Scholar]
- 23. Martens RJH, van Adrichem AJ, Mattheij NJA, et al. Hemocytometric characteristics of COVID-19 patients with and without cytokine storm syndrome on the Sysmex XN-10 hematology analyzer. Clin Chem Lab Med 2021;59:783–93. 10.1515/cclm-2020-1529 [DOI] [PubMed] [Google Scholar]
- 24. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-ncov) by real-time RT-PCR. Euro Surveill 2020;25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nkuipou-Kenfack E, Engel H, Fakih S, et al. Improving efficiency of viability-PCR for selective detection of live cells. J Microbiol Methods 2013;93:20–4. 10.1016/j.mimet.2013.01.018 [DOI] [PubMed] [Google Scholar]
- 26. RIVM, t.N . Variants of the coronavirus SARS-cov-2 (dutch: varianten van het coronavirus SARS-cov-2); 2022.
- 27. Gorgels KMF, Dingemans J, van der Veer B, et al. Linked nosocomial COVID-19 outbreak in three facilities for people with intellectual and developmental disabilities due to SARS-cov-2 variant B.1.1.519 with spike mutation T478K in the netherlands. BMC Infect Dis 2022;22:139. 10.1186/s12879-022-07121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. R.C.T. R . A language and environment for statistical computing R foundation for statistical computing. 2022. 2020. Available: http://www.r-project.org/index.html
- 29. Wickham H, Averick M, Bryan J, et al. Welcome to the tidyverse. JOSS 2019;4:1686. 10.21105/joss.01686 [DOI] [Google Scholar]
- 30. Bates D. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- 31. Buuren van S, Groothuis-Oudshoorn K. MICE: multivariate imputation by chained equations in R. J Stat Soft 2011;45:1–67. 10.18637/jss.v045.i03 [DOI] [Google Scholar]
- 32. Stekhoven DJ, Bühlmann P. MissForest -- non-parametric missing value imputation for mixed-type data. Bioinformatics 2012;28:112–8. 10.1093/bioinformatics/btr597 [DOI] [PubMed] [Google Scholar]
- 33. Kuhn M. Building predictive models in R using the caret package. J Stat Software 2008;28:1–26. 10.18637/jss.v028.i05 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2022-069455supp001.pdf (49.9KB, pdf)