Abstract
Knowledge of an underlying genetic predisposition to cancer allows the use of personalised prognostic, preventive and therapeutic strategies for the patient and carries clinical implications for family members. Despite great progress, we identified six challenging areas in the management of patients with hereditary cancer predisposition syndromes and suggest recommendations to aid in their resolution. These include the potential for finding unexpected germline variants through somatic tumour testing, optimal risk management of patients with hereditary conditions involving moderate-penetrance genes, role of polygenic risk score in an under-represented Asian population, management of variants of uncertain significance, clinical trials in patients with germline pathogenic variants and technology in genetic counselling. Addressing these barriers will aid the next step forward in precision medicine in Singapore. All stakeholders in healthcare should be empowered with genetic knowledge to fully leverage the potential of novel genomic insights and implement them to provide better care for our patients.
Keywords: Cancer genetics, genetic testing, precision medicine, Singapore
INTRODUCTION
The application of germline genetic testing is becoming more common in cancer treatment selection and hereditary cancer risk evaluation and management in Singapore, with a significant increase in the number of cases referred for genetic counselling.[1] This is likely aided by the rising genetics awareness and literacy amongst both healthcare professionals and the Singaporean public,[2] partly as a result of sustained efforts from the genetics community.[3] Cancer has often been dubbed an aberration of the genome. Even with complete sequencing of the human genome in 2003, we have only begun to scratch the surface in understanding the interconnected pathways of carcinogenesis.[4] Multigene assays allow comprehensive genetic analysis at low cost and improve accessibility to germline testing for patients and at-risk relatives because of more expansive testing criteria. Knowing the sequence is the first step; interpreting the data and translating this into clinical practice is the next major hurdle. As we begin to uncover the multitude of genomic pathways and their role in cancer development, we are beginning to find novel therapeutics with pan-tumour indications. This heralds the onset of precision medicine, which aims to deliver the right drug to the right patient at the right time.[5]
Tumour molecular profiling is the bedrock of precision medicine,[6] having originated in the germline arena. Germline and somatic sequencing in the management of cancer can identify pathogenic variants that affect patient management in several ways. Firstly, knowledge of an underlying cancer predisposition syndrome may impact surgical decision. For example, BRCA1/2 pathogenic variant carriers may opt for upfront risk-reducing bilateral mastectomy and salpingo-oophorectomy to reduce the lifetime risk of breast and ovarian cancers, respectively. Secondly, clinicians may fine-tune patient surveillance with prior knowledge of a cancer predisposition syndrome. As an example, Li-Fraumeni patients with a TP53 pathogenic variant on surveillance should consider minimising radiation exposure and avoid regular surveillance with computed tomography scans because of the increased risk of carcinogenesis.[7,8] Patients with germline pathogenic variants are at increased risk for certain cancers, and gene-directed surveillance is starting to make inroads in this area. The use of whole-body magnetic resonance imaging in patients with Li-Fraumeni syndrome, as demonstrated by the Toronto protocol, has been shown to improve cancer detection and survival rates.[9,10] This intervention amongst high-risk individuals aims to identify cancers early, so that curative surgical resection can be initiated when cancer is at its earliest and most treatable stage. Cancer prevention is a highly attractive option for policymakers because it can reduce healthcare costs and the demand for existing healthcare resources [Figure 1].[11,12]
Figure 1.
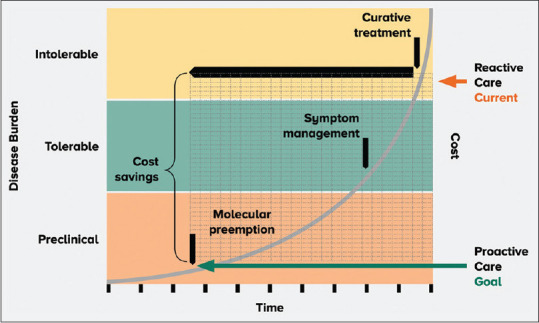
Diagram shows how germline genetic testing allows clinicians to identify high-risk patients early, which translates to proactive care and cost savings for the healthcare system.
The cancer genetics community has had some success in advancing care and broadening knowledge, but we are still facing barriers in other areas. We have chosen six challenging areas to highlight in this review and suggest recommendations to aid in their resolution. We believe that addressing these barriers will be important for the next step forward in precision medicine in Singapore and globally.
POTENTIAL FOR IDENTIFYING GERMLINE VARIANTS THROUGH SOMATIC TUMOUR TESTING
Most molecular diagnostic laboratories have replaced single gene assays with massively parallel high-throughput sequencing, termed next-generation sequencing (NGS), to analyse tumour samples for clinically relevant somatic variants.[13] This technique has changed the sequencing landscape because it allows simultaneous interrogation of multiple genes in several samples with limited amounts of DNA. This improvement in diagnostics saves both time and cost and increases diagnostic yield with its massive sequencing capacity.[13] It is possible to sequence a large number of genes and even whole genomes even though the turnaround time is shorter and the cost is more affordable.[14,15] The challenge is no longer sequencing, but the evaluation and interpretation of the read-out to guide diagnostic, prognostic and treatment decisions.
Increasingly, oncologists have turned to these multigene somatic solid tumour panels to enhance detection of actionable pathogenic variants, using these results to ascertain potential targeted therapies for the specific molecular profile of the cancer.[16,17] Various commercial and academic laboratories provide these NGS capabilities and generate a detailed report of somatic variants found. A joint consensus recommendation from various American medical societies has proposed a four-tiered system to categorise these somatic sequence variations based on their clinical significance: variants with strong clinical significance (tier I), variants with potential clinical significance (tier II), variants of unknown clinical significance (tier III), and variants deemed benign or likely benign (tier IV).[18] However, there is a lack of uniform classification of somatic variants[18,19,20] and variable selection of genes,[21] and the clinical significance of a large proportion of the variants is still unknown. Laboratories may have differing classifications, depending on the subjective analysis of each laboratory. This subjectivity can result in varied classifications and clinical interpretation of the same variant. Consequently, this leads to a heterogeneous mix of data in precision medicine clinical trials, which makes generalisability and application of findings difficult. Molecular tumour boards, comprising oncologists, pathologists, geneticists and scientists, have thus been convened to discuss and pair patients’ unique tumour profiles with appropriately selected targeted interventions to enhance treatment outcomes. One such example is the ongoing genomically matched IMPACT trial at the National Cancer Centre Singapore.[22]
Increased use of NGS in tumour samples not only uncovers somatic genetic defects, but it may also suggest a germline cause that may not be expected by the patient or healthcare provider. The American College of Medical Genetic and Genomics (ACMG) has introduced three iterations of its recommendations for significant incidental findings (ACMG secondary findings v3.0), which should be explained to the patient when returning the somatic test results [Table 1].[23] These findings ought to trigger a discussion with a genetics professional in view of the possibility of an underlying hereditary cancer predisposition syndrome. It is important for clinicians ordering somatic tumour panels to be cognisant of findings that may herald an underlying hereditary cancer predisposition syndrome, inform the patient of this possibility and refer the patient to a cancer genetics clinic for evaluation when this is suspected.[1]
Table 1.
Genes related to cancer phenotypes in the American College of Medical Genetics and Genomics secondary findings v3.0 gene list.
| Phenotype | Gene | Inheritance |
|---|---|---|
| Familial adenomatous polyposis | APC | AD |
|
| ||
| Multiple endocrine neoplasia type 1, familial medullary thyroid cancer | RET | AD |
|
| ||
| Hereditary breast and/or ovarian cancer | BRCA1, BRCA2, PALB2 | AD |
|
| ||
| Hereditary paraganglioma-phaeochromocytoma syndrome | SDHD, SDHA, SDHAF2, SDHC, SDHB, MAX, TMEM127 | AD |
|
| ||
| Juvenile polyposis syndrome | BMPR1A, SMAD4 | AD |
|
| ||
| Li-Fraumeni syndrome | TP53 | AD |
|
| ||
| Lynch syndrome | MLH1, MSH2, MSH6, PMS2, EPCAM | AD |
|
| ||
| Multiple endocrine neoplasia type 1 | MEN1 | AD |
|
| ||
| MUTYH-associated polyposis | MUTYH | AR |
|
| ||
| Neurofibromatosis type 2 | NF2 | AD |
|
| ||
| Peutz-Jeghers syndrome | STK11 | AD |
|
| ||
| PTEN hamartoma tumour syndrome | PTEN | AD |
|
| ||
| Retinoblastoma | RB1 | AD |
|
| ||
| Tuberous sclerosis syndrome | TSC1, TSC2 | AD |
|
| ||
| Von Hippel-Lindau syndrome | VHL | AD |
|
| ||
| WT1-related Wilms tumour | WT1 | AD |
AD: autosomal dominant, AR: autosomal recessive
A germline pathogenic variant confirms a hereditary aetiology and has implications for both the patient and his/her family. Thus, individuals considering germline testing should undergo pretest genetic counselling to understand the significance of this.[1] Genetic counselling involves more than just relay of information — it addresses the psychosocial impact of hereditary conditions.[24] With the availability of pretest genetic counselling, time is spent with patients and their families to conduct assessments, provide information, address doubts and concerns and promote informed decision-making, whilst providing the necessary emotional support. This improves patient outcomes, including empowerment, positive behavioural change and decisional satisfaction.[25–28] Furthermore, this triggers a cascade effect where patients reported to have a germline pathogenic variant confirming a hereditary cause can share the genetics knowledge gained from the consult with their extended families to help with downstream risk management of at-risk relatives.[29–31] The overall goal of genetic testing is to promote preventive medicine for the patient and his/her family by tailoring an individualised management plan based on his/her cancer risk.[32] Germline genetic testing allows clinicians to identify high-risk patients early, which translates to proactive care and cost savings for the healthcare system [Figure 1]. Pretest and posttest genetic counselling ensure that accurate information is relayed and both time and space are provided for patients and their families to make an informed decision on germline testing.[27] In a multilingual country such as Singapore, English may be the working language, but not necessarily the native language of the patient. Healthcare providers should seek to explain genetics concepts at a level appropriate to patients’ understanding and in a language that the patient is comfortable in, supplementing with visual aids such as charts and diagrams to improve information retention. In addition, cultural sensitivity requires healthcare professionals to understand the strong impact of language and culture on health beliefs and decisions,[33] tailoring each consult while keeping in mind nuanced unwritten cultural norms. The importance of pretest counselling by trained healthcare providers prior to germline genetic testing cannot be overstated.
OPTIMAL MANAGEMENT OF MODERATE-PENETRANCE GENES
Multigene NGS panels are commonly used to test for germline pathogenic variants in the cancer genetics clinic. The use of multigene panels in germline evaluation has added to the body of literature on moderate-penetrance genes. There have been concerns regarding the clinical validity and actionability of these moderate-penetrance genes because of limited understanding of the cancer risks involved.[34] The appropriate management of individuals harbouring these moderate-penetrance genes is unclear because the cancer risk estimates are lower than those for high-penetrance genes such as BRCA1 and BRCA2, but remain elevated compared to the general population. Recent publications of large consortiums[35,36] have refined risk estimates of breast cancer risk in genes other than BRCA1/2. This has helped to give patients a more accurate picture of relative risk with regard to moderate-penetrance breast genes [Figure 2]. Armed with this information, patients can make a more informed choice in deciding whether to include these moderate-penetrance breast genes in their panel testing, which often raises cancer risk in tandem with environmental and other exogenous risk factors. The availability of more data has helped to refine management guidelines for individuals with variants in moderate-penetrance cancer-susceptibility genes, instead of extrapolating management from high-penetrant genes, which could bring about substantial harm as a result of needless risk-reducing surgery and overly intensive surveillance protocols. Clinical guidelines for these moderate-penetrance genes are beginning to emerge, an example being the PALB2 management guidelines released by the ACMG.[37]
Figure 2.
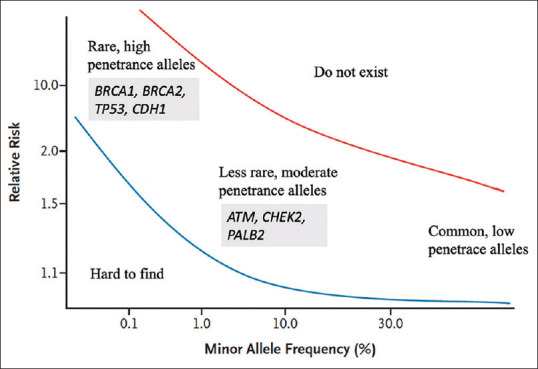
Graph shows breast cancer susceptibility loci and genes.
Guidelines for germline genetic testing are constantly evolving and expanding. The National Comprehensive Cancer Network (NCCN) has issued five revisions to the ‘Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic’ in the past 2 years.[38] There have been recent discussions to re-frame our focus from ‘who to test’ to ‘who not to test’.[39] This is testament to the potential impact that precision medicine has in stall for the oncology community. Yet this statement is fraught with controversy.[40] A major limitation of genetic studies is that they are specific to populations and generalisability cannot be assumed.[41,42] Partly as a result of their lack of generalisability, indications for germline testing can differ from region to region, as evidenced by the differing European Society of Medical Oncology (ESMO) and NCCN guidelines on germline testing in pancreas[43,44] and prostate cancer.[45,46]
A method of addressing this uncertainty in NGS panel selection is to actively involve the patient in a shared decision-making approach.[25] Shared decision-making is a process with three phases of communication: (1) choice talk, where the patient is made aware that choice exists; (2) option talk, where patients are informed about the NGS panel options in detail; and (3) decision talk, where patient preferences are discussed and the decision supported by exploring ‘what matters most’ to the patient.[47] During this process, patients journey from their initial preferences to informed preferences through deliberation. The healthcare provider shares information with the patient on possible panel selections, based on indication and utility. The testing options, potential benefits and limitations are presented and discussed whilst considering the patient's own values and preferences. With this approach, decisions regarding medical interventions are made collaboratively between the clinician and patient. Shared decision-making can empower the patient in making a choice that he/she deems optimal.
There is space for our own local recommendations,[48] yet this must take into account the speed of knowledge generation in genetics. There may be a need for regular revisions to keep the local guidelines up to date, because the last update to our local guidelines was in 2015.[48] It is prudent for clinicians to be cognisant of new data and keep abreast of the latest development in genetics and its impact on their patients.
ROLE OF POLYGENIC RISK SCORES IN CANCER RISK MANAGEMENT
Polygenic risk scores (PRSs) are a matter of great interest in the genetics community. A PRS is calculated from the total effect size of all relevant genetic variants estimated from a genome-wide association study (GWAS) and pertains to an individual's risk for disease based on a combination of genetic factors drawn from the population it is studied in. The promise of personalised risk for an individual is exciting and holds great potential, yet several key barriers remain unaddressed. PRSs are ancestry specific, and the reference population is critical in its interpretation. This is especially relevant for a diverse population such as Singapore, where our ancestors hail from different parts of Asia with differing ancestral genetic backgrounds. A previous study has shown that PRSs are not portable across populations because there is limited concordance between different ancestries.[49] Differing patterns of linkage disequilibrium will result in different tagging of causal variants.[50] Different genetic architectures with different causal variants and different effect sizes in different populations all add to the complexity of PRS interpretation in our heterogeneous Singaporean population. It will take more time and data to establish a reliable population reference before PRS can be brought to the frontline in multi-ethnic Singapore.
Currently, flagship population databases such as the gnomAD under-represent the Asian population, with South and East Asians combined constituting only 17.9%.[51] There is no category for South-East Asians. Similarly, there is a marked European bias in individuals in the GWAS catalogue,[50] with 78% Europeans and only 10% Asians included. Other reference databases such as 1000 Genomes show a similarly small representation of the Asian population. The lack of global representation hampers our ability to understand the genetic background of carcinogenesis in our Asian population, which results in inaccurate evaluation of pathogenic variants in clinical genetic studies. In turn, this leads to suboptimal and even potentially incorrect development of public health policy. Any attempts to use PRS from European-based studies in non-European populations may result in inaccurate risk assessment and inappropriate interventions. Because most data come from studies in populations of European descent, it is vital to collect our own data in Singapore to understand the risks in the local population and to tailor population guidelines. Furthermore, PRS may be oversimplifying the complexities of carcinogenesis. The role of endogenous genomic alterations may not paint the full picture in oncogenesis. Environmental and external influences such as diet and lifestyle may play an equal, if not greater, role in cancer development. Thus, more data need to be accumulated and evaluated before PRS is ready for mainstream practice.
MANAGEMENT OF VARIANTS OF UNCERTAIN SIGNIFICANCE
The optimal management of variants of uncertain significance (VUS) is a difficult area for both clinicians and patients. Local data have shown that up to 40% of patients who undergo genetic testing will receive a VUS result.[52] This is largely a result of the lack of Asian representation in the reference database, which was derived primarily from studies on populations of European descent, resulting in a higher proportion of VUS results compared to that in the UK and US, which have a VUS rate of approximately 20%. VUS management is challenging because of its uncertain nature. Even though the majority of VUS are reclassified to benign/likely benign variants, the number of families, and thus individuals, who are affected by reclassification is not insignificant. In a previous study, our group has shown that up to one third of patients with reclassified variants have actionable results that necessitate a change in medical management.[52] This not only affects that patient but has a cascade effect on all family members with the reclassified variant.
Variants are classified from pathogenic to benign based partly on information from reference genomic data. Because Asians have limited representation in the reference database, we identify fewer pathogenic and benign variants and instead get more VUS results, where the clinical significance and implications for disease have yet to be resolved. It is certainly hoped that with more data, the clinical relevance of all encountered genomic variants will be readily predictable, rendering the diagnostic designation ‘VUS’ obsolete and helping to clarify risk management.[53] Individual genomes have VUS mainly because of limited ability to interpret missense variants. Currently, computational predictions can only predict possible outcomes. Functional evidence of an effect on protein function is needed to conclude if a variant is pathogenic or benign. This has sparked off the entire field of proteomics, but the data set in proteins is vast. What we need are high-throughput assays to interrogate the effect of every possible variant in a gene of interest. Novel gene editing and synthesis methods[54] have now made this possible for BRCA1 and may pave the way in other genes to determine if a VUS is responsible for a hereditary cancer predisposition syndrome.
VUS can be challenging to interpret and manage for both the patient and healthcare provider. A clinical report of a VUS that is given back to the patient may state that ‘there is not enough information to determine if the variant contributes to disease or is benign’. A BRCA1/2 VUS may cause great unease in a patient with a personal history of breast cancer, for example, and she may opt for a risk-reducing mastectomy even if there is no medical need to do so. VUS often cause patients distress, anxiety and worry. Additionally, if the clinical significance of the VUS is incorrectly interpreted by the healthcare provider as disease causing, this may result in misdiagnosis and mismanagement.
From the clinician perspective, it is important to follow up selected patients with VUS results and ensure that they are empowered to understand the intricate interpretation and management of a VUS result.[52] It is a matter of debate whether the responsibility of reporting a VUS reclassification lies with the genetics service or the patient. In our local study, the median time to reclassification was found to be 1 year, though some reclassifications took up to 4 years.[52] We recommend that the genetics service puts in place a system to regularly review VUS and recall patients if reclassification occurs.[52] This will ensure that any updates in management are communicated in a timely manner.
GERMLINE GENE-DIRECTED TRIALS
Beyond cancer prevention, knowledge of an underlying hereditary cancer predisposition syndrome can help clinicians initiate the most effective therapy and inform on reproductive risks. Therapeutic needs have not only increased the demand for somatic testing, but have also similarly driven requests for germline testing as well. The number of patients who underwent germline genetic testing has grown locally and globally,[55] with rising demand for genetic counsellors.[3,56] Since 2014, there has been a significant increase in the number of cases referred for genetic counselling in Singapore as a result of the demand for genetic testing for treatment-based decisions.[1] More genetics-trained personnel will be needed in the years to come to meet the demand for services.
Drugs such as poly (ADP-ribose) polymerase (PARP) inhibitors have demonstrated improved survival in individuals with germline BRCA1/2 pathogenic variants with ovarian[57,58] and breast cancer,[59,60] with expanding indications from the metastatic to the adjuvant setting. There are also a range of tumour site–agnostic molecular features, such as microsatellite instability, which may select for patients who have the potential for dramatic benefit with immune checkpoint inhibitors such as pembrolizumab.[61] In addition, germline pathogenic variants can introduce potential vulnerabilities to the cancers that arise as part of the hereditary cancer syndrome. These intrinsic vulnerabilities may be targeted by novel drug classes in clinical trials. Examples include mitogen-activated protein kinase kinase (MEK) inhibitors, selumetinib[62,63] and trametinib,[64] which have demonstrated response in patients with neurofibromatosis type 1 (NF1)-associated tumours, such as progressive low-grade gliomas and inoperable plexiform neurofibromas. MEK inhibition may represent a new therapeutic option in a disease population with previously limited treatment options. In addition, patients with Von Hippel–Lindau (VHL) disease have a high incidence of renal cell carcinoma owing to VHL gene inactivation and constitutive activation of the transcription factor hypoxia-inducible factor 2α (HIF-2α). The HIF-2α inhibitor belzutifan has shown responses in VHL patients with renal cell carcinoma, pancreatic neuroendocrine tumours and central nervous system hemangioblastomas.[65]
TECHNOLOGY-ENABLED GENETIC COUNSELLING
A well-documented pedigree takes centre stage for determining which individuals might be at higher risk for a cancer predisposition syndrome. It is essentially free, save for manpower and time. Clinicians should be aware of red flags, including rare cancers such as adrenocortical carcinoma, cancers at a young age, synchronous or metachronous malignancies or a strong family history of cancer that require a genetics referral to assess if there is a hereditary component to their personal and/or family history of cancer. Electronic medical records can trigger alerts to remind the clinician to make a referral to the cancer genetics clinic when a diagnosis worrisome for a cancer predisposition syndrome is found. However, the absence of a family history of malignancy does not rule out the possibility of an underlying cancer predisposition syndrome because of the possibility of de novo variants, moderate-penetrance genes, recessive and other inheritance patterns, lack of information and variable expressivity. Unfortunately, current clinical platforms are not designed to capture family history. It is difficult to succinctly illustrate a pedigree on the available electronic healthcare platforms in Singapore, resulting in hardcopy pedigrees that are in opposition to the overall drive for digitisation of healthcare records. A possible solution is for chatbots, which can collect family history, to provide educational information on genetic testing and answer frequently asked questions before the genetics consult. This frees up time and leads to greater accessibility of genetic counselling services. In Singapore, family history is sporadically captured and typically distributed across multiple individuals and providers without linkage information. Tools to compile pedigree data, and make it accessible and usable during a clinic consult, are essential, so primary care providers and specialists can identify a potential patient at risk for cancer predisposition syndromes and refer the individual to a genetics service for a discussion on genetic testing. Even though Singapore is a small island state, family members may be seen at different hospitals depending on their household address, and we lack an integrated platform to link pedigrees and determine if a prior pathogenic germline variant has been found in a family member or distinguish which family members have been seen for testing and who needs follow-up. The lack of a centralised genetic registry means patients may be overlooked, leading to missed opportunities for intervention and the potential to prevent disease.
The COVID-19 pandemic has changed the way we deliver care. We need to review our current clinical care models to include genetic counselling using video consultation, which has shown encouraging uptake rates.[66] Remote genetic counselling using video or telephone has brought convenience to patients, and a prior study showed that patients are receptive towards this novel service model in Singapore.[66] Telephone consultation was shown to be non-inferior to face-to-face genetic counselling.[67] Remote counselling using video or telephone reduces travelling and wait time in the hospital for the patient, results in cost savings for the patient and the healthcare system, encourages family members to participate in the discussion when required[66,67] and provides access of care to underserved populations. Furthermore, remote genetic counselling opens up the possibility of group genetic counselling for the family, which has been shown to reduce time spent by the provider and increase patient satisfaction.[68] However, advances in technology need to be reflected with parallel innovations in healthcare reimbursement.
CONCLUSION
The regular use of genomic information will transition from boutique to mainstream in all clinical settings, making genomic testing routine.[53] We look forward to an established electronic database to capture the wealth of somatic and germline information and using this information to further advance patient care. National efforts are ongoing to set up a centralised genomic medicine platform to integrate local efforts to ensure coordination amongst the multiple stakeholders. All stakeholders along the healthcare journey must be equipped and empowered to bring genomic intelligence to the patient. This means that providers such as family physicians, specialists, ministry and healthcare administrators all play an equally important role to alter the trajectory of cancer. This is especially relevant in cancer care where early detection and treatment leads to improved outcomes.
To fully leverage these breakthroughs, the healthcare community must be ready to accommodate these new findings, workflows and data sets from novel genomic insights and implement them in daily clinics and practice.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Chiang J, Ngeow J. The management of BRCA1 and BRCA2 carriers in Singapore. Chin Clin Oncol. 2020;9:62. doi: 10.21037/cco-20-104. [DOI] [PubMed] [Google Scholar]
- 2.Chin T-M, Tan S-H, Lim S-E, Iau P, Yong W-P, Wong S-W, et al. Acceptance, motivators, and barriers in attending breast cancer genetic counseling in Asians. Cancer Detect Prev. 2005;29:412–8. doi: 10.1016/j.cdp.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 3.Abacan M, Alsubaie L, Barlow-Stewart K, Caanen B, Cordier C, Courtney E, et al. The global state of the genetic counseling profession. Eur J Hum Genet. 2019;27:183–97. doi: 10.1038/s41431-018-0252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams E. Right Drug—Right Patient—Right Time: Personalized medicine coalition. Clin Transl Sci. 2008;1:11–2. doi: 10.1111/j.1752-8062.2008.00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dugger SA, Platt A, Goldstein DB. Drug development in the era of precision medicine. Nat Rev Drug Discov. 2018;17:183–96. doi: 10.1038/nrd.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frebourg T, Bajalica Lagercrantz S, Oliveira C, Magenheim R, Evans DG European Reference Network GENTURIS. Guidelines for the Li-Fraumeni and heritable TP53-related cancer syndromes. Eur J Hum Genet. 2020;28:1379–86. doi: 10.1038/s41431-020-0638-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mai PL, Best AF, Peters JA, DeCastro RM, Khincha PP, Loud JT, et al. Risks of first and subsequent cancers among TP53 mutation carriers in the National Cancer Institute Li-Fraumeni syndrome cohort. Cancer. 2016;122:3673–81. doi: 10.1002/cncr.30248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villani A, Shore A, Wasserman JD, Stephens D, Kim RH, Druker H, et al. Biochemical and imaging surveillance in germline TP53 mutation carriers with Li-Fraumeni syndrome: 11 year follow-up of a prospective observational study. Lancet Oncol. 2016;17:1295–305. doi: 10.1016/S1470-2045(16)30249-2. [DOI] [PubMed] [Google Scholar]
- 10.Ballinger ML, Best A, Mai PL, Khincha PP, Loud JT, Peters JA, et al. Baseline surveillance in Li-Fraumeni syndrome using whole-body magnetic resonance imaging: A meta-analysis. JAMA Oncol. 2017;3:1634–9. doi: 10.1001/jamaoncol.2017.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray F, Jemal A, Torre LA, Forman D, Vineis P. Long-term realism and cost-effectiveness: Primary prevention in combatting cancer and associated inequalities worldwide. J Natl Cancer Inst. 2015;107:djv273. doi: 10.1093/jnci/djv273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soerjomataram I, Bray F. Planning for tomorrow: Global cancer incidence and the role of prevention 2020-2070. Nat Rev Clin Oncol. 2021;18:663–72. doi: 10.1038/s41571-021-00514-z. [DOI] [PubMed] [Google Scholar]
- 13.Luthra R, Chen H, Roy-Chowdhuri S, Singh RR. Next-generation sequencing in clinical molecular diagnostics of cancer: Advantages and challenges. Cancers. 2015;7:2023–36. doi: 10.3390/cancers7040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S-T, Yuen J, Zhou K, Ishak NDB, Chen Y, Met-Domestici M, et al. Impact of subsidies on cancer genetic testing uptake in Singapore. J Med Genet. 2017;54:254–9. doi: 10.1136/jmedgenet-2016-104302. [DOI] [PubMed] [Google Scholar]
- 15.Courtney E, Chok AK-L, Ang ZLT, Shaw T, Li S-T, Yuen J, et al. Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom Med. 2019;4:22. doi: 10.1038/s41525-019-0096-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31:1491–505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
- 17.Colomer R, Mondejar R, Romero-Laorden N, Alfranca A, Sanchez-Madrid F, Quintela-Fandino M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine. 2020;25:100487. doi: 10.1016/j.eclinm.2020.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jennings LJ, Arcila ME, Corless C, Kamel-Reid S, Lubin IM, Pfeifer J, et al. Guidelines for validation of next-generation sequencing-based oncology panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19:341–65. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froyen G, Le Mercier M, Lierman E, Vandepoele K, Nollet F, Boone E, et al. Standardization of somatic variant classifications in solid and haematological tumours by a two-level approach of biological and clinical classes: An initiative of the Belgian ComPerMed expert panel. Cancers. 2019;11:2030. doi: 10.3390/cancers11122030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giri VN, Knudsen KE, Kelly WK, Cheng HH, Cooney KA, Cookson MS, et al. Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol Off J Am Soc Clin Oncol. 2020 Aug;20(38):2798–811. doi: 10.1200/JCO.20.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seet AOL, Tan AC, Tan TJ, Ng MCH, Tai DWM, Lam JYC, et al. Individualized molecular profiling for allocation to clinical trials Singapore study—An Asian tertiary cancer center experience. JCO Precis Oncol. 2021;5 doi: 10.1200/PO.20.00261. PO.20.00261. doi: 10.1200/PO.20.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller DT, Lee K, Chung WK, Gordon AS, Herman GE, Klein TE, et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23:1381–90. doi: 10.1038/s41436-021-01172-3. [DOI] [PubMed] [Google Scholar]
- 24.Austin J, Semaka A, Hadjipavlou G. Conceptualizing genetic counseling as psychotherapy in the era of genomic medicine. J Genet Couns. 2014;23:903–9. doi: 10.1007/s10897-014-9728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Courtney E, Li S-T, Shaw T, Chen Y, Allen JC, Ngeow J. Predictors of next-generation sequencing panel selection using a shared decision-making approach. NPJ Genom Med. 2018;3:1–7. doi: 10.1038/s41525-018-0050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuen J, Lee SY, Courtney E, Lim J, Soh H, Li ST, et al. Evaluating empowerment in genetic counseling using patient-reported outcomes. Clin Genet. 2020;97:246–56. doi: 10.1111/cge.13646. [DOI] [PubMed] [Google Scholar]
- 27.Yuen J, Fung SM, Sia CL, Venkatramani M, Shaw T, Courtney E, et al. An in-depth exploration of the post-test informational needs of BRCA1 and BRCA2 pathogenic variant carriers in Asia. Hered Cancer Clin Pract. 2020;18:22. doi: 10.1186/s13053-020-00154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun S, Li S-T, Ngeow J. Factors shaping at-risk individuals’ decisions to undergo genetic testing for cancer in Asia. Health Soc Care Community. 2020;28:1569–77. doi: 10.1111/hsc.12981. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman S, Lahad A, Tomer A, Koka S, BenUziyahu M, Raz A, et al. Familial communication and cascade testing among relatives of BRCA population screening participants. Genet Med. 2018;20:1446–54. doi: 10.1038/gim.2018.26. [DOI] [PubMed] [Google Scholar]
- 30.Li S-T, Sun S, Lie D, Met-Domestici M, Courtney E, Menon S, et al. Factors influencing the decision to share cancer genetic results among family members: An in-depth interview study of women in an Asian setting. Psychooncology. 2018;27:998–1004. doi: 10.1002/pon.4627. [DOI] [PubMed] [Google Scholar]
- 31.Chiang J, Yuen J, Shaw T, Goh HX, Li S-T, Courtney E, et al. Predictive testing for tumor predisposition syndromes in pediatric relatives: An Asian experience. Front Pediatr. 2020;8:568528. doi: 10.3389/fped.2020.568528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Courtney E, Chin XW, Yuen J, Li S-T, Chen Y, Allen JC, et al. Risk management adherence following genetic testing for hereditary cancer syndromes: A Singaporean experience. Fam Cancer. 2018;17:621–6. doi: 10.1007/s10689-018-0071-9. [DOI] [PubMed] [Google Scholar]
- 33.Shaw T, Ishak D, Lie D, Menon S, Courtney E, Li S-T, et al. The influence of Malay cultural beliefs on breast cancer screening and genetic testing: A focus group study. Psychooncology. 2018;27:2855–61. doi: 10.1002/pon.4902. [DOI] [PubMed] [Google Scholar]
- 34.Tung N, Domchek SM, Stadler Z, Nathanson KL, Couch F, Garber JE, et al. Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol. 2016;13:581–8. doi: 10.1038/nrclinonc.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu C, Hart SN, Gnanaolivu R, Huang H, Lee KY, Na J, et al. A population-based study of genes previously implicated in breast cancer. N Engl J Med. 2021;384:440–51. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, et al. Breast Cancer Association Consortium. Breast cancer risk genes-association analysis in more than 113,000 women. N Engl J Med. 2021;384:428–39. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tischkowitz M, Balmaña J, Foulkes WD, James P, Ngeow J, Schmutzler R, et al. Management of individuals with germline variants in PALB2: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG) Genet Med. 2021;23:1416–23. doi: 10.1038/s41436-021-01151-8. [DOI] [PubMed] [Google Scholar]
- 38.Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Cancer Netw. 2021;19:77–102. doi: 10.6004/jnccn.2021.0001. [DOI] [PubMed] [Google Scholar]
- 39.Lincoln SE, Nussbaum RL, Kurian AW, Nielsen SM, Das K, Michalski S, et al. Yield and utility of germline testing following tumor sequencing in patients with cancer. JAMA Netw Open. 2020;3:e2019452. doi: 10.1001/jamanetworkopen.2020.19452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yadav S, Couch FJ. Germline genetic testing for breast cancer risk: The past, present, and future. Am Soc Clin Oncol Educ Book Am Soc Clin Oncol Annu Meet. 2019;39:61–74. doi: 10.1200/EDBK_238987. [DOI] [PubMed] [Google Scholar]
- 41.van Rooij JGJ, Jhamai M, Arp PP, Nouwens SCA, Verkerk M, Hofman A, et al. Population-specific genetic variation in large sequencing data sets: Why more data is still better. Eur J Hum Genet. 2017;25:1173–5. doi: 10.1038/ejhg.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu D, Dou J, Chai X, Bellis C, Wilm A, Shih CC, et al. Large-scale whole-genome sequencing of three diverse asian populations in Singapore. Cell. 2019;179:736–49.e15. doi: 10.1016/j.cell.2019.09.019. [DOI] [PubMed] [Google Scholar]
- 43.Tempero MA. NCCN Guidelines Updates: Pancreatic cancer. J Natl Compr Cancer Netw. 2019;17:603–5. doi: 10.6004/jnccn.2019.5007. [DOI] [PubMed] [Google Scholar]
- 44.Stjepanovic N, Moreira L, Carneiro F, Balaguer F, Cervantes A, Balmaña J, et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann Oncol. 2019;30:1558–71. doi: 10.1093/annonc/mdz233. [DOI] [PubMed] [Google Scholar]
- 45.Parker C, Castro E, Fizazi K, Heidenreich A, Ost P, Procopio G, et al. Prostate cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol Off J Eur Soc Med Oncol. 2020;31:1119–34. doi: 10.1016/j.annonc.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 46.Schaeffer E, Srinivas S, Antonarakis ES, Armstrong AJ, Bekelman JE, Cheng H, et al. NCCN Guidelines Insights: Prostate cancer, Version 1.2021: Featured updates to the NCCN Guidelines. J Natl Compr Canc Netw. 2021;19:134–43. doi: 10.6004/jnccn.2021.0008. [DOI] [PubMed] [Google Scholar]
- 47.Elwyn G, Frosch D, Thomson R, Joseph-Williams N, Lloyd A, Kinnersley P, et al. Shared decision making: A model for clinical practice. J Gen Intern Med. 2012;27:1361–7. doi: 10.1007/s11606-012-2077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singapore Cancer Network (SCAN) Cancer Genetics Workgroup. Singapore Cancer Network (SCAN) Guidelines for Referral for Genetic Evaluation of Common Hereditary Cancer Syndromes. Ann Acad Med Singapore. 2015;44:492–510. [PubMed] [Google Scholar]
- 49.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51:584–91. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sirugo G, Williams SM, Tishkoff SA. The missing diversity in human genetic studies. Cell. 2019;177:26–31. doi: 10.1016/j.cell.2019.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiang J, Chia TH, Yuen J, Shaw T, Li S-T, Binte Ishak ND, et al. Impact of variant reclassification in cancer predisposition genes on clinical care. JCO Precis Oncol. 2021;5:577–84. doi: 10.1200/PO.20.00399. [DOI] [PubMed] [Google Scholar]
- 53.Green ED, Gunter C, Biesecker LG, Di Francesco V, Easter CL, Feingold EA, et al. Strategic vision for improving human health at The Forefront of Genomics. Nature. 2020;586:683–92. doi: 10.1038/s41586-020-2817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Findlay GM, Daza RM, Martin B, Zhang MD, Leith AP, Gasperini M, et al. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–22. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurian AW, Ward KC, Abrahamse P, Bondarenko I, Hamilton AS, Deapen D, et al. Time trends in receipt of germline genetic testing and results for women diagnosed with breast cancer or ovarian cancer, 2012-2019. J Clin Oncol Off J Am Soc Clin Oncol. 2021;39:1631–40. doi: 10.1200/JCO.20.02785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bamshad MJ, Magoulas PL, Dent KM. Genetic counselors on the frontline of precision health. Am J Med Genet C Semin Med Genet. 2018;178:5–9. doi: 10.1002/ajmg.c.31610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–84. doi: 10.1016/S1470-2045(17)30469-2. [DOI] [PubMed] [Google Scholar]
- 58.Moore K, Colombo N, Scambia G, Kim B-G, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 59.Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–33. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 60.Johnston SRD, Harbeck N, Hegg R, Toi M, Martin M, Shao ZM, et al. Abemaciclib combined with endocrine therapy for the adjuvant treatment of HR+, HER2-, node-positive, high-risk, early breast cancer (monarchE) J Clin Oncol Off J Am Soc Clin Oncol. 2020;38:3987–98. doi: 10.1200/JCO.20.02514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dombi E, Baldwin A, Marcus LJ, Fisher MJ, Weiss B, Kim A, et al. Activity of selumetinib in neurofibromatosis Type 1-related plexiform neurofibromas. N Engl J Med. 2016;375:2550–60. doi: 10.1056/NEJMoa1605943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gross AM, Wolters P, Baldwin A, Dombi E, Fisher MJ, Weiss BD, et al. SPRINT: Phase II study of the MEK 1/2 inhibitor selumetinib (AZD6244, ARRY-142886) in children with neurofibromatosis type 1 (NF1) and inoperable plexiform neurofibromas (PN) J Clin Oncol. 2018;36(15 Suppl):10503. [Google Scholar]
- 64.McCowage GB, Mueller S, Pratilas CA, Hargrave DR, Moertel CL, Whitlock J, et al. Trametinib in pediatric patients with neurofibromatosis type 1 (NF-1)–associated plexiform neurofibroma: A phase I/IIa study. J Clin Oncol. 2018;36(15 Suppl):10504. [Google Scholar]
- 65.Jonasch E, Donskov F, Iliopoulos O, Rathmell WK, Narayan VK, Maughan BL, et al. Belzutifan for renal cell carcinoma in von Hippel–Lindau disease. N Engl J Med. 2021;385:2036–46. doi: 10.1056/NEJMoa2103425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sim J, Shaw T, Li S-T, Courtney E, Yuen J, Chiang J, et al. Understanding patients’ views and willingness toward the use of telehealth in a cancer genetics service in Asia. J Genet Couns. 2021;30:1658–70. doi: 10.1002/jgc4.1432. [DOI] [PubMed] [Google Scholar]
- 67.Schwartz MD, Valdimarsdottir HB, Peshkin BN, Mandelblatt J, Nusbaum R, Huang A-T, et al. Randomized noninferiority trial of telephone versus in-person genetic counseling for hereditary breast and ovarian cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2014;32:618–26. doi: 10.1200/JCO.2013.51.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hynes J, MacMillan A, Fernandez S, Jacob K, Carter S, Predham S, et al. Group plus “mini” individual pre-test genetic counselling sessions for hereditary cancer shorten provider time and improve patient satisfaction. Hered Cancer Clin Pract. 2020;18:3. doi: 10.1186/s13053-020-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]