CRISPR technology has revolutionized the biological research world, making animals heretofore recalcitrant to genetic manipulation, accessible to analysis of specific gene functions. Building upon the demonstration of targeted gene mutations in the sea urchin (CRISPR knock-out) (Fleming et al., 2021; Lin et al., 2019; Lin & Su, 2016; Liu et al., 2019; Vyas et al., 2022), investigators may now be able to insert exogenous DNA into specific locations in the genome (CRISPR knock-in). Such Cas9-mediated knock-ins will reveal sites of gene expression, and function. By judicious selection of exogenously encoded tags e.g. a fluorescent reporter, an investigator may then follow specific gene activities and cell lineages throughout development in live embryos. This tag can also be used for protein pull-down without requiring an antibody for the targeted protein. Here we describe a procedure for CRISPR-based knock-in DNA in the sea urchin Strongylocentrotus purpuratus.
Sea urchin larvae produce echinochrome pigments that require several gene functions including the enzyme polyketide synthase 1 (PKS) (Barsi et al., 2015; Calestani et al., 2003; Calestani & Wessel, 2018; Perillo et al., 2020; Wessel et al., 2020). Sp PKS1 expression is restricted to a small population of ~50 cells of the Veg2 lineage of the animal (Calestani et al., 2003; Barsi et al., 2015). We realized that using PKS1 to evaluate CRISPR knock-in success was highly stringent since the insertion must occur within that small lineage, and be expressed by yet a smaller population of the lineage. Mutations of the gene encoding PKS1 by CRISPR knock-out resulted in albino larvae, an easy phenotype to assess with simple brightfield microscopy (Oulhen & Wessel, 2016a). A single gRNA was previously shown to mutate PKS1 by Cas9 activity, nearly 100% of the time in embryos from S. purpuratus and Hemicentrotus pulcherrimus (Liu et al., 2019; Oulhen et al., 2022; Oulhen & Wessel, 2016a). We took advantage of this highly efficient gRNA to test and to optimize Cas9-mediated methodology in the sea urchin Strongylocentrotus purpuratus.
We tested three different donor templates for their efficacy in selectively knocking-in exogenous DNA encoding a fluorescent protein: plasmid DNA, linear double stranded DNA, single stranded DNA. The key for this test is a highly efficient gRNA against the target gene, and a DNA repair template that contains homologous regions to the target sequence for homology directed repair (SFigure.1).
Investigators have previously injected linear DNA into sea urchin eggs/early embryos, which results in rapid and extensive concatenation (McMahon et al., 1985) that appears to be detrimental to high-fidelity insertion (data not shown). To counter this concern, we tested circular plasmid – based strategies. Here the DNA repair template targeting the cleaved genomic locus was contained within a plasmid and was accessible for insertion before or following CRISPR-Cas9 cutting of the same flanking sequence in the plasmid as in the targeted genomic locus. This strategy resulted in GFP insertions into PKS1, but was inconsistent for reasons not yet clear (SFigures 2 and 3).
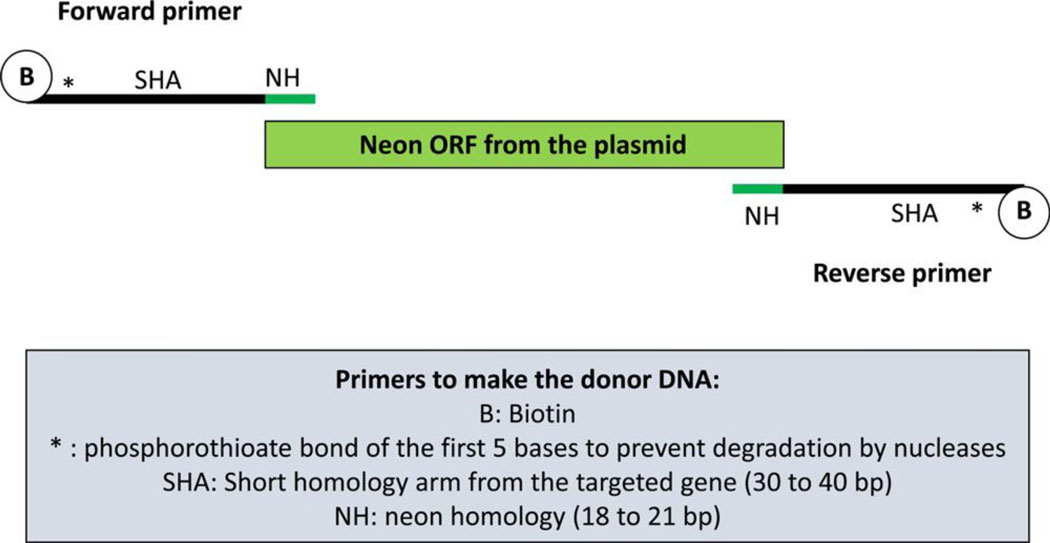
We obtained the best results using a different strategy based on a double strand PCR product as the DNA repair template whose termini were blocked from concatenation (Gutierrez-Triana et al., 2018; Kimura et al., 2014; Paix et al., 2017; Paix et al., 2019; Seleit et al., 2021; Winkler et al., 1991). We provide a detailed protocol in the Supplemental document explaining how to insert a Neon fluorescent tag in the gene Sp PKS1 using this strategy. This approach relies on a double-stranded DNA donor (single stranded DNA did not yield insertions, SFigure 1) containing short homology arms from the Sp PKS1 gene, flanking a fluorescent protein reporter sequence (Figures 1 and 2). The fluorescent protein Neon was selected for this task because of its intense fluorescence properties (Addgene 98877). Forward and reverse primers were designed that contain 30 to 40 bp of the targeted gene (longer homologous arms e.g. 200 bp were not successful), followed by 18 to 21 bp of the fluorescent protein sequence for annealing with the plasmid template. The 5’ end of the forward and reverse primers were biotinylated to prevent concatenation of the DNA once it had been amplified and injected into the embryo. The first five bases of the primers were also modified (phosphorothioate) to reduce exo-DNA degradation (Figure. 2). Development of the method presented here was guided by a similarly successful protocol for use in the medaka (Seleit et al., 2021). This same method has been used successfully in the teleost Medaka and the sea anemone, Nematostella. The percentage of success depends on the gene targeted. For example, in Medaka, 11% (for mapre1b) to 59% (for g3bp1) of embryos expressed the fluorescent reporter (Seleit et al., 2021). In Nematostella a recent study reported that between 2.2% (for lamin) and 37.7% (for cdh1) of injected embryos were successfully fluorescent (Paix et al., 2023).
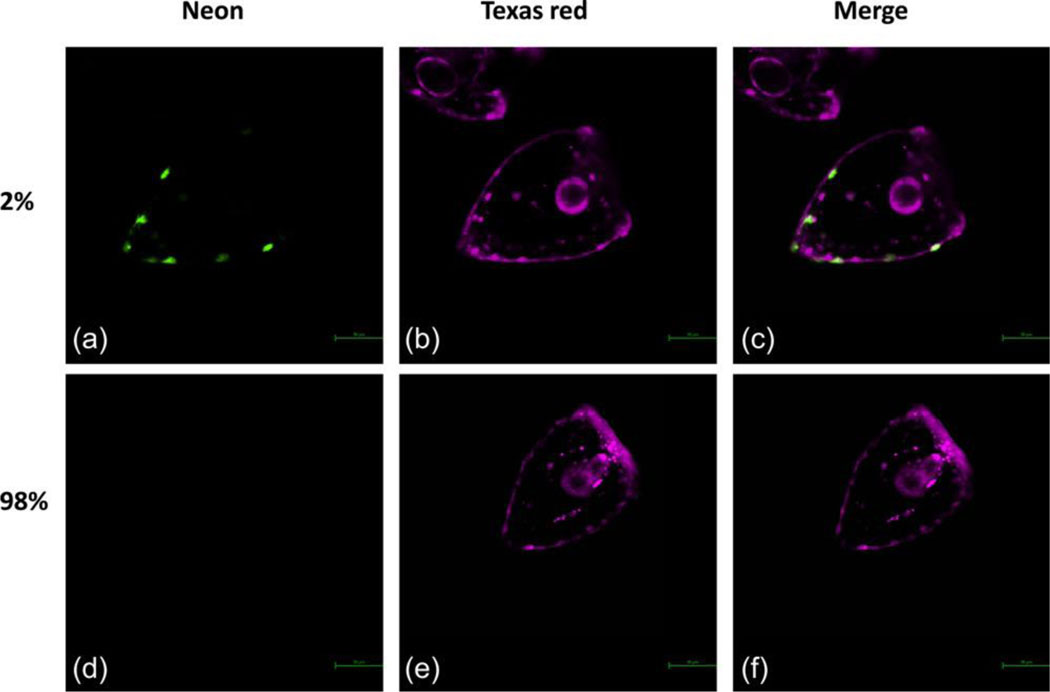
Figure 1: Sp PKS1 Crispr knock-in.
Two percent of embryos express Sp PKS1 Neon in their pigment cells (A). Ninety-eight percent of embryos didn’t show any detectable expression of Sp PKS1 Neon (D). The Texas red dye was co-injected in the zygotes with the knock-in components. This fluorescent dye is used to visualize and sort embryos after injections.
Figure 2:
Description of the method used to design the primers for successful CRISPR/Cas9 knock-in (double stranded DNA, 40 bp of homologous arms from the Sp PKS1 gene).
We obtained 2% of successful and consistent CRISPR PKS1 knock-in larvae presenting fluorescent pigment cells. All of these successful knock-in larvae were mosaic (not every cell of these larvae contained the insert). A low frequency of visual neon insertion actually underestimates the efficiency of the method since PKS1 is expressed in a small population of cells. By genotyping the knock-in locus using PCR and sequencing of the resulting amplicon, we found actually that 10% of the injected larvae contained the correct insert (SFigures 4,5 and 6). The resulting fluorescent larvae were albino, indicating that the Neon insertion into the ketosynthase domain of PKS1, resulted in a nonfunctional PKS1 enzyme not based on sequence but based on structure (Li et al., 2022). Moving forward, we will test other domains in the large PKS1 protein for their essential functionality, and to broaden the utility of the procedure by testing the targeting of ubiquitously expressed genes. We will also test smaller, non-fluorescent tags that may enhance efficiency of insertion. Additional protocol variations will be tested in the future to attempt to achieve a higher knock-in efficiency. It is possible that homologous recombination is not highly efficient in the sea urchin. Some protocols rely instead of non-homologous end joining (NHEJ) (He et al., 2016). In zebrafish, due to the inefficiency of homologous recombination and the error-prone nature of the integrations in this animal, researchers have developed a creative approach to insert their PCR donors by taking advantages of the non-coding regions of the targeted genes (Levic et al., 2021).
CRISPR/Cas9 knock-in will enable researchers to follow the expression of their favorite genes in live cells and embryos, even over multiple generations (Vyas et al., 2022) if the insertion is transmitted to the germline. Fundamental processes such as the biology of the pigment cells (using Sp PKS1 gene) and the biology of the germ cells (using Sp Nanos2 (Oulhen & Wessel, 2016b)), can now be explored in live embryos with this method.
Supplementary Material
Supplemental Figure S1: Summary of the different methods tested to optimize Crispr knock-in in the sea urchin Strongylocentrotus purpuratus. The targeted gene is Sp PKS1. The inserted DNA was the neon sequence, except for the plasmid strategy in which the GFP sequence was used. The best methods are represented in red. Five hundred larvae were injected for each condition.
Supplemental Figure S2: Description of the plasmid method. Double strand break was induced at the Sp Pks1 locus specified by Sp PKS1_547 gRNA. The knock-in construct (KI construct) containing GFP ORF and 1kb of homologous arms was linearized by cutting at the Sp PKS1_547 gRNA target sites located at the both ends of KI-construct. The linearized KI construct is integrated into the second exon of Pks1 locus by homology-directed repair (HDR).
Supplemental Figure S3: CRISPR knock-in obtained with the plasmid method. Cas9 mRNA (A–Á´), Cas9 mRNA and gRNA (B–B´´), KI construct (C–Ć´), Cas9 mRNA, gRNA and KI construct (D–D´´) and Cas9 mRNA, gRNA, KI construct and DN-dnl4 (E–É´) were injected into fertilized eggs. All these zygotes were co-injected with a mCherry mRNA as a control, the mCherry protein was detected in all of them (A–E). GFP expression was observed in animals injected with Cas9, gRNA and KI construct (D–D´´ and E–É´). Proportions of GFP-expressing larvae are indicated in A’’ to E’’.
Supplemental Figure S4: Genomic DNA of the injected embryos was extracted and PKS1 was amplified using primers surrounded the insert. This figure represents an example of PCR showing that some embryos contain the insert at the correct size (expected PCR product at 1245 bp, white arrow). Each lane represents an individual embryo. The embryos labelled 1 to 16 represent the embryos that were injected with the Crispr knock-in mix. The embryos C1 to C8 are the control embryos that were injected with the Cas9 and the DNA donor, but no gRNA. No insertion is detected in the control embryos.
Supplemental Figure S5: The DNA donor was inserted in the correct location and the correct frame. For example, this sequence was obtained from the band presented in Supplemental Figure S2 (embryo 6, band pointed by the right arrow). Primers used for the PCR are in blue. gRNA sequence is in red. Neon ORF is in green.
Supplemental Figure S6: Sp PKS1 and Sp PKS1 neon partial protein sequences resulting from the translation of the DNA sequences obtained by PCR from injected control larvae and injected knock-in larvae. Neon protein is in light green. The pink and orange colors indicate the location of the neon insertion.
Acknowledgments:
The authors thank the National Institutes of Health (1R35GM140897, G.W.; 1P20GM119943, N.O.; R15 GM139113-01A1, J.F.W.) and the National Science Foundation (IOS-1923445, G.W.).
References
- Barsi JC, Tu Q, Calestani C, & Davidson EH (2015). Genome-wide assessment of differential effector gene use in embryogenesis. Development, 142(22), 3892–3901. 10.1242/dev.127746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calestani C, Rast JP, & Davidson EH (2003). Isolation of pigment cell specific genes in the sea urchin embryo by differential macroarray screening. Development, 130(19), 4587–4596. 10.1242/dev.00647 [DOI] [PubMed] [Google Scholar]
- Calestani C, & Wessel GM (2018). These Colors Don’t Run: Regulation of Pigment-Biosynthesis in Echinoderms. Results Probl Cell Differ, 65, 515–525. 10.1007/978-3-319-92486-1_22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming TJ, Schrankel CS, Vyas H, Rosenblatt HD, & Hamdoun A. (2021). CRISPR/Cas9 mutagenesis reveals a role for ABCB1 in gut immune responses to Vibrio diazotrophicus in sea urchin larvae. J Exp Biol, 224(7). 10.1242/jeb.232272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Triana JA, Tavhelidse T, Thumberger T, Thomas I, Wittbrodt B, Kellner T, Anlas K, Tsingos E, & Wittbrodt J. (2018). Efficient single-copy HDR by 5’ modified long dsDNA donors. Elife, 7. 10.7554/eLife.39468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Tan C, Wang F, Wang Y, Zhou R, Cui D, You W, Zhao H, Ren J, & Feng B. (2016). Knock-in of large reporter genes in human cells via CRISPR/Cas9-induced homology-dependent and independent DNA repair. Nucleic Acids Res, 44(9), e85. 10.1093/nar/gkw064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, & Higashijima S. (2014). Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci Rep, 4, 6545. 10.1038/srep06545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levic DS, Yamaguchi N, Wang S, Knaut H, & Bagnat M. (2021). Knock-in tagging in zebrafish facilitated by insertion into non-coding regions. Development, 148(19). 10.1242/dev.199994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lin Z, Torres JP, Hill EA, Li D, Townsend CA, & Schmidt EW (2022). Sea Urchin Polyketide Synthase SpPks1 Produces the Naphthalene Precursor to Echinoderm Pigments. J Am Chem Soc, 144(21), 9363–9371. 10.1021/jacs.2c01416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Oulhen N, Wessel G, & Su YH (2019). CRISPR/Cas9-mediated genome editing in sea urchins. Methods Cell Biol, 151, 305–321. 10.1016/bs.mcb.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, & Su YH (2016). Genome editing in sea urchin embryos by using a CRISPR/Cas9 system. Dev Biol, 409(2), 420–428. 10.1016/j.ydbio.2015.11.018 [DOI] [PubMed] [Google Scholar]
- Liu D, Awazu A, Sakuma T, Yamamoto T, & Sakamoto N. (2019). Establishment of knockout adult sea urchins by using a CRISPR-Cas9 system. Dev Growth Differ, 61(6), 378–388. 10.1111/dgd.12624 [DOI] [PubMed] [Google Scholar]
- McMahon AP, Flytzanis CN, Hough-Evans BR, Katula KS, Britten RJ, & Davidson EH (1985). Introduction of cloned DNA into sea urchin egg cytoplasm: replication and persistence during embryogenesis. Dev Biol, 108(2), 420–430. 10.1016/0012-1606(85)90045-4 [DOI] [PubMed] [Google Scholar]
- Oulhen N, Pieplow C, Perillo M, Gregory P, & Wessel GM (2022). Optimizing CRISPR/Cas9-based gene manipulation in echinoderms. Dev Biol, 490, 117–124. 10.1016/j.ydbio.2022.07.008 [DOI] [PubMed] [Google Scholar]
- Oulhen N, & Wessel GM (2016a). Albinism as a visual, in vivo guide for CRISPR/Cas9 functionality in the sea urchin embryo. Mol Reprod Dev, 83(12), 1046–1047. 10.1002/mrd.22757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N, & Wessel GM (2016b). Differential Nanos 2 protein stability results in selective germ cell accumulation in the sea urchin. Dev Biol, 418(1), 146–156. 10.1016/j.ydbio.2016.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Basu S, Steenbergen P, Singh R, Prevedel R, & Ikmi A. (2023). Endogenous tagging of multiple cellular components in the sea anemone Nematostella vectensis. Proc Natl Acad Sci U S A, 120(1), e2215958120. 10.1073/pnas.2215958120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, & Seydoux G. (2017). Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods, 121-122, 86–93. 10.1016/j.ymeth.2017.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Rasoloson D, Folkmann A, & Seydoux G. (2019). Rapid Tagging of Human Proteins with Fluorescent Reporters by Genome Engineering using Double-Stranded DNA Donors. Curr Protoc Mol Biol, 129(1), e102. 10.1002/cpmb.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perillo M, Oulhen N, Foster S, Spurrell M, Calestani C, & Wessel G. (2020). Regulation of dynamic pigment cell states at single-cell resolution. Elife, 9, e60388. 10.7554/eLife.60388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seleit A, Aulehla A, & Paix A. (2021). Endogenous protein tagging in medaka using a simplified CRISPR/Cas9 knock-in approach. Elife, 10, e75050. 10.7554/eLife.75050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas H, Schrankel CS, Espinoza JA, Mitchell KL, Nesbit KT, Jackson E, Chang N, Lee Y, Warner J, Reitzel A, Lyons DC, & Hamdoun A. (2022). Generation of a homozygous mutant drug transporter (ABCB1) knockout line in the sea urchin Lytechinus pictus. Development, 149(11). 10.1242/dev.200644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Kiyomoto M, Shen TL, & Yajima M. (2020). Genetic manipulation of the pigment pathway in a sea urchin reveals distinct lineage commitment prior to metamorphosis in the bilateral to radial body plan transition. Sci Rep, 10(1), 1973. 10.1038/s41598-020-58584-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C, Vielkind JR, & Schartl M. (1991). Transient expression of foreign DNA during embryonic and larval development of the medaka fish (Oryzias latipes). Mol Gen Genet, 226(1–2), 129–140. 10.1007/BF00273596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Summary of the different methods tested to optimize Crispr knock-in in the sea urchin Strongylocentrotus purpuratus. The targeted gene is Sp PKS1. The inserted DNA was the neon sequence, except for the plasmid strategy in which the GFP sequence was used. The best methods are represented in red. Five hundred larvae were injected for each condition.
Supplemental Figure S2: Description of the plasmid method. Double strand break was induced at the Sp Pks1 locus specified by Sp PKS1_547 gRNA. The knock-in construct (KI construct) containing GFP ORF and 1kb of homologous arms was linearized by cutting at the Sp PKS1_547 gRNA target sites located at the both ends of KI-construct. The linearized KI construct is integrated into the second exon of Pks1 locus by homology-directed repair (HDR).
Supplemental Figure S3: CRISPR knock-in obtained with the plasmid method. Cas9 mRNA (A–Á´), Cas9 mRNA and gRNA (B–B´´), KI construct (C–Ć´), Cas9 mRNA, gRNA and KI construct (D–D´´) and Cas9 mRNA, gRNA, KI construct and DN-dnl4 (E–É´) were injected into fertilized eggs. All these zygotes were co-injected with a mCherry mRNA as a control, the mCherry protein was detected in all of them (A–E). GFP expression was observed in animals injected with Cas9, gRNA and KI construct (D–D´´ and E–É´). Proportions of GFP-expressing larvae are indicated in A’’ to E’’.
Supplemental Figure S4: Genomic DNA of the injected embryos was extracted and PKS1 was amplified using primers surrounded the insert. This figure represents an example of PCR showing that some embryos contain the insert at the correct size (expected PCR product at 1245 bp, white arrow). Each lane represents an individual embryo. The embryos labelled 1 to 16 represent the embryos that were injected with the Crispr knock-in mix. The embryos C1 to C8 are the control embryos that were injected with the Cas9 and the DNA donor, but no gRNA. No insertion is detected in the control embryos.
Supplemental Figure S5: The DNA donor was inserted in the correct location and the correct frame. For example, this sequence was obtained from the band presented in Supplemental Figure S2 (embryo 6, band pointed by the right arrow). Primers used for the PCR are in blue. gRNA sequence is in red. Neon ORF is in green.
Supplemental Figure S6: Sp PKS1 and Sp PKS1 neon partial protein sequences resulting from the translation of the DNA sequences obtained by PCR from injected control larvae and injected knock-in larvae. Neon protein is in light green. The pink and orange colors indicate the location of the neon insertion.