Summary
Cellular exposure to free fatty acids (FFA) is implicated in the pathogenesis of obesity-associated diseases. However, studies to date have assumed that a few select FFAs are representative of broad structural categories, and there are no scalable approaches to comprehensively assess the biological processes induced by exposure to diverse FFAs circulating in human plasma. Furthermore, assessing how these FFA-mediated processes interact with genetic risk for disease remains elusive. Here we report the design and implementation of FALCON (Fatty Acid Library for Comprehensive ONtologies) as an unbiased, scalable and multimodal interrogation of 61 structurally diverse FFAs. We identified a subset of lipotoxic monounsaturated fatty acids (MUFAs) with a distinct lipidomic profile associated with decreased membrane fluidity. Furthermore, we developed a new approach to prioritize genes that reflect the combined effects of exposure to harmful FFAs and genetic risk for type 2 diabetes (T2D). Importantly, we found that c-MAF inducing protein (CMIP) protects cells from exposure to FFAs by modulating Akt signaling and we validated the role of CMIP in human pancreatic beta cells. In sum, FALCON empowers the study of fundamental FFA biology and offers an integrative approach to identify much needed targets for diverse diseases associated with disordered FFA metabolism.
Keywords: Type 2 diabetes, obesity, transcriptomics, lipidomics, pancreatic beta cell, high-throughput screen, GWAS, CMIP, monounsaturated fatty acid, erucic acid
Introduction
Obesity promotes and exacerbates a growing burden of chronic diseases including type 2 diabetes (T2D), cancer, cardiovascular disease and neurodegeneration (Pillon et al. 2021; Rathmell 2021; Yoon et al. 2021). Obesity interacts with underlying genetic risk to drive this unprecedented health crisis; however, at present we do not understand the molecular mechanisms underpinning the complex interplay between genetic and environmental factors (Pillon et al. 2021). Genome wide association studies (GWAS) are powerful in identifying genetic risk factors but are limited by challenges in prioritizing disease-relevant loci (Pillon et al. 2021; Uffelmann et al. 2021). Metabolomic and lipidomic studies have linked aberrant levels of circulating lipids to metabolic diseases (Musunuru and Kathiresan 2019, 2016). However, the contribution of individual lipid species (i.e. triacylglycerols, lipoprotein particles, phospholipids, diacylglycerols and fatty acids) remains an area of active investigation (Farese et al. 2012; Jha et al. 2018; Imamura et al. 2017). Cellular models of fatty acid overload have been used to define “lipotoxicity” as the harmful effects of prolonged and increased exposure to specific lipids (Lytrivi, Castell, et al. 2020; Sharma and Alonso 2014), but the link between cellular exposure to diverse FFA species circulating in human blood and toxicity phenotypes remains poorly understood.
T2D, one of the most prevalent obesity-associated diseases, is characterized by insulin resistance in peripheral tissues and progressive pancreatic beta cell failure. Although liver, adipose and muscle tissue play a central role in insulin resistance (da Silva Rosa et al. 2020), T2D GWAS to date have highlighted genes critical to beta cell function (Petrie, Pearson, and Sutherland 2011; Pasquali et al. 2014; Udler et al. 2018; Parker et al. 2013). In vitro, beta cells are strongly affected by lipid overload; excess chronic fatty acid exposure disrupts their metabolic homeostasis and their capacity to secrete insulin, pushing them towards ER stress and cell death (Prentki and Madiraju 2012; Randle et al. 1963). Therefore, FFA studies in beta cells are centrally important to our understanding of the combined effects of excess FFA exposure and genetic risk for T2D.
Not all fatty acids produce the same cellular effects, and distinctions are often made based on their saturation level. Saturated fatty acids (SFAs) contain no double bonds in their carbon chains. In contrast, monounsaturated fatty acids (MUFAs) contain one double bond and polyunsaturated fatty acids (PUFAs) contain more than one double bond. At the cellular level, studies to date have primarily relied on the effects of a single SFA, palmitic acid (PA) (Palomer et al. 2018; Piccolis et al. 2019) to explore the role of FFAs in lipotoxicity. However, large-scale epidemiological studies, including the Framingham Heart Study, have shown an association between the abundance of TAG species composed of a wide spectrum of structurally diverse FFAs and the severity of metabolic diseases (Rhee et al. 2011), hinting at the importance of largely unexplored and clinically relevant FFA biology. Considering that (i) TAGs are hydrolyzed by lipases into FFAs (and glycerol) before uptake into peripheral cells (Saponaro et al. 2015), and (ii) there is a strong association between TAG composition and obesity-associated diseases such as T2D (Al-Sulaiti et al. 2018; Rhee et al. 2011), there is an urgent need to thoroughly interrogate the biological effects of FFAs across their structural spectrum.
Here we reveal FALCON (Fatty Acid Library for Comprehensive ONtologies), a cell-based platform for the unbiased, multimodal investigation of structurally diverse FFAs found in human plasma (Abdelmagid et al. 2015; Aubourg et al. 1993; Lust et al. 2021) by integrating transcriptomics, cell morphological features, lipidomics, FFA structural characterization, and functional profiling. Out of 61 FFAs, FALCON identified 20 FFAs that were toxic to beta cells, grouped into a cluster marked by a distinct transcriptomic signature. These lipotoxic FFAs could not be classified based on traditional structural designations such as degree of saturation. More than half (12/20) were MUFAs whose toxicity is not fully understood on a mechanistic level. Exposure to these MUFAs was associated with a distinct lipidomic profile and decreased membrane fluidity. Aiming to understand the combined effects of environmental exposure and genetic risk for T2D, we prioritized 25 genes that emerged from the overlap between genes differentially regulated after exposure to lipotoxic FFAs and genes nominated in a large-scale T2D GWAS. We thus identified CMIP, a gene with no prior mechanistic links to metabolic disease, and showed that it protects mouse and human pancreatic beta cells from excess fatty acid exposure. In sum, FALCON empowered a comprehensive, multiplexed query of FFA biology and revealed CMIP as a previously unrecognized suppressor of lipotoxicity.
Results
A systematic approach to study FFA biology
To enable the systematic study of FFAs, we needed to (i) curate a comprehensive set of FFAs beyond those traditionally used in targeted experiments (mostly PA and oleic acid (OA)), (ii) engineer an approach that would solve the difficulty of working with compounds of varying levels of hydrophobicity, and (iii) assess the biological effects of these FFAs in an unbiased fashion. FALCON fulfilled all three criteria, and we used it to systematically interrogate 61 structurally diverse FFAs (Table S1) using multiple high-content assays in a manner widely applicable to any cell type of choice. Most of the analyzed FFAs were readily detectable in both human and mouse blood (Table S1).
We performed proof of concept studies in MIN6 cells, a widely used mouse insulinoma pancreatic beta cell line that displays robust glucose-sensing and insulin secretion, and shares well-conserved molecular programs with human beta cells (Fang et al. 2019; Skelin, Rupnik, and Cencic 2010). We generated several datasets to deeply phenotype the effects of treating cells with each of the 61 FFAs including measures of cell viability/function, lipidomics, transcriptomics and unbiased cell morphology measurements (Fig. 1A, tier 1). We analyzed and integrated these diverse datasets using (i) Gene Set Enrichment Analysis (GSEA (Subramanian et al. 2005)) to identify alterations in cellular pathway activity; (ii) Molecular Operating Environment (MOE) software (Vilar, Cozza, and Moro 2008) to correlate FFA chemical structures with biological features, and (iii) Multimarker Analysis of GenoMic Annotation (MAGMA (de Leeuw et al. 2015)) ranking to incorporate genetic predisposition to human metabolic disease (Fig. 1A, Tier 2). Importantly, we validated our results from MIN6 cells in two human beta cell systems: iPSC derived pancreatic beta cells and acutely isolated human pancreatic islets from donors. Collectively, this multimodal approach allowed us to (i) group FFAs in an unbiased manner (i.e. remaining agnostic to structural features) solely based on similarity of biological readouts, (ii) identify previously unrecognized lipotoxic FFAs, and (iii) define toxicity phenotypes representative of these lipotoxic FFAs.
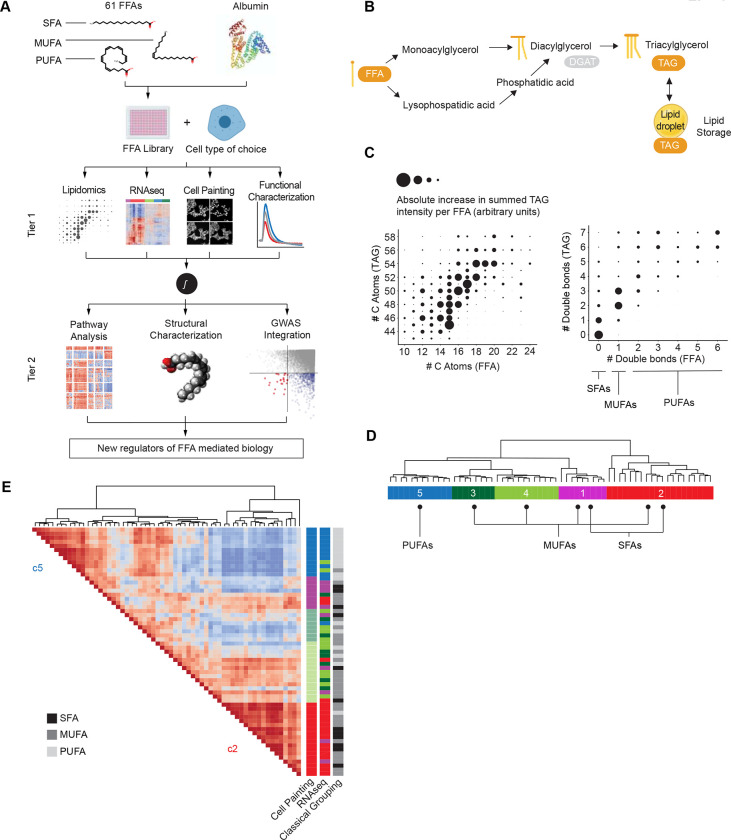
Fig. 1. FALCON, a multiplexed platform for the systematic interrogation of structurally diverse FFAs defines 5 FFA clusters.
(A) Analysis workflow for FALCON. SFA, saturated fatty acid; MUFA, monounsaturated fatty acid; PUFA, polyunsaturated fatty acid. In Tier 1, we use several methods, as listed, to characterize the cellular effects of each of 61 FFAs. In Tier 2, the Tier 1 datasets are integrated to reveal mechanistic insights. (B) Schematic of triacylglyceride (TAG) synthesis from FFAs. Shown here are the monoacylglycerol (top) and the glycerol-3-phosphate pathway (bottom). For simplicity, the acylation of dihydroxyacetone phosphate is not shown. (C) Qualitative correlation of structural features (number of C atoms, number of double bonds) of externally applied FFAs from the library (x-axis) versus structural features of endogenous TAGs (y-axis) measured by lipidomics. Distinct TAG profiles detected in cells treated with SFAs, MUFAs or PUFAs. (D) Five FFA clusters (c1-c5) were identified after hierarchical clustering of transcriptomic profiles derived from exposure to each of 61 FFAs (methods). (E) Cell Painting analysis of immunofluorescence images from cells exposed to each of 61 FFAs (methods) independently clustered together the FFAs transcriptomically assigned to c2 and separately the FFAs assigned to c5.
The crucial gating step in establishing FALCON was the ability to reproducibly and reliably deliver solvent-free BSA-bound FFAs to cells in multiwell plates at scale, using concentrations of magnitude similar to human blood (Abdelmagid et al. 2015; Aubourg et al. 1993; Tsoukalas et al. 2019; Mir et al. 2015; Mogilenko et al. 2019; Chen et al. 2020; Ubellacker et al. 2020; Lytrivi, Ghaddar, et al. 2020)). To achieve this goal, FFAs were dissolved in DMSO and non-covalently bound to FFA-free BSA at a 7:1 ratio (there are 7 FFA binding sites in human albumin (Petitpas et al. 2001; Curry et al. 1998; Bhattacharya, Grüne, and Curry 2000; Fujiwara and Amisaki 2008)). A centrifugal evaporation/concentration step removed water and DMSO to generate solvent-free crystals of BSA-bound FFAs (Fig. S1A). The crystals were dissolved in culture medium, which was then applied to cells (Fig. S1A). We confirmed FFA binding to BSA by differential scanning calorimetry (Michnik 2003)(Fig. S1B).
To assess the effects of FFA treatment on cellular lipid composition, we performed mass spectrometry-based lipidomics of cells exposed to each of the 61 FFAs (Fig. S1C). TAGs, which are synthesized from FFAs available within a cell (Coleman and Mashek 2011) (Fig. 1B), constituted the majority of detected lipid species and thus became the focus of our initial lipidomic analysis. We concluded that FFAs were likely incorporated into cellular TAGs, as evidenced by the correlation between FFA and TAG structural features, such as chain length and number of double bonds (Fig. 1C, Fig. S1D). In summary, FALCON assessed the biological effects of structurally diverse FFAs in a reliable, high-throughput, and quantitative manner.
Identification of non-canonical FFA clusters
To comprehensively define FFA-induced cell states without prior assumptions about biological effects or FFA structural features, we generated transcriptomic profiles of MIN6 cells in response to each of 61 FFAs in the library (Fig. S1E). We detected induction of carnitine palmitoyltransferase 1 expression (CPT1A), the rate-limiting enzyme of FFA beta-oxidation (Thumelin et al. 1997), across the entire library, demonstrating successful FFA delivery and intracellular metabolism (Fig. S1F). Hierarchical clustering of FFA-induced transcriptomes revealed 5 distinct “clusters” (c1-c5)(Fig. 1D, Fig. S2A,B). At the extremes, FFAs segregated by saturation and chain length: SFAs were major constituents of cluster 1 (c1) and also well represented in cluster 2 (c2; 8/20 c2 FFAs were SFAs), whereas cluster 5 (c5) was exclusively composed of PUFAs. In line with the traditional paradigm (Palomer et al. 2018), palmitic acid (lipotoxic FFA) and oleic acid (OA; lipotoxicity-protective FFA), clustered separately, in c2 and c3, respectively. Strikingly, MUFAs were distributed across four clusters c1 to c4 (Fig. 1D), indicating that the transcriptomic signatures classified some MUFAs as functionally similar to SFAs, and not to other MUFAs. These data argue against a simple grouping of FFAs based on saturation (SFA, MUFA, and PUFA) (Palomer et al. 2018) and suggest instead that traditional criteria do not adequately capture the observed transcriptomically-defined heterogeneity (Fig. S2C).
To probe cellular responses to FFA exposure using an independent method, we profiled the entire FFA library using Cell Painting, a high-content image analysis method that simultaneously measures several hundred cellular imaging features (Bray et al. 2016) (Fig. S2D). This morphologic analysis independently clustered together the SFAs and MUFAs transcriptomically assigned to c2 and separated them from the PUFAs in c5 (Fig. 1E). We thus concluded that the algorithms analyzing the cell imaging data were in agreement with the FFA clustering derived from transcriptomics with regard to the two most prominent clusters: c2 and c5 (Fig. 1D–E).
Cellular transcriptomes define key biological responses to FFAs
To understand the basis behind the FFA clusters identified by FALCON, we applied Gene Set Enrichment Analysis (GSEA) (Subramanian et al. 2005) to each of the 61 FFA-induced transcriptomes using multiple gene set collections (MSigDB (Liberzon et al. 2015)). By looking at how multiple gene sets were enriched across the 5 clusters, we were able to categorize them into modules of gene expression (Fig. 2A; Methods). Crucially, this analysis ensured that our gene set annotation was comprehensive and not reliant on any single database of curated gene lists.
Fig. 2. Transcriptomic analysis identifies key biological responses to FFAs, and functional assays validate novel FFA clustering.
(A) Hierarchical clustering of a gene set enrichment matrix (based on normalized enrichment scores of gene sets, NES) revealed gene set modules of interest. Representative leading edge genes from each module are listed on the right. (B, C) Scatter plots of two independent replicates of cell viability (B) and ER Ca2+ level measurements (C)(n=5–7 replicates / FFA / screen). Closed dots represent FFAs that showed significant difference (p < 0.05, Bonferroni) from controls in both replicates, open dots represent non-significant FFAs in at least one replicate. Colors indicate corresponding FFA cluster membership (Fig. 1D). (D) Summary of functional assays. Top bar represents FFA clusters derived from transcriptomic analysis. Second color bar represents clusters derived from Cell Painting (cellular morphology) analysis (methods). Bar in grayscale indicates classical grouping of FFAs based on saturation level. The next two bars are heat maps displaying log2 fold changes of ER Ca2+ levels and cell viability, respectively. X-axis labels show FFA structure (in Simplified Molecular Input Line Entry System, SMILES). The box highlights the 20 FFAs in cluster 2 (c2) identified as the lipotoxicity cluster. Highlighted FFAs were chosen as cluster representatives for further downstream studies. PA, palmitic acid; EA, erucic acid; PSA, petroselenic acid; OA, oleic acid; AA, arachidonic acid; GLA: gamma-linoleic acid.
Since the identified gene modules served as the basis for the separation of diverse FFAs into five discrete clusters, we next sought to better understand the biological processes these modules represented. Consistent with exposure of cells to FFAs, a key module shared among all clusters represented pathways regulating FFA transport and metabolism, including upregulation of SLC25A20 and CPT1A (core genes that encode the mitochondrial carnitine shuttle (Tonazzi et al. 2021; Longo, Frigeni, and Pasquali 2016). A second module, whose induction (positive normalized enrichment score (NES)) was detected specifically in c1 and c2, consisted of ER stress and unfolded protein response (UPR) genes (PPP1R15A, ATF6, ERN1 (IRE1) and XBP1; (Schröder and Kaufman 2005)) (Fig. 2A). A third gene module comprised stress response pathways, NFkB signaling and inflammation (Fig. 2A) consistent with lipotoxicity (Hotamisligil 2010; Eguchi et al. 2012; Donath et al. 2013). Genes in the third module such as PAM, CCK, OXTR, RGS4 and the chemokine, CXCL12 were specifically upregulated in the c2 FFA cluster. Module four comprised pathways related to cholesterol metabolism (including key genes PCSK9, LDLR, HMGCR and SQLE; (Abifadel et al. 2003; Hobbs, Brown, and Goldstein 1992; Howe et al. 2017)), and module five consisted of programs related to proteasome activity and responses to reactive oxygen species (ROS) (upregulated genes DLL1, GCLC, GSTP1 in the c5 cluster) (Fig. 2A). A sixth gene module was associated with MAPK signaling, a crucial regulator of cell proliferation and apoptosis that directly impacts insulin secretion in beta cells (Khoo et al. 2004). The MAPK signaling module was specifically upregulated in the c2 cluster (as reflected by the induction of genes such as DUSP4, SPRED2 and NUPR1 (Fig. 2A)). In sum, FALCON defined the biological processes that served as the basis for the separation of diverse FFAs into five previously unrecognized clusters. More specifically, the second, third and sixth modules pointed to the c2 cluster as a putative mediator of cell stress and lipotoxicity.
To explore the biological relevance and reproducibility of the newly defined c2 transcriptomic signature, we asked if differentially expressed genes from published lipotoxicity datasets in other systems might overlap with genes differentially expressed upon exposure to c2 FFAs. We found a distinct overlap of genes upregulated by c2 FFAs in MIN6 cells with (i) genes upregulated in the transcriptome of pancreatic beta cells isolated from mice fed a high fat diet (Dusaulcy et al. 2019) and (ii) genes upregulated in human islets exposed to PA (Lytrivi, Ghaddar, et al. 2020) (Fig. S2E,F; Methods). These analyses indicated that cellular responses to c2 FFAs were conserved and shared between mouse MIN6 cells and human beta cells in vitro, as well as mouse beta cells in vivo.
Functional validation of FFA clustering
To functionally validate the transcriptome-derived clusters, we first measured cell viability in MIN6 cells exposed to each of the 61 FFAs (Fig. 2B). The c2 FFAs induced the most consistent and significant reduction in cell numbers among all clusters. Based on the observation that FFA-induced ER stress is associated with decreased ER Ca2+ levels and apoptosis (Marmugi et al. 2016; Fu et al. 2011; Orrenius, Zhivotovsky, and Nicotera 2003), we measured ER Ca2+ in a high-throughput fluorometric assay after exposure to each of the 61 FFAs (Fig. S3A, Methods). We detected decreased ER Ca2+ levels in cells treated with c2 FFAs and, to a smaller extent, c1 FFAs, thus functionally validating the ER stress signature derived from transcriptomics (Fig. 2C). In contrast, we detected a consistent increase in ER Ca2+ levels for cells exposed to c5 FFAs, while c3 and c4 FFAs did not alter ER Ca2+ levels (Fig. 2C). Collectively, these data showed that, in addition to SFAs and in contrast to c3 and c4 MUFAs, a specific subset of 12 MUFAs in the c2 cluster caused ER Ca2+ deficits and cell death (Fig. 2D). These results enhanced our interest into further understanding c2 FFAs, including c2 MUFAs, as mediators of lipotoxicity. More broadly, these data served as functional validation for FALCON in pancreatic beta cells.
FALCON is applicable at scale to different cell types
Exposure to excess circulating FFAs has been implicated in diseases affecting many cells and organs in addition to pancreatic beta cells, including the kidney (Sun et al. 2020; Jang et al. 2020) and the brain (Yin 2022; Guttenplan et al. 2021; Ioannou et al. 2019). To assess the general versatility of FALCON, we tested two additional disease-relevant cell types: human kidney tubular epithelial cells, associated with kidney disease (Sun et al. 2020; Jang et al. 2020), and human iPSC-derived microglia, associated with neurodegenerative disease (Yin 2022; Tcw et al. 2022). In both cell types, c2 cluster FFAs were the most toxic, consistent with the results in beta cells (Fig. 3A–E). Of note, there were also some cell-type specific differences. In human kidney epithelial cells, c3–4 FFAs universally increased cell numbers, indicating that these FFAs may promote cell proliferation (Fig. 3A,D). In microglia, c5 PUFAs were more toxic than in beta cells and kidney epithelial cells, on par with c2 FFAs (Fig. 3A). High levels of PUFAs are associated with increased levels of ROS-induced damage due to lipid peroxidation leading to ferroptosis (W. S. Yang et al. 2016; Dixon et al. 2012; Ryan et al. 2023). Our data, the first comprehensive snapshot of the effect of diverse FFAs on human microglia, suggest that these critical neuro-immune cells may be particularly susceptible to PUFA-mediated injury (Fig. 3A), an intriguing finding with implications for neurodegenerative diseases (Bachiller et al. 2018). In sum, these studies demonstrated the utility of FALCON for multiple cell types of interest, and its potential to yield fundamental insights into FFA biology.
Fig. 3. FALCON is applicable at scale to different cell types.
(A) Heatmap showing comparison of viability changes across 3 different cell types: MIN6 pancreatic beta cells, human iPSC-derived microglia and human kidney tubular epithelial cells. c2 FFAs are toxic for all three cell types studied. (B) Representative images from all 3 cell types highlighting the toxicity of c2 FFAs including erucic acid (EA). EA induces cell death in beta cells (green, Hoescht), in microglia (green, GFP) and in kidney tubular epithelial cells (red, propidium iodide). Scale bars: 100 μm. (C-E) Bar plots indicate change in cell viability relative to BSA induced by OA, PA, or EA in each cell type after exposure to FFAs at 500 μM for 72 h (MIN6) (C), 500 μM for 15 h (epithelial cells) (D), or 250 μM for 24 h (microglia) (E). PA and EA consistently induce cell death across all three cell types as assessed by 1-way ANOVA followed by Dunnett’s test (*p < 0.05, ****p < 0.0001). Data are mean ± SD.
Cell biological hallmarks of lipotoxicity characterize the newly-defined c2 FFA cluster
To better understand the mechanisms underlying c2 FFA-induced toxicity, which was universally observed in beta cells, kidney epithelial cells and microglia, we took a closer look at the 8 SFAs and 12 MUFAs comprising this cluster (Fig. 2D, 3A) and focused on erucic acid (EA), a 22-carbon c2 MUFA. Although there is evidence that long chain MUFAs such as EA are toxic (Plötz et al. 2019), the mechanisms behind this toxicity are only beginning to be explored (Chen et al. 2020). In addition, previous in vivo studies had not clarified whether exposure to EA is harmful or not (Vogtmann et al. 1975; Aubourg et al. 1993). In our studies, despite its classification as a MUFA with a similar length to various c5 PUFAs, EA was distinctly clustered with lipotoxic c2 FFAs. To more deeply characterize EA, we examined it in comparison to (i) palmitic acid, a saturated c2 FFA, (ii) oleic acid (OA) and petroselinic acid (PSA), two MUFAs in the non-toxic c3 cluster, and (iii) arachidonic acid (AA) and gamma-linoleic acid (GLA), two long chain FFAs in the c5 PUFA cluster. Given the high disease burden from T2D and the availability of well-characterized cellular assays (as described for example in Figs. 1, 2), we focused our analysis on beta cells. Out of the six FFAs studied, only EA and PA consistently induced cell death (Fig. 4A).
Fig. 4. Cell biological hallmarks of lipotoxicity characterize the c2 FFA cluster.
(A) Quantification of fluorescence imaging of cells treated with representative FFAs for 48 h. Apoptotic cells (positive y-axis) are measured by caspase activity. Dead cells are measured by propidium iodide positive nuclei (n = 5 wells). Reduction in cell viability (negative y-axis) is defined as the fraction of caspase positive and propidium iodide positive cells. Data are mean ± SD. Student’s t-test (two-sided) ****p < 0.0001, corrected for multiple testing (Bonferroni). (B) Dose-response curve of live cell numbers after 65 h of EA or OA treatment compared to BSA. Live cell number is assessed by total number of Hoechst-positive nuclei that were negative for propidium iodide and for cleaved caspase 3/7. EA is toxic in a concentration dependent manner; OA is not toxic. (C) Western blots show ATF4 and CHOP induction by PA and EA (lipotoxic c2 FFAs). CPT1A induction is a control for intracellular FFA delivery. BSA, negative control. (D) Quantification of peak amplitude as readout for ER Ca2+ levels, relative to negative control (BSA). Data are mean ± SD. Student’s t-test (two-sided) *p < 0.05, ****p < 0.0001, corrected for multiple testing (Benjamini-Hochberg, entire FFA library). (E) Quantification of RELA translocation as percentage of total number of cells (t = 18 h, n = 5 replicates). Data are mean ± SD. Student’s t-test (two-sided) ****p < 0.0001, corrected for multiple testing (Bonferroni). (F) Glucose stimulated insulin secretion (GSIS) after FFA exposure ([FFA] = 500 μM, t = 24 h, n = 6 wells) was measured by ELISA. (G) Autophagosome formation was assessed by imaging LC3B puncta normalized to the number of total cells (t = 48 h, n = 4 wells). Data are mean ± SD. One-way ANOVA followed by Dunnett’s test. For A, D, E, G and H: bar color represents cluster identity (green, c3; red, c2; blue, c5; Fig. 1D). (H) Representative images of LC3B immunofluorescence (gray) in MIN6 cells treated with FFAs, BSA as negative control, or autophagy modulating drugs. Nuclei were detected by Hoechst (blue). c2 FFAs induce autophagosomes. Scale bars: 25 μm. (I) Number of iPSC-derived beta cells after exposure to EA or OA for 24, 48, or 72 h at 250 μM, 500 μM, or 750 μM. Only EA decreased cell count in a dose and time dependent manner. All EA conditions are significant per two-way ANOVA (p < 0.0001); all OA conditions are not significant. Data are mean ± SD, n = 4–6 wells/timepoint/FFA. (J) Representative images of iPSC-derived beta cells after treatment with BSA, EA, or OA for 48 h at 500 μM. Nuclei are marked by Hoechst and beta cell identity is marked by C-peptide staining (orange). Scale bars: 100 μm. (K) Quantification of C-peptide positive cells human beta cells dissociated from cadaveric primary islets normalized to the BSA control after exposure to each of 10 FFAs at 3 different concentrations (t = 5 days, n = 6 wells). c2 cluster MUFAs decreased human beta cell viability in a dose-dependent manner. Data are mean ± SD. Multiple t-test with Bonferroni correction (gold: p<0.05, blue: p<0.01, green: p<0.001, red: p<0.0001). (L) Representative images of human islets after treatment with BSA, EA, or OA. Nuclei are marked by Hoechst, and beta cell identity is marked by C-peptide staining (orange). Scale bars: 100 μm. PA, palmitic acid; EA, erucic acid; PSA, petroselenic acid; OA, oleic acid; AA, arachidonic acid; GLA: gamma-linoleic acid.
Next we sought to explore the dose-dependent toxicity of EA, an FFA whose abundance in human plasma may be 10–200 times lower than PA (one of the most well-studied and abundant FFAs) (Abdelmagid et al. 2015; Aubourg et al. 1993; Lust et al. 2021). Recognizing that (1) plasma concentrations are not indicative of tissue concentrations (D. Zhang et al. 2019) and (2) FFA concentrations have not been measured in any tissue, including the pancreas, we tested a range of EA concentrations. We found that as low as ~75–100 μM EA, a concentration previously measured in human plasma (Aubourg et al. 1993; Lust et al. 2021), induced high levels of cell death (Fig. 4B). These dose-response experiments confirmed that the biological effects we measured were physiologically relevant for PA and toxic MUFAs such as EA.
Sustained activation of the UPR has been linked to the toxicity induced by prolonged exposure to SFAs. We assessed ER stress upon exposure to FFAs by probing the cell death-inducing PERK/ATF4/CHOP arm of the UPR (Oyadomari and Mori 2004; Chan et al. 2015). Only treatment with EA or PA increased ATF4 and CHOP protein abundance, pointing to lipotoxicity-induced activation of the UPR (Fig. 4C). An increase in CPT1A protein abundance across all conditions showed successful intracellular delivery of all FFAs (Fig. 4C) (Thumelin et al. 1997). In line with previous work demonstrating that depletion of ER calcium is associated with lipotoxicity-induced ER stress (Oyadomari and Mori 2004; Marmugi et al. 2016), we observed significant reduction in ER Ca2+ levels only in cells treated with PA or EA (Fig. 4D). In contrast, cells treated with OA and PSA remained near-baseline, and cells treated with AA and GLA had increased ER Ca2+ levels (Fig. 4D).
Lipotoxic inflammation and recruitment of immune cells into pancreatic islets plays a central role in beta cell dysfunction and death in the context of T2D (Hotamisligil 2017). Based on the transcriptomic analysis (Fig. 2A) and previous work with PA (Eguchi et al. 2012), we focused on inflammatory signaling through NFkB measured by nuclear translocation of RELA (p65), a major component of NFkB-mediated transcription (Hayden and Ghosh 2008). Robust RELA nuclear translocation was noted after treatment with EA (Fig. 4E, S3B) and PA (Fig. 4E). In line with the transcriptomic analysis (Fig. 2A), PSA and OA triggered only modest RELA translocation (Fig. 4E) without affecting cell viability (Fig. 4A).
Next, we measured glucose stimulated insulin secretion (GSIS), a function specific to beta cells. Exposure to EA caused significant GSIS impairment (Fig. 4F). PA had a similarly deleterious effect (Fig. 4F)(Oh et al. 2018). In contrast, OA, PSA, AA and GLA had no effect on GSIS (Fig. 4F). Since impaired insulin secretion has been linked to disrupted autophagy (Ebato et al. 2008), and lipotoxicity increases autophagosome number (Mir et al. 2015), we quantified LC3B-positive autophagosomes in cells exposed to FFAs. Similar to PA, EA increased autophagosome numbers, whereas OA, PSA, AA and GLA had no effect (Fig. 4G–H). In sum, despite structural similarities with other MUFAs or very long chain FFAs, EA functionally behaved like the lipotoxic saturated FFAs (as summarized in Fig. 2D). Overall, these results reinforced the FFA clustering derived from systematic analyses, assigned lipotoxic effects to EA, and more broadly, to a previously unrecognized subset of 12 c2 MUFAs (Fig. 2D).
c2 MUFAs are toxic to human cells
We took two additional approaches to assess the effects of toxic MUFAs such as EA in human pancreatic beta cells. First, taking advantage of advances enabling the differentiation of human induced pluripotent stem cells (iPSCs) into pancreatic beta cells (Pagliuca et al. 2014; K. Yang et al. 2020), we generated human iPSC-derived beta cells (Maxwell and Millman 2021; Hogrebe et al. 2020), and treated them with either EA (putative toxic MUFA) or OA (putative benign MUFA). Exposure to EA induced cell death in a dose and time-dependent manner, whereas OA had no effect on cell viability (Fig. 4I,J). In a complementary approach, we studied human pancreatic islets acutely isolated from cadaveric donors. We treated these islets with MUFAs representing all three MUFA-containing clusters c2, c3, and c4 (Fig. 4K,L). Similar to MIN6 cells and iPSC-derived human beta cells, c2 MUFAs (7 FFAs including EA) were toxic to human islet beta cells in a dose-dependent manner, whereas c3 (oleic and petroselenic acid) and c4 MUFAs (10(Z)-nonadecenoic acid) had no effect on cell viability (Fig. 4K,L). We concluded that the c2 MUFAs can induce significant injury, making them highly relevant to human beta cell biology.
Structural characterization of c2 MUFAs
After confirming the toxicity of c2 MUFAs in both mouse and human beta cells, we next asked whether molecular or chemical features could predict and explain this newly defined FFA grouping. Recognizing that double bond number alone did not account for transcriptome-based clustering, we generated a matrix of all 2D FFA structural features (Methods). A random forest classifier trained on the pre-processed structural feature matrix successfully predicted assignment to transcriptome-based predefined clusters c1, c2 and c5 with high sensitivity and specificity based on leave-one-out cross validation (Fig. S3C; Methods). This classifier showed that while the number of double bonds was an important distinguishing feature (Fig. 5A, b_double), the longest chain of single bonds (b_max1len) and bond rotation (b_rotR) were highly predictive for c2 cluster assignment even amongst the MUFAs (c1-c4; Fig. 5A). The longest chain of single bonds also reliably captured the single PUFA (13(Z), 16(Z), 19(Z)-docosatrienoic acid) that was transcriptomically assigned to c2, and predicted its separation from the rest of the PUFAs in the c5 cluster (Fig. 5A).
Fig. 5. Long single bond chain in MUFA (EA) induces a distinct lipidomic profile that is associated with increased membrane rigidity.
(A) Selected features from the decision tree analysis based on meta-features of highest importance (Mean Decrease Accuracy; methods). The longest single bond chain is the meta-feature that predicts the inclusion of 13(Z), 16(Z), 19(Z)-docosatrienoic acid as the only PUFA in the c2 cluster, and distinguishes between toxic EA and non-toxic OA. (B) Accumulation of longer unsaturated acyl chains found by lipidomic analysis of MIN6 cells treated for 24 h with 500 μM EA (left). A network analysis of the biochemical relationship (lines) between significantly enriched lipid species in the EA-induced lipidome highlighted the accumulation of EA (22:1)-containing triglyceride (TAG) species (right). (C) Membrane rigidity as measured by the GP index of Laurdan dye in INS1E beta cells after 12 h treatment with 3–6 different concentrations of BSA, PA, EA, or OA. PA increases membrane rigidity proportionally to its concentration; EA decreases membrane rigidity at low concentration similar to OA, but at higher, toxic levels, EA increases membrane rigidity similar to PA. Data are mean ± SEM; n=6 wells.
Erucic acid induces a lipidome distinct from palmitic acid that is associated with changes in membrane fluidity
Exposure to PA leads to an increase in the abundance of saturated acyl chains of more complex lipid species (Piccolis et al. 2019). This in turn is linked to the activation of the UPR, which can sense changes in lipid composition or lipid bilayer stress (Olzmann and Carvalho 2018; Promlek et al. 2011; Volmer, van der Ploeg, and Ron 2013; Shen et al. 2017; Ho et al. 2020). Since c2 MUFAs caused significant cellular injury, similar to PA, we hypothesized that exposure to c2 MUFAs with long single-bond chains (such as EA) may induce specific changes to the cellular lipidome that could explain their lipotoxic effect. We performed a lipidomics analysis and found that exposure of cells to EA led to the accumulation of longer, unsaturated acyl chains in multiple lipid classes (DAGs, PCs, PEs), and especially in triglycerides (TAGs)(Fig. 5B, S3D). This profile was consistent with the integration of the very long-chain unsaturated EA into more complex lipids, and represented a lipidome distinct from that induced by PA (Fig. 5B, Table S2).
To probe the functional consequences of incorporation of long-chain FFAs into the lipidome, we measured membrane fluidity using a Laurdan dye that fluoresces at different wavelengths in accordance with lipid order/disorder, following a protocol previously used to demonstrate maladaptive PA-induced membrane rigidity (Golfetto, Hinde, and Gratton 2013; Pérez-Martí et al. 2022). Notably, EA had a unique biphasic effect on membrane fluidity. At the lowest concentrations, EA was similar to OA with a trend towards increasing membrane fluidity (Fig. 5C). At higher concentrations, similar to those measured in plasma from patients on FFA-rich diet (Aubourg et al. 1993; Lust et al. 2021) (and at which we found that EA is toxic (Fig. 4B)), membrane fluidity was decreased (Fig. 5C). Of interest, the membrane regions that were most rigid in EA treated cells differed from those in PA treated cells. Exposure to PA led to linear regions of increased rigidity reminiscent of the ER (Ruiz et al. 2019, 2021)(Fig. S3E). In contrast, EA induced distinct spherical regions of increased rigidity reminiscent of lipid droplet morphology (Fig. S3E). These observations are consistent with robust EA incorporation into TAGs (Fig. 5B). In sum, we found that exposure to EA generated a distinct cellular lipidome characterized by species with long unsaturated acyl chains that induced toxicity by decreasing cellular membrane fluidity.
Identification of genes at the intersection of FFA exposure and genetic risk for metabolic disease
Complex diseases arise from the interaction of genetic risk and environmental exposures (Dempfle et al. 2008). GWAS studies have greatly contributed to our understanding of the genomic architecture of complex diseases (Claussnitzer et al. 2020), including T2D (Mahajan et al. 2014; Petrie, Pearson, and Sutherland 2011; Fuchsberger et al. 2016). While several T2D genomic studies have shown a strong association with loci linked to beta cell function (Pasquali et al. 2014; Flannick et al. 2014; Dwivedi et al. 2019; Thomsen et al. 2018), it remains challenging to annotate variants and prioritize genes for mechanistic functional follow-up studies (www.icda.bio).
In our experiments, beta cell exposure to 20 c2 FFAs significantly up- or downregulated specific genes (Fig. 2A, Fig. S2A). We hypothesized that a subset of these genes could also be implicated in genetic risk for T2D. Based on this hypothesis, we sought to prioritize disease-relevant genes by investigating the overlap between an annotated large-scale T2D GWAS dataset (Mahajan et al. 2018) and genes highlighted by our transcriptomic analysis (Fig. 2A, Fig. S2A).
Taking advantage of the Multimarker Analysis of GenoMic Annotation tool (MAGMA; (de Leeuw et al. 2015); Methods), we generated a ranked list of T2D genes. We used the MAGMA gene set analysis (GSA) function (de Leeuw et al. 2015) to test whether the beta cell c2 FFA transcriptomic signature was enriched in the MAGMA-ranked T2D genes. Overlapping of the two datasets revealed that the top 5% of significantly differentially expressed lipotoxicity genes were enriched among the T2D GWAS ranked genes (FDR < 0.001, Fig. 6A). This enrichment was specific to T2D as compared to GWAS datasets for type 1 diabetes, an autoimmune disease (Onengut-Gumuscu et al. 2015) or schizophrenia, an unrelated neurodevelopmental disorder (Schizophrenia Working Group of the Psychiatric Genomics Consortium 2014); Fig. 6A). T2D GWAS genes were specifically enriched in the signature of the c2 cluster but not in the other four clusters (Fig. 6B). These results independently validated the utility and human relevance of FALCON for gaining new insights into FFA-associated metabolic diseases such as T2D (Fig. 1A, tier 2). Next, we plotted the genes that drove this enrichment according to their MAGMA rank on the x-axis (Table S3) and their lipotoxicity rank on the y-axis (based on differential expression p-value, Table S4, Fig. 6C). To highlight genes that drove the observed enrichment, we conservatively chose the top 5% lipotoxicity gene set as the y-axis boundary (464 genes) and the top 600 genes of the ranked MAGMA list as the x-axis boundary. We thus identified 25 genes at the intersection of lipotoxicity and T2D (Fig. 6C,D).
Fig 6. Integration of lipotoxicity transcriptomic signature with T2D GWAS dataset identifies CMIP as mediator of genetic and environmental risk for disease.
(A) Gene set analysis (GSA) results based on the c2 lipotoxicity signature. T2D GWAS genes were ranked based on MAGMA score. A T1D and a schizophrenia GWAS dataset served as negative controls. Lipotoxicity gene sets are defined as top DE genes (1, 5, 10%) in transcriptomic profiles from the c2 cluster ranked by p-value and log2 fold change (LFC). GSA showed significant enrichment (FDR<0.05) for the top 5% (boxed) and 10% lipotoxicity gene sets. (B) Enrichment analysis for all FFA clusters (top 5% gene set) revealed that the c2 cluster gene set is uniquely enriched in the T2D GWAS dataset. (C) Scatter plot of genes based on T2D MAGMA rank (x-axis) and lipotoxicity rank (y-axis). Horizontal boundary defines top 5% lipotoxicity signature genes, vertical boundary defines top 600 MAGMA ranked T2D genes. As demonstrated in the schematic (top), genes located in the left (blue) quadrants are associated with T2D genetic risk, genes located in the bottom (yellow) quadrants are associated with lipotoxicity environmental risk, and genes located in the bottom left (green) quadrant are associated with both genetic and environmental risk. Genes of interest (red) drove the enrichment of the lipotoxicity signature in the GWAS dataset. (D) Expression pattern for the top 25 overlapping T2D-lipotoxicity genes across all FFA clusters. Size of dots represents the percentage of FFAs/cluster that induce significant differential expression of corresponding gene (p<0.05, Benjamini-Hochberg), colors represent strength and directionality of transcriptional changes (log2 fold change). (E) Venn diagram comparing the results of the analysis using the PA-induced signature alone compared with using the c2 lipotoxicity signature derived from all 20 lipotoxic FFAs. 19/25 genes, including GLP1R and CMIP, would have been missed if the analysis was limited to the PA-induced signature alone.
The sensitivity by which we could detect genes of interest was greatly enhanced by the fact that the lipotoxicity signature used in our analysis was based on an integrated transcriptome derived from all 20 lipotoxic c2 FFAs (including the newly defined lipotoxic MUFAs). Accordingly, the transcriptomic signature generated by PA treatment alone could predict only 6 of the 25 T2D-lipotoxicity genes, thus missing 19 important gene candidates, among them GLP1R, SLC30A8, ADCY5, and CMIP (Fig. 6E). GLP1R is a G-protein coupled receptor, the drug target for a class of medicines known as incretin mimetics (GLP1R agonists) approved for the management of obesity and T2D (Winzell and Ahrén 2008; Doyle-Delgado et al. 2020; Blonde et al. 2006; Wadden et al. 2013; Wilding et al. 2021). SLC30A8 encodes the zinc transporter ZnT8 which plays an important role in insulin storage and glucose tolerance (Lemaire et al. 2009; Pound et al. 2009; Nicolson et al. 2009; Wijesekara et al. 2010). Variants in SLC30A8 modify T2D risk (Flannick et al. 2014; Dwivedi et al. 2019). T2D risk alleles are also associated with decreased ADCY5 expression in human islets (Roman et al. 2017). The identification of biologically and therapeutically relevant targets such as GLP1R directly validated our approach of using an integrated lipotoxicity signature derived from all 20 c2 FFAs to identify genes of interest at the intersection of genetic and lipotoxic risk for disease. Of the T2D-lipotoxicity genes revealed by our analysis, we focused our next set of experiments on CMIP because (i) it had not been previously implicated in beta cell biology (despite its poorly understood association with the Maf transcription factor family that bears connections to islet biology (Kataoka et al. 2004)) and (ii) it had no known role in lipotoxicity, thus offering opportunities for new biological insights.
CMIP suppresses lipotoxicity in beta cells
C-MAF inducing protein (CMIP) has been implicated in kidney disease (Moktefi et al. 2016; Bouachi et al. 2018) and cancer (J. Zhang et al. 2017), but it has not been studied in beta cells. Of interest, CMIP-associated risk loci are linked to alterations in body mass index (Cao et al. 2018) and dyslipidemia (Mo et al. 2018). In immune cells, CMIP interacts with RELA and reduces NFkB activation (Kamal et al. 2009). A genome-wide interaction analysis with the insulin secretion locus MTNR1B identified an interaction with a CMIP intronic SNP affecting T2D risk (Keaton et al. 2018). To study CMIP in beta cells, we generated MIN6 Cmip knockout cell lines using CRISPR/Cas9, and isolated one Cmip knockout (CMIP KO) clone with a complete deletion of the major CMIP isoform (Fig. S4A). At baseline, this CMIP KO line and the non-targeting guide WT control displayed (i) similar morphology (Fig. S4B), (ii) expressed key beta cell markers (qPCR; Fig. S4C), (iii) responded to high glucose by a 2-fold increase in insulin secretion (Fig. S4D), and (iv) showed similar doubling rates in culture (Fig. 7A). These data suggested that at baseline CMIP is not required for beta cell survival or insulin secretion. In contrast, CMIP deletion significantly increased sensitivity to lipotoxic stress and cell death (Fig. 7B,C). Specifically, CMIP deletion exacerbated the lipotoxic effects of EA, and it even converted the typically non-toxic c3 MUFAs oleic and petroselenic acid into toxic FFAs (Fig. 7B). Functionally, CMIP deletion increased NFkB signaling (Fig. 7D,E)(Kamal et al. 2009). In CMIP KO cells, EA shifted the peak of RELA nuclear translocation from 18 hours to 3 hours while increasing the overall peak and baseline signal (Fig. 7D). Exposure to PA or EA, but not PSA or OA, increased RELA nuclear translocation in CMIP KO cells more than 4-fold compared to WT controls (Fig. 7E). Thus, CMIP deletion specifically increased inflammatory signaling in beta cells in response to lipotoxic FFAs. CMIP deletion also worsened the EA- or PA-induced reduction in insulin secretion (Fig. 7F). Overall, CMIP deletion rendered beta cells more vulnerable to FFA exposure.
Fig 7. CMIP deletion sensitizes beta cells to FFA-mediated injury and cell death.
(A) CMIP deletion does not affect cell proliferation. No difference in the doubling rate between WT and CMIP KO cells (Two-sided t-test, n=19 splits). (B) After exposure to lipotoxic PA or EA, cell death is increased in CMIP KO compared to WT cells. Non-toxic OA and PSA are rendered toxic in CMIP KO cells. Cell death was measured as number of viable (caspase 3/7 and propidium iodide negative) cells compared to the non-treated control for WT and CMIP KO cells after 72 h exposure to FFAs (500 μM, n=21 wells). Data are mean ± SD. Two-way ANOVA with multiple comparisons (Šídák correction,****p < 0.0001). (C) Representative images of the cell death assay showing increased susceptibility to EA in CMIP-deleted cells. Nuclei were marked by Hoechst and apoptosis was measured by a caspase 3/7 dye (green). Dead cells were stained with propidium iodide (red). Scale bars: 100 μm. (D) Percentage of cells with RELA nuclear translocation after exposure to EA or BSA (500 μM) at 3 h intervals from 0 to 21 h after subtraction of baseline signal from non-treated cells. Data are mean ± SD, n = 7 wells. Two-way ANOVA with multiple comparisons (Šídák correction,****p < 0.0001). (E) Percentage of cells with RELA nuclear translocation after 3 h exposure to FFAs (500 μM) or TNFα (50 ng/ml; positive control) after subtraction of baseline signal from non-treated cells. CMIP KO increased the percentage of RELA translocation after PA or EA treatment. Data are mean ± SD. Two-way ANOVA with multiple comparisons (Šídák correction, ****p < 0.0001). (F) Insulin secretion at baseline normalized to BSA control after 24 h treatment with FFA (500 μM). Insulin secretion was reduced in CMIP deleted cells upon exposure to lipotoxic FFAs. Data are mean ± SD. Two-way ANOVA with multiple comparisons (Holm-Šídák correction, *p < 0.05). PA, palmitic acid; EA, erucic acid; PSA, petroselenic acid; OA, oleic acid.
To confirm that the observed phenotypes could be attributed to the loss of CMIP, we next tested whether reintroducing CMIP in CMIP KO cells could reverse its deleterious effects. Restoring CMIP abundance in CMIP KO cells (as quantified in Fig. S4F) led to a partial rescue from EA-induced cell death (Fig. 8A) and to a faster decline in EA-induced inflammatory NFkB signaling (Fig. 8B; no effect on NFkB signaling in the absence of EA (Fig. S4G)). Similarly, glucose stimulated insulin release was improved upon restoring CMIP expression, especially in EA-treated cells (Fig. 8C). Taken together, these data revealed that CMIP, initially found among thousands of loci in a T2D GWAS, could now be prioritized as a putative suppressor of lipotoxic injury in beta cells.
Fig 8. CMIP protects beta cells from FFAs by regulating Akt activity, and human beta cells lacking CMIP are vulnerable to FFA-induced cell death.
(A) Reintroduction of CMIP in CMIP deleted cells (CMIP rescue) attenuates the toxic effects of EA. Percentage of cell death as measured by decreases in MIN6 viable cells compared to the non-treated control after 24 h exposure to FFAs (250 μM, n=9 wells). Data are mean ± SD. Two-way ANOVA with multiple comparisons (Tukey correction,****p < 0.0001). (B) CMIP rescue attenuates inflammatory NFkB signaling. Percentage of cells with RELA nuclear translocation upon treatment with EA (500 μM) for the indicated times after subtraction of baseline signal from non-treated cells. Data are mean ± SD, n = 6–9 wells. (C) CMIP rescue partially restores insulin secretion in cells exposed to c2 FFAs. Insulin secretion upon glucose stimulation normalized to BSA control for WT and CMIP KO cells after 24 h treatment with FFA (500 μM). Data are mean ± SD. Two way ANOVA with Šídák multiple comparison test (*p < 0.05, n=3 wells). (D) PI3K p85 immunoprecipitates with CMIP in beta cells. Western blot displaying lysate input (left) or co-IP with a CMIP or IgG control antibody (right) stained for CMIP (top) or PI3K (bottom) (n=3 blots). (E) Phosphorylated Akt (pAkt) abundance is increased in CMIP KO cells compared to WT controls. EA exposure increases pAkt in WT cells, but not in CMIP KO cells indicating that CMIP deletion maximizes pAkt levels at baseline. Western blot for pAkt and total Akt after 24 h treatment with 500 μM FFAs (GAPDH, loading control; n=3 blots). (F) In human iPSC-derived beta cells, CMIP deletion promotes cell death, in agreement with experiments in MIN6 beta cells (Fig. 7B). Cell death in human iPSC-derived beta cells as measured by decreases in cell count compared to BSA-treated control. Cells were treated with BSA, EA, or OA at 500 μM for 24 h (n=24 wells). (****p<0.0001, two-way ANOVA with Bonferroni multiple comparison test). (G) Representative images of cell death assay in human iPSC-derived beta cells as measured by number of nuclei (Hoechst). Scale bars: 100 μm. PA, palmitic acid; EA, erucic acid; OA, oleic acid.
To gain insights into CMIP’s function in beta cells, we analyzed transcriptomic data from human islets (Taneera et al. 2012). CMIP gene expression correlated with several pathways including PI3K-Akt signaling, insulin secretion, FFA metabolism, and AMPK signaling (Fig. S4H,I), suggesting that CMIP may play a role in metabolic regulation. Intriguingly, consistent with prior work in peripheral blood mononuclear cells (Kamal et al. 2010), we found that that the regulatory subunit of PI3K (p85) co-immunoprecipitated with CMIP in beta cells (Fig. 8D), suggesting that this interaction may connect CMIP to metabolic signaling pathways (Fig. S4H,I).
To experimentally probe these pathways, we assessed the activity of three critical metabolic sensors in beta cells: AMPK, Akt, and FOXO1. AMPK and FOXO1 signaling were unchanged after CMIP deletion (Fig. S4J). In contrast, CMIP deletion resulted in increased pAkt protein abundance (with no effect on total Akt, Fig. 8E). Interestingly, pAkt in WT cells exposed to EA was at the same level as pAkt in CMIP KO cells at baseline (Fig. 8E). When exposed to EA, CMIP KO cells could not increase pAkt further (Fig. 8E). Since modulation of Akt signaling in response to metabolic stress is thought to promote beta cell survival (Camaya, Donnelly, and O’Brien 2022; Elghazi and Bernal-Mizrachi 2009), and PI3K activity increases the presence of phosphoinositide signaling molecules (PIP2 and PIP3) in the plasma membrane to recruit Akt for activation and downstream signaling (Long et al. 2021), the simplest explanation for our data is that CMIP modulates Akt activity through its interaction with PI3K. Upon CMIP deletion, cells lose the ability to dynamically regulate Akt, making them more vulnerable to FFA-induced cell death compared to WT beta cells (Fig. 7B).
As a final test, and to probe for the human relevance of CMIP, we generated human iPSC-derived beta cells in which CMIP was deleted. In these human cells, CMIP deletion reduced cell viability after treatment with either EA or OA (Fig. 8F,G). In sum, exposure to excess FFAs was particularly deleterious to human beta cells lacking CMIP, validating the key role of CMIP in a relevant model for human health and disease.
Discussion
Lipids, including FFAs, are ubiquitous in living organisms and essential for life, and yet, significant knowledge gaps remain in our understanding of FFA biology. In this study we pioneered FALCON, a multimodal, systematic approach to functionally characterize structurally diverse FFAs. Our approach was comprehensive both in input (number of FFAs tested) and output (multimodal readouts). The added dimensionality - a direct consequence of studying the effects of 61 diverse FFAs - provided the necessary power to uncover biological features across the entire spectrum of FFAs tested. Our studies led to several important conclusions.
First, 20 structurally diverse FFAs defined the toxic (c2) cluster, a group of FFAs united solely by the fact that they mediated similar functional outcomes. The identification of MUFAs in the c2 cluster (12/20 FFAs) suggests a shift in how we interpret the toxicity of FFAs, because we show that saturation alone is not sufficient to predict the lipotoxic potential of a given FFA. MUFAs like OA have been proposed to have harmless or even beneficial effects (Palomer et al. 2018). However, our experiments, including studies in human islets, kidney epithelial cells and microglia showed that OA is not representative of the entire MUFA class, and highlighted several MUFAs, such as EA, that were highly toxic. Our classifier (Fig. S3C) showed that the longest chain of single bonds was predictive of c2 MUFA toxicity, correlating with toxic SFAs rather than non-toxic MUFAs in other clusters (c3 and c4). These findings suggest that a double-bond containing MUFA with a long stretch of carbons linked by single bonds is functionally similar to SFAs, at least in terms of cellular toxicity. Despite induction of similar transcriptomic responses, EA induced a lipidome that was distinct from that induced by PA. In contrast to the well-known accumulation of saturated acyl chains after PA treatment (Piccolis et al. 2019), the EA lipidome was characterized by an accumulation of long-chain unsaturated acyl chains. The distinct lipidomic profiles induced by EA versus PA prompted us to investigate why exposure to these FFAs leads to similar biological outcomes in beta cells (Fig. 4). The membrane fluidity studies hint at the mechanism behind this unique MUFA toxicity; at low concentrations, EA behaves like unsaturated FFAs (e.g. OA), but at higher concentrations, EA becomes harmful because the incorporation of its long unsaturated chains into complex lipid species changes the properties of lipid membranes in a manner similar to SFAs (e.g. PA). The resulting increase in membrane rigidity likely contributes to lipid bilayer stress culminating in activation of cell death pathways (Volmer, van der Ploeg, and Ron 2013). Future work may test whether increased membrane rigidity at high concentrations of EA is directly related to the predicted impact of incorporating its long chain of single bonds (Fig. S3C) into membrane lipids. In sum, we identified and characterized the biological effects of a previously unrecognized set of toxic MUFAs, a discovery enabled by our FALCON platform, and provided mechanistic insight into their particular behavior compared to toxic but structurally dissimilar SFAs.
Second, the comprehensive interrogation of many FFAs with FALCON allowed us to gain new biological insights. Due to the large number of FFAs studied simultaneously (e.g. 20 c2 toxic FFAs), we gained power well beyond that achievable by interrogating the effects of a single FFA alone (such as PA, Fig. 6E). Accordingly, we identified a set of 25 genes that are transcriptionally responsive to lipotoxic stress and are also associated with variants that confer genetic risk for T2D. Our analysis was internally validated by the identification of GLP1R, a well-known obesity (Wilding et al. 2021) and T2D drug target (Scott et al. 2016), and SLC30A8, a gene in which coding variants have been shown to modify risk for T2D (Krentz and Gloyn 2020; Flannick et al. 2014; Dwivedi et al. 2019). Importantly, we identified CMIP as a previously unrecognized suppressor of lipotoxicity, and we confirmed that these findings are relevant to humans using iPSC-derived beta cells. Future studies will focus on the precise molecular mechanism by which CMIP senses FFAs and modulates Akt and related pathways. Nevertheless, this proof-of-concept study illustrates FALCON’s ability to prioritize genes of high mechanistic value that may have otherwise gone unnoticed based on genomics data alone, and offers a method for prioritizing targets that reflect the combined effects of environmental exposure and genetic risk for disease.
Third, FALCON can serve as a valuable tool for exploring fundamental lipid biology in different cell types and tissues. Lipotoxicity and alterations in lipid metabolism have been implicated in numerous disorders including kidney disease (Morgado-Pascual and Opazo-Ríos 2020; Sidhom et al. 2021), neuropathy (Durán et al. 2019), cancer (Munir et al. 2019; Ringel et al. 2020; Ubellacker et al. 2020), liver disease (Liangpunsakul and Chalasani 2019), and Alzheimer’s disease (de la Monte and Tong 2014; Cutuli et al. 2020; Desale and Chinnathambi 2020; Chausse et al. 2019; Madore et al. 2020; Snowden et al. 2017). However, many of the fundamental mechanisms involving lipotoxic FFAs in these diseases have yet to be fully elucidated (Yoon et al. 2021). For example, metabolic alterations in cancer cells that increase fatty acid synthesis and uptake have long been correlated with cancer progression (Munir et al. 2019; German et al. 2016). In other tissues, such as the heart, studies exploring the mechanisms underlying lipotoxic cardiomyopathy have explicitly called for investigations into the contributions of individual fatty acids to disease pathogenesis (Nakamura et al. 2019). FALCON provides the fatty acid level resolution necessary to begin to tackle these complex questions across many cell types and diseases of interest.
Finally, an important translational implication of our study is the notion that the precise FFA profiles in human blood or tissue may carry valuable information about personalized disease risk and progression. Although extensive work is required to fully assess the implications of these findings in vivo, our results motivate the measurement of FFA species in serum samples from large cohorts of well-phenotyped patients. These patient-derived FFA profiles may generate a nuanced view of FFAs as an environmental risk factor, and integrate with the mechanistic insights generated from FALCON. We speculate that integration of polygenic risk scores for complex metabolic diseases (such as obesity, T2D and NAFLD) with serum or tissue-derived “lipotoxicity risk scores’’ for individual patients could become a useful tool for patient risk assessment and stratification in clinical trials.
Limitations of the Study
Since some FFAs in our platform are protective while others are harmful, the study of FFA combinations may be of interest. We note that the large number of potential FFA combinations (>1800) currently limits the feasibility of such a systematic study, but future work may prioritize some FFA combinations for follow-up work.
While the work described in this manuscript was largely focused on c2 FFAs, we uncovered many additional biological processes that merit further study, for example, the putative role of ferroptosis induced by c5 PUFAs in microglia. By making all of our datasets publicly available, we hope that many colleagues in the scientific community will be empowered to explore them to gain fundamental insights into FFA biology in different cell types.
Materials and Methods
Cell Culture
MIN6 cells were purchased (Addex Bio, #C0018008) and cultured as described previously (Burns et al. 2015). In short, cells were maintained in DMEM with 4.5 g/L glucose, supplemented with 10% FBS (fetal bovine serum, Life Technologies, #26140079), 100 U/ml penicillin and 100 μg/ml streptomycin (#15140 Invitrogen) and 55 μM beta-mercaptoethanol (Sigma, #M6250). MIN6 cells were cultured and used for experiments up to passage 30.
Immortalized human kidney epithelial cells used in this study were maintained at 37°C with 5% CO2 in RenaLife Renal Basal Medium supplemented with RenaLife LifeFactors® (Lifeline Cell Technology), with the exclusion of gentamicin and amphotericin B. The cells were generated with informed consent under WFUHS IRB00014033.
INS-1E cells were grown in RPMI 1640 media, supplemented with 10% FBS, 1% penicillin and streptomycin, 1% sodium pyruvate, and 50 μM β-mercaptoethanol (all from Life Technologies). Cells were maintained in flasks pre-coated with supernatant from rat 804G cell line (804G matrix) as previously described (Parnaud et al. 2008).
Cell lines were routinely checked and were negative for mycoplasma.
FFA Preparation
Enzo SCREEN-WELL® Fatty Acid library (#BML-2803-0100) containing FFAs dissolved in DMSO ([FFA]stock = 10 μM) was stored in glass vials at −20°C in the compound management facility of the Broad Institute. Template plates for High Throughput Screens were stored up to 4 weeks at 4°C. To prepare compound plates, small volumes of DMSO dissolved FFAs were transferred into microplates containing fatty acid free BSA (Sigma #A8806) solutions in ddH2O in a molecular ratio of 1:6.67 (BSA:FFA, [FFA]final = 500 μM) with an automated simultaneous pipettor (Analytik Jena CyBio® Well Vario). Plates were incubated overnight for 24 h at 37 °C to ensure complete binding of FFAs to BSA. Next, DMSO and ddH2O were completely removed with the GeneVac HT-12 evaporator for 12 h with full vacuum at 37 °C and continuous centrifugation at 400g. Plates with dry FFA bound BSA crystals in the wells were resuspended in MIN6 culture medium at room temperature for 4–8 h on an orbital plate shaker. After resuspension, compound plates were spun down at 5000g for 10 min and manually transferred to 384 MultiScreenHTS HV Filter Plates (0.45 μm, Millipore, #MZHVN0W10) and spun down again for 1 min at 500g into an empty compound plate. Resulting filtered compound plates were transferred into assay plates of the same format as the CyBio® Well Vario simultaneous pipettor. Representative FFAs were ordered from Nu-Chek Prep, Inc., manually dissolved in DMSO ([FFA]stock = 10 μM) and prepared in glass vials according to the same protocol.
Differential Scanning Calorimetry
All differential scanning calorimetry (DSC) measurements were performed with a MicroCal VP-Capillary DSC Automated system (Malvern Panalytical). Selected FFAs were bound to BSA in microplates according to the protocol described above and resuspended in PBS to a final concentration of [BSA]final = 50 μM. Sample measurements included one measurement of PBS vs. PBS to record a baseline reference curve at the scan rate of 200 °C/h. The samples were heated from Tstart = 10 °C up to Tend = 90 °C at the same scan rate. The melting temperature Tm was determined from the resulting single-peak melting curve using FFA-free BSA as a control.
Lipid Profiling
40,000 MIN6 cells/well were seeded in 96 well plates 24 h prior to treatment in three replicates. FFA library compound plates were transferred into assay plates which were then incubated for 24 h. The lipid fraction of cells was isolated with isopropanol after washing the plates 3 times with ice cold PBS. After the addition of isopropanol, plates were incubated for 1 h at 4 °C. IPA extracts were then manually transferred to autosampler vials (Waters), capped, and stored at −80 °C until analysis. Lipid profiling was done as previously described (Paynter et al. 2018). Briefly, non-targeted liquid chromatography mass spectrometry (LC-MS) data were acquired using a system composed of a Nexera X2 U-HPLC system (Shimadzu Scientific Instruments; Marlborough, MA) coupled to a Q Exactive Focus Orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Cell extracts (2 μL) were injected directly onto a 100 × 2.1 mm, 1.7 μm ACQUITY BEH C8 column (Waters; Milford, MA). The column was then eluted isocratically with 80% mobile phase A (95/5/0.1; vol/vol/vol 10mM ammonium acetate/methanol/formic acid) for 1 min followed by a linear gradient to 80% mobile-phase B (99.9/0.1; vol/vol methanol/formic acid) over 2 min, a linear gradient to 100% mobile phase B over 7 min, then 3 min at 100% mobile-phase B. Mass spectrometry analyses were carried out using ESI in the positive ion mode using full scan analysis over 220–1100 m/z. Raw data were processed and visually inspected using TraceFinder 3.3 software (Thermo Fisher Scientific; Waltham, MA) and Progenesis QI (Nonlinear Dynamics; Newcastle upon Tyne, UK). The identity of individual metabolites and lipid families was confirmed by matching their retention time to that of authentic reference standards.
For the comparison between the PA and EA-induced lipidomes, MIN6 were seeded at 800,000 per well in a 6-well plate and grown for 3 days (n=4). After the cells were treated with the respective FFAs at 500 μM for 24h, cells were washed 3x with 1ml of room temperature PBS. Cells were scraped in 500ul room temperature PBS, pelleted at 13,000×g, and flash frozen in liquid nitrogen for storage after the supernatant was removed. Cell pellets were lysed in double distilled water following three cycles of freeze-thawing in a water bath sonicator. Subsequently, lipids were extracted according to Folch’s Method. The organic phase of each sample, normalized by tissue weight, were then separated using ultra high performance liquid chromatography coupled to tandem mass spectrometry (UHPLC-MSMS) (Narváez-Rivas and Zhang 2016). UHPLC analysis was performed employing a C30 reverse-phase column (Thermo Acclaim C30, 2.1 × 250 mm, 3 μM, operated at 55° C; Thermo Fisher Scientific) connected to a Dionex UltiMate 3000 UHPLC system and a Q-Exactive Orbitrap high resolution mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI-II) probe. Extracted lipid samples were dissolved in 2:1 methanol:chloroform (v/v) and 5 μl of each sample was analyzed separately using positive and negative ionization modes, respectively. Mobile phase A consisted of 60:40 water/acetonitrile (v/v), 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of 90:10 isopropanol/acetonitrile (v/v), 10 mM ammonium formate and 0.1% formic acid. Lipids were separated over a 90 min gradient; during 0–7 minutes, elution starts with 40% B and increases to 55%; from 7 to 8 min, increases to 65% B; from 8 to 12 min, elution is maintained with 65% B; from 12 to 30 min, increase to 70% B; from 30 to 31 min, increase to 88% B; from 31 to 51 min, increase to 95% B; from 51 to 53 min, increase to 100% B; during 53 to 73 min, 100% B is maintained; from 73 to 73.1 min, solvent B was decreased to 40% and then maintained for another 16.9 min for column re-equilibration. The flow-rate for chromatographic separation was set to 0.2 mL/min. The column oven temperature was set at 55° C, and the temperature of the autosampler tray was set to 4° C. The spray voltage was set to 4.2 kV, and the heated capillary and the HESI were held at 320° C and 300° C, respectively. The S-lens RF level was set to 50, and the sheath and auxiliary gas were set to 35 and 3 units, respectively. These conditions were held constant for both positive and negative ionization mode acquisitions. External mass calibration was performed using the standard calibration mixture every 7 days. MS spectra of lipids were acquired in full-scan/data-dependent MS2 mode. For the full-scan acquisition, the resolution was set to 70,000, the AGC target was 1e6, the maximum injection time was 50 msec, and the scan range was m/z = 133.4–2000. For data-dependent MS2, the top 10 precursor selection in each full scan were isolated with a 1.0 Da window, fragmentation using stepped normalized collision energy of 15, 25, and 35 units, and analyzed at a resolution of 17,500 with an AGC target of 2e5 and a maximum injection time of 100 msec. The underfill ratio was set to 0. The selection of the top 10 precursors was subject to isotopic exclusion with a dynamic exclusion window of 5.0 sec. All data were analyzed using the LipidSearch version 5.0 SP (Thermo Fisher Scientific) and all identified species (grade A, B) were reported.
RNASeq / SmartSeq 2
40,000 MIN6 cells/well were seeded in 96-well plates 24 h prior to treatment in three replicates. FFA library compound plates were transferred into assay plates which were then incubated for 24 h (n=6). RNA was extracted from cells using TCL buffer (#1031576, Qiagen) with 1% beta-mercaptoethanol followed by an RNA clean-up with Agencourt RNA cleanup XP (#A63987, Beckman Coulter). Bulk RNA (1 μl) was added to a 3 step cDNA synthesis reaction with 3’RT (5’-AGCAGTGGTATCAACGCAGAGTAC(T30)VN-3’, IDT), template switching (5’-AAGCAGTGGTATCAACGCAGAGTACrGrG+G-3’, Qiagen), and ISPCR (5’-AAGCAGTGGTATCAACGCAGAGT-3’, IDT) oligos from the SMART-seq2 protocol (Picelli et al. 2014). cDNA was purified using AMPure XP Agencourt (#100609, Beckman Coulter) and quantified using Qubit dsDNA High Sensitivity (#102689, Life Technologies). Samples were diluted to 0.2 ng/ul in TE and tagmented (Nextera XT DNA Library Preparation Kit (#FC-131–1096, Illumina). Indexing was performed using the Nextera XT Index Kit (#FC-131-1001, Illumina). Final libraries were QCed using the Qubit dsDNA High Sensitivity kit and Bioanalyzer High Sensitivity DNA Kit (#5067–4627, Agilent). Libraries were sequenced at a concentration of 1.8 pM on a NextSeq with a 75 cycle v2 kit (#TG-160-2002, Illumina) with a read structure of Read 1 37bp, Read 2 37bp, Index 1 8bp, and Index 2 8bp. Each sample had approximately 4 million reads.
Cell Painting
MIN6 cells were seeded at a density of 15,000 per well in a 384-well plate (Perkin Elmer, CellCarrier Ultra, #6057300). After 24 h recovery, cells were treated with the FFA library and incubated for 24 h. Cell staining and fixation was performed according to previous protocols (Bray et al. 2016). Images were acquired with the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer) with a 63X/1.15NA water immersion lens. Image quality control was carried out using CellProfiler 2.2.0 (Kamentsky et al. 2011) and CellProfiler Analyst (Dao et al. 2016) according to a prior machine-learning based protocol (Bray et al. 2012). Image illumination correction and analysis were performed in CellProfiler (pipelines available upon request). After analysis, the data were compiled and normalized in Cytominer (code available at https://github.com/cytomining/cytominer) as described previously (Bray et al. 2016). Briefly, single cell level features were aggregated per well by computing averages. The mean values and dispersion of the 2202 features measured in all samples were normalized to negative controls (BSA). Features with near-zero variance were removed, and a non-redundant feature set was created by inspecting pairwise correlations. The remaining 1222 features comprise the morphological profile of a given well.
Data Analysis
Unless otherwise stated all computational and statistical analysis in this study has been performed in Python and R.
(1). Lipidomics
A blocked experimental design with one replicate of each FFA in the library, together with multiple BSA controls per 96-well plate, was chosen (n=3). Raw lipidomic profiles received from the Metabolomics Platform at the Broad Institute were filtered for samples with strongly deviating sample medians (manual cutoff, 7 out of 280 or 3% of the samples were discarded). Lipid metabolites with more than 30% of missing data points were removed, otherwise missing values were substituted with 50% of the minimum value of the respective metabolite’s intensity. To account for variations in total amount of captured metabolites, samples were scaled towards the global sample median. Only annotated lipid metabolites were used for further differential abundance analysis. We sought to understand the relationship between structural features of externally added FFAs and changes in the triglyceride fraction of the cells (Fig. 1C). For each externally added FFA, triglyceride intensity deviations from the BSA control were summed based on the structural feature of interest (number of C-atoms, number of double bonds). Then, triglyceride profiles of externally added FFAs were summarized based on the structural feature of interest of the FFAs (number of C-atoms, number of double bonds) and normalized to the number of FFAs making up each group. For assessment of the global lipidome in response to erucic and palmitic acid, lipid metabolites were filtered as described and subsequently imported into lipidR (Mohamed, Molendijk, and Hill 2020). Differential analysis of lipid abundance was calculated using the empirical Bayes procedure. Fold change in lipid abundance (EA vs. PA) was then normalized based on noted structural features (number of C-atoms, number of double bonds) and visualized in R. Network analysis of the biochemical relationship between differentially abundant lipid species was performed using the Lipid Network Explorer with default settings (Köhler et al. 2021).
(2). RNASeq Pipeline and Gene Set Enrichment Analysis
A blocked experimental design with one replicate of each FFA in the library together with multiple BSA controls per 96-well plate was chosen (n=6). Raw data from NextSeq runs were de-multiplexed and converted to sample specific fastq files. Alignment was performed with STAR (Dobin et al. 2013), reads were counted with HTSeq (Anders, Pyl, and Huber 2015) and QC metrics were generated with RNA-SeQC (DeLuca et al. 2012).
The resulting count matrix was filtered by column for samples with more than 103 detected genes (counts > 0) and by row for coding genes (as defined by the MGI database) with a row sum across all samples > 500 counts (with a total number of 500 samples). The resulting normalized and filtered count matrix was then variance stabilized using the vst method from the DESeq2 R package (Love, Huber, and Anders 2014). Surrogate variable analysis (SVA, R package)(Leek et al. 2012) was performed on the vst count matrix to account for linear batch effects. In addition, we performed differential expression analysis with DESeq2 for each sample, including derived surrogate variables to the linear model. To cluster the samples, we chose the top 500 most common significant differentially expressed genes (padj < 0.05) across the entire dataset. Samples were either transformed to z-scores or replicates were collapsed by calculating their signal to noise ratio (with respect to the BSA control) before performing hierarchical clustering based on Euclidean distance and Ward’s linkage method. Clusters were extracted with the Dynamic Tree Cut function (Langfelder, Zhang, and Horvath 2008). After assigning each FFA to a cluster, we performed differential expression analysis based on cluster labels and BSA controls (based on the vst count matrix) and calculated adjusted p-values for each gene (Mann–Whitney U, Bonferroni). For gene set enrichment analysis (GSEA), gene lists for each FFA transcriptome were ranked by log2 fold changes as compared to BSA control to weigh genes according to their differential expression vs control. MsigDB H: HALLMARK gene sets, C2: KEGG and REACTOME gene sets and C5: GO BP gene sets were tested for enrichment (Liberzon et al. 2011; Subramanian et al. 2005). 250 Genesets with the most common, significant differential enrichment across the whole FFA library were selected and transformed into a normalized enrichment score (NES) matrix. Hierarchical clustering of those genesets resulted in the presented gene modules presented in Fig. 2A. Pearson correlation analysis of human islet gene expression (Taneera et al. 2012) was performed using MATLAB 2020a (MathWorks), as previously described (Brown et al. 2021). Genes significantly correlated with CMIP (FDR < 0.01,Benjamini-Hochberg) were subjected to KEGG pathway analysis using Metascape and visualized using Cytoscape.
(3). Rank-rank hypergeometric overlap (RRHO).
The R package RRHO2 (Cahill et al. 2018) was used to perform RRHO analysis to evaluate the cutoff-free overlap in differential expression results from FFA Clusters (C1-C5) versus human islets (Lytrivi, Ghaddar, et al. 2020) and mice pancreatic beta cells (Dusaulcy et al. 2019). RRHO2 plots show a heatmap with four quadrants to display the overlap between expression list comparisons. This includes: downregulated observations in both data sets (top right, Fig, S2E,F), downregulated in the comparative data and upregulated observations in our data (bottom right, Fig. S2E,F), upregulated in the comparative data and downregulated observations in our data (top left, Fig. S2E,F) and upregulated observations in both data sets (bottom left, Fig. S2E,F). For each comparison, one-sided enrichment tests were used on −log(p-values) with default step size for each quadrant.
(4). Structural analysis of FFAs
Molecular structural features were generated with the MOE software. A complete list of generated features are summarized in Table S5. Detailed descriptions of these features are available in the MOE user manual (v2016.08). The molecular feature matrix was filtered for non-constant features across the FFA library and transformed into z-scores. To decrease the linear dependence between features, meta-features were extracted from the original molecular features by hierarchical clustering based on Pearson correlation and Ward’s linkage method (cutreeDynamic, WGCNA, R package)(Langfelder, Zhang, and Horvath 2008). Clustering was performed iteratively until the maximum Pearson r correlation coefficient between any two meta-features was less than 0.8 (n=3 iterations). The first principal component of molecular feature clusters were used to define meta-features.
The random forest classifier (RFC) was performed (randomForest; R package)(Breiman 2001). Optimal values for ntree (number of trees to grow) and mtry (number of variables randomly sampled as candidates at each split) were determined empirically based on the classification accuracy of the RFC run on the entire dataset. The RFC was then run with leave-one-out cross validation. Meta-feature importance measures for RFC prediction were calculated (importance).
(5). Morphological feature analysis
Confocal images were acquired as described for Cell Painting using the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer)(Bray et al. 2016). Image analysis was performed using Harmony software (PerkinElmer). Single nuclei were first identified using Hoechst staining. Associated cell bodies around each nuclei were identified by the “find cytoplasm function” method A (individual cutoff 0.1) from the Harmony software in the 488 nm channel. ER and mitochondrial regions were identified (Find Image Region) based on their respective channels (stained with Concanavalin A and MitoTracker® respectively) For each cell, the standard morphological features (area and roundness); as well as advanced STAR (Symmetry, Threshold compactness, Axial or Radial) and SER (Spots, Edges, Ridges) morphology for ER and mitochondria were calculated. A full list of extracted features is summarized in Table S6. Data was exported per cell object, and downstream analysis was performed in R.
(6). MAGMA analysis pipeline, gene functional readout correlations
The MAGMA software (de Leeuw et al. 2015) (v 1.07) was used to perform SNP annotation, gene analysis to generate ranked lists of genes from GWAS summary statistics and gene set analysis (GSA) according to the instructions provided in the user manual. For multiple hypothesis testing we used a permutation-based approach to generate an empirical Null Hypothesis to account for the enrichment of beta cell genes in previous T2D GWAS analyses. For each gene set, we generated 1000 randomly sampled gene sets based on the MIN6 transcriptome of the same size and calculated the FDR accordingly.
iPSC-beta Cell Differentiation
The iPSC-derived beta cells were differentiated as described previously (Maxwell and Millman 2021; Hogrebe et al. 2020) from an episomal reprogrammed iPSC line (Gibco, #A18945). The factors added each day of differentiation can be found in Table S7. After the conclusion of the 28 day differentiation process, these cells were maintained in enriched serum-free media (ESFM) with media changes every other day. On day 30, the cultures were dissociated with TrypLE incubation for a maximum of 5 min, followed by neutralization with ESFM, TrypLE removal through centrifugation at 200×g for 3 min, filtration through a 40 μm filter to remove large clumps, and seeding onto HTB-9 ECM coated 96-well plates (Perkin Elmer, CellCarrier Ultra, #6055308) at 10,000/well.
iPSC-microglia Cell Differentiation
AICS-0036-006 from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research was used in this study. iPSCs were cultured in Essential 8 (E8) (Thermo Fisher Scientific) media on Matrigel (Corning) coated 6-well plates. Media was changed daily until confluence. iMGLs were differentiated as previously described (Abud et al. 2017; Dolan et al. 2022). When confluent, iPSCs were dissociated using Accutase (Stem Cell technologies), centrifuged for 5 mins at 300xg and counted using trypan blue (Thermo Fisher Scientific). 200,000 cells/well were resuspended in E8 containing 10 μM Y27632 ROCK inhibitor (Selleckchem) in low adherence 6-well plates (Corning). For the first 10 days, cells were cultured in HPC medium [50% IMDM (Thermo Fisher Scientific), 50% F12 (Thermo Fisher Scientific), ITSG-X 2% v/v (Thermo Fisher Scientific), L-ascorbic acid 2-Phosphate (64 ug/ml, Sigma), monothioglycerol (400mM, Sigma), Poly(vinyl) alcohol (PVA) (10mg/ml, Sigma), Glutamax (1X, Thermo Fisher Scientific), chemically-defined lipid concentrate (1X, Thermo Fisher Scientific) and non-essential amino acids (Thermo Fisher Scientific)]. At day 0, embryoid bodies (EB) were gently collected, centrifuged at 100xg and resuspended in HPC medium supplemented with 1 μM ROCK inhibitor, FGF2 (50 ng/ml, Thermo Fisher Scientific), BMP4 (50ng/ml, Thermo Fisher Scientific), Activin-A (12.5ng/ml, Thermo Fisher Scientific) and LiCL (2 mM, Sigma), then incubated in a hypoxic incubator (5% O2, 5% CO2, 37 °C). On day 2, cells were gently collected and the media changed to HPC medium supplemented with FGF2 (50ng/ml, Thermo Fisher Scientific) and VEGF (50 ng/ml, PeproTech) and returned to the hypoxic incubator. On day 4, cells were collected and media changed to HPC medium supplemented with FGF2 (50ng/ml, Thermo Fisher Scientific), VEGF (50 ng/ml, PeproTech), TPO (50 ng/ml, PeproTech), SCF (10ng/ml, Thermo Fisher Scientific), IL6 (50ng/ml, PeproTech) and IL3 (10ng/ml, PeproTech) and incubated in a normoxic incubator (20% O2, 5% CO2, 37C). At day 6 and 8, 1 ml of day 4 media was added in each well. On day 10, cells were collected, counted using trypan blue and frozen in Cryostor (Sigma Aldrich) in aliquots of 300,000–500,000 cells.
For iMGL differentiation, cells were thawed, washed 1x with PBS and plated at 200,000 cells per well in 6-well plates coated with matrigel in iMGL media [DMEM/F12 (Thermo Fisher Scientific), ITS-G (2% v/v, Thermo Fisher Scientific), B27 (2% v/v, Thermo Fisher Scientific), N2 (0.5% v/v, Thermo Fisher Scientific), monothioglycerol (200 mM, Sigma), Glutamax (1X, Thermo Fisher Scientific), non-essential amino acids (1X, Thermo Fisher Scientific)] supplemented with M-CSF (25 ng/ml, PeproTech ), IL-34 (10 ng/ml, PeproTech) and TGFB-1 (50ng/ml, PeproTech). Cells were fed every 2 days and replated at day 22. On day 30, cells were collected and replated in iMGL media supplemented with M-CSF (25 ng/ml, PeproTech), IL-34 (10 ng/ml, PeproTech), TGFB-1 (50ng/ml, PeproTech), CD200 (100ng/ml, VWR) and CX3CL1 (100ng/ml, PeproTech).
CMIP KO Clone Selection
Lentivirus containing a plasmid programmed to express either CMIP-specific sgRNA (ACGTCTTCAATGGCGCTGTAGG, Millipore Sigma, Sanger Clone MM5000005403) or non-targeting control sgRNA (Millipore Sigma, CRISPR20–1EA) was obtained through transfecting HEK293T cells with these plasmids in combination with a second generation CMV lentiviral packaging system and FuGENE transfection agent (E2311, Promega).
MIN6 cells stably expressing Cas9 were infected with this lentivirus at half volume with 8 μg/ml polybrene (TR-1003-G, Sigma) overnight. The media was changed 12 h later, and 48 h after the first media change, 1 μg/ml of puromycin was added to the media. After one week of puromycin selection, the cultures were dispersed into single cells, seeded into 96-well plates for expansion, and sequenced for CMIP mutations. One WT clone and one possessing a truncation within the first 15% of the longest CMIP isoform were used for the experiments described.
To derive CMIP KO cells from the human iPSC-beta cell line, we delivered Cas9-RNP complexes with either CMIP-specific sgRNA or non-targeting sgRNA (same as Min6 above) to the parental iPSC cells using a Lonza nucleofector kit. Upon confirmation of successful transfection and generation of a stable knockout line, single clones were isolated and sequenced to ultimately identify the clone used in the studies described.
CMIP Overexpression for Rescue Studies
To re-express CMIP back into the MIN6 CMIP KO line, we obtained lentivirus from VectorBuilder containing a mouse CMIP ORF (NM_001163262.1) with a modified PAM site under a CMV promoter and accompanied by a neomycin resistance gene. We used the same vector with amino acids 2–83 of E. coli beta-galactosidase as a substitute for the CMIP ORF for our control line. 600,000 cells were seeded into each well of a 6-well plate and one week later were infected with up to 100ul virus per well (and 8 ug/ml polybrene (TR-1003-G, Sigma)) overnight. The media was changed 12 h later, and 48 h after the first media change, 800 μg/ml of G418 was added to the media. After selection for one week, the cells were used for the experiments described.
Cell Viability
For the high throughput cell viability assay, cells were seeded in 384-well plates (Perkin Elmer, CellCarrier Ultra, #6057300) and treated for 24, 48 and 72 h with the FFA library (n=7 / FFA). Just before readout, cell nuclei were stained with Hoechst (Thermo Fisher Scientific) for 1 h at 37 °C and imaged with the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer). Number of counted nuclei was determined with the image analysis software Harmony (PerkinElmer) and used as a proxy for cell viability. For validation experiments, cells were treated for 48 h with representative FFAs in CellCarrier-384 Ultra Microplates. Caspase 3/7 (Thermo Fisher Scientific, #C10423) activation and propidium iodide (Thermo Fisher Scientific, #P3566) staining were used to calculate the fraction of apoptotic cells and dead cells, respectively. Single cells were identified and counted after staining their nuclei with Hoechst. Fluorescence intensities were measured and the threshold for caspase 3/7 and propidium iodide positive staining was determined manually. Cell viability was calculated as the fraction of cells that were neither caspase 3/7 nor propidium iodide positive. For the EA dose-response curve, MIN6 cells were grown as described and then treated for 65 hours with BSA, EA or OA. The concentrations were (in mM): 0.7, 2.1, 6.2, 18.5, 55.6, 166.7, 500, 1000 and were prepared by serial dilution from a stock concentration. Cell viability was assessed as described.
iPSC-derived microglia
On day 30 of the differentiation, iMGLs were plated in 96-well plates (Perkin Elmer, CellCarrier) at a density of 20,000 cells per well. One day before treatment, FFA and BSA were reconstituted in iMGL media and gently rocked overnight at room temperature. At day 40, iMGL were treated with a final concentration 250 μM of each FFA or BSA for 24 h followed by live imaging. Cells were imaged using the Opera Phenix confocal system using a 20X objective, temperature was maintained at 37 °C and CO2 at 5% during the imaging period. One plane was taken per picture and 20 images were taken per well. Harmony software was used for analysis, flatfield correction (Basic Method) and cell identification using EGFP signal (common Threshold 0.02; area >50 μm2). Total number of cells per well was used for quantification with an n=4 for all conditions.
Kidney tubular epithelial cells
Prior to treatment, BSA-bound FFAs were reconstituted and incubated overnight in RenaLife media at room temperature. Cells were seeded in CellCarrier-384 Ultra Microplates and maintained at 37°C with 5% CO2. Cells were treated with 500 μM FFA or BSA for 15 h followed by live imaging using the Opera Phenix High Content Screening System. Number of cells and number of dead cells was determined on Harmony image analysis software using digital phase contrast and propidium iodide staining (Thermo Fisher Scientific, #P3566), respectively. Viability was assessed by calculating the number of propidium iodide-negative cells per total number of cells per well (n=4).
Western Blot
MIN6 cells were lysed (#9803, Cell Signaling Technology) in the presence of protease inhibitors (#05892791001, Roche) and phosphatase inhibitors (#04906837001, Roche). Protein concentrations were quantified with the Pierce BCA Protein Assay Kit (#23225, Thermo Fisher Scientific). NuPAGE LDS sample buffer (#NP0008, Thermo Fisher Scientific) was added to normalized protein lysates together with NuPAGE reducing agent (#NP0004, Thermo Scientific). Lysates were heated to 95 °C for 5 min prior to SDS-PAGE gel electrophoresis (NuPAGE MES SDS running buffer, Thermo Fisher Scientific, #NP0002). Proteins were transferred to a nitrocellulose membrane (#1704158, BioRad) with the Trans-Blot® Turbo TM Blotting System (#1704155, BioRad) according to the manufacturer’s protocol. Membranes were blocked in 5% Nonfat Dry Milk (#9999S, Cell Signaling Technology) in PBS with 0.1% Tween® 20 (PBS-T). AKT and pAKT blots were blocked instead in 5% Bovine Serum Albumin (#1900–0016, LGC Clinical Diagnostics) in PBS-T. Primary antibodies were incubated at 4 °C overnight, secondary antibodies were incubated at room temperature for 1 h. Super Signal West Dura (#34076, Thermo Fisher Scientific) or SuperSignal West Pico (#34087, Thermo Fisher Scientific) were used to visualize immunoreactive bands imaged by G:BOX Chemi XT4 (BOX-CHEMI-XT4, Syngene). Primary antibodies used in this study were CPT1A: (#ab128568, Abcam), ATF4: (#11815, Cell Signaling Technology), CHOP: (#2895, Cell Signaling Technology), CMIP: (NBP2–58180, Novus Bio), AKT: (#9272, Cell Signaling Technology), pAKT: (#4060, Cell Signaling Technology), AMPK : (#2532, Cell Signaling Technology), pAMPK : (#2535, Cell Signaling Technology), FOXO1: (#2880, Cell Signaling Technology), pFOXO1: (#9464, Cell Signaling Technology), GAPDH-HRP: (#3683, Cell Signaling Technology). Secondary antibodies used were: anti-rabbit IgG-HRP (#7074, Cell Signaling Technology), anti-mouse IgG-HRP (#7076, Cell Signaling Technology).
Coimmunoprecipitation (co-IP)
Two 10cm dishes of Min6 cells were lysed with 1ml of co-IP lysis buffer (100mM NaCl, 5mM EDTA, 50mM Tris-HCl pH 7.5, 1% NP-40, protease inhibitor tablet, phosphatase inhibitor tablet) after being washed once with ice cold PBS. The lysate was rotated at 4°C for 30m, spun at 13,000xg for 20m at 4°C, and the supernatant was collected. 1mg of protein was combined with 10ug of either CMIP antibody (12851-1-AP, Proteintech) or rabbit IgG control antibody (10500C, Thermo Fisher) and rotated overnight at 4°C. The following day, Pierce Protein A/G Magnetic Beads (88802, Thermo Fisher) were warmed to room temperature, washed twice with 1ml TBS-T (150mM NaCl, 50mM Tris-HCl pH 7.5, 0.05% Tween-20) on a magnet, and resuspended in the original volume of co-IP lysis buffer. 25ul of beads were added to each lysate tube and the solution was rotated at 4°C for 1h. The beads were then washed 3x with 1ml TBS-T, once with 1ml distilled water, and eluted with 30ul of 2X NuPAGE LDS Sample Buffer and 1X NuPAGE Sample Reducing Agent (DTT). The beads were incubated at room temperature for 10m with occasional flicking of the tube, and then the supernatant was extracted on the magnet. The supernatant was incubated at 95°C for 10m and then the samples were run on a gel as detailed in the Western Blot section above. The primary antibodies used for staining were CMIP: (12851-1-AP, Proteintech) and PI3K p85: (#4292, Cell Signaling Technology).
Immunofluorescence (IF) staining
MIN6 NFkB Immunofluorescence
MIN6 cells grown on 384-well CellCarrier Ultra microplates (#6057308, PerkinElmer) were fixed for 10 min in PBS containing 4% PFA (Electron Microscopy Sciences), permeabilized for 15 min in 0.5% Triton X-100 (Sigma-Aldrich), blocked for 1 h in blocking reagent (100 mM Tris HCL pH 8; 150 mM NaCL; 5 g/L Blocking Reagent (#11096176001, Roche)) and treated for 1.5 h with primary antibody diluted in blocking reagent (NF-κB p65/RELA, Rabbit monoclonal antibody, 1:200, #8242, Cell Signaling Technology). Cells were washed three times in PBS and incubated for 0.5 h with fluorescent-labeled secondary antibody in blocking solution (1:500, Alexa Fluor 568 Goat anti-Rabbit IgG, (#A11036, Thermo Fisher Scientific)). Cytoplasmic actin filaments were stained with Phalloidin conjugated with Alexa 647 (1:40, #A22287, Thermo Fisher Scientific) and nuclei were counterstained with Hoechst (1:2000, #H3570, Thermo Fisher Scientific). Cells were washed three times in PBS and imaged using the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer). A minimum of nine fields were acquired per well using 20x water immersion objectives in confocal mode. Image analysis was performed using the Harmony software (PerkinElmer). Cell nuclei were first identified using Hoechst staining and a nuclear region was defined for each cell. Phalloidin staining was then used to detect and define the cytoplasmic region of the cell. RELA fluorescent intensity was measured separately in the nuclear and cytoplasmic regions and a threshold for a nuclear translocation was defined using negative (BSA) and positive (TNF) controls. For each well, the fraction of cells identified for RELA nuclear translocation was calculated.
MIN6 LC3B Immunofluorescence
MIN6 cells grown on 384-well CellCarrier Ultra microplates (#6057308, PerkinElmer) were treated for 48 h with 500 μM FFAs or 25 nM of rapamycin (Sigma-Aldrich, R8781) or bafilomycin A1 (Sigma-Aldrich SML1661). Cells were fixed for 20 min in ice-cold methanol (Sigma-Aldrich, 154903), washed twice with PBS, permeabilized for 15 min in 0.5% Triton X-100 (Sigma-Aldrich, 10789704001), washed twice with PBS, blocked for 1 h in 5% BSA in PBS (SeraCare, 1900–0016) and incubated for 1.5 h with primary antibody diluted in blocking reagent (Lc3b (D11) XP® Rabbit mAb, 1:500, #3868, Cell Signaling Technology). Cells were washed four times in PBS and incubated for 45 min with fluorescent-labeled secondary antibody in blocking solution (1:500, Alexa Fluor 568 Donkey anti-Rabbit IgG, (#A10042, Thermo Fisher Scientific)). Nuclei were counterstained with Hoechst (1:2000, #H3570, Thermo Fisher Scientific). Cells were washed four times in PBS and imaged using the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer). A minimum of nine fields were acquired per well using 63x water immersion objectives in confocal mode. Image analysis was performed using the Harmony software (PerkinElmer). Cell nuclei were first identified using Hoechst staining and a nuclear region was defined for each cell. LC3B puncta were then detected based on intensity and size with rapamycin and bafilomycin treatments serving as controls.
Primary Human Islet Immunofluorescence
Primary human islets from three independent cadaveric donors were received from the Integrated Islet Distribution Program. Intact islets were dissociated using accutase followed by trituration and stained using Trypan Blue to confirm viability. They were seeded at 15,000/well into 384-well Cell Carrier Ultra microplates (Perkin Elmer, CellCarrier Ultra, #6057300) coated with HTB-9-derived ECM, and treated two days later with FFAs at 250 μM, 500 μM, and 1 mM. After treatment for 5 days, the islets were fixed for 20 min in 3% PFA followed by permeabilization for 20 min with 0.2% TritonX-100. Blocking with 2% BSA in PBS (SeraCare, AP-45100-80) was conducted for 1 h at room temperature, followed by primary incubation with c-peptide antibody (Developmental Studies Hybridoma Bank at the University of Iowa, GN-ID4) at 1:100 overnight at 4 °C. The plate was washed 3x with PBS followed by secondary incubation with 568 goat anti-rat (Life technologies, A11077) at 1:1000 and Hoechst (1:2000, #H3570, Thermo Fisher Scientific) for 1 h at room temperature. The plate was then washed 5x with PBS and imaged on the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer). A minimum of nine fields were acquired per well using a 20x water immersion objective in confocal mode. Image analysis was performed using the Harmony software (PerkinElmer). Beta cells were identified and counted by c-peptide positive staining. All three donors displayed similar trends in c2 toxicity; the data presented in Fig. 5C are representative of all donors.
iPSC-Derived Beta Cell Immunofluorescence
IPSC-derived beta cells after 28 days of differentiation as per the protocol listed above were dissociated and seeded at 10,000/well into 96-well Cell Carrier Ultra microplates (Perkin Elmer, CellCarrier Ultra, #6055308) coated with HTB-9-derived ECM. These cells were treated three days later with FFAs at 250 μM, 500 μM, and 750 μM for 24, 48, and 72 h followed by fixation with 4% PFA for 30 min at room temperature. The plates were washed twice with PBS, permeabilized for 15 min with 0.5% Triton-X 100, washed twice again with PBS, and blocked with 5% BSA in PBS for 1 h at room temperature. Hoechst was added (1:2000, #H3570, Thermo Fisher Scientific) and incubated for 1 h at room temperature. The plate was then washed thrice with PBS and imaged on the Opera Phenix High Content Screening System (#HH14000000, Perkin Elmer). A minimum of nine fields were acquired per well using a 20x water immersion objective in confocal mode. Image analysis was performed using the Harmony software (PerkinElmer) and the cells were counted.
ER Calcium Levels
MIN6 cells were plated in 384-well plates (Aurora, Black 384 SQ Well 188 micron Film, #1022–10110) and treated with the FFA library for 24 h prior to readout (n=5 / FFA). Cells were carefully washed three times with HBSS (with calcium, Thermo Fisher Scientific, #14025076) using an automated simultaneous pipettor (analytikjena CyBio® Well vario) and incubated with the fluorescent calcium indicator Fluo4 (2 μM, Life Technologies, #F14202) in DMEM without supplementation for 1 h at room temperature. Then, cells were washed again in HBSS (with calcium) and incubated for another 30 min at room temperature in DMEM without supplementation. Just before the readout, cells were washed in calcium free assay buffer solution (140 mM NaCl, 5 mM KCl, 10 mM HEPES, 2 mM MgCl2, 10 mM EGTA, 10 mM Glucose) and left with 25 μl assay volume per well. Assay plates were immediately transferred to the FLIPR Tetra® High-Throughput Cellular Screening System. The plate was recorded with a frequency of 1Hz for 10 min. Baseline was recorded for 30 s before the automated liquid transfer system of the FLIPR added the SERCA inhibitor Thapsigargin (final concentration 10 μM) in calcium free assay buffer. The resulting passive efflux of calcium from the ER induced a transient cytosolic fluorescence signal and the peak amplitudes were used to indirectly quantify ER calcium levels (See Fig. S3A). The resulting trajectories were corrected for a pipetting artifact and baseline normalized. Log2 Fold changes were calculated according to plate location specific negative (BSA) controls. We allowed for the exclusion of one outlier / FFA / plate (n=5) based on a 3-sigma cutoff. P-values were calculated with Student’s t-test (two-sided) and corrected for multiple testing (Benjamini & Hochberg).
Glucose Stimulated Insulin Secretion (GSIS)
To measure GSIS, cells were plated in 96-well plates (Perkin Elmer, CellCarrier Ultra, #6055308) and treated with the FFA library for 24 h prior to readout (n=6 / FFA). First, cells were pre-incubated in Krebs Ringer Buffer (KRB) with 2.8 mM glucose for 1 h and then stimulated with fresh KRB containing 16.7 mM glucose for another hour. The supernatant was then transferred to a mouse insulin ELISA (Thermo Fisher, EMINS) at a dilution of 1:150. The cells were imaged on the Opera Phenix High Content Screening System, (#HH14000000, Perkin Elmer) and subsequently counted via Hoechst nuclear staining using Harmony software (PerkinElmer). A minimum of four fields were acquired per well using a 10x air objective in a confocal mode. The insulin concentrations measured via ELISA were normalized to the number of cells in each respective well.
Membrane Fluidity Assay
This assay has been well established and optimized in INS-1E cells, a rat beta cell line. INS-1E cells were seeded at 20,000 cells per well in a CellCarrier Ultra 96-well plate (Perkin Elmer). Cells were incubated for 24 h, washed once with PBS, treated with BSA-bound fatty acids in a serum-free media and further incubated for 18 h at 37°C. Cells were washed twice with PBS and stained with Laurdan dye (6-dodecanoyl-2-dimethylaminoaphthalene) (Thermofisher) at 10 μM for 45 min. Cells were washed twice with PBS and images were acquired using an Opera Phenix High-Content Screening system (Perkin Elmer). Temperature (37°C) and carbon dioxide (5%) were controlled during live-cell imaging. Cells were excited with a 405 nm laser and the emission recorded between 435 and 480 nm (ordered phase) and between 500 and 550 nm (disordered phase). Images were analyzed using Harmony High-Content Imaging and Analysis Software (Perkin Elmer) and data represented as general polarization (GP) index as calculated by GP = (intensityorder - intensitydisorder)/(intensityorder + intensitydisorder) (Harris, Best, and Bell 2002; Parasassi et al. 1991).
Supplementary Material
Highlights.
FALCON (Fatty Acid Library for Comprehensive ONtologies) enables multimodal profiling of 61 free fatty acids (FFAs) to reveal 5 FFA clusters with distinct biological effects
FALCON is applicable to many and diverse cell types
A subset of monounsaturated FAs (MUFAs) equally or more toxic than canonical lipotoxic saturated FAs (SFAs) leads to decreased membrane fluidity
New approach prioritizes genes that represent the combined effects of environmental (FFA) exposure and genetic risk for disease
C-Maf inducing protein (CMIP) is identified as a suppressor of FFA-induced lipotoxicity via Akt-mediated signaling
Acknowledgements
We thank Katie Liguori for excellent graphic design work. We are also grateful to colleagues in the Center for the Development of Therapeutics for their assistance in the preparation of the FFA library. This work was supported by a seed grant from the Broad Institute of MIT and Harvard (Bn10 to AG), who is also supported by DK095045, DK099465, and Cure Alzheimer’s Fund. NW was supported by a Deutsche Forschungsgesellschaft Fellowship (WI 4612/1-1) and is a participant in the BIH-Charité Junior Digital Clinician Scientist Program funded by the Charité –Universitätsmedizin Berlin and the Berlin Institute of Health, JCF was supported by the National Institute of General Medical Sciences (T32GM007753, T32GM007726), E-H.S. was supported by the National Institutes of Health (F30DK112477), C.K. was supported by the National Institutes of Health (F31DK126252), J.R. was supported by the National Institute of General Medical Sciences (T32GM007753), M.R.B was supported by the National Institutes of Health (K00DK123834), M.D.-L. was supported by a Broad–Israeli Science Foundation Fellowship, E.S. was supported by NIH 1-R01-DK126855-01 and an American Surgical Association Foundation Fellowship, MC was supported by FNIH AMP-T2D RFB8b, NIDDK UM1 DK126185, AEC and SS were supported by the National Institutes of Health (NIH MIRA R35 GM122547 to AEC), and RD was supported by Knut and Alice Wallenberg Foundation, Sweden (KAW 2019.0580). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing Interests
NW, JCF and AG are co-inventors of a patent on the composition, method and use for FFA screening, application No.: 52199-550P01US. A.G. serves as a founding advisor to a new company launched by Atlas Ventures, an agreement reviewed and managed by Brigham and Women’s Hospital, Mass General Brigham, and the Broad Institute of MIT and Harvard in accordance with their conflict of interest policies.
References
- Abdelmagid Salma A., Clarke Shannon E., Nielsen Daiva E., Badawi Alaa, El-Sohemy Ahmed, Mutch David M., and Ma David W. L.. 2015. “Comprehensive Profiling of Plasma Fatty Acid Concentrations in Young Healthy Canadian Adults.” PLOS ONE. 10.1371/journal.pone.0116195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abifadel Marianne, Varret Mathilde, Rabès Jean-Pierre, Allard Delphine, Ouguerram Khadija, Devillers Martine, Cruaud Corinne, et al. 2003. “Mutations in PCSK9 Cause Autosomal Dominant Hypercholesterolemia.” Nature Genetics 34 (2): 154–56. [DOI] [PubMed] [Google Scholar]
- Abud Edsel M., Ramirez Ricardo N., Martinez Eric S., Healy Luke M., Nguyen Cecilia H. H., Newman Sean A., Yeromin Andriy V., et al. 2017. “iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases.” Neuron 94 (2): 278–93.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sulaiti Haya, Diboun Ilhame, Banu Sameem, Al-Emadi Mohamed, Amani Parvaneh, Harvey Thomas M., Dömling Alex S., Latiff Aishah, and Elrayess Mohamed A.. 2018. “Triglyceride Profiling in Adipose Tissues from Obese Insulin Sensitive, Insulin Resistant and Type 2 Diabetes Mellitus Individuals.” Journal of Translational Medicine 16 (1): 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders Simon, Pyl Paul Theodor, and Huber Wolfgang. 2015. “HTSeq--a Python Framework to Work with High-Throughput Sequencing Data.” Bioinformatics 31 (2): 166–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubourg Patrick, Adamsbaum Catherine, Lavallard-Rousseau Marie-Claude, Rocchiccioli Francis, Cartier Nathalie, Jambaque Isabelle, Jakobezak Christine, et al. 1993. “A Two-Year Trial of Oleic and Erucic Acids (‘Lorenzo’s Oil’) as Treatment for Adrenomyeloneuropathy.” The New England Journal of Medicine 329 (11): 745–52. [DOI] [PubMed] [Google Scholar]
- Bachiller Sara, Jiménez-Ferrer Itzia, Paulus Agnes, Yang Yiyi, Swanberg Maria, Deierborg Tomas, and Boza-Serrano Antonio. 2018. “Microglia in Neurological Diseases: A Road Map to Brain-Disease Dependent-Inflammatory Response.” Frontiers in Cellular Neuroscience 12 (December): 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A. A., Grüne T., and Curry S.. 2000. “Crystallographic Analysis Reveals Common Modes of Binding of Medium and Long-Chain Fatty Acids to Human Serum Albumin.” Journal of Molecular Biology 303 (5): 721–32. [DOI] [PubMed] [Google Scholar]
- Blonde L., Klein E. J., Han J., Zhang B., Mac S. M., Poon T. H., Taylor K. L., Trautmann M. E., Kim D. D., and Kendall D. M.. 2006. “Interim Analysis of the Effects of Exenatide Treatment on A1C, Weight and Cardiovascular Risk Factors over 82 Weeks in 314 Overweight Patients with Type 2 Diabetes.” Diabetes, Obesity & Metabolism 8 (4): 436–47. [DOI] [PubMed] [Google Scholar]
- Bouachi Khedidja, Moktefi Anissa, Zhang Shao-Yu, Oniszczuk Julie, Sendeyo Kelhia, Remy Philippe, Audard Vincent, Pawlak Andre, Ollero Mario, and Sahali Djillali. 2018. “Expression of CMIP in Podocytes Is Restricted to Specific Classes of Lupus Nephritis.” PloS One 13 (11): e0207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray Mark-Anthony, Fraser Adam N., Hasaka Thomas P., and Carpenter Anne E.. 2012. “Workflow and Metrics for Image Quality Control in Large-Scale High-Content Screens.” Journal of Biomolecular Screening 17 (2): 266–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray Mark-Anthony, Singh Shantanu, Han Han, Davis Chadwick T., Borgeson Blake, Hartland Cathy, Kost-Alimova Maria, Gustafsdottir Sigrun M., Gibson Christopher C., and Carpenter Anne E.. 2016. “Cell Painting, a High-Content Image-Based Assay for Morphological Profiling Using Multiplexed Fluorescent Dyes.” Nature Protocols 11 (9): 1757–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman Leo. 2001. “Random Forests.” Machine Learning 45 (1): 5–32. [Google Scholar]
- Brown Matthew R., Holmes Heather, Rakshit Kuntol, Javeed Naureen, Her Tracy K., Stiller Alison A., Sen Satish, et al. 2021. “Electrogenic Sodium Bicarbonate Cotransporter NBCe1 Regulates Pancreatic β Cell Function in Type 2 Diabetes.” The Journal of Clinical Investigation 131 (17). 10.1172/JCI142365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns Sean M., Vetere Amedeo, Walpita Deepika, Dančík Vlado, Khodier Carol, Perez Jose, Clemons Paul A., Wagner Bridget K., and Altshuler David. 2015. “High-Throughput Luminescent Reporter of Insulin Secretion for Discovering Regulators of Pancreatic Beta-Cell Function.” Cell Metabolism. 10.1016/j.cmet.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Cahill Kelly M., Huo Zhiguang, Tseng George C., Logan Ryan W., and Seney Marianne L.. 2018. “Improved Identification of Concordant and Discordant Gene Expression Signatures Using an Updated Rank-Rank Hypergeometric Overlap Approach.” Scientific Reports 8 (1): 9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camaya Inah, Donnelly Sheila, and O’Brien Bronwyn. 2022. “Targeting the PI3K/Akt Signaling Pathway in Pancreatic β-Cells to Enhance Their Survival and Function: An Emerging Therapeutic Strategy for Type 1 Diabetes.” Journal of Diabetes 14 (4): 247–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Yaying, Wang Tao, Wu Yiqun, Juan Juan, Qin Xueying, Tang Xun, Wu Tao, and Hu Yonghua. 2018. “Opposite Genetic Effects of CMIP Polymorphisms on the Risk of Type 2 Diabetes and Obesity: A Family-Based Study in China.” International Journal of Molecular Sciences 19 (4). 10.3390/ijms19041011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Jeng Yie, Luzuriaga Jude, Maxwell Emma L., West Phillip K., Bensellam Mohammed, and Laybutt D. Ross. 2015. “The Balance between Adaptive and Apoptotic Unfolded Protein Responses Regulates β-Cell Death under ER Stress Conditions through XBP1, CHOP and JNK.” Molecular and Cellular Endocrinology 413 (September): 189–201. [DOI] [PubMed] [Google Scholar]
- Chausse Bruno, Kakimoto Pamela A., Caldeira-da-Silva Camille C., Chaves-Filho Adriano B., Yoshinaga Marcos Y., da Silva Railmara Pereira, Miyamoto Sayuri, and Kowaltowski Alicia J.. 2019. “Distinct Metabolic Patterns during Microglial Remodeling by Oleate and Palmitate.” Bioscience Reports 39 (4). 10.1042/BSR20190072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Xiaocui, Shang Lin, Deng Senwen, Li Ping, Chen Kai, Gao Ting, Zhang Xiao, Chen Zhilan, and Zeng Jia. 2020. “Peroxisomal Oxidation of Erucic Acid Suppresses Mitochondrial Fatty Acid Oxidation by Stimulating Malonyl-CoA Formation in the Rat Liver.” The Journal of Biological Chemistry 295 (30): 10168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer Melina, Cho Judy H., Collins Rory, Cox Nancy J., Dermitzakis Emmanouil T., Hurles Matthew E., Kathiresan Sekar, et al. 2020. “A Brief History of Human Disease Genetics.” Nature 577 (7789): 179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman Rosalind A., and Mashek Douglas G.. 2011. “Mammalian Triacylglycerol Metabolism: Synthesis, Lipolysis, and Signaling.” Chemical Reviews 111 (10): 6359–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry Stephen, Mandelkow Hendrik, Brick Peter, and Franks Nick. 1998. “Crystal Structure of Human Serum Albumin Complexed with Fatty Acid Reveals an Asymmetric Distribution of Binding Sites.” Nature Structural Biology 5 (9): 827–35. [DOI] [PubMed] [Google Scholar]
- Cutuli Debora, Landolfo Eugenia, Nobili Annalisa, Paola De Bartolo Stefano Sacchetti, Chirico Doriana, Marini Federica, et al. 2020. “Behavioral, Neuromorphological, and Neurobiochemical Effects Induced by Omega-3 Fatty Acids Following Basal Forebrain Cholinergic Depletion in Aged Mice.” Alzheimer’s Research & Therapy. 10.1186/s13195-020-00705-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao David, Fraser Adam N., Hung Jane, Ljosa Vebjorn, Singh Shantanu, and Carpenter Anne E.. 2016. “CellProfiler Analyst: Interactive Data Exploration, Analysis and Classification of Large Biological Image Sets.” Bioinformatics. 10.1093/bioinformatics/btw390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca David S., Levin Joshua Z., Sivachenko Andrey, Fennell Timothy, Nazaire Marc-Danie, Williams Chris, Reich Michael, Winckler Wendy, and Getz Gad. 2012. “RNA-SeQC: RNA-Seq Metrics for Quality Control and Process Optimization.” Bioinformatics. 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempfle Astrid, Scherag André, Hein Rebecca, Beckmann Lars, Chang-Claude Jenny, and Schäfer Helmut. 2008. “Gene–environment Interactions for Complex Traits: Definitions, Methodological Requirements and Challenges.” European Journal of Human Genetics: EJHG 16 (10): 1164–72. [DOI] [PubMed] [Google Scholar]
- Desale Smita Eknath, and Chinnathambi Subashchandrabose. 2020. “Role of Dietary Fatty Acids in Microglial Polarization in Alzheimer’s Disease.” Journal of Neuroinflammation. 10.1186/s12974-020-01742-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon Scott J., Lemberg Kathryn M., Lamprecht Michael R., Skouta Rachid, Zaitsev Eleina M., Gleason Caroline E., Patel Darpan N., et al. 2012. “Ferroptosis: An Iron-Dependent Form of Nonapoptotic Cell Death.” Cell 149 (5): 1060–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin Alexander, Davis Carrie A., Schlesinger Felix, Drenkow Jorg, Zaleski Chris, Jha Sonali, Batut Philippe, Chaisson Mark, and Gingeras Thomas R.. 2013. “STAR: Ultrafast Universal RNA-Seq Aligner.” Bioinformatics. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan Michael-John, Therrien Martine, Jereb Saša, Kamath Tushar, Atkeson Trevor, Marsh Samuel E., Goeva Aleksandrina, et al. 2022. “A Resource for Generating and Manipulating Human Microglial States in Vitro.” bioRxiv. 10.1101/2022.05.02.490100. [DOI] [Google Scholar]
- Donath Marc Y., Dalmas Élise, Sauter Nadine S., and Böni-Schnetzler Marianne. 2013. “Inflammation in Obesity and Diabetes: Islet Dysfunction and Therapeutic Opportunity.” Cell Metabolism 17 (6): 860–72. [DOI] [PubMed] [Google Scholar]
- Doyle-Delgado Kacie, Chamberlain James J., Shubrook Jay H., Skolnik Neil, and Trujillo Jennifer. 2020. “Pharmacologic Approaches to Glycemic Treatment of Type 2 Diabetes: Synopsis of the 2020 American Diabetes Association’s Standards of Medical Care in Diabetes Clinical Guideline.” Annals of Internal Medicine. 10.7326/m20-2470. [DOI] [PubMed] [Google Scholar]
- Durán Alfonso M., Salto Lorena M., Câmara Justin, Basu Anamika, Paquien Ivette, Beeson W. Lawrence, Firek Anthony, Cordero-MacIntyre Zaida, and De León Marino. 2019. “Effects of Omega-3 Polyunsaturated Fatty-Acid Supplementation on Neuropathic Pain Symptoms and Sphingosine Levels in Mexican-Americans with Type 2 Diabetes.” Diabetes, Metabolic Syndrome and Obesity: Targets and Therapy 12 (January): 109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusaulcy Rodolphe, Handgraaf Sandra, Visentin Florian, Howald Cedric, Dermitzakis Emmanouil T., Philippe Jacques, and Gosmain Yvan. 2019. “High-Fat Diet Impacts More Changes in Beta-Cell Compared to Alpha-Cell Transcriptome.” PloS One 14 (3): e0213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi Om Prakash, Lehtovirta Mikko, Hastoy Benoit, Chandra Vikash, Krentz Nicole A. J., Kleiner Sandra, Jain Deepak, et al. 2019. “Loss of ZnT8 Function Protects against Diabetes by Enhanced Insulin Secretion.” Nature Genetics 51 (11): 1596–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebato Chie, Uchida Toyoyoshi, Arakawa Masayuki, Komatsu Masaaki, Ueno Takashi, Komiya Koji, Azuma Kosuke, et al. 2008. “Autophagy Is Important in Islet Homeostasis and Compensatory Increase of Beta Cell Mass in Response to High-Fat Diet.” Cell Metabolism 8 (4): 325–32. [DOI] [PubMed] [Google Scholar]
- Eguchi Kosei, Manabe Ichiro, Oishi-Tanaka Yumiko, Ohsugi Mitsuru, Kono Nozomu, Ogata Fusa, Yagi Nobuhiro, et al. 2012. “Saturated Fatty Acid and TLR Signaling Link β Cell Dysfunction and Islet Inflammation.” Cell Metabolism 15 (4): 518–33. [DOI] [PubMed] [Google Scholar]
- Elghazi Lynda, and Bernal-Mizrachi Ernesto. 2009. “Akt and PTEN: Beta-Cell Mass and Pancreas Plasticity.” Trends in Endocrinology and Metabolism: TEM 20 (5): 243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Zhou, Weng Chen, Li Haiyan, Tao Ran, Mai Weihua, Liu Xiaoxiao, Lu Leina, et al. 2019. “Single-Cell Heterogeneity Analysis and CRISPR Screen Identify Key β-Cell-Specific Disease Genes.” Cell Reports 26 (11): 3132–44.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farese Robert V., Zechner Rudolf Jr, Newgard Christopher B., and Walther Tobias C.. 2012. “The Problem of Establishing Relationships between Hepatic Steatosis and Hepatic Insulin Resistance.” Cell Metabolism 15 (5): 570–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannick Jason, Thorleifsson Gudmar, Beer Nicola L., Jacobs Suzanne B. R., Grarup Niels, Burtt Noël P., Mahajan Anubha, et al. 2014. “Loss-of-Function Mutations in SLC30A8 Protect against Type 2 Diabetes.” Nature Genetics 46 (4): 357–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsberger Christian, Flannick Jason, Teslovich Tanya M., Mahajan Anubha, Agarwala Vineeta, Gaulton Kyle J., Ma Clement, et al. 2016. “The Genetic Architecture of Type 2 Diabetes.” Nature 536 (7614): 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Shin-Ichi, and Amisaki Takashi. 2008. “Identification of High Affinity Fatty Acid Binding Sites on Human Serum Albumin by MM-PBSA Method.” Biophysical Journal 94 (1): 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Suneng, Yang Ling, Li Ping, Hofmann Oliver, Dicker Lee, Hide Winston, Lin Xihong, Watkins Steven M., Ivanov Alexander R., and Hotamisligil Gökhan S.. 2011. “Aberrant Lipid Metabolism Disrupts Calcium Homeostasis Causing Liver Endoplasmic Reticulum Stress in Obesity.” Nature 473 (7348): 528–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German Natalie J., Yoon Haejin, Yusuf Rushdia Z., Murphy J. Patrick, Finley Lydia W. S., Laurent Gaëlle, Haas Wilhelm, et al. 2016. “PHD3 Loss in Cancer Enables Metabolic Reliance on Fatty Acid Oxidation via Deactivation of ACC2.” Molecular Cell 63 (6): 1006–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golfetto Ottavia, Hinde Elizabeth, and Gratton Enrico. 2013. “Laurdan Fluorescence Lifetime Discriminates Cholesterol Content from Changes in Fluidity in Living Cell Membranes.” Biophysical Journal 104 (6): 1238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan Kevin A., Weigel Maya K., Prakash Priya, Wijewardhane Prageeth R., Hasel Philip, Rufen-Blanchette Uriel, Münch Alexandra E, et al. 2021. “Neurotoxic Reactive Astrocytes Induce Cell Death via Saturated Lipids.” Nature 599 (7883): 102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris Faith M., Best Katrina B., and Bell John D.. 2002. “Use of Laurdan Fluorescence Intensity and Polarization to Distinguish between Changes in Membrane Fluidity and Phospholipid Order.” Biochimica et Biophysica Acta 1565 (1): 123–28. [DOI] [PubMed] [Google Scholar]
- Hayden Matthew S., and Ghosh Sankar. 2008. “Shared Principles in NF-kappaB Signaling.” Cell 132 (3): 344–62. [DOI] [PubMed] [Google Scholar]
- Hobbs H. H., Brown M. S., and Goldstein J. L.. 1992. “Molecular Genetics of the LDL Receptor Gene in Familial Hypercholesterolemia.” Human Mutation 1 (6): 445–66. [DOI] [PubMed] [Google Scholar]
- Hogrebe Nathaniel J., Augsornworawat Punn, Maxwell Kristina G., Velazco-Cruz Leonardo, and Millman Jeffrey R.. 2020. “Targeting the Cytoskeleton to Direct Pancreatic Differentiation of Human Pluripotent Stem Cells.” Nature Biotechnology 38 (4): 460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Nurulain, Yap Wei Sheng, Xu Jiaming, Wu Haoxi, Koh Jhee Hong, Goh Wilson Wen Bin, George Bhawana, Chong Shu Chen, Taubert Stefan, and Thibault Guillaume. 2020. “Stress Sensor Ire1 Deploys a Divergent Transcriptional Program in Response to Lipid Bilayer Stress.” The Journal of Cell Biology 219 (7). 10.1083/jcb.201909165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil Gökhan S. 2010. “Endoplasmic Reticulum Stress and the Inflammatory Basis of Metabolic Disease.” Cell 140 (6): 900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil Gökhan S. 2017. “Inflammation, Metaflammation and Immunometabolic Disorders.” Nature 542 (7640): 177–85. [DOI] [PubMed] [Google Scholar]
- Howe Vicky, Sharpe Laura J., Prabhu Anika V., and Brown Andrew J.. 2017. “New Insights into Cellular Cholesterol Acquisition: Promoter Analysis of Human HMGCR and SQLE, Two Key Control Enzymes in Cholesterol Synthesis.” Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids 1862 (7): 647–57. [DOI] [PubMed] [Google Scholar]
- Imamura Fumiaki, Sharp Stephen J., Koulman Albert, Schulze Matthias B., Kröger Janine, Griffin Julian L., Huerta José M., et al. 2017. “A Combination of Plasma Phospholipid Fatty Acids and Its Association with Incidence of Type 2 Diabetes: The EPIC-InterAct Case-Cohort Study.” PLoS Medicine 14 (10): e1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou Maria S., Jackson Jesse, Sheu Shu-Hsien, Chang Chi-Lun, Weigel Aubrey V., Liu Hui, Pasolli H. Amalia, et al. 2019. “Neuron-Astrocyte Metabolic Coupling Protects against Activity-Induced Fatty Acid Toxicity.” Cell 177 (6): 1522–35.e14. [DOI] [PubMed] [Google Scholar]
- Jang Hee-Seong, Noh Mi Ra, Kim Jinu, and Padanilam Babu J.. 2020. “Defective Mitochondrial Fatty Acid Oxidation and Lipotoxicity in Kidney Diseases.” Frontiers of Medicine 7 (March): 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha Pooja, McDevitt Molly T., Halilbasic Emina, Williams Evan G., Quiros Pedro M., Gariani Karim, Sleiman Maroun B., et al. 2018. “Genetic Regulation of Plasma Lipid Species and Their Association with Metabolic Phenotypes.” Cell Systems 6 (6): 709–21.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal Maud, Pawlak Andre, BenMohamed Fatima, Valanciuté Asta, Dahan Karine, Candelier Marina, Lang Philippe, Guellaën Georges, and Sahali Djillali. 2010. “C-Mip Interacts with the p85 Subunit of PI3 Kinase and Exerts a Dual Effect on ERK Signaling via the Recruitment of Dip1 and DAP Kinase.” FEBS Letters 584 (3): 500–506. [DOI] [PubMed] [Google Scholar]
- Kamal Maud, Valanciute Asta, Dahan Karine, Ory Virginie, Pawlak Andre, Lang P., Guellaen Georges, and Sahali Djillali. 2009. “C-Mip Interacts Physically with RelA and Inhibits Nuclear Factor Kappa B Activity.” Molecular Immunology 46 (5): 991–98. [DOI] [PubMed] [Google Scholar]
- Kamentsky Lee, Jones Thouis R., Fraser Adam, Bray Mark-Anthony, Logan David J., Madden Katherine L., Ljosa Vebjorn, Rueden Curtis, Eliceiri Kevin W., and Carpenter Anne E.. 2011. “Improved Structure, Function and Compatibility for CellProfiler: Modular High-Throughput Image Analysis Software.” Bioinformatics. 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K., Shioda S., Ando K., Sakagami K., Handa H., and Yasuda K.. 2004. “Differentially Expressed Maf Family Transcription Factors, c-Maf and MafA, Activate Glucagon and Insulin Gene Expression in Pancreatic Islet Alpha- and Beta-Cells.” Journal of Molecular Endocrinology. 10.1677/jme.0.0320009. [DOI] [PubMed] [Google Scholar]
- Keaton Jacob M., Gao Chuan, Guan Meijian, Hellwege Jacklyn N., Palmer Nicholette D., Pankow James S., Fornage Myriam, et al. 2018. “Genome-Wide Interaction with the Insulin Secretion Locus MTNR1B Reveals CMIP as a Novel Type 2 Diabetes Susceptibility Gene in African Americans.” Genetic Epidemiology 42 (6): 559–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo Shih, Gibson Tara Beers, Arnette Don, Lawrence Michael, January Bridgette, McGlynn Kathleen, Vanderbilt Colleen A., Griffen Steven C., German Michael S., and Cobb Melanie H.. 2004. “MAP Kinases and Their Roles in Pancreatic Beta-Cells.” Cell Biochemistry and Biophysics 40 (3 Suppl): 191–200. [DOI] [PubMed] [Google Scholar]
- Köhler Nikolai, Rose Tim Daniel, Falk Lisa, and Pauling Josch Konstantin. 2021. “Investigating Global Lipidome Alterations with the Lipid Network Explorer.” Metabolites 11 (8). 10.3390/metabo11080488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz Nicole A. J., and Gloyn Anna L.. 2020. “Insights into Pancreatic Islet Cell Dysfunction from Type 2 Diabetes Mellitus Genetics.” Nature Reviews. Endocrinology 16 (4): 202–12. [DOI] [PubMed] [Google Scholar]
- Langfelder Peter, Zhang Bin, and Horvath Steve. 2008. “Defining Clusters from a Hierarchical Cluster Tree: The Dynamic Tree Cut Package for R.” Bioinformatics 24 (5): 719–20. [DOI] [PubMed] [Google Scholar]
- Leek Jeffrey T., Johnson W. Evan, Parker Hilary S., Jaffe Andrew E., and Storey John D.. 2012. “The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments.” Bioinformatics 28 (6): 882–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw Christiaan A. de, Mooij Joris M., Heskes Tom, and Posthuma Danielle. 2015. “MAGMA: Generalized Gene-Set Analysis of GWAS Data.” PLoS Computational Biology 11 (4): e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire K., Ravier M. A., Schraenen A., Creemers J. W. M., Van de Plas R., Granvik M., Van Lommel L., et al. 2009. “Insulin Crystallization Depends on Zinc Transporter ZnT8 Expression, but Is Not Required for Normal Glucose Homeostasis in Mice.” Proceedings of the National Academy of Sciences of the United States of America 106 (35): 14872–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liangpunsakul Suthat, and Chalasani Naga. 2019. “Lipid Mediators of Liver Injury in Nonalcoholic Fatty Liver Disease.” American Journal of Physiology-Gastrointestinal and Liver Physiology 316 (1): G75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon Arthur, Birger Chet, Thorvaldsdóttir Helga, Ghandi Mahmoud, Mesirov Jill P., and Tamayo Pablo. 2015. “The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection.” Cell Systems 1 (6): 417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon Arthur, Subramanian Aravind, Pinchback Reid, Thorvaldsdóttir Helga, Tamayo Pablo, and Mesirov Jill P.. 2011. “Molecular Signatures Database (MSigDB) 3.0.” Bioinformatics 27 (12): 1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Hui-Zhi, Cheng Yan, Zhou Zi-Wei, Luo Hong-Yu, Wen Dan-Dan, and Gao Li-Chen. 2021. “PI3K/AKT Signal Pathway: A Target of Natural Products in the Prevention and Treatment of Alzheimer’s Disease and Parkinson’s Disease.” Frontiers in Pharmacology. 10.3389/fphar.2021.648636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo Nicola, Frigeni Marta, and Pasquali Marzia. 2016. “Carnitine Transport and Fatty Acid Oxidation.” Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1863 (10): 2422–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love Michael I., Huber Wolfgang, and Anders Simon. 2014. “Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2.” Genome Biology. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lust Cody A. C., Bi Xinyan, Henry Christiani Jeyakumar, and Ma David W. L.. 2021. “Development of Fatty Acid Reference Ranges and Relationship with Lipid Biomarkers in Middle-Aged Healthy Singaporean Men and Women.” Nutrients 13 (2). 10.3390/nu13020435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytrivi Maria, Castell Anne-Laure, Poitout Vincent, and Cnop Miriam. 2020. “Recent Insights Into Mechanisms of β-Cell Lipo- and Glucolipotoxicity in Type 2 Diabetes.” Journal of Molecular Biology. 10.1016/j.jmb.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytrivi Maria, Ghaddar Kassem, Lopes Miguel, Rosengren Victoria, Piron Anthony, Yi Xiaoyan, Johansson Henrik, et al. 2020. “Combined Transcriptome and Proteome Profiling of the Pancreatic β-Cell Response to Palmitate Unveils Key Pathways of β-Cell Lipotoxicity.” BMC Genomics 21 (1): 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madore C., Leyrolle Q., Morel L., Rossitto M., Greenhalgh A. D., Delpech J. C., Martinat M., et al. 2020. “Essential Omega-3 Fatty Acids Tune Microglial Phagocytosis of Synaptic Elements in the Mouse Developing Brain.” Nature Communications. 10.1038/s41467-020-19861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan Anubha, Min Jin Go Weihua Zhang, Below Jennifer E., Gaulton Kyle J., Ferreira Teresa, Horikoshi Momoko, et al. 2014. “Genome-Wide Trans-Ancestry Meta-Analysis Provides Insight into the Genetic Architecture of Type 2 Diabetes Susceptibility.” Nature Genetics 46 (3): 234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan Anubha, Taliun Daniel, Thurner Matthias, Robertson Neil R., Torres Jason M., Rayner N. William, Payne Anthony J., et al. 2018. “Fine-Mapping Type 2 Diabetes Loci to Single-Variant Resolution Using High-Density Imputation and Islet-Specific Epigenome Maps.” Nature Genetics 50 (11): 1505–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmugi Alice, Parnis Julia, Chen Xi, Carmichael Leanne, Hardy Julie, Mannan Naila, Marchetti Piero, et al. 2016. “Sorcin Links Pancreatic β-Cell Lipotoxicity to ER Ca2+ Stores.” Diabetes 65 (4): 1009–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell Kristina G., and Millman Jeffrey R.. 2021. “Applications of iPSC-Derived Beta Cells from Patients with Diabetes.” Cell Reports Medicine 2 (4): 100238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnik A. 2003. “Thermal Stability of Bovine Serum Albumin DSC Study.” Journal of Thermal Analysis and Calorimetry 71 (2): 509–19. [Google Scholar]
- Mir Shakeel U. R., George Nicholas M., Zahoor Lubna, Harms Robert, Guinn Zachary, and Sarvetnick Nora E.. 2015. “Inhibition of Autophagic Turnover in β-Cells by Fatty Acids and Glucose Leads to Apoptotic Cell Death.” The Journal of Biological Chemistry 290 (10): 6071–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilenko Denis A., Haas Joel T., L’homme Laurent, Fleury Sébastien, Quemener Sandrine, Levavasseur Matthieu, Becquart Coralie, et al. 2019. “Metabolic and Innate Immune Cues Merge into a Specific Inflammatory Response via the UPR.” Cell 178 (1): 263. [DOI] [PubMed] [Google Scholar]
- Mohamed Ahmed, Molendijk Jeffrey, and Hill Michelle M.. 2020. “Lipidr: A Software Tool for Data Mining and Analysis of Lipidomics Datasets.” Journal of Proteome Research 19 (7): 2890–97. [DOI] [PubMed] [Google Scholar]
- Moktefi Anissa, Zhang Shao-Yu, Vachin Pauline, Ory Virginie, Henique Carole, Audard Vincent, Rucker-Martin Catherine, et al. 2016. “Repression of CMIP Transcription by WT1 Is Relevant to Podocyte Health.” Kidney International 90 (6): 1298–1311. [DOI] [PubMed] [Google Scholar]
- Mo Man-Qiu, Pan Ling, Lu Qing-Mei, Li Qiu-Lin, and Liao Yun-Hua. 2018. “The Association of the CMIP rs16955379 Polymorphism with Dyslipidemia and the Clinicopathological Features of IgA Nephropathy.” International Journal of Clinical and Experimental Pathology 11 (10): 5008–23. [PMC free article] [PubMed] [Google Scholar]
- Monte Suzanne M. de la, and Ming Tong. 2014. “Brain Metabolic Dysfunction at the Core of Alzheimer’s Disease.” Biochemical Pharmacology 88 (4): 548–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado-Pascual J. L., and Opazo-Ríos L.. 2020. “Pathogenic Pathways and Therapeutic Approaches Targeting Inflammation in Diabetic Nephropathy.” International Journal of. https://www.mdpi.com/1422-0067/21/11/3798. [DOI] [PMC free article] [PubMed]
- Munir Rimsha, Lisec Jan, Swinnen Johannes V., and Zaidi Nousheen. 2019. “Lipid Metabolism in Cancer Cells under Metabolic Stress.” British Journal of Cancer 120 (12): 1090–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru Kiran, and Kathiresan Sekar. 2016. “Surprises From Genetic Analyses of Lipid Risk Factors for Atherosclerosis.” Circulation Research 118 (4): 579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2019. “Genetics of Common, Complex Coronary Artery Disease.” Cell 177 (1): 132–45. [DOI] [PubMed] [Google Scholar]
- Nakamura Michinari, Liu Tong, Husain Seema, Zhai Peiyong, Warren Junco S., Hsu Chiao-Po, Matsuda Takahisa, et al. 2019. “Glycogen Synthase Kinase-3α Promotes Fatty Acid Uptake and Lipotoxic Cardiomyopathy.” Cell Metabolism 29 (5): 1119–34.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narváez-Rivas Mónica, and Zhang Qibin. 2016. “Comprehensive Untargeted Lipidomic Analysis Using Core-Shell C30 Particle Column and High Field Orbitrap Mass Spectrometer.” Journal of Chromatography. A 1440 (April): 123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson Tamara J., Bellomo Elisa A., Wijesekara Nadeeja, Loder Merewyn K., Baldwin Jocelyn M., Gyulkhandanyan Armen V., Koshkin Vasilij, et al. 2009. “Insulin Storage and Glucose Homeostasis in Mice Null for the Granule Zinc Transporter ZnT8 and Studies of the Type 2 Diabetes–Associated Variants.” Diabetes 58 (9): 2070–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Yoon S., Bae Gong D., Baek Dong J., Park Eun-Young, and Jun Hee-Sook. 2018. “Fatty Acid-Induced Lipotoxicity in Pancreatic Beta-Cells During Development of Type 2 Diabetes.” Frontiers in Endocrinology 9 (July): 384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann James A., and Carvalho Pedro. 2018. “Dynamics and Functions of Lipid Droplets.” Nature Reviews. Molecular Cell Biology 20 (3): 137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onengut-Gumuscu Suna, Chen Wei-Min, Burren Oliver, Cooper Nick J., Quinlan Aaron R., Mychaleckyj Josyf C., Farber Emily, et al. 2015. “Fine Mapping of Type 1 Diabetes Susceptibility Loci and Evidence for Colocalization of Causal Variants with Lymphoid Gene Enhancers.” Nature Genetics 47 (4): 381–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrenius Sten, Zhivotovsky Boris, and Nicotera Pierluigi. 2003. “Regulation of Cell Death: The Calcium–apoptosis Link.” Nature Reviews. Molecular Cell Biology 4 (7): 552–65. [DOI] [PubMed] [Google Scholar]
- Oyadomari S., and Mori M.. 2004. “Roles of CHOP/GADD153 in Endoplasmic Reticulum Stress.” Cell Death and Differentiation 11 (4): 381–89. [DOI] [PubMed] [Google Scholar]
- Pagliuca Felicia W., Millman Jeffrey R., Mads Gürtler Michael Segel, Alana Van Dervort Jennifer Hyoje Ryu, Peterson Quinn P., Greiner Dale, and Melton Douglas A.. 2014. “Generation of Functional Human Pancreatic β Cells In Vitro.” Cell 159 (2): 428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomer Xavier, Pizarro-Delgado Javier, Barroso Emma, and Vázquez-Carrera Manuel. 2018. “Palmitic and Oleic Acid: The Yin and Yang of Fatty Acids in Type 2 Diabetes Mellitus.” Trends in Endocrinology & Metabolism. 10.1016/j.tem.2017.11.009. [DOI] [PubMed] [Google Scholar]
- Parasassi T., De Stasio G., Ravagnan G., Rusch R. M., and Gratton E.. 1991. “Quantitation of Lipid Phases in Phospholipid Vesicles by the Generalized Polarization of Laurdan Fluorescence.” Biophysical Journal 60 (1): 179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker Stephen C. J., Stitzel Michael L., Taylor D. Leland, Orozco Jose Miguel, Erdos Michael R., Akiyama Jennifer A., van Bueren Kelly Lammerts, et al. 2013. “Chromatin Stretch Enhancer States Drive Cell-Specific Gene Regulation and Harbor Human Disease Risk Variants.” Proceedings of the National Academy of Sciences of the United States of America 110 (44): 17921–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnaud G., Bosco D., Berney T., Pattou F., Kerr-Conte J., Donath M. Y., Bruun C., Mandrup-Poulsen T., Billestrup N., and Halban P. A.. 2008. “Proliferation of Sorted Human and Rat Beta Cells.” Diabetologia 51 (1): 91–100. [DOI] [PubMed] [Google Scholar]
- Pasquali Lorenzo, Gaulton Kyle J., Rodríguez-Seguí Santiago A., Mularoni Loris, Miguel-Escalada Irene, Akerman İldem, Tena Juan J., et al. 2014. “Pancreatic Islet Enhancer Clusters Enriched in Type 2 Diabetes Risk-Associated Variants.” Nature Genetics 46 (2): 136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter Nina P., Balasubramanian Raji, Giulianini Franco, Wang Dong D., Tinker Lesley F., Gopal Shuba, Deik Amy A., et al. 2018. “Metabolic Predictors of Incident Coronary Heart Disease in Women.” Circulation 137 (8): 841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martí Albert, Ramakrishnan Suresh, Li Jiayi, Dugourd Aurelien, Molenaar Martijn R., De La Motte Luigi R., Grand Kelli, et al. 2022. “Reducing Lipid Bilayer Stress by Monounsaturated Fatty Acids Protects Renal Proximal Tubules in Diabetes.” eLife 11 (May). 10.7554/eLife.74391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitpas I., Grüne T., Bhattacharya A. A., and Curry S.. 2001. “Crystal Structures of Human Serum Albumin Complexed with Monounsaturated and Polyunsaturated Fatty Acids.” Journal of Molecular Biology 314 (5): 955–60. [DOI] [PubMed] [Google Scholar]
- Petrie John R., Pearson Ewan R., and Sutherland Calum. 2011. “Implications of Genome Wide Association Studies for the Understanding of Type 2 Diabetes Pathophysiology.” Biochemical Pharmacology 81 (4): 471–77. [DOI] [PubMed] [Google Scholar]
- Piccolis Manuele, Bond Laura M., Kampmann Martin, Pulimeno Pamela, Chitraju Chandramohan, Jayson Christina B. K., Vaites Laura P., et al. 2019. “Probing the Global Cellular Responses to Lipotoxicity Caused by Saturated Fatty Acids.” Molecular Cell 74 (1): 32–44.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli Simone, Faridani Omid R., Björklund Asa K., Winberg Gösta, Sagasser Sven, and Sandberg Rickard. 2014. “Full-Length RNA-Seq from Single Cells Using Smart-seq2.” Nature Protocols 9 (1): 171–81. [DOI] [PubMed] [Google Scholar]
- Pillon Nicolas J., Loos Ruth J. F., Marshall Sally M., and Zierath Juleen R.. 2021. “Metabolic Consequences of Obesity and Type 2 Diabetes: Balancing Genes and Environment for Personalized Care.” Cell 184 (6): 1530–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plötz T., von Hanstein A. S., Krümmel B., Laporte A., Mehmeti I., and Lenzen S.. 2019. “Structure-Toxicity Relationships of Saturated and Unsaturated Free Fatty Acids for Elucidating the Lipotoxic Effects in Human EndoC-βH1 Beta-Cells.” Biochimica et Biophysica Acta, Molecular Basis of Disease 1865 (11): 165525. [DOI] [PubMed] [Google Scholar]
- Pound Lynley D., Sarkar Suparna A., Benninger Richard K. P., Wang Yingda, Suwanichkul Adisak, Shadoan Melanie K., Printz Richard L., et al. 2009. “Deletion of the Mouse Slc30a8 Gene Encoding Zinc Transporter-8 Results in Impaired Insulin Secretion.” Biochemical Journal 421 (3): 371–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki Marc, and Murthy Madiraju S. R.. 2012. “Glycerolipid/free Fatty Acid Cycle and Islet β-Cell Function in Health, Obesity and Diabetes.” Molecular and Cellular Endocrinology 353 (1–2): 88–100. [DOI] [PubMed] [Google Scholar]
- Promlek Thanyarat, Ishiwata-Kimata Yuki, Shido Masahiro, Sakuramoto Mitsuru, Kohno Kenji, and Kimata Yukio. 2011. “Membrane Aberrancy and Unfolded Proteins Activate the Endoplasmic Reticulum Stress Sensor Ire1 in Different Ways.” Molecular Biology of the Cell 22 (18): 3520–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randle P. J., Garland P. B., Hales C. N., and Newsholme E. A.. 1963. “The Glucose Fatty-Acid Cycle. Its Role in Insulin Sensitivity and the Metabolic Disturbances of Diabetes Mellitus.” The Lancet 1 (7285): 785–89. [DOI] [PubMed] [Google Scholar]
- Rathmell Jeffrey C. 2021. “Obesity, Immunity, and Cancer.” The New England Journal of Medicine 384 (12): 1160–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Eugene P., Cheng Susan, Larson Martin G., Walford Geoffrey A., Lewis Gregory D., McCabe Elizabeth, Yang Elaine, et al. 2011. “Lipid Profiling Identifies a Triacylglycerol Signature of Insulin Resistance and Improves Diabetes Prediction in Humans.” The Journal of Clinical Investigation 121 (4): 1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel Alison E., Drijvers Jefte M., Baker Gregory J., Catozzi Alessia, García-Cañaveras Juan C., Gassaway Brandon M., Miller Brian C., et al. 2020. “Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti-Tumor Immunity.” Cell 183 (7): 1848–66.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman Tamara S., Cannon Maren E., Vadlamudi Swarooparani, Buchkovich Martin L., Wolford Brooke N., Welch Ryan P., Morken Mario A., et al. 2017. “A Type 2 Diabetes–Associated Functional Regulatory Variant in a Pancreatic Islet Enhancer at the ADCY5 Locus.” Diabetes 66 (9): 2521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Mario, Devkota Ranjan, Panagaki Dimitra, Bergh Per-Olof, Kaper Delaney, Henricsson Marcus, Nik Ali, et al. 2022. “Sphingosine 1-Phosphate Mediates Adiponectin Receptor Signaling Essential For Lipid Homeostasis and Embryogenesis.” Nature Communications 13 (1): 7162. doi: 10.1038/s41467-022-34931-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz Mario, Ståhlman Marcus, Borén Jan, and Pilon Marc. 2019. “AdipoR1 and AdipoR2 Maintain Membrane Fluidity in Most Human Cell Types and Independently of Adiponectin.” Journal of Lipid Research 60 (5): 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan Sean K., Zelic Matija, Han Yingnan, Teeple Erin, Chen Luoman, Sadeghi Mahdiar, Shankara Srinivas, et al. 2023. “Microglia Ferroptosis Is Regulated by SEC24B and Contributes to Neurodegeneration.” Nature Neuroscience. 10.1038/s41593-022-01221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponaro Chiara, Gaggini Melania, Carli Fabrizia, and Gastaldelli Amalia. 2015. “The Subtle Balance between Lipolysis and Lipogenesis: A Critical Point in Metabolic Homeostasis.” Nutrients 7 (11): 9453–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. 2014. “Biological Insights from 108 Schizophrenia-Associated Genetic Loci.” Nature 511 (7510): 421–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder Martin, and Kaufman Randal J.. 2005. “The Mammalian Unfolded Protein Response.” Annual Review of Biochemistry 74: 739–89. [DOI] [PubMed] [Google Scholar]
- Scott Robert A., Freitag Daniel F., Li Li, Chu Audrey Y., Surendran Praveen, Young Robin, Grarup Niels, et al. 2016. “A Genomic Approach to Therapeutic Target Validation Identifies a Glucose-Lowering GLP1R Variant Protective for Coronary Heart Disease.” Science Translational Medicine 8 (341): 341ra76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma Rohit B., and Alonso Laura C.. 2014. “Lipotoxicity in the Pancreatic Beta Cell: Not Just Survival and Function, but Proliferation as Well?” Current Diabetes Reports 14 (6): 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Yihui, Zhao Zhilun, Zhang Luyuan, Shi Lingyan, Shahriar Sanjid, Chan Robin B., Paolo Gilbert Di, and Min Wei. 2017. “Metabolic Activity Induces Membrane Phase Separation in Endoplasmic Reticulum.” Proceedings of the National Academy of Sciences of the United States of America 114 (51): 13394–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhom Eriene-Heidi, Kim Choah, Maria Kost-Alimova May Theng Ting, Keller Keith, Avila-Pacheco Julian, Watts Andrew Jb, et al. 2021. “Targeting a Braf/Mapk Pathway Rescues Podocyte Lipid Peroxidation in CoQ-Deficiency Kidney Disease.” The Journal of Clinical Investigation 131 (5). 10.1172/JCI141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa Silva, da Simone C., Nayak Nichole, Caymo Andrei Miguel, and Gordon Joseph W.. 2020. “Mechanisms of Muscle Insulin Resistance and the Cross-Talk with Liver and Adipose Tissue.” Physiological Reports 8 (19): e14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skelin Masa, Rupnik Marjan, and Cencic Avrelija. 2010. “Pancreatic Beta Cell Lines and Their Applications in Diabetes Mellitus Research.” ALTEX 27 (2): 105–13. [DOI] [PubMed] [Google Scholar]
- Snowden Stuart G., Ebshiana Amera A., Hye Abdul, An Yang, Pletnikova Olga, O’Brien Richard, Troncoso John, Legido-Quigley Cristina, and Thambisetty Madhav. 2017. “Association between Fatty Acid Metabolism in the Brain and Alzheimer Disease Neuropathology and Cognitive Performance: A Nontargeted Metabolomic Study.” PLoS Medicine 14 (3): e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian Aravind, Tamayo Pablo, Mootha Vamsi K., Mukherjee Sayan, Ebert Benjamin L., Gillette Michael A., Paulovich Amanda, et al. 2005. “Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles.” Proceedings of the National Academy of Sciences of the United States of America 102 (43): 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Yue, Ge Xin, Li Xue, He Jinrong, Wei Xinzhi, Du Jie, Sun Jian, et al. 2020. “High-Fat Diet Promotes Renal Injury by Inducing Oxidative Stress and Mitochondrial Dysfunction.” Cell Death & Disease 11 (10): 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneera Jalal, Lang Stefan, Sharma Amitabh, Fadista Joao, Zhou Yuedan, Ahlqvist Emma, Jonsson Anna, et al. 2012. “A Systems Genetics Approach Identifies Genes and Pathways for Type 2 Diabetes in Human Islets.” Cell Metabolism 16 (1): 122–34. [DOI] [PubMed] [Google Scholar]
- Tcw Julia, Qian Lu, Pipalia Nina H., Chao Michael J., Liang Shuang A., Shi Yang, Jain Bharat R., et al. 2022. “Cholesterol and Matrisome Pathways Dysregulated in Astrocytes and Microglia.” Cell 185 (13): 2213–33.e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen Soren K., Raimondo Anne, Hastoy Benoit, Sengupta Shahana, Dai Xiao-Qing, Bautista Austin, Censin Jenny, et al. 2018. “Type 2 Diabetes Risk Alleles in PAM Impact Insulin Release from Human Pancreatic β-Cells.” Nature Genetics 50 (8): 1122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumelin Stéphane, Roche Enrique, Esser Victoria, McGarry J. Denis, Prentki Marc, and Others. 1997. “Fatty Acids Rapidly Induce the Carnitine Palmitoyltransferase I Gene in the Pancreatic β-Cell Line INS-1.” The Journal of Biological Chemistry 272 (3): 1659–64. [DOI] [PubMed] [Google Scholar]
- Tonazzi Annamaria, Giangregorio Nicola, Console Lara, Palmieri Ferdinando, and Indiveri Cesare. 2021. “The Mitochondrial Carnitine Acyl-Carnitine Carrier (SLC25A20): Molecular Mechanisms of Transport, Role in Redox Sensing and Interaction with Drugs.” Biomolecules 11 (4). 10.3390/biom11040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoukalas Dimitris, Fragoulakis Vassileios, Sarandi Evangelia, Docea Anca Oana, Papakonstaninou Evangelos, Tsilimidos Gerasimos, Anamaterou Chrysanthi, et al. 2019. “Targeted Metabolomic Analysis of Serum Fatty Acids for the Prediction of Autoimmune Diseases.” Frontiers in Molecular Biosciences 6 (November): 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubellacker Jessalyn M., Tasdogan Alpaslan, Ramesh Vijayashree, Shen Bo, Mitchell Evann C., Martin-Sandoval Misty S., Gu Zhimin, et al. 2020. “Lymph Protects Metastasizing Melanoma Cells from Ferroptosis.” Nature 585 (7823): 113–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udler Miriam S., Kim Jaegil, von Grotthuss Marcin, Bonàs-Guarch Sílvia, Cole Joanne B., Chiou Joshua, Anderson Christopher D. on behalf of METASTROKE and the ISGC, et al. 2018. “Type 2 Diabetes Genetic Loci Informed by Multi-Trait Associations Point to Disease Mechanisms and Subtypes: A Soft Clustering Analysis.” PLoS Medicine 15 (9): e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uffelmann Emil, Huang Qin Qin, Munung Nchangwi Syntia, de Vries Jantina, Okada Yukinori, Martin Alicia R., Martin Hilary C., and Lappalainen Tuuli. 2021. “Genome-Wide Association Studies.” Nature Reviews Methods Primers 1 (1): 1–21. [Google Scholar]
- Vilar Santiago, Cozza Giorgio, and Moro Stefano. 2008. “Medicinal Chemistry and the Molecular Operating Environment (MOE): Application of QSAR and Molecular Docking to Drug Discovery.” Current Topics in Medicinal Chemistry 8 (18): 1555–72. [DOI] [PubMed] [Google Scholar]
- Vogtmann H., Christian R., Hardin R. T., and Clandinin D. R.. 1975. “The Effects of High and Low Erucic Acid Rapeseed Oils in Diets for Rats.” International Journal for Vitamin and Nutrition Research. Internationale Zeitschrift Fur Vitamin- Und Ernahrungsforschung. Journal International de Vitaminologie et de Nutrition 45 (2): 221–29. [PubMed] [Google Scholar]
- Volmer Romain, van der Ploeg Kattria, and Ron David. 2013. “Membrane Lipid Saturation Activates Endoplasmic Reticulum Unfolded Protein Response Transducers through Their Transmembrane Domains.” Proceedings of the National Academy of Sciences of the United States of America 110 (12): 4628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadden T. A., Hollander P., Klein S., Niswender K., Woo V., Hale P. M., Aronne L., and NN8022–1923 Investigators. 2013. “Weight Maintenance and Additional Weight Loss with Liraglutide after Low-Calorie-Diet-Induced Weight Loss: The SCALE Maintenance Randomized Study.” International Journal of Obesity 37 (11): 1443–51. [DOI] [PubMed] [Google Scholar]
- Wijesekara N., Dai F. F., Hardy A. B., Giglou P. R., Bhattacharjee A., Koshkin V., Chimienti F., Gaisano H. Y., Rutter G. A., and Wheeler M. B.. 2010. “Beta Cell-Specific Znt8 Deletion in Mice Causes Marked Defects in Insulin Processing, Crystallisation and Secretion.” Diabetologia 53 (8): 1656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding John P. H., Batterham Rachel L., Calanna Salvatore, Davies Melanie, Van Gaal Luc F., Lingvay Ildiko, McGowan Barbara M., et al. 2021. “Once-Weekly Semaglutide in Adults with Overweight or Obesity.” The New England Journal of Medicine 384 (11): 989. [DOI] [PubMed] [Google Scholar]
- Winzell Maria Sörhede, and Ahrén Bo. 2008. “Durable Islet Effects on Insulin Secretion and Protein Kinase A Expression Following Exendin-4 Treatment of High-Fat Diet-Fed Mice.” Journal of Molecular Endocrinology 40 (2): 93–100. [DOI] [PubMed] [Google Scholar]
- Yang Kisuk, Lee Miseon, Peter Anthony Jones Sophie S. Liu, Zhou Angela, Xu Jun, Sreekanth Vedagopuram, et al. 2020. “A 3D Culture Platform Enables Development of Zinc-Binding Prodrugs for Targeted Proliferation of β Cells.” Science Advances 6 (47): eabc3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Wan Seok, Kim Katherine J., Gaschler Michael M., Patel Milesh, Shchepinov Mikhail S., and Stockwell Brent R.. 2016. “Peroxidation of Polyunsaturated Fatty Acids by Lipoxygenases Drives Ferroptosis.” Proceedings of the National Academy of Sciences 113 (34): E4966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Fei. 2022. “Lipid Metabolism and Alzheimer’s Disease: Clinical Evidence, Mechanistic Link and Therapeutic Promise.” The FEBS Journal, January. 10.1111/febs.16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon Haejin, Shaw Jillian L., Haigis Marcia C., and Greka Anna. 2021. “Lipid Metabolism in Sickness and in Health: Emerging Regulators of Lipotoxicity.” Molecular Cell 81 (18): 3708–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Donglu, Hop Cornelis E. C., Patilea-Vrana Gabriela, Gampa Gautham, Seneviratne Herana Kamal, Unadkat Jashvant D., Kenny Jane R., et al. 2019. “Drug Concentration Asymmetry in Tissues and Plasma for Small Molecule–Related Therapeutic Modalities.” Drug Metabolism and Disposition: The Biological Fate of Chemicals 47 (10): 1122–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Jianlin, Huang Jin, Wang Xingyu, Chen Weidong, Tang Qinqing, Fang Maoyong, and Qian Yeben. 2017. “CMIP Is Oncogenic in Human Gastric Cancer Cells.” Molecular Medicine Reports 16 (5): 7277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.