Abstract
Overexpression and inhibitor studies have suggested that the c-Myc target gene for ornithine decarboxylase (ODC), the enzyme which converts ornithine to putrescine, plays an important role in diverse biological processes, including cell growth, differentiation, transformation, and apoptosis. To explore the physiological function of ODC in mammalian development, we generated mice harboring a disrupted ODC gene. ODC-heterozygous mice were viable, normal, and fertile. Although zygotic ODC is expressed throughout the embryo prior to implantation, loss of ODC did not block normal development to the blastocyst stage. Embryonic day E3.5 ODC-deficient embryos were capable of uterine implantation and induced maternal decidualization yet failed to develop substantially thereafter. Surprisingly, analysis of ODC-deficient blastocysts suggests that loss of ODC does not affect cell growth per se but rather is required for survival of the pluripotent cells of the inner cell mass. Therefore, ODC plays an essential role in murine development, and proper homeostasis of polyamine pools appears to be required for cell survival prior to gastrulation.
Overexpression of the c-Myc, N-Myc, or L-Myc members of the Myc oncogene family is a common event in human tumors. This selection likely reflects Myc's ability to provide continuous proliferative and angiogenic signals under growth-limiting conditions, such as those that occur in the tumor microenvironment (38). However, Myc's propensity to induce continuous proliferation also blocks terminal differentiation (11) and triggers the apoptotic program (2). c-Myc is a basic helix-loop-helix leucine zipper protein that exhibits sequence-specific DNA binding to CACGTG or CACATG elements when dimerized with its obligate basic helix-loop-helix leucine zipper partner Max (8). Despite Myc's well-defined function as a transcriptional transactivator, the numbers of its ascribed targets that have been proven to be direct are relatively few, but they do include ornithine decarboxylase (ODC) (6), α-prothymosin (15), eIF-4E (44), carbamoyl-phosphate synthase-aspartate carbamoyltransferase-dihydroorotase (cad) (28), a DEAD box-related gene (MrDb) (20), the ubiquitin E2 ligase Cul1 (32), and ECA39 (7). A compelling example of a target gene that contributes to c-Myc's biological effects is ODC (34, 35), which is activated by growth factors and by c-Myc through two conserved CACGTG sites present in the first intron of vertebrate ODC genes (6). ODC is the key regulator of the polyamine biosynthetic pathway and decarboxylates l-ornithine to form putrescine (50). ODC expression is highly regulated by changes in its transcription, translation, and RNA and protein half-life (40). Furthermore, ODC enzyme activity is tightly controlled and shows biphasic induction during late G1 and at G2/M (5). Inhibition of ODC by difluoromethylornithine (DFMO) compromises cell growth and transformation (3) and induces cell cycle arrest in G1 (35, 43). ODC inhibition results in marked reductions in the intracellular levels of the polyamines putrescine, spermidine, and spermine, which appear essential for fundamental processes such as stabilization of chromatin and cytoskeletal structure (4), translation (37), transcription (10), semiconservative DNA replication (42), and the protection of cells from DNA damage (25). Chronic reductions in polyamine levels have also been reported to lead to apoptosis, especially following exposure to oxidative stress (14). Paradoxically, ODC overexpression, which upregulates putrescine levels, can also trigger the apoptotic program (34). Overall, these findings strongly support the concept that proper homeostasis of polyamine pools is a critical determinant of cell fate.
In eukaryotes, loss-of-function mutations in ODC have been created in Saccharomyces cerevisiae and Leishmania donovani, and a naturally occurring ODC mutant has been identified for the nematode Caenorhabditis elegans. Loss of ODC in haploid yeast results in a cessation of growth (45), whereas deletion of ODC in L. donovani (23) and C. elegans (26) is lethal, unless these animals are supplied with exogenous putrescine or polyamines in their diets. However, the cause of the lethality of ODC deficiency in these lower organisms is not resolved, and relatively little is known about the role of ODC during vertebrate embryogenesis. To address this issue directly, we examined the biological role of ODC in the mouse by gene targeting, and we demonstrate a critical in vivo role for ODC in promoting cell survival prior to gastrulation.
MATERIALS AND METHODS
Construction of the ODC targeting vector.
Genomic clones of the murine ODC gene were isolated from a 129/SVE mouse genomic library in λEMBL3 using a full-length murine ODC cDNA probe (kindly provided by Daniel Nathans). Positive clones were restriction mapped, subcloned into pBluescript SK(+), and sequenced. Standard recombinant techniques were used to design the targeting vector schematically shown in Fig. 1A. A 304-bp SalI-SstI fragment that included most of exon 2 and all of exon 3, including the translation start site of the ODC gene, was replaced by an internal ribosome entry site-linked LacZ-neomycin cassette (31), which allowed the positive selection of recombinant clones. A herpes simplex virus thymidine kinase cassette mediating negative selection was inserted in the 3′ end of the construct at the HindIII site within ODC (Fig. 1A).
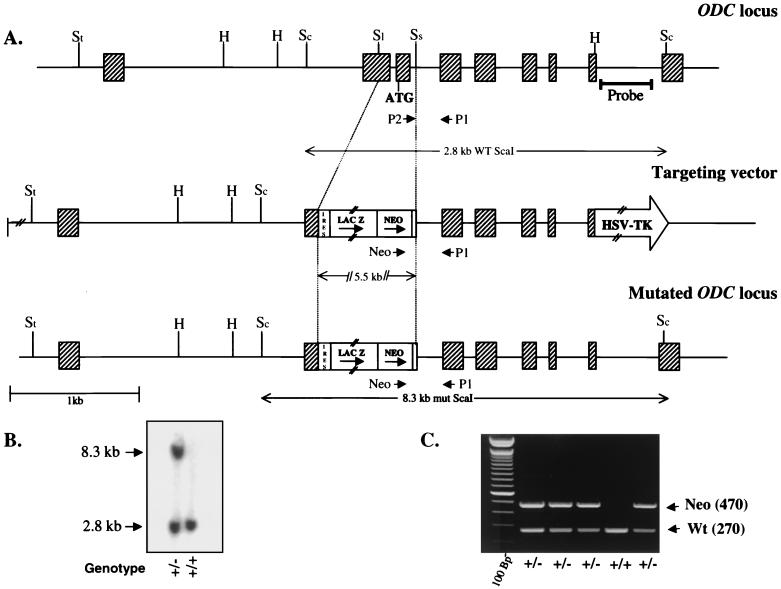
FIG. 1.
(A) Targeting strategy of the ODC genomic locus. Schematics of the wild-type locus (top), targeting vector (middle), and recombined locus (bottom) are shown. Exons are indicated by hatched boxes, and the arrows correspond to the three primers used for PCR genotyping. Abbreviations: St, StuI; H, HindIII; Sc, ScaI; Sl, SalI; Ss, SstI. (B) Southern blot analysis of genomic DNA isolated from ES cell clones. Digestion of the targeted ODC locus with ScaI and hybridization to a 3′ external probe show a 2.8- and an 8.3-kb band specific to the wild-type and targeted alleles, respectively, confirming homologous recombination. (C) Transmission of the ODC mutant allele to the progeny was determined by PCR amplification of tail DNAs using the primers described in Materials and Methods. PCR products were resolved on a 2% agarose gel in Tris-acetate-EDTA buffer. WT and Wt, wild type; mut, mutant; HSV-TK, herpes simplex virus thymidine kinase; IRES, internal ribosome entry site.
Targeting of ES cells and generation of ODC-deficient mice.
Linearized vector DNA was introduced by electroporation into W9.5 embryonic stem (ES) cells cultured as described previously (36). Following selection with G418 (250 μg/ml) and 1-(2'-deoxy-2'-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU) (1 μM), eight correctly targeted homologous recombinants were identified by PCR and Southern blotting (see below). To generate chimeric mice, C57BL/6J blastocysts injected with two independent ODC+/− ES clones were implanted into pseudopregnant foster mothers. Several chimeric males, identified by their agouti coat color, gave germ line transmission, and their offspring were screened for the presence of the disrupted ODC gene. All animal experiments performed fully complied with federal and institutional guidelines.
PCR assays.
Genotyping of mice and embryos older than E8.5 was performed on tail DNA and visceral yolk sac DNA, respectively, lysed at 55°C in 400 μl of lysis buffer (500 mM KCl, 100 mM Tris-HCl [pH 8.3], 0.1 mg of gelatin/ml, 1% NP-40, 1% Tween 20, 500 μg of proteinase K/ml) for 3 h. The proteinase K was inactivated by boiling for 10 min, and 3 μl from each sample was used for standard PCR using the Taq PCR Core kit (Qiagen). For embryos younger than E8.5, and for blastocyst outgrowths, different buffers (described in reference 51) were used. In all cases, a mixture of three PCR primers was used to detect wild-type and mutant alleles: P1 (5′-CGAGGTCCGCAACATAGAACG-3′), P2 (5′-CTCTGTAAGTACGGGAAGCCC-3′), and NEO (5′-CCCACACCTCCCCCTGAACC-3′), which amplified 270-bp (wild-type) and 470-bp (knockout) fragments. The PCR cycle profile was as follows: 1 cycle of 94°C for 4 min, followed by 34 cycles (standard) or 39 cycles (blastocysts) of 94°C for 1 min, 64°C for 1 min, and 72°C for 1 min, and finally 1 cycle of 72°C for 6 min. PCR products were analyzed by standard agarose gel electrophoresis.
In vitro culture of blastocyst outgrowths.
Natural matings between male and female ODC+/− mice were used to obtain embryos of all genotypes (wild type, ODC+/−, and ODC−/−). Embryos were staged according to the detection of vaginal plugs resulting from the crosses (noon of day 1 of plugging equals E0.5). Blastocysts at E3.5 were flushed from the uterine horns, extensively washed in phosphate-buffered saline (PBS), and cultured individually in ES cell medium lacking leukemia inhibitory factor on gelatin-coated 96-well plates for 5 to 6 days. Morphology of the outgrowths was assessed at specific intervals, photographs were taken with an inverted microscope (Olympus), and the genotype of the blastocyst cultures was determined by PCR.
Whole-mount LacZ staining.
Free-floating blastocysts were fixed in 0.2% (vol/vol) glutaraldehyde and 1% (vol/vol) formaldehyde in PBS for 10 min at room temperature (RT) and washed extensively. LacZ activity was assessed by incubation of the embryos at 37°C in the histochemical reaction mixture [1 mg of 4-chloro-5-bromo-3-indolyl-β-galactosidase (X-Gal)/ml, 4 mM K4Fe(CN)6 · 3H2O, 4 mM K3Fe(CN)6, and 2 mM MgCl2 in PBS] for 20 h in a humidified chamber.
TUNEL assays.
E3.5 to E4.0 embryos resulting from heterozygous intercrosses were collected, washed in PBS, and fixed in freshly prepared 4% paraformaldehyde in PBS for 15 min at RT. After permeabilization in PBS containing 0.1% (vol/vol) Na citrate–0.1% (vol/vol) Triton X-100 for 10 min at RT, they were washed twice in PBS and tested for evidence of apoptosis by TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling), according to the manufacturer's instructions (in situ cell death detection kit, fluorescein; Roche). At the end of the assay, blastocysts were washed, individually transferred to a drop of PBS on a depression slide, and examined by confocal laser scanning microscopy.
Immunofluorescence.
The first steps of the procedure were the same as for the TUNEL assays described above. Following permeabilization and washing, the embryos were stained overnight at 4°C with an antibody specific to phosphohistone H3 at Ser10 (1:200 dilution; Upstate Biotechnology) in a drop of PBS containing 0.7% bovine serum albumin and 10% goat serum, under mineral oil. After three washes in PBS for 5 min, anti-phosphohistone H3 antibody binding was detected with Cy3-conjugated secondary antibody to rabbit immunoglobulin G (1:200; Jackson Laboratories). Embryos were then washed in PBS and immediately analyzed for immunofluorescence by confocal microscopy.
For ODC whole-mount immunofluorescence, the blastocysts were first incubated in blocking solution (1% goat serum in PBS) for 20 min and then stained with anti-ODC antibody (B250-1; Accurate Chemical & Scientific Corp.) at a 1:100 dilution in blocking solution for 2 h at RT. After three washes in PBS (10 min each), they were incubated with the secondary antibody Alexa-488 anti-rabbit immunoglobulin G (Molecular Probes) in blocking solution for 1 h at RT. The embryos were then treated for 30 min at RT with RNase A and stained with propidium iodide for 5 min. They were finally washed in PBS and mounted on slides for analysis by confocal microscopy.
Histological analysis.
Deciduae at E5.5 were isolated, fixed in 10% buffered formalin overnight at 4°C, processed, and embedded in paraffin using standard procedures. Four-micrometer-thick sections were prepared and stained with hematoxylin and eosin. Coverslips were removed from sections to be microdissected by immersion in xylene, and the sections were air dried. Embryonic tissue was microdissected using a PALM laser microscope system (PALM Microlaser Technologies, Bernied, Germany). Microdissected tissue was catapulted into the caps of microcentrifuge tubes, and the DNA was subsequently extracted and subjected to PCR amplification.
Putrescine rescue experiments.
Intact uteri were removed at E6.5 and E7.5 from heterozygous pregnant females which had been administered putrescine (Sigma) at a concentration of 0.1 mM in their drinking water from the day on which a vaginal plug was detected. Water was changed daily to ensure putrescine stability and effectiveness. Embryos were dissected from the decidua and genotyped as described above.
RESULTS
Gene targeting of murine ODC results in embryonic lethality.
We constructed a targeting vector in which ODC sequences encompassing most of ODC exon 2 and all of exon 3, including the ATG initiation codon, were deleted by replacement with an internal ribosome entry site-LacZ-Neo (β-geo) selection cassette. The herpes simplex virus thymidine kinase gene was used as a negative selection marker (Fig. 1A). The choice of a promoterless targeting strategy was based on the observations made by Northern blotting that the ODC gene is actively transcribed in ES cells (data not shown). The insertion of the β-geo expression cassette deletes the first 35 amino acids of ODC. This targeting construct was introduced into W9.5 ES cells by electroporation, and cells that had undergone homologous recombination were enriched by selection with G418 and FIAU and identified by PCR and Southern blotting (Fig. 1B). Eight independent correctly targeted clones were identified, and two having a normal karyotype were microinjected into C57BL/6 blastocysts and transplanted into pseudopregnant females. High- and medium-chimeric mice were obtained and subsequently transmitted the mutated ODC allele to their progeny. The validity of the ODC mutation was confirmed by performing ODC enzyme assays using whole-cell extracts isolated from wild-type and heterozygous (ODC+/−)-derived MEFs. ODC enzyme activity was reduced by half in MEFs prepared from ODC+/− mice compared to the activity in those derived from wild-type littermates, clearly showing a dosage effect (data not shown). Male and female ODC+/− mice appeared phenotypically normal, were fertile, and were indistinguishable in size or growth from their wild-type littermates. However, genotyping the litters of mice arising from ODC+/− intercrosses failed to show any homozygous null ODC mice, indicating a prenatal lethality (Fig. 1C), although numbers of wild-type and ODC+/− mice were as expected.
To precisely pinpoint the time of embryonic death, we dissected embryos from heterozygous intercrosses at different days of gestation from E6.5 to E12.5 and genotyped them by PCR. As shown in Table 1, of the 233 deciduae tested, 178 contained morphologically normal fetuses, of which 118 were ODC+/− and 60 were wild type. No ODC−/− embryos were detected at these embryonic stages. In accord with this finding, embryos were not found in about 25% of the deciduae at E6.5, and attempts to genotype the residual tissues from these empty implantation sites were not successful. These data indicate that ODC-deficient embryos cannot survive to gastrulation.
TABLE 1.
Genotype analysis of ODC+/− intercross progeny
| Age | No. with genotypea
|
Unknown | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Neonate | 453 | 951 | 0 | NAb |
| E12.5 | 4 | 8 | 0 | 3c |
| E10.5 | 8 | 21 | 0 | 10c |
| E9.5 | 6 | 15 | 0 | 8c |
| E8.5 | 6 | 11 | 0 | 5c |
| E7.5 | 12 | 20 | 0 | 10d |
| E6.5 | 24 | 43 | 0 | 19d |
The ODC genotype of embryos was determined by PCR as described in Materials and Methods.
NA, not applicable.
Resorptions.
Residues of embryonic material.
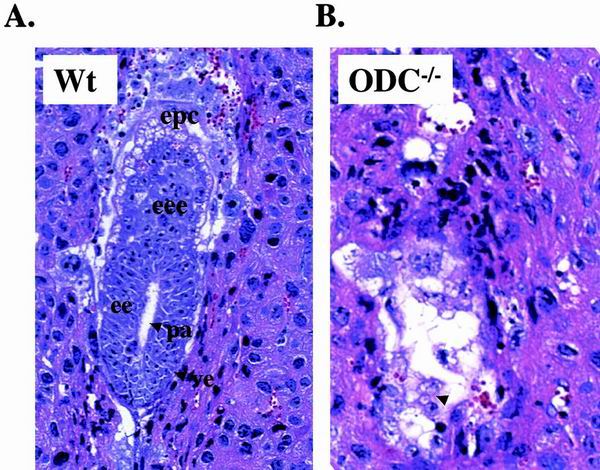
To assess at what stage ODC-deficient embryos died, more timed matings were initiated and E5.5 deciduae obtained from ODC+/− intercrosses were fixed, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Histological sections revealed obvious abnormalities that distinguished normal from mutant conceptuses following implantation (Fig. 2). All wild-type and ODC+/− embryos showed normal growth patterns, with the typical structure of early egg cylinders. By contrast, at this stage of development, almost one-quarter of decidual swellings were virtually empty, with decreased numbers of cells and small and poorly organized embryos (Fig. 2). Embryos having these abnormalities in organization and cellular morphology were confirmed to be ODC−/− by genotyping residual tissue isolated by laser microdissection. Overall, the data suggested that ODC−/− embryos were able to implant but quickly expired thereafter.
FIG. 2.
Histological sections of wild-type (Wt) and ODC−/− mutant embryos in utero. (A) Transverse section through the decidua of a normal embryo at early egg cylinder stage (E5.5). Note the appearance of the proamniotic cavity and the clearly differentiated embryonic and extraembryonic ectoderms. (B) Transverse section through a decidua of a degenerating E5.5 ODC−/− embryo (arrow). No discernible structure can be distinguished. pa, proamniotic cavity; ee, embryonic ectoderm; epc, ectoplacental cone; eee, extraembryonic ectoderm; ve, visceral endoderm.
Exogenous sources of putrescine can rescue developmental defects in ODC-deficient C. elegans and L. donovani (23, 26). Furthermore, putrescine supplementation in drinking water effectively blocks the inhibitory effects of DFMO on tumor formation in a mouse model of skin tumors (39). We therefore addressed whether providing similar levels of putrescine in the drinking water of pregnant female mice could extend the development of ODC-deficient embryos. Although 0.5 to 1 mM putrescine in the drinking water was nontoxic to adult female mice, these doses of putrescine totally compromised pregnancy and this was associated with a marked necrosis of the uterus (data not shown). Presumably, this is due to high levels of diamine oxidase that are present in the placenta and which oxidize putrescine to produce hydrogen peroxide (9). Decreasing doses of putrescine to 0.1 mM in the drinking water allowed mice to carry litters to term. Plugged heterozygous mice were therefore treated daily with this dose of putrescine from day 1 of pregnancy, and at various intervals of gestation, litters were harvested, examined morphologically, and genotyped. Although we failed to detect ODC-deficient embryos at day E7.5 and beyond (n = 41), ODC-deficient embryos were detected at E6.5 by PCR genotyping that appeared phenotypically normal in terms of development and size (data not shown). However, the expected Mendelian ratio was not obtained, as only two E6.5 ODC-null embryos were observed out of a total of 68, indicating that this dose of putrescine can only partially rescue the deleterious effects of ODC deficiency.
Zygotic ODC is dispensable prior to implantation.
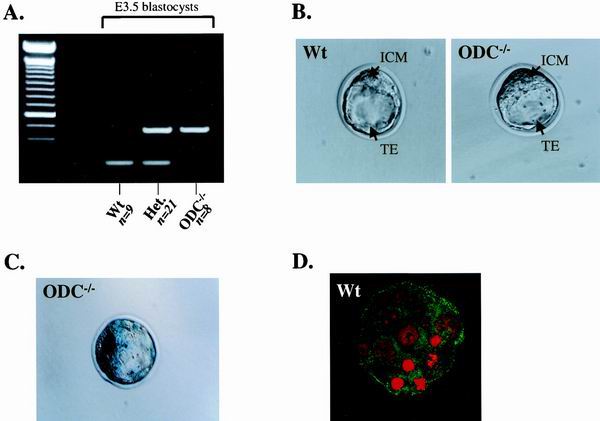
To further address the developmental stage at which the defect of ODC-null embryos was manifest, we analyzed blastocysts (E3.5) generated from ODC+/− intercrosses after flushing these from the uteral lumen. PCR genotyping of the blastocysts revealed that ODC−/− blastocysts were present at nearly an expected Mendelian ratio (21% [Fig. 3A]) and that they appeared morphologically identical to their wild-type and heterozygous counterparts, with a distinct inner cell mass (ICM) and an outer layer of trophoectoderm (TE) cells (Fig. 3B).
FIG. 3.
Zygotic ODC is expressed in blastocysts but is dispensable for blastocyst formation. (A) Representative genotypic analysis of E3.5 embryos from ODC+/− breedings. DNA from individual blastocysts was isolated as described in Materials and Methods, and primers were used to amplify fragments from the wild-type and/or ODC mutant allele in each sample. ODC−/− embryos were found at nearly an expected Mendelian ratio (21%). (B) Phase-contrast photographs of wild-type (left panel) and ODC-null (right panel) blastocysts. ODC-deficient blastocysts appear morphologically normal. (C) Expression of zygotic ODC, as detected by LacZ staining. An intense signal is observed in both the ICM and the TE of an ODC-deficient blastocyst. (D) A similar pattern of ODC expression (green) was detected with an ODC-specific antibody in a wild-type blastocyst. Blastocysts were also stained with propidium iodide (red), which detects DNA. Wt, wild type; Het., heterozygous.
ODC expression has been detected by reverse transcription-PCR in two-cell embryos (13), and yet it is unclear whether ODC is zygotic or maternal in origin. Analysis of ODC and β-galactosidase expression in heterozygous embryos by immunohistochemistry at later stages of development (E10.5 to E12.5) confirmed coincident expression of the β-galactosidase allele and the remaining ODC wild-type allele (data not shown). We therefore determined the pattern of zygotic ODC expression in E3.5 blastocysts. In agreement with ODC expression in ES cells, analysis of β-galactosidase activity demonstrated that the LacZ-Neo-marked ODC allele was strongly expressed in the ICM and yet was also detected in the TE (Fig. 3C). β-Galactosidase expression can be maternally inherited. However, β-galactosidase staining was also evident in embryos resulting from crosses between ODC+/− males and wild-type females, proving that the expression detected is zygotic. Immunostaining with an ODC-specific antibody confirmed that ODC was expressed in both the ICM and TE of wild-type blastocysts (Fig. 3D). Reduced staining of ODC protein was evident in ODC−/− blastocysts (data not shown), and we presume that some maternally derived ODC protein persists to this stage. Thus, reductions in ODC do not impair development prior to implantation, and the lethality of ODC-deficient embryos indeed occurs after E3.5.
ODC is required for the expansion of the ICM in vitro.
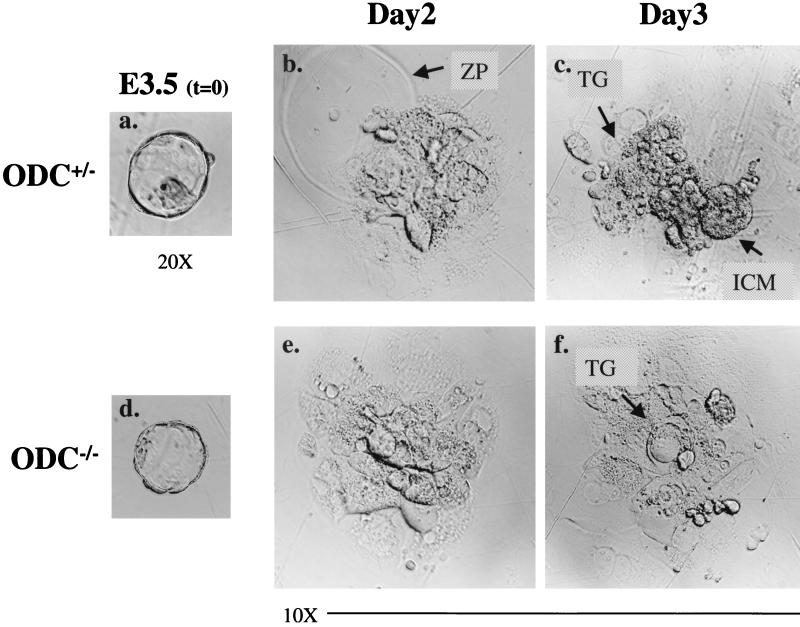
To address whether the abortive development of ODC-deficient embryos was due to some generalized defect in cell growth, or to lineage-specific defects, blastocysts isolated at E3.5 were individually cultured in vitro on gelatin-coated plates for a period of 5 days and then genotyped. During the early phases of these cultures, most blastocysts obtained from three separate litters (8 wild-type, 12 heterozygous, and 5 null, as genotyped after 3 days in culture) attached to the substratum, hatched from the zona pellucida, and started to expand their ICMs and to form outgrowths having the classical appearance of migrating trophoblastic cells (Fig. 4). However, by 18 h, the ICMs of the five ODC-null blastocysts stopped proliferating, and they degenerated soon thereafter (Fig. 4). This phenotype became fully manifest over the next 2 days, and only ODC-deficient trophoblastic giant cells persisted in long-term culture (Fig. 4). Two embryos of unknown genotype were able to attach but quickly collapsed and regressed inside the zona pellucida, suggesting that some ODC-deficient blastocysts had more severe defects. A range of concentrations of putrescine (1 to 100 μM) that are known to override a DFMO-induced cell cycle arrest (35) failed to rescue the growth of ODC-deficient blastocysts, suggesting that losses in polyamines prior to this stage compromise the capacity of ODC-deficient ICMs to expand. Overall, these results demonstrate that the expansion of the ICM is strictly dependent upon ODC.
FIG. 4.
ODC-deficient E3.5 blastocysts are compromised in their expansion in vitro. Photographs of the same heterozygous E3.5 blastocyst (a) at 2 (b) and 3 (c) days in culture and an ODC-deficient blastocyst (d) at 2 (e) and 3 (f) days in culture are shown. A well-developed ICM is evident in the ODC+/− embryo after 3 days, whereas by this time the ICM of the ODC-deficient blastocyst has already completely degenerated. Only the nondividing trophoblastic giant cells derived from ODC−/− blastocysts remained on the plates. ZP, zona pellucida; TG, trophoblastic giant cells.
Loss of ODC results in increased apoptosis of the ICM.
The in vitro growth deficits of ODC-deficient ICMs could be due to defects in cell cycle progression and/or survival. Polyamines appear required for chromatin structure and semiconservative DNA replication but are also required for proper transit through G2/M (5). To address whether ODC-null blastocysts had proliferative defects, we immunostained 17 blastocysts with an antibody that specifically recognizes histone H3 phosphorylated at Ser10, which selectively detects cells in mitosis (1). No striking differences were observed in any of the embryos analyzed, and ODC-deficient blastocysts (n = 3) contained similar numbers of mitotic cells as their wild-type littermates (Fig. 5A). Although we cannot formally exclude an ODC-dependent proliferative defect after this particular stage of development, these data suggest that E3.5 ODC-null embryos are not ostensibly impaired in their proliferation.
FIG. 5.
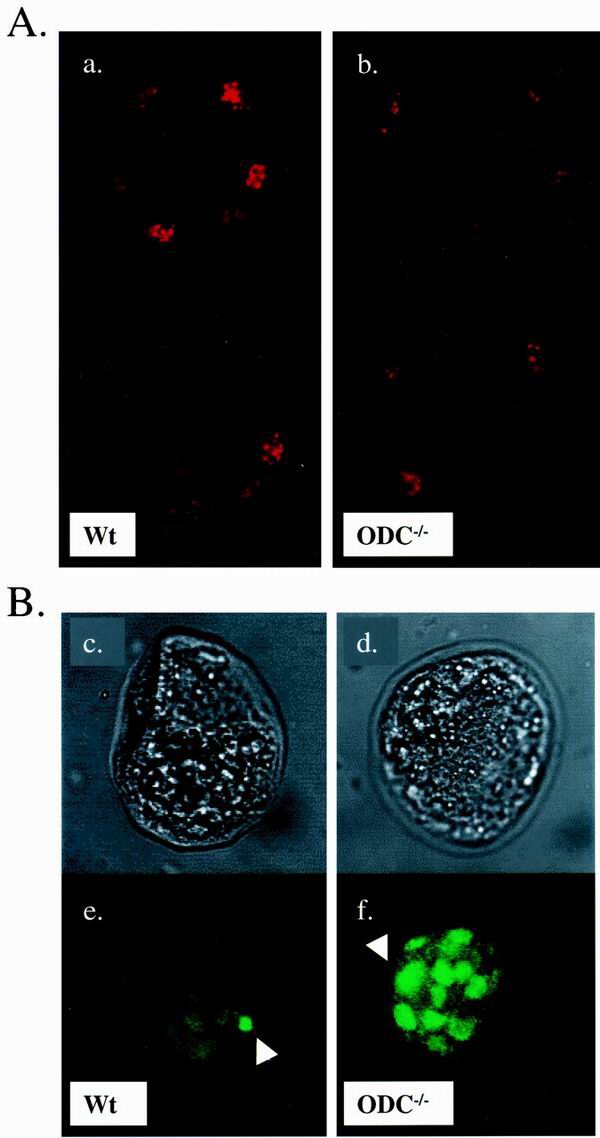
ODC is required for cell survival in the ICM. (A) Comparison of the proliferation index between wild-type (a) and ODC−/− (b) E3.5 embryos. Blastocysts were isolated, fixed, and stained with an antibody specific to phosphohistone H3 at Ser10. Two serial sections captured by confocal imaging of each blastocyst are shown. Mitotic cells are readily detected in embryos from both genotypes. (B) TUNEL analysis of E3.5 blastocysts. The upper panels show phase-contrast photographs of wild-type (c) and ODC−/− (d) blastocysts, and the lower panels correspond to immunodetection of TUNEL labeling (e and f). The wild-type embryo displays minimal apoptosis (e). In contrast, the ODC-deficient embryo exhibits massive cell death confined to the ICM (f). The arrowheads indicate fluorescent dots corresponding to fragmented DNA. Wt, wild type.
To address whether ODC-deficient blastocysts had defects in survival, TUNEL assays were performed on E3.5 blastocysts isolated from five litters (n = 45). We observed evidence of increased chromosomal DNA breakage associated with apoptosis in about 15% (n = 7) of the blastocysts. Although we could genotype only some of these embryos, ODC-deficient embryos displayed many fluorescent dots compared to the small number of TUNEL-positive cells found in their littermates (Fig. 5B). Strikingly, this massive cell death occurs in the ICM, suggesting that the failure of ODC-deficient ICMs to expand is due to their intrinsically high rates of apoptosis.
DISCUSSION
ODC is a well-defined target gene for c-Myc and other oncogenes (6), and ODC activity is necessary and sufficient to promote transformation (3). Genetic studies of lower organisms such as C. elegans and L. donovani have demonstrated that ODC plays an essential developmental role, but the cause of lethality in these ODC-deficient organisms is not resolved (23, 26). In vertebrates, there is little evidence for a developmental role for ODC other than inhibitor studies. For example, in vitro depletion of spermine and spermidine by an inhibitor of S-adenosyl-l-methionine decarboxylase arrests growth before the eight-cell or morula stage of fertilized murine embryos, whereas DFMO, a potent inhibitor of ODC, arrests development at the morula-to-blastocyst transition (54). Furthermore, chicken embryos injected with DFMO have marked reductions in their polyamine levels and fail to develop past gastrulation (22). Similarly, scheduled administration of DFMO during pregnancy in mice induces resorption of embryos when introduced at gestational days 7 and 8 (17). However, these inhibitors may have other targets, and these experiments fail to discriminate whether arrest in embryonic development is due to a lack of maternal ODC or zygotic ODC. Here we have demonstrated that eliminating ODC function by gene targeting compromises early mouse embryonic development. ODC-heterozygous mice exhibit no visible pathology, at least by 1 year of age. By contrast, embryos that lack ODC develop normally to the blastocyst stage and implant but die shortly thereafter, before the onset of gastrulation.
In spite of the severe phenotype found later, the cleavage stages, compaction, differentiation of TE, and blastocoel formation can occur independently of zygotic ODC. This is perhaps not surprising if one posits that maternal stores of ODC and/or polyamines are present in sufficient quantities in the embryo until E3.5. In support of this concept, ODC transcripts are detected in the oocyte by reverse transcription-PCR and continue to increase in abundance until the morula and blastocyst stages (13). In addition, DFMO treatment arrests development in vitro at the morula-to-blastocyst transition (54), whereas ODC-null embryos develop to the blastocyst stage. Thus, it appears likely that a maternal component(s) contributes to the rescue of preimplantation ODC-deficient embryos.
Zygotic ODC expression appears to be crucial when the decidual reaction takes place in the uterus. This hormonally regulated process follows the attachment of the hatched blastocyst and involves an increase in the numbers and permeability of local capillaries. At the time of implantation, the first differentiation event within the blastocyst is the proliferation and invasion of trophoblastic cells through the uterine epithelium. This ensures tight connections between the conceptus and maternal tissue. Concurrent with this process, the ICM undergoes rapid proliferation that extends into the blastocoel cavity to form a structure known as the egg cylinder.
Our work has demonstrated that zygotic ODC is expressed in both embryonic (ICM) and extraembryonic (TE) compartments (Fig. 3C). ODC-deficient embryos are able to initiate implantation in vivo, and trophoblast outgrowths appear to occur normally in culture (Fig. 2). Thus, at face value, these data would suggest that an extraembryonic defect is not the cause of the death of ODC-deficient embryos. However, at this juncture other interpretations of these results are plausible, and it will be important to perform chimeric studies to evaluate the developmental fate of ODC-deficient TE. The data are most consistent with the hypothesis that ODC is critical for the survival of cells within the ICM compartment, as ODC-deficient ICMs exhibit a marked increase in their apoptotic index (Fig. 5B). This most likely prevents the expansion of ODC-deficient ICMs during ex vivo culture, as there appears to be no defect in the growth of ODC-deficient cells as judged by phosphorylation status of H3. However, we cannot exclude the possibility that there might be later defects in cell growth. The onset of the ODC-deficient phenotype is stochastic, as some ODC-deficient embryos can implant whereas others have high rates of apoptosis at the blastocyst stage. In part, these differences could reflect the ability of maternal products to maintain viability of some ODC-deficient embryos. The partial effects of putrescine rescue in promoting survival of ODC-deficient embryos to E6.5 could similarly be due to differences in damage suffered by blastocysts and/or to maternal effects. Alternatively, a more trivial interpretation is that the putrescine rescue actually works but that it is compromised by maternal drinking behavior. To test this hypothesis, we are currently trying other ways to supplement the ODC+/− pregnant females with putrescine.
A hallmark of the ODC-deficient phenotype is the degeneration of the embryo at the early egg cylinder stage. The knockouts of two other Myc targets, Cul1 and H-ferritin, also result in embryonic lethal phenotypes, although the lethalities appear to occur somewhat later (12, 16, 52). The deletion of Max, the required dimerization partner of Myc, is associated with defects in the ICM, but this has been attributed previously to alterations in proliferation rather than survival (46). In contrast, similar to ODC-deficient embryos, ATR- and Chk-1-deficient blastocysts have marked defects in their survival. These observations, along with the null phenotypes of the oxidative regulators thioredoxin and H-ferritin (16, 27), suggest interesting connections between ODC and the DNA damage pathway. Polyamines stabilize chromatin structure, and this may account for their ability to prevent DNA damage (48, 49). Furthermore, cells depleted of polyamines are deficient in their ability to repair X-ray-induced DNA strand breaks, suggesting that they also play a role in stimulating DNA repair (47). Finally, an emerging concept in the field is that polyamines play a proactive role in circumventing DNA damage by acting as antioxidants (25). Indeed, it has been shown previously that spermine is an endogenous scavenger of reactive oxygen species (ROS), which directly induce DNA damage and chromosomal breakage (21). Paradoxically, the catabolism of polyamines leads to the production of ROS (30), and this has been proposed previously to play a physiological role in regulating programmed cell deaths in the blastocyst, where 10% of the cells of the ICM undergo apoptosis (19). This process eliminates ICM cells having TE potential, to prevent the formation of TE within the germ layers during gastrulation (41). By contrast, our data indicate that loss of ODC, and thus depletion of polyamines, leads to massive apoptosis in the ICM.
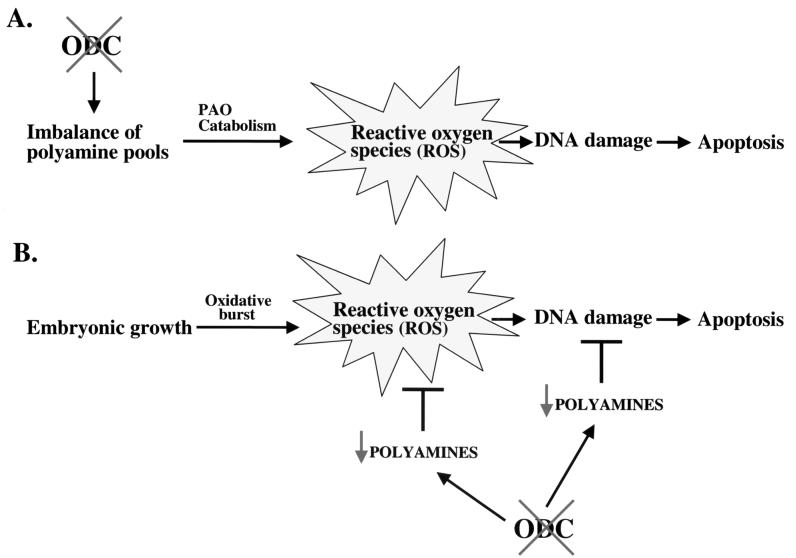
Based on these observations, we propose two models that may explain the apoptotic phenotype of ODC deficiency (Fig. 6), and these are not necessarily mutually exclusive. Firstly, imbalances in polyamine pools may result in inappropriate polyamine catabolism and lead to excessive levels of ROS, which would result in DNA damage and cell death. Secondly, rapid embryonic growth is always accompanied by an oxidative burst, and the levels of ROS generated must be detoxified to prevent cytotoxicity (53). Reductions in intracellular polyamines would compromise one line of defense, as polyamines directly scavenge ROS, and this would place cells of the ICM at risk of DNA damage and death (Fig. 6).
FIG. 6.
Two models of apoptosis caused by loss of ODC. (A) Loss of ODC would be predicted to lead to reductions and an imbalance in polyamine pools. Polyamine catabolism by polyamine oxidase (PAO) would continue and result in the production of ROS, which result in DNA damage and cell death. (B) The rapidly dividing cells of the ICM are at risk of damage due to ROS from the oxidative burst, and these are normally countered by polyamines, which can act as direct scavengers of ROS and/or protect DNA or stimulate DNA repair (21, 47, 48). Loss of ODC would lead to reductions of polyamine pools and thus place these cells at high risk of death from ROS.
Taking into account the pleiotropic roles played by polyamines in cellular processes, additional alternatives causing lethality in ODC-deficient embryos cannot be excluded, and the drastic phenotype observed may be a combination of simultaneous defects. In addition to accepted effects on transcription, translation, replication, and growth that are difficult to reconcile, reductions in polyamines could also affect DNA methylation. DNA methylation is required for early embryonic development, and this presumably reflects the demethylation of DNA at the blastula stage and subsequent de novo methylation of the entire genome at the time of implantation (29). There are three DNA methyltransferase genes that play essential roles during development, Dnmt1, Dnmt3A, and Dnmt3B. Dnmt1-deficient embryos die between gastrulation and E9.5 (24), and Dnmt3A/3B double-null embryos die before E11.5 (33), both substantially later than ODC-deficient embryos. However, the triple Dnmt knockout has not been created, and it is formally possible that these embryos will display an earlier lethality. The substrate for DNA methyltransferases is S-adenosylmethionine (AdoMet), which is also a precursor in the synthesis of the polyamines (50). Conversely, decarboxylated AdoMet acts as a competitive inhibitor of DNA methyltransferase (22). Depletions of putrescine and spermidine typically lead to dramatic increases of decarboxylated AdoMet (18). In the case of the ODC-deficient embryo, we propose that reductions in putrescine should result in the accumulation of decarboxylated AdoMet and that this may inhibit methylation of the genome. Further evaluation of the ODC knockout should allow us to distinguish between these alternatives.
ACKNOWLEDGMENTS
We are grateful to G. Oliver, C. Kelley, J. Ihle, and M. Donohoe for advice and stimulating discussions during the course of this work and to E. White, C. Yang, K. Barnes, and C. Beard for excellent technical assistance. We thank O. Lagutin and J. Swift for help in isolating blastocysts, J. Raucci for microinjection, and B. Lorsbach for assistance with the histological laser microdissection. We offer special thanks to the personnel of SJCRH's animal resource center and art department for their efficient and helpful work.
H.P. was a holder of a doctoral fellowship from the FRIA (Belgian government). This work was supported by grants DK44158 and CA76379 (to J.L.C.), by Cancer Center Core grant CA-21765, and by the American Lebanese Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital.
REFERENCES
- 1.Ajiro K, Yoda K, Utsumi K, Nishikawa Y. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J Biol Chem. 1996;271:13197–13201. doi: 10.1074/jbc.271.22.13197. [DOI] [PubMed] [Google Scholar]
- 2.Askew D S, Ashmun R A, Simmons B C, Cleveland J L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991;6:1915–1922. [PubMed] [Google Scholar]
- 3.Auvinen M, Paasinen A, Andersson L C, Holtta E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360:355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 4.Balasundaram D, Tabor C W, Tabor H. Spermidine or spermine is essential for the aerobic growth of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:5872–5876. doi: 10.1073/pnas.88.13.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balasundaram D, Tyagi A K. Polyamine-DNA nexus: structural ramifications and biological implications. Mol Cell Biochem. 1991;100:129–140. doi: 10.1007/BF00234162. [DOI] [PubMed] [Google Scholar]
- 6.Bello-Fernandez C, Cleveland J L. c-myc transactivates the ornithine decarboxylase gene. Curr Top Microbiol Immunol. 1992;182:445–452. doi: 10.1007/978-3-642-77633-5_56. [DOI] [PubMed] [Google Scholar]
- 7.Benvenisty N, Leder A, Kuo A, Leder P. An embryonically expressed gene is a target for c-Myc regulation via the c-Myc-binding sequence. Genes Dev. 1992;6:2513–2523. doi: 10.1101/gad.6.12b.2513. [DOI] [PubMed] [Google Scholar]
- 8.Blackwood E M, Eisenman R N. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 9.Bruun L, Houen G. In situ detection of diamine oxidase activity using enhanced chemiluminescence. Anal Biochem. 1996;233:130–136. doi: 10.1006/abio.1996.0017. [DOI] [PubMed] [Google Scholar]
- 10.Celano P, Baylin S B, Giardiello F M, Nelkin B D, Casero R A. Effect of polyamine depletion on c-myc expression in human colon carcinoma cells. J Biol Chem. 1988;263:5491–5494. [PubMed] [Google Scholar]
- 11.Coppola J A, Cole M D. Constitutive c-myc oncogene expression blocks mouse erythroleukaemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 12.Dealy M J, Nguyen K V, Lo J, Gstaiger M, Krek W, Elson D, Arbeit J, Kipreos E T, Johnson R S. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat Genet. 1999;23:245–248. doi: 10.1038/13886. [DOI] [PubMed] [Google Scholar]
- 13.Domashenko A D, Latham K E, Hatton K S. Expression of myc-family, myc-interacting, and myc-target genes during preimplantation mouse development. Mol Reprod Dev. 1997;47:57–65. doi: 10.1002/(SICI)1098-2795(199705)47:1<57::AID-MRD8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 14.Dypbukt J M, Ankarcrona M, Burkitt M, Sjoholm A, Strom K, Orrenius S, Nicotera P. Different prooxidant levels stimulate growth, trigger apoptosis, or produce necrosis of insulin-secreting RINm5F cells. The role of intracellular polyamines. J Biol Chem. 1994;269:30553–30560. [PubMed] [Google Scholar]
- 15.Eilers M, Schirm S, Bishop J M. The MYC protein activates transcription of the alpha-prothymosin gene. EMBO J. 1991;10:133–141. doi: 10.1002/j.1460-2075.1991.tb07929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira C, Bucchini D, Martin M E, Levi S, Arosio P, Grandchamp B, Beaumont C. Early embryonic lethality of H ferritin gene deletion in mice. J Biol Chem. 2000;275:3021–3024. doi: 10.1074/jbc.275.5.3021. [DOI] [PubMed] [Google Scholar]
- 17.Fozard J R, Part M L, Prakash N J, Grove J. Inhibition of murine embryonic development by alpha-difluoromethylornithine, an irreversible inhibitor of ornithine decarboxylase. Eur J Pharmacol. 1980;65:379–391. doi: 10.1016/0014-2999(80)90342-8. [DOI] [PubMed] [Google Scholar]
- 18.Frostesjo L, Holm I, Grahn B, Page A W, Bestor T H, Heby O. Interference with DNA methyltransferase activity and genome methylation during F9 teratocarcinoma stem cell differentiation induced by polyamine depletion. J Biol Chem. 1997;272:4359–4366. doi: 10.1074/jbc.272.7.4359. [DOI] [PubMed] [Google Scholar]
- 19.Gramzinski R A, Parchment R E, Pierce G B. Evidence linking programmed cell death in the blastocyst to polyamine oxidation. Differentiation. 1990;43:59–65. doi: 10.1111/j.1432-0436.1990.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 20.Grandori C, Mac J, Siebelt F, Ayer D E, Eisenman R N. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 21.Ha H C, Sirisoma N S, Kuppusamy P, Zweier J L, Woster P M, Casero R A. The natural polyamine spermine functions directly as a free radical scavenger. Proc Natl Acad Sci USA. 1998;95:11140–11145. doi: 10.1073/pnas.95.19.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heby O, Emanuelsson H. Role of the polyamines in germ cell differentiation and in early embryonic development. Med Biol. 1981;59:417–422. [PubMed] [Google Scholar]
- 23.Jiang Y, Roberts S C, Jardim A, Carter N S, Shih S, Ariyanayagam M, Fairlamb A H, Ullman B. Ornithine decarboxylase gene deletion mutants of Leishmania donovani. J Biol Chem. 1999;274:3781–3788. doi: 10.1074/jbc.274.6.3781. [DOI] [PubMed] [Google Scholar]
- 24.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 25.Lovaas E. Antioxidative and metal-chelating effects of polyamines. Adv Pharmacol. 1997;38:119–149. doi: 10.1016/s1054-3589(08)60982-5. [DOI] [PubMed] [Google Scholar]
- 26.MacRae M, Kramer D L, Coffino P. Developmental effect of polyamine depletion in Caenorhabditis elegans. Biochem J. 1998;333(Part 2):309–315. doi: 10.1042/bj3330309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsui M, Oshima M, Oshima H, Takaku K, Maruyama T, Yodoi J, Taketo M M. Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev Biol. 1996;178:179–185. doi: 10.1006/dbio.1996.0208. [DOI] [PubMed] [Google Scholar]
- 28.Miltenberger R J, Sukow K A, Farnham P J. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 30.Morgan D M. Polyamines. An overview. Mol Biotechnol. 1999;11:229–250. doi: 10.1007/BF02788682. [DOI] [PubMed] [Google Scholar]
- 31.Mountford P S, Smith A G. Internal ribosome entry sites and dicistronic RNAs in mammalian transgenesis. Trends Genet. 1995;11:179–184. doi: 10.1016/S0168-9525(00)89040-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Hagan R C, Ohh M, David G, de Alboran I M, Alt F W, Kaelin W G, Jr, DePinho R A. Myc-enhanced expression of Cul1 promotes ubiquitin-dependent proteolysis and cell cycle progression. Genes Dev. 2000;14:2185–2191. doi: 10.1101/gad.827200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano M, Bell D W, Haber D A, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 34.Packham G, Cleveland J L. Ornithine decarboxylase is a mediator of c-Myc-induced apoptosis. Mol Cell Biol. 1994;14:5741–5747. doi: 10.1128/mcb.14.9.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Packham G, Porter C W, Cleveland J L. c-Myc induces apoptosis and cell cycle progression by separable, yet overlapping, pathways. Oncogene. 1996;13:461–469. [PubMed] [Google Scholar]
- 36.Parganas E, Wang D, Stravopodis D, Topham D J, Marine J C, Teglund S, Vanin E F, Bodner S, Colamonici O R, van Deursen J M, Grosveld G, Ihle J N. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 37.Park M H. The essential role of hypusine in eukaryotic translation initiation factor 4D (eIF-4D). Purification of eIF-4D and its precursors and comparison of their activities. J Biol Chem. 1989;264:18531–18535. [PubMed] [Google Scholar]
- 38.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 39.Peralta S A, Gilliard G, Megosh L, George K, O'Brien T G. Polyamines regulate expression of the neoplastic phenotype in mouse skin. Cancer Res. 1998;58:1654–1659. [PubMed] [Google Scholar]
- 40.Persson L, Wallstrom E L, Nasizadeh S, Dartsch C, Jeppsson A, Wendt A, Holmgren J. Regulation of mammalian ornithine decarboxylase. Biochem Soc Trans. 1998;26:575–579. doi: 10.1042/bst0260575. [DOI] [PubMed] [Google Scholar]
- 41.Pierce G B, Gramzinski R A, Parchment R E. Amine oxidases, programmed cell death, and tissue renewal. Philos Trans R Soc Lond B Biol Sci. 1990;327:67–74. doi: 10.1098/rstb.1990.0043. [DOI] [PubMed] [Google Scholar]
- 42.Pohjanpelto P, Holtta E. Phosphorylation of Okazaki-like DNA fragments in mammalian cells and role of polyamines in the processing of this DNA. EMBO J. 1996;15:1193–1200. [PMC free article] [PubMed] [Google Scholar]
- 43.Ray R M, Viar M J, Yuan Q, Johnson L R. Polyamine depletion delays apoptosis of rat intestinal epithelial cells. Am J Physiol Cell Physiol. 2000;278:C480–C489. doi: 10.1152/ajpcell.2000.278.3.C480. [DOI] [PubMed] [Google Scholar]
- 44.Rosenwald I B, Rhoads D B, Callanan L D, Isselbacher K J, Schmidt E V. Increased expression of eukaryotic translation initiation factors eIF-4E and eIF-2 alpha in response to growth induction by c-myc. Proc Natl Acad Sci USA. 1993;90:6175–6178. doi: 10.1073/pnas.90.13.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwartz B, Hittelman A, Daneshvar L, Basu H S, Marton L J, Feuerstein B G. A new model for disruption of the ornithine decarboxylase gene, SPE1, in Saccharomyces cerevisiae exhibits growth arrest and genetic instability at the MAT locus. Biochem J. 1995;312(Part 1):83–90. doi: 10.1042/bj3120083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen-Li H, O'Hagan R C, Hou H, Horner J W, Lee H W, DePinho R A. Essential role for Max in early embryonic growth and development. Genes Dev. 2000;14:17–22. [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder R D. Inhibition of X-ray-induced DNA strand break repair in polyamine-depleted HeLa cells. Int J Radiat Biol. 1989;55:773–782. doi: 10.1080/09553008914550821. [DOI] [PubMed] [Google Scholar]
- 48.Snyder R D. Polyamine depletion is associated with altered chromatin structure in HeLa cells. Biochem J. 1989;260:697–704. doi: 10.1042/bj2600697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snyder R D, Lachmann P J. Hyperthermia, polyamine depletion, and inhibition of X-ray-induced DNA strand break repair. Radiat Res. 1989;120:121–128. [PubMed] [Google Scholar]
- 50.Tabor C W, Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- 51.van Deursen J, Boer J, Kasper L, Grosveld G. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 1996;15:5574–5583. [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Penfold S, Tang X, Hattori N, Riley P, Harper J W, Cross J C, Tyers M. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr Biol. 1999;9:1191–1194. doi: 10.1016/S0960-9822(00)80024-X. [DOI] [PubMed] [Google Scholar]
- 53.Winn L M, Wells P G. Phenytoin-initiated DNA oxidation in murine embryo culture, and embryo protection by the antioxidative enzymes superoxide dismutase and catalase: evidence for reactive oxygen species-mediated DNA oxidation in the molecular mechanism of phenytoin teratogenicity. Mol Pharmacol. 1995;48:112–120. [PubMed] [Google Scholar]
- 54.Zwierzchowski L, Czlonkowska M, Guszkiewicz A. Effect of polyamine limitation on DNA synthesis and development of mouse preimplantation embryos in vitro. J Reprod Fertil. 1986;76:115–121. doi: 10.1530/jrf.0.0760115. [DOI] [PubMed] [Google Scholar]