Abstract
Genomic structural variation (SV) can have profound effects on an organism’s evolution, often serving as a novel source of genetic variation. Gene copy number variation (CNV), a specific form of SV, has repeatedly been associated with adaptive evolution in eukaryotes, especially to biotic and abiotic stresses. Resistance to the most widely used herbicide, glyphosate, has evolved through target-site CNV in many weedy plant species, including the economically important cosmopolitan grass, Eleusine indica (goosegrass); however, the origin and mechanisms of these resistance CNVs remain elusive in many weed species due to limited genetic and genomics resources. In order to study the target site CNV in goosegrass, we generated high-quality reference genomes for both glyphosate-susceptible and -resistant individuals, fine assembled the duplication of glyphosate’s target site gene enolpyruvylshikimate-3-phosphate synthase (EPSPS), and revealed a novel rearrangement of EPSPS into the subtelomeric region of the chromosomes, ultimately leading to herbicide resistance evolution. This discovery adds to the limited knowledge of the importance of subtelomeres as rearrangement hotspots and novel variation generators as well as provides an example of yet another unique pathway for the formation of CNVs in plants.
1. Introduction
Eleusine indica (Indian goosegrass) is one of the most economically important weed species in tropical and sub-tropical regions globally, commonly infesting cereals (especially rice), legumes, cotton, and vegetable crops, and it is also a common weed of turf systems. Decades of using the herbicide glyphosate for goosegrass control has applied enormous selective pressure for glyphosate resistance evolution. Glyphosate is a non-selective herbicide that targets the protein 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS)1, an essential enzyme for aromatic amino acid synthesis in plants. In some cases, glyphosate resistance in goosegrass is caused by EPSPS target site mutations such as the Pro106 single mutations and the Thr102Ile and Pro106Ser (i.e. TIPS) double mutation2; however, increased EPSPS copy number variation (CNV)3, a type of genomic structural variation (SV), is the more frequent mechanism3 in this species. With both mechanisms present in goosegrass, some populations, or even individuals, can have both EPSPS CNV and target-site mutations4, with some, most, or all the copies of EPSPS having one or multiple mutations.
Genomic SV can have profound effects on an organism’s evolution5. As opposed to other forms of genetic variation, such as single nucleotide polymorphisms (SNPs) or insertions/deletions (InDels), SV does not always occur at a constant rate6. Instead, SV formation is punctuated and depends on several factors including the environment, transposable element activity, genetics, hybridization events, and the state of the epigenome. In plants, SV is a broad category and may include smaller-scale events like trans/cis-duplications, tandem duplications, and inversions as well as large scale events like chromosome arm inversions and even polyploidy7. SV that varies gene copy number (i.e., CNV) can have a direct impact on gene expression without changing the nucleotide sequence of the gene itself. Furthermore, additional gene copies often diverge over time and can eventually neo- or sub-functionalize, resulting in increased genetic diversity8.
Some regions of the genome, as well as some gene families, are especially prone to generating SVs and CNVs. Chief among these are regions of highly repetitive sequences where unequal crossing over happens frequently due to misalignment of sister chromatids, homologous chromosomes, and even non-homologous chromosomes. Chromosomes are often highly repetitive at their ends in the telomeres and subtelomeres. The subtelomeres are loosely defined and vary across taxa but are typically the next 50kb to 100kb of genome adjacent to the telomeres. Subtelomeres, while generally gene poor, can be sources of novel CNV events as crossing over can happen frequently. For instance, it has been previously shown in Phaseolus vulgaris that certain pathogen resistance genes exist in or near the subtelomere and that due to their proximity to the subtelomeres, these resistance genes are highly duplicated leading to certain pathogen resistance phenotypes9. This phenomenon is also relevant in monocotyledonous species. In allohexaploid bread wheat (Triticum aestivum), there is generally less synteny of genic regions among subgenomes in the subtelomeres compared to interstitial regions between the centromeres and subtelomeres, partially from high levels of CNV10. CNVs in subtelomeres are not limited to plants; in human subtelomeres, CNVs constitute around 80% of the most distal 100kb of the chromosomes11.
In plants, we are only now beginning to understand the importance of SV as a novel source of genetic variation due to the advent of ubiquitous and inexpensive genome sequencing technologies12. Projects like the 1001 Genomes Project13 are thoroughly investigating SV and its importance in Arabidopsis thaliana. Additionally, there have been other examples demonstrating the importance of SVs in the evolution of crops and non-crops and the massive effects that they can have on phenotypes6,14–16. One of the most striking examples of SV in action has been the evolution of glyphosate (Roundup) resistance in weedy species that effect row crop production. In cases where CNVs cause glyphosate resistance, the plant over-produces the EPSPS enzyme, to a degree that an enormous amount of glyphosate is needed to inhibit the entire EPSPS protein pool.
At least nine divergent, monocot and eudicot weed species have independently evolved glyphosate resistance via EPSPS CNV, an astounding example of convergent evolution17. Furthermore, each species has evolved these CNVs uniquely. The first EPSPS CNV was discovered in Amaranthus palmeri (palmer amaranth)18. It was eventually discovered that the EPSPS gene is being duplicated by a novel, extra-chromosomal, circular piece of DNA named “the replicon”19,20. The replicon independently replicates from palmer’s core genome and tethers itself to the core genome during cellular division20,21. Other weed species have duplicated EPSPS in more familiar ways; for example, EPSPS is duplicated in Bassia scoparia (kochia) in tandem and is thought to be the result of a combination of transposable element activity and unequal crossing over22. Efforts to identify mechanisms of CNV formation have been concentrated on dicot species in the Americas, although globally, weedy monocot species are more problematic.
Despite E. indica’s global economic importance, molecular biology and genomics research has remained difficult due to the lack of a quality reference genome and other molecular tools23,24. In this work, we generated a nearly end-to-end assembly of a glyphosate susceptible (GS) goosegrass individual and a near complete assembly of a glyphosate resistant (GR) individual. Furthermore, we use genomic resequencing, comparative genomics, and transcriptomics to identify the complete genomic region involved in goosegrasses EPSPS CNV event and provide insight in the mechanism driving increased copy numbers. This is the first in-depth investigation into the nature of EPSPS CNV in a grass species and is the first description of subtelomeric repeat driven herbicide resistance in any species.
2. Results and Discussion
2.1. Genome assembly, annotation, and overview
We assembled a chromosome-scale genome of a GS E. indica plant using PacBio HiFi and long-range interaction (Hi-C) datasets. The assembly consists of nine chromosomes spanning 509,878,742 base pairs estimated to be approximately 98.0% complete by Benchmarking Universal Single-Copy Orthologs (BUSCO)25 analysis. Gene models for this species were called using a de novo Isoseq dataset and predicted genes were prescribed function using homology and other protein domain predictive software. Ultimately 27,487 gene models were predicted in the GS E. indica genome. Additionally, we assembled another, near-chromosome level (>99% in 15 contigs), 541,164,105 base pair long genome using a separate PacBio HiFi dataset from a single GR individual, also estimated to be 98.0% complete by BUSCO. Using the same annotation pipeline, 29,090 genes were predicted in the GR E. indica genome. This resistant individual was confirmed to have increased EPSPS gene copy-number as its major glyphosate resistance mechanism in previous work via qPCR of EPSPS and EPSPS sanger sequencing4.
On average, gene density and transposable element density vary inversely in both GR and GS genomes; gene density decreases near the centromeres but increases in the distal parts of the chromosome arms, with the opposite being true for transposable element density (Fig. 1). There are higher numbers of LTR transposons (i.e. Copia, Gypsy, and all other TEs), as well as other transposable elements, clustering at the centromeres and the most distal arms of each chromosome near the telomeres. The Gypsy superfamily is especially prevalent, having the highest transposable element density (Fig. 1). The GS genome contains 59.89% total repetitive sequence, including 2.53% DNA transposons and 40.21% retro-elements (38.29% of which are long terminal repeat elements). The GR genome contains 58.30% total repetitive sequence, including 2.08% DNA transposons and 35.71% retro-elements (33.91% of which are long terminal repeat elements). The most dominant transposon family in both genomes were classified as Gypsy transposons (~25%) in both genome assemblies, with Copia transposons being the second most abundant (~11%). All other transposons make up less than 2% of each genome. There were no large differences at the superficial scale in which we annotated repeat content between GS and GR.
Figure 1: Overview of the glyphosate susceptible Eleusine indica genome.
The Circos plot shows (a.) the length (Mb) of chromosomes one through nine as an index with corresponding (b.) gene-rich (blue) and gene-poor (yellow) genomic regions, (c.) Gypsy (red), Copia (blue), and other (black) transposable element family coverage across the genome (scale: 0–50%), (d.) transposable element rich (red) and transposable element poor (yellow) genomic regions, and the location of the EPSPS cassette (EPSPS-A & EPSPS-B).
E. indica has Arabidopsis-type telomere sequence (TTTAGGGn)26 that are tandemly repeated at the terminal ends of the chromosomes in up to 39,800bp long stretches. In the GS genome, chromosomes two, three, and four have these tandem telomeric repeats at one terminal end and the remaining chromosomes, one, five, six, seven, eight, and nine do not contain any terminal tandem telomeric repeats (Fig. 2). In the GR genome, chromosomes one, two, four, seven and eight begin and end with tandem telomeric repeats, while chromosomes three, five, six, and nine only have tandem telomeric repeats at one end of the chromosome (Fig. 2). This indicates we have captured most but not all the full-length chromosomes in the GR genome. The increase in telomere-to-telomere coverage in the GR genome compared to the GS may be explained by biological factors, such as increased homozygosity of the resistant line, or computational factors, such as different amounts of input PacBio or PacBio read N50 size. Most likely the highly repetitive chromosome ends and redundancy between the telomeric repeats makes further refinement difficult without the implementation of novel techniques specifically designed to resolve these regions.
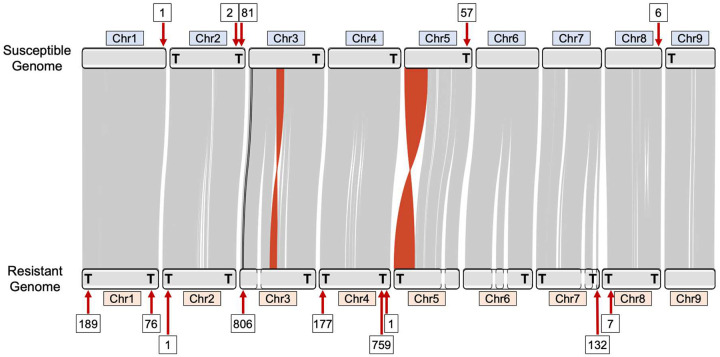
Figure 2: An ideogram of glyphosate susceptible and resistant genome alignment.
Grey links indicate shared synteny between chromosome pairs. Red links indicate large inversions of synteny between the genomes. Black links represent Region-A and Region-B of the EPSPS-cassette in their native locations. Numbers in boxes above and below the ideogram indicate the number of copies of sub-telomeric repeats at each locus. A bold letter “T” on the karyotype represents ends of the chromosomes where the assembly reaches the telomeres.
The GS and GR assemblies are highly syntenic, as indicated by the linear arrangement of large (>10kb) lengths of nearly identical sequences that remain in order over entire chromosomes (Fig. 2). Due to the high amount of synteny between the assemblies, the slightly more fragmented GR genome has been manually ordered and named to maximize alignment with the GS genome. In the GR assembly, chromosomes three and five are composed of two contigs each while chromosomes six, and seven are comprised of three contigs, with the remaining chromosomes being in single contigs. Two large inversions were assembled in the GR genome at the ends of chromosomes five and six; and without Hi-C or optical mapping, we cannot say for certain how supported these inversions are or if they have any impact on the resistance phenotype.
In both the GS and GR genomes, EPSPS, glyphosate’s target, was located approximately 1.6Mb from the telomere of chromosome three. In the GR genome, EPSPS was also identified in 23 short unscaffolded contigs, but always co-assembled in these un-scaffolded contigs with a copy of a sequence normally 2.4Mb from the telomere of chromosome three, about 1Mb downstream of EPSPS. We call the original location of EPSPS in the genome “Region-A,” and the region co-assembled with Region-A, “Region-B” for ease of discussion and clarity (Fig. 1; Fig. 3). Contrary to the tandem EPSPS duplications conferring glyphosate resistance in the weed Bassia scoparia (kochia)22, only one copy of Region-A was assembled at the native location on chromosome three in the GR genome, suggesting at least one initial trans-duplication event of the EPSPS must have occurred before subsequent duplications.
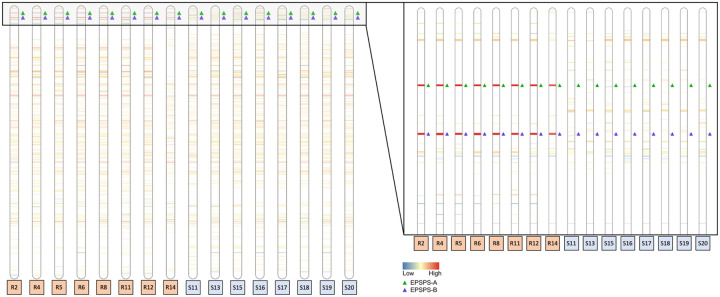
Figure 3: Copy number variation in chromosome three across eight GR and eight GS Eleusine indica individuals.
The ideogram shows deletions below 0.25x of average read depth (blue color spectrum), copy number variation above 0.25 of average read depth and below 4x of average read depth and with a p-value greater than 0.01 (white), and duplications above 4x of average read depth (red color spectrum) across chromosome three in eight glyphosate-resistant (GR: R) versus eight glyphosate-susceptible (GS: S) E. indica individuals at a scale of (a.) full chromosome length (63,742,515 base pairs) and (b.) the first 5,000,000 base pairs of chromosome three. Band thickness is proportional to the length of the genomic region exhibiting copy number variation. The EPSPS cassette, duplicated consistently around 32x compared to average read depth in GR (R) individuals but is not duplicated in any of the GS (S) individuals, is marked by region: EPSPS-A with a green triangle and EPSPS-B with a purple triangle.
2.2. EPSPS structural variation
Eight GS and eight GR E. indica genomes were re-sequenced using PacBio HiFi to determine both the uniformity of the EPSPS CNV event in terms of structure but also in terms of other polymorphisms such as SNPs and InDels. This resequencing data was aligned to the GS genome and analyzed for 1) changes in read depth (indicating duplications or deletions) of certain regions that were uniform across the GR population and divergent from GS individuals, and 2) breaks in read alignment that describe rearrangements, translocations, inversions, and duplications. There were 34 shared predicted CNV duplication events among all GR individuals, 15 in the pseudomolecules and 19 in the unscaffolded contigs. CNV2 (Region-A; average read depth: 22.03) and CNV3 (Region-B; average read depth: 22.46) had nearly identical read depth in all individuals (Table 1; Fig. 3). Seven of the eight GR individuals had EPSPS read depths of approximately 25–29x above background except for sample R14 which only had 8x (Fig. 3). From this observation we believe this sample was a heterozygote, while the other seven GR individuals had copies in both the paternal and maternal genome. Additionally, these results indicate both that Region-A and Region-B are co-duplicated in every copy of the EPSPS CNV, and that Region-A and Region-B were translocated and fused prior to CNV proliferation. Four other CNV events (CNV21, CNV26, CNV 29, and CNV30) that were predicted to have high copy number (>20x) but are solely located in unscaffolded contigs that are mainly composed of highly repetitive DNA and/or transposons.
Table 1:
Table of all duplicated CNVs shared in glyphosate resistant (GR) individuals that do not appear in any glyphosate-susceptible (GS) individuals.
| CNV Event Number | Chromosome or Scaffold | Start | Stop | Length | Average Read Depth in GR |
|---|---|---|---|---|---|
| CNV1 | Chrl | 54,792,251 | 54,809,000 | 16,749 | 2.39 |
| CNV2 (A) | Chr3 | 1,666,751 | 1,701,750 | 34,999 | 22.03 |
| CNV3 (B) | Chr3 | 2,719,751 | 2,767,250 | 47,499 | 22.46 |
| CNV4 | Chr4 | 30,001,501 | 30,024,250 | 22,749 | 3.31 |
| CNV5 | Chr4 | 32,703,001 | 32,731,250 | 28,249 | 9.19 |
| CNV6 | Chr5 | 10,538,001 | 10,552,000 | 13,999 | 2.23 |
| CNV7 | Chr5 | 13,291,251 | 13,298,500 | 7,249 | 2.22 |
| CNV8 | Chr6 | 120,001 | 124,750 | 4,749 | 2.18 |
| CNV9 | Chr6 | 142,751 | 153,250 | 10,499 | 3.40 |
| CNV10 | Chr7 | 27,384,251 | 27,401,750 | 17,499 | 2.03 |
| CNV11 | Chr7 | 35,146,001 | 35,150,750 | 4,749 | 2.14 |
| CNV12 | Chr7 | 40,076,251 | 40,112,750 | 36,499 | 3.68 |
| CNV13 | Chr8 | 42,437,251 | 42,510,250 | 72,999 | 2.06 |
| CNV14 | Chr9 | 11,015,251 | 11,024,750 | 9,499 | 5.86 |
| CNV15 | Chr9 | 18,044,251 | 18,072,250 | 27,999 | 2.80 |
| CNV16 | Scaffold12 | 46,751 | 94,500 | 47,749 | 3.29 |
| CNV17 | Scaffold12 | 121,751 | 135,250 | 13,499 | 4.66 |
| CNV18 | Scaffold26 | 99,251 | 118,500 | 19,249 | 4.71 |
| CNV19 | Scaffold29 | 32,251 | 58,000 | 25,749 | 3.04 |
| CNV20 | Scaffold29 | 88,251 | 124,750 | 36,499 | 4.28 |
| CNV21 | Scaffold30 | 13,251 | 123,250 | 109,999 | 32.46 |
| CNV22 | Scaffold35 | 10,001 | 120,500 | 110,499 | 2.93 |
| CNV23 | Scaffold36 | 1 | 38,500 | 38,499 | 3.04 |
| CNV24 | Scaffold36 | 52,001 | 88,750 | 36,749 | 2.16 |
| CNV25 | Scaffold36 | 104,751 | 117,250 | 12,499 | 5.38 |
| CNV26 | Scaffold44 | 1 | 93,250 | 93,249 | 47.34 |
| CNV27 | Scaffold45 | 1 | 50,250 | 50,249 | 2.07 |
| CNV28 | Scaffold45 | 68,001 | 91,000 | 22,999 | 5.88 |
| CNV29 | Scaffold47 | 1,251 | 80,000 | 78,749 | 45.20 |
| CNV30 | Scaffold49 | 1 | 77,750 | 77,749 | 65.48 |
| CNV31 | Scaffold51 | 1 | 23,250 | 23,249 | 2.25 |
| CNV32 | Scaffold55 | 1 | 16,250 | 16,249 | 3.84 |
| CNV33 | Scaffold56 | 37,251 | 65,500 | 28,249 | 6.15 |
| CNV34 | Scaffold58 | 48,501 | 64,000 | 15,499 | 4.42 |
Region-A is approximately 35kb-long at the coordinates Chr3:1,666,751–1,701,750 in GS and Chr3:2,163,092–2,198,095 in GR. Region-A contains glyphosate’s target, EPSPS, as well as four other predicted genes: A390, A400, A410, and A440. Region-B is approximately 41kb-long at coordinates Chr3:2,719,751–2,760,750 in GS and Chr3:3,206,579–3,247,578 in GR with four predicted genes: B510, B520, B560, and B570 (Table 2). The TIPS double amino acid substitution of EPSPS was only found in sample R14. In R14, T102I was present in 12% of the 583 cDNA reads that mapped to EPSPS and P106S was present in 12% of 562 reads, each in approximately one out of the eight predicted copies. This indicates that as mentioned previously, this individual is heterozygous with one haplotype containing the EPSPS CNV and the other containing the TIPS mutation. Possibly associated with the TIPS mutation, a synonymous alanine substitution (GCA to GCG) was also found 28 amino acid residues upstream from T102I on all reads containing the TIPS mutations in the same 12% read coverage. The co-occurrence of TIPS and EPSPS CNV in this individual is not unexpected because it has been shown previously that E. indica individuals can have stacked resistance mechanisms and fitness penalties associated with mutations can be compensated for with unmutated copies of EPSPS. Such scenarios were predicted when the TIPS double mutation was first identified2 and later discovered in goosegrass4. Furthermore, it indicated that SNPs should also be carefully looked for in GR weeds with low EPSPS CNV frequency as a potential primary source of glyphosate resistance.
Table 2:
RNA-Seq data of genes contained within the EPSPS-Cassette
| Gene ID | Label | Annotation | Coordinates | RNA-Seq | ||
|---|---|---|---|---|---|---|
| Glyphosate susceptible Glyphosate resistant |
Start GS Start GR |
Stop GS Stop GR |
logFC | P-value | ||
| Region A | ||||||
| EleInSChr3g081370 EleInRChr3_2g092340 |
EPSPS | EPSPS | 1,669,076 2,165,535 |
1,673,225 2,169,359 |
4.6 | 1.6e-11 |
| EleInSChr3g081410 EleInRChr3_2g092360 |
A410 | Ribosomal subunit protein | 1,673,439 2,169,409 |
1,675,161 2,171,640 |
5.8 | 2.8e-11 |
| EleInSChr3g081390 EleInRChr3_2g092350 |
A390 | tRNA 2’-phosphotransferase 1 | 1,675,833 2,172,099 |
1,680,441 2,176,989 |
4.6 | 4.9e-13 |
| EleInSChr3g081400** None. |
A400 | Unknown protein | 1,682,414 2,178,757 |
1,687,866 2,184,208 |
1.2 | 0.42 |
| EleInSChr3g081440 EleInRChr3_2g080580 |
A440 | Unknown protein of E. coracana | 1,695,565 2,192,031 |
1,701,288 2,198,135 |
5.2 | 3e-14 |
| Region B | ||||||
| EleInSChr3g082510 EleInRChr3_2g221600 |
B510 | DNA repair protein RadA-like | 2,724,808 3,211,487 |
2,735,415 3,224,071 |
5.2 | 6.9e-11 |
| EleInSChr3g082560 EleInRChr3_2g 119210 |
B560 | 6-phosphofructokinase 1 (PFK) gene, complete CDS | 2,742,394 3,229,211 |
2,747,873 3,234,785 |
0.41 | 0.78 |
| EleInSChr3g082520** Not Annotated in R |
B520 | Putative dual specificity protein phosphatase DSP8 | 2,748,924 3,235,752 |
2,752,196 3,239,024 |
1.1 | 0.03 |
| EleInSChr3g082570* EleInRChr3_2g091630 |
B570 | E3 ubiquitin-protein ligase 1 | 2,758,205 3,245,033 |
2,759,136 3,245,964 |
- | - |
Filtered from differential expression plot during edgeR processing
Not annotated in GR genome. Coordinates from BLAST of GS gene against GR genome.
In GR individuals, Region-A and Region-B are fused and associated with a 3,396bp region of unknown origin inserted at the beginning of Region-B, labeled ‘Region-I’ (Figures 4a and 4b). Region-I contains no predicted genes or features of significance we can decern. Together, Region-A, Region-B, and Region-I makeup the entire ‘EPSPS-Cassette’, a structure not found in any of the glyphosate-susceptible individuals (Fig. 3a). Flanking the EPSPS-Cassette on both sides is a 452bp subtelomeric sequence that can also be found in the most distal part on various chromosomes in the susceptible and resistant E. indica genomes as well as to the subtelomere from other grass species. On one side of the cassette the subtelomeric sequence is repeated 12 times in the forward and 31 times in the reverse directions and serves to invert the EPSPS cassette. On the other side is a much larger array of repeats consisting of a minimum of 43 repeats in the forward and 294 repeats in the reverse directions (Fig. 3a). We were able to span and assemble across the small array of repeats; however, the larger array became ambiguous. Given previous fluorescent in situ hybridization (FISH) experiments EPSPS exists on two chromosomes in GR E. indica27. This information indicates that the EPSPS-Cassette is not scattered or widely dispersed but located in tandem on the ends of one or two chromosomes.
Figure 4. A self-alignment of the EPSPS-Cassette assembled from the GR genome.
A) The cassette consists of several domains. The A-region is approximately 35kb and corresponds to Chr3:1666751–1701750 and contains the EPSPS gene itself. The B-region is approximately 41kb and corresponds to Chr3: 2719751–2760750. The I-region is a small, 450bp sequence inserted into the beginning of the B-region from an unknown origin. The entire cassette is assembled in reverse orientation, and it is denoted as A’, B’, and I’ in reverse orientation. A shorter stretch of subtelomeric repeats (472bp tandem repeat units) separates the forward and reverse copy of the cassette, and a larger stretch of repeats flanks the two cassettes on either end. B) PacBio reads from the resistant genome were aligned to the forward copy of the cassette to validate the junctions of each domain (STs-A, A-B, B-I, I-B, and B-ST) and to quantify their abundance. All junctions were confirmed to be present and assembled correctly. Furthermore, we confirmed that the inversion point of the cassette in the small subtelomeric repeat region was half as abundant (set at 1x coverage/265 reads). All other junctions were shown to be approximately twice as abundant (461–556 reads).
To verify the EPSPS-Cassette model presented here and specifically confirm each region-junction PacBio resequencing data were aligned to the EPSPS-Cassette (explicitly junctions checked are: Subtelomere-A, A-B, B-I, I-B, and B-Subtelomere). Around 545 PacBio reads were aligned to the manually assembled EPSPS-cassette model to support the Subtelomere-A and B-Subtelomere junctions, 466 reads support the A-B, B-I, and I-B junctions, and 265 reads support the subtelomere junction between the reverse and forward EPSPS-Cassettes (Fig. 3b). The number of reads supporting the inversion is almost exactly half of the number of reads that support all other junctions, indicating that the inversion exists half as much as the others. That is to say, the full-length cassette consists of one forward and one reverse copy of the A-B fusion, joined by the inverted, shorter subtelomeric repeat and flanked by much larger subtelomeric sequences on both sides.
In addition, an RNA-seq experiment was performed to investigate gene expression changes driven by the EPSPS CNV. Although the entire EPSPS-Cassette and all genes within it are co-duplicated in GR individuals, four of the five genes in Region-A are significantly overexpressed (p-value<0.01 and fold-change>2), while only one out of four genes in Region-B is significantly overexpressed (Table 2, Fig. 5). Genes overexpressed other than EPSPS from Region-A include A410: A ribosomal subunit protein, A390: a tRNA-2’phosphotransferase, and A440: a protein of unknown function. Interestingly, homologs of A410 and A390 are also co-duplicated with EPSPS in Bassia scoparia, a eudicot weed with a tandemly duplicated EPSPS CNV22. Given the annotation of these proteins, it is unlikely they are directly involved in the EPSPS CNV formation. B510 is the only significantly overexpressed (log fold-change: 5.2, p-value: 6.9e-11) gene in Region-B (Fig. 7; Table 2). Gene B510 encodes a RadA-like protein, a type of protein known to be associated with EPSPS in glyphosate resistance in other prominent weed species such as kochia. This gene involvement in the formation of the EPSPS CNV is unknown; however, its overexpression indicates it is currently active. RadA proteins are DNA-dependent ATPase that process DNA recombination intermediates and are therefore involved in repairing DNA breaks28. These proteins are particularly interesting in the case of EPSPS duplication due to their role in catalyzing homologous recombination. Whether or not RadA is directly involved in the duplication of the EPSPS loci from various weed species or merely coincidental is open for examination.
Figure 5: Differential expression of eight GR versus eight GS Eleusine indica individuals.
The plot from RNA-Seq data shows over-expressed (red) and under-expressed (blue) genes in GR E. indica individuals with labels for all identified genes within the EPSPS cassette. Gene labels with a non-integer numerical value represent splice variants of the same gene. Genes below a p-value of 0.01 or a fold change value below two were considered not differentially expressed (grey) between the two treatment groups.
2.3. Subtelomeres in Eleusine indica
Whole genome alignment reveals highly conserved subtelomeric repeat sequence from the EPSPS-Cassette near the ends of many of the assembled chromosomes in both the GS and GR goosegrass genomes (Fig. 2). Interestingly, over twice as many subtelomeres at higher average copy number in each region was assembled in the GR genome (Fig. 2). There is an especially high copy number of subtelomeric repeats in chromosomes one, three, four, and seven of the GR genome that could be sites of meiotic recombination and subtelomere rearrangement in and between chromosomes (Fig. 2)29. It has been previously shown in Phaseolus vulgaris that similar subtelomeric sequences are predisposed to unequal intra-strand homologous recombination30 commonly resulting in large duplications of whole pathogen resistance gene pathways.
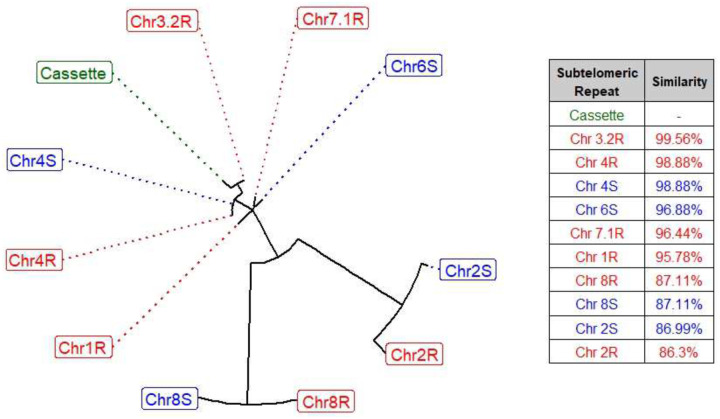
The 452bp-long subtelomeric repeat unit that flanks the EPSPS-Cassette is most like the subtelomeric repeat region of second contig that comprises chromosome three from the GR genome (99.556%), indicating chromosome three is likely the location of the EPSPS-Cassette in GR plants. EPSPS is natively on chromosome three in both assembled genomes. The subtelomeric sequence of chromosome four in both the GS and GR genomes are also highly similar to the EPSPS-Cassette subtelomeric repeat region (98.884%). The subtelomeric repeats found on chromosomes one and seven of the GR genome and chromosome six of the GS genome are 95.778%−96.882% similar to the subtelomeric region of the EPSPS-Cassette, while chromosomes eight and two of both the GS and GR genomes are the least related (86.301%−87.113%). The subtelomeric repeats found on chromosomes four and eight of both genomes are identical (Fig. 6). Work by other researchers using FISH cytometry have shown that the EPSPS CNVs in goosegrass on one or possibly two chromosomes27. Given the sequence similarity of the subtelomeric repeat in the EPSPS-Cassette and the subtelomeres on chromosomes three and four, translocation of the EPSPS-Cassette between these two regions through non-homologous recombination seems feasible; however, we assembled chromosome four of the GR genome from telomere to telomere, completely through the subtelomeric region, and found no evidence of the EPSPS-Cassette on chromosome four in this population.
Figure 6: Relatedness of EPSPS-Cassette subtelomere sequence to chromosomal subtelomeric sequences of the GR and GS Eleusine indica genomes.
The below plot shows the relatedness of subtelomeric sequences found on the glyphosate-resistant (GR: R, red) and glyposate-susceptible (GS: S, blue) E. indica genomes to the subtelomeric sequence found on the EPSPS-Cassette (green). Chromosomes at branch tips further from Cassette are less related to Cassette than chromsomes closer to Cassette. Branch distance is based on BLAST similarity. The sequence with the highest relatedness to the EPSPS-Cassette subtelomere sequence on each chromsome were used as representative sequences to make a tree.
2.4. Subtelomeres in plant evolution
Subtelomeres of eukaryotic organisms are hotspots for adaptive evolution due to frequent, error-prone recombination events during meiosis that lead to rapidly changing genes. In common bean (Phaseolus vulgaris), segmental duplications, sometimes up to 100kb-long, of disease resistance genes located in the subtelomeres are facilitated by non-homologous end joining and frequent interchromosomal recombination30. The large duplications of these distal disease resistance genes allow for divergence of homologous genes into paralogs to allow novel resistance that accounts for the rapid evolution of pathogens. Analogous to the generation of novel disease resistance genes in the subtelomeres of common bean, CNV increases variation and diversity of virulence genes in the subtelomeres of several eukaryotic pathogens including Plasmodium falciparum, Trypanosoma brucei and cruzi, and Pneumocystis carinii and sugar metabolism genes in yeast31. Similarly, the translocation of genomic regions associated with herbicide or other abiotic stress resistance, like the EPSPS-Cassette to the subtelomeric region, may allow for novel abiotic resistance to develop from the rapid accumulation of variation between duplicate loci in addition to an increase in transcript abundance.
Duplications of the EPSPS-Cassette in the subtelomeres, especially in a grass species such as goosegrass, may not be unusual given subtelomeres propensity for generating and selectively maintaining novel variation in other grasses. In Oryza sativa (rice) the subtelomeres are theorized to be associated with high rates of transcription, recombination, and novel gene generation because they contain a large amount of highly similar paralogs, including some stress-response genes32. Despite the apparent stochastic nature of some rice subtelomeric regions, some subtelomeric regions in Sorghum haplensis (sorghum), Brachypodium distachyon (purple false brome), and several species of the Oryza genus exhibit duplications of the same genomic regions and preferential gene conversions within these duplicated regions, suggesting continuous concerted evolution and selective conservation of certain genes in subtelomeric regions33. Such precise selectivity is also evident in several Avena species (oats), where a subtelomeric, 12-gene cluster developed since the evolutionary divergence of Aveneae is maintained with gene order colinear to the biosynthetic pathway34. Given the importance of subtelomeres in other monocots for generating novel genetic variation and our findings, we have strong evidence that E. Indica EPSPS copy number variation is due to unequal crossing over in the subtelomeres of this species after an initial translocation event.
3. Conclusion
The large population size, plastic genomes, and large selection pressures exerted on weed species makes weeds an excellent system to study adaptive genome evolution. As we explore these genomes, we continuously find new ways that plant genomes innovate and overcome various stresses and discover exactly what makes weedy species like goosegrass such a successful survivor. The goals of this research were to both obtain high quality genomic resources for E. indica, a major, global weed, and to use those resources to investigate the genomic rearrangements and mechanism(s) that perpetrated EPSPS gene duplication and therefore glyphosate resistance in this species. By assembling both a GS and GR E. indica genome, genomic resequencing eight individuals of both populations, performing RNA-seq on eight individuals from each population, and manually curating the assembly surrounding the EPSPS locus, we have discovered that the EPSPS gene in GR E. indica has been duplicated, fused with another part of the genome, and inserted in one or more of the subtelomeric regions of the genome. We hypothesize that after this initial translocation and fusion, EPSPS duplication has carried on through unequal crossing over of the subtelomeres, facilitated by the high amounts of similarity between the subtelomeric sequence on the ends of chromosomes three. This is the first report of an herbicide resistance trait brought about by this type of genomic rearrangement and adds to both our knowledge of herbicide resistance evolution, and to the relatively limited information we have about the importance of subtelomeres as rearrangement hotspots and novel variation generators.
4. Methods
4.1. Eleusine indica tissue generation
Glyphosate-susceptible (GS) and glyphosate-resistant (GR) populations of E. indica that have been characterized in previous studies collected from Guangdong Province, China4. These populations were purified and self-pollinated for increased homozygosity and consistency of glyphosate susceptibility or resistance phenotypes. The GR population was confirmed to have EPSPS copy number variation by DNA quantitative PCR before purification.
Seeds of purified GS and GR biotypes were sown on wet filter paper in Petri dishes in a climate chamber at 28–30 °C, with 12h/12h light/dark period and 70% relative humidity. The two-leaf stage seedlings were transplanted into 28 × 54 cm trays (50 plants per tray) filled with potting soil and grown in a glasshouse. At the tillering stage, about ten individuals were randomly selected each from the GS and GR E. indica population and characterized. Three tillers of each plant were separated and repotted (one tiller per pot, 60 pots in total). One tiller of each plant was used for glyphosate resistance and susceptibility phenotyping, one for EPSPS CNV estimation and one for subsequent sequencing.
For glyphosate resistance and susceptibility phenotyping, one regrowth tiller (three days after tiller cloning) was treated with commercial glyphosate (41% glyphosate isopropylamine salt, 400 g ai ha-1 for GS and 1600 g ai ha-1 for GR), and GR (i.e., survivors) and GS (i.e., killed) phenotypes were determined three weeks after treatment. EPSPS CNV was again assessed in the resistant population to ensure the CNV event was still present before genomics work began. Leaf material from untreated tillers of corresponding resistant and susceptible plants was used for genomic DNA isolation using the Plant Genomic DNA kit (Trans Gen Biotech Beijing Co., LTD). Quantitative PCR was performed using published primer pairs and methods where EPSPS copy number was compared to the single copy acetolactate synthase (ALS) gene as the internal reference. The untreated tiller of a confirmed GS plant was used for genomic sequencing performed at Shanghai OE Biotech Co., Ltd (Shanghai, China).
4.2. Susceptible genome assembly.
The initial GS genome was generated from 112.2 Gb (223x) of raw PacBio Sequel II sequencing data and assembled using Falcon35. The resulting initial genome was 518,672,752 base pairs long in 239 contigs with a contig N50 of 18.8 Mb. Error corrections were conducted using the Arrow36 algorithm and a hi-coverage Illumina dataset. The error corrected assembly was compared to the NCBI37 nucleotide database using BLAST to identify possible bacterial or mammalian contamination, of which none was found. After polishing and removing contamination from the assembly, the assembly was 519,224,895 base pairs long in 239 contigs with a contig N50 of 18.8 Mb.
To further increase the continuity of the assembly and fix any possible large-scale errors, Hi-C data was obtained. The Hi-C library utilized the DpnII restriction enzyme (GATC cut sites) to generate sufficient digestion of the fixed DNA38. The final library was sequenced with Illumina HiSeq resulting in 150 base pair long paired end reads. Fastp (version 0.20; Chen et al., 2018) was used to clean the Hi-C paired end reads of adaptors and redundant PCR reads and perform analysis on the cleaned reads. Linker sequences and reads with greater than or equal to 5 N (not AGCT) bases were also removed. Sliding window (window size of 4 base pairs) was performed to excise windows with an average base quality score below 20. Filtered reads less than 75 base pairs long or with an average base quality score below 15 were removed. The resulting clean Hi-C reads had a total yield (G) of 93.48, 641,488,988 read pairs, a Q20% of 97.41%, a Q30% of 92.69%, and a GC content of 44.28%.
Juicer39 was used with the default parameters of both bwa-mem40 and 3D-DNA41 to scaffold the contigs of the PacBio only assembly. Contigs were then clustered by contact point proximity and sorted to generate a Hi-C interaction matrix that was imported into juicebox42 for visualization and manual inspection. The resulting matrix presented no abnormalities and contigs were able to be clustered into 9 chromosome scale scaffolds and 154 much smaller scaffolds. Gaps of 500 Ns were added between each contig to link the chromosomes for filling later. The scaffolded assembly was 519,302,895 base pairs long with an N50 of 57.27 Mb.
Finally, PBjelly43 was used to gap filling the assembly by aligning the original PacBio sequencing data to the Hi-C assembled genome. The gap filled assembly was now 522,502,607 base pairs long with a scaffold N50 of 57.37 Mb and a contig N50 of 42.10 Mb. Arrow36 was then used for self-comparison and another round of error corrections. Last, second-generation sequencing data was used for two rounds of Pilon44 error correction, resulting in a final assembly that was 522,557,097 base pairs long with a scaffold N50 of 57.37 Mb, a contig N50 of 42.10 Mb, and 62 total gaps. The final assembly was benchmarked for gene content using BUSCO)25of 1375 single-copy genes from the embryonic plants database the 1348 (98.0%) were identified as either single or multi copy.
4.3. Resistant genome assembly
The initial GR genome was generated from 28Gb (~53x) of PacBio HiFi45 sequencing data, assembled using HiCanu46 with a predicted genome size of 492Mb. The resulting initial genome was 541,164,105 base pairs long in 2014 contigs with a contig N50 of 47.4 Mb. Per the instruction of HiCanu, post-assembly error corrections were not conducted to avoid introducing errors and dropping below the 99.99% accuracy rating. A BUSCO assessment rated the resulting assembly as 98.0% complete. Contigs were aligned to the glyphosate-susceptible genome to identify chromosomes.
4.4. Genome annotation
Assembled GS and GR E. indica genomes were both annotated using a custom genome annotation pipeline developed by the International Weed Genomics Consortium genome annotation pipeline, as described below (https://www.weedgenomics.org/). First, repeat regions were annotated using RepeatModeler47 (version 2.0.2) and then masked using RepeatMasker48 (version 4.1.2) and bedtools49 (version 2.30.0) as a measure of data reduction before further annotation. Isoseq reads were then mapped to both repeat-masked genomes using Minimap250 (version 2.24) to determine sites of transcription. The resulting Sequence Alignment Map (SAM) files were converted into Binary Alignment Map (BAM) files using the SAMtools51 (version 1.11) view command before being collapsed using cDNA Cupcake52 (version 28.0). The genomes, collapsed cDNA Cupcake outputs, repeat libraries from RepeatModeler, and a protein FASTA file from a close relative, Eleusine corocana (Phytozome genome ID: 560), were fed into MAKER53 (version) to predict the genomic coordinates of putative gene models. Genes that produced proteins under 32 amino acids long were removed from further annotation with only the longest proteins from each gene and unique untranslated regions (UTRs) used for functional annotation.
Functional annotation began by first selecting the longest isoforms from each gene using AGAT54 (version 0.8.0) and gffread55 (version 0.12.7). Longest isoforms sequence similarity searches were conducted using MMseqs256 (version 4.1) with NCBI, UniRef 5057, and the InterPro58 database using InterProScan 559 (version 5.47–82.0) locally. Protein localization was predicted using MultiLoc260 (version 1.0). Using this pipeline, 27,487 genes in GS (BUSCO: 90.5%) and 29,090 genes in GR (BUSCO: 90.4%) were predicted.
4.5. Genome-resequencing and transcriptomics
Illumina 150bp paired-end sequences were generated from the DNA of eight GS and eight GR individuals from the above-named populations. DNA was extracted using leaf material from untreated tillers of corresponding GS and GR plants using the Plant Genomic DNA kit (TIANGEN, Beijing, China). DNA was sequenced using the Illumina HiSeq X Ten sequencing platform (Illumina Inc., San Diego, CA, USA) with an average of 30x coverage. Illumina reads were cleaned using FASTQ and aligned to the susceptible genome using HiSat261 (version 2.1.0) using standard options for paired-end reads. CNVnator62 (version 0.4.1) was used to scan the read depth from the alignments in 5kb windows to roughly call regions of the genome that deviated significantly from the average read depth. CNVnator outputs were intersected using bedtools49 (version 2.30.0) intersect so that regions that were amplified in all eight GR individuals but none of the GS individuals were identified. Only two such regions were discovered. The region containing EPSPS on chromosome three was labeled “Region-A”, while the other, was labeled as “Region-B” for further analysis.
Illumina 150bp paired-end sequences were generated from the RNA of the same eight GS and eight GR individuals that were used for genome resequencing. RNA was extracted from the leave sheath material using the mirVana miRNA Isolation Kit (Ambion). Illumina reads were cleaned and aligned to the GS genome predicted transcriptome using HiSat2 (version 2.1.0) using standard options for paired-end reads. Alignments were converted into count tables using SAMtools (version 1.11). The count table was loaded into R (version 4.2.0) and differential gene expression was calculated using package edgeR63 (version 3.38.1) quasi-likelihood F-tests.
4.6. Investigation of the EPSPS-Cassette and subtelomeres
The EPSPS-Cassette model was resolved by first using BLAST to identify all contigs containing EPSPS, Region-A, and Region-B. Contigs of interest were self-aligned in YASS64 to visualize macrostructure, especially repeat structure. Contigs were manually assembled into putative models based on their repeat macrostructure. Contig junctions of the putative models were confirmed by aligning genomic reads to them using HiSat2 (version 2.1.0). The large subtelomeric repeat regions flanking the reverse-forward EPSPS-Cassette duplications were not able to be assembled completely using this method. The locations and relatedness of sequences similar to the 452bp subtelomeric repeat unit were found using Minimap2 (version 2.24) and BLAST.
4.7. Plot generation
Circos65 (version 0.69–9) was used to visually summarize the overall E. indica genome (Fig. 1). Coverage windows used to make the Circos tracks were generated using bedtools (version 2.30.0; Fig. 1). RIdeogram66 (version 0.2.2) was used to visualize duplications and deletions of EPSPS on chromosome three detected using CNVnator (version 0.4.1; Fig. 2). Synteny plots were made by aligning both genomes using MiniMap2 (version 2.24) and visualized using RIdeogram (version 0.2.2) in R (version 4.2.2; Fig. 3 & Fig. 5). The EPSPS Cassette was visualized using YASS (Fig. 4a & Fig. 4b). Differential expression between eight GS and eight GR E. indica individuals was visualized using ggplot267 (version 3.4.0) in R (version 3.6.0). The subtelomere relatedness tree was generated using RAXML-NG68 (version 1.1.0), ggtree69 (version 3.6.2), cowplot70 (version 1.1.1), and ggplot2 (version 3.4.0) in R (version 4.2.2; Fig. 7).
Acknowledgements
This work is supported by National Science Foundation of China (project nos: 32272568 and 31871984) and the Department of Science and Technology foundation of Guangdong Province, China (2019B121201003) to Dr Chun Zhang and Prof Xingshan Tian, and by Michigan State University and the National Science Foundation Research Traineeship Program (DGE-1828149) and a fellowship from Michigan State University under the Training Program in Plant Biotechnology for Health and Sustainability (T32-GM110523) to Nicholas A. Johnson. Funding for Dr Qin Yu by Australian Grains Research and Development Corporation (GRDC) is acknowledged.
Footnotes
Code availability.
All code used in this research is available upon request to the corresponding authors.
Additional Declarations: There is NO Competing Interest.
Supplementary Files
Data availability.
The assembled genomes, associated GFF annotation files, and all functional annotation information are publicly available through the International Weed Genomics Consortium online database, ‘Weedpedia’ (https://weedpedia.weedgenomics.org/). Raw sequencing data has been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA); Genome resequencing data with the accession numbers SRR23364316-SRR23364331 and RNA-seq data with the accession numbers SRR23372273-SRR23372288.
References
- 1.Sammons R. D. & Gaines T. A. Glyphosate resistance: State of knowledge. Pest Management Science vol. 70 Preprint at 10.1002/ps.3743 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu Q. et al. Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance. Plant Physiol 167, 1440–1447 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J., Mei Y., Zhang L., Hao L. & Zheng M. The Resistance Levels and Target-Site Based Resistance Mechanisms to Glyphosate in Eleusine indica from China. Agronomy 12, 2780 (2022). [Google Scholar]
- 4.Zhang C. et al. Evolution of multiple target-site resistance mechanisms in individual plants of glyphosate-resistant Eleusine indica from China. Pest Manag Sci 77, 4810–4817 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Wellenreuther M., Mérot C., Berdan E. & Bernatchez L. Going beyond SNPs: The role of structural genomic variants in adaptive evolution and species diversification. Mol Ecol 28, 1203–1209 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Debolt S. Copy number variation shapes genome diversity in Arabidopsis over immediate family generational scales. Genome Biol Evol 2, 441–453 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Y., Bayer P. E., Batley J. & Edwards D. Current status of structural variation studies in plants. Plant Biotechnology Journal vol. 19 2153–2163 Preprint at 10.1111/pbi.13646 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marroni F., Pinosio S. & Morgante M. Structural variation and genome complexity: Is dispensable really dispensable? Current Opinion in Plant Biology vol. 18 31–36 Preprint at 10.1016/j.pbi.2014.01.003 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Calderón M. D. C., Rey M. D., Cabrera A. & Prieto P. The subtelomeric region is important for chromosome recognition and pairing during meiosis. Sci Rep 4, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salina E. A.;, Numerova Ozkan, M. ; & Feldman H. Alterations in subtelomeric tandem repeats during early stages of allopolyploidy in wheat. Genome vol. 47 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Riethman H. Human subtelomeric copy number variations. Cytogenetic and Genome Research vol. 123 244–252 Preprint at 10.1159/000184714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francia E., Pecchioni Nicola, Policriti Alberto & Scalabrin Simone. ‘CNV and Structural Variation in Plants: Prospects of NGS Approaches’ in Advances in the Understanding of Biological Sciences Using Next Generation Sequencing (NGS) Approaches. Springer 211–232 (2015). [Google Scholar]
- 13.Weigel D. & Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol 10, 107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolatabadian A., Patel D. A., Edwards D. & Batley J. Copy number variation and disease resistance in plants. Theoretical and Applied Genetics vol. 130 2479–2490 Preprint at 10.1007/s00122-017-2993-2 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Zmieńko A., Samelak A., Kozłowski P. & Figlerowicz M. Copy number polymorphism in plant genomes. Theoretical and Applied Genetics vol. 127 1–18 Preprint at 10.1007/s00122-013-2177-7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook D. E. et al. Copy Number Variation of Multiple Genes at Rhg1 Mediates Nematode Resistance in Soybean. www.soystats.com (2012). [DOI] [PubMed]
- 17.Patterson E. L., Pettinga D. J., Ravet K., Neve P. & Gaines T. A. Glyphosate resistance and EPSPS gene duplication: Convergent evolution in multiple plant species. Journal of Heredity 109, 117–125 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Gaines T. A. et al. Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci U S A 107, 1029–1034 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koo D. H. et al. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. Proc Natl Acad Sci U S A 115, 3332–3337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molin W. T., Yaguchi A., Blenner M. & Saski C. A. The EccDNA replicon: A heritable, extranuclear vehicle that enables gene amplification and glyphosate resistance in Amaranthus palmeri. Plant Cell 32, 2132–2140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaines T. A., Patterson E. L. & Neve P. Molecular mechanisms of adaptive evolution revealed by global selection for glyphosate resistance. New Phytologist vol. 223 1770–1775 Preprint at 10.1111/nph.15858 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Patterson E. L. et al. The Draft Genome of Kochia scoparia and the Mechanism of Glyphosate Resistance via Transposon-Mediated EPSPS Tandem Gene Duplication. Genome Biol Evol 11, 2927–2940 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravet K. et al. The power and potential of genomics in weed biology and management. Pest Manag Sci 74, 2216–2225 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Patterson E. L., Saski C., Küpper A., Beffa R. & Gaines T. A. Omics potential in herbicide-resistant weed management. Plants vol. 8 Preprint at 10.3390/plants8120607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. v. & Zdobnov E. M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Richards E. J. & Ausubel F. M. Isolation of a Higher Eukaryotic Telomere from Arabidopsis thaliana. Cell vol. 53 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Chen J., Cui H., Ma X., Ma Y. & Li X. Distribution differences in the EPSPS gene in chromosomes between glyphosate-resistant and glyphosate-susceptible goosegrass (Eleusine indica). Weed Sci 68, 33–40 (2019). [Google Scholar]
- 28.Seitz E. M., Brockman J. P., Sandler S. J., Clark A. J. & Kowalczykowski S. C. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange possesses the characteristics of a DNA strand exchange protein: The RadA protein is a DNA-dependent ATPase, forms a nucleoprotein filament on DNA, and catalyzes DNA pairing and strand exchange. www.genesdev.org (1998). [DOI] [PMC free article] [PubMed]
- 29.Lambing C., Franklin F. C. H. & Wang C. J. R. Understanding and manipulating meiotic recombination in plants. Plant Physiol 173, 1530–1542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen N. W. G. et al. Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front Plant Sci 9, 1–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baird D. M. Telomeres and genomic evolution. Philosophical Transactions of the Royal Society B: Biological Sciences vol. 373 Preprint at 10.1098/rstb.2016.0437 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan C. et al. The subtelomere of Oryza sativa chromosome 3 short arm as a hot bed of new gene origination in rice. Mol Plant 1, 839–850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacquemin J., Laudié M. & Cooke R. A recent duplication revisited: Phylogenetic analysis reveals an ancestral duplication highly-conserved throughout the Oryza genus and beyond. BMC Plant Biol 9, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y. et al. Subtelomeric assembly of a multi-gene pathway for antimicrobial defense compounds in cereals. Nat Commun 12, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin C. S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chin C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10, 563–569 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Sayers E. W. et al. Database resources of the national center for biotechnology information. Nucleic Acids Res 50, D20–D26 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durand N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments HHS Public Access. Cell Syst 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. (2013). [Google Scholar]
- 41.Dudchenko O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science (1979) 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudchenko O. et al. The Juicebox Assembly Tools module facilitates de novo assembly of mammalian genomes with chromosome-length scaffolds for under $1000. doi: 10.1101/254797. [DOI]
- 43.English A. C., Salerno W. J. & Reid J. G. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics vol. 15 http://www.biomedcentral.com/1471-2105/15/180 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker B. J. et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hon T. et al. Highly accurate long-read HiFi sequencing data for five complex genomes. Sci Data 7, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nurk S. et al. HiCanu: accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flynn Jullien M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. PNAS 117, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smit A., Hubley R. & Green P. RepeatMasker Open-4.0. http://www.repeatmasker.org/ (2015). [Google Scholar]
- 49.Quinlan A. R. & Hall I. M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Danecek P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tseng E. cDNA_Cupcake. Github (2022). [Google Scholar]
- 53.Cantarel B. L. et al. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18, 188–196 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dainat J. AGAT: Another Gff Analysis Toolkit to handle annotations in any GTF/GFF format. Zenodo (2022) doi: 10.5281/zenodo.3552717. [DOI] [Google Scholar]
- 55.Pertea M. & Pertea G. GFF Utilities: GffRead and GffCompare. F1000Res 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinegger M. & Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nature Biotechnology vol. 35 1026–1028 Preprint at 10.1038/nbt.3988 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Suzek B. E., Huang H., McGarvey P., Mazumder R. & Wu C. H. UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Paysan-Lafosse T. et al. InterPro in 2022. Nucleic Acids Res (2022) doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P. et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blum T., Briesemeister S. & Kohlbacher O. MultiLoc2: Integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10, 274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D., Paggi J. M., Park C., Bennett C. & Salzberg S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abyzov A., Urban A. E., Snyder M. & Gerstein M. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21, 974–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson M. D., McCarthy D. J. & Smyth G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Noé L. & Kucherov G. YASS: Enhancing the sensitivity of DNA similarity search. Nucleic Acids Res 33, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krzywinski M. et al. Circos: An information aesthetic for comparative genomics. Genome Res 19, 1639–1645 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hao Z. et al. RIdeogram: Drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci 6, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wickham H. ggplot2. ggplot2 (Springer; New York, 2009). doi: 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- 68.Kozlov A. M., Darriba D., Flouri T., Morel B. & Stamatakis A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu G., Smith D. K., Zhu H., Guan Y. & Lam T. T. Y. ggtree: an r package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8, 28–36 (2017). [Google Scholar]
- 70.Wilke C. O. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’. CRAN-R (2020). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled genomes, associated GFF annotation files, and all functional annotation information are publicly available through the International Weed Genomics Consortium online database, ‘Weedpedia’ (https://weedpedia.weedgenomics.org/). Raw sequencing data has been submitted to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA); Genome resequencing data with the accession numbers SRR23364316-SRR23364331 and RNA-seq data with the accession numbers SRR23372273-SRR23372288.