Abstract
Omega-3 (n-3) and omega-6 (n-6) polyunsaturated fatty acids (PUFAs) play critical roles in human health. Prior genome-wide association studies (GWAS) of n-3 and n-6 PUFAs in European Americans from the CHARGE Consortium have documented strong genetic signals in/near the FADS locus on chromosome 11. We performed a GWAS of four n-3 and four n-6 PUFAs in Hispanic American (n = 1454) and African American (n = 2278) participants from three CHARGE cohorts. Applying a genome-wide significance threshold of P < 5 x 10−8, we confirmed association of the FADS signal and found evidence of two additional signals (in DAGLA and BEST1) within 200 kb of the originally reported FADS signal. Outside of the FADS region, we identified novel signals for arachidonic acid (AA) in Hispanic Americans located in/near genes including TMX2, SLC29A2, ANKRD13D and POLD4, and spanning a >9 Mb region on chromosome 11 (57.5Mb ~ 67.1 Mb). Among these novel signals, we found associations unique to Hispanic Americans, including rs28364240, a POLD4 missense variant for AA that is common in CHARGE Hispanic Americans but absent in other race/ancestry groups. Our study sheds light on the genetics of PUFAs and the value of investigating complex trait genetics across diverse ancestry populations.
Introduction
Omega-3 (n-3) and omega-6 (n-6) polyunsaturated fatty acids (PUFAs) are critical structural components of cell membranes, which can influence cellular activities by promoting the fluidity, flexibility, and the permeability of a membrane.1-3 Additionally, PUFAs affect a variety of other biological processes and molecular pathways, including modulating membrane channels and proteins, regulating gene expression through nuclear receptors and transcription factors, and conversion of the PUFAs themselves into bioactive metabolites.4 Levels of circulating PUFAs and long chain (≥ 20 carbons) PUFAs (LC-PUFAs) are associated with reduced risk of cardiovascular disease5,6, type 2 diabetes mellitus7, cognitive decline8, Alzheimer's disease9, metabolic syndrome10 and breast cancer11, as well as all-cause mortality.12
PUFAs and LC-PUFAs are characterized by the position of the first double bond from the methyl terminal (omega; ω; or n – FAs) and fall into two primary families, n-3 and n-6. The most abundant n-3 PUFAs are alpha-linolenic acid (ALA), eicosapentaenoic acid (EPA), docosapentaenoic acid (DPA) and docosahexaenoic acid (DHA), while the primary n-6 PUFAs are linoleic acid (LA), gamma-linolenic acid (GLA), dihomo-γ-linolenic acid (DGLA) and arachidonic acid (AA). ALA and LA are essential n-3 and n-6 PUFAs consumed from the diet and these then can be converted to more unsaturated LC-PUFAs through a set of desaturation and elongation enzymatic steps. For example, DGLA and AA can be synthesized from LA, while EPA, DPA and DHA can be produced from ALA (Fig. 1). The precursors LA and ALA are essential fatty acids that must be provided by the diet. Due to the lower abundance of ALA in Western diets and the inefficiency of conversion of ALA to longer chain n-3 LC-PUFAs such as EPA and DHA, dietary intake of these via fatty fish or marine oil supplementation is often recommended.13,14
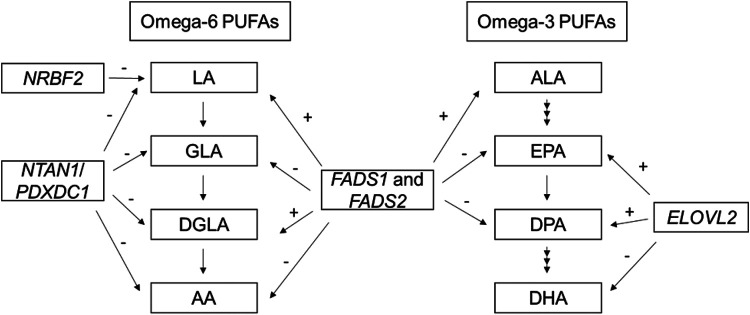
Figure 1. PUFAs metabolic pathway and summary of genome-wide association from previous CHARGE GWAS of n-3 and n-6 PUFAs in European Americans.
Figure 1 shows the summary of genome-wide association from previous CHARGE GWAS of n-3 and n-6 PUFAs in European Americans. + and − signs indicate the direction of the associations for the minor allele of most significant SNP at each locus. The SNPs used to determine the directions of effect at each locus:
FADS1 and FADS2: rs174547 (ALA, DPA, LA, GLA, DGLA and AA); rs174538 (EPA)
ELOVL2: rs780094 (DPA); rs3798713 (EPA); rs2236212 (DHA)
NTAN1/PDXDC1: rs16966952 (LA, GLA, DGLA and AA)
NRBF2: rs10740118 (LA).
Previous studies have shown that African ancestry populations have higher circulating levels of LC-PUFAs compared to European Americans.15 These large differences can be explained in part by variation in the allele frequencies of FADS variants associated with different biosynthetic efficiencies in these two populations.16 Mathias et al. also revealed that African Americans have significantly higher levels of AA and lower levels of the AA precursor DGLA, and that FADS1 variants were significantly associated with AA, DGLA and the AA/DGLA ratio in a sample of fewer than 200 African Americans from the GeneSTAR study.15 In addition, African ancestry populations have higher frequencies of the “derived” FADS haplogroup (represented by the variant rs174537 allele G)17 that is associated with more efficient conversion for PUFAs.16 In contrast, Amerind ancestry Hispanic populations have higher frequencies of the “ancestral” FADS haplogroup (represented by rs174537 allele T) that has a reduced capacity to synthesize PUFAs. Accordingly, we demonstrated that higher global proportions of Amerind ancestry are associated with lower levels of PUFAs in Hispanic populations.17
Genome-wide association studies (GWAS) of n-3 and n-6 PUFAs were performed by the CHARGE consortium in European ancestry (EUR) participants.18-20 The CHARGE GWAS of n-3 PUFAs in 8,866 European Americans identified genetic variants in/near FADS1 and FADS2 associated with higher levels of ALA and lower levels of EPA and DPA, as well as SNPs in ELOVL2 associated with higher EPA and DPA and lower DHA. The CHARGE GWAS of n-6 PUFAs in 8,631 European Americans confirmed that variants in the FADS gene cluster were associated with LA and AA, and it revealed that variants near NRBF2 were associated with LA and those in NTAN1 were associated with LA, GLA, DGLA, and AA (Fig. 1). In the Framingham Heart Offspring Study, variants in/near PCOLCE2, LPCAT3, DHRS4L2, CALN1 FADS1/2, and ELOVL2 were associated with PUFAs in European ancestry participants.21,22 Collectively, these studies played an important role in identifying the genetic associations of n-3 and n-6 PUFAs in European ancestry populations.
To address the paucity of GWAS of PUFAs in non-European ancestry cohorts, we performed a metaanalysis of genome-wide association studies for n-3 and n-6 PUFAs for Hispanic American (HIS) and African American (AFA) participants from three CHARGE consortium cohorts: the Multi-Ethnic Study of Atherosclerosis (MESA), the Cardiovascular Health Study (CHS) and the Framingham Heart Study (FHS) Omni cohort. The major goals of the study were (1) to examine whether the major loci identified in European Americans are shared across race/ancestry groups, and (2) to examine evidence for genetic association unique to HIS and AFA populations. As GWAS approaches are not sufficient to identify the causal variants and determine the number of independent signals, especially in the context of long stretches of linkage disequilibrium (LD) within the FADS locus15,23, we conducted statistical fine-mapping24 to identify the most likely causal variants within each n-3 and n-6 PUFA-associated locus. We performed cross-ancestry replication analysis in CHARGE and MESA, with validation using the multiancestry GWAS of lipids from the Global Lipids Genetics Consortium (GLGC).25 Subsequently, we performed integrative analysis leveraging gene expression data from MESA26,27 and the Genotype-Tissue Expression (GTEx) project28 to identify genes that could contribute to our identified genetic association results. Finally, we examined open chromatin defined by ATAC-seq to determine the impact and physical contact of the identified variants with nearby genes. Our study demonstrates the vital importance of diverse ancestry genetic studies for the study of complex traits, and particularly for metabolites that have been subject to evolutionary pressures and are closely regulated by specific protein-coding genes.
Results
Participant characteristics
The participants in the meta-analysis of GWAS for PUFAs included 1,454 HIS and 2,278 AFA unrelated participants (Table 1; fatty acid levels are expressed as the percentage of total fatty acids throughout the entire manuscript). There were some differences in the distributions of fatty acid levels observed across cohorts, which were likely due to the sources of biospecimens for the assays (plasma phospholipids for MESA and CHS versus erythrocytes for FHS). For example, mean levels of DPA varied from 0.85% (CHS: plasma phospholipids) to 2.54% of total fatty acids (FHS: erythrocytes) in AFA and AA from 11.01% (MESA: plasma phospholipids) to 16.56% (FHS: erythrocytes) in HIS (Table 1). In addition, n-6 PUFAs, especially LA and AA, have relatively higher mean levels than n-3 PUFAs in all cohorts (Table 1).
Table 1.
CHARGE cohorts descriptives
| MESA/Hispanic Americans |
FHS/Hispanic Americans |
MESA/African Americans |
CHS/African Americans |
FHS/African Americans |
|
|---|---|---|---|---|---|
| Participant characteristics | |||||
| No. subjects | 1243 | 211 | 1472 | 603 | 203 |
| Women | 629 (50.6) | 129 (61.1) | 788 (53.5) | 390 (64.7) | 130 (64.0) |
| Age, years | 61 [53, 69] | 53 [44, 60] | 63 [53, 70] | 74 [71, 79] | 58 [50, 67] |
| n-3 Polyunsaturated Fatty Acids | |||||
| ALA (% of total fatty acids) | 0.16 [0.12, 0.20] | 0.21 [0.16, 0.27] | 0.15 [0.12, 0.19] | 0.13 [0.11, 0.17] | 0.18 [0.15, 0.23] |
| EPA | 0.53 [0.37, 0.74] | 0.57 [0.47, 0.78] | 0.68 [0.51, 0.98] | 0.53 [0.39, 0.67] | 0.68 [0.48, 1.01] |
| DPA | 0.86 [0.73, 1.00] | 2.49 [2.13, 2.79] | 0.93 [0.80, 1.07] | 0.85 [0.75, 0.97] | 2.54 [2.25, 2.89] |
| DHA | 2.96 [2.29, 3.77] | 4.21 [3.45, 5.13] | 4.05 [3.25, 4.95] | 3.46 [2.87, 4.17] | 5.23 [4.21, 6.47] |
| n-6 Polyunsaturated Fatty Acids | |||||
| LA | 20.92 [18.87, 23.07] | 14.32 [12.24, 16.76] | 18.88 [17.12, 20.84] | 17.84 [16.46, 19.40] | 12.53 [10.88, 15.16] |
| GLA | 0.11 [0.08, 0.14] | 0.15 [0.10, 0.18] | 0.10 [0.08, 0.13] | 0.07 [0.05, 0.09] | 0.10 [0.07, 0.15] |
| DGLA | 3.57 [3.04, 4.13] | 1.95 [1.63, 2.35] | 2.89 [2.47, 3.33] | 2.76 [2.39, 3.24] | 1.51 [1.32, 1.78] |
| AA | 11.01 [9.37, 12.84] | 16.56 [15.17, 17.74] | 13.21 [11.65, 14.82] | 12.64 [11.57, 13.86] | 17.17 [15.95, 18.48] |
Table 1 shows the participant characteristics of the Hispanic Americans and African Americans from each cohort (MESA, CHS and FHS). Data are presented as n (%) for binary measures or median [IQR] for continuous measures. Summary statistics are reported for the subset of individuals with data available for at least one of the fatty acid traits examined in genetic analyses. Fatty acids were measured in plasma phospholipids in MESA and CHS and in erythrocytes in FHS.
Regardless of whether the fatty acids were measured in plasma phospholipids or erythrocytes, AFA populations had higher levels of AA and elevated ratios of AA to DGLA and AA to LA relative to Hispanic populations. This result would be expected given the frequency differences in the derived (efficient) to ancestral (inefficient) FADS haplogroups between these two populations. As expected, due to the lower levels of dietary ALA relative to LA entering the biosynthetic pathway, levels of n-3 LC-PUFAs including EPA, DPA and DHA were significantly lower than the n-6 LC-PUFA, AA. Additionally, African Americans had higher levels of n-3 LC-PUFAs than Hispanic Americans, again likely due to differences in the ratio of the derived to ancestral FADS haplogroups. These differences are similar to those observed examining the same PUFAs and LC-PUFAs and ratios when comparing African Americans and European Americans.15,29
Confirmation Of Top Variants Identified In Prior Charge Eur Gwas Of Pufas
We began by examining associations of seven known PUFA-associated signals from CHARGE EUR in our current study of CHARGE HIS and AFA. Multiple variants identified by previous CHARGE EUR GWAS metaanalyses were also identified in CHARGE HIS (FADS1/2 region: rs174547 and rs174538, PDXDC1 variant: rs16966952 and GCKR variant: rs780094) and AFA (FADS1/2 region: rs174547, PDXDC1 variant: rs16966952, GCKR variant: rs780094 and ELOVL2 variant: rs3734398) after adjusting for multiple testing for the number of variants examined across the eight PUFAs (P < 0.05/8 = 0.006) (Table S1). The directions of effect observed in HIS and AFA for these variants were consistent with those reported for European ancestry populations in prior CHARGE GWAS meta-analyses of n-3 and n-6 PUFAs (Table S1).
Gwas And Fine-mapping Identify Novel Pufa-associated Genetic Signals In Charge His And Afa
Based on a genome-wide significance threshold of P < 5 x 10−8, our complete GWAS of n-3 and n-6 PUFAs identified associations on chromosomes 11, 15 and 16 in CHARGE HIS (Table 2) and chromosomes 6, 7, 10 and 11 in CHARGE AFA (Table 3). For regions with more than one genome-wide significant variant, we applied statistical fine-mapping to identify the independent putative causal signals (credible sets) for each genome-wide significant locus. We carried out these analyses separately for our CHARGE HIS and CHARGE AFA GWAS meta-analysis results.
Table 2.
Genome-wide significant signals (Credible sets) for PUFAs in CHARGE Hispanic Americans.
| Variants (Chr:Pos:EFF:OTH) |
EAF | Zscore | Pvalue | Cluster | #Of SNP |
Novel/ Known |
Nearest Gene |
|
|---|---|---|---|---|---|---|---|---|
| AA | rs102274 (11:61557826:C:T) | 0.506 | −24.26 | 5.1E-130 | 1 | 7 | Known | TMEM258 |
| rs142068305 (11:67065755:T:G) | 0.196 | −7.06 | 1.63E-12 | 2 | 1 | Novel | ANKRD13D | |
| rs28364240 (11:67120530:G:C) | 0.204 | −7.04 | 1.88E-12 | 3 | 1 | Novel | POLD4 | |
| rs2668898 (11:61725498:G/A) | 0.402 | −5.83 | 5.32E-09 | 4 | 1 | Known | BEST1 | |
| rs180792704 (11:67325239:C:G) | 0.199 | −7.56 | 3.81E-14 | 5 | 1 | Novel | NA | |
| rs198434 (11:61483417:A:G) | 0.710 | −8.97 | 2.80E-19 | 6 | 1 | Known | DAGLA | |
| rs518804 (11:57494487:C:A) | 0.420 | −7.73 | 1.01E-14 | 7 | 1 | Novel | TMX2 | |
| rs3177514 (11:66130358:G:T) | 0.699 | −5.60 | 2.06E-08 | 8 | 1 | Novel | SLC29A2 | |
| ALA | rs174562 (11:61585144:G:A) | 0.503 | 7.84 | 4.30E-15 | 1 | 23 | Known | FADS1 |
| DGLA | rs174538 (11:61560081:A:G) | 0.488 | 14.70 | 6.03E-49 | 1 | 1 | Known | TMEM258 |
| rs174585 (11:61611694:A:G) | 0.274 | 9.82 | 8.72E-23 | 2 | 1 | Known | FADS2 | |
| rs198434 (11:61483417:A:G) | 0.710 | 6.27 | 3.57E-10 | 3 | 1 | Known | DAGLA | |
| rs198461 (11:61524366:C:A) | 0.363 | −5.95 | 2.54E-09 | 4 | 1 | Known | MYRF | |
| rs57112407 (15:78088914:T:C) | 0.255 | −5.86 | 4.46E-09 | NA | NA | Novel | LINGO1 | |
| rs4985155 (16:15129459:G:A) | 0.524 | −7.72 | 1.16E-14 | 1 | 25 | Known | PDXDC1 | |
| DPA | rs1535 (11:61597972:G:A) | 0.520 | −11.31 | 1.07E-29 | 1 | 18 | Known | FADS2 |
| rs198434 (11:61483417:A:G) | 0.710 | −6.26 | 3.67E-10 | 2 | 1 | Known | DAGLA | |
| EPA | rs102274 (11: 61557826:C:T) | 0.506 | −11.56 | 6.18E-31 | 1 | 17 | Known | TMEM258 |
| GLA | rs174576 (11: 61603510:A:C) | 0.546 | −7.73 | 1.07E-14 | 1 | 19 | Known | FADS2 |
| LA | rs174564 (11:61588305:G:A) | 0.520 | 15.11 | 1.23E-51 | 1 | 10 | Known | FADS2 |
| rs10751002 (11:63617634:G:T) | 0.664 | 6.06 | 1.36E-09 | 2 | 1 | Novel | MARK2 | |
| rs2668898 (11:61725498:G:A) | 0.402 | 5.54 | 2.99E-08 | 3 | 1 | Known | BEST1 | |
| rs28364240 (11:67120530:G:C) | 0.204 | 5.90 | 3.44E-09 | 4 | 1 | Novel | POLD4 | |
| rs11039018 (11:46909524:A:C) | 0.67 | −6.10 | 1.01E-09 | 5 | 1 | Novel | LRP4 | |
| rs518804 (11:57494487:C:A) | 0.420 | 6.03 | 1.62E-09 | 6 | 1 | Known | TMX2 |
Table 3.
Genome-wide significant signals (Credible sets) for PUFAs in CHARGE African Americans.
| Variants (Chr:Pos:EFF:OTH) |
EAF | Zscore | Pvalue | Cluster | #Of SNP |
Novel/ Known |
Nearest Gene |
|
|---|---|---|---|---|---|---|---|---|
| AA | rs174585 (11:61611694:A:G) | 0.060 | −9.32 | 1.08E-20 | 1 | 1 | Known | FADS2 |
| rs174607 (11:61627321:C:G) | 0.078 | −6.49 | 8.47E-11 | 2 | 1 | Known | FADS2 | |
| rs174564 (11:61588305:G:A) | 0.133 | −14.85 | 6.43E-50 | 3 | 1 | Known | FADS2 | |
| rs174559 (11:61581656:A:G) | 0.078 | −13.68 | 1.27E-42 | 4 | 1 | Known | FADS1 | |
| rs17161592 (7:9388418:C:G) | 0.085 | −6.31 | 2.75E-10 | 1 | 2 | Novel | NA | |
| DGLA | rs174560 (11:61581764:C:T) | 0.216 | 9.12 | 7.51E-20 | 1 | 1 | Known | FADS1 |
| rs1136001 (16:15131974:T:G) | 0.220 | −6.11 | 9.69E-10 | 2 | 17 | Known | PDXDC1 | |
| DPA | rs717894 (6:22119292:A:G) | 0.250 | −5.48 | 4.11E-08 | 1 | 1 | Novel | CASC15 |
| rs9295741 (6:10997166:T:C) | 0.223 | 5.54 | 2.89E-08 | 2 | 2 | Known | ELOVL2 | |
| DHA | rs114622288 (10:14663844:A:G) | 0.050 | −5.71 | 1.16e-08 | NA | NA | Novel | FAM107B |
| LA | rs28456 (11:61597972:G:A) | 0.163 | 7.88 | 3.14E-15 | 1 | 2 | Known | FADS2 |
Table 2 shows the signals (credible sets) of putative causal variants identified at each chromosome for each PUFAs from SuSiE in the HIS. All variant positions are presented based on Human Genome Build 37. Variants previously documented in the CHARGE GWAS meta-analysis of n-3 and n-6 PUFAs were considered known prior to the current meta-analysis. Additionally, those variants demonstrating linkage disequilibrium (LD) R-squared > 0.2 with one or more previously reported GWAS variants were considered known. The remaining variants that were not in LD with known GWAS variants were considered novel in the current study. There was only one genome-wide significant variant on chromosome 15 for DGLA (rs57112407) in HIS, this signal was not carried forward for fine-mapping.
Table 3 shows the signals (credible sets) of putative causal variants identified at each chromosome for each PUFAs from SuSiE in AFA. All variant positions are presented based on Human Genome Build 37. Variants previously documented in the CHARGE GWAS meta-analysis of n-3 and n-6 PUFAs were considered known at the current meta-analysis. Additionally, those variants demonstrating linkage disequilibrium (LD) R-squared > 0.2 with one or more previously reported GWAS variants were considered known. The remaining variants that were not in LD with known GWAS variants were considered novel in the current study. There was only one genome-wide significant variant on chromosome 10 for DHA (rs114622288) in AFA, this signal was not carried forward for fine-mapping.
We identified multiple independent putative causal signals for the PUFA traits [AA: 8 signals (credible sets); ALA: 1; DGLA: 5, DPA: 2; EPA: 1; GLA: 1; LA: 6] in HIS and [AA: 5; DGLA: 2, DPA: 2, LA: 1] in AFA (Table 2, Table 3, Table S2 and Table S3). We examined the overlap of signals identified from fine-mapping in HIS versus AFA. We observed that the credible sets were generally smaller in AFA (average number of variants in credible set: HIS:3.4; AFR:2.2) possibly driven by the lower average LD in AFA.
Among the independent credible sets identified, most were novel associated signals within a +/− 5 Mb region of the previously reported FADS signal on chromosome 11 (Tables 2-3). Examining all the signals for PUFAs in HIS and AFA, we observed that the lead signal (reflecting the strongest evidence of association) on chromosome 11 represents the FADS signal reported in the previous GWAS.20 For example, rs174547, the FADS1 variant reported in the previous CHARGE EUR GWAS, is one of the variants in the first credible set for AA in HIS.19,20 In addition to the known FADS signals, we also observed multiple novel independent signals at other regions of chromosome 11 for PUFAs [AA: 5 novel signals (credible sets) and LA: 3] in HIS, for example, in/near ANKRD13D, TMX2, POLD4 and SLC29A2 and spanning a long range (57.5Mb ~ 67.1 Mb) on chromosome 11 for AA in HIS (Table 2). Additionally, we observed several novel independent signals on other chromosomes showing associations with the PUFA traits in AFA [AA: 1 novel signal on chromosome 7 and DPA: 1 on chromosome 6] (Table 3).
Additional independent PUFA-associated signals on chromosome 11 demonstrate chromatin contacts with FADS and other genes
While prior studies have represented the FADS signal as primarily just one signal,19,20 our study demonstrates numerous independent signals within the FADS region (+/−1 Mb of the top variant, rs107724) (Fig. 2A). We examined this region to identify the subset of variants that may affect cis-regulatory elements in physical contact with nearby genes. Four variants within the credible sets in this region were located in regions of open chromatin defined by ATAC-seq and were in contact with gene promoters defined by Promoter Capture C in multiple metabolic-relevant cell types (human mesenchymal stem cells [hMSC], adipocytes derived from in vitro from the hMSC [hMSC_Adipocytes], induced pluripotent stem cell derived Hepatocytes [iPSC_Hepatocytes], embryonic stem cell derived Hypothalamic Neurons [hESC_HypothalamicNeurons], Enteroids, and HepG2s). Almost all of the interactions we detected were bait-to-bait interactions, meaning that they reflected physical contact between promoters of two different genes (Table S4). For example, the region surrounding rs2668898 near BEST1 showed evidence of physical contact with the TMEM258, FADS1 and FADS2 region in multiple cell types and TMEM258 region also showed evidence of physical contact with the FADS1 and FADS2 region (Fig. 3A and Table S4). Besides the FADS region, we further found evidence of physical contact between POLD4 and ANKRD13D(Fig. 3B and Table S4), corresponding to the regions surrounding two signals identified in fine-mapping of AA in HIS (Fig. 2A).
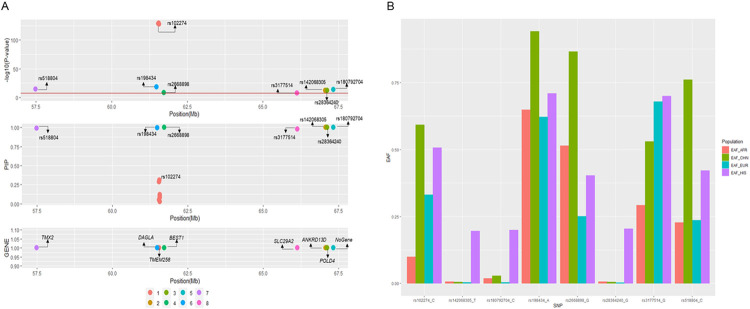
Figure 2. Summary of signals (credible sets) identified in association with AA on chromosome 11 in Hispanic Americans.
Figure 2 shows the information (Panel A: GWAS_Pvalue, Posterior Inclusion Probability, Nearest genes and Panel B: Allele effect frequencies) of the putative causal variants of each signal (credible set) showing the association with AA on chromosome 11 in Hispanic Americans. In panel A, the upper panel shows the P-value of the putative causal variants of each signal (credible set) on chromosome 11 from GWAS; middle panel shows the Posterior Inclusion Probability (PIP) of the putative causal variants from statistical fine-mapping using SuSIE; bottom panel shows the Gene near/in the putative causal variants of each signal. Panel B shows the effect allele frequencies across four race/ethnic groups (African American, European, Hispanics and Chinese) of the most significant putative causal variant from each signal (credible set).
Figure 3. Chromatin contact analysis of selected genome-wide significant variants identified on Chromosome 11.
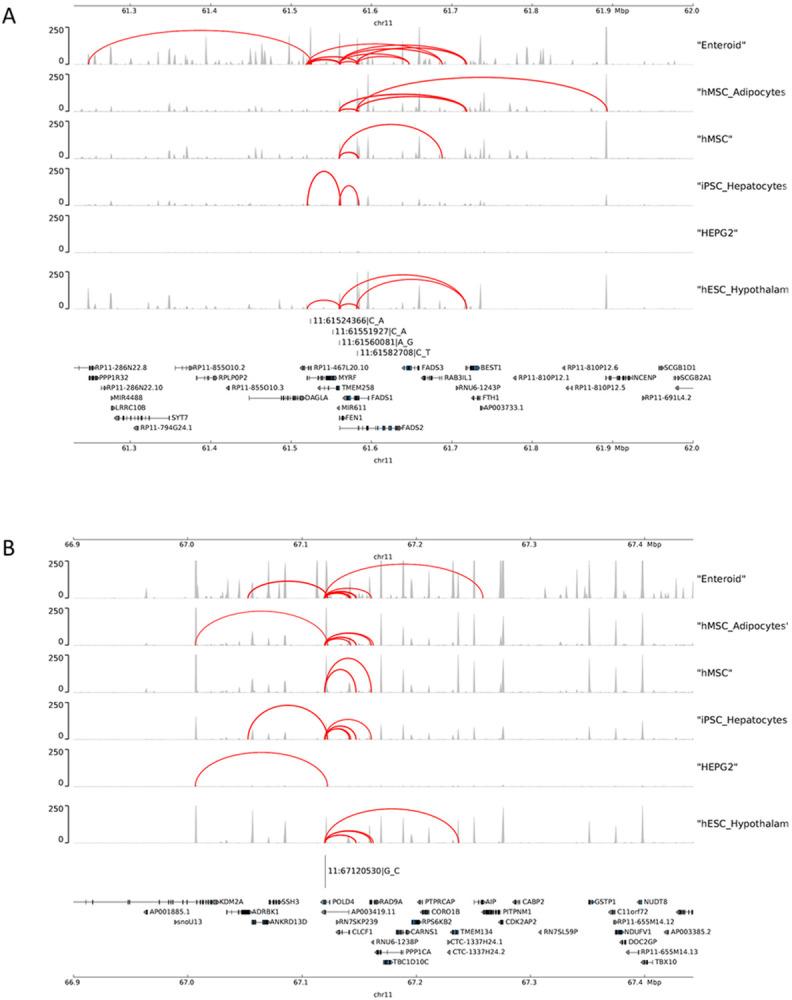
Figure 3 shows the chromatin contact between the causal variants within the signals (Panel A: FADS region and Panel B: POLD4region) located in open chromatin defined by ATAC-seq with gene promoters defined by Promoter Capture C in multiple metabolic-relevant cell types, including: human mesenchymal stem cells (hMSC), which were also differentiated in vitro to adipocytes (hMSC_Adipocytes), induced pluripotent stem cell derived Hepatocytes (iPSC_Hepatocytes), embryonic stem cell derived Hypothalamic Neurons (hESC_HypothalamicNeurons), Enteroids, and HepG2s.
Three novel signals on chromosome 11 identified in HIS show evidence of cross-ancestry replication or validation
We examined evidence of cross-ancestry replication for signals identified in our present GWAS of CHARGE HIS and AFA by examining evidence of genetic association in European Americans (CHARGE EUR and MESA EUR), African Americans (CHARGE AFA), Hispanic Americans (CHARGE HIS) and Chinese Americans (MESA CHN). Replication analysis was performed with multiple testing correction (HIS: P < 0.05/19 signals = 0.0026 and AFA: P < 0.05/11 signals = 0.004).
As noted previously, the first credible set identified in our present GWAS of HIS and AFA for each trait (reflecting the strongest evidence of association) generally coincided with the region of chromosome 11 reported in prior CHARGE GWAS efforts. These signals showed evidence of genetic association in European Americans, as well as across race/ancestry groups. For example, rs102274 for AA was replicated in the MESA EUR, CHARGE AFA and MESA CHN groups (MESA EUR: P = 1.04 x 10−151, CHARGE AFA: P = 2.36 x 10−47, MESA CHN: P = 8.75 x 10−92) (Table S5).
Additionally, one novel signal was also replicated across race/ancestry groups (Table 4). LRP4 variant rs11039018 in credible set 5 for LA was replicated in the CHARGE AFA (CHARGE AFA: P = 1.90 x 10−13).
Table 4.
Novel PUFA-associated signals (credible sets) from analysis of HIS with external replication or validation evidence.
| Traits | Variants (chr:pos:effect:other) |
Replication | Validation | Direction | Nearest Gene |
|---|---|---|---|---|---|
| AA | rs518804 (11:57494487:C:A) | NS | HDL: P = 1.96E-06 logTG: P = 0.001 |
HDL: (−) LDL: (−) logTG: (+) |
TMX2 |
| LA | rs10751002 (11:63617634:G:T) | NS | LDL: P = 3.31E-12 TC: P = 5.74E-09 |
LDL: (+) TC: (+) |
MARK2 |
| rs11039018 (11:46909524:A:C) | AFA: P = 1.90E-13 | HDL: P = 2.85E-74 logTG: P = 4.5E-43 |
AFA: (+) HDL: (+) logTG: (−) |
LRP4 |
Table 4 shows the novel putative causal variants in each signal (credible set) identified from Fine-mapping for PUFAs with replication and validation evidence in HIS. Variants that weren’t previously documented in the CHARGE GWAS meta-analysis of n-3 and n-6 PUFAs and weren’t in LD with known GWAS variants were considered novel in the current study.
Some of the novel signals without cross-ancestry replication demonstrated large differences in allele frequencies across groups. For example, the effect allele frequency of rs28364240, a POLD4 missense variant in credible set 3 for AA in Hispanics, is 0.204 in our CHARGE HIS group, but close to zero in the other race/ancestry groups examined (EUR: 0.003, AFR: 0.007, CHN: 0.005) (Fig. 2B and Table S5) and the effect allele frequency of rs142068305, a ANKRD13D intron variant, is 0.196 in our CHARGE HIS group while 0.007, 0.004 and 0.005 in AFR, EUR and CHN, respectively. These results suggest evidence of genetic association signals unique to HIS or other groups carrying Amerindian ancestry or admixture.
As some variants could not be interrogated using independent GWAS of PUFA traits, given those studies’ focus on specific race/ancestry groups which may not include our variants of interest and/or limited sample sizes, we performed validation analyses using the results of multi-ancestry GWAS of lipid levels from the GLGC including ~ 1.65 million individuals from five genetic ancestry groups (admixed African or African, East Asian, European, Hispanic and South Asian). We focused on the most significant putative causal variants from each credible set and applied multiple testing correction for the number of validated variants (HIS: P < 0.05/19 = 0.0026 and AFA: P < 0.05/11 = 0.004). Interestingly, we observed that two novel signals without cross-ancestry replication did demonstrate association with one or more lipid levels. For example, the AA associated TMX2 intron variant rs518804 was validated based on its association with HDL and Triglycerides (HDL: 1.96 x 10−06 and Triglycerides: 0.001), while the LA associated MARK2 intron variant rs10751002 was validated based on its association with LDL and Total Cholesterol (LDL: 3.31 x 10−12 and Total Cholesterol: 5.74 x 10−09) (Table 4, Table S7 and Table S8).
Integrative Analyses Identify Putative Causal Genes For The Pufa Loci
Using colocalization with eQTL resources, we identified candidate genes underlying the genetic association signals for the PUFA traits. In HIS, we found colocalization with expression of the genes MED19, TMEM258, PACS1, RAD9A, C11orf24, CTTN on chromosome 11 and PDXDC1 on chromosome 16 based on MESA multi-ancestry eQTL resources26 (Table 5 and Table S9). In further analysis using eQTL resources from GTEx whole blood, we confirmed colocalization with TMEM258 and MED19 identified using the MESA multi-ancestry eQTLs, and also identified colocalization with FADS1, RPS4XP13, AP001462.2, PGA5, PGA5, TPCN2, MEN1 on chromosome 11 and RP11-156C22.5 on chromosome 16. (Table 5 and Table S10).
Table 5.
Integrative analysis (Colocalization and PrediXcan) in the Hispanic Americans using multi-ancestry resources from MESA and GTEx.
| Colocalization Analysis | PrediXcan | |||
|---|---|---|---|---|
| MESA multi- ethnics eQTLs |
GTEx eQTLs |
MESA | GTEx | |
| AA | Chromosome 11 | |||
| MED19, TMEM258, PACS1, RAD9A | RPS4XP13, AP001462.6 | TMEM258, TMEM109, ZBTB3, TTC9C, FERMT3, MED19, POLD4, CLCF1, INCENP, MADD, SSH3, C11orf24, PRPF19, TBC1D10C, BANF1, CCDC86, NXF1, MS4A6E, CCS, COX8A, CCDC88B, ACP2, MAP4K2 | TMEM258, TMEM223, NXF1, INCENP, MUS81, C11orf84, MED19, MEN1, BBS1, NEAT1, DPP3, SSH3, ACP2, ASRGL1, RNASEH2C | |
| ALA | Chromosome 11 | |||
| TMEM258, MED19 | MED19, PGA5, TMEM258 | TMEM258, TMEM109 | TMEM258 | |
| DGLA | Chromosome 11 | |||
| TMEM258 | TMEM258, ZBTB3 | TMEM258, FADS1, FADS2 | ||
| Chromosome 16 | ||||
| PDXDC1 | RP11-426C22.5 | PDXDC1 | NPIPA2 | |
| DPA | Chromosome 11 | |||
| TMEM258, C11orf24, RAD9A | PGA5 | TMEM258, TMEM109 | TMEM258, SSH3, TMEM223 | |
| EPA | Chromosome 11 | |||
| TMEM258 | TPCN2 | TMEM258, FERMT3, TMEM109 | TMEM258, SSH3, TMEM223 | |
| GLA | Chromosome 11 | |||
| TMEM258 | MEN1 | TMEM258 | TMEM258 | |
| Chromosome 11 | ||||
| LA | MED19, CTTN, C11orf24, RAD9A | MED19, TPCN2, FADS1, RPS4XP13, AP001462.6 | TMEM258, TMEM109, FERMT3, ZBTB3, COX8A, MADD, POLD4, TBC1D10C, INCENP, TTC9C, MED19, CLCF1, SSH3, ACP2 | TMEM258, INCENP, SSH3, C11orf84, TMEM223, GIF, NXF1, MED19, MUS81, ACP2 |
Table 5 shows the results of integrative analysis including Colocalization analysis and PrediXcan in the HIS by using MESA data and GTEx data. For Colocalization analysis, eQTL resources include MESA multiethnic eQTL from purified monocytes and GTEx European ancestry whole blood tissue eQTL. GWAS signals with posterior colocalization probability of hypothesis 4 (PP.H4) > 0.80, or PP.H4 > 0.50 and the ratio of PP.H4 / PP.H3 > 5 were considered colocalized with eQTL. For PrediXcan, reference gene expression prediction models include MESA purified monocytes and GTEx European ancestry whole blood. Multiple testing correction of PrediXcan was applied for all genes (MESA: P < 0.05/4470 = 0.00001 and GTEx: P < 0.05/4350 = 0.00001).
We also performed complementary integrative analysis using PrediXcan, identifying significant associations for predicted expression of TMEM258 with AA, ALA, DGLA, DPA, EPA, GLA and LA (after multiple testing correction for all genes examined: P < 0.05/4470 = 0.00001), based on integration with eQTL from both MESA and GTEx. PrediXcan also identified TMEM109, ZBTB3, TTC9C, POLD4, INCENP and FERMT3 on chromosome 11 and PDXDC1 on chromosome 16 as putative genes associated with PUFAs in HIS (Table 5, Table S11 and Table S12). For AFA, colocalization and PrediXcan analyses did not identify any genes of interest that met our pre-specified thresholds for statistical significance.
Incorporating the prior chromatin contacts identified (Table S4), we found that several of our GWAS regions had physical contact with one or more genes identified by integration with eQTL resources. For example, RAD9A was supported by colocalization with MESA eQTL and also showed chromatin contact with POLD4 in nearly all cell types examined (Fig. 3B). In addition, INCENP was supported by PrediXcan using both MESA and GTEx resources and also showed chromatin contact with TMEM258, FADS1 and FADS2 in nearly all cell types examined (Fig. 3A). We further observed that CLCF1, RAD9A, FADS2, TMEM258, INCENP, FADS1 identified from colocalization or PrediXcan were additionally supported by chromatin contacts analyses (Table S4, Fig. 3A and 3B).
To follow-up on the genes of interest identified by colocalization and PrediXcan analyses, we examined their co-expression with FADS1 using GTEx whole blood gene expression with multiple testing correction for the number of genes under consideration (HIS: P < 0.05/39 = 0.0012). In both unadjusted and age/sex-a djusted regression models, multiple genes showed statistically significant co-expression with FADS1, for example, TMEM258, MED19, POLD4, RAD9A and SSH3 (Table S13), suggesting these genes have shared patterns of expression.
Discussion
To address the relative lack of prior studies examining genetics of PUFA levels in non-European ancestry populations, we carried out a meta-analysis of GWAS of n-3 and n-6 PUFAs in HIS and AFA across three cohorts: MESA, CHS and FHS. Examining genetic variants identified in prior CHARGE GWAS of the same traits in European Americans, we demonstrated evidence of association with n-3 and n-6 PUFAs for the signals in/near FADS1/2 on chromosome 11, PDXDC1 on chromosome 16, and GCKR on chromosome 2 in both HIS and AFA from our current CHARGE GWAS, as well as for ELOVL2 on chromosome 6 in AFA only.
Through genome-wide analysis and subsequent statistical fine-mapping of our ancestry-specific results, we demonstrated evidence of multiple independent novel signals within the FADS1/2 locus in both HIS and AFA, and in/near ELOVL2 in AFA. Among these independent novel signals, we found one of the novel signals for LA identified in HIS demonstrated evidence of replication in AFA based on association with the same PUFA traits in MESA and CHARGE (HIS: rs11039018 intronic to LRP4 [LDL receptor related protein 4]). This finding is supported by animal studies showing that deficiency of Lrp4 in adipocytes increased glucose and insulin tolerance and reduced serum fatty acids.30 Additionally, multiple novel signals without cross-ancestry replication did show evidence of validation based on association with lipid levels in GLGC. For example, rs518804, a TMX2 (thioredoxin related transmembrane protein 2) intron variant associated with AA and LA was validated based on its association with HDL and Triglycerides, while a MARK2 (microtubule affinity regulating kinase 2) intron variant rs10751002 associated with LA was validated based on its association with LDL and Total Cholesterol.
While we identified one signal in HIS with evidence of cross-ancestry replication, we also found a large number of signals in HIS that could not be replicated across race/ancestry groups (European, African American and Chinese), in part to differences in allele frequencies. For example, the chromosome 11 POLD4 (DNA polymerase delta 4, accessory subunit) missense variant rs28364240 and ANKRD13D (ankyrin repeat domain 13D) intron variant rs142068305 identified in association with AA have minor allele frequencies of 0.204 and 0.196 in HIS, compared to frequencies close to zero in other race/ancestry groups.
Examining the distance between the putative causal variants in different credible sets identified in HIS, we observed that the signals on chromosome 11 span a long range (57.5Mb ~ 67.1 Mb). The extended physical distance covered by these independent PUFA-associated variants, combined with their subsequent validation in association with selected lipid traits, suggests there may be long-range chromatin interactions or other forms of physical interaction that may have yielded distinct genetic associations across this region.31 Interestingly, prior studies have reported the FADS signal on chromosome 11 as primarily just one genetic signal.19,20 However, our study provides evidence of two more independent signals (BEST1 and DAGLA) within this FADS region. In order to understand the chromatin interactions of the FADS region on chromosome 11, we used ATAC-seq peaks and chromatin loops to perform the chromatin contact analyses. We identified multiple genes from colocalization or PrediXcan also supported by chromatin contacts, including CLCF1, RAD9A, FADS2, TMEM258, INCENP and FADS1, providing support for the role of our identified genetic signals in regulating these genes. In addition, we observed evidence of chromatin contacts among multiple distinct credible sets identified based on our fine-mapping of genetic signals on chromosome 11. For example, the region surrounding rs2668898 near BEST1 also showed evidence of physical contact with the TMEM258, FADS1 and FADS2 region in multiple cell types and TMEM258 also showed evidence of physical contact with the FADS1 and FADS2 region. This support for physical contact among some of the multiple independent signals within the FADS region opens the possibility of coordinated regulation among these distinct genetic signals. Besides the FADS region, POLD4 also showed evidence of physical contact with the ANKRD13D region in multiple cell types. The cell types examined for chromatin interaction correspond to pancreas, liver, and other cell types that could play a role in synthesis and regulation of fatty acids. While the cell types used to examine chromatin interactions are distinct from those used for our integrative eQTL analyses, the chromatin interaction results do provide support for the plausible role of the genes identified by colocalization and PrediXcan.
Through integrative analyses including colocalization analysis and PrediXcan and overlapping our GWAS of PUFA levels with selected eQTL resources, we identified putative candidate genes that may shed light on the functional mechanisms of our identified genetic association signals. On chromosome 11 containing the FADS genes, we identified overlap with eQTL for multiple other genes including MED19 (Mediator Complex Subunit 19), TMEM258 (Transmembrane Protein 258), PACS1 (Phosphofurin Acidic Cluster Sorting Protein 1), RAD9A (RAD9 Checkpoint Clamp Component A) and CTTN (Cortactin) suggesting additional complexity within this region beyond the FADS genes. For the signals on chromosome 16 identified based on analyses of DGLA in HIS and AFA, in/near NTAN1 and PDXDC1, our integrative PrediXcan analyses identified PDXDC1 (Pyridoxal Dependent Decarboxylase Domain Containing 1) (but not NTAN1) as a putative gene for DGLA. Additionally, having identified association with AA in HIS for the POLD4 missense variant rs28364240, our subsequent identification of POLD4 (DNA Polymerase Delta 4, Accessory Subunit) based on the PrediXcan analyses brings additional support for this gene. To follow-up on the genes of interest identified by colocalization and PrediXcan analyses, we examined their co-expression with FADS1 using GTEx whole blood gene expression. Multiple genes on chromosome 11 identified in our integrative analyses combining the GWAS of PUFAs with whole blood expression from GTEx showed evidence of co-expression with FADS1, for example, TMEM258, POLD4, TMEM109 and ZBTB3. This finding suggests some genomic regions at a considerable distance from FADS1 may play a role in regulating its expression, and ultimately influence circulating PUFA levels.
While our genetic association study of PUFA levels in HIS and AFA provides novel insights, our work has several limitations. First, while we have combined data from multiple CHARGE cohorts, the overall sample size of our study is still relatively small for a GWAS. Second, as we began this GWAS effort some years ago, our work makes use of older imputation panels based on the 1000 Genomes. We expect future work could leverage newer resources including imputation based on the Trans-omics for Precision Medicine (TOPMed) reference panel or newer whole genome sequence data from TOPMed32. Third, the circulating PUFA levels examined in this study are derived from heterogeneous sources (plasma phospholipids in MESA and CHS vs. erythrocytes in FHS), which could have resulted in heterogeneity of genetic associations across the included studies and overall loss of power. Finally, while our integration of GWAS with eQTL proved useful in some cases, our efforts were driven in part by the available resources. We made use of multi-ancestry eQTL resources based on purified monocytes in MESA, as we knew these resources were well-matched with our GWAS cohorts in terms of LD structure, although purified monocytes were likely not the most relevant cell type for our study. We complemented those efforts with whole blood eQTL from GTEx through which we were able to capture colocalization of FADS1 that was not observed in MESA due to the lack of significant cis-eQTL for FADS1. This limitation underscores the need for more diverse ancestry eQTL resources across a wider range of tissues and cell types.
In summary, working with the CHARGE Consortium, we conducted the first consortium-based GWAS of circulating PUFA levels in HIS and AFA cohorts. Our study demonstrated evidence of shared genetic influences on PUFA levels across race/ancestry groups, and demonstrated for the first time the large number of distinct genetic association signals within a neighborhood of the well documented FADS region on chromosome 11.19,20 Our findings provide new insight into the complex genetics of circulating PUFA levels that reflect, in part, their response to evolutionary pressures across the course of human history.33,34 Overall, our study demonstrates the value of investigating complex trait genetics in diverse ancestry populations and highlights the need for continued efforts for expanded genetic association efforts in cohorts with genetic ancestry that reflects that of the general population within the United States and worldwide.
Methods
Study participants
The participants in this study were recruited from three population-based cohorts: the Multi-Ethnic Study of Atherosclerosis (MESA)35, the Cardiovascular Health Study (CHS) and the Framingham Heart Study (FHS). This manuscript focuses on HIS participants from MESA (N = 1,243) and FHS (N = 211) and AFA participants from MESA (N = 1472), CHS (N = 603) and FHS (N = 203).
Ethical Review
All cohort participants gave written informed consent, including consent to participate in genetic studies. All studies received approval from local ethical oversight committees.
Fatty Acid Measurements
Circulating PUFA levels were quantified from plasma phospholipids in MESA and CHS, and from erythrocytes in FHS. Details on measurement of the PUFAs are provided in the Supplementary Methods.
Genotyping And Imputation
Each of the participating cohorts had genome-wide genotype data based on a GWAS array, followed by imputation based on the 1000 Genomes Phase 1 v3 (for CHS) or Phase 3 (for MESA and FHS) Cosmopolitan reference panel.36 Details on genotyping, quality control and imputation are provided in the Supplementary Methods.
Data transformation and detection of outliers in measured PUFA Levels.
After examining the raw phenotype distributions for each of the phenotypes of interest, we applied variable transform for traits exhibiting deviation from normality. Log-transformation was applied for ALA, EPA and GLA. In addition, outliers for all of the PUFA levels were identified by the limits of median +/− 3.5 * MAD’, where MAD’ is computed with a scale factor constant of 1.4826 [default for the mad() function in R]. The value of MAD’ = 1.4826 * MAD0 where MAD0 is the raw value of median absolute deviation (MAD). For all the PUFAs, outliers were winsorized to the value of (median +/− 3.5 * MAD’).
Meta-analysis Of Genome-wide Association Study
Genome-wide association analysis was carried out separately in each cohort and stratified by race/ancestry with covariate adjustment for age, sex, study site and principal components of ancestry. Cohort-specific GWAS results were filtered using EasyQC based on minor allele count (MAC) > 6 and imputation R-squared > 0.3. Cohort-specific results were combined using weighted sum of z-score metaanalysis in METAL37 and filtered using Effective Heterozygosity Filter (effHET) > 60. A threshold of P < 5 x 10−8 was applied to identify genome-wide significant loci.
Identification Of Novel Versus Previously Reported Variants
Variants previously documented in the CHARGE GWAS meta-analysis of n-3 (n = 8,866)19 and n-6 (n = 8,631)20 PUFAs in European ancestry cohorts were considered known for the current meta-analysis. Additionally, those variants demonstrating linkage disequilibrium (LD) R-squared > 0.2 with one or more previously reported GWAS variants were considered known. The remaining variants were considered novel in the current study.
Statistical Fine-mapping Using Susie
For each chromosome with more than one genome-wide significant variant (at P < 5 x 10−8), we carried out statistical fine-mapping to identify the putative causal variants and estimate the number of independent signals. We used Sum of Single Effect model (SuSiE)24 to identify the credible set of putative causal variants, providing as input all variants with P < 5 x 10−8 from the meta-analysis results. For fine-mapping of signals identified in our meta-analysis of HIS and AFA, we used imputed genotype dosage for the same set of variants in MESA HIS and AFA, respectively. To select the parameter L (prior number of independent signals) for fine-mapping in SuSiE, DAP-G (Deterministic Approximation of Posteriors)38 was conducted to provide a starting value for L based on the number of credible sets that the threshold of posterior inclusion probability was greater than 0.95.
Follow-up Replication And Validation Analyses
Following statistical fine-mapping, cross-ancestry replication analyses were conducted for the most highly supported putative causal variant from each credible set using data on n-3 and n-6 PUFAs from other race/ancestry groups. The resources for replication analyses included the following:
European Americans (EUR): 2344 self-reported European American participants from MESA (using 1000 Genomes Phase 3 imputation, for comparison with the current study), as well as summary statistics from the previously published CHARGE GWAS meta-analysis of n-3 (n = 8,866)19 and n-6 (n = 8,631)20 PUFAs based on imputation from the HapMap Phase I and II,
African Americans (AFA): summary statistics from the present GWAS of PUFAs in AFA to examine cross-ancestry replication of variants identified in the present GWAS of HIS,
Hispanic Americans (HIS): summary statistics from the present GWAS of PUFAs in HIS to examine cross-ancestry replication of variants identified in the present GWAS of AFA, and
Chinese Americans (CHN): 649 self-reported Chinese American participants from MESA (using 1000 Genomes Phase 3 imputation, for comparison with the current study).
Given the limited number of cohorts available for ethnic-specific and cross-ethnic replication of PUFA traits, additional validation analyses were conducted for the same set of variants using multi-ancestry genetic association with lipid traits (HDL, LDL, total cholesterol and triglycerides) from the Global Lipids Genetics Consortium (GLGC).25 Multiple testing correction was applied to account for the number of variants examined in cross-ethnic replication (HIS: P < 0.05/19 = 0.0026 and AFA: P < 0.05/11 = 0.004).
Bayesian Colocalization Analysis
Bayesian colocalization analysis has proven an effective approach for identification of downstream genes underlying GWAS loci.35 We used the R/coloc package to conduct Bayesian colocalization analysis39 to identify the putative gene(s) corresponding to each credible set of variants using MESA multi-ancestry eQTL data from purified monocytes26 and GTEx multi-ancestry whole blood tissue eQTL data.40 Bayesian colocalization analysis tested the following hypotheses: H0. neither GWAS of PUFAs nor eQTL has a genetic association in the region (within 1 Mb of the transcription start site); H1. only GWAS of PUFAs has a genetic association in the region; H2. only eQTL has a genetic association in the region; H3. both GWAS of PUFAs and eQTL are associated, but with different causal variants; H4. both GWAS of PUFAs and eQTL are associated and share a single causal variant. Colocalization for variants in credible sets was defined by (1) a posterior colocalization probability of hypothesis 4 (PP.H4) > 0.80, or (2) a PP.H4 > 0.50 and the ratio of PP.H4 / PP.H3 > 5.
PrediXcan model.
PrediXcan, a gene-based association method focused on identifying trait-associated genes by quantifying the effect of gene expression on the phenotype on interest.41 In this study, we applied summary-statistics based PrediXcan (S-PrediXcan)42 using reference gene expression prediction models from MESA purified monocytes26 and GTEx multi-ancestry whole blood.43 S-PrediXcan associations were considered genomewide significant if they passed the multiple testing correction for all genes (MESA: P < 0.05/4470 = 0.00001 and GTEx: P < 0.05/4350 = 0.00001).
Chromatin Contact Analysis
To identify variants located in open chromatin regions in contact gene promoters, we used GenomicRanges (v. 1.46.1 ; R version 4.1.1) to intersect the genomic coordinates (hg19) of the variants contained in the credible sets with the open chromatin peaks (called using the ENCODE pipeline) in significantly enriched contact with gene promoter determined by Promoter Capture C (Chicago Score > 5). We queried chromatin accessibility and promoter contacts in human mesenchymal stem cells (hMSC) and Adipocytes differentiated in vitro from these (hMSC_Adipocytes), embryonic stem cell derived hypothalamic neurons (hESC Hypothalamic Neurons), induced pluripotent-dervived Heptocytes (IPS-Hepatocytes), Enteroids, and the hepatic carcinoma HepG2 cell line.44-49 Details on Promoter Capture C and ATAC-seq library generation and analyses have been previously described.44
Gene Co-expression Analysis.
We used the GTEx whole blood gene expression version 8 TPM dataset to examine co-expression with FADS1 for genes identified by integrative analyses, including colocalization and PrediXcan. Two models for gene co-expression analysis were used for the trait of interest,
an unadjusted model FADS1 ~ gene expression; and
a covariate adjusted model FADS1 ~ age + gender + gene expression.
Gene co-expression associations were considered statistically significant if they passed the multiple testing correction for all genes examined from colocalization and PrediXcan (P < 0.05/39 = 0.0012).
Acknowledgments
This work was supported by NCCIH R01 AT008621 (to FHC) and by the Daniel B. Burke Endowed Chair for Diabetes Research (to SFG).
The Multi-Ethnic Study of Atherosclerosis: MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts HHSN268201500003I, N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420, UL1-TR-001881, and DK063491. Funding for SHARe genotyping was provided by NHLBI Contract N02-HL-64278. Genotyping was performed at Affymetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affymetrix Genome-Wide Human SNP Array 6.0.
The Cardiovascular Health Study: This research was supported by contracts HHSN268201200036C, HHSN268200800007C, HHSN268201800001C, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, 75N92021D00006, and grants U01HL080295 and U01HL130114 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.. Additional support for infrastructure was provided by R01HL105756 and additional genotyping among the African-American cohort was supported in part by R01HL085251. DNA handling and genotyping at Cedars-Sinai Medical Center was supported in part by National Center for Research Resources grant UL1RR033176, now at the National Center for Advancing Translational Technologies CTSI grant UL1TR000124; in addition to the National Institute of Diabetes and Digestive and Kidney Diseases grant P30DK063491 to the Southern California Diabetes Endocrinology Research Center.
The Framingham Heart Study: This research was partially supported by the National Human Genome Research Institute (R15HG006915; R01HL152215; NT). The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195, HHSN268201500001I and 75N92019D00031). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Data for this project was obtained via dbGaP (phs000007.v32.p1). Funding to support the Omni cohort recruitment, retention and examination was provided by NHLBI Contract N01-HC-25195, HHSN268201500001I and 75N92019D00031, as well as NHLBI grants R01-HL070100, R01-HL076784, R01-HL49869, and U01-HL-053941. “Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding for Affymetrix genotyping of the FHS Omni cohorts was provided by Intramural NHLBI funds from Andrew D. Johnson and Christopher J. O’Donnell. Funding support for the Framingham Red Blood Cell Fatty Acids dataset was provided by NHLBI grant R01 HL089590.
The authors acknowledge Research Computing at The University of Virginia for providing computational resources and technical support that have contributed to the results reported within this publication.
Contributor Information
Chaojie Yang, University of Virginia.
Jenna Veenstra, Dordt University.
Traci Bartz, University of Washington.
Matthew Pahl, Children's Hospital of Philadelphia.
Brian Hallmark, University of Arizona.
Yii-Der Ida Chen, Lundquist Institute for Biomedical Innovation at Harbor-UCLA Medical Center.
Jason Westra, Dordt University.
Lyn Steffen, University of Mineesota.
Christopher Brown, Perelman School of Medicine, University of Pennsylvania.
David Siscovick, 42. The New York Academy of Medicine, New York, NY.
Michael Tsai, University of Minneosta.
Alexis Wood, Baylor College of Medicine.
Stephen Rich, University of Virginia.
Caren Smith, Tufts University.
Timothy O'Connor, University of Maryland, Baltimore.
Dariush Mozaffarian, Tufts University.
Struan Grant, Children's Hospital of Philadelphia Research Institute.
Floyd Chilton, University of Arizona.
Nathan Tintle, Fatty Acid Research Institute.
Rozenn Lemaitre, Cardiovascular Health Research Unit, University of Washington.
Ani Manichaikul, University of Virginia School of Medicine.
References
- 1.Calder P. C., Yaqoob P., Harvey D. J., Watts A. & Newsholme E. A. Incorporation of fatty acids by concanavalin A-stimulated lymphocytes and the effect on fatty acid composition and membrane fluidity. Biochemical Journal 300, 509–518 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Los D. A. & Murata N. Structure and expression of fatty acid desaturases. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism 1394, 3–15 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Stubbs C. D. & Smith A. D. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochimica et Biophysica Acta (BBA) - Reviews on Biomembranes 779, 89–137 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Mozaffarian D. & Wu J. H. Y. Omega-3 fatty acids and cardiovascular disease: effects on risk factors, molecular pathways, and clinical events. J Am Coll Cardiol 58, 2047–2067 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Aung T. et al. Associations of Omega-3 Fatty Acid Supplement Use With Cardiovascular Disease Risks: Meta-analysis of 10 Trials Involving 77917 Individuals. JAMA Cardiology 3, 225–233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Importance of the Omega-6/Omega-3 Fatty Acid Ratio in Cardiovascular Disease and Other Chronic Diseases - Artemis P. Simopoulos, 2008. https://journals.sagepub.com/doi/abs/10.3181/0711-MR-311. [DOI] [PubMed]
- 7.Krachler B. et al. Fatty acid profile of the erythrocyte membrane preceding development of Type 2 diabetes mellitus. Nutrition, Metabolism and Cardiovascular Diseases 18, 503–510 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Conquer J. A., Tierney M. C., Zecevic J., Bettger W. J. & Fisher R. H. Fatty acid analysis of blood plasma of patients with alzheimer’s disease, other types of dementia, and cognitive impairment. Lipids 35, 1305–1312 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Söderberg M., Edlund C., Kristensson K. & Dallner G. Fatty acid composition of brain phospholipids in aging and in Alzheimer’s disease. Lipids 26, 421 (1991). [DOI] [PubMed] [Google Scholar]
- 10.Warensjö E., Sundström J., Lind L. & Vessby B. Factor analysis of fatty acids in serum lipids as a measure of dietary fat quality in relation to the metabolic syndrome in men. Am J Clin Nutr 84, 442–448 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Pizer E. S. et al. Inhibition of Fatty Acid Synthesis Induces Programmed Cell Death in Human Breast Cancer Cells. Cancer Res 56, 2745–2747 (1996). [PubMed] [Google Scholar]
- 12.Harris W. S. et al. Blood n-3 fatty acid levels and total and cause-specific mortality from 17 prospective studies. Nat Commun 12, 2329 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenna J. T. Efficiency of conversion of alpha-linolenic acid to long chain n-3 fatty acids in man. Curr Opin Clin Nutr Metab Care 5, 127–132 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Plourde M. & Cunnane S. C. Extremely limited synthesis of long chain polyunsaturates in adults: implications for their dietary essentiality and use as supplements. Appl Physiol Nutr Metab 32, 619–634 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Mathias R. A. et al. The impact of FADS genetic variants on ω6 polyunsaturated fatty acid metabolism in African Americans. BMC Genet. 12, 50 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris D. N. et al. Evolution of Hominin Polyunsaturated Fatty Acid Metabolism: From Africa to the New World. Genome Biol Evol 11, 1417–1430 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Impact of Amerind ancestry and FADS genetic variation on omega-3 deficiency and cardiometabolic traits in Hispanic populations ∣ Communications Biology. https://www.nature.com/articles/s42003-021-02431-4?proof=t%29. [DOI] [PMC free article] [PubMed]
- 18.Psaty B. M. et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet 2, 73–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemaitre R. N. et al. Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium. PLOS Genetics 7, e1002193 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan W. et al. Genome-Wide Association Study of Plasma N6 Polyunsaturated Fatty Acids within the CHARGE Consortium. Circ Cardiovasc Genet 7, 321–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tintle N. L. et al. A genome-wide association study of saturated, mono- and polyunsaturated red blood cell fatty acids in the Framingham Heart Offspring Study. Prostaglandins Leukot Essent Fatty Acids 94, 65–72 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalsbeek A. et al. A genome-wide association study of red-blood cell fatty acids and ratios incorporating dietary covariates: Framingham Heart Study Offspring Cohort. PLoS One 13, e0194882 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckley M. T. et al. Selection in Europeans on Fatty Acid Desaturases Associated with Dietary Changes. Mol Biol Evol 34, 1307–1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G., Sarkar A., Carbonetto P. & Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 82, 1273–1300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graham S. E. et al. The power of genetic diversity in genome-wide association studies of lipids. Nature 600, 675–679 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mogil L. S. et al. Genetic architecture of gene expression traits across diverse populations. PLoS Genet 14, e1007586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y. et al. Methylomics of gene expression in human monocytes. Hum Mol Genet 22, 5065–5074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sergeant S. et al. Differences in arachidonic acid levels and fatty acid desaturase (FADS) gene variants in African Americans and European Americans with diabetes or the metabolic syndrome. Br J Nutr 107, 547–555(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S. P. et al. Lrp4 expression by adipocytes and osteoblasts differentially impacts sclerostin’s endocrine effects on body composition and glucose metabolism. J Biol Chem 294, 6899–6911 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fadason T., Schierding W., Lumley T. & O’Sullivan J. M. Chromatin interactions and expression quantitative trait loci reveal genetic drivers of multimorbidities. Nat Commun 9, 5198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taliun D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathias R. A. et al. Adaptive evolution of the FADS gene cluster within Africa. PLoS ONE 7, e44926 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ameur A. et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet 90, 809–820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bild D. E. et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am. J. Epidemiol. 156, 871–881 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer C. J., Li Y. & Abecasis G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen X., Lee Y., Luca F. & Pique-Regi R. Efficient Integrative Multi-SNP Association Analysis via Deterministic Approximation of Posteriors. Am J Hum Genet 98, 1114–1129 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giambartolomei C. et al. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet 10, e1004383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lonsdale J. et al. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamazon E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbeira A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun 9, 1825 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbeira A. N. et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol 22, 49 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chesi A. et al. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun 10, 1260 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pahl M. C. et al. Cis-regulatory architecture of human ESC-derived hypothalamic neuron differentiation aids in variant-to-gene mapping of relevant complex traits. Nat Commun 12, 6749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lasconi C. et al. Variant-to-Gene-Mapping Analyses Reveal a Role for the Hypothalamus in Genetic Susceptibility to Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol 11, 667–682 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond R. K. et al. Biological constraints on GWAS SNPs at suggestive significance thresholds reveal additional BMI loci. Elife 10, e62206 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Çalıçkan M. et al. Genetic and Epigenetic Fine Mapping of Complex Trait Associated Loci in the Human Liver. Am J Hum Genet 105, 89–107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramdas S. et al. A multi-layer functional genomic analysis to understand noncoding genetic variation in lipids. http://biorxiv.org/lookup/doi/10.1101/2021.12.07.470215 (2021) doi: 10.1101/2021.12.07.470215. [DOI] [PMC free article] [PubMed] [Google Scholar]