The Rh blood group system is the most complex blood group system known.1 Currently, 48 antigens are distinguished. Even this number does not fully reflect the serologic diversity of the Rh blood group, because this list of antigens disregards the complexity of the D antigen revealed by monoclonal antibodies binding to different epitopes2,3 and by the anti-D formed in carriers of partial D phenotypes.4 This review presents an overview of the molecular structures shaping the serologic complexity of the Rh blood group system. We outline the general principles underlying the relationship of Rh molecular structure and phenotype.
Rh proteins
The antigens of the Rh blood group system are located on two proteins.5,6 RhD carries the D (Rh1) antigen, and RhCE carries the C, c, E, and e (Rh2 to Rh5) antigens. Both proteins are composed of 417 amino acids.5,7 Current structural models predict 6 extracellular loops and 12 transmembranous and 7 intracellular protein segments.8,9 Both C- and N-terminal protein ends are intracellular (Fig.1).Depending on the RHCE allele considered, RhD and RhCE differ in 34 to 37 amino acids. These differences are dispersed throughout the amino acid sequence of the protein. Only a limited number of these differences are located exofacially; such exofacial differences are restricted to loop 3 encoded by exon 4, loop 4 encoded by exon 5, and loop 6 encoded by exon 7. In loop 2 encoded by exon 2, the c allele but not the C allele of RHCE differs from RhD (Fig. 1).
Fig. 1.
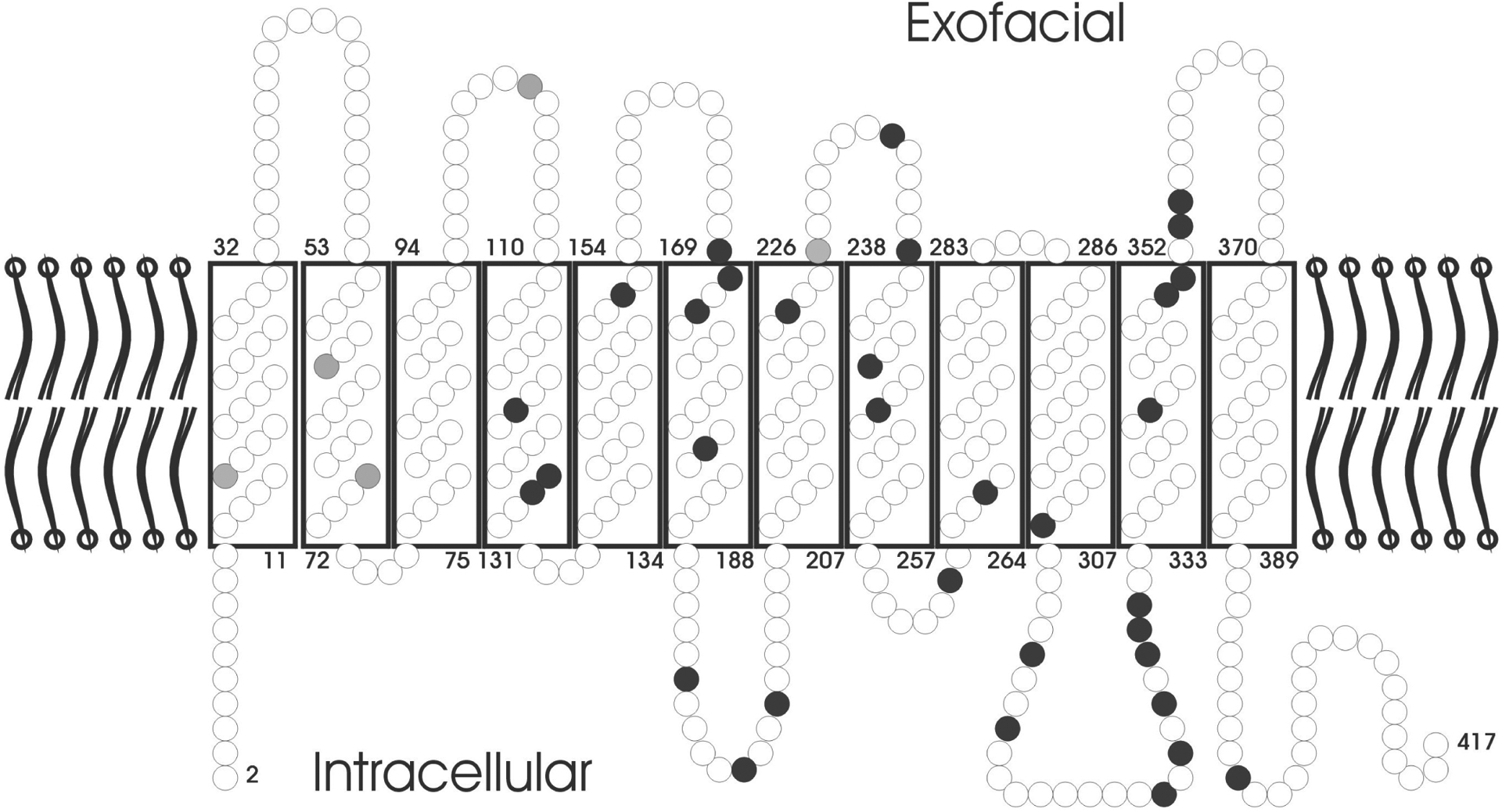
Rh topology in the RBC membrane. The protein is assumed to possess 12 transmembranous segments, 6 extracellular loops, and 5 intracellular loops. Both the C- and N-terminal ends of the protein are intracellular. Each amino acid is depicted by a circle; black circles indicate positions that differ between RhD and RhCE in all frequent alleles, grey circles indicate positions that differ between RhD and RhCE only in some alleles. Most differences are located in transmembranous or intracellular segments; among the extracellular loops, only loops 3, 4, and 6 differ between RhD and RhCE. The latter fact is most important for D antigen expression and is discussed in detail in the text.
In the RBC membrane, the Rh proteins form a complex with Rh-associated glycoprotein (RhAG), previously known as RH50.10 This “Rh complex” is tightly linked to the cytoskeleton.11 Several additional proteins, such as CD47, LW, and the Duffy glycoprotein, are associated with the Rh complex but not necessary for Rh expression. The membrane expression of Rh depends on functional RhAG: mutations in RhAG could be shown to underly the “regulator form” of the Rhnull phenotype characterized by lack of all Rh antigens.12
The RhAG and Rh proteins share homologies with ammonia transport proteins and have been shown to transport ammonia.13–15 Currently, it is unknown whether this represents their sole function; indirect data16 feed speculation on transport functions for other gases more relevant to the RBCs, such as CO2 or O2. Furthermore, the distorted RBC shape in the Rhnull phenotype indicates the importance of the correct interaction of the Rh complex with the cytoskeleton.11
RH gene locus
The two RH genes, RHD and RHCE, are each composed of 10 exons17 and are spread along about 60,000 bp genomic sequence each. The genes have opposite orientation,18,19 face each other by their 3′ ends, and are separated by only about 30,000 bp.18,19 A third gene, SMP1, is interspersed between RHD and RHCE.18 There is no indication that SMP1 is functionally related to RH or expressed on the RBC surface; rather SMP1 is considerably more conserved throughout evolution than RH20 and mainly expressed in the cytoplasm.21 Based on a comparison of the RH loci of man and mice, RHD is the duplicated gene,20 whereas RHCE with its close proximity to SMP1 represents the ancestral position (Fig. 2).
Fig. 2.
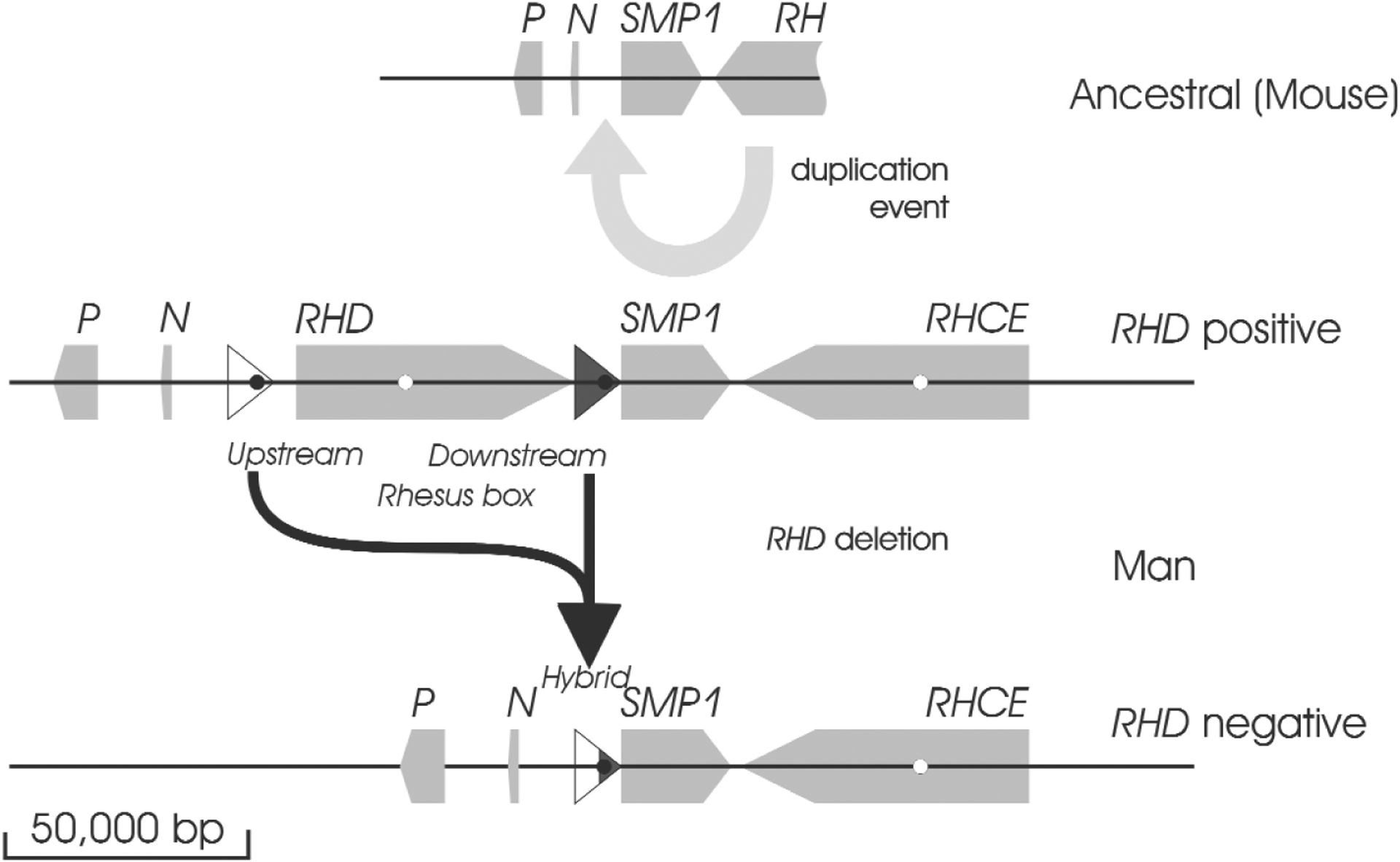
RH duplication and deletion. In the ancestral state (deduced from the mouse RH locus), a single RH gene is in close proximity to the SMP1 gene. Two other genes, the P29-associated protein (P) and NPD014 (N) are upstream of SMP1. In the duplication event, an inversed RH gene is introduced between NPD014 and SMP1. At the insertion point, a 9000-bp DNA segment is duplicated, resulting in the formation of upstream and downstream Rhesus boxes, which flank the RHD gene. The RHD deletion occurred by a recombination of the upstream and downstream Rhesus boxes and led to a RH locus, which closely resembles the ancestral state before the RH duplication occurred.
The RHD gene is flanked by two DNA segments of 9000 bp, called Rhesus boxes.18 The D− phenotype in Whites is usually caused by the homozygous presence of a haplotype in which all the RHD gene is deleted.22 This deletion occurred in the Rhesus boxes,18 probably by an unequal crossing over. Hence, the RH locus of the RHD negative haplotype is almost identical to the ancestral RH locus before the duplication event.
The characterization of the deletion site was instrumental for the specific detection of the RHD deletion.18 Since then, the RHD deletion may be detected even in heterozygous form, i.e., if it occurs in trans to the normal RHD allele. Thus, it became possible to distinguish D+ individuals with two RHD genes from D+ individuals with one RHD gene and one RHD deletion. For instance, it is important to predict the probability of a D+ pregnancy if the mother is D−.23 Such determination cannot be achieved by serology. However, even current molecular methods for the detection of the RHD deletion are not yet reliable in people of African descent.24,25
Mechanisms contributing to the molecular variability of the RH locus
Several different mechanisms contributed to the large number of RH alleles. Many of these mechanisms are shared by most other genes and contributed to the complexity of other blood groups, like KEL and LU. A single nucleotide substitution may cause a change of the encoded amino acid (missense mutation), leading to a single amino acid substitution; introduce a premature stop codon (nonsense mutation), leading to a truncated protein; or destroy the splice consensus sequence (splice site mutation), preventing the correct splicing of the allele. Insertions or deletions of one or a few nucleotides usually lead to a frameshift resulting in completely aberrant amino acid sequences. Finally, recombinations between different alleles lead to alleles sharing peculiarities of both parent alleles.
Two mechanisms that are favored by the structure of the RH locus are rare in other blood group systems. The two highly similar Rhesus boxes flanking the RHD gene18 allowed the deletion of RHD by an unequal crossing over (Fig. 2) in the common RHD negative haplotype. Obviously, a similar mechanism is not possible for RHCE, and there is no common RHCE negative haplotype.
The high similarity of both genes and their opposite orientation favored gene conversions in cis (Fig. 3), in which internal parts of one gene are replaced by the corresponding parts of the other gene.26 The results were RHD-CE-D or RHCE-D-CE hybrid alleles. Some haplotypes (e.g., DCw–27) are composed of two single hybrid alleles of type RHD-CE and RHCE-D.28,29 Such haplotypes are best explained by gene conversions involving more than one gene (Fig. 3).
Fig. 3.
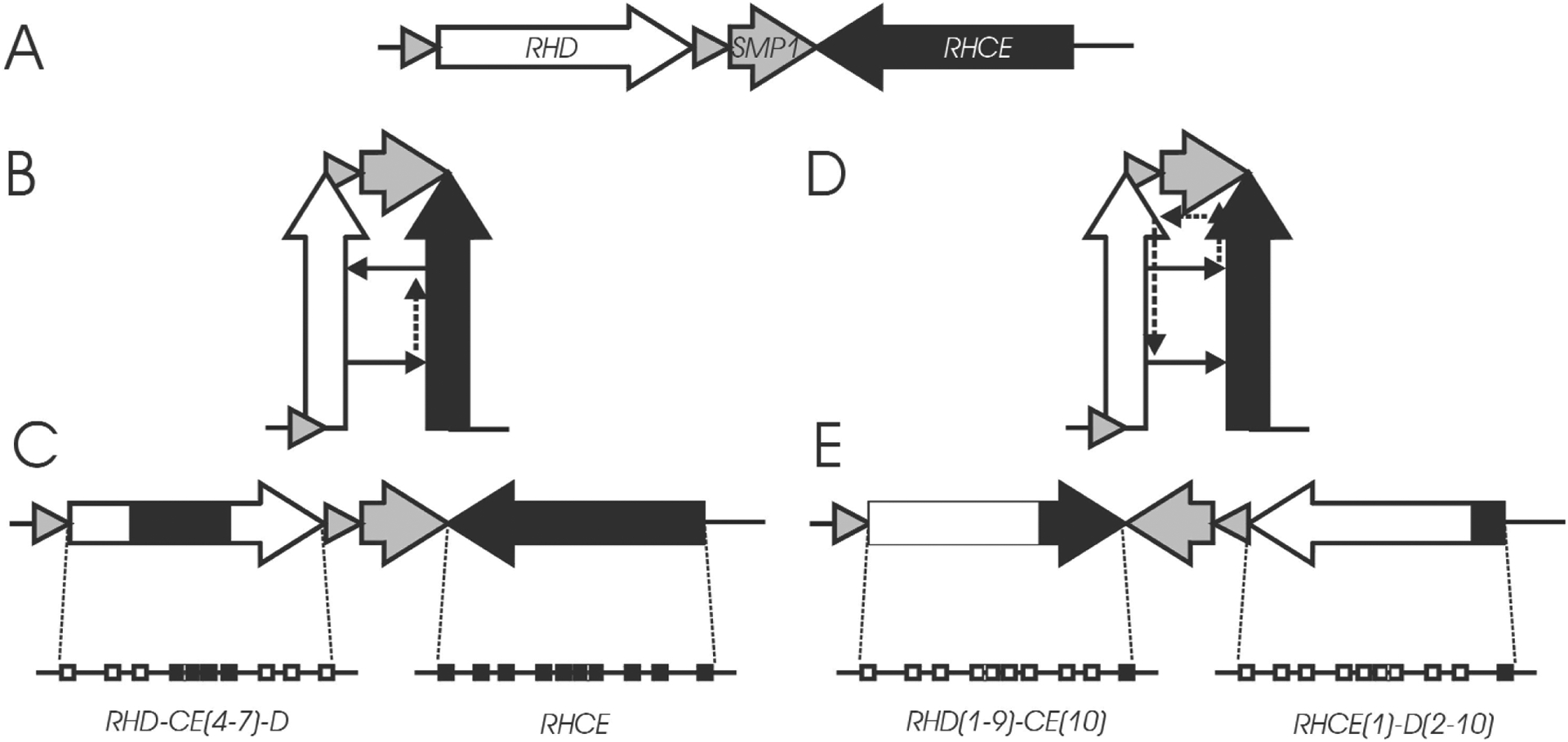
Gene conversions in cis. Panel A:The two RH genes have opposite orientation. Panel B:A gene conversion in cis might be favored by a hairpin-like structure. Panel C:As a result of the gene conversion, part of one RH gene is replaced by the corresponding segments of the other gene. Panel D: If transcription continues on the wrong template throughout SMP1, it may return to the correct template when it enters the second RH gene. Panel E:The result of this multi-gene-conversion are haplotypes with two “single hybrid” alleles, as described for CWD−,27 EKH,28 and some D· ·.29
Molecular basis of the antigen D
Independent of the exact binding site, any anti-RBC antibody binding to an Rh protein is considered an anti-D if the antibody binds to RhD but not to RhCE. With a few exceptions, the presence of D antigen may be equated to the presence of RhD-specific amino acids in any of the exofacial loops 3, 4, or 6. These three loops are the only exofacial protein segments that differ between RhD and RhCE. Monoclonal antibodies binding to different parts of RhD usually differ in their ability to bind to aberrant forms of RhD; this phenomenon was instrumental for establishing the serologic classification of D epitopes.30
Aberrant RHCE alleles encoding D-specific amino acids in the extracellular loops 3, 4, and 6 often express some D epitopes. The best known example is DHAR caused by an RHCE-D(5)-CE hybrid allele31 that encodes D epitope 6 (epD6)2 and is often typed as D+ by using commercial, licensed monoclonal anti-D.
Recently, the observation of the RHCE allele ceRT32 has added to the complexity of D antigen expression: the ceRT allele encodes part of epD6 without encoding any D-specific amino acid. This observation provided direct evidence that the Ser to Thr missense mutation at codon 154 forms structural features that are present in RhD but not in standard RhCE. This conclusion is in congruence with the notion that most D epitopes represent three-dimensional structures rather than linear protein segments.33
Molecular basis of the D− phenotype and of Del
The D− phenotype may be caused by the lack of functional RhD protein or by the presence of aberrant forms of RhD that do not express D antigen.
The most frequent cause for the absence of a functional RhD protein is a deletion of the whole RHD gene.18,22 This D− haplotype represents 40 percent of all haplotypes in Whites, and even in Africans, it may be more often the cause of D− than all other causes combined.18,25 The RHD deletion resulted from an unequal crossing over of the Rhesus boxes and is characterized by the presence of a “hybrid Rhesus box”18 (Fig. 2). The absence of the RHD gene in most D− Whites was a fortunate coincidence fostering the analyses of the molecular basis of D antigen: Whites with weak D or partial D often carry the RHD deletion in trans, a fact that much simplified the analyses of their single aberrant RHD alleles. Likewise, almost any RHD-specific polymorphism is a good predictor of D antigen in Whites.
Other observed sources of non-functional RhD proteins are nonsense mutations leading to premature stop codons, insertions and deletions leading to frameshifts, and splice site mutations that prevent the correct splicing of the RhD mRNA. These nonfunctional alleles are important for any genotyping approach34 including RHD genotyping by PCR: unless the mutation is detected specifically, RHD PCR will give a falsely positive prediction for D antigen.26 The most important allele of this type is RHDΨ,35 which is about as frequent as the RHD deletion among some African populations.35 Therefore, a correct D antigen prediction by PCR in Africans became possible only after the characterization of the structure of RHDΨ,35 which harbors a 37-bp insertion at the intron 3/exon 4 junction, a stop codon in exon 6, and several missense mutations. Today, a specific detection of RHDΨ is considered mandatory for any D antigen prediction by PCR.26
Some splice site mutations are permissive for the expression of minute traces of D antigen resulting in a Del phenotype, in which D antigen may only be demonstrated by adsorption and elution of anti-D. The most important example is RHD(G1227A),26 the most frequent Del allele in Japanese and Chinese,36 and the second most frequent Del allele among Whites.26
Other forms of aberrant RHD alleles can express RhD proteins that however do not carry D antigen. Such RhD proteins are encoded by RHD-CE-D hybrid alleles in which the RHCE segments must encompass at least exons 4 to 7.26 This causes exofacial loops 3 to 6 to resemble their RhCE counterparts. Since loops 1 and 2 do not differ between RhD and RhCE, the whole exofacial protein part of these hybrid proteins is identical to RhCE, and hence these aberrant RhD proteins cannot express D antigen. The two most important examples of such alleles are dCces,37 a RHD-CE(4–7)-D hybrid allele with some additional substitutions, and RHD-CE(2–9)-D.27 The dCces allele is the third most common cause of D− phenotypes among Africans,25 and RHD-CE(2–9)-D is a major cause for D− phenotypes in Chinese.36 It should be kept in mind that, although these hybrid alleles do not express D antigen, they are present in the RBC membrane and may carry other antigens, albeit often in modified, e.g., weakened, forms. For example, the RhD protein of dCces carries C antigen.38
Partial D caused by hybrid proteins
What happens if some, but not all, RhD-specific extracellular loops are replaced by their RhCE counterparts? Depending on their exact binding site on the RhD protein, some monoclonal anti-D antibodies will be able to bind to such hybrid proteins, other anti-D will not be able to bind. Thus, these hybrid RhD proteins present themselves serologically as a partial D phenotype whose hallmark is the lack of some, but not all, distinct D epitopes (epD), which are defined by monoclonal anti-D. Using polyclonal anti-D, these partial D RBCs are typed as D+, but carriers of these alleles may form anti-D antibodies, which bind to those parts of normal RhD that are lacking in the aberrant RhD protein.
Similar to the RHD-CE-D hybrids represented by dCces, the partial D phenotype is mainly determined by the RhD- or RhCE-specificity of the exofacial loops 3, 4, and 6 and influenced by the RhE/Rhe origin of loop 4.39 There are six possible combinations of the segments encoding exofacial loops 3, 4, and 6 (Table 1). Substitutions in nonexofacial protein segments have generally minor effects on the phenotype.40 Alleles that differ in the exact extent of the substitution but express the same exofacial protein segments form clusters of alleles that share similar phenotypes.39–41 Three of these clusters (DIVb, DVa, and DVI) were recognized early on and form three major groups of the D category classification.42,43 The other possible combinations are represented by DFR,43 DHAR,31 and DBT44 and have been recognized only since the mid 1980s.
Table 1.
Phenotype of RHD-CE-D and RHCE-D-CE hybrid alleles with segmental substitutions in exofacial loops 3 to 6
| Loop 3 (exon 4*) |
Loop 4 (exon 5) |
Loop 6 (exon 7) |
Phenotype | Antigens expressed | |
|---|---|---|---|---|---|
| D epitopes | Low-frequency antigens | ||||
| RhCE† | RhD | RhD | DFR | (1),(2),3,4,(5),(6/7),9 | FPTT (Rh50) |
| RhD | RhCE | RhD | DVa‡ | 2,3,4,6/7,8,9 | DW (Rh23) |
| RhD | RhD | RhCE | DIVb | 5,6/7,8 | Evans (Rh37) |
| RhCE | RhCE | RhD | DVI | 3,4,9 | BARC (Rh52) |
| RhCE | RhD | RhCE | DHAR | (5),(6/7) | RH33, FPTT (Rh50) |
| RhD | RhCE | RhCE | DBT | (6/7),8 | Rh32 |
The polymorphic parts of loops 3, 4, and 6 are encoded by exons 4, 5, and 7, respectively.
RhD indicates RhD-like sequence in the loop, RhCE RhCE-like sequence.
For the expression of a DVa phenotype, the RhD-specific alanine at position 226 must be retained (donor allele e). A proline at 226 (donor allele E) leads to a different phenotype, e.g. in DBS.
The unique combinations of RhD- and RhCE-specific extracellular loops found in hybrid RHD-CE-D alleles often are antigenic and may explain the low-frequency antigens expressed by those alleles, e.g., FPTT in DFR45 and BARC in D category VI.46
Of course, there are a few exceptions to this general outline, which are exemplified by two known partial D phenotypes that involve segmental substitutions other than loops 3 to 6: (1) DIIIb is caused by a substitution of exon 2 corresponding to loop 247 and (2) DIIIc by a substitution of exon 348 that presumably does not affect any exofacial amino acid. Although these phenotypes may become immunized to normal D, the alterations of the D antigen are much more limited than in the other hybrids, and their serologic detection using monoclonal antibodies is difficult.
Partial D caused by missense mutations affecting the exofacial protein segments
Missense mutations affecting the exofacial protein segments of RhD generally lead to a partial D phenotype. Because there are many more possible missense mutations than segmental substitutions, the phenotypic changes are much more diverse than those observed in partial D caused by hybrid proteins (Table 2). However, the phenotypic changes are often limited, correlating with an often low anti-D immunization risk and a difficult serologic detection. Based on the frequency of anti-D immunization events,49 DNB50 and DVII51 may be the two most important partial D of this type.
Table 2.
Partial D caused by missense mutations
| Loop involved | Position involved | Substitution | Trivial name |
|---|---|---|---|
| 1* | 54 | Leu to Pro | DMH81 |
| 2 | 103 | Ser to Pro | (G negative RhD)63 |
| 110 | Leu to Pro | DVII51 | |
| 3 | 166 | His to Pro | DFW82 |
| 4 | 229 | Arg to Lys | DHR83 |
| 233 | Glu to Lys | DHK28 | |
| 234 | Arg to Trp | DYU† | |
| 235 | Lys to Thr | DHO53 | |
| 5 | 283 | Thr to Ile | DHMi84 |
| 284 | Ser to Leu | “Sample A”85 | |
| 285 | Cys to Tyr | DIM52 | |
| 6 | 353 | Gly to Arg | DNU86 |
| 354 | Ala to Asp | DII86 | |
| 355 | Gly to Ser | DNB50 | |
| 358 | Met to Thr | DWI87 |
Depending on the model used, this amino acid position is considered transmembranous closely adjacent to the RBC surface.
GenBank entry AJ557827
Similar to hybrid alleles, the presence of aberrant amino acids in the RhD sequence may cause the expression of low-frequency antigens. The most important example is the Tar (RH40) antigen accompanying DVII46 that is caused by a leucine to a proline substitution at position 110.
Weak D caused by missense mutations affecting the nonexofacial protein segments
If a missense mutation affects a nonexofacial segment of the RhD protein, the influence on the D antigen is limited. However, such mutations tend to interfere with membrane integration of the RhD protein and are causing the vast majority of the weak D phenotypes.9 Although each allele has a distinct phenotype,52 a purely serologic discrimination of the many alleles is almost impossible, because the phenotypic differences are most often minute. The weak D types are designated by numbers according to their molecular structure; the lowest numbers were given to the most frequent types in the initial study,9 which has been proved since then to be representative for White populations in general (Table 3).53–55 No allo-anti-D immunization has been reported for weak D type 1 to type 3.49,52 Caution should be applied when considering the transfusion strategy for the less frequent weak D types, because, at the moment, it is not possible to exclude the possibility that changes in nonexofacial protein segments may allow allo-anti-D immunization.
Table 3.
Examples of clinically important and well defined weak D types*
| Weak D type | Position involved | Substitution | Intracellular/transmembranous |
|---|---|---|---|
| 1 | 270 | Val to Gly9 | transmembranous |
| 2 | 385 | Gly to Ala9 | transmembranous |
| 3 | 3 | Ser to Cys9 | intracellular |
| 4.0 | 201 | Thr to Arg9 | transmembranous |
| 223 | Phe to Val9 | transmembranous | |
| 4.1 | 16 | Trp to Cys50 | transmembranous |
| 201 | Thr to Arg | transmembranous | |
| 223 | Phe to Val | transmembranous | |
| 4.2† | 201 | Thr to Arg50 | transmembranous |
| 223 | Phe to Val | transmembranous | |
| 342 | Ile to Thr | transmembranous | |
| 5 | 149 | Ala to Asp9 | transmembranous |
| 11 | 295 | Met to Ile9 | transmembranous |
| 15 | 282 | Gly to Asp9 | transmembranous |
| 20 | 417 | Phe to Ser88 | intracellular |
A regularly updated list can be found at the RhesusBase: http://www.uniulm.de/~wflegel/Rh/.
The protein sequence of weak D type 4.2 is identical to the partial D DAR.59
The mechanisms underlying reduced RhD expression by weak D alleles are not completely understood and may differ depending on the allele concerned.56 Almost all the involved single point mutations relate to amino acids conserved throughout species.57 The missense mutations seem to occur in clusters9 (Fig.4), which might hint to regions important for the correct integration of the Rh proteins in the membrane or the correct interaction with RhAG.
Fig. 4.
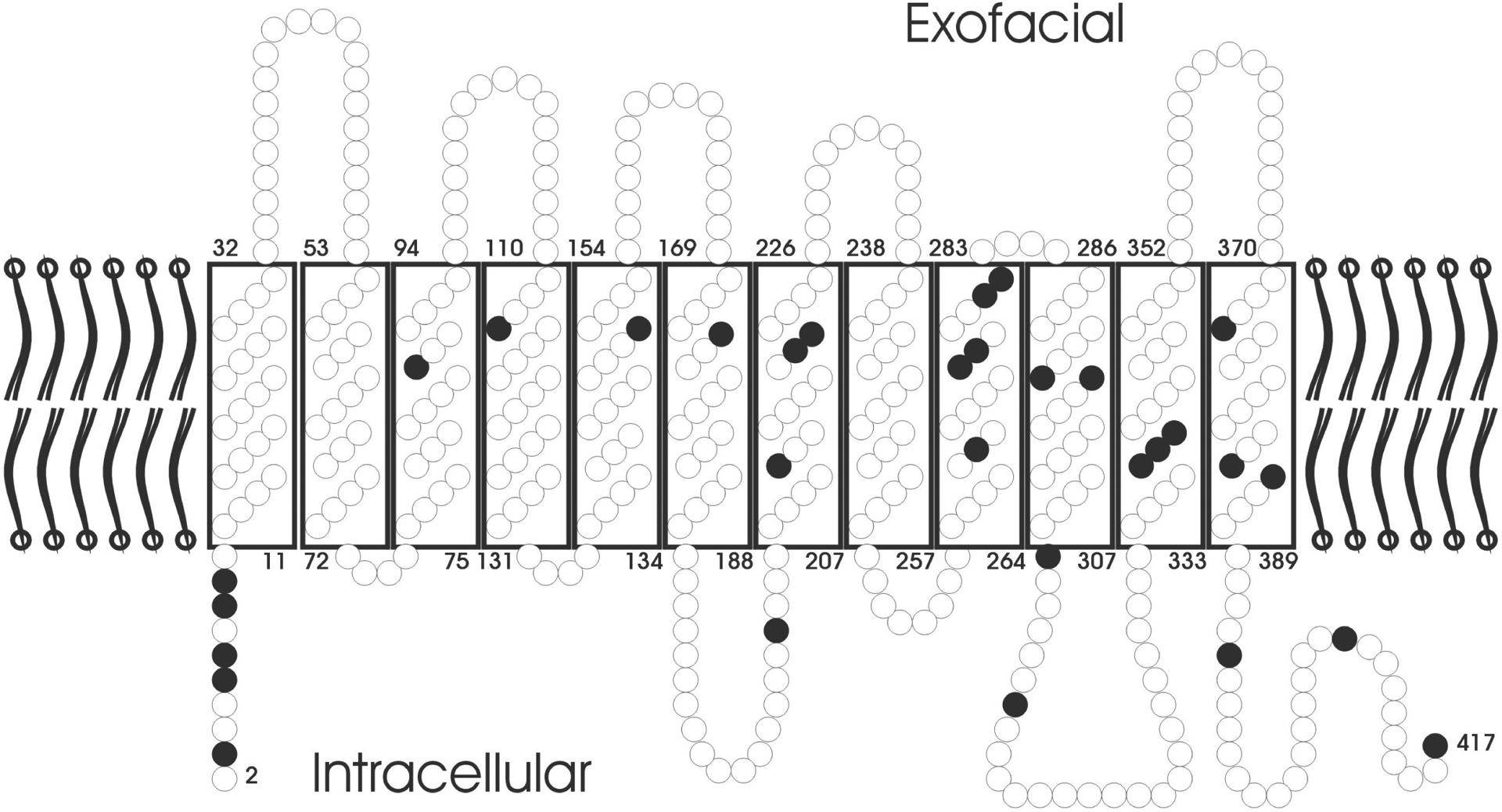
The predicted topology of RhD in the RBC membrane is shown. Amino acids are depicted as circles. Black circles indicate amino acid substitutions, each of which was correlated with a molecularly distinct weak D type.
Dispersed mutations, “African” alleles and the phylogeny of RHD alleles
A few partial D43,49,52,58–60 first detected among individuals of African descent are characterized by a multitude of missense mutations dispersed throughout the amino acid sequence of the RhD protein. The substitutions often are typical for RhCE but do not form a continuous stretch of RhCE sequence within RhD. The observation of these alleles is best explained assuming an RHD phylogeny in which almost all RHD alleles detected in Eurasians form just one of four branches.60 The multitude of missense mutations in “African” alleles simply reflects a longer phylogenetic distance from standard RhD (Fig. 5). The lack of “African” alleles in Eurasians is probably the result of a bottleneck during the migration out of Africa. Only a few “African” alleles, such as weak D type 49 and DAU-0,60 are occasionally observed among Whites. It is currently unknown whether these alleles were present during the primary bottleneck or entered the Eurasian allele pool by secondary migrations. Among Africans, alleles of all four clusters, including the alleles of the “Eurasian” cluster, are frequent.
Fig. 5.
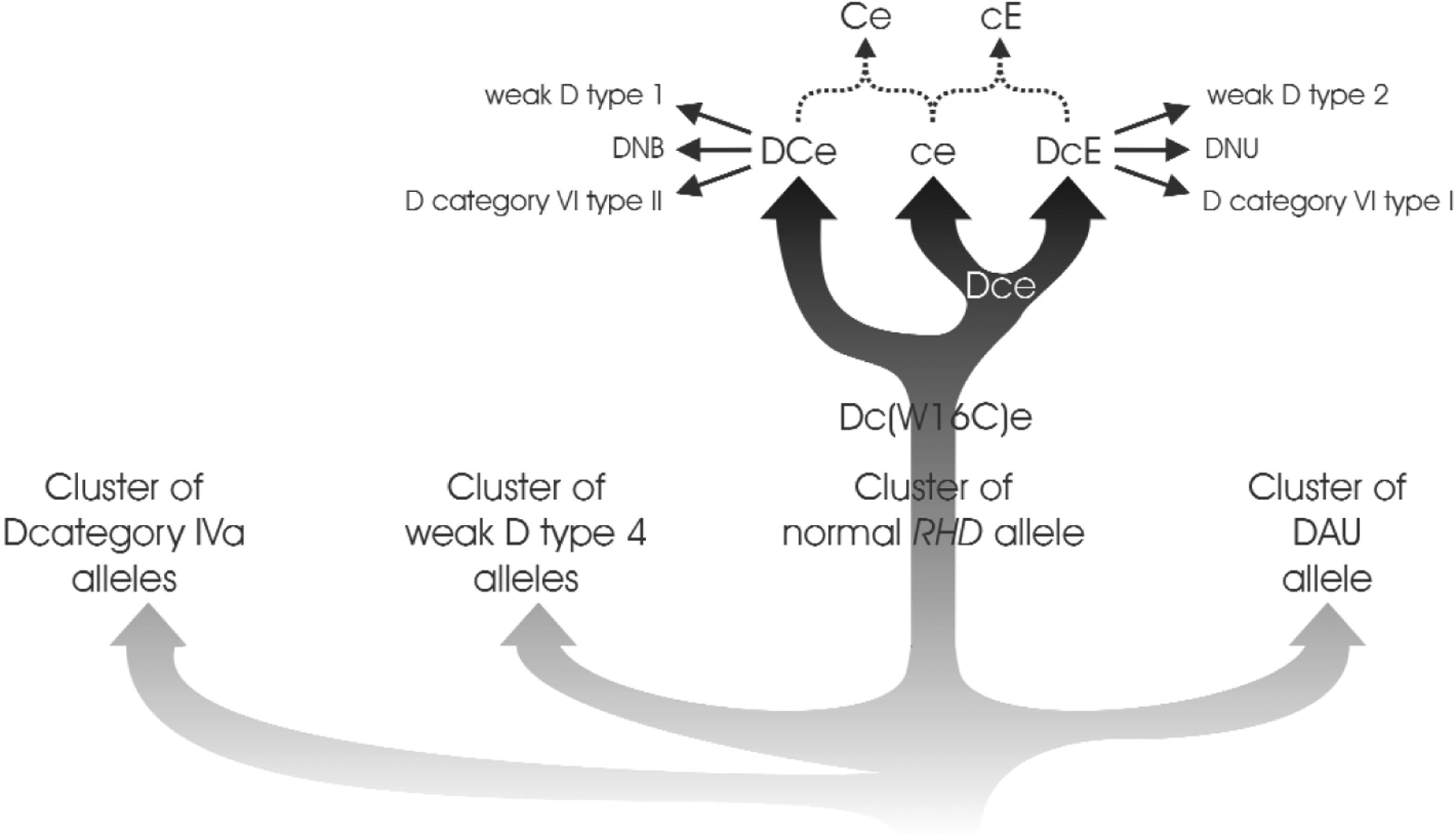
Phylogenetic tree of RHD. There are four independent branches of RHD alleles: The D category IVa cluster, the weak D type 4 cluster, the Eurasian D cluster, and the DAU cluster. The alleles of the D category IVa, weak D type 4, and DAU clusters are largely confined to individuals of African ancestry and generally occur in Dce haplotypes. The alleles of the Eurasian D cluster are predominant in Eurasian populations and most often occur in DCe and DcE haplotypes. The vast majority of aberrant alleles detected in individuals of Eurasian populations belong to the Eurasian D cluster and may be derived from “standard” RHD by a single molecular event (i.e., single nucleotide substitution or single gene conversion). In contrast, most alleles of the “African” clusters differ from “standard” (Eurasian) RHD by more than a single molecular event.
The phylogenic relationship between the four clusters is not completely resolved, the depicted topology is just one possibility. Likewise, it should be noted that the RHCE variation present among “African” haplotypes is not depicted.
Molecular basis of c, E, and G antigens
These three antigens are strongly correlated with the presence of a specific amino acid in an Rh protein: a c antigen is determined by Pro at position 103,61,62 G antigen by Ser at position 103,63 and E antigen by Pro at position 226.61 Ser at position 103 is present both in RhD and in the C allele of RhCE, therefore, both C+ and D+ haplotypes generally also express G antigen.
The mechanisms leading to partial D, weak D, and D− phenotypes may also occur in RhCE and lead to aberrant RhCE alleles carrying partial antigens, weakened antigens, or low-frequency antigens or not expressing the specific antigen despite PCR prediction of the corresponding allele (Table 4). However, these phenomena are recognized less frequently, because the antigens are less immunogenic than D and there are fewer monoclonal antibodies that could unravel lacking epitopes.
Table 4.
Examples of partial and weak antigens caused by aberrant RHCE alleles
| Mechanism | Example | “Parent allele” | Mutation | Antigens lost | Antigens expressed |
|---|---|---|---|---|---|
| Gene conversion | E category II 89 | cE | Ce-D(2–3)-ce | ||
| 90 | Ce | RHCE-D(4)-CE | Rh46 | Rh32 | |
| Ce | RHCE-D(3 partial-4)-CE | ||||
| R 0 Har 31 | ce * | RHCE-D(5)-CE | Rh33, Rh50 | ||
| CeVA 91 | Ce | RHCE-D(5)-CE | Rh33, Rh50 | ||
| Missense mutation (exofacial) | C X 92 | Ce | A36T | MAR | CX |
| C W 92 | Ce | Q41R | MAR | CW | |
| RH:-26 93 | ce | G96S | Rh26 | ||
| CeMA | Ce | R114W | (Har*) | ||
| E cat V = EHK 28 | cE | R154T | E epitopes | ||
| ceRT 32 | ce | R154T | D epitope 6 | ||
| E cat I 89 | E | M167K | E epitopes | ||
| E cat III 89 | E | Q233E, M238V | E epitopes | ||
| Missense mutation (non-exofacial) | E cat IV 89 | E | R201T | E epitopes ? | |
| VS 70 | ce(W16C) † | L245V | VS | ||
| In-frame-deletion | e U 95 | Del 229 | Very weak e |
RBC samples harboring this allele react with human serum Har. It is unknown whether the target antigen of this serum is identical to a known low-frequency antigen of the Rh blood group
Molecular basis of e and C antigens
The molecular bases of e and C antigens are a bit more complicated, because the amino acids characterizing these alleles of RhCE are also present in RhD. While the e antigen is associated with the presence of alanine at position 226,61 which is also found in RhD, e antigen is only expressed if its typical alanine occurs in an “RhCE context.” The minimal necessary “RhCE” context is unknown: RHCED(5)-CE alleles like R0Har lack RHCE exon 5 but express some e antigen. These alleles may cause a falsely negative e antigen prediction by PCR.64
The hallmark of alleles expressing C antigen is the presence of RHD exon 2 (encoding exofacial loop 2) in an RHCE context.61 Two mechanisms have led to such proteins: the “standard” C allele (Ce) differs from “standard” ce by an RHD-like type exon 2 that resulted from a gene conversion with RHD65; a small duplication at the insertion point was found to be the most reliable molecular polymorphism to predict this mechanism of C antigen expression.66 In addition, in this type of C+ allele, a cysteine must be present at position 16 encoded by exon 1. This cysteine is necessary for expression of C antigen,67 although it is not C-specific as it is shared by many ce alleles of the Dce haplotype.68,69 A different mechanism leads to the C antigen of dCces: this C antigen is carried by an aberrant RhD protein38 encoded by an RHD-CE(4–7)-D hybrid allele with several additional substitutions,70 including an RHCE-like threonine at position 152.The dCces haplotype codes for both C and c, because the accompanying RhCE protein carries c antigen.
Large RHCE-D-CE hybrids and the D– – phenotype
Similar to the loss of D antigen in RHD-CE-D hybrid alleles with a CE segment ranging from exons 4 to 7, substitution of all or almost all RhCE-specific extracellular protein segments with RhD sequence results in RhCE proteins that do not express CE antigens or parts thereof.71 Depending on the extent of the substitution, three forms of RhCE proteins lacking CE antigens, dubbed “CE-silent haplotypes,”72 result: substitution of exons 2 to 7 leads to the absence of all CE antigens in D– –.73,74 If exon 7 retains RHCE, the resulting phenotype is D··,75 which differs from D– – by the presence of the low-frequency antigen Evans (Rh37) and the high-frequency antigen Dav (RH47). Third, if a c-type exon 2, which encodes loop 2, is present, a cD− phenotype will result.76,77 In the CWD−phenotype,27,77 exon 2 is RHD but the CW-specific mutation in exon 1 is present. There is no frequent CE-negative haplotype, therefore CE-silent haplotypes are only detected if they occur in homozygous or compound heterozygous form or become apparent by family studies. The often enhanced expression of D antigen probably derives from the additional expression of D antigen in the aberrant RhCE protein.
“African” RHCE alleles
In analogy to the RHD allele of African origin, several RHCE alleles are frequent in Africans but rare in Europeans and often differ by a multitude of mutations (Table 5).37,59,70,78 These alleles code for RhCE proteins that lack some high-frequency antigens. Immunizations to these antigens may pose serious logistic problems, especially if they occur in individuals depending on chronic transfusion support because of inherited anemias.78
Table 5.
“African” RHCE alleles*
| Allele type | Allele name | Antigens lacking | “New” antigens | Molecular bases† |
|---|---|---|---|---|
| RH:-18,-19 (HrS-,hrS-) | ceEK78 | RH18, RH19 (HrS, hrS) | Trp16Cys, Met238Val, Arg263Gly, Met267Lys | |
| ceBI78 | RH18, RH19 (HrS, hrS) | Trp16Cys, Met238Val, Ala273Val, Leu378Val | ||
| ceAR59 | RH18, RH19 (HrS, hrS) | Trp16Cys, Met238Val, Leu245Val, Arg263Gly, Met267Lys, Ile306Val | ||
| RH: −19 (hrS-), Partial e | ceMO96 | RH19 (hrS) | Trp16Cys, Val223Phe | |
| RH:−34 (HrB-) | Ccde s 37 | RH34 (HrB) | RH20 (VS) | dCce S |
| Partial e | ces(340CT)78 | Arg114Trp, Leu245Val | ||
| RH:32,−46 | 90 | RH46 | RH32† | RHCE-D(4)-CE |
| 90 | RH46 | RH32 | RHCE-D(3 partial-4)-CE | |
| Weak e | ce(W16C)94 | Trp16Cys | ||
| ces 70 | RH10, RH20 (V,VS) | Trp16Cys, Leu245Val | ||
| ceCF97 | RH20, RH43 (VS, Crawford) | Trp16Cys, Gln233Glu, Leu245Val |
Only important examples relevant to specific antigens are given. Several additional RHCE alleles have been described in individuals of African ancestry but often still await a full serologic characterization.
Among Africans, R0Har appears to be linked to a D+ allele.
Rhnull of the amorph type
Carriers of the “amorph” type of Rhnull were shown to lack any functional RHCE and any functional RHD genes. Generally, they are caused by nonsense mutations in RHCE in an RHD negative background.79,80
Unresolved issues of RH genotype and phenotype
The models presented in this review correlate the Rh phenotype with the type of the extracellular protein segments and missense, nonsense, and splice site mutations. These models are powerful in explaining much of the allelic and antigenic variation observed in vivo (Table 6). However, it is important to realize that the exact relationship of D epitopes (epD) and RhD structures as well as the molecular basis of several Rh antigens remains unresolved. In addition, there may be some RHD alleles that lack D antigen without obvious changes in the RHD gene.55 Detection of the RHD deletion by PCR is still hampered by falsely negative and falsely positive results in Africans, indicating a variability in Rhesus boxes in Africans that may well surpass the variation observed among Whites. Finally, while the function of Rh proteins in the Rh complex is only emerging, the functional role of different Rh variants and a possible selection pressure generating and maintaining the astounding Rh antigenic variability is a remaining mystery and poses an opportunity for continuing research.
Table 6.
Most likely explanations for Rh antigens using a loop-centered approach*
| Antigen Number | Antigen symbol | Molecular basis (protein) | Molecular basis (DNA/Codons) |
|---|---|---|---|
| 1 | D | RhD exofacial loops 3, 4, and 6 | RHD exon 4, 5, 7 |
| 2 | C | RhD-like loop 2 in RhCE with Cys 16 | RHD exon 2 in RHCE |
| 3 | E | Pro 226 in loop 4 | 226 Pro in exon 5 |
| 4 | c | Pro 103 in loop 2 | 103 Pro in exon 2 |
| 5 | e | Ala 226 in RhCE loop 4 | 226 Ala in RHCE exon 5 |
| 6 | f | Pro103 in loop 2 and Ala 226 in loop 4 | 103 Pro in exon 2 and 226 Ala in exon 5 |
| 7 | Ce | RhD-like loop 2 in RhCE and Ala 226 in loop 4 and Leu 245 (transmembranous) | RHD exon 2 and 226 Ala, 245 Leu in exon 5 RHCE |
| 8 | CW | Arg 41 in loop 1 | 41 Arg in exon 1 |
| 9 | CX | Thr 36 in loop 1 | 36 Thr in exon 1 |
| 10 | V | Val 245 and Gly 336 (transmembranous location) | 245 Val in exon 5 and 336 Gly in exon 7 |
| 11 | EW | ||
| 12 | G | Ser 103 in loop 2 | 103 Ser in exon 2 (both RHD and RHCE possible) |
| 17 | Hr0 | RhCE exofacial loops 3, 4 and 6 | RHCE exons 4,5,7 |
| 18 | Hr | Met 238 in RhCE loop 4 | 238 Met in RHCE exon 5 |
| 19 | hrs | Ala 226 and Met 238 in RhCE loop 4 | 226 Ala and 238 Met in RHCE exon 5 |
| 20 | VS | Val 245 (transmembranous location?) | 245 Val in exon 5 |
| 21 | CG | Ser 103 in RhCE loop 2 | 103 Ser in RHCE exon 2 |
| 22 | CE | Ser 103 in loop 2 and Pro 226 in loop 4 | 103 Ser in exon 2 and 226 Pro in exon 5 (RHCE) |
| 23 | DW | RhCE loop 4 (Gln 233) and RhD loop 3 and 6 | RHCE exon 5 and RHD exons 4 and 7 |
| 26 | c-like | Gly 96 and Pro 103 in RhCE loop 2 | 96 Gly and 103 Pro in RHCE exon 2 |
| 27 | cE | Pro 103 in loop 2 and Pro 226 in loop 4 | 103 Pro in exon 2 and 226 Pro in exon 5 |
| 28 | hrH | ||
| 29 | Total RH | Any Rh protein | Any expressed RH gene |
| 30 | Goa | Carried by DIVa† | Carried by DIVa |
| 31 | hrB | ||
| 32 | Rh32 | RhD loop 3 together with RhCE loop 4 and 6 | RHD exon 4 together with RHCE exon 5 and 7 |
| 33 | Har | Probably RhD loop 4 together with RhCE loop 3 and 6 | RHD exon 5 together with RHCE exon 4 and 7 |
| 34 | HrB | Cys 336 in RhDE | 336 Cys in RHDE |
| 35 | 1114 | ||
| 36 | Bea | ||
| 37 | Evans | RhD loops 3 and 4 with RhCE loop 6 | RHD exon 4 and 5 together with RHCE exon 7 |
| 39 | C-like | ||
| 40 | Tar | Pro 110 in RhDE loop 2 | 110 Pro in RHDE exon 2 |
| 41 | Ce-like | RhD-like loop 2 in RhCE and 226 Ala in loop 4 and normal loop 1 (Gln 41) | RHD exon 2,226 Ala in exon 5, normal exon 1 |
| 42 | Ces | Carried by dCces† | |
| 43 | Crawford | Gln 233 in loop 4 in VS-like allele | 233 Gln in exon 5 of VS-like allele |
| 44 | Nou | ||
| 45 | Riv | Carried by DIVa(C)-† | |
| 46 | Sec | RhCE loop 3 | RHCE exon 4 |
| 47 | Dav | Probably RhCE loop 6 | RHCE exon 7 |
| 48 | JAL | ||
| 49 | STEM | ||
| 50 | FPTT | RhCE loop 3 together with RhD loop 4 | RHCE exon 4 together with RHD exon 5 |
| 51 | MAR | Normal RhCE loop 1 (Ala 37 and Gln 41) | Normal RHCE exon 1 (37 Ala and 41 Gln) |
| 52 | BARC | RhCE loop 3 and 4 (with Ala 226) together with RhD loop 6; | RHCE exon 4 and 5 (with 226 Ala) together with RHD exon 7 |
| 53 | JAHK | RhD loop 2 in RhCE without Cys 16 | RHD exon 2 in RHCE without 16 Cys |
| 54 | DAK | Carried by DIIIa, DOL, and | |
| 55 | LOCR |
All explanations are tentative and based on the published distribution of the antigens. These interpretations may need modifications, if additional haplotypes encoding or not encoding the antigens are identified.
The region of Rh relevant for the antigen cannot be deduced from the published distribution of the antigen among RH haplotypes.
Contributor Information
F. F. Wagner, Priv.-Doz. Dr. Med., DRK Blutspendedienst NSTOB, Zentralinstitut Springe, Eldagsener Str. 38, D-31830 Springe, Germany
W. A. Flegel, Institut für klinische Transfusionsmedizin und Immungenetik Ulm, Ulm, Germany.
References
- 1.Daniels GL, Cartron JP, Fletcher A, et al. International Society of Blood Transfusion Committee on terminology for red cell surface antigens: Vancouver Report. Vox Sang 2003;84: 244–7. [DOI] [PubMed] [Google Scholar]
- 2.Scott M. Rh serology—coordinator’s report. Transfus Clin Biol 1996;3:333–7. [DOI] [PubMed] [Google Scholar]
- 3.Scott M. Section 1A: Rh serology. Coordinator’s report. Transfus Clin Biol 2002;9:23–9. [DOI] [PubMed] [Google Scholar]
- 4.Tippett P, Sanger R. Observations on subdivisions of the Rh antigen D. Vox Sang 1962;7:9–13. [DOI] [PubMed] [Google Scholar]
- 5.Le van Kim C, Mouro I, Cherif-Zahar B, et al. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci USA 1992;89:10925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arce MA, Thompson ES, Wagner S, Coyne KE, Ferdman BA, Lublin DM. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood 1993;82: 651–655 [PubMed] [Google Scholar]
- 7.Cherif-Zahar B, Bloy C, Le Van Kim C, et al. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci USA 1990;87:6243–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avent ND, Ridgwell K, Tanner MJ, Anstee DJ. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J 1990;271:821–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood 1999;93:385–93. [PubMed] [Google Scholar]
- 10.Ridgwell K, Spurr NK, Laguda B, MacGeoch C, Avent ND, Tanner MJ. Isolation of cDNA clones for a 50 kDa glycoprotein of the human erythrocyte membrane associated with Rh (rhesus) blood-group antigen expression. Biochem J 1992;287: 223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas V, Le Van Kim C, Gane P, et al. Rh-RhAG/ankyrin-R, a new interaction site between the membrane bilayer and the red cell skeleton, is impaired by Rh(null)-associated mutation. J Biol Chem 2003;278:25526–33. [DOI] [PubMed] [Google Scholar]
- 12.Cherif-Zahar B, Raynal V, Gane P, et al. Candidate gene acting as a suppressor of the RH locus in most cases of Rh-deficiency. Nat Genet 1996;12:168–73. [DOI] [PubMed] [Google Scholar]
- 13.Marini AM, Matassi G, Raynal V, Andre B, Cartron JP, Cherif-Zahar B. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat Genet 2000;26:341–4. [DOI] [PubMed] [Google Scholar]
- 14.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem 2002;277:12499–502. [DOI] [PubMed] [Google Scholar]
- 15.Hemker MB, Cheroutre G, van Zwieten R, et al. The Rh complex exports ammonium from human red blood cells. Br J Haematol 2003;122:333–40. [DOI] [PubMed] [Google Scholar]
- 16.Soupene E, King N, Feild E, et al. Rhesus expression in a green alga is regulated by CO(2). Proc Natl Acad Sci USA 2002;99:7769–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherif-Zahar B, Le Van Kim C, Rouillac C, Raynal V, Cartron JP, Colin Y. Organization of the gene (RHCE) encoding the human blood group RhCcEe antigens and characterization of the promoter region. Genomics 1994;19:68–74. [DOI] [PubMed] [Google Scholar]
- 18.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood 2000;95:3662–8. [PubMed] [Google Scholar]
- 19.Suto Y, Ishikawa Y, Hyodo H, Uchikawa M, Juji T. Gene organization and rearrangements at the human Rhesus blood group locus revealed by fiber-FISH analysis. Hum Genet 2000;106:164–71. [DOI] [PubMed] [Google Scholar]
- 20.Wagner FF, Flegel WA. RHCE represents the ancestral RH position, while RHD is the duplicated gene. Blood 2002;99:2272–3. [DOI] [PubMed] [Google Scholar]
- 21.Kumada M, Iwamoto S, Kamesaki T, Okuda H, Kajii E. Entire sequence of a mouse chromosomal segment containing the gene Rhced and a comparative analysis of the homologous human sequence. Gene 2002;299:165–72. [DOI] [PubMed] [Google Scholar]
- 22.Colin Y, Cherif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood 1991;78:2747–52. [PubMed] [Google Scholar]
- 23.Chiu RW, Murphy MF, Fidler C, Zee BC, Wainscoat JS, Lo YM. Determination of RhD zygosity: comparison of a double amplification refractory mutation system approach and a multiplex real-time quantitative PCR approach. Clin Chem 2001;47: 667–72. [PubMed] [Google Scholar]
- 24.Matheson KA, Denomme GA. Novel 3’Rhesus box sequences confound RHD zygosity assignment. Transfusion 2002;42:645–50. [DOI] [PubMed] [Google Scholar]
- 25.Wagner FF,Moulds JM,Tounkara A,Kouriba B,Flegel WA. RHD allele distribution in Africans of Mali. BMC Genet 2003;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet 2001;2:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang CH. Alteration of RH gene structure and expression in human dCCee and DCW– red blood cells: phenotypic homozygosity versus genotypic heterozygosity. Blood 1996;88:2326–33. [PubMed] [Google Scholar]
- 28.Kashiwase K, Ishikawa Y, Hyodo H, et al. E variants found in Japanese and c antigenicity alteration without substitution in the second extracellular loop. Transfusion 2001;41:1408–12. [DOI] [PubMed] [Google Scholar]
- 29.Cheng GJ, Chen Y, Reid ME, Huang CH. Evans antigen: a new hybrid structure occurring on background of D·· and D– – Rh complexes. Vox Sang 2000;78:44–51. [DOI] [PubMed] [Google Scholar]
- 30.Lomas C, Tippett P, Thompson KM, Melamed MD, Hughes-Jones NC. Demonstration of seven epitopes on the Rh antigen D using human monoclonal anti-D antibodies and red cells from D categories. Vox Sang 1989;57:261–4. [DOI] [PubMed] [Google Scholar]
- 31.Beckers EAM, Faas BHW, von dem Borne AEGK, Overbeeke MAM, van Rhenen DJ, van der Schoot CE. The R0Har Rh:33 phenotype results from substitution of exon 5 of the RHCE gene by the corresponding exon of the RHD gene. Br J Haematol 1996;92:751–7. [DOI] [PubMed] [Google Scholar]
- 32.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion 2003;43:1248–54. [DOI] [PubMed] [Google Scholar]
- 33.Liu W, Avent ND, Jones JW, Scott ML, Voak D. Molecular configuration of Rh D epitopes as defined by site-directed mutagenesis and expression of mutant Rh constructs in K562 erythroleukemia cells. Blood 1999;94:3986–96. [PubMed] [Google Scholar]
- 34.Wagner FF, Flegel WA. Polymorphism of the h allele and the population frequency of sporadic nonfunctional alleles. Transfusion 1997;37:284–90. [DOI] [PubMed] [Google Scholar]
- 35.Singleton BK, Green CA, Avent ND, et al. The presence of an RHD pseudogene containing a 37 base-pair duplication and a nonsense mutation in Africans with the Rh D− blood group phenotype. Blood 2000;95:12–8. [PubMed] [Google Scholar]
- 36.Shao CP, Maas JH, Su YQ, Kohler M, Legler TJ. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang 2002;83:156–61. [DOI] [PubMed] [Google Scholar]
- 37.Faas BH, Beckers EA, Wildoer P, et al. Molecular background of VS and weak C expression in blacks. Transfusion 1997;37:38–44. [DOI] [PubMed] [Google Scholar]
- 38.Blunt T, Daniels G, Carritt B. Serotype switching in a partially deleted RHD gene. Vox Sang 1994;67:397–401. [DOI] [PubMed] [Google Scholar]
- 39.Wagner FF, Ernst M, Sonneborn HH, Flegel WA. A D(V)-like phenotype is obliterated by A226P in the partial D DBS. Transfusion 2001;41:1052–8. [DOI] [PubMed] [Google Scholar]
- 40.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Three molecular structures cause Rhesus D category VI phenotypes with distinct immunohematologic features. Blood 1998;91:2157–68. [PubMed] [Google Scholar]
- 41.Omi T, Okuda H, Iwamoto S, et al. Detection of Rh23 in the partial D phenotype associated with the D(Va) category. Transfusion 2000;40:256–8. [DOI] [PubMed] [Google Scholar]
- 42.Mouro I, Le Van Kim C, Rouillac C, et al. Rearrangements of the blood group RhD gene associated with the DVI category phenotype. Blood 1994;83:1129–35. [PubMed] [Google Scholar]
- 43.Rouillac C, Colin Y, Hughes-Jones NC, et al. Transcript analysis of D category phenotypes predicts hybrid Rh D-CE-D proteins associated with alteration of D epitopes. Blood 1995;85:2937–44. [PubMed] [Google Scholar]
- 44.Beckers EA, Faas BH, Simsek S, et al. The genetic basis of a new partial D antigen: DDBT. Br J Haematol 1996;93:720–7. [DOI] [PubMed] [Google Scholar]
- 45.Lomas C, Grassmann W, Ford D, et al. FPTT is a low-incidence Rh antigen associated with a “new” partial Rh D phenotype, DFR. Transfusion 1994;34:612–6. [DOI] [PubMed] [Google Scholar]
- 46.Tippett P, Lomas-Francis C, Wallace M. The Rh antigen D: partial D antigens and associated low incidence antigens. Vox Sang 1996;70(3):123–31. [DOI] [PubMed] [Google Scholar]
- 47.Rouillac C, Le Van Kim C, Blancher A, Roubinet F, Cartron JP, Colin Y. Lack of G blood group antigen in DIIIb erythrocytes is associated with segmental DNA exchange between RH genes. Br J Haematol 1995;89:424–6. [DOI] [PubMed] [Google Scholar]
- 48.Beckers EA, Faas BH, Ligthart P, et al. Characterization of the hybrid RHD gene leading to the partial D category IIIc phenotype. Transfusion 1996;36:567–74. [DOI] [PubMed] [Google Scholar]
- 49.Flegel WA. Rhesus Immunisierungsregister (RIR) [The Rhesus Immunization Surveillance]. Ulm:DRK Blutspendedienst Baden-Württemberg-Hessen. <http://www.uni-ulm.de/~wflegel/RH/RIR/>. [Google Scholar]
- 50.Wagner FF, Eicher NI, Jorgensen JR, Lonicer CB, Flegel WA. DNB: a partial D with anti-D frequent in Central Europe. Blood 2002;100:2253–6. [DOI] [PubMed] [Google Scholar]
- 51.Rouillac C, Le Van Kim C, Beolet M, Cartron JP, Colin Y. Leu110Pro substitution in the RhD polypeptide is responsible for the DVII category blood group phenotype. Am J Hematol 1995;49:87–8. [DOI] [PubMed] [Google Scholar]
- 52.Wagner FF, Frohmajer A, Ladewig B, et al. Weak D alleles express distinct phenotypes. Blood 2000;95:2699–708. [PubMed] [Google Scholar]
- 53.Müller TH, Wagner FF, Trockenbacher A, et al. PCR screening for common weak D types shows different distributions in three Central European populations. Transfusion 2001;41:45–52. [DOI] [PubMed] [Google Scholar]
- 54.Cowley NM, Saul A, Hyland CA. RHD gene mutations and the weak D phenotype: an Australian blood donor study. Vox Sang 2000;79:251–2. [DOI] [PubMed] [Google Scholar]
- 55.Döscher A, Ladewig B, Das Gupta C, et al. Molecular genetic RHD characterization of 577 cases with serologic suspect for weak D (abstract). Transfus Med Hemother 2003;30S1:B2.03. [Google Scholar]
- 56.Kamesaki T, Iwamoto S, Kumada M, et al. Molecular characterization of weak D phenotypes by site-directed mutagenesis and expression of mutant Rh-green fluorescence protein fusions in K562 cells. Vox Sang 2001;81:254–8. [DOI] [PubMed] [Google Scholar]
- 57.Wagner FF. Die molekulare Basis der RH-Haplotypen mit schwacher Expression des Antigens D [The molecular basis of RH haplotypes expressing weak D antigens]. Habilitationsschrift. Universität Ulm; 1999. <http://vts.uni-ulm.de/query/longview.meta.asp?document_id=584 > [Google Scholar]
- 58.Huang CH, Chen Y, Reid M. Human D(IIIa) erythrocytes: RhD protein is associated with multiple dispersed amino acid variations. Am J Hematol 1997;55:139–45. [DOI] [PubMed] [Google Scholar]
- 59.Hemker MB, Ligthart PC, Berger L, van Rhenen DJ, van der Schoot CE, Wijk PA. DAR, a new RhD variant involving exons 4, 5, and 7, often in linkage with ceAR, a new rhce variant frequently found in African blacks. Blood 1999;94:4337–42. [PubMed] [Google Scholar]
- 60.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood 2002;100:306–11. [DOI] [PubMed] [Google Scholar]
- 61.Mouro I, Colin Y, Cherif-Zahar B, Cartron JP, Le Van Kim C. Molecular genetic basis of the human Rhesus blood group system. Nat Genet 1993;5:62–5. [DOI] [PubMed] [Google Scholar]
- 62.Faas BH, Beuling EA, Ligthart PC, van Rhenen DJ, van der Schoot CE. Partial expression of RHc on the RHD polypeptide. Transfusion 2001;41:1136–42. [DOI] [PubMed] [Google Scholar]
- 63.Faas BH, Beckers EA, Simsek S, et al. Involvement of Ser103 of the Rh polypeptides in G epitope formation. Transfusion 1996;36:506–11. [DOI] [PubMed] [Google Scholar]
- 64.Hundhausen T, Petershofen EK, Doescher A, Bauerfeind U, Muller TH, Schunter F. RHCE-D-CE hybrid genes can cause false-negative DNA typing of the Rh e antigen. Vox Sang 2002. Oct;83(3):268–72. [DOI] [PubMed] [Google Scholar]
- 65.Carritt B, Kemp TJ, Poulter M. Evolution of the human RH (rhesus) blood group genes: a 50 year old prediction (partially) fulfilled. Hum Mol Genet 1997. Jun;6(6):843–50. [DOI] [PubMed] [Google Scholar]
- 66.Poulter M, Kemp TJ, Carritt B. DNA-based Rhesus typing: simultaneous determination of RHC and RHD status using the polymerase chain reaction. Vox Sang 1996;7:164–8. [DOI] [PubMed] [Google Scholar]
- 67.Mouro I, Colin Y, Gane P, et al. Molecular analysis of blood group Rh transcripts from a rGr variant. Br J Haematol 1996;93:472–4. [DOI] [PubMed] [Google Scholar]
- 68.Wolter LC, Hyland CA, Saul A. Refining the DNA polymorphisms that associate with the rhesus c phenotype. Blood 1994;84:985–6. [PubMed] [Google Scholar]
- 69.Gassner C, Schmarda A, Kilga-Nogler S, et al. RHD/CE typing by polymerase chain reaction using sequence-specific primers. Transfusion 1997; 37:1020–6. [DOI] [PubMed] [Google Scholar]
- 70.Daniels GL, Faas BH, Green CA, et al. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion 1998;38:951–8. [DOI] [PubMed] [Google Scholar]
- 71.Avent ND, Reid ME. The Rh blood group system: a review. Blood 2000;95:375–87. [PubMed] [Google Scholar]
- 72.Blumenfeld OO, Reid ME, Huang CH. Blood group antigen gene mutation database: rare alleles of RH loci. New York, accessed 2003: <http://www.bioc.aecom.yu.edu/bgmut/rh_rare.htm> [Google Scholar]
- 73.Huang CH, Reid ME, Chen Y. Identification of a partial internal deletion in the RH locus causing the human erythrocyte D– – phenotype. Blood 1995; 86:784–90. [PubMed] [Google Scholar]
- 74.Cherif-Zahar B, Raynal V, Cartron JP. Lack of RHCE-encoded proteins in the D– –phenotype may result from homologous recombination between the two RH genes. Blood 1996;88:1518–20. [PubMed] [Google Scholar]
- 75.Huang CH, Chen Y, Reid M, Ghosh S. Genetic recombination at the human RH locus: a family study of the red-cell Evans phenotype reveals a transfer of exons 2–6 from the RHD to the RHCE gene. Am J Hum Genet 1996;59:825–33. [PMC free article] [PubMed] [Google Scholar]
- 76.Cotorruelo CM, Biondi CS, Borras SE, Di Monaco RA, Racca A. A Dc– phenotype encoded by an RHCE-D(5–7/8)-CE hybrid allele. Vox Sang 2003; 85:102–8. [DOI] [PubMed] [Google Scholar]
- 77.Cherif-Zahar B, Raynal V, D’ Ambrosio AM, Cartron JP, Colin Y. Molecular analysis of the structure and expression of the RH locus in individuals with D– –, Dc–, and DCw– gene complexes. Blood 1994;84:4354–60. [PubMed] [Google Scholar]
- 78.Noizat-Pirenne F, Lee K, Pennec PY, et al. Rare RHCE phenotypes in Black individuals of Afro-Caribbean origin: identification and transfusion safety. Blood 2002;100:4223–31. [DOI] [PubMed] [Google Scholar]
- 79.Huang CH, Chen Y, Reid ME, Seidl C. Rhnull disease: the amorph type results from a novel double mutation in RhCe gene on D-negative background. Blood 1998;92:664–71. [PubMed] [Google Scholar]
- 80.Cherif-Zahar B, Matassi G, Raynal V, et al. Molecular defects of the RHCE gene in Rh-deficient individuals of the amorph type. Blood 1998;92:639–46. [PubMed] [Google Scholar]
- 81.Avent ND, Poole J, Singleton B, et al. Studies of two partial Ds: DMH and DOL (abstract). Transfus Med 9, 1999;33(suppl):33. [Google Scholar]
- 82.Wagner FF, Gassner C, Eicher NI, Lonicer C, Flegel WA. Characterization of D category IV type IV, DFW, and DNB (abstract). Transfusion 1998; 38(suppl): 63S. [Google Scholar]
- 83.Jones JW, Finning K, Mattock R, et al. The serological profile and molecular basis of a new partial D phenotype, DHR. Vox Sang 1997;73:252–6. [DOI] [PubMed] [Google Scholar]
- 84.Liu W, Jones JW, Scott ML, Voak D, Avent ND. Molecular analysis of two D-variants, DHMi and DHMii (abstract). Transfus Med 6, 1996; 21(suppl):21.8696444 [Google Scholar]
- 85.Döscher A, Ladewig B, Gerdes I, et al. Six new RHD-alleles with previously unknown polymorphisms (abstract). Transfus Med Hemother 2003;30(suppl 1):B2.04. [Google Scholar]
- 86.Avent ND, Jones JW, Liu W, et al. Molecular basis of the D variant phenotypes DNU and DII allows localization of critical amino acids required for expression of Rh D epitopes epD3, 4 and 9 to the sixth external domain of the Rh D protein. Br J Haematol 1997;97:366–71. [DOI] [PubMed] [Google Scholar]
- 87.Körmöczi GF, Legler TJ, Daniels GL, et al. A novel partial RhD with highly retained epitope composition in an individual with alloanti-D (abstract). Transfus Med Hemother 2003;30(suppl 1):B2.07. [Google Scholar]
- 88.Witter B. Die Verteilung von Antigendichten und weak D-Allelen im Rhesusphänotyp ccD.Ee Dissertationsschrift. Universität Ulm; 2000. <http://vts.uni-ulm.de/query/longview.meta.asp?document_id=765> [Google Scholar]
- 89.Noizat-Pirenne F, Mouro I, Gane P, et al. Heterogeneity of blood group RhE variants revealed by serological analysis and molecular alteration of the RHCE gene and transcript. Br J Haematol 1998;103:429–36. [DOI] [PubMed] [Google Scholar]
- 90.Rouillac C, Gane P, Cartron J, Le Pennec PY, Cartron JP, Colin Y. Molecular basis of the altered antigenic expression of RhD in weak D(Du) and RhC/e in Rh phenotypes. Blood 1996;87:4853–61. [PubMed] [Google Scholar]
- 91.Noizat-Pirenne F, Le Pennec PY, Mouro I, et al. Molecular background of D(C)(e) haplotypes within the white population. Transfusion 2002;42: 627–33. [DOI] [PubMed] [Google Scholar]
- 92.Mouro I, Colin Y, Sistonen P, Le Pennec PY, Cartron JP, Le Van Kim C. Molecular basis of the RhCW (Rh8) and RhCX (Rh9) blood group specificities. Blood 1995;86:1196–201. [PubMed] [Google Scholar]
- 93.Faas BH, Ligthart PC, Lomas-Francis C, Overbeeke MA, von dem Borne AE, van der Schoot CE. Involvement of Gly96 in the formation of the Rh26 epitope. Transfusion 1997;37:1123–30. [DOI] [PubMed] [Google Scholar]
- 94.Westhoff CM, Silberstein LE, Wylie DE, Skavdahl M, Reid ME. 16Cys encoded by the RHce gene is associated with altered expression of the e antigen and is frequent in the RO haplotype. Br J Haematol 2001;113:666–71. [DOI] [PubMed] [Google Scholar]
- 95.Huang CH, Reid ME, Chen Y, Novaretti M. Deletion of Arg 229 in RhCE polypeptide alters expression of Rhe and ce-associated Rh6 antigen (abstract). Blood 1992;90(suppl):272a. [Google Scholar]
- 96.Noizat-Pirenne F, Mouro I, Le Pennec PY, et al. Two new alleles of the RHCE gene in Black individuals: the RHce allele ceMO and the RHcE allele cEMI. Br J Haematol 2001;113:672–9. [DOI] [PubMed] [Google Scholar]
- 97.Schlanser G, Moulds MK, Flegel WA, Wagner FF, Frame T. Crawford (Rh43), a low-incidence antigen, is associated with a novel RHCE variant RHce allele, ceCF (abstract). Transfusion 2003;43(suppl): 35A. [Google Scholar]


