Abstract
The assembly of newly synthesized DNA into chromatin is essential for normal growth, development, and differentiation. To gain a better understanding of the assembly of chromatin during DNA synthesis, we identified, cloned, and characterized the 180- and 105-kDa polypeptides of Drosophila chromatin assembly factor 1 (dCAF-1). The purified recombinant p180+p105+p55 dCAF-1 complex is active for DNA replication-coupled chromatin assembly. Furthermore, we have established that the putative 75-kDa polypeptide of dCAF-1 is a C-terminally truncated form of p105 that does not coexist in dCAF-1 complexes containing the p105 subunit. The analysis of native and recombinant dCAF-1 revealed an interaction between dCAF-1 and the Drosophila anti-silencing function 1 (dASF1) component of replication-coupling assembly factor (RCAF). The binding of dASF1 to dCAF-1 is mediated through the p105 subunit of dCAF-1. Consistent with the interaction between dCAF-1 p105 and dASF1 in vitro, we observed that dASF1 and dCAF-1 p105 colocalized in vivo in Drosophila polytene chromosomes. This interaction between dCAF-1 and dASF1 may be a key component of the functional synergy observed between RCAF and dCAF-1 during the assembly of newly synthesized DNA into chromatin.
In the nucleus, DNA is packaged into a nucleoprotein structure known as chromatin. The basic repeating unit of chromatin, the nucleosome, consists of approximately two turns of DNA wrapped around an octamer of core histone proteins (22). The structure and dynamics of chromatin have far-ranging consequences for many nuclear processes, such as DNA replication, transcription, recombination, and repair (44).
Chromatin assembly accompanies the synthesis of DNA and is required for the growth and maintenance of cells (for reviews, see references 1, 9, 11, 16, 25, 40, and 43). It has been found that the assembly of chromatin involves the initial deposition of a heterotetramer of histones H3 and H4 onto the DNA and the subsequent incorporation of two heterodimers of histones H2A and H2B to complete the nucleosome. Chromatin assembly is mediated by factors that function to deliver the core histones to the sites of DNA synthesis, such as chromatin assembly factor 1 (CAF-1), replication-coupling assembly factor (RCAF), nucleosome assembly protein-1 (NAP-1), and nucleoplasmin, as well as by an ATP-dependent motor protein, such as ATP-utilizing chromatin assembly and remodeling factor (ACF), which catalyzes the assembly of histones into periodic nucleosome arrays.
CAF-1 was identified as a protein that participates in the assembly of newly synthesized DNA into chromatin during simian virus 40 (SV40) DNA replication in vitro (32, 36). CAF-1 binds to histones H3 and H4 (15, 33), and the protein can be isolated as a complex with histones H3 and (acetylated) H4 (42). CAF-1 also appears to be involved in the assembly of chromatin during the repair of DNA damage (7, 8, 17). In addition, the phenotypes of Saccharomyces cerevisiae lacking CAF-1 activity are consistent with a function of CAF-1 as a chromatin assembly factor (5, 6, 17, 18, 27). In Arabidopsis thaliana, CAF-1 is important for the stable maintenance of gene expression states at shoot and root apical meristems (19). CAF-1 has been found to be localized to sites of DNA replication (20, 23, 37), and a specific interaction has been observed between CAF-1 and the PCNA component of the DNA replication and repair machinery (26, 30, 45). CAF-1 has also been observed to bind to heterochromatin protein 1 and to be localized to heterochromatin (28).
In contrast to CAF-1 from yeast and humans, which consists of three subunits (15, 17), dCAF-1 preparations from Drosophila embryos contained four predominant polypeptides with apparent molecular masses of 180, 105, 75, and 55 kDa (hereafter referred to as p180, p105, p75, and p55) (14, 39). The nature of the additional subunit in dCAF-1 is not known. The p55 component of dCAF-1 is highly conserved among eukaryotes and is found in other protein complexes that are involved in chromatin remodeling, histone acetylation, and histone deacetylation (for examples, see references 24 and 39). The p180, p105, and p75 components of dCAF-1 are likely to be unique to the dCAF-1 complex, yet they remain uncharacterized.
The analysis of factors that are required in addition to CAF-1 for DNA replication-coupled chromatin assembly led to the identification of RCAF (41). RCAF comprises the Drosophila homologue of the yeast anti-silencing function 1 protein (dASF1) and histones H3 and H4 (21, 31, 41). The specific acetylation pattern of H3 and H4 in RCAF is identical to that of newly synthesized histones that are assembled onto newly replicated DNA (35, 41). RCAF functions synergistically with CAF-1 in the assembly of chromatin in DNA replication-chromatin assembly reactions. The study of yeast strains that are lacking CAF-1 and/or RCAF further suggested that CAF-1 and RCAF have both common and unique functions in the cell (41). RCAF-mediated chromatin assembly appears to be essential for normal progression through the cell cycle, gene expression, DNA replication, and DNA repair (41). Furthermore, it appears that the checkpoint kinase Rad53 may regulate the chromatin assembly function of ASF1 during DNA replication and repair (4).
In this study, we investigated the composition and function of Drosophila CAF-1 (dCAF-1). To this end, we have cloned the p180 and p105 subunits of dCAF-1, and we found that dCAF-1 p75 is encoded by the p105 gene. In addition, we have discovered and characterized a novel interaction between dCAF-1 and ASF1 that is mediated through the p105 subunit of dCAF-1. This interaction is likely to coordinate the CAF-1-dependent assembly of newly replicated DNA into chromatin with ASF1.
MATERIALS AND METHODS
Protein microsequencing of dCAF-1 p180, p105, and p75.
Native dCAF-1 was purified from nuclear extracts derived from 0- to 12-h Drosophila embryos, as described previously (14). The peak material from the glycerol gradient purification step was subjected to electrophoresis on a 10% polyacrylamide–sodium dodecyl sulfate (SDS) gel and stained with Coomassie brilliant blue G (Sigma). The p180, p105, and p75 bands were excised, and the proteins were digested with a lysylendopeptidase (Achromobacter protease I; Wako Chemicals). The resulting peptides were purified by high-performance liquid chromatography with a C18 column (Vydac) and sequenced by automated Edman degradation (Applied Biosystems). The following peptide sequence information was obtained (where X = an unidentified residue and brackets indicate a lower confidence of accura-cy): dCAF-1 p180 subunit—FVETRLPFK, GSPAPIQIK, NDQATIDLFMG(Q), LAEERRLK, DEEDDDDVQVIDYLSPAGLP(E)(I)VEQQ(K), YLHFADNRRPPYYG, SSSISARRPLAQDK, LQVLQQEFAQEMK, TQATAEANQTTLPSK, FQLPDLQLQNQWNYTLTP(K); dCAF-1 p105 subunit—EVWLTLK, LLLTPSGITDYDGVVK, PINTSYGF, (V)LXG(H)REDIYDLSXAPNSQF(L)V(S)(G)(S)XX(N)XA; dCAF-1 p75 subunit—PINTSYGFSR(H)(D)(L)(S), V(N)TEAVPPAETSQPALAVIPVFE, and VLRGHREDIYDLS(S)P.
Isolation of cDNAs that encode dCAF-1 p105 and p180.
Degenerate primers that corresponded to the expected coding sequences of the p180 and p105 peptides (and incorporated the preferred codon usage for Drosophila [2]) were used to generate partial cDNA fragments of the dCAF-1 p180 and p105 cDNAs by PCR. By screening a Drosophila embryo cDNA library (0- to 4-h embryos) in λZAPII (Stratagene) with radiolabeled cDNA fragments, we isolated eight independent full-length cDNAs for p105, six independent cDNAs that contain the majority of p180 but lack the 5′ end, and five independent cDNAs that contain the 5′ end of p180. For p105, two full-length cDNA clones were sequenced on both strands to determine the open reading frame. The p180 open reading frame was reconstructed by sequencing both strands of two overlapping cDNA clones. By in situ hybridization to Drosophila polytene chromosomes, the dCAF-1 p180 and dCAF-1 p105 loci were mapped to regions 8A1-2 and 47A7-8, respectively (T. Laverty, unpublished data).
Purification of recombinant dCAF-1.
Recombinant baculoviruses that express the p180, p105, and p55 dCAF-1 subunits were prepared with the BaculoGold system in conjunction with a pAcUW51-based baculovirus transfer vector (PharMingen). The p180-FLAG recombinant baculovirus encodes the full-length p180 protein with a carboxyl-terminal extension of DYKDDDK, which is the FLAG peptide. The p105-His6 recombinant baculovirus encodes the full-length p105 protein with a carboxyl-terminal extension of 6 His residues. The p55 recombinant baculovirus encodes the full-length p55 protein without any extraneous sequences or tags.
dCAF-1 subcomplexes that included p180-FLAG were purified by anti-FLAG (M2) affinity chromatography as follows. Sf9 cells were infected with the appropriate recombinant baculoviruses (at a multiplicity of infection of 5) for 72 h at 26°C. The cells were collected and washed with phosphate-buffered saline (3.5 mM sodium phosphate [dibasic], 1.5 mM potassium phosphate [monobasic], 137 mM NaCl, 2.7 mM KCl) and resuspended in 1 ml (per 150-mm-diameter plate of cells) of Buffer A (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.1% Nonidet P-40, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium metabisulfite, 0.5 mM benzamidine, and 10 mM 2-glycerophosphate) containing 500 mM NaCl. The cell suspension was homogenized with 20 strokes in a glass Dounce homogenizer, incubated on ice for 15 min, and subjected to centrifugation at 10,000 rpm for 10 min in a SS-34 rotor. The supernatant was diluted by the addition of an equal volume of Buffer A and incubated with 50 μl of M2 agarose resin (Sigma) for 2 h at 4°C. The resin was washed five times with 200 volumes of Buffer A containing 150 mM NaCl. Then, the purified recombinant dCAF-1 was eluted by incubation of the resin with 50 μl of Buffer A containing 150 mM NaCl and 100 μg of FLAG peptide (Sigma) per ml for 5 min on ice followed by brief microcentrifugation at 1,000 rpm. The elution process was repeated three times, and the amounts of the purified proteins were estimated by 7.5% polyacrylamide–SDS gel electrophoresis with bovine serum albumin (BSA) protein standards.
dCAF-1 subcomplexes that included p105-His6 were purified by Ni(II) affinity chromatography as follows. Sf9 cells were infected with the appropriate recombinant baculoviruses (at a multiplicity of infection of 5) for 72 h at 26°C. The cells were collected and washed with phosphate-buffered saline and resuspended in 1 ml (per 150-mm-diameter plate of cells) of Buffer A containing 500 mM NaCl and 20 mM imidazole. The cell suspension was homogenized with 20 strokes in a glass Dounce homogenizer, incubated on ice for 15 min, and subjected to centrifugation at 10,000 rpm for 10 min in an SS-34 rotor. The supernatant was diluted by the addition of an equal volume of Buffer A and incubated with 100 μl of Ni-nitrilotriacetic acid (NTA) agarose resin (Qiagen) for 2 h at 4°C. The resin was washed five times with 200 volumes of Buffer A containing 150 mM NaCl. The purified recombinant dCAF-1 was eluted by incubation of the resin with 100 μl of Buffer A containing 150 mM NaCl and 250 mM imidazole for 5 min on ice followed by brief microcentrifugation at 1,000 rpm. The elution process was repeated three times, and the amounts of the purified proteins were estimated by 7.5% polyacrylamide–SDS gel electrophoresis with BSA protein standards.
Purification of recombinant dASF1.
Escherichia coli HMS174 cells were transformed with plasmid pETdASF1-His6, which contains the entire dASF1 open reading frame (41) with a C-terminal His6 tag. The cells (500 ml) were grown to an A600 of 0.6, and the synthesis of dASF1-His6 was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside for 16 h at 14°C. The bacterial pellet was suspended in 5 ml of RCAF buffer (10 mM HEPES [K+], pH 7.6, 10% glycerol, 1 mM EDTA, 0.1% Nonidet P-40, 0.2 mM phenylmethylsulfonyl fluoride, 0.5 mM sodium metabisulfite, 0.5 mM benzamidine, 10 mM 2-glycerophosphate) containing 0.4 M NaCl and 20 mM imidazole. The cells were lysed by sonication, and the insoluble material was removed by centrifugation at 10,000 rpm for 10 min in an SS-34 rotor. The supernatant was incubated for 4 h at 4°C with 0.1 ml of Ni-NTA silica resin (Qiagen). Then, the resin was washed five times with 100 volumes of RCAF buffer containing 0.4 M NaCl and 20 mM imidazole. The purified dASF1-His6 protein was eluted by incubation of the resin with 100 μl of RCAF buffer containing 0.4 M NaCl and 250 mM imidazole for 5 min on ice followed by brief microcentrifugation at 1,000 rpm. The elution process was repeated three times, and the amounts of the purified protein were estimated by 15% polyacrylamide–SDS gel electrophoresis with BSA protein standards.
SV40 DNA replication-chromatin assembly assays.
SV40 DNA replication reactions were performed and analyzed as described previously (32, 36, 41), except that the SV40 origin-containing plasmid pSVL-ΨCRK was used instead of pSV011. [α-32P]dATP was included in the reaction mixtures to label the newly replicated DNA. The reaction mixtures were deproteinized, and the DNA species were resolved by 1% agarose gel electrophoresis. The gels were stained with ethidium bromide to visualize the bulk DNA and subjected to autoradiography to visualize the newly replicated DNA. Assembly of chromatin was observed by the generation of negative supercoils into the plasmid DNA.
Antibodies and Western blot analyses.
Polyclonal antibodies that recognize dCAF-1 p180 were raised against a purified, bacterially synthesized fragment of p180 (amino acids 756 to 915). Polyclonal antibodies that recognize both dCAF-1 p105 and p75 were raised against a purified, bacterially synthesized fragment of p105 (amino acids 440 to 539). Polyclonal antibodies that recognize dCAF-1 p105, but not p75, were raised against a purified, bacterially synthesized fragment of p105 (amino acids 610 to 709). All antibodies were affinity purified with their purified recombinant antigens prior to use. For Western blotting, protein samples were subjected to polyacrylamide-SDS gel electrophoresis and transferred to nitrocellulose membranes. The proteins were detected by using horseradish peroxidase-coupled secondary antibody and a chemiluminescence reagent (Amersham). The Western blot analysis of Drosophila proteins at different stages of development was performed as described previously (12).
Coimmunoprecipitation of native dCAF-1 and dASF1.
S190 extract (300 μl) derived from Drosophila embryos (13) was incubated with 10 μg of the appropriate preimmune sera or affinity-purified antibodies for 2 h at 4°C. A 50:50 slurry of protein A-Sepharose (30-μl volume in RCAF buffer containing 0.5 M NaCl) was added to the mixture, which was incubated for an additional 1 h at 4°C. The immobilized proteins were washed extensively with RCAF buffer containing 0.5 M NaCl and then were subjected to polyacrylamide-SDS gel electrophoresis and Western blot analysis. We additionally found that the presence of 0.5 mg of ethidium bromide/ml did not affect the results of these coimmunoprecipitation experiments (data not shown) and, thus, it is unlikely that the protein interactions are mediated through DNA.
Protein-protein interaction analyses.
dCAF-1 proteins were immobilized on anti-FLAG (M2) agarose resin or Ni-NTA resin, as described above. After washing the unbound proteins from the resin, the immobilized dCAF-1 was incubated with either 300 μl of S190 extract from Drosophila embryos (13) or 10 μg of purified recombinant dASF1-His6 for 2 h at 4°C. The immobilized proteins were washed five times with 100 volumes of RCAF buffer containing 150 mM NaCl and then subjected to polyacrylamide-SDS gel electrophoresis and staining with Coomassie blue and/or Western blot analysis.
Chromosomal localization of CAF-1 and RCAF subunits.
Chromosome spreads were prepared from salivary glands of wandering third-instar larvae and stained with polyclonal primary antibodies and fluorescein isothiocyanate (FITC)-labeled or Texas Red-labeled secondary antibodies, as previously described (3, 29, 38). In some cases, chromosomes were counterstained with propidium iodide (PI) to allow visualization of chromosome morphology and cytological mapping. For the determination of colocalization of p105 and dASF1, both of which were detected with rabbit antibodies, simultaneous immunostaining was carried out as follows. Incubation with the first primary antibody (anti-p105) and then biotinylated goat anti-rabbit secondary antibody was followed by extensive washing before incubation with the second primary antibody (anti-dASF1) and a second secondary antibody (Texas Red-labeled anti-rabbit). Finally, after an extensive final washing, incubation with avidin-FITC was carried out to allow detection of p105 with minimal photobleaching from the extended processing time of the procedure. To control for the undesired possibility that the second secondary antibody might detect residual first primary antibody that remained unbound by the first secondary, the entire procedure was also carried out in parallel in the absence of the second primary antibody. No red signal from the second secondary antibody was detected in these control experiments. Images were obtained with a Zeiss/Bio-Rad confocal microscope as previously described (29).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the p180 and p105 cDNAs are AF367177 and AF367178, respectively.
RESULTS AND DISCUSSION
Cloning of the p180 and p105 subunits of dCAF-1.
To gain a better understanding of the function of dCAF-1, we isolated the cDNAs that encode its p180 and p105 subunits. To this end, we purified the dCAF-1 complex from Drosophila embryos and obtained partial amino acid sequences of several peptides of each subunit by protein microsequencing. With the amino acid sequence data, we used reverse transcription-PCR techniques to generate fragments of the p180 and p105 cDNAs. These cDNA fragments were then used to isolate full-length cDNAs for p180 and p105.
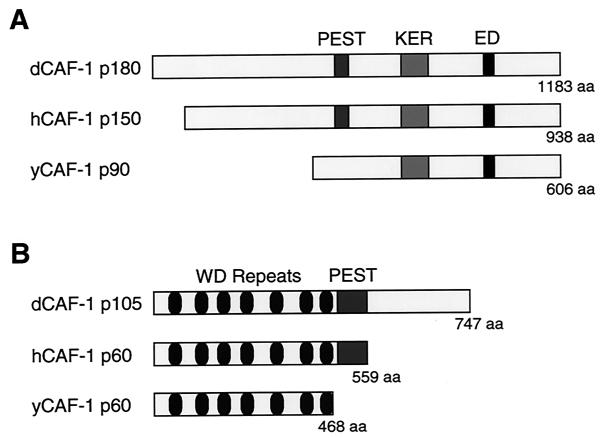
The dCAF-1 p180 cDNA encodes a polypeptide of 1,183 amino acid residues that includes 12 peptide sequences that were obtained from the p180 protein (Fig. 1A). Like its human counterpart, dCAF-1 p180 possesses a PEST degradation consensus sequence (amino acid residues 392 to 403). In addition, dCAF-1 p180 has an acidic C terminus that includes several repetitive stretches of charged amino acids that are referred to as the KER and ED regions (15). The dCAF-1 p180 protein is encoded by a single gene, as determined by Southern blot analysis, hybridization to polytene chromosomes, and analysis of the Drosophila genome sequence. By Western blot analysis, we observed that the dCAF-1 p180 protein is present throughout Drosophila development (data not shown).
FIG. 1.
The p180 and p105 subunits of dCAF-1 share homology with components of human and yeast CAF-1. (A) Schematic diagrams of the predicted open reading frames of the largest CAF-1 subunits from Drosophila, humans, and yeast. There is approximately 26% amino acid identity between dCAF-1 p180 and hCAF-1 p150 (15) as well as between dCAF-1 p180 and yCAF-1 p90 (17). (B) Schematic diagrams of the predicted open reading frames of the middle CAF-1 subunits from Drosophila, humans, and yeast. dCAF-1 p105 exhibits approximately 38% identity with hCAF-1 p60 (15) and about 32% identity with yCAF-1 p60 (17).
The dCAF-1 p105 cDNA encodes a protein of 747 amino acids (Fig. 1B). The N-terminal portion of the dCAF-1 p105 polypeptide comprises seven WD-repeat sequences that are likely to fold into a β-propeller structure (34). dCAF-1 p105 has two PEST protein degradation consensus sequences (amino acid residues 443 to 466). The presence of PEST consensus sequences in the dCAF-1 p180 and p105 proteins suggests that there is regulation of the levels of dCAF-1 in the cell.
The p105 and p75 subunits of Drosophila CAF-1 are closely related.
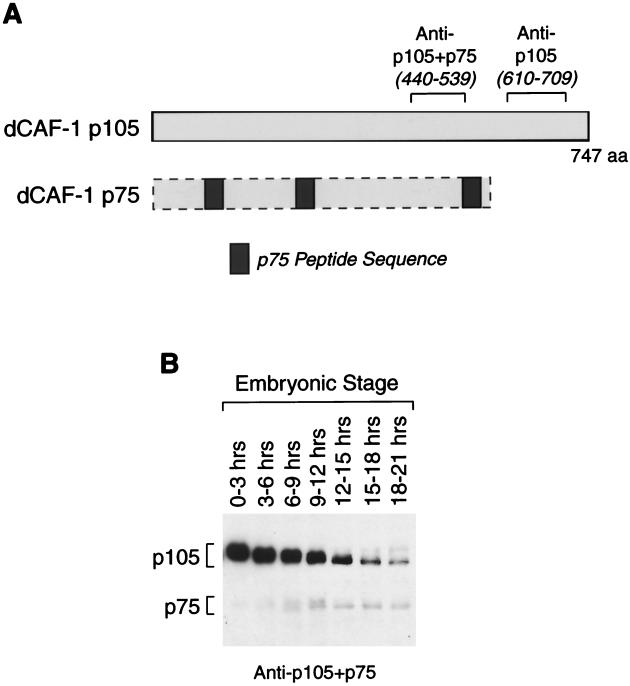
We also sought to determine the identity of the p75 subunit of dCAF-1. The available evidence suggests that p75 is an N-terminal fragment of p105. First, we carried out protein microsequencing of p75 and found that the amino acid sequences of three p75 peptides were nearly identical to regions in the predicted amino acid sequence of p105 (Fig. 2A). Second, polyclonal antibodies against amino acid residues 440 to 539 of p105 strongly cross-reacted with both p105 and p75 proteins, whereas polyclonal antibodies against amino acid residues 610 to 709 of p105 were able to recognize p105 but not p75 (Fig. 2A; also see Fig. 3). Third, exhaustive screening of Drosophila cDNA libraries with DNA probes or anti-p105+p75 antibodies yielded only the p105 cDNA. Lastly, analysis of the Drosophila genome sequence revealed that the dCAF-1 p105 gene is the only known Drosophila gene that is capable of encoding the p75-derived peptides. Thus, it appears that the p75 protein is encoded by the p105 gene.
FIG. 2.
The dCAF-1 p75 subunit appears to be related to dCAF-1 p105. (A) Schematic showing the apparent relationship between the p105 and p75 proteins. The positions of peptide sequences that were identified by microsequencing of p75 are indicated by the gray boxes and are shown relative to the positions of the corresponding sequences in the p105 open reading frame. The regions of the p105 protein that were used as antigens to raise the dCAF-1 p105+p75 and dCAF-1 p105 antisera are also indicated. (B) Western blot analysis of the p105 and p75 proteins during Drosophila embryogenesis. The relative intensities of the p105 bands versus the p75 bands do not reflect their abundance in vivo.
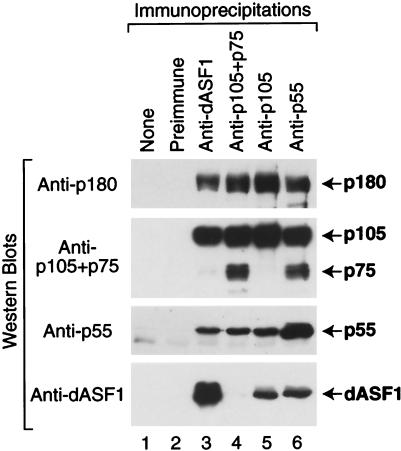
FIG. 3.
Coimmunoprecipitation analysis of native dCAF-1 and dASF1. The antibodies indicated above each lane were used to immunoprecipitate proteins from a crude Drosophila embryo extract. The resulting immunoprecipitates were then subjected to Western blot analysis with the antibodies indicated at the left.
Analysis of the developmental expression of the dCAF-1 p105 and p75 proteins indicated that both p105 and p75 are present throughout Drosophila embryogenesis (Fig. 2B). The dCAF-1 p105 protein is most abundant in early embryogenesis and has an expression pattern that is similar to that of the dCAF-1 p55, dCAF-1 p180, ACF, nucleoplasmin, and NAP-1 chromatin assembly proteins (10, 12, 39). In contrast to the dCAF-1 p105 protein, the dCAF-1 p75 protein is most abundant from 9 to 21 h after egg deposition (Fig. 2B). The time lag between the peak of the p105 protein and the appearance of the p75 protein raises the possibility that the p75 protein is derived from the p105 protein via posttranslational processing. Furthermore, expression of the dCAF-1 p105 cDNA in rabbit reticulocyte lysates or in Sf9 cells with baculovirus vectors yields p105 but not p75. Therefore, the generation of the dCAF-1 p75 protein from the p105 gene may be a consequence of specific events that occur during Drosophila embryogenesis. We also performed Northern blot analyses of poly(A)+ RNA from embryos at different times throughout embryogenesis and observed only a single p105 mRNA species (data not shown). Thus, there was no apparent alternate splicing of the p105 transcript.
It is also appropriate to consider the formal possibility that the p75 protein is generated by proteolysis of p105 subsequent to lysis of the embryos. As seen in Fig. 2B, however, when extracts are prepared by lysis of whole embryos directly into SDS sample buffer followed immediately by boiling prior to application to the gel, there is only a trace of p75 observed in early embryos (such as 0 to 3 h or 3 to 6 h after egg deposition) and, thus, there is almost no detectable postlysis conversion of p105 into p75 under these conditions. Then, in contrast with older embryos under otherwise identical conditions, we do observe significant levels of p75. Based on these results, it appears likely that the ratio of p105 to p75 species in the lysates reflects the distribution of these proteins in the embryos.
The p180, p105, and p55 proteins comprise a distinct form of the dCAF-1 complex.
dCAF-1 from Drosophila embryos consists of four polypeptides (p180, p105, p75, and p55), whereas CAF-1 from a human cell line (293 cells) or S. cerevisiae consists of three polypeptides. As noted above, the primary amino acid sequences of these polypeptides suggest that the largest and smallest polypeptides are homologous, whereas dCAF-1 p105 and p75 are homologous to the middle-sized polypeptides of yeast and human CAF-1. We therefore sought to determine whether dCAF-1 exists as a single four-polypeptide complex or as multiple smaller (e.g., three subunits) complexes.
To address this question, we performed coimmunoprecipitation analyses from Drosophila embryo extracts (Fig. 3). These experiments revealed that all four subunits of dCAF-1 (p180, p105, p75, and p55) were coimmunoprecipitated by antibodies against p55. Similarly, all four subunits of dCAF-1 were coimmunoprecipitated by the anti-p105+p75 antibodies. In contrast, p180, p105, and p55, but not p75, were immunoprecipitated by the anti-p105 antibodies, which recognize p105 but not p75. These results indicate that p105 and p75 are not present in the same complex. It thus appears that there is a distinct dCAF-1 complex that comprises p180, p105, and p55.
Due to the lack of p75-specific reagents, we have not been able to test directly the existence of a dCAF-1 complex that comprises p180, p75, and p55. It is likely, however, that such a complex exists, because p75 is closely related to p105, p75 copurifies with the other dCAF-1 subunits through multiple purification steps (14), and p75 coimmunoprecipitates with the antibodies against p55 (Fig. 3, lane 6). Hence, these findings suggest that there are two distinct forms of dCAF-1 in Drosophila embryos that consist of p180+p105+p55 and p180+p75+p55 proteins.
The dCAF-1 p180 subunit interacts with the p105 and p55 subunits.
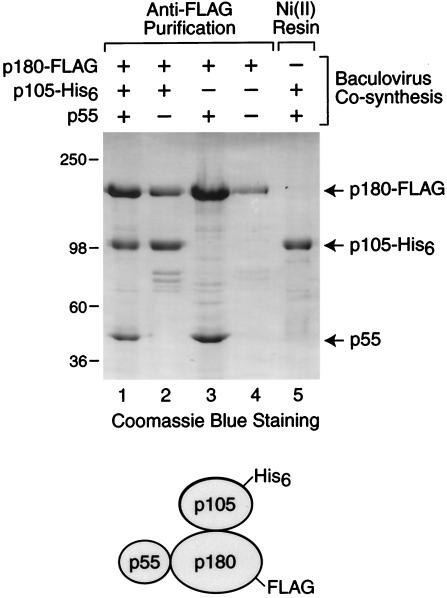
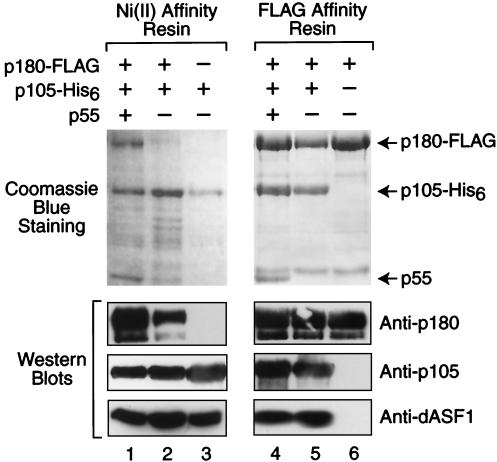
To analyze the biochemical properties of dCAF-1, we synthesized the p180, p105, and p55 proteins in Sf9 cells by using baculovirus expression vectors. The p180 subunit contained a C-terminal FLAG epitope tag and was thus designated as p180-FLAG. The p105 subunit contained a C-terminal His6 tag and was therefore termed p105-His6. As shown in Fig. 4, different combinations of dCAF-1 subunits were synthesized and purified by either anti-FLAG or Ni(II) affinity chromatography. When p180-FLAG, p105-His6, and p55 were cosynthesized and subjected to anti-FLAG immunoaffinity chromatography, the purified p180+p105+p55 dCAF-1 complex was obtained. Similarly, cosynthesis of p180-FLAG with either p105-His6 or p55 yielded p180+p105 and p180+p55 subcomplexes. Although the three-subunit p180+p105+p55 complex can be purified by Ni(II) affinity chromatography via p105-His6 (see, for example, Fig. 7), cosynthesis of p105-His6 and p55 and subsequent Ni(II) affinity chromatography yielded only p105. Hence, these findings indicate that dCAF-1 p180 interacts with both p105 and p55, but that p105 and p55 do not interact with one another, as depicted at the bottom of Fig. 4.
FIG. 4.
The p180 subunit of dCAF-1 interacts with the p55 and p105 subunits. Sf9 cells were coinfected with the indicated recombinant baculoviruses, and the proteins were purified by either anti-FLAG or Ni(II) affinity chromatography. The resulting protein preparations were subjected to SDS-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue R-250. The diagram at the bottom of the figure depicts the arrangement of the dCAF-1 subunits, as deduced from these experiments.
FIG. 7.
The p105 subunit of dCAF-1 interacts with dASF1. Purified recombinant dCAF-1 subunits, as indicated, were immobilized onto either a Ni-NTA resin via the p105-His6 subunit (left panel) or an anti-FLAG M2 affinity resin via the p180-FLAG subunit (right panel). The resins were each incubated with a crude Drosophila embryo extract, washed to remove nonspecifically bound proteins, and analyzed by SDS-polyacrylamide gel electrophoresis and staining with Coomassie blue (upper panels). The presence of dCAF-1 p180, dCAF-1 p105, and dASF1 in these protein preparations was detected by Western blot analysis (lower panels).
The p105 and p180 subunits are essential for dCAF-1-mediated chromatin assembly.
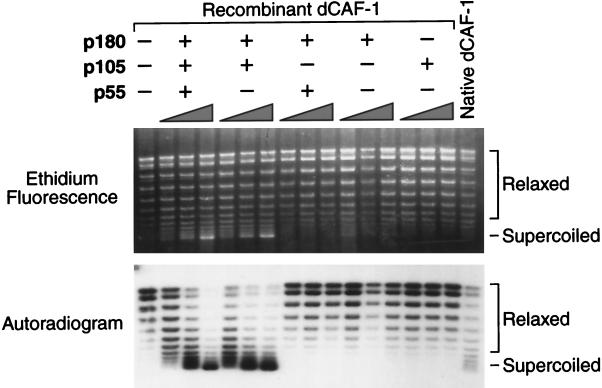
To test whether the p180, p105, and p55 subunits are required for chromatin assembly, we performed DNA replication-chromatin assembly reactions with partial and complete (i.e., p180+p105+p55) dCAF-1 complexes (Fig. 5). These experiments revealed that the purified recombinant p180+p105+p55 dCAF-1 complex possesses a specific activity for DNA replication-coupled chromatin assembly that is comparable to that of native dCAF-1, as demonstrated by plasmid supercoiling analysis (Fig. 5). We have further confirmed that dCAF-1-mediated plasmid supercoiling was a consequence of chromatin assembly by using micrococcal nuclease digestion analysis (data not shown). In addition, the two-subunit p180+p105 subcomplex is fully active for chromatin assembly. In contrast, neither the p180 subunit alone nor the p105 subunit alone is sufficient for chromatin assembly. These results thus indicate that the p180 and p105 subunits are each essential for DNA replication-coupled chromatin assembly by dCAF-1.
FIG. 5.
The p180 and p105 subunits of dCAF-1 are essential for the assembly of newly replicated DNA into chromatin. DNA replication-chromatin assembly reactions were performed in the presence or absence of purified recombinant dCAF-1 proteins, as indicated, and the resulting DNA products were resolved by agarose gel electrophoresis. The upper panel shows the total DNA, as visualized by ethidium bromide staining, whereas the lower panel shows the newly replicated DNA, as detected by autoradiography of the same gel (the newly replicated DNA was radiolabeled by the inclusion of [α-32P]dATP in the replication reaction medium). The positions of relaxed and supercoiled DNA species are shown. The supercoiling of the DNA indicates chromatin assembly.
It is relevant that the DNA replication extract used in these experiments contains significant amounts of hCAF-1 p60 and hCAF-1 p48 (also known as RbAp48) (15), which are homologous to dCAF-1 p105 and dCAF-1 p55, respectively. Based on the requirement of dCAF-1 p105 for chromatin assembly, it appears that the hCAF-1 p60 subunit cannot function with the Drosophila CAF-1 polypeptides. On the other hand, the lack of a requirement for dCAF-1 p55 may be due to the ability of the hCAF-1 p48 subunit, which is about 87% identical to dCAF-1 p55 (39), to function with the dCAF-1 p180 and p105 subunits in lieu of dCAF-1 p55. It is also possible, however, that the dCAF-1 p180+p105 subcomplex has the intrinsic ability to mediate chromatin assembly. We have not been able to immunodeplete the hCAF-1 p48 protein from the DNA replication extract to differentiate between these possibilities. It is noteworthy, however, that the Arabidopsis equivalent of dCAF-1 p55 is required for DNA replication-coupled chromatin assembly with the same assay (19).
dCAF-1 p55, p105, and p180 are associated with chromatin in vivo.
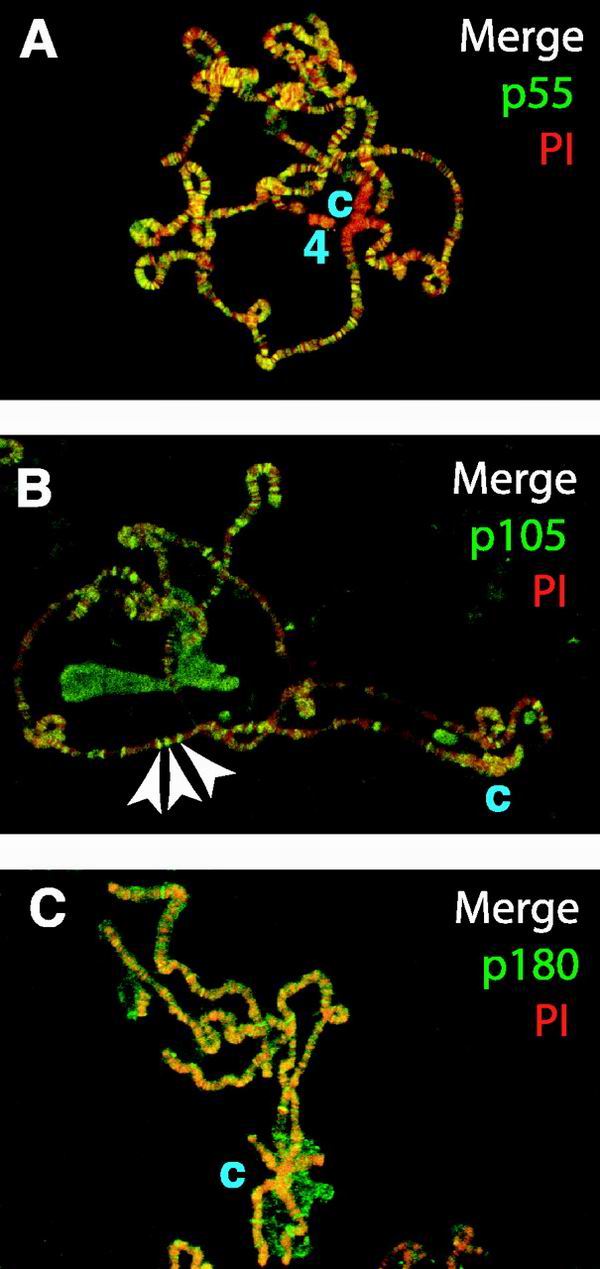
To observe interactions between dCAF-1 and native chromatin, we used immunofluorescence microscopy to examine the distribution of the dCAF-1 subunits on salivary gland polytene chromosomes from third-instar Drosophila larvae (Fig. 6). As noted previously (24), the dCAF-1 p55 protein is broadly localized over the Drosophila genome (Fig. 6A). In addition, it appears that p55 is largely excluded from the heterochromatic chromosome 4 and the chromocenter. In contrast to p55, neither p105 nor p180 is excluded from the heterochromatic chromocenter (Fig. 6B and C). The p180 protein is distributed somewhat generally throughout the polytene chromosomes (Fig. 6C). In contrast, p105 has a more specific pattern of staining that correlates with the counterstaining of DNA with PI (Fig. 6B). (Note that the anti-p105+p75 antibodies yielded a staining pattern that is similar to that obtained with anti-p105 [data not shown].) The staining of p180 and p105, but not p55, to heterochromatin as well as the staining of p105 and p75, but not p55, to distinct foci in chromatin suggest that there are functions of p180, p105, and p75 that do not involve the p55 protein. Note, however, that the polytene chromosomes are not actively undergoing DNA replication. Thus, the observed association of dCAF-1 subunits to chromatin may be due to the prior role of dCAF-1 in chromatin assembly during S phase, or to a function of dCAF-1 outside of S phase.
FIG. 6.
Localization of dCAF-1 subunits to Drosophila polytene chromosomes. Indirect immunofluorescent staining of dCAF-1 polypeptides was performed on Drosophila polytene chromosomes with an anti-rabbit FITC secondary antibody (green). PI counterstaining is shown in red. Yellow indicates coincidence of FITC and PI. The letter c specifies the location of the chromocenter. For all of the dCAF-1 subunits, protein A-purified antibodies and affinity-purified antibodies generated apparently identical polytene localization results. (A) dCAF-1 p55 protein. Chromosome 4 is indicated as “4.” (B) dCAF-1 p105 protein. A few representative bands that stain intensely for both p105 protein and DNA are denoted by arrows. (C) dCAF-1 p180 protein.
dCAF-1 interacts with the ASF1 component of RCAF chromatin assembly factor.
We previously observed that the assembly of newly replicated DNA into chromatin requires both dCAF-1 and the RCAF chromatin assembly factor, which comprises Drosophila ASF1 (dASF1) and specifically acetylated histones H3 and H4 (41). To investigate this effect further, we performed coimmunoprecipitation analyses with a crude Drosophila embryo extract (Fig. 3). In these experiments, we observed that immunoprecipitation with anti-dASF1 results in the coimmunoprecipitation of dCAF1 p180, p105, and p55, but not dCAF-1 p75. Conversely, we found that immunoprecipitation with anti-p105 or with anti-p55 results in the coimmunoprecipitation of dASF1. Thus, these findings indicate that native dASF1 interacts with the native p180+p105+p55 form of dCAF-1 but not with the p75-containing form of dCAF-1. We also found that immunoprecipitation of dCAF-1 with anti-p105+p75 did not result in the coimmunoprecipitation of dASF1, which suggests that the anti-p105+p75 antibodies destabilize the interaction between dASF1 and the p180+p105+p55 form of dCAF-1.
The p105 subunit of dCAF-1 mediates the interaction with ASF1.
To characterize further the interaction between dCAF-1 and dASF1, we sought to identify the component of dCAF-1 that mediates its interaction with dASF1. To this end, we purified different recombinant dCAF-1 complexes and subcomplexes by anti-FLAG or Ni(II) affinity chromatography and then incubated the immobilized proteins with a crude Drosophila embryo extract. The resulting bound proteins were washed, and the presence of native dASF1 (from the extract) associated with the immobilized recombinant dCAF-1 proteins was detected by Western blot analysis. These experiments revealed that dCAF-1 p105 alone can bind to dASF1 and that the binding of dASF1 to dCAF-1 proteins does not occur in the absence of p105 (Fig. 7). In addition, the apparent destabilization of the interaction between dCAF-1 and dASF1 with the anti-p105+p75 antibodies (Fig. 3, lane 4) further indicates a key role for p105 in the binding of dCAF-1 to dASF1. Thus, based on these data, we conclude that the dCAF-1 p105 subunit mediates the interaction between dCAF-1 and dASF1.
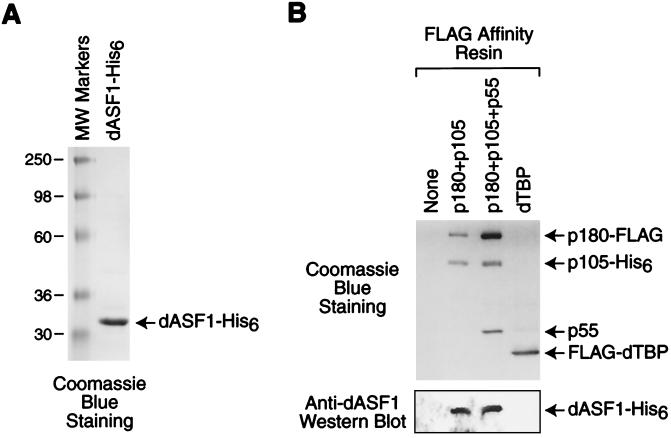
We next sought to test whether the interaction between ASF1 and dCAF-1 is direct or mediated by other proteins in the crude Drosophila extracts that were used as the source for native dASF1. To this end, we synthesized recombinant dASF1-His6 in bacteria and purified the protein by Ni(II) affinity chromatography (Fig. 8A). We then incubated the purified dASF1-His6 with immobilized dCAF-1 complexes, washed the complexes, and then detected the presence of bound dASF1-His6 by Western blot analysis. These experiments revealed that dASF1 binds to dCAF-1 as well as to a p180+p105 subcomplex, but not to a TATA-binding protein (TBP) control protein (Fig. 8B). These results therefore suggest that the interaction between dASF1 and dCAF-1 is direct.
FIG. 8.
dASF1 binds directly to dCAF-1. (A) Purification of recombinant dASF1. The protein was analyzed by SDS-polyacrylamide gel electrophoresis and staining with Coomassie blue. (B) Purified recombinant dASF1, as shown in panel A, was incubated with purified, immobilized dCAF-1 proteins or dTBP (as a control for nonspecific binding), as indicated (upper panel). The resins were washed, and the remaining proteins were subjected to Western blot analysis with anti-dASF1 (bottom panel).
Colocalization of dCAF-1 p105 and dASF1 in Drosophila polytene chromosomes.
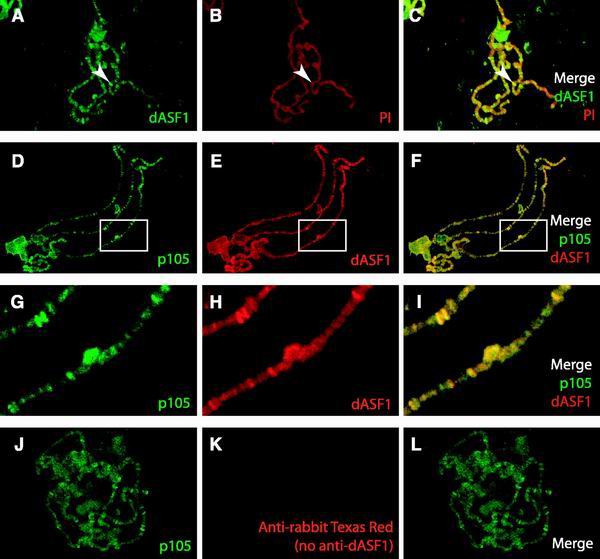
To test whether there is an interaction between dCAF-1 and dASF1 in vivo, we compared the localization of dCAF-1 and dASF1 in polytene chromosomes (Fig. 9). Immunolocalization of the dASF1 protein demonstrated that dASF1 is broadly localized over the chromosomes (Fig. 9A to C). The pattern of ASF1 localization closely follows the DNA staining, and many intense bands of ASF1 protein are present that colocalize with bands that counterstain strongly with PI. This pattern of dASF1 localization is similar to that of the dCAF-1 p105 protein (Fig. 6B). Accordingly, simultaneous immunolocalization of dCAF-1 p105 and dASF1 demonstrates that dCAF-1 and ASF1 are colocalized on Drosophila polytene chromosomes (Fig. 9D to F). The coincidence of the p105 and ASF1 proteins is particularly apparent in the magnified view of the polytene chromosomes (Fig. 9G to I). As a control, we additionally showed that this coincident staining of the p105 and dASF1 proteins is not due to cross-reaction of the anti-rabbit Texas Red secondary antibody with the anti-p105 rabbit primary antibodies (Fig. 9J to L). Like p105, dASF1 is not excluded from the heterochromatic regions of the Drosophila polytene chromosomes. Based on these observations, we thus conclude that dASF1 and dCAF-1 p105 interact in vivo.
FIG. 9.
Colocalization of dCAF-1 p105 and dASF1 in polytene chromosomes. (A) Indirect immunofluorescent staining of dASF1 with an anti-rabbit FITC secondary antibody (green). (B) PI counterstaining (red) of the chromosomes in panel A. (C) Merge of the images in panels A and B. Yellow indicates coincidence of DNA and dASF1. A representative region of intense, coincident staining of dASF1 protein and DNA is denoted by an arrow. (D) Indirect immunofluorescent staining of dCAF-1 p105 protein with FITC. The methodology used for the results shown in panels D to L is described in Materials and Methods. (E) Indirect immunofluorescent staining with Texas Red of dASF1 in the same chromosomes shown in panel D. (F) Merge of the images in panels D and E, which shows the extent of p105 and dASF1 colocalization. (G, H, and I) Enlargements of the regions indicated by the white boxes in panels D, E, and F, respectively. (J, K, and L) To test whether there is binding of the second secondary antibody (Texas Red-labeled anti-rabbit) to the first primary antibody (anti-p105), the entire staining procedure, such as that in panels D, E, and F, was performed in parallel in the absence of the second primary antibody (anti-dASF1). As shown in panels K and L, no red signal from the second secondary antibody (anti-rabbit Texas Red) was detected in these control experiments.
Summary and perspectives.
In this study, we have described the isolation of the cDNAs encoding the p180 and p105 subunits of dCAF-1. We found that the p75 subunit of dCAF-1 appears to be a C-terminally truncated form of p105 and that there are distinct forms of dCAF-1 that contain either the p105 subunit or the p75 subunit. The p105-containing form of dCAF-1 comprises the p180, p105, and p55 proteins. The purified recombinant p180+p105+p55 dCAF-1 complex is as active for DNA replication-coupled chromatin assembly as native dCAF-1. Both the p180 and p105 subunits are essential for chromatin assembly. We have discovered a preexisting interaction between dCAF-1 and the dASF1 chromatin assembly factor in crude extracts. This dCAF-1-ASF1 interaction occurs via the dCAF-1 p105 subunit, and this interaction appears to be direct. We additionally observed that dASF1 and dCAF-1 p105 colocalize in vivo in Drosophila polytene chromosomes. These results suggest that there is physical cooperation between dCAF-1 and dASF1 during chromatin assembly.
CAF-1 from S. cerevisiae and from a human cultured cell line (293 cells) consists of three polypeptides (15, 17, 32, 42), whereas dCAF-1 isolated from Drosophila embryos comprises four polypeptides (14, 39). In this study, we have found that the p105 and p75 subunits of dCAF-1 are closely related, and that dCAF-1 is not a single four-subunit complex but rather a three-subunit p180+p105+p55 complex and a presumed p180+p75+p55 complex. Thus, the basic three-subunit structure of CAF-1 is conserved among yeast, Drosophila, and humans. The presence of multiple forms of dCAF-1 is of particular interest. Because dCAF-1 was isolated from whole embryos instead of a specific cell line, there is potential for considerable diversity in the range of functions that may be performed by the different forms of dCAF-1. It is possible, for instance, that the p105-containing form of dCAF-1 functions in ASF1-dependent processes, whereas the p75-containing form of dCAF-1 may function in ASF1-independent processes. Alternatively, the activity of dCAF-1 may be regulated during embryogenesis by processing the p105 polypeptide into p75.
This physical interaction between dCAF-1 and dASF1 may be a key component of the functional synergy observed between RCAF and dCAF-1 during the assembly of newly synthesized DNA into chromatin (41). The coupling of DNA synthesis and chromatin assembly appears to require a specific interaction between CAF-1 and PCNA (26, 30, 45). The results presented in this work further extend this model to include the binding of ASF1 to CAF-1. It is possible, for instance, that a complex of RCAF and CAF-1 is recruited to sites of DNA synthesis via the interaction of CAF-1 with PCNA. In the future, it will be interesting to study how RCAF and CAF-1 mediate the formation of nucleosomes in conjunction with the other components of the chromatin assembly machinery.
ACKNOWLEDGMENTS
We thank Dmitry Fyodorov, Jennifer Butler, Vassili Alexiadis, Buyung Santoso, Mark Levenstein, and Tom Boulay for critical reading of the manuscript. We are grateful to Todd Laverty and Gerry Rubin for determination of the cytological location of the dCAF-1 p180 and p105 genes. Tara Dobson provided excellent technical assistance.
This work was supported by a grant from the National Institutes of Health (GM58272) to J.T.K. J. K. Tyler was supported by a Leukemia and Lymphoma Society Special Fellowship and a Howard Hughes Institutional Award from the University of Colorado.
REFERENCES
- 1.Adams C R, Kamakaka R T. Chromatin assembly: biochemical identities and genetic redundancy. Curr Opin Genet Dev. 1999;9:185–190. doi: 10.1016/S0959-437X(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 2.Aota S, Gojobori T, Ishibashi F, Maruyama T, Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1988;16(Suppl.):R315–R402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinwalla V, Jane E P, Harte P J. The Drosophila trithorax protein binds to specific chromosomal sites and is co-localized with Polycomb at many sites. EMBO J. 1995;14:2056–2065. doi: 10.1002/j.1460-2075.1995.tb07197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emili A, Schieltz D M, Yates III J R, Hartwell L H. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 5.Enomoto S, McCune-Zierath P D, Gerami-Nejad M, Sanders M A, Berman J. RLF2, a subunit of yeast chromatin assembly factor-I, is required for telomeric chromatin function in vivo. Genes Dev. 1997;11:358–370. doi: 10.1101/gad.11.3.358. [DOI] [PubMed] [Google Scholar]
- 6.Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaillard P-H, Martini E M, Kaufman P D, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- 8.Game J C, Kaufman P D. Role of Saccharomyces cerevisiae chromatin assembly factor-I in repair of ultraviolet radiation damage in vivo. Genetics. 1999;151:485–497. doi: 10.1093/genetics/151.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Bulger M, Kobayashi R, Kadonaga J T. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly-spaced nucleosomal arrays. Mol Cell Biol. 1996;16:3112–3124. doi: 10.1128/mcb.16.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Tyler J K, Kadonaga J T. Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells. 1997;2:593–600. doi: 10.1046/j.1365-2443.1997.1500348.x. [DOI] [PubMed] [Google Scholar]
- 12.Ito T, Levenstein M E, Fyodorov D V, Kutach A K, Kobayashi R, Kadonaga J T. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamakaka R T, Bulger M, Kadonaga J T. Potentiation of RNA polymerase II transcription by Gal4-VP16 during but not after DNA replication and chromatin assembly. Genes Dev. 1993;7:1779–1795. doi: 10.1101/gad.7.9.1779. [DOI] [PubMed] [Google Scholar]
- 14.Kamakaka R T, Bulger M, Kaufman P D, Stillman B, Kadonaga J T. Post-replicative chromatin assembly by Drosophila and human chromatin assembly factor-1. Mol Cell Biol. 1996;16:810–817. doi: 10.1128/mcb.16.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman P D, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman P D. Nucleosome assembly: the CAF and the HAT. Curr Opin Cell Biol. 1996;8:369–373. doi: 10.1016/s0955-0674(96)80012-3. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman P D, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman P D, Cohen J L, Osley M A. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaya H, Shibahara, Taoka K-I, Iwabuchi M, Stillman B, Araki T. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell. 2001;104:131–142. doi: 10.1016/s0092-8674(01)00197-0. [DOI] [PubMed] [Google Scholar]
- 20.Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- 21.Le S, Davis C, Konopka J B, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Luger K, Mäder A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 23.Marheineke K, Krude T. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J Biol Chem. 1998;273:15279–15286. doi: 10.1074/jbc.273.24.15279. [DOI] [PubMed] [Google Scholar]
- 24.Martínez-Balbás M A, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello J A, Almounzi G. The ins and outs of nucleosome assembly. Curr Opin Genet Dev. 2001;11:136–141. doi: 10.1016/s0959-437x(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 26.Moggs J G, Grandi P, Quivy J-P, Jónsson Z O, Hübscher U, Becker P B, Almouzni G. A CAF-1–PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monson E K, de Bruin D, Zakian V A. The yeast Cac1 protein is required for the stable inheritance of transcriptionally repressed chromatin at telomeres. Proc Natl Acad Sci USA. 1997;94:13081–13086. doi: 10.1073/pnas.94.24.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 29.Orlando V, Jane E P, Chinwalla V, Harte P J, Paro R. Binding of trithorax and Polycomb proteins to the bithorax complex: dynamic changes during early Drosophila embryogenesis. EMBO J. 1998;17:5141–5150. doi: 10.1093/emboj/17.17.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- 31.Singer M S, Kahana A, Wolf A J, Meisinger L L, Peterson S E, Goggin C, Mahowald M, Gottschling D E. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150:613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 33.Smith S, Stillman B. Stepwise assembly of chromatin during DNA replication in vitro. EMBO J. 1991;10:971–980. doi: 10.1002/j.1460-2075.1991.tb08031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith T F, Gaitatzes C, Saxena K, Neer E J. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 35.Sobel R E, Cook R G, Perry C A, Annunziato A T, Allis C D. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 37.Taddei A, Roche D, Sibarita J-B, Turner B M, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tie F, Furuyama J T, Prasad-Sinha J E, Harte P J. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development. 2001;128:275–286. doi: 10.1242/dev.128.2.275. [DOI] [PubMed] [Google Scholar]
- 39.Tyler J K, Bulger M, Kamakaka R T, Kobayashi R, Kadonaga J T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol Cell Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyler J K, Kadonaga J T. The “dark side”of chromatin remodeling: repressive effects on transcription. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 41.Tyler J K, Adams C R, Chen S-R, Kobayashi R, Kamakaka R T, Kadonaga J T. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 42.Verreault A, Kaufman P D, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- 43.Verreault A. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 2000;14:1430–1438. [PubMed] [Google Scholar]
- 44.Wolffe A P. Chromatin: structure and function. 3rd ed. San Diego, Calif: Academic Press; 1998. [Google Scholar]
- 45.Zhang Z, Shibahara K, Stillman B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature. 2000;408:221–225. doi: 10.1038/35041601. [DOI] [PubMed] [Google Scholar]