Abstract
Background
Immunotherapy for malignant tumors has made great progress, but many patients do not benefit from it. The complex intratumoral heterogeneity (ITH) hindered the in-depth exploration of immunotherapy. Conventional bulk sequencing has masked intratumor complexity, preventing a more detailed discovery of the impact of ITH on treatment efficacy. Hence, we initiated this study to explore ITH at the multi-omics spatial level and to seek prognostic biomarkers of immunotherapy efficacy considering the presence of ITH.
Methods
Using the segmentation strategy of digital spatial profiling (DSP), we obtained differential information on tumor and stromal regions at the proteomic and transcriptomic levels. Based on the consideration of ITH, signatures constructed by candidate proteins in different regions were used to predict the efficacy of immunotherapy.
Results
Eighteen patients treated with a bispecific antibody (bsAb)-KN046 were enrolled in this study. The tumor and stromal areas of the same samples exhibited distinct features. Signatures consisting of 11 and 18 differentially expressed DSP markers from the tumor and stromal areas, respectively, were associated with treatment response. Furthermore, the spatially resolved signature identified from the stromal areas showed greater predictive power for bsAb immunotherapy response (area under the curve=0.838). Subsequently, our stromal signature was validated in an independent cohort of patients with non-small cell lung cancer undergoing immunotherapy.
Conclusion
We deciphered ITH at the spatial level and demonstrated for the first time that genetic information in the stromal region can better predict the efficacy of bsAb treatment.
Trial registration number
Keywords: biomarkers, tumor; immunotherapy; lung neoplasms
WHAT IS ALREADY KNOWN ON THIS TOPIC
The presence of heterogeneity within the tumor can affect the efficacy of immunotherapy and confuse prognostic biomarkers of cancer, but the specific mechanism is still unknown.
WHAT THIS STUDY ADDS
In this study, spatial segmentation through digital spatial profiling (DSP) was used to discover intratumoral heterogeneity (ITH) in advanced non-small cell lung cancer and enabled us to distinguish the efficacy-related biomarkers that from tumor or stromal regions. After segmentation, the stromal regions showed more genetic information, which can better predict the efficacy of immunotherapy.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Through DSP, we fully considered the ITH and overcame the limitations of previous bulk sequencing. This study provided 18 protein candidates as the efficacy predictive biomarkers of bispecific antibody immunotherapy targeting programmed death ligand-1 and cytotoxic T lymphocyte-associated antigen-4.
Background
A tumor can be conceived as an ecosystem in which tumor cells are interconnected with other cells such as immune cells in the tumor microenvironment (TME).1 Intratumoral heterogeneity (ITH) has a significant effect on the efficacy of various immunotherapies and confuses clinically prognostic biomarkers; thus, understanding ITH is the key to measuring clinical efficacy and developing new targets for cancer immunotherapy.2–4 Conventional bulk RNA or protein sequencing was largely limited to process thousands of cells at a time and get an average level of variation, which masked the intratumor complexity.4 5 It is difficult to distinguish whether the identified efficacy-related biomarkers are from tumor or stromal cells.
Immunotherapy has changed the treatment landscape of advanced non-small cell lung cancer (aNSCLC) and markedly increased overall survival (OS). However, immunotherapy is still confronted with many challenges, particularly in overcoming drug resistance and lack of accurate predictive biomarkers.6–10 With the development of precise tumor therapy, treatment using multitarget antitumor drugs takes on a development direction.11 12 KN046 is a bispecific antibody (bsAb) from Alphamab Oncology targeting programmed death ligand-1 (PD-L1) and cytotoxic T lymphocyte-associated antigen-4 (CTLA-4). The safety and clinical effectiveness of KN046 in patients with aNSCLC were evaluated in a phase II multicenter clinical trial. The median progression-free survival (mPFS) showed a beneficial advantage compared with the historical data of anti-PD-1/PD-L1 monotherapy.13 Nevertheless, not all patients with aNSCLC could benefit from KN046. The existence of ITH and its impact on the efficacy of bsAB remains unknown.
To explore ITH at the multi-omics spatial level in patients with aNSCLC, we used the GeoMx digital spatial profiling (DSP) technology (NanoString Biotechnology), combined with next-generation sequencing (NGS) and Nanostring nCounter Analysis System, to quantify RNAs and proteins in both tumor and stomal areas from spatially distinct regions.14 15 We also conducted a multiomics analysis in patients from different efficacy groups treated with KN046 to look for differential information related to efficacy in different spaces.
Methods
Patient and sample selection
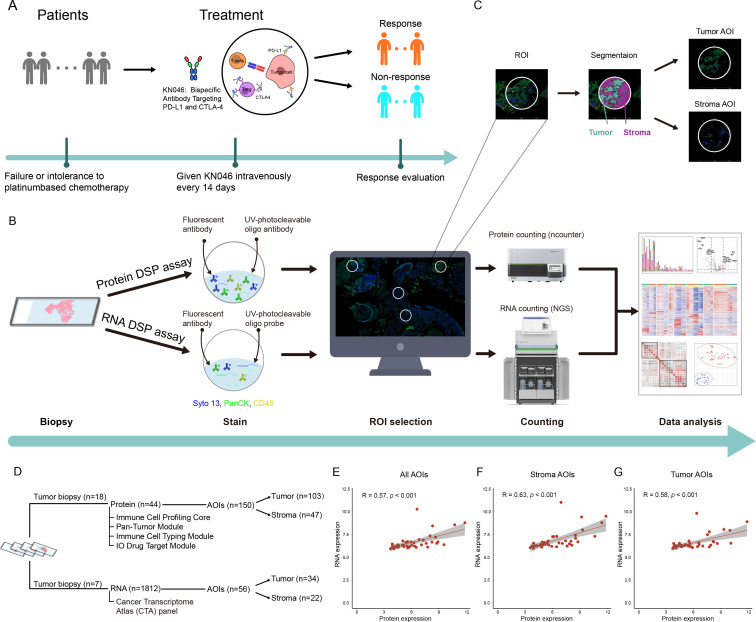
All participants were enrolled in a phase II multicenter clinical trial. Available tissues from the Shanghai Pulmonary Hospital Center were selected. Enrolled patients with aNSCLC who progressed after first-line platinum-based chemotherapy were administered bsAb-KN046 intravenously every 14 days until disease progression or intolerable toxicity (figure 1A).13 The inclusion criteria were 18–75 years of age, stage IV NSCLC confirmed by histology or pathology, negative epidermal growth factor receptor mutation or anaplastic lymphoma kinase rearrangement, and patients who failed or did not tolerate first-line platinum-based chemotherapy. Tumor response was evaluated according to the Response Evaluation Criteria in Solid Tumors V.1.1.16 To meet the requirements of DSP, samples were analyzed by DSP protein assay and DSP RNA assay.
Figure 1.
Overview of methods used to investigate spatial heterogeneity. (A) Schematic of the study participants. (B) Schematic of the DSP workflow. (C) ROI selection and segmentation. (D) AOIs collected from different regions from FFPE. (E) Correlation analysis of all regions between proteome and transcriptome data. (F) Correlation analysis of stromal regions between proteome and transcriptome data. (G) Correlation analysis of tumor regions between proteome and transcriptome data. AOI, area of interest; CTLA-4, cytotoxic T lymphocyte-associated antigen-4; DSP, digital spatial profiling; NGS, next-generation sequencing; PD-L1, programmed death ligand-1; ROI, region of interest; UV, ultraviolet.
Sample preparation for nanostring GeoMx DSP protein assay
Formalin-fixed paraffin-embedded (FFPE) slides (4 µm) were baked at 60°C for 1.5 hours, and then deparaffinized and rehydrated as follows: 3×5 min in CitriSolv, 2×5 min in 100% ethanol, 2×5 min in 95% ethanol, and 2×5 min in double-distilled water. For antigen retrieval, slides were placed in a staining jar containing 1× citrate buffer with pH 6 at 25°C. The staining jar containing the slides was placed in a preheated pressure cooker and run at high pressure and temperature for 15 min. After carefully releasing the pressure, transferring the staining jar to the lab bench, removing the lid, and letting it stand for 25 min, the slides were then washed with 1× tris-buffered saline with Tween-20 (TBS-T) for 5 min. Blocking was performed by placing the slide in a humidity chamber in a horizontal position and covering it with sufficient Buffer W (NanoString). The slides were then incubated with Buffer W for 1 hour at 25°C in a humidity chamber. Ultraviolet (UV)-photocleavable oligo antibody sets (Immune Cell Profiling Core, Immuno-oncology (IO) Drug Target Module, Immune Cell Typing Module, and Pan-Tumor Module), containing 44 targets, were used for protein detection. A mixture of UV-photocleavable oligo antibody sets and morphological markers panCK and CD45 (NanoString, 121300301) was diluted in Buffer W. The slides were removed from the humidity chamber and Buffer W was discarded then placed back into the humidity chamber and covered with diluted antibody solution. The humidity chamber was then transferred to a 4°C freezer and incubated overnight. Postfix was performed by removing the slide from the humidity chamber and carefully aspirating the antibody solution from the slide. The slides were washed for 3×10 min in TBS-T. The samples were covered with 4% paraformaldehyde and incubated for 30 min at 25°C in a humidity chamber. After incubation, the slides were washed for 2×5 min in TBS-T. For nuclear staining, the slides were incubated with SYTO 13 (NanoString, 121300301) for 15 min at 25°C in a humidity chamber and rinsed with 1× TBS-T. Finally, the slides were loaded onto the GeoMx instrument.
Sample preparation for nanostring GeoMx DSP RNA assay
FFPE slides (4 µm) were baked at 60°C for 1.5 hours, deparaffinized and rehydrated as follows: 3×5 min in CitriSolv, 2×5 min in 100% ethanol, 1×5 min in 95% ethanol, and 1×5 min in phosphate-buffered saline (PBS). For antigen retrieval, the slides were dipped in diethylpyrocarbonate-treated water for 10 s to increase the temperature of the slides to 99°C, then quickly transferred to 1× tris-EDTA at 99°C and incubated for 20 min. The slides were immediately moved to 25°C and washed in 1×PBS for 5 min. The RNA target was exposed by incubating the slides in 1 µg/mL proteinase K solution at 37°C for 15 min and washed in 1×PBS for 5 min. After exposure, the slides were postfixed as follows: 1×5 min in 10% neutral buffered formalin (NBF), 2×5 min in NBF stop buffer, and 1×5 min in PBS. UV-photocleavable oligo probes (Cancer Transcriptome Atlas, CTA) containing 1812 gene probes were used for RNA detection. In situ hybridization was performed by adding hybridization solution to the slides. The slides were placed in a hybridization oven and incubated at 37°C overnight (16–24 hours). To remove off-target probes, the slides were dipped in 2× saline sodium citrate (SSC) buffer and washed 2×25 min in stringent wash at 37°C, washed 2×2 min in 2× SSC, and removed from the 2× SSC, excess liquid was aspirated, returned to the humidity chamber, covered with Buffer W, and stored at 25°C for 30 min in the dark. Buffer W was then removed and the slides were placed back in the humidity chamber. The slides were covered with morphological markers panCK and CD45, and nuclear stain SYTO 13 (NanoString, 121300310) at 25°C for 1 hour in a humidity chamber. After staining, the slides were washed for 2×5 min in 2× SSC. Finally, the slides were loaded onto the GeoMx instrument.
Digital spatial profiling
Region of interest (ROI) selection was performed on the immunofluorescence images. Compartment-specific areas of interest (AOI) was assigned from the sequential masks as the stroma (panCK-negative staining and tumor-inverse segment) and tumor (panCK-positive staining and tumor-enriched segment) compartments. Each ROI was selected and independently determined by two participating authors, pathologists at the Department of Pathology of Shanghai Pulmonary Hospital. ROIs ranging from 1‒17 in per sample were selected for the analysis. Spatially indexed barcode cleavage and collection were performed using a GeoMx Digital Spatial Profiling instrument (NanoString). The barcodes were collected for each AOI. For protein quantification, the aspirate was dried and then rehydrated in a DSP collection plate. The transferring samples and GeoMx Hyb Code Pack reagents were transferred to a new plate for hybridization. Hybridization was performed overnight, and the products were pooled through the column into strip tubes. The pooled samples were loaded onto the nCounter FLEX (NanoString) to count oligonucleotides collected by GeoMx DSP. Once the counting was complete, the reporter code count (RCC) files were uploaded to the GeoMx DSP system to integrate the oligonucleotide counts with spatial data, followed by data analysis. For RNA quantification, the collected aspirate was dried and rehydrated in a DSP collection plate. Samples, 5× PCR master mix, and primer mix were transferred to a new plate for library preparation. During PCR amplification, i5 and i7 indexing sequences were added to the GeoMx Seq Code primers. The PCR products were purified, assessed for quality, quantified, and then sequenced on an Illumina NextSeq 550AR instrument. Once sequencing was complete, the FASTQ files were converted to digital count conversion (DCC) files. The DCC files were uploaded to the GeoMx DSP system to integrate the oligonucleotide counts with the spatial data, followed by data analysis.
Spatial proteomic data analysis
The RCC files were uploaded to the GeoMx DSP system. Analysis was performed on the GeoMx DSP Control Center using the Data Analysis module V.2.4.0.421. Quality control (QC) of the protein data was performed using an nCounter assay, which included six positive and six negative control probes. The QC content contains the field of view (FOV) detection percentage, binding density, nuclei count, and surface area (online supplemental table S1A). FOV recording was used to remove the AOIs when the FOV detection percentage was <75%. AOIs with binding density between 0.1 and 2.25 were retained. For each AOI, the number of nuclei was >20, or the surface area was >1600 square microns. The raw digital counts from the barcodes corresponding to particular probes were normalized. The sizes of the different ROIs were adjusted by area normalization and different cell numbers to avoid variations across the ROIs. Data that met the QC criteria were normalized across samples based on the background of immunoglobin G.
jitc-2022-006234supp001.xlsx (336.4KB, xlsx)
The normalized counts were used to compare differences between groups and to construct the signatures of pan-tumor, immune cell profiling, immune cell typing, and IO drug with the corresponding proteins (online supplemental table S2). The signature scores were calculated using the equation reported in a previous study17: , where Xi is the normalized expression value of each protein, and n is the number of proteins included in the signature. Wilcoxon test was used to analyze the differences between groups, and the significant difference was determined with a p value of <0.05.
jitc-2022-006234supp002.xlsx (10.5KB, xlsx)
To calculate the simulated bulk sequencing data for the corresponding proteins in each sample, the following equation was used to calculate the protein expression of all AOIs, stromal AOIs, and tumor AOIs in each sample: average value=(X1+X2…+Xn)/N (N is the number of corresponding AOIs in each sample). The average value is the simulated bulk sequencing expression value of the corresponding protein in the target region.
Spatial transcriptomic data analysis
GeoMx NGS Pipeline Software V2.0.0.16 was used to convert the sequenced FASTQ files to DCC files. The DCC files were then uploaded to the GeoMx DSP system. Analysis was performed on the GeoMx DSP Control Center using the Data Analysis module V.2.4.0.421. The QC of the transcriptome data included technical signal, technical background, probe, and normalization. Technical signal QC was used to remove the ROIs when the alignment rate of the reads compared with the template sequence was <80%. The technical background included three indicators: no template control (NTC) count, negative probe count, and parameters of ROI. NTC count was used to detect template contamination during library building. ROIs with an NTC of >1000 were removed. Negative probe count was used to measure the overall technical signal level in the CTA experiment. The threshold of the negative probe count was four counts. Parameters of the ROI included nuclei counts (>100) and surface area (>8000 square microns) (online supplemental table S1B). The sizes of the different ROIs were adjusted by area normalization and different cell numbers to avoid variations across the ROIs. Data that met QC were normalized across samples using quantile 3 normalization.18
The gene expression matrix was used to compare differences between groups and to calculate the different signatures, including cell function, immune response, metabolism, and signaling pathways (online supplemental table S3). The 14 immune cell signature scores were calculated by SpatialDecon V.1.2.0, which used deconvolution to estimate the number of immune and stromal cells in the TME.19 The other signature score was also obtained by calculating with the equation mentioned previously: , where Xi is the normalized expression value of the gene corresponding to the cells, and n is the number of genes included in the signature. The ‘filterByExpr’ function with parameter ‘min.count=3’ in edegR V.3.34.0 package was applied to remove low expression genes. Subsequently, the common functions with default parameters in edgeR package were used to identify the differentially expressed genes (DEGs), including glmFit, glmLRT, estimateGLMCommonDisp, estimateGLMTrendedDisp, and estimateGLMTagwiseDisp.20 DEGs with |log2FoldChange| of >1 and false discovery rate of <0.05 were considered as significant DEGs. Subsequently, these significant DEGs were used to perform Kyoto Encyclopedia of Genes and Genomes pathway enrichment using the KOBAS-i web tool, which incorporates seven functional class scoring tools and two pathway topology tools into a single ensemble score.21
jitc-2022-006234supp003.xlsx (92.9KB, xlsx)
To calculate the simulated bulk sequencing data for the corresponding RNAs in each sample, the following equation was used to average the RNA expression of all AOIs, stromal AOIs, and tumor AOIs in each sample: average value=(X1+X2…+Xn)/N (N is the number of corresponding AOIs in each sample). The average value was the simulated bulk sequencing expression value of the corresponding RNA in the target region.
Construction of the predictive signature
The arithmetic means of all different proteins between the two groups (p<0.05) were used to make up the signature [Σ(log2(Xi+1))−Σ(log2(Yk+1))]/N where Xi is the normalized expression value of the protein significantly expressed in the response group, while Yk is the normalized expression value of the protein significantly expressed in the non-responding group, and N is the number of proteins included in the signature.
Calculation of the predictive signature score from the validation cohort
Raw sequencing data for 65 patients with NSCLC treated with immune checkpoint inhibitors (ICIs) were obtained from three previously reported studies.22–24 The trim galore V.0.6.7 was used to filter out low-quality reads and adapters. The read counts and transcripts per million values were calculated using Kallisto V.0.46.2, which pseudoaligned sequencing reads to the reference transcripts downloaded from the Gencode V.38 database.25 The signature score was calculated using the equation [Σ(log2(Xi+1))- Σ(log2(Yk+1))]/N. The median score was used as the cut-off value to divide the patients into the high-core and low-score groups. According to the response of the patients, complete response and partial response (PR) were divided into the response groups, and stable disease (SD) and progressive disease (PD) were the non-response groups.
Multiplex immunohistochemistry (mIHC)
mIHC was performed by staining 4 μm FFPE whole tissue sections with standard, primary antibodies sequentially and paired with a tyramide signal amplification (TSA) six-color kit (abs50014-100T; Absinbio, Shanghai). The cells were then stained with 4′,6-diamidino-2-phenylindole (DAPI). To clearly distinguish tumor areas from stromal areas, we selected panCK consistent with DSP. Positive areas of panCK were defined as tumor areas, while negative areas were defined as stromal areas. Deparaffinized slides were incubated with anti-panCK antibody (4545, CST) for 30 min and then treated with an anti-rabbit/mouse horseradish peroxidase-conjugated secondary antibody (abs50015-02, Absinbio) for 10 min. Labeling was then developed for a strictly observed 10 min using TSA 570 per manufacturer’s direction. The slides were washed in TBS-T buffer and transferred to preheated citrate solution (90°C) before being heat-treated using a microwave set at 20% maximum power for 15 min. The slides were then cooled to 25°C in the same solution. Between all steps, the slides were washed with tris buffer. The same process was repeated for the following antibodies and fluorescent dyes: CD11c antibody (45581, CST)/TSA 570, anti-Tim3 (ab241332, Abcam)/TSA 520, anti-CD4 (ab133616, Abcam)/TSA 690, and anti-CD45 (13917, CST)/TSA 780. Each slide was then treated with two drops of DAPI (abs47047616, Absinbio), washed in distilled water, and manually cover-slipped. The slides were air-dried and photographed with a Pannoramic MIDI II (3DHISTECH). Images were analyzed using the Indica Halo software.
PD-L1 testing
PD-L1 quantitative detection was performed using the DSP protocol described previously. We also detected PD-L1 expression using traditional automatic IHC. The prepared FFPE sections were placed into a Ventana BenchMark XT automatic immunohistochemical staining instrument platform (Ventana PD-L1 SP263). The expression was evaluated according to the coloring ratio. PD-L1 positivity was defined according to the Tumor Proportion Score (TPS) staining on tissue: TPS=(number of PD-L1 membrane staining positive tumor cells/number of total tumor cells)×100%. TPS of <1% was negative; 1%‒49% was weakly positive; and ≥50% was strongly positive.
Tumor mutation burden (TMB) analysis
Tumor samples were extracted from FFPE specimens for DNA isolation and genomic DNA sequencing. DNA fragments of 200–400 bp were isolated from the tissue after shearing, and DNA libraries were constructed using KAPA Hyper Prep kits. The prepared library was hybridized with two different hybridization reagents and blockers in the SureSelectXT Target Enrichment System. The enrichment library was then amplified using P5/P7 primers. Finally, the library was sequenced on the NovaSeq6000 platform (Illumina) after identification with a 2200 bioanalyzer and was quantified by Qbit3 and quantitative real-time PCR NGS library quantitative kits. A panel of 825 cancer-related genes was compared with an average coverage depth of >700 (Genetron Health, Beijing, China). TMB=number of non-synonymous mutations/panel size (mut/Mb).
Statistical analysis
Heatmaps were drawn using the R V.4.1.0 package of ComplexHeatmap V.2.8.0.26 Dimension reduction analysis was performed using umap V.0.2.8.0. Wilcoxon test was used to compare the differences between two groups. Correlation analysis was conducted using Pearson correlation analysis. Receiver operating characteristic (ROC) curves were used to evaluate the predictive power of the model. The area under the curve (AUC) was calculated to measure the discriminatory ability using pROC V.1.18.0. When comparing different classification models, the ROC of each model can be drawn, and the AUC can be compared as an indicator of the merits and disadvantages of the model. The significance of OS and PFS was analyzed using Kaplan-Meier (K-M) curve analysis. The K-M curve was compared using the log-rank test. The median follow-up time was calculated using the reverse K-M method. The effect size was expressed as the standardized mean difference with a 95% CI. Statistical significance was set at a p value of <0.05.
Results
Spatial transcriptomic and proteomic analysis of aNSCLC
After sample collection and quality detection, 18 patients from pretreatment of KN046 were selected in our discovery cohort. The baseline characteristics of the patients are shown in table 1.
Table 1.
Clinical characteristics
| Characteristics | All cases | PR group | PD group | SD group |
| Total, n | 18 | 8 | 9 | 1 |
| Median age, years (range) | 65 (48–72) | 63.0 (48–70) | 60.1 (50–72) | 66 |
| Age group (years), n (%) | ||||
| <65 | 10 (55.6) | 4 (50.0) | 6 (66.7) | 0 (0.0) |
| ≥65 | 8 (44.4) | 4 (50.0) | 3 (33.3) | 1 (100.0) |
| Gender, n (%) | ||||
| Male | 15 (83.3) | 7 (87.5) | 8 (88.9) | 0 (0.0) |
| Female | 3 (16.7) | 1 (12.5) | 1 (11.1) | 1 (100.0) |
| Smoking history, n (%) | ||||
| Never smoker | 5 (27.8) | 2 (25.0) | 2 (22.2) | 1 (100.0) |
| Former/current smoker | 13 (72.2) | 6 (75.0) | 7 (77.8) | 0 (0.0) |
| ECOG performance status, n (%) | ||||
| 0 | 2 (11.1) | 1 (12.5) | 0 (0.0) | 1 (100.0) |
| 1 | 16 (88.9) | 7 (87.5) | 9 (100.0) | 0 (0.0) |
| Pathological classification, n (%) | ||||
| Adenocarcinoma | 13 (72.2) | 6 (75.0) | 6 (66.7) | 1 (100.0) |
| Squamous carcinoma | 5 (27.8) | 2 (25.0) | 3 (33.3) | 0 (0.0) |
| Biopsy location, n (%) | ||||
| Lung | 17 (94.4) | 8 (100.0) | 8 (88.9) | 1 (100.0) |
| Lymph node | 1 (5.6) | 0 (0.0) | 1 (11.1) | 0 (0.0) |
| One-line treatment regimen, n (%) | ||||
| AC/AP | 8 (44.4) | 3 (37.5) | 4 (44.4) | 1 (100.0) |
| AC/AP+Bev | 3 (16.7) | 1 (12.5) | 2 (22.2) | 0 (0.0) |
| GC/GP | 6 (33.3) | 3 (37.5) | 3 (33.3) | 0 (0.0) |
| PC | 1 (5.6) | 1 (12.5) | 0 (0.0) | 0 (0.0) |
| PD-L1 expression, n (%) | ||||
| <1% | 13 (72.2) | 6 (75.0) | 6 (66.7) | 1 (100.0) |
| 1%–49% | 4 (22.2) | 2 (25.0) | 2 (22.2) | 0 (0.0) |
| ≥50% | 1 (5.6) | 0 (0.0) | 1 (11.1) | 0 (0.0) |
| TMB, n (%) | ||||
| ≥10 mut/Mb | 13 (72.2) | 5 (62.5) | 8 (88.9) | 0 (0.0) |
| <10 mut/Mb | 1 (5.6) | 0 (0.0) | 1 (11.1) | 0 (0.0) |
| NA | 4 (22.2) | 3 (37.5) | 0 (0.0) | 1 (100.0) |
AC/AP, pemetrexed+cisplatin/carboplatin; Bev, bevacizumab; ECOG, Eastern Cooperative Oncology Group; GC/GP, gemcitabine+cisplatin/carboplatin/nedaplatin; NA, not available; PC, paclitaxel+cisplatin; PD, progressive disease; PD-L1, programmed death ligand-1; PR, partial response; SD, stable disease; TMB, tumor mutation burden.
Until 30 July 2022, the median follow-up time calculated by reverse K-M method was 33.51 months. The cohort was balanced in clinical response, with nine patients with PR and SD and nine patients with PD. Samples from two patients were available from both pretreatment and post treatment and used to further evaluate the tumor and immune microenvironment changes during therapy. Overall, 20 tumor samples from 18 patients with aNSCLC were used for subsequent analyses.
GeoMx DSP technology was used to quantitate the expression of mRNAs and proteins in spatially defined tumor sample regions. To measure mRNA expression, we used the CTA panel which was designed to profile over 1800 transcripts. To measure protein expression, we used the DSP panel that included 44 proteins and 6 internal references (figure 1B).
Because of the cellular heterogeneity within each ROI, it was difficult to untangle the mRNA and protein expression signals of the target cells from those in their surrounding cells. Thus, complex heterogeneity was masked if all ROIs were adopted for analysis. To address this issue, we used a masking and segmentation strategy to separate the tumor or stroma cells from their surrounding cells by combining the staining patterns of morphological markers with digital optical barcoding technology, which enabled us to dissect the crosstalk between tumor cells and their microenvironment (figure 1C). In this way, primary ROIs were further subdivided into predominant tumor and stromal AOIs. Therefore, 206 AOIs were obtained including 136 tumor AOIs and 70 stromal AOIs. Among them, 150 and 56 AOIs were subjected to the protein DSP and RNA DSP assays, respectively (figure 1D).
After selecting ROIs, the DNA oligonucleotides were UV-cleaved and sequenced on an Illumina sequencer for mRNA quantitation and nCounter Analysis System for protein quantitation. All ROI/AOI met the QC criterion (online supplemental figures S1 and S2). Comparison analysis revealed medium positive correlations between RNA and protein expression in all AOIs (R=0.57, p<0.001), stromal AOIs (R=0.63, p<0.001), and tumor AOIs (R=0.58, p<0.001) (figure 1E–G).
jitc-2022-006234supp004.pdf (42.3KB, pdf)
jitc-2022-006234supp005.pdf (38.6KB, pdf)
Distinct expression patterns of intratumoral spatially AOIs identified by DSP
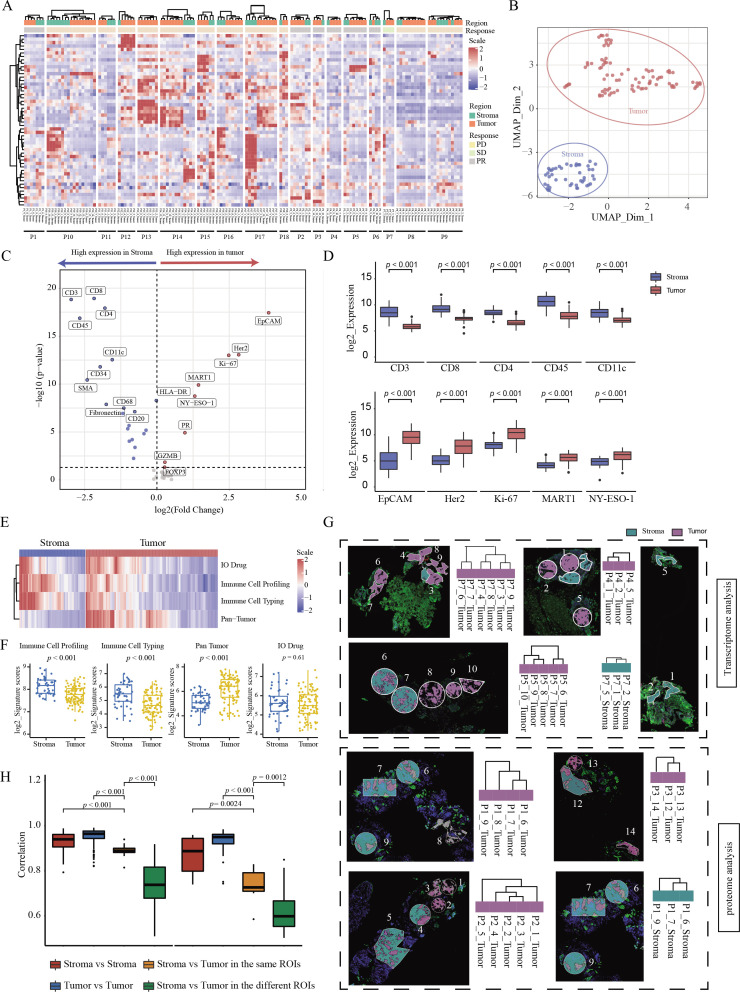
To assess ITH, we analyzed the expression patterns of both proteins and RNAs among spatially distinct AOIs for each sample. First, significant differences were observed between tumor and stromal AOIs from the same sample at both the protein and RNA levels (figure 2A and online supplemental figure S3A). Specifically, tumor and stromal AOIs from the same sample were grouped into separate clusters in most patient samples. For example, in patient 1 (P1), the DSP protein results showed that four tumor AOIs and three stromal AOIs were independently clustered into two categories. At the same time, we observed that the stromal and tumor AOIs of the two patients (P6 and P9) clustered together. Such distinct patterns could be observed more clearly using dimensionality reduction analysis (figure 2B and online supplemental figure S3B). Subsequently, we used DSP to investigate specific molecular markers associated with the tumor and stromal regions. The protein markers that were most associated with the tumor phenotype were epithelial cell adhesion molecule, human epidermal growth factor receptor 2, and Ki-67 (all p<0.001). Immune cell markers (CD45), including T-cell markers (CD3, CD8, and CD4) and dendritic cells (DCs) (CD11c), were significantly increased in the stromal areas (all p<0.001) (figure 2C, D, and online supplemental table S4). Consistent with the findings based on individual proteins, the signature scores of the Immune Cell Profiling Core and Immune Cell Typing Module were significantly higher in stromal AOIs (both p<0.001), while the scores of the Pan-Tumor Module were significantly higher in tumor AOIs (p<0.001) (figure 2E, F, and online supplemental table S5). To confirm the findings derived from the protein data, we also used DSP CTA data to perform the analysis, and similar results were obtained at the RNA level (online supplemental figure S3C, D and table S6).
Figure 2.
Distinct expression patterns of intratumoral spatially distinct AOIs identified by DSP (A) Heatmap of target proteins in different spatial regions of each patient. The region of AOIs for each patient is shown in the top track. The clinical response for each patient is shown under the track of the region. The protein expression from each AOI is shown in the central heatmap. The corresponding region and patient for each AOI are shown under the central heatmap. (B) UMAP of dimensionality reduction analysis at the protein level. (C) Volcanic map of differentially expressed proteins in different regions. (D) Box plot of top five differential proteins within different spatial regions. (E) Heatmap of DSP protein panels about pan-tumor, immune cell profiling, immune cell typing, and IO drug in different regions. (F) Scatter plot of DSP protein panels in different regions. (G) Comparison between spatial distance of different ROIs in one sample and molecular clustering results from transcriptome and proteome images. The top four and bottom four images with spatially adjacent AOIs are from the DSP RNA and protein assay, respectively. The cluster maps of AOIs on the side of the images were based on the expression level of RNA or proteins, respectively. (H) Correlation comparisons of stroma or tumor regions in the same or different ROIs. AOI, area of interest; DSP, digital spatial profiling; PD, progressive disease; PR, partial response; ROI, region of interest; SD, stable disease.
jitc-2022-006234supp006.pdf (828KB, pdf)
jitc-2022-006234supp007.xlsx (13.3KB, xlsx)
jitc-2022-006234supp008.xlsx (12.6KB, xlsx)
jitc-2022-006234supp009.xlsx (13.8KB, xlsx)
To further investigate ITH at the spatial level, we characterized the molecular profiles of geographically disparate regions in each sample. According to the process of tumor evolution, we speculate that spatially adjacent tumor regions will have higher similarity at the molecular level. To prove this, we selected 12 tumor samples with a clear positional relationship between different AOIs in the spatial distance for analysis. Among the 12 DSP samples, the spatial distances of the tumor AOIs in the 8 samples were consistent with the molecular clustering patterns (figure 2G). For example, in patient 7 (P7), the spatially closed tumor AOIs (06‒07, 04‒08, and 03‒09) clustered together at the RNA expression level; in P1, a pair of tumor AOIs with a relatively close spatial distance (06‒07), their protein expression patterns clustered together. Moreover, the inferred relationships among disparate regions by molecular clustering may reflect the path of tumor development. Accordingly, based on the cluster results of five geographically disparate regions (01‒05) in patient 2, we could infer that the development of the tumor may originate from AOI 02 and 03, and a similar phenomenon was also observed among stroma AOIs (figure 2G).
Since tumor cells are closely connected with their surrounding stromal cells in each ROI, we speculated that the molecular profiles between the tumor and stromal regions in the same ROI were more similar than those in different regions. To address this issue, we used paired tumor and stromal AOIs that were segmented from the same region for further analysis. We found that the correlation between the tumor and stromal regions segmented from the same ROI was significantly higher than that between different ROI (all p≤0.0024). Meanwhile, the correlation among tumor AOIs was strongest at both protein and RNA levels (figure 2H). The correlation analysis for each patient is shown in online supplemental figure 4A, B.
jitc-2022-006234supp010.pdf (74.1KB, pdf)
Characterization of immune cell types and pathways in tumor and stromal AOIs
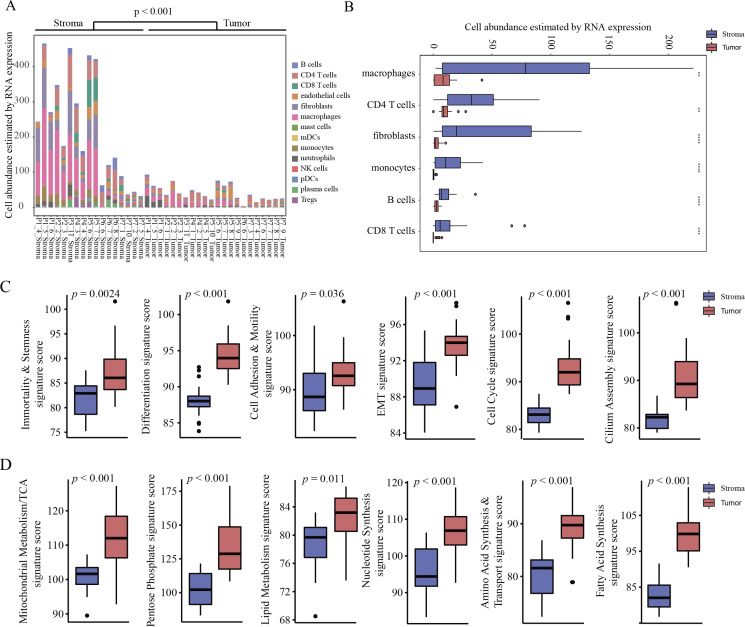
The immune cell repertoire and signature of various cancer signaling pathways have not been well-characterized in NSCLC at the spatial level. To evaluate lymphocyte infiltration, we used DSP to quantify transcripts that marked different immune cell types (online supplemental table S7). Because the tumor AOIs captured in this study focused on areas enriched for neoplastic cells, which were largely devoid of immune cells, we observed significantly lower lymphocyte abundance in the tumor AOIs than in the stromal AOIs (p<0.001) (figure 3A and online supplemental table S8). Macrophages, CD4 T cells, fibroblasts, monocytes, B cells, and CD8 T cells were the most highly detected cell types in stromal AOIs (figure 3B). The immune response-related cell signature scores between different regions were then analyzed. We found that lymphocyte trafficking and lymphocyte regulation signature scores were associated with stromal AOIs (both p<0.001), which further supported the results of the lymphocyte abundance analysis. In addition, the interleukin (IL)-2 signaling, type II interferon, and phagocytosis signature scores were significantly higher in stromal AOIs, while IL-1 and IL-17 signatures exhibited a significant increase in tumor AOIs (all p<0.001) (online supplemental figure S5A).
Figure 3.
Characterization of immune cell types and cancer pathways in tumor and stroma AOIs. (A) Lymphocyte infiltration abundance estimated by RNA expression between stromal and tumor regions. (B) Box plot of abundance for macrophages, CD4 T cells, fibroblasts, monocytes, B cells, and CD8 T cells estimated by RNA expression between stromal and tumor regions. (C) Box plot of signatures about cell function and signaling pathway related to proliferation and metastasis. (D) Box plot of signature scores related to energy metabolism and macromolecule synthesis. AOI, area of interest; EMT, epithelial–mesenchymal transition; mDCs, myeloid dendritic cells; NK, natural killer; pDCs, plasmacytoid dendritic cells; TCA, tricarboxylic acid.
jitc-2022-006234supp011.xlsx (9.7KB, xlsx)
jitc-2022-006234supp012.xlsx (19.9KB, xlsx)
jitc-2022-006234supp013.pdf (140.5KB, pdf)
Subsequently, we evaluated the expression programs of AOIs within each patient, indicating cancer-related cell function, signaling pathways, and metabolism. The scores for 76.3% of cancer-related programs were significantly higher for tumor AOIs than for stromal AOIs (online supplemental figure S5B). For example, signatures of cell function and signaling pathways related to proliferation and metastasis were significantly increased in tumor AOIs (all p≤0.036) (figure 3C and online supplemental figure S5C), which is consistent with the ability of tumor cells to proliferate indefinitely and metastasize. In the tumor regions, the normal functions of cells are often dysregulated (figure 3D and online supplemental figure S5D). In addition, the signaling scores related to energy metabolism and macromolecule synthesis were also significantly higher in the tumor AOIs (all p≤0.011), consistent with the observation that the rapid proliferation of tumor cells requires more energy and biological macromolecules (figure 3D).27 As a pivotal molecule regulating cell growth and metabolism, the mammalian target of rapamycin and its upstream and downstream signaling pathways was also highly expressed in the tumor areas (all p≤0.014) (online supplemental figure S5E).28 Those results demonstrated that DSP could assign biological observations to specific phenotypic cell types within the complex tumor ecosystem, which would be masked or obfuscated using bulk sequencing.
Identification of DSP markers revealed the impact of ITH on finding candidate biomarkers related to treatment efficacy
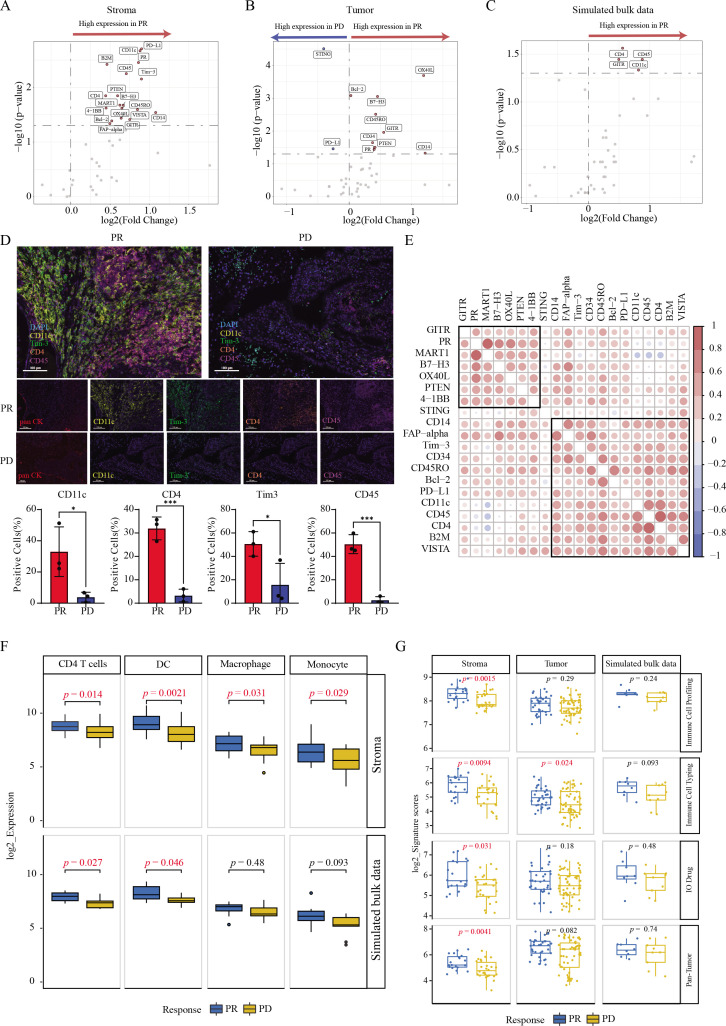
To assess the effect of ITH on finding treatment efficacy related biomarkers, DSP data were used to investigate the efficacy of bsAb-KN046 therapy. Owing to the limited sample size for biopsy samples, we averaged all AOIs per tumor sample to derive a composite value of simulated bulk sequencing data and compared it with the results after spatial segmentation. A comparative analysis revealed a high positive correlation (R=0.81, p<0.001) between the DSP simulated bulk sequencing data and bulk RNA-seq data (online supplemental figure S6A). A total of 18 (B7-H3, B-cell lymphoma 2 (Bcl-2), beta-2-microglobulin, CD11c, CD14, CD4, CD45, CD45RO, fibroblast activation protein alpha (FAP-alpha), glucocorticoid-induced TNFR-related (GITR), melanocyte antigen (MART1), OX40L, PD-L1, progesterone receptor, phosphatase and tensin homolog (PTEN), T-cell immunoglobulin and mucin domain-containing protein 3 (Tim-3), V-domain Ig-containing suppressor of T-cell activation (VISTA), and 4-1BB) and 11 (OX40L, Bcl-2, B7-H3, CD45RO, GITR, CD34, PTEN, progesterone receptor, CD14, stimulator of interferon genes (STING), and PD-L1) efficacy-associated DSP protein markers were identified in the stromal and tumor areas, respectively (figure 4A, B, and online supplemental table S9), whereas only 4 proteins including CD45, CD4, GITR, and CD11c were identified using simulated bulk sequencing data (figure 4C). DSP protein data and simulated bulk sequencing data consistently showed that the expression of these four proteins increased in the PR group. The fold changes of PR/PD in most protein markers in the stromal regions, tumor regions, and simulated bulk sequencing data were consistent with the results above, several proteins showed discrepancies (online supplemental figure S6B). For example, PD-L1 was highly expressed in the stromal of patients with PR but decreased in tumor and simulated bulk sequencing data. This is probably because PD-L1 is not only expressed on the tumor surface, but is also expressed in antigen-presenting cells under the stimulation of interferon gamma. However, previous bulk detection often missed the expression of cells in the stromal region, while DSP could be accurately detected.
Figure 4.
Identification of DSP markers revealed the impact of ITH on treatment efficacy. (A) Volcanic map of differential proteins in stromal regions between different response groups. (B) Volcanic map of differential proteins in tumor regions between different response groups. (C) Volcanic map of differential proteins through simulated bulk sequencing in whole areas. (D) Expression of CD11c, Tim-3, CD45, and CD4 in stromal regions from patients with PR and PD detected by mIHC. (E) Correlation analysis of DSP protein markers in different regions. (F) Abundance of immune-related cell types among different groups was demonstrated by differential genes corresponding to proteins. (G) Scatter plot of four target protein signatures in different response groups. DSP, digital spatial profiling; ITH, intratumoral heterogeneity; mIHC, multiplex immunohistochemistry; PD, progressive disease; PR, partial response; Tim-3, T-cell immunoglobulin and mucin domain-containing protein 3.
jitc-2022-006234supp014.pdf (1.8MB, pdf)
jitc-2022-006234supp015.xlsx (13.7KB, xlsx)
To further verify the expression of proteins in different regions, three samples from PR group and three from PD group were selected for mIHC verification with four common proteins in the stromal regions including CD11c, Tim-3, CD45, and CD4. We selected panCK consistent with DSP to identify tumor cells. The positive areas of panCK were defined as tumor areas, while the negative areas were stroma areas. We calculated the proportion of cells that were positive for each marker. The proportion of the above four proteins in the stromal region of PR group were significantly higher than that in the PD group (all p<0.05) (figure 4D and online supplemental figure S6C). In addition, we found that the correlation between tumor and immune DSP markers was distinguishable, suggesting different actions of tumor and immune cell populations on treatment efficacy (figure 4E).
Subsequently, we evaluated the immune infiltration in the stromal region of patients in different response groups and found that the abundance of four types of immune cells, including CD4 T cells (p=0.014), DCs (p=0.0021), macrophages (p=0.031), and monocytes (p=0.029), was significantly higher in the PR group, but only CD4 T cells and DCs could be identified by simulated bulk sequencing data (figure 4F and online supplemental table S10). Meanwhile, immune cell profiling (p=0.0015), immune cell typing (p=0.0094), IO drug (p=0.031), pan-tumor signature scores (p=0.0041) from stroma, and immune cell typing signature scores (p=0.024) from tumor regions were significantly higher in patients with PR, none of which could be observed in simulated bulk data (figure 4G).
jitc-2022-006234supp016.xlsx (13.1KB, xlsx)
How the tumor ecosystem changes during bsAb treatment is still poorly understood, prompting us to further analyze tumor and stromal changes across treatments. In our study, the tissues from two patients were sampled at the time of pretreatment and post-treatment with bsAb-KN046 therapy. The results revealed a significant increase in activated leukocyte cell adhesion molecule (ALCAM) and keratin five levels with the treatment of KN046 (online supplemental figure S7A and table S11). High ALCAM expression was associated with poor prognosis in NSCLC.29 In addition, B cells were significantly decreased after disease progression (p=0.037) (online supplemental figure S7B). Subsequently, we used related genes to construct a panel to explore the differences in immune exhaustion-related pathways before and after treatment and found that the immune exhaustion signature was significantly decreased after resistance to bsAb-KN046 (p<0.001) (online supplemental figure S7C).
jitc-2022-006234supp017.pdf (447.4KB, pdf)
jitc-2022-006234supp018.xlsx (12KB, xlsx)
Spatially resolved signature of stromal regions has greater clinical response relevance than that of tumor regions
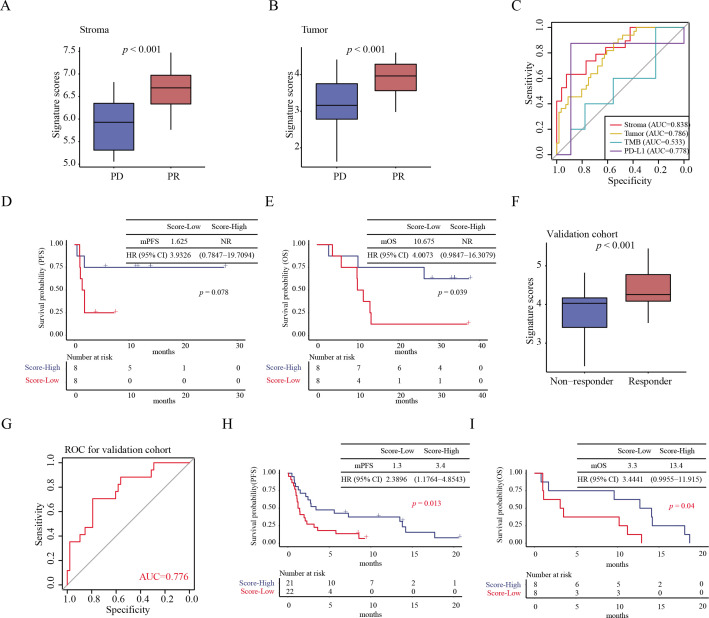
Given the ITH in response to immunotherapy, the clinical relevance of spatially resolved biomarkers from both stromal and tumor regions was evaluated. We calculated the scores of spatially resolved signatures based on efficacy-related DSP protein markers (protein markers from tumor areas: OX40L, Bcl-2, B7-H3, CD45RO, GITR, CD34, PTEN, progesterone receptor, CD14, STING, and PD-L1; protein markers from stromal areas: B7-H3, Bcl-2, Beta-2-microglobulin, CD11c, CD14, CD4, CD45, CD45RO, FAP-alpha, GITR, MART1, OX40L, PD-L1, progesterone receptor, PTEN, Tim-3, VISTA, and 4-1BB) (online supplemental table S12A, B). Significant difference in signature scores were observed between patients with PD and PR patients in both tumor and stromal areas (both p<0.001) (figure 5A, B, and online supplemental table S13A, B). To further investigate the role of stromal and tumor signatures in predicting the efficacy of KN046, the effects of age, sex, smoking history, Eastern Cooperative Oncology Group performance status, pathological classification, PD-L1 expression, and response signatures on the clinical outcomes of ROIs were analyzed using multi-factor Cox regression. Stromal and tumor signatures were favorable factors for clinical outcomes (online supplemental figure S8). Moreover, the AUC values in the stromal and tumor regions were 0.838 and 0.786, respectively. Compared with the tumor signature and traditional biomarkers, such as PD-L1 and TMB, the signature from the stromal showed a stronger clinical relevance (figure 5C). Furthermore, patients with high stromal signature scores had longer median OS (p=0.039) than those with low scores, but mPFS did not reach statistical significance (p=0.078) (figure 5D, E).
Figure 5.
Construction of a spatially resolved signature to predict clinical response to immunotherapy. Boxplot of signature scores from stromal (A) and tumor regions (B) between different response groups. (C) ROC curves of efficacy prediction of constructed signatures in the stromal, tumor regions, as well as traditional biomarkers PD-L1 and TMB. (D) Survival curves of the PFS prognostic value of the stroma signature. (E) Survival curves of the OS prognostic value of the stroma signature. (F) Box plot of efficacy prediction of signature constructed by genes corresponding to differential proteins between different response groups in a validation cohort. (G) ROC of gene signature in the validation cohort. (H) Survival curves showed the PFS prognostic value of gene signature in validation cohort. (I) Survival curves showed the OS prognostic value of gene signature in validation cohort. mPFS, median progression-free survival; NR, not reached; OS, overall survival; PD, progressive disease; PD-L1, programmed death ligand-1; PFS, progression-free survival; PR, partial response; ROC, receiver operating characteristic; TMB, tumor mutation burden.
jitc-2022-006234supp019.xlsx (11.1KB, xlsx)
jitc-2022-006234supp020.xlsx (14.6KB, xlsx)
jitc-2022-006234supp021.pdf (287KB, pdf)
To determine whether the signature of stromal regions remained effective in predicting the efficacy of general immunotherapy, we validated the spatially resolved signature constructed with relevant genes identified from the stromal area in an independent set of 65 patients with NSCLC treated with ICIs from public datasets (online supplemental table S14). Similar to the results of our cohort, in the validation datasets, we observed a significant increase in gene signature scores in the response group (p<0.001) (figure 5F). The AUC value of the spatially resolved signature for predicting clinical response was 0.776 (figure 5G). Moreover, the PFS (p=0.013) and OS (p=0.040) were significantly longer in the group with high signature scores than in those with low scores (figure 5H, I). These results suggested that molecules from the stromal region may have a greater potential in evaluating the clinical efficacy of immunotherapy.
jitc-2022-006234supp022.xlsx (12.7KB, xlsx)
Discussion
In this study, we characterized ITH and its effect on bsAb immunotherapy efficacy in patients with aNSCLC. To this end, we used DSP, NGS, and nCounter to profile the transcriptomic and proteomic information of more than 100 ROIs at different spatial locations and different efficacy groups in patients with NSCLC. We found that the tumor and stromal areas from the same tumor sample exhibited the distinct features, and multiple gene programs uniquely identified from tumor or stromal areas were associated with treatment response, most of which could not be identified using simulated bulk sequencing. Furthermore, we compared the spatially resolved signature to predict clinical response to immunotherapy between stromal and tumor areas and found that the signature in the stromal areas showed a greater predictive power for treatment response.
We observed spatial heterogeneity in the biopsy samples. The differences in genetic information caused by ITH correlated with the prediction of efficacy. Previous studies on the relationship between efficacy and ITH have reported that ITH is one of the major factors leading to resistance to antitumor therapy.30 31 High ITH is associated with poor prognosis in multiple cancer types including lung cancer.32–34 However, most past studies confirmed ITH and clonal evolution by collecting surgical samples and conducting multiregional sampling for detection.4 34 35 In recent studies on the heterogeneity of lung cancer, Wu et al suggested that the proteome was spatially heterogeneous within tumors in dozens of regions within the same tissue sample, and they described the spatially heterogeneous genome of individual tumor cells in lung adenocarcinoma.36 Since the heterogeneity of tumors has been widely verified, and numerous studies have proven that it is related to the therapeutic effect, it is worth further exploration whether ITH promotes a biomarker that can predict the therapeutic effect more accurately, or whether ITH affects the accuracy of biomarkers. Kazdal et al confirmed through TruSight Oncology 500 targeted sequencing panel that tumor amount and spatial diversity spectrum can affect the estimation of the traditional biomarker TMB, thus revealing that tumor subclonal development and subsequent ITH can influence TMB values. Therefore, TMB as a predictive biomarker has certain limitations.37 In a subsequent study on the exploration of immunotherapeutic biomarkers in NSCLC, Moutafi et al applied DSP technology and found that CD44 could serve as a novel indicative biomarker to achieve optimal patient stratification.38 However, in this study, the researchers took each sample as an ROI, which failed to consider the problem of ITH. To find more accurate biomarkers under the influence of ITH, we performed spatial segmentation of tumor tissues. In addition, owing to the different timing of tissue acquisition and different immunotherapy drugs, a significant relationship between CD44 and efficacy was not found in our study.
An interesting phenomenon in our study is that in the global analysis, PD-L1 expression was slightly higher in the PR group, but after spatial segmentation, PD-L1 expression in stromal regions was significantly higher in the PR group. However, the expression of PD-L1 in the tumor area was slightly higher in the PD group. Through spatial segmentation, some traditional biomarkers will be shown more detailed. This further demonstrated the influence of ITH on accurate predictive biomarkers of efficacy. PD-L1, as a traditional biomarker, reflects the impact of ITH on biomarker interpretation. Hwang et al used the 22C3 IHC pharmDx assay and found that in many PD-L1 negative small biopsy samples, ITH may lead to the misclassification of PD-L1 status.39 Another reason is that the patients enrolled in our study were treated with PD-L1 and CTLA-4 bsAb, which may be different from the previous immunomab in the discovery of biomarkers. The results of the CheckMate 227 study showed that the benefits of nivolumab (NIVO) plus ipilimumab (IPI) in PD-L1≥1% and < 1% groups were basically the same, and a similar phenomenon was observed in the subgroup of patients who survived 5 years and completed 2 years of immunotherapy. In addition, especially in the population with PD-L1<1%, the efficacy of NIVO+IPI combined regimen was superior to that of NIVO / IPI+chemotherapy in all aspects.40 Therefore, these results also indicated that PD-L1 expression could not be a single indicator for predicting the efficacy of dual immunotherapy. For patients treated with bsAbs, it is more important to accurately identify biomarkers in the exclusion of ITH, rather than just using the traditional detection technique.
KN046 is a novel recombinant humanized bsAb that can simultaneously block the PD-1/PD-L1 and CTLA-4 pathways and restore the immune response of T cells.41 By blocking the CTLA-4 target, the regulatory T cells (Treg) function of tumor invasion is blunted, thereby enhancing the immune response.42 However, in the detection of lymphocyte infiltration, we did not find a correlation between the expression level of Treg cells and the efficacy of KN046 because we marked Treg cells only by identifying forkhead box P3 (FOXP3). Other Treg markers, such as CD4, CD45, and GITR, were significantly increased in the stromal region of the PR group. In addition, since OX40 was mainly expressed in Tregs, its ligand OX40L was highly expressed in both the tumor and stromal regions in the PR group. This further verified the Treg recognition function of KN046 and suggested that Treg-related markers in immunotherapy targeting CTLA-4 might be related to the therapeutic response. As we hope to find more efficacy prediction biomarkers associated with bsAbs, we used DSP technology to find more candidates. In the field of immunotherapy companion diagnostics, Gupta et al and Zugazagoitia et al confirmed that DSP seems to have quantitative potential compared with IHC, and the technology can perform concomitant diagnostic tests for immunotherapy.43 44 Another study successfully identified 20 biomarkers in patients with melanoma, in which the expression of PD-L1 in CD68+ cells rather than tumor cells was an important factor in determining PFS, OS, and treatment response.45 DSP has the potential to become an accurate technique for determining the therapy prognosis of immunotherapy. We found that the predictive signature of stromal regions, including 18 proteins, had a good efficacy prediction with an AUC of 0.838. This suggested that bsAb immunotherapy may alter the initial TME and induce greater changes in protein expression in stromal regions. The combination of spatially different proteins may ameliorate some drawbacks of the traditional biomarkers. It also provides a new method and idea for exploring the predictive efficacy predictive biomarkers of other bsAb drugs.
Despite our earlier findings, this study also had its limitations. First, we analyzed tumor samples collected from a single-center retrospective immunotherapy cohort. Retrospective studies have limitations, such as the possibility of selection bias. Therefore, although ITH was found in the biopsy samples and high signature scores constructed in the stroma region were associated with the efficacy of the bsAbs targeting PD-L1 and CTLA-4 in two independent cohorts, these results need to be interpreted with caution. However, this can only be regarded as a hypothesis. In addition, because of the lack of data on this type of bsAb in the public database, only patients undergoing immunotherapy were used in our verification cohort, which may indicate that our findings are universal in predicting the efficacy of other immunotherapies. However, the verification cohort cannot accurately verify our findings in bsAb. Another limitation is that not all patients underwent DSP CTA assay because of the strict requirements of paraffin section time, which may have resulted in some transcriptomic results not matching the proteomic results individually. Finally, we found that some protein expressions may show a trend that is inconsistent with previous studies, which may be because the enrolled patients were in an advanced stage and had received other therapies such as chemotherapy in the early stage. Moreover, the situation of bsAb therapy is different from that of immunotherapy alone, which may lead to abnormal results. However, owing to the small clinical sample size and lack of verifiable data, larger and well-powered studies are needed. In the future, our bsAb-KN046 signature needs to be further validated in subsequent prospective studies and clinical trials involving multiple centers. We will continue to focus on this subset of patients and expand the sample size. We plan to add other techniques such as single-cell sequencing and spatial transcriptome sequencing to further verify our findings.
Conclusion
ITH is present in patients with aNSCLC. For the different therapeutic groups treated with bsAb immunotherapy, the stromal region showed more differential genetic information, and the signature constructed by the stromal region had better predictive efficiency. Furthermore, simple bulk sequencing may miss some key information, failing to accurately screen the beneficiaries. It may be more accurate and effective to search for efficacy-related biomarkers after the spatial segmentation due to the presence of ITH.
Acknowledgments
We thank the enrolled patients and their families for participating in the study
and their physicians for submitting the clinical samples. We also express great gratitude to Alphamab Biopharmaceuticals for providing the drug and the data from the clinical trial KN046-201.
Footnotes
Contributors: XS, HL, and CZho conceived the research and designed the experiments. XS, AX, LC, and CZha collected the clinical samples. XS, ZZ, and HL made the formalin-fixed paraffin-embedded slices and selected the field of view. JW supervised the study. XS, XZ, CP, and XC participated in the data analysis. FW, XL, TJ, PC, and ZW reviewed and edited the manuscript. CZho was responsible for the overall content as the guarantor. All the authors contributed to the review of the manuscript.
Funding: This study was supported by Shanghai Pulmonary Hospital Innovation Research Group Project (FKCX1903), The Science and Technology Innovation Action Plan of Shanghai Science and Technology Commission (19411950300), and Shanghai Collaborative Innovation Project (2020CXJQ02).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
All data relevant to the study are included in the article or uploaded as supplementary information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by the ethics committee of Shanghai Pulmonary Hospital (ID 18205ZL). The participants gave informed consent to participate in the study before taking part.
References
- 1. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (time) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546–58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Patel AP, Tirosh I, Trombetta JJ, et al. Single-Cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396–401. 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang J, Fujimoto J, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science 2014;346:256–9. 10.1126/science.1256930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sharma A, Merritt E, Hu X, et al. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep 2019;29:2164–74. 10.1016/j.celrep.2019.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science 2011;331:1565–70. 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- 7. Azoury SC, Straughan DM, Shukla V. Immune checkpoint inhibitors for cancer therapy: clinical efficacy and safety. Curr Cancer Drug Targets 2015;15:452–62. 10.2174/156800961506150805145120 [DOI] [PubMed] [Google Scholar]
- 8. Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443–54. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xia L, Liu Y, Wang Y. PD-1/PD-L1 blockade therapy in advanced non-small-cell lung cancer: current status and future directions. Oncologist 2019;24:S31–41. 10.1634/theoncologist.2019-IO-S1-s05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tumeh PC, Harview CL, Yearley JH, et al. Pd-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brinkmann U, Kontermann RE. The making of bispecific antibodies. MAbs 2017;9:182–212. 10.1080/19420862.2016.1268307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Labrijn AF, Janmaat ML, Reichert JM, et al. Bispecific antibodies: a mechanistic review of the pipeline. Nat Rev Drug Discov 2019;18:585–608. 10.1038/s41573-019-0028-1 [DOI] [PubMed] [Google Scholar]
- 13. Zhou C, Xiong A, Li W, et al. P77.03 a phase II study of KN046 (bispecific anti-PD-L1/CTLA-4) in patients (PTS) with metastatic non-small cell lung cancer (NSCLC). Journal of Thoracic Oncology 2021;16:S636. 10.1016/j.jtho.2021.01.1161 [DOI] [Google Scholar]
- 14. Merritt CR, Ong GT, Church S, et al. High multiplex, digital spatial profiling of proteins and RNA in fixed tissue using genomic detection methods. Pathology [Preprint] 2019. 10.1101/559021 [DOI] [PubMed]
- 15. Merritt CR, Ong GT, Church SE, et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat Biotechnol 2020;38:586–99. 10.1038/s41587-020-0472-9 [DOI] [PubMed] [Google Scholar]
- 16. Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 17. Ghatalia P, Gordetsky J, Kuo F, et al. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J Immunother Cancer 2019;7:139. 10.1186/s40425-019-0621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao Y, Wong L, Goh WWB. How to do quantile normalization correctly for gene expression data analyses. Sci Rep 2020;10:15534. 10.1038/s41598-020-72664-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Danaher P, Kim Y, Nelson B, et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat Commun 2022;13:385. 10.1038/s41467-022-28020-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Robinson MD, McCarthy DJ, Smyth GK. EdgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bu D, Luo H, Huo P, et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 2021;49:W317–25. 10.1093/nar/gkab447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung H, Kim HS, Kim JY, et al. Dna methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat Commun 2019;10:4278. 10.1038/s41467-019-12159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee JS, Nair NU, Dinstag G, et al. Synthetic lethality-mediated precision oncology via the tumor transcriptome. Cell 2021;184:2487–502. 10.1016/j.cell.2021.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cho J-W, Hong MH, Ha S-J, et al. Genome-Wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer. Exp Mol Med 2020;52:1550–63. 10.1038/s12276-020-00493-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bray NL, Pimentel H, Melsted P, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 26. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- 27. Abdel-Wahab AF, Mahmoud W, Al-Harizy RM. Targeting glucose metabolism to suppress cancer progression: prospective of anti-glycolytic cancer therapy. Pharmacol Res 2019;150:104511. 10.1016/j.phrs.2019.104511 [DOI] [PubMed] [Google Scholar]
- 28. Kim J, Guan KL. Mtor as a central hub of nutrient signalling and cell growth. Nat Cell Biol 2019;21:63–71. 10.1038/s41556-018-0205-1 [DOI] [PubMed] [Google Scholar]
- 29. Münsterberg J, Loreth D, Brylka L, et al. Alcam contributes to brain metastasis formation in non-small-cell lung cancer through interaction with the vascular endothelium. Neuro Oncol 2020;22:955–66. 10.1093/neuonc/noaa028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 2017;168:613–28. 10.1016/j.cell.2017.01.018 [DOI] [PubMed] [Google Scholar]
- 31. Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 2018;15:81–94. 10.1038/nrclinonc.2017.166 [DOI] [PubMed] [Google Scholar]
- 32. Li S, Garrett-Bakelman FE, Chung SS, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med 2016;22:792–9. 10.1038/nm.4125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ding X, He M, Chan AWH, et al. Genomic and epigenomic features of primary and recurrent hepatocellular carcinomas. Gastroenterology 2019;157:1630–45. 10.1053/j.gastro.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 34. Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med 2017;376:2109–21. 10.1056/NEJMoa1616288 [DOI] [PubMed] [Google Scholar]
- 35. de Bruin EC, McGranahan N, Mitter R, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science 2014;346:251–6. 10.1126/science.1253462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu H-J, Temko D, Maliga Z, et al. Spatial intra-tumor heterogeneity is associated with survival of lung adenocarcinoma patients. Cell Genom 2022;2:100165. 10.1016/j.xgen.2022.100165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kazdal D, Endris V, Allgäuer M, et al. Spatial and temporal heterogeneity of panel-based tumor mutational burden in pulmonary adenocarcinoma: separating biology from technical artifacts. J Thorac Oncol 2019;14:1935–47. 10.1016/j.jtho.2019.07.006 [DOI] [PubMed] [Google Scholar]
- 38. Moutafi MK, Molero M, Martinez Morilla S, et al. Spatially resolved proteomic profiling identifies tumor cell CD44 as a biomarker associated with sensitivity to PD-1 axis blockade in advanced non-small-cell lung cancer. J Immunother Cancer 2022;10:e004757. 10.1136/jitc-2022-004757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hwang DM, Albaqer T, Santiago RC, et al. Prevalence and heterogeneity of PD-L1 expression by 22C3 assay in routine population-based and reflexive clinical testing in lung cancer. J Thorac Oncol 2021;16:1490–500. 10.1016/j.jtho.2021.03.028 [DOI] [PubMed] [Google Scholar]
- 40. Brahmer JR, Lee J-S, Ciuleanu T-E, et al. Five-year survival outcomes with nivolumab plus ipilimumab versus chemotherapy as first-line treatment for metastatic non-small-cell lung cancer in checkmate 227. J Clin Oncol 2023;41:1200–12. 10.1200/JCO.22.01503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 2015;27:450–61. 10.1016/j.ccell.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Redmond WL, Linch SN, Kasiewicz MJ. Combined targeting of costimulatory (OX40) and coinhibitory (CTLA-4) pathways elicits potent effector T cells capable of driving robust antitumor immunity. Cancer Immunol Res 2014;2:142–53. 10.1158/2326-6066.CIR-13-0031-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gupta S, Zugazagoitia J, Martinez-Morilla S, et al. Digital quantitative assessment of PD-L1 using digital spatial profiling. Lab Invest 2020;100:1311–7. 10.1038/s41374-020-0424-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zugazagoitia J, Gupta S, Liu Y, et al. Biomarkers associated with beneficial PD-1 checkpoint blockade in non-small cell lung cancer (NSCLC) identified using high-plex digital spatial profiling. Clin Cancer Res 2020;26:4360–8. 10.1158/1078-0432.CCR-20-0175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Toki MI, Merritt CR, Wong PF, et al. High-plex predictive marker discovery for melanoma immunotherapy-treated patients using digital spatial profiling. Clin Cancer Res 2019;25:5503–12. 10.1158/1078-0432.CCR-19-0104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-006234supp001.xlsx (336.4KB, xlsx)
jitc-2022-006234supp002.xlsx (10.5KB, xlsx)
jitc-2022-006234supp003.xlsx (92.9KB, xlsx)
jitc-2022-006234supp004.pdf (42.3KB, pdf)
jitc-2022-006234supp005.pdf (38.6KB, pdf)
jitc-2022-006234supp006.pdf (828KB, pdf)
jitc-2022-006234supp007.xlsx (13.3KB, xlsx)
jitc-2022-006234supp008.xlsx (12.6KB, xlsx)
jitc-2022-006234supp009.xlsx (13.8KB, xlsx)
jitc-2022-006234supp010.pdf (74.1KB, pdf)
jitc-2022-006234supp011.xlsx (9.7KB, xlsx)
jitc-2022-006234supp012.xlsx (19.9KB, xlsx)
jitc-2022-006234supp013.pdf (140.5KB, pdf)
jitc-2022-006234supp014.pdf (1.8MB, pdf)
jitc-2022-006234supp015.xlsx (13.7KB, xlsx)
jitc-2022-006234supp016.xlsx (13.1KB, xlsx)
jitc-2022-006234supp017.pdf (447.4KB, pdf)
jitc-2022-006234supp018.xlsx (12KB, xlsx)
jitc-2022-006234supp019.xlsx (11.1KB, xlsx)
jitc-2022-006234supp020.xlsx (14.6KB, xlsx)
jitc-2022-006234supp021.pdf (287KB, pdf)
jitc-2022-006234supp022.xlsx (12.7KB, xlsx)
Data Availability Statement
All data relevant to the study are included in the article or uploaded as supplementary information.