Abstract
The relative stiffness of naked DNA is evident from measured values of longitudinal persistence length (∼150 bp) and torsional persistence length (∼180 bp). These parameters predict that certain arrangements of eukaryotic transcription activator proteins in gene promoters should be much more effective than others in fostering protein-protein interactions with the basal RNA polymerase II transcription apparatus. Thus, if such interactions require some kind of DNA looping, DNA loop energies should depend sensitively on helical phasing of protein binding sites, loop size, and intrinsic DNA curvature within the loop. Using families of artificial transcription templates where these parameters were varied, we were surprised to find that the degree of transcription activation by arrays of Gal4-VP1 transcription activators in HeLa cell nuclear extract was sensitive only to the linear distance separating a basal promoter from an array of bound activators on DNA templates. We now examine the hypothesis that this unexpected result is due to factors in the extract that act to enhance apparent DNA flexibility. We demonstrate that HeLa cell nuclear extract is rich in a heat-resistant activity that dramatically enhances apparent DNA longitudinal and torsional flexibility. Recombinant mammalian high-mobility group 2 (HMG-2) protein can substitute for this activity. We propose that the abundance of HMG proteins in eukaryotic nuclei provides an environment in which DNA is made sufficiently flexible to remove many constraints on protein binding site arrangements that would otherwise limit efficient transcription activation to certain promoter geometries.
DNA looping allows juxtaposition of distant DNA sequences and is thought to play a role in numerous cellular processes, including transcription, DNA replication, and recombination (17, 26). In eukaryotic transcription, activators bound upstream of a basal promoter facilitate transcription initiation by binding to limiting components of the transcription apparatus and delivering these components to the promoter, perhaps via DNA looping (6, 22, 33). The resulting protein-DNA complex recruits RNA polymerase. The thermodynamic stabilities of complexes involving a DNA loop should be dependent on loop size, the intrinsic shape and flexibility of the DNA, and the relative helical orientation of the sites being juxtaposed (2, 23, 26).
Using in vitro DNA ligation assays, the physical properties of naked DNA have been carefully examined, and the energetic costs of DNA bending and twisting have been estimated (reviewed in reference 8). Cyclization rates have been shown to be highest for DNA sequences of approximately 500 bp, with cyclization rates dropping for both shorter and longer DNA fragments (23, 28). Based on these experiments, it has been predicted that for DNA sequences less than ∼500 bp in length, both intrinsic DNA curvature (14) and the relative helical orientation (27, 28) of the sites being juxtaposed should significantly affect the rate of loop formation. Such curvature and phasing effects should increase as DNA length decreases.
In prokaryotic transcription, evidence of such helical phasing (2, 16) and DNA curvature (5, 12; but see reference 1) effects has been detected in vivo. However, in eukaryotes, the effects of DNA phasing and curvature are less clear. The precise helical relationship between individual transcription factor binding sites within the human beta interferon and T cell receptor α gene enhancers has been shown to affect the strength of enhancement (13, 32). Similarly, the helical alignment of simian virus 40 early promoter elements has been shown to be important for transcription initiation in HeLa cells over a very short distance (31). However, examination of a variety of other promoters has revealed that activators often function independently of helical alignment relative to the basal promoter (7, 25, 35). Furthermore, while some evidence suggests that DNA curvature increases synergy between bound activator proteins (15), it is unclear if insertion of intrinsically curved DNA sequences between activators and a basal promoter affects the strength of activation.
Previously members of our group designed a set of 32 eukaryotic transcription templates with different intrinsic shapes in order to examine the effects of DNA curvature and helical orientation on transcription activation (24). The templates were based on the adenovirus E4 basal promoter. We inserted five phased Gal4 binding sites upstream of the promoter and varied the DNA length and shape (number of phased A5 tracts) between the promoter and activator binding sites. We then measured the efficiency of transcription activation in cell-free transcription experiments using HeLa cell nuclear extract (NE) supplemented with Gal4-VP1 activator. Surprisingly, for both linearized and supercoiled templates, spacing between the activator binding sites and the promoter was the primary determinant of the activated level of transcription, with little or no effect of binding site phasing or intrinsic template curvature (24). These results differ substantially from predictions based on looping models of activation by direct protein-protein contacts (23).
Our data suggested the possibility that factors present in NE enhance DNA flexibility, thereby masking the expected differences between the templates predicted based on the behavior of naked DNA in dilute solution. We have therefore performed cyclization assays with transcription templates to examine the flexibility of DNA in the presence and absence of NE. In the absence of NE, we found that DNA length, curvature, and helical phasing all affected cyclization rates as expected. However, apparent DNA flexibility is enhanced in the presence of NE. Consistent with the results of previous transcription experiments by members of our group (24), DNA cyclization rates became independent of DNA curvature and helical phasing and were dependent only on DNA length. We show that recombinant rat high-mobility group 2 (rHMG-2) protein substitutes for NE in promoting cyclization and overcoming DNA curvature and helical phasing effects on cyclization rates.
MATERIALS AND METHODS
DNA multimerization-cyclization assay.
NE was prepared by standard methods (9). rHMG-2 was kindly provided by Dean Edwards. All enzymes were obtained from New England Biolabs. Where indicated, NE or rHMG-2 was heat treated by incubation at 95°C for 5 min, followed by incubation on ice for 5 min and clarification by brief centrifugation. Radiolabeled DNA duplexes (1 μM) were mixed with the indicated amount of NE in 10-μl reactions containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg of bovine serum albumin (BSA)/ml, and 400 U of T4 DNA ligase. Ligation reactions were incubated at 22°C for 30 min. Ten microliters of each ligation reaction was then mixed with 1 μl of water, 1 U of BAL 31 exonuclease, and 3 μl of 5× Nuclease BAL-31 buffer and incubated at 30°C for 30 min. Sodium dodecyl sulfate was then added to 1.5%, followed by incubation at 37°C for 20 min, phenol-chloroform extraction, and ethanol precipitation. DNA circles were resolved on 5% native polyacrylamide gels (acrylamide/bisacrylamide ratio, 75:1), 12 V/cm, at 22°C. Gels were exposed to storage phosphor screens and analyzed using a Molecular Dynamics Storm 840 phosphorimager, and bands were quantitated with ImageQuant software.
Cyclization assay data were quantitated by measuring the storage phosphor signal for each DNA circle. Data were expressed as the signal for a given DNA circle, relative to the total signal for all DNA circles in the lane, and further normalized to account for intrinsic differences in signal among DNA circles of different size, according to the formula
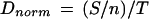 |
1 |
where Dnorm gives the normalized DNA circle yield, S is the storage phosphor signal for a given gel band in arbitrary units, n is the number of ligated duplexes present in that species, and T is the total storage phosphor signal in all DNA circles for a given DNA duplex. In plots of Dnorm versus circle size, areas under the curves are not equal because the numerator in equation 1 is corrected for the number of 21-bp units, while the denominator is not.
Probes for DNA ring closure assay.
Phasing, curved, and uncurved DNA cyclization probes were generated from the promoters of previously studied transcription templates (24). Templates were restricted to remove the five phased Gal4 sites, and a DNA duplex containing two phased Gal4 sites was inserted. PCR products of the indicated series were generated using the promoters from each template, incorporating Acc65I restriction sites at each terminus. These fragments were then subcloned and sequenced. Probe designations indicate base-pair spacing between sequence elements.
To construct longer DNA probes, irrelevant sequences of 154, 254, and 355 bp from plasmid pFW11-null (34) were inserted into the initial series of phasing constructs.
To generate radiolabeled cyclization probes, plasmids were digested with Acc65I and treated with calf intestinal alkaline phosphatase. Restriction fragments were purified by native 5% polyacrylamide gel electrophoresis and radiolabeled using T4 polynucleotide kinase and [γ-32P]ATP. The radiolabeled probes were purified by spermine precipitation.
DNA ring closure assay.
Fifty-microliter reaction mixtures were prepared with 0.1 nM DNA, 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM dithiothreitol, 1 mM ATP, 25 μg of BSA/ml, and 0.08 U of T4 DNA ligase. Where indicated, 0.1 mg of heat-treated NE/ml or 3 μg of heat treated rHMG-2/ml was added. Reactions were incubated at room temperature. At various times, 10-μl aliquots were removed, added to 10 μl of 50 mM EDTA, and treated with sodium dodecyl sulfate. After phenol-chloroform extraction, samples were electrophoresed on native polyacrylamide gels. The fraction cyclized at each time point (Ft) was quantitated by measuring the storage phosphor signal of the circular products and of the unligated linear monomer and applying the formula
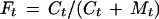 |
2 |
where Ct is the storage phosphor signal for the circular products (arbitrary units) at time t, and Mt is the storage phosphor signal for the unligated linear monomer gel band (arbitrary units). Estimation of J factors (effective concentration of intramolecular DNA termini) was not possible because, in many cases, a significant fraction of starting material was depleted over these experimental time scales in the presence of NE or rHMG-2, violating a required assumption in the derivation of J factors from such assays.
RESULTS
NE contains heat-resistant activities that overcome effects of intrinsic DNA curvature.
To examine whether NE could overcome shape-dependent differences in DNA looping, we studied the multimerization and cyclization of 21-bp DNA duplexes by T4 DNA ligase. As multimer ligation products accumulate, cyclization begins to compete with linear growth. The probability of cyclization is dependent on the local concentration of the termini of a growing multimer, and therefore the distribution of circle sizes reflects both the apparent shape and the flexibility of the individual duplexes.
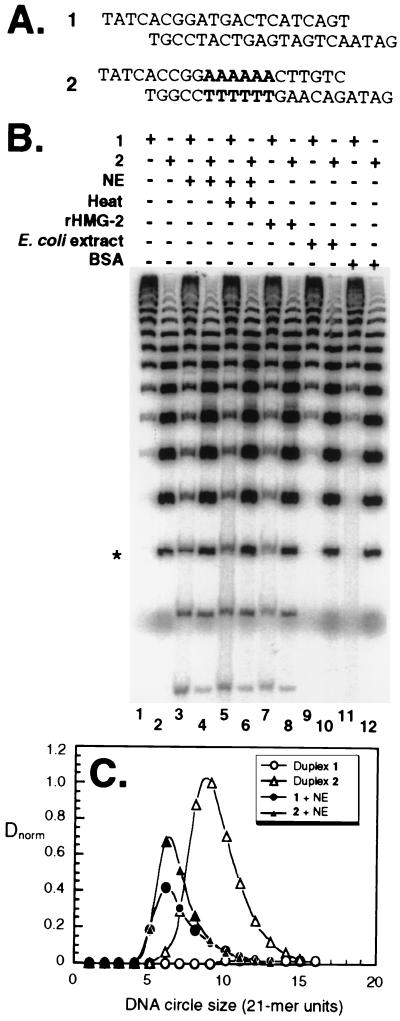
The DNA duplexes used in this experiment are shown in Fig. 1A. Duplex 1 is predicted to be relatively linear. Duplex 2 contains an A6 tract that contributes ∼18° of curvature. Figure 1B displays the circular ligation products obtained from multimerization-cyclization of DNA duplexes 1 and 2 under various conditions. For the naked DNA duplexes, the expected shape-dependent differences are observed (Fig. 1B, compare lanes 1 and 2). Duplex 1 is linear and cyclizes poorly, forming only large circles (Fig. 1B, lane 1). Duplex 2 (Fig. 1B, lane 2) is curved, cyclizes more readily, and forms circles as small as 147 bp.
FIG. 1.
DNA cyclization is enhanced in the presence of NE and rHMG-2. (A) DNA duplexes (21 bp) used in this study. Duplex 1 is predicted to have a linear geometry. Duplex 2 contains an A6 tract (boldface) that induces ∼18° of axial curvature. (B) DNA multimerization-cyclization assay. Gel electrophoretic analysis of circular ligation products obtained after incubation of either duplex 1 or duplex 2 under the indicated conditions. The 147-bp (seven 21-mer units) DNA circle is indicated (*). (C) Quantitation of DNA cyclization. Data are plotted as the normalized circle yield, Dnorm, versus the size of the DNA circle (21-mer units) in the absence (open) or presence (filled) of NE for duplex 1 (circles) and duplex 2 (triangles).
We found NE to be a rich source of an activity that enhanced DNA flexibility. Fifteen nanograms of NE induced DNA cyclization at much smaller sizes than in the absence of NE (Fig. 1B, compare lanes 1 and 2 and lanes 3 and 4). Remarkably, in the presence of NE, duplexes 1 and 2 displayed similar apparent shapes and flexibilities, forming circles as small as 105 bp (Fig. 1B, lanes 3 and 4). Even after heat treatment, NE enhanced DNA flexibility (Fig. 1B, lanes 5 and 6). This observation suggested the involvement of HMG proteins (19, 20). We therefore tested rHMG-2 protein and found it to substitute for NE in enhancing the formation of small circles, diminishing the difference between curved and uncurved templates (Fig. 1B, lanes 7 and 8). In contrast, 15 ng of Escherichia coli extract or bovine serum albumin did not detectably enhance DNA flexibility (Fig. 1B, lanes 9 to 12).
Characterization of DNA looping by transcription templates.
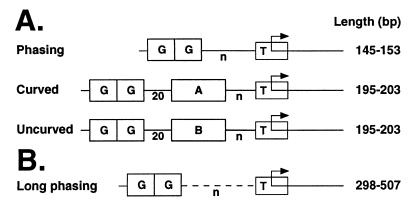
We then tested how NE would affect the flexibility of transcription templates. DNA cyclization, like transcription activation, requires DNA looping. We generated DNA fragments from the promoters of some of our group's previous transcription templates (24) (Fig. 2A) and employed these probes in cyclization assays. We hypothesized that when ligations were performed in the absence of NE, cyclization rates would depend on length, intrinsic DNA curvature, and the precise helical alignment (phasing) of the restriction fragment termini. In contrast, based on our previous transcription results, we predicted that cyclization rates in the presence of NE would be dependent only on DNA probe length. We tested three sets of probes (Fig. 2A). Each set contained four probes that varied in length by a total of 8 bp. All three sets contained two Gal4 DNA binding sites upstream of an adenovirus E4 basal promoter. Phasing probes (Fig. 2A) were designed primarily to test the effects of helical phasing due to DNA torsional stiffness, as measured by DNA cyclization. Curved probes (Fig. 2A) contained 72° of intrinsic curvature due to the presence of four phased A5 tracts. In the uncurved set, intrinsic DNA curvature was replaced with an uncurved sequence of the same length. The curved and uncurved sets of templates were designed to test directly the effect of intrinsic DNA curvature on looping.
FIG. 2.
Schematic design of cyclization probes. Probes were based on transcription templates and contain two tandem 20-bp sequences carrying Gal4 binding sites (G) and the TATA box derived from the adenovirus E4 promoter (T). Probes have Acc65I restriction sites at each terminus. (A) Phasing probes contain a variable-length spacer (n). Curved probes contain four phased A5 tracts (A). This curvature was replaced with a scrambled sequence of identical length in the uncurved templates. (B) Long phasing probes.
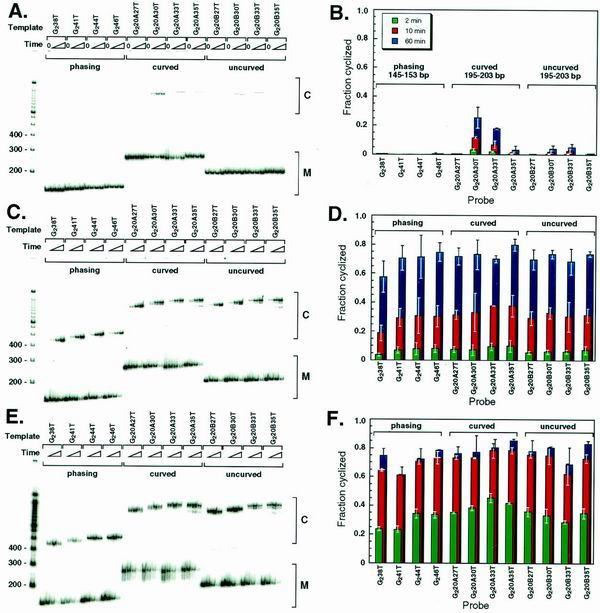
In the absence of NE, the predicted differences in cyclization rates were strikingly observed (Fig. 3A). The set of short phasing probes did not cyclize regardless of the phasing of the termini (Fig. 3A and B), whereas some cyclization was detected for longer uncurved probes with properly phased termini (Fig. 3A and B, probes G220B30T and G220B33T). The curved probes cyclized much more efficiently than uncurved probes of the same lengths (Fig. 3A and B, compare G220A30T to G220A33T and G220B30T to G220B33T). These results confirm expectations for naked DNA. Over short distances, DNA torsional and longitudinal stiffnesses dictate accessible loop geometries such that only properly preconfigured geometries loop appreciably. This result is exactly what had been predicted, but not observed, in our previous studies of in vitro transcription activation (24).
FIG. 3.
Cyclization of curved, uncurved, and phasing probes. (A) Electrophoretic analysis of the DNA probes shown in Fig. 2A after ligation for 0, 2, 10, and 60 min in the absence of NE. (B) Quantitation of panel A. Error bars represent standard deviations after two to five repeats. (C) Electrophoretic analysis of DNA probes in Fig. 2A after ligation for 0, 2, 10, and 60 min in the presence of 0.1 mg of heat-treated NE/ml. (D) Quantitation of panel C. (E) Electrophoretic analysis of the DNA probes shown in Fig. 2A after ligation for 0, 2, 10, and 60 min in the presence of 3 μg of heat-treated rHMG-2/ml. (F) Quantitation of panel E.
In the presence of NE, cyclization is independent of DNA curvature and helical phasing.
In dramatic contrast to the results for naked DNA probes (Fig. 3A and B), all of the probes cyclized with nearly identical efficiency in the presence of NE (Fig. 3C and D). This result indicates a profound change in the physical properties of the DNA: neither helical phasing nor DNA curvature significantly affected cyclization efficiency.
rHMG-2 can substitute for NE.
Based on the ability of rHMG-2 to substitute for NE in promoting cyclization in the multimerization-cyclization assay (Fig. 1), we tested whether rHMG-2 protein could substitute for NE in promoting cyclization and overcoming effects of DNA curvature and helical phasing among the transcription template derivatives shown in Fig. 2A. rHMG-2 protein alone could substitute for NE in enhancing apparent DNA flexibility and mitigating the effects of DNA curvature and helical phasing (Fig. 3E and F).
Longer DNA probes show length-dependent cyclization with and without added NE.
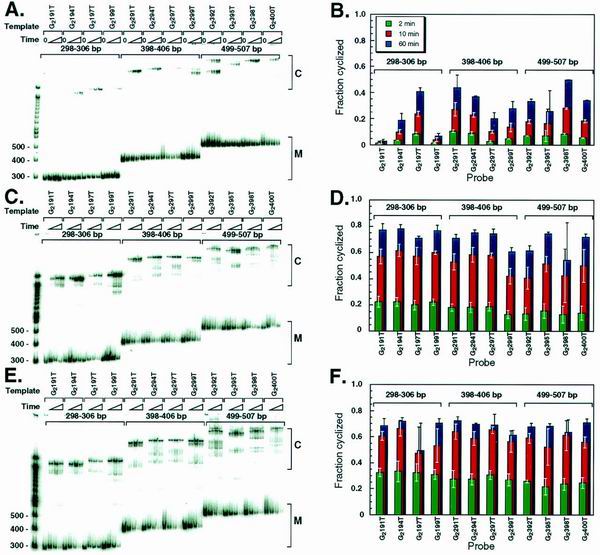
Over the length range so far tested (150 to 203 bp), DNA length did not significantly affect looping efficiency in the presence of NE or rHMG-2. We hypothesized that in the presence of NE or rHMG-2, the length range tested may be near the optimum length for DNA cyclization, and we therefore predicted that longer probes should cyclize less efficiently. To test this notion, we constructed three additional sets of long phasing probes (Fig. 2B). These three longer probe sets were ∼300, ∼400, and ∼500 bp in length, respectively. Each set comprised four probes that spanned a length range of approximately one helical turn.
In the absence of NE, a helical phasing effect was still observed for the long phasing probes (Fig. 4A and B). This effect gradually decreased as the probe length increased (Fig. 4B, compare 298- to 306-bp probes with 499- to 507-bp probes). By contrast, no significant phasing effect was observed in the presence of NE (Fig. 4C and D) or rHMG-2 (Fig. 4E and F). Furthermore, increased torsional flexibility is revealed in multiple topoisomeric forms of circular monomers (e.g., G2395T in Fig. 4A versus C).
FIG. 4.
Cyclization with longer DNA probes. (A) Electrophoretic analysis of the DNA probes shown in Fig. 2B after ligation for 0, 2, 10, and 60 min in the absence of NE. (B) Quantitation of panel A. Error bars represent standard deviations after 2 to 5 repeats. (C) Electrophoretic analysis of the DNA probes shown in Fig. 2B after ligation for 0, 2, 10, and 60 min in the presence of 0.1 mg of heat-treated NE/ml. (D) Quantitation of panel C. (E) Electrophoretic analysis of the DNA probes in shown Fig. 2B after ligation for 0, 2, 10, and 60 min in the presence of 3 μg of heat-treated rHMG-2/ml. (F) Quantitation of panel E.
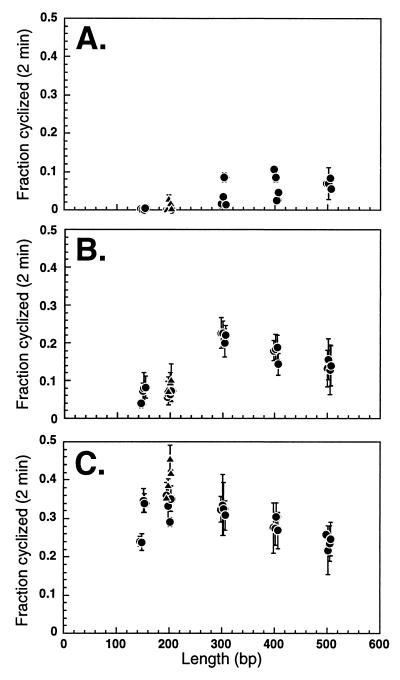
The effects of NE and rHMG-2 are particularly apparent when the fraction of each probe cyclized after 2 min is plotted against the probe length (Fig. 5). Three major effects of NE and rHMG-2 are observed. First, the optimal length for cyclization is shifted. For naked DNA, cyclization rates increased with increasing DNA length up to a length of ∼400 bp (Fig. 5A). These results are consistent with previously reported DNA J factor values, which peak at ∼500 bp (23). However, in the presence of NE, the optimal length for cyclization was shifted to ∼300 bp (Fig. 5B). rHMG-2 further shifted the optimal cyclization length to ∼200 bp (Fig. 5C). Second, NE and rHMG-2 both significantly enhanced cyclization rates (Fig. 5). Finally, the vertical data scatter for cyclization of naked DNA (Fig. 5A) was reduced by NE (Fig. 5B) and rHMG-2 (Fig. 5C), emphasizing the capacity of these factors to overcome the torsional rigidity of DNA.
FIG. 5.
NE and rHMG-2 change the optimal DNA length for cyclization. Fractions of DNA probes cyclized in 2-min ligation reactions (from Fig. 3 and 4) are plotted as a function of probe length. Curved templates are indicated with triangles, while all other probes are indicated with circles. (A) Naked DNA. (B) DNA in the presence of NE. (C) DNA in the presence of rHMG-2.
NE enhances DNA cyclization but not intermolecular dimerization.
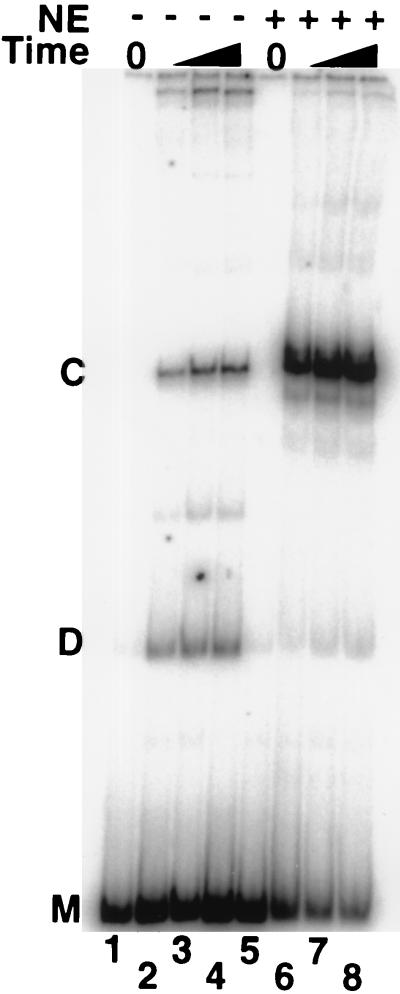
We considered the possibility that NE or rHMG-2 protein affected the overall ligation rate rather than specifically the cyclization rate. We therefore tested the effect of NE on intermolecular dimerization of DNA probes. When ligation reactions were performed at higher DNA concentrations such that DNA cyclization and dimerization occurred at comparable rates, the cyclization rate, but not the dimerization rate, was increased by NE (Fig. 6). In the absence of NE, time-dependent ligation under these conditions yields both monomer circles and linear dimers in comparable yields (Fig. 6, lanes 1 to 4). In contrast, upon addition of NE, cyclic products are dramatically enhanced at the expense of linear dimers (Fig. 6, lanes 5 to 8). If NE enhanced cyclization by a general stimulation of DNA end-joining activity, the yield of both circular and linear products should have increased. Instead, the results indicate that NE specifically facilitates cyclization and does not contribute to the intermolecular ligation rate, consistent with enhancement of DNA flexibility. In control experiments where DNA ligase was omitted, heat-treated NE did not contribute detectable ligase activity (data not shown).
FIG. 6.
NE specifically enhances DNA cyclization rather than intermolecular ligation. Electrophoretic analysis of the uncurved DNA probe G220B33T (200 bp) after ligation for 0, 2, 10, and 60 min in the absence or presence of NE. The DNA concentration was increased to enhance intermolecular dimer ligation products (D) to a yield comparable to intramolecular circular ligation products (C). M indicates unligated linear probe precursor. Upon addition of NE, product C is enhanced at the expense of product D, consistent with increased DNA flexibility rather than increased total end-joining activity.
Enhancement of DNA flexibility by NE and rHMG-2.
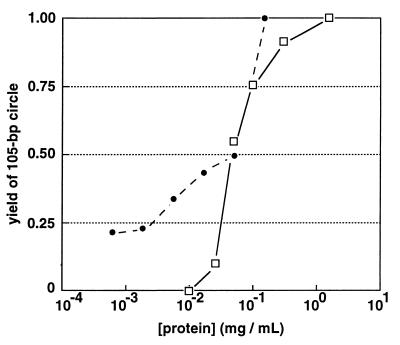
It is of interest to know whether the level of free nuclear HMG activity in vivo is sufficient to eliminate DNA phasing and curvature effects over the distances tested here. Although it is not possible to estimate what fraction of HMG activity is free in vivo or what fraction of bulk DNA is accessible to HMG binding, we used a DNA multimerization-cyclization assay comparable to that shown in Fig. 1 to titrate the activities of heat-treated NE and rHMG-2 in the presence of 1 μM DNA (Fig. 7). Enhancement of DNA flexibility was monitored as the yield of 105-bp DNA circles upon addition of DNA ligase to a 1 μM solution of DNA duplex 1 (Fig. 1). No 105-bp circular product is detected in the absence of HMG activity. Yields were normalized to the amount of 105-bp circle obtained at saturation. Dilution of both NE and rHMG-2 preparations resulted in decreased DNA flexibility, as anticipated. Remarkably, the activity of crude NE per unit mass of protein was roughly comparable to that of purified rHMG-2 (Fig. 7). rHMG-2 retained 50% of its maximal DNA flexibility enhancement activity at a concentration of ∼2 μM, corresponding to a 2:1 rHMG-2/DNA ratio. This result suggests that NE must contain a variety of HMG activities, some of them more potent than rHMG-2.
FIG. 7.
Quantitation of HMG activities. A DNA multimerization-cyclization assay (see Materials and Methods and Fig. 1) was employed to monitor the ability of HMG activity to promote formation of a 105-bp circular product upon addition of DNA ligase to a 1 μM solution of 21-bp DNA duplex 1 (see Fig. 1 for the sequence) over a range of NE (□) or rHMG-2 (●) dilutions. No 105-bp circle could be detected under these conditions upon ligation in the absence of HMG activity. Yield of 105-bp circle was normalized to the yield at the highest protein concentration tested, which appeared to be saturating for both NE and rHMG-2. In the case of rHMG-2, a 0.5 fractional yield of 105-bp circle was observed at a ∼2 μM concentration of protein, corresponding to a 1:2 DNA/protein stoichiometry.
DISCUSSION
High mobility group proteins 1 and 2 (HMG1/2) are abundant, highly conserved, chromatin-associated proteins (11). Previous reports have suggested that HMG1/2 may play roles in numerous processes including transcription (29, 30) and nucleosome assembly (4, 18). HMG1/2 are capable of low-affinity binding to a wide variety of DNA sequences with little sequence specificity (19) and display a high level of affinity for bent structures, such as cruciforms (3) and cisplatin modified DNA (21). These proteins bind in the DNA minor groove and cause significant DNA bending. How might the random binding of HMG proteins enhance apparent DNA flexibility? Repeated transient nonspecific binding by HMG1/2 will be accompanied by strong transient bending and a slight unwinding of the DNA with each binding event. Because these bending and unwinding events will be distributed randomly, the average properties of the DNA chain will be drastically altered. Rapid fluctuations due to random bending and unwinding events will reduce longitudinal and torsional persistence lengths, increasing DNA longitudinal and torsional flexibility, as we observed. Such a process will permit the DNA to sample randomly more extreme conformations with each HMG binding event than would otherwise be probable. In effect, the energy of the protein-DNA complex is thus used to overcome DNA stiffness.
The present studies involve DNA transcription templates incubated in the presence of nuclear extract but not deliberately assembled into chromatin. The remarkable ability of HMG proteins to facilitate DNA looping and overcome helical phasing effects would have obvious physiological significance in cases where nucleosomes are disrupted by chromatin remodeling prior to looping of the unencumbered DNA to allow protein-protein interactions at a distance. It remains unclear whether such DNA unpacking typically precedes direct recruitment of the basal transcription machinery (presumably by DNA looping) during the process of transcription activation. In cases where nucleosomal packing might participate directly in facilitating protein-protein interactions at a distance, the relevance of the activities we demonstrate for HMG proteins is less clear. Thus, it is not known to what extent DNA constrained on histone octamers is a substrate for transient HMG binding, bending, and unwinding. Local effects of HMG binding in the context of chromatin are unlikely to propagate over distances as they would for naked DNA. The ability of HMG proteins to alter the properties of exposed linker DNA could remain of profound significance in fostering protein-protein interactions in the context of chromatin.
While the ability of HMG proteins to promote DNA looping has been previously reported (19), what is particularly striking about our results is the ability of these factors to virtually eliminate the effects of intrinsic DNA curvature and helical phasing over the DNA lengths tested (Fig. 3 to 5). The cyclization results in the presence of NE or rHMG-2 parallel our previous observations in cell-free transcription experiments. In both cases, DNA curvature, length, and helical phasing were predicted to have significant effects on the reaction rates. However, in both cases, only DNA length had a measured effect. We propose that these results should be interpreted as consistent with (i) a role of DNA looping in transcription activation in vitro and (ii) a prominent role for HMG proteins in promoting such looping.
While the effect of NE on cyclization persisted even in the presence of 2,000-fold-excess nonspecific competitor plasmid DNA (data not shown), it is difficult to accurately estimate either the concentration of free HMG-2 in the nucleus or the fraction of chromosomal DNA available at any given time for binding by HMG proteins. Therefore, it remains unclear to what extent the profound activities of HMG proteins in our in vitro experiments reflect activities expected in the nuclei of living eukaryotic cells.
Nevertheless, the relative abundance of HMG1/2 (105 to 106 molecules/nucleus [10]) suggests that the helical phasing and DNA curvature effects observed in traditional in vitro cyclization assays with naked DNA may not be relevant to DNA in eukaryotic nuclei. HMG proteins have the potential to relieve constraints on promoter geometry that would otherwise limit interactions of proteins acting at a distance. These results are consistent with previous reports that HMG1/2 can act as general transcription factors (30). Our experiments show that even acting alone, HMG proteins profoundly alter the apparent physical properties of DNA, endowing this otherwise locally stiff polymer with enhanced longitudinal and torsional flexibility. We argue that deployment of HMG proteins fundamentally alters the behavior of DNA and promotes interactions between proteins tethered to DNA far beyond what would otherwise be predicted (23, 26).
ACKNOWLEDGMENTS
We thank Michael Carey, Rod Hori, Jason Kahn, and members of the Maher Lab for materials, helpful discussion, and assistance. We acknowledge Dean Edwards for providing rHMG-2, M. Doerge in the Mayo Foundation Molecular Biology Core Facility for providing excellent oligonucleotide synthesis services, and the National Cell Culture Center for HeLa cells.
This work was supported by the Mayo Foundation and NIH grant GM54411 to L.J.M.
REFERENCES
- 1.Aiyar S E, Gourse G L, Ross W. Upstream A-tracts increase bacterial promoter activity through interactions with RNA polymerase α subunit. Proc Natl Acad Sci USA. 1998;95:14652–14657. doi: 10.1073/pnas.95.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellomy G R, Mossing M C, Record M T., Jr Physical properties of DNA in vivo as probed by the length dependence of the lac operator looping process. Biochemistry. 1988;27:3900–3906. doi: 10.1021/bi00411a002. [DOI] [PubMed] [Google Scholar]
- 3.Bianchi M E, Beltrame M, Paonessa G. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 4.Bonne-Andrea C, Harper F, Sobczak J, De Recondo A M. Rat liver HMG1: a physiological nucleosome assembly factor. EMBO J. 1984;3:1193–1199. doi: 10.1002/j.1460-2075.1984.tb01950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracco L, Kotlarz D, Kolb A, Diekmann S, Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989;8:4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 7.Chodosh L A, Carthew R W, Morgan J G, Crabtree G R, Sharp P A. The adenovirus major late transcription factor activates the rat gamma-fibrinogen promoter. Science. 1987;238:684–688. doi: 10.1126/science.3672119. [DOI] [PubMed] [Google Scholar]
- 8.Crothers D M, Drak J, Kahn J D, Levene S D. DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol. 1992;212:3–29. doi: 10.1016/0076-6879(92)12003-9. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duguet M, de Recondo A M. A deoxyribonucleic acid unwinding protein isolated from regenerating rat liver. Physical and functional properties. J Biol Chem. 1978;253:1660–1666. [PubMed] [Google Scholar]
- 11.Falciola L, Spada F, Calogero S, Langst G, Voit R, Grummt I, Bianchi M E. High mobility group 1 protein is not stably associated with the chromosomes of somatic cells. J Cell Biol. 1997;137:19–26. doi: 10.1083/jcb.137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gartenberg M R, Crothers D M. Synthetic DNA bending sequences increase the rate of in vitro transcription initiation at the Escherichia coli lac promoter. J Mol Biol. 1991;219:217–230. doi: 10.1016/0022-2836(91)90563-l. [DOI] [PubMed] [Google Scholar]
- 13.Giese K, Kingsley C, Kirshner J R, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 14.Kahn J D, Crothers D M. DNA bending in transcription initiation, p. 115–122. Cold Spring Harbor Symp Quant Biol. 1993;58:115–122. doi: 10.1101/sqb.1993.058.01.015. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Klooster S, Shapiro D J. Intrinsically bent DNA in a eukaryotic transcription factor recognition sequence potentiates transcription activation. J Biol Chem. 1995;270:1282–1288. doi: 10.1074/jbc.270.3.1282. [DOI] [PubMed] [Google Scholar]
- 16.Lee D H, Schleif R F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci USA. 1989;86:476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews K S. DNA looping. Microbiol Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nightingale K, Dimitrov S, Reeves R, Wolffe A P. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996;15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 19.Paull T T, Haykinson M J, Johnson R C. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–1534. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 20.Pil P M, Chow C S, Lippard S J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc Natl Acad Sci USA. 1993;90:9465–9469. doi: 10.1073/pnas.90.20.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pil P M, Lippard S J. Specific binding of chromosomal protein HMG1 to DNA damaged by the anticancer drug cisplatin. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 22.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 23.Rippe K, von Hippel P H, Langowski J. Action at a distance: DNA-looping and initiation of transcription. Trends Biol Sci. 1995;20:500–506. doi: 10.1016/s0968-0004(00)89117-3. [DOI] [PubMed] [Google Scholar]
- 24.Ross E D, Keating A M, Maher L J. DNA constraints on transcription activation in vitro. J Mol Biol. 2000;297:321–334. doi: 10.1006/jmbi.2000.3562. [DOI] [PubMed] [Google Scholar]
- 25.Ruden D M, Ma J, Ptashne M. No strict alignment is required between a transcriptional activator binding site and the “TATA box” of a yeast gene. Proc Natl Acad Sci USA. 1988;85:4262–4266. doi: 10.1073/pnas.85.12.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schleif R. DNA looping. Annu Rev Biochem. 1992;61:199–223. doi: 10.1146/annurev.bi.61.070192.001215. [DOI] [PubMed] [Google Scholar]
- 27.Shore D, Baldwin R L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983;170:957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- 28.Shore D, Langowski J, Baldwin R L. DNA flexibility studied by covalent closure of short fragments into circles. Proc Natl Acad Sci USA. 1981;78:4833–4837. doi: 10.1073/pnas.78.8.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shykind B M, Kim J, Sharp P A. Activation of the TFIID-TFIIA complex with HMG-2. Genes Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- 30.Singh J, Dixon G H. High mobility group proteins 1 and 2 function as general class II transcription factors. Biochemistry. 1990;29:6295–6302. doi: 10.1021/bi00478a026. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K, Vigneron M, Matthes H, Wildeman A, Zenke M, Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986;319:121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- 32.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 33.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy clues. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 34.Whipple F W. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in Escherichia coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirth T, Staudt L, Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987;329:174–178. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]