ABSTRACT
The hallmark of cellular events observed upon macroautophagic/autophagic induction is the conjugation of LC3B, one of the mammalian Atg8 homologs, with phosphatidylethanolamine. This conversion from LC3B-I (an unconjugated form) to LC3B-II (a conjugated form) is essential for phagophore expansion and formation of autophagosomes. Our recent study revealed that LC3B binds to RNAs with a preference for the consensus AAUAAA motif and recruits the CCR4-NOT deadenylase complex. Consequently, LC3B elicits rapid degradation of mRNAs, which we have termed as LC3B-mediated mRNA decay (LMD). LMD requires the conversion of LC3B-I to LC3B-II and occurs before the formation of autolysosomes. Furthermore, we identified PRMT1 mRNA, which encodes a protein that functions as a negative regulator of autophagy, as an LMD substrate. A failure of rapid degradation of PRMT1 mRNA via LMD results in inefficient autophagy. Thus, our study unravels an important role of LC3B in autophagy as an RNA-binding protein for efficient mRNA decay.
KEYWORDS: ATG8, autophagy, CCR4-NOT deadenylase, LC3B, mRNA decay, PRMT1
The network of interactions between RNAs and RNA-binding proteins (RBPs) is pivotal in RNA biology. Recent advances in the large-scale identification of RBPs have revealed that human cells express more than 1,000 different such proteins. Many of these contain one or more canonical RNA-binding domains, such as the RNA recognition motif, K-homology domain, and zinc-finger domain. However, hundreds of proteins identified as RBPs lack any previously characterized RNA-binding domains, implicating the existence of a vast number of unexplored RBPs, possibly with their own biological and molecular functions.
RNA biology is now being connected with autophagy. During autophagy, various cellular components are eliminated through lysosomes in either a selective or nonselective manner. Autophagy takes place in the following sequential manner: nucleation of a double membrane called a phagophore, sequestration of cellular materials within double-membrane autophagosomes, fusion of autophagosomes with lysosomes to form autolysosomes, and degradation of cellular materials within autolysosomes. These stepwise processes are tightly coordinated by ATG (autophagy related) proteins.
In our recent study [1] , we demonstrated that LC3B, a mammalian LC3 isoform involved in the formation of phagophores, has RNA-binding ability. Transcriptome-wide analysis using cross-linking immunoprecipitation coupled with high-throughput sequencing (CLIP-seq) revealed that LC3B is an RNA-binding protein with a preference for an AAUAAA consensus motif. The direct binding of LC3B to the AAUAAA motif was corroborated by an electrophoretic mobility shift assay and fluorescence polarization assay using purified recombinant LC3B and an in vitro-synthesized RNA probe harboring the AAUAAA motif.
What molecular events are affected by LC3B binding to RNA? Although the AAUAAA motif is a well-known polyadenylation signal found in the 3′-untranslated region (3′ UTR) of most eukaryotic mRNAs, the frequency of alternative polyadenylation was not affected by LC3B downregulation. We next analyzed changes in mRNA abundance and half-life under the conditions wherein cells were treated with either rapamycin (a potent inhibitor of MTOR kinase and thus an inducer of autophagy) or siRNA against LC3B. The results showed that rapamycin treatment considerably reduces both the abundance and half-life of LC3B-bound mRNAs (mRNAs harboring peaks in LC3B CLIP-seq) when compared to LC3B-unbound mRNAs (mRNAs lacking peaks in LC3B CLIP-seq). Of note, we observe the most drastic reduction in the abundance and half-life when mRNAs have an LC3B peak at the AAUAAA motif in the 3′ UTR. Intriguingly, the reduction is significantly reversed by LC3B downregulation. These observations indicate that the direct interaction between LC3B and target mRNAs triggers rapid mRNA degradation, which we refer to as LC3B-mediated mRNA decay (LMD).
Among the top-ranked potential LMD substrates obtained from our transcriptome analysis, we chose PRMT1 mRNA, the protein encoded by which functions as a negative regulator of autophagy. Our CLIP-seq results showed that PRMT1 mRNA contains a single LC3B peak at the AAUAAA motif of its 3′ UTR. From a series of experiments using PRMT1 mRNA as an LMD reporter, we uncovered several molecular features of LMD (Figure 1). First, we found that autophagic induction promotes the binding of LC3B to LMD substrates. During this process, RNA secondary or tertiary structures upstream of the AAUAAA motif facilitate the efficient loading of LC3B onto the AAUAAA motif. Although most eukaryotic mRNAs harbor the AAUAAA motif for polyadenylation, LC3B does not interact with all the mRNAs harboring this motif. Therefore, it is most likely that the RNA structure itself and/or the associated RBPs may communicate with LC3B and provide additional binding specificity and strength of LC3B toward LMD substrates. Second, we observed that efficient LMD depends on the conjugation of LC3B with phosphatidylethanolamine (conversion of LC3B-I to LC3B-II), which is required for the formation of autophagosomes. In support of these observations, treatment of cells with 3-methyladenine, a chemical inhibitor that blocks autophagosome formation, abolishes LMD. In contrast, LMD efficiency is not significantly affected upon treatment with a chemical inhibitor (bafilomycin A1 or chloroquine) that blocks the fusion of autophagosomes with lysosomes, indicating that LMD occurs concomitantly with autophagosome formation and occurs before the formation of autolysosomes. Third, LC3B is associated with the CCR4-NOT deadenylase complex. Notably, autophagic activation causes an increased association between LC3B and the CCR4-NOT complex both outside and within the autophagic puncta. These data indicate that LMD may initiate mRNA degradation even before the complete formation of autophagic puncta. Finally, the efficient degradation of PRMT1 mRNA via LMD facilitates autophagy.
Figure 1.
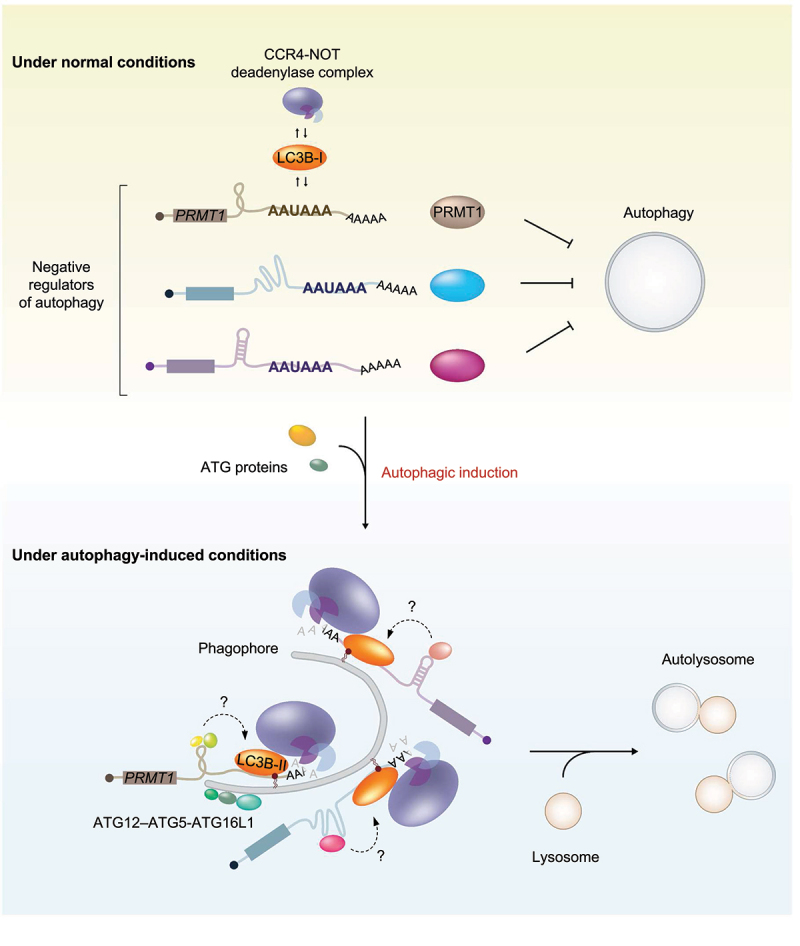
A proposed model illustrating the dual role of LC3B upon autophagic induction. Under normal conditions, LC3B binds to the AAUAAA motif in the target mRnas; however, LMD is not efficient because of a weak interaction between LC3B and its target mRnas, a lack of LC3B conversion, and inefficient association with LC3B and the CCR4-NOT complex. Immediately after the autophagic induction, LC3B-I is conjugated to phosphatidylethanolamine and converted into LC3B-II at the phagophore membrane causing an expansion to form an autophagosome. Concomitantly, an interaction between LC3B and LMD substrates becomes stronger and recruits the CCR4-NOT deadenylase complex. Consequently, LMD substrates (such as mRnas encoding proteins that function as negative regulators of autophagy) are destabilized before the formation of autolysosomes, facilitating efficient autophagy. During this process, RNA secondary or tertiary structures upstream of the AAUAAA motif and the RBPs associating with the structures may crosstalk with the downstream LC3B-II complex for efficient LMD.
These observations lead to an important question regarding why autophagy employs LMD. Considering that (i) LMD is a target-specific mRNA degradation pathway, (ii) efficient autophagy requires degradation of PRMT1 mRNA, an LMD substrate, and (iii) PRMT1 protein acts as a negative regulator of autophagy, it is plausible that immediately after autophagic activation, LMD may shape and orchestrate the cellular transcriptome to prepare an appropriate environment for efficient autophagy. Indeed, our transcriptome analysis suggests another LMD substrate, mRNA encoding DDIT4 (DNA damage inducible transcript 4), whose induction causes impaired autophagy in dry eye.
In summary, we have demonstrated that LC3B plays dual roles in autophagy, namely as a cellular factor for phagophore formation and as an RBP for rapid mRNA degradation. These two distinct functions of LC3B may be independent or interconnected. It is also interesting to know how the conversion of LC3B-I to LC3B-II drives the recruitment of the CCR4-NOT complex to LMD substrates. In addition, further studies are required to understand the additional factors that may contribute to the specificity and affinity of LC3B binding to RNA.
Funding Statement
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korea Government (Ministry of Science, ICT, and Future Planning [NRF-2015R1A3A2033665 and NRF-2018R1A5A1024261].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Reference
- [1].Hwang HJ, Ha H, Lee BS, et al. LC3B is an RNA-binding protein to trigger rapid mRNA degradation during autophagy. Nature Commun. 2022 Mar 17;13:1436. DOI: 10.1038/s41467-022-29139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


