Abstract
The Ty5 retrotransposons of Saccharomyces cerevisiae integrate preferentially into regions of silent chromatin at the telomeres and silent mating loci (HMR and HML). We define a Ty5-encoded targeting domain that spans 6 amino acid residues near the C terminus of integrase (LXSSXP). The targeting domain establishes silent chromatin when it is tethered to a weakened HMR-E silencer, and it disrupts telomeric silencing when it is overexpressed. As determined by both yeast two-hybrid and in vitro binding assays, the targeting domain interacts with the C terminus of Sir4p, a structural component of silent chromatin. This interaction is abrogated by mutations in the targeting domain that disrupt integration into silent chromatin, suggesting that recognition of Sir4p by the targeting domain is the primary determinant in Ty5 target specificity.
The long terminal repeat (LTR) retrotransposons are a large and ubiquitous class of mobile genetic elements. Like their cousins the retroviruses, they replicate by reverse transcribing an element mRNA and then integrating the cDNA product into their host's chromosomes. LTR retrotransposons are typically abundant components of nuclear genomes, constituting a few percentages of the Saccharomyces cerevisiae genome to over 50% of the genomes of some plants such as maize (31, 45). As the genome sequencing projects progress, it is apparent that most retrotransposons are not randomly distributed on chromosomes. In Drosophila melanogaster and Arabidopsis thaliana, for example, retrotransposons are highly enriched in pericentromeric heterochromatin (19, 44). This nonrandom distribution may be the result of preferential integration to these sites. It has been suggested that the low gene density of heterochromatin may offer a safe haven for transposition, which ensures persistence of retrotransposons by avoiding the harmful consequences of mutations that might occur if integration were random (5). Because repetitive sequences can form heterochromatin in some species, the accumulation of retrotransposons in certain regions of the genome may, in turn, contribute to the formation of chromatin domains (18).
How is it that retrotransposons identify certain chromosomal regions during integration? One model suggests that the integration apparatus recognizes specific chromatin states or DNA-bound protein complexes (7). This interaction tethers the integration machinery to target sites and results in the observed target site biases. This model is best supported by studies of the S. cerevisiae retrotransposons. Over 90% of native Ty1, Ty2, Ty3, and Ty4 insertions are located upstream of genes transcribed by RNA polymerase III (RNAP III) (31). These regions are often gene poor and, like heterochromatin, may provide a safe haven for transposition within the streamlined S. cerevisiae genome (5). For Ty1 and Ty3, the association with sites of RNAP III transcription is due to targeted integration. Targeting requires assembly of the RNAP III transcription complex, and promoter mutations in target genes that prevent transcription complex assembly render them inefficient targets (8, 9, 13). In vitro targeted-transposition assays have been developed for Ty3 in which binding of TFIIIB and TFIIIC to tRNA gene templates is sufficient for targeting (32). The critical factors within these complexes appear to be the TATA binding protein and Brf (also called TFIIIB70) (50). These data support the model that targeting results when the retrotransposon preintegration complex recognizes specific DNA-bound proteins.
In contrast to the other S. cerevisiae retrotransposons, native Ty5 elements are not located at sites of RNAP III transcription. Rather, like retrotransposons in many other organisms, Ty5 insertions are predominantly found within the heterochromatin-like domains of the S. cerevisiae genome, such as at the telomeres and silent mating loci (HMR and HML) (57). The chromatin at these sites is referred to as silent chromatin, because it represses transcription of genes located in these regions. Silent chromatin is made up of a large number of proteins that assemble at specific DNA sequences (reviewed in references 35 and 36). Several proteins or protein complexes bind the E and I silencers that flank HMR and HML, including the origin recognition complex (ORC), the transcription factor Abf1, and the repressor activator protein Rap1p. These proteins also bind to sequences near the telomeres: ORC and Abf1 bind to the subtelomeric X repeat, and Rap1p binds to the telomeric repeat sequences (TG1–3). These DNA-bound proteins recruit additional components of silent chromatin, including the well-studied Sir proteins. Sir2p is a histone deacetylase (21, 33, 47), and Sir3p and Sir4p are considered structural components of silent chromatin. All three proteins interact with each other, and they nucleate at the silencers and spread outward along the chromosome (17, 40).
Ty5 integrates preferentially into regions of silent chromatin. Over 95% of de novo Ty5 transposition events occur within a 3-kb window on either side of the HM silencers or the subtelomeric X repeat (54, 55). Silent chromatin is required for this target choice, because mutations in HMR-E that prevent its assembly abolish targeting to this locus (56). Targeting decreases by approximately 50% in sir2Δ strains, whereas it is virtually abolished in sir3Δ and sir4Δ strains (53). An allele of SIR4 (sir4-42) causes a dramatic change in the chromosomal distribution of the Sir complex (29). In sir4-42 strains, Sir3p and Sir4p move from the telomeres and silent mating loci to the ribosomal DNA (rDNA) (30). This change in Sir protein distribution is related to mother cell aging, and sir4-42 strains are long-lived (29). Ty5 target specificity changes with the chromosomal distribution of the Sir complex in sir4-42 strains, and over 25% of the insertions occur within the rDNA (53). These results suggest that the Sir complex, particularly Sir3p and Sir4p, determines Ty5 target choice.
Ty5-encoded proteins are also important for target site selection. A Ty5 missense mutation decreases targeting more than 20-fold and provides the first direct evidence that retroelements encode their own targeting determinants (14). In this paper, we further define Ty5-encoded factors required for targeting and describe a short targeting domain (TD) near the integrase C terminus (INC). This TD interacts with silent chromatin because, when tethered to a defective HMR-E silencer, reporter genes at HMR are transcriptionally silenced in a Sir-dependent fashion. Overexpression of the TD disrupts telomeric silencing, likely by titrating away critical silencing components. We show that the TD interacts with Sir4p in both yeast two-hybrid and in vitro binding assays. Sir4p, therefore, appears to be the primary host determinant mediating Ty5 target specificity, and the interaction between the Ty5-encoded TD and Sir4p appears to determine target choice.
MATERIALS AND METHODS
Mutagenesis of Ty5 elements.
A BspEI-PflMI fragment of Ty5 (Fig. 1) was mutagenized by PCR, using two different protocols to minimize mutation biases. The first used the nucleoside triphosphate analogue dPTP, which pairs with both A and G and thereby increases the mutation spectrum (51). A typical 20-μl reaction mixture included 5 ng of template DNA (pXW27, a plasmid containing the BspEI-PflMI fragment), 2.5 U of Taq polymerase, 2 μl of 10× buffer, 0.5 μl of the universal and reverse primers (20 pM), 1.6 μl of 25 mM MgCl2, 4 μl of deoxynucleoside triphosphates (dNTPs) (2.5 mM each), and 2.5 μl of dPTP (400 μM). The PCR was carried out for five cycles as follows: 92°C for 1 min, 50°C for 1.5 min, and 72°C for 5 min. An aliquot of the reaction mixture (0.5 μl) was then used for PCR amplification without dPTP (3). The second mutagenesis method used Mn2+ (rather than Mg2+) and biased amounts of dNTPs (46). A typical 50-μl reaction mixture included 5 ng of template DNA, 2.5 U of Taq polymerase, 5 μl of 10× buffer, 0.5 μl of each primer (20 pM), 14 μl of 25 mM MgCl2, 0.75 μl of 10 mM MnCl2, 4 μl of dNTPs (2.5 mM each), 4 μ l of dCTP (10 mM), and 4 μl of dTTP (10 mM). The PCR was carried out for 13 cycles as follows: 94°C for 30 s, 50°C for 45 s, and 72°C for 3 min. An aliquot of the reaction mixture (0.5 μl) was used as a template in a standard PCR amplification (3). The mutant Ty5 library was constructed by replacing the mutagenized BspEI-PflMI fragment with the corresponding fragment in a wild-type Ty5 element on pNK254 (27).
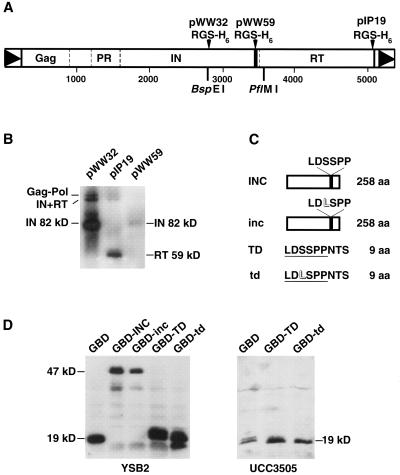
FIG. 1.
The Ty5 TD is located at the C terminus of integrase. (A) Ty5 is 5,375 bp in length. It expresses a full-length protein of 182 kDa, which is processed by protease (PR) into Gag, IN, and RT (22). The cleavage sites, based on the mobilities of mature proteins by SDS-PAGE, are shown by dashed lines. The black bar marks the position of TD. The BspEI and PflMI sites define the region of IN used in the mutagenesis experiment. pIP19, pWW32, and pWW59 carry Ty5 elements that were modified by an RGS-H6 tag. The tag replaced TD (pWW59) or was inserted either into the middle of IN (pWW32) or at the end of RT (pIP19) (22). (B) The modified Ty5 elements were expressed in yeast, and an anti-RGS-H6 antibody was used to identify IN or RT on immunoblots. The partially processed and mature protein species are indicated. (C) The Ty5 IN fragments used throughout this paper are shown. INC and inc are 258 aa long (the small letters indicate the version with the S1094L mutation). TD and td represent the wild-type and mutant TD plus 3 flanking aa from Ty5. (D) Western blots demonstrating that the wild-type and mutant GBD fusion proteins are expressed at comparable levels in the test strains (YSB2 and UCC3505). GBD-INC and GBD-TD have molecular masses of approximately 47 and 19 kDa, respectively.
Mutations were made by PCR-based site-directed mutagenesis to define the TD. pNK254 was PCR amplified using the reverse primer and a mutagenic primer. The amplification product was digested with EcoRI and then inserted into the EcoRI site of pWW37, a Ty5 subclone containing an HpaI-SacI fragment. The BspEI-PflMI fragment of the recombinant plasmid was used to replace the corresponding fragment in pNK254. The mutagenic primers were as following: DVO754 for mutant pWW39 (5′-GGA-ATT-CAA-TCG-AAT-CTC-CTC-CAT-CGG-TGG-ATT-CAT-C), DVO755 for mutant pWW40 (5′-GGA-ATT-CAA-TCG-AAT-CTC-CTC-CAT-CGT-TGG-CTT-CAT-CGC-C), DVO756 for mutant pWW41 (5′-GGA-ATT-CAA-TCG-AAT-CTC-CTC-CAT-CGT-TGG-ATT-CAT-CGG-CTC-CAA-ATA-C), DVO757 for mutant pWW42 (5′-GGA - ATT - CAA - TCG - AAT - CTC - CTC - CAT - CGT - TGG - ATT - CAT - CGC - CTC - CAG-CTA-CCT-CAT-TT), DVO631 for mutant pXW198 [5′-GGA-ATT-CAA-TCG-AA(T/G)-CTC-CTC-CA(T/G)-CGT-TGG-AT], DVO632 for mutants pXW199, pXW200, and pXW201 [5′-GGA-ATT-CAA-TCG-AAT-CT(C/G)-CT(C/G)-CAT-CGT-TG], and DVO634 for mutant pXW202 [5′-GGA-ATT-CGG - TCG - AAT - CTC - CTC - CAT - CGT - TGG - ATT - CAT - CGC - CTC - CAA - AT(A/G)-CC(T/G)-CAT-TTA-AC].
The Ty5 mutants rut3, rut15, rut31, rut38, rut41, and rut46 were constructed by replacing an EcoRI-PflMI fragment of the wild-type Ty5 element on pNK254 with the corresponding fragment from mutants ut3, ut15, ut31, ut38, ut41, and ut46. Because of the multiple EcoRI sites in Ty5, this was accomplished in two steps: first, an EcoRI-EcoRI fragment from the original mutant element was inserted into the EcoRI site of plasmid pWW37, which contains an HpaI-SacI fragment of Ty5; second, the BspEI-PflMI fragment of the resulting plasmid was used to replace the corresponding fragment of pNK254. For all mutant elements tested in this study, targeting was measured using our plasmid-based targeting assay described in detail in our previous study (14).
Tethered silencing.
Gal4p DNA binding domain (GBD) fusion proteins were generated using pGBD plasmids, which contain the GAL4 DNA binding domain under the control of the ADH1 promoter (24). The plasmid expressing GBD-INC (pXW140) contains amino acids (aa) 879 to 1136 of Ty5 inserted between the BamHI and PstI sites of pGBDU; the GBD-inc construct (pXW158) is identical, except that it carries the S1094L mutation (14). The plasmid expressing TD fused to GBD (GBD-TD) (pXW205) was generated by inserting into the EcoRI and BglII sites of pGBDU a short DNA fragment created from two complementary oligonucleotides, DVO690 (5′-AAT-TCT-TGG-ATT-CAT-CGC-CTC-CAA-ATA-CCT-CA) and DVO691 (5′-GAT-CTG-AGG-TAT-TTG-GAG-GCG-ATG-AAT-CCA-AG); the GBD-td construct (pXW213) is identical, except that it has the S1094L mutation. We generated versions of these plasmids (pWW48, GBD; pWW49, GBD-TD; pWW50, GBD-td) in which the TRP1 marker gene of pGBD was replaced with HIS3. This was accomplished by replacing the EcoRV-XbaI fragment with a HIS3-containing NruI-XbaI fragment. LEU2-based expression plasmids (pWW44, GBD; pWW45, GBD-TD; pWW46, GBD-td) were generated by swapping the Gal4p activation domain (GAD)-encoding SphI fragment from pGAD (which has the LEU2 marker [24]) with an SphI fragment encoding the various GBD fusion proteins.
To test silencing of the HMR reporter gene, the above-described expression plasmids were transformed into strains with different HMR-E mutations (10). These strains include YSB1 (aeB but no UASg), YSB2 (aeB::3×UASg) and YSB35 (Aeb::3×UASg). sir derivatives of YSB2 and YSB35 include RS1072 and RS112 (sir1::URA3), RS1042 and RS1132 (sir2::URA3), RS1061 and RS1133 (sir3::URA3), and RS1067 (sir4::URA3) (1). The strain used to assess telomeric silencing was UCC3505 (kind gift of D. Gottschling) (16). Complementation of GBD-TD-induced loss of telomeric silencing was tested by introducing SIR genes (kind gift of J. Rine) on a 2μm plasmid (pRS424 [11]). The SIR2 plasmid (pSZ270) carries a NotI-XhoI fragment from pRS315-SIR2, the SIR3 plasmid (pSZ282) carries a BamHI-SalI fragment from pJR104, and the SIR4 plasmid (pSZ269) carries a SacII-ClaI fragment from pRS316-SIR4. To measure silencing, an overnight culture was grown to saturation for each strain and adjusted to an optical density at 600 nm of 1. Tenfold serial dilutions were made; 10 μl of each dilution was spotted onto both the test plate and the control plate. The plates were incubated at 30°C for 2 days.
Two-hybrid assays.
Two-hybrid assays were performed using yeast strain L40 (20), which has HIS3 and lacZ reporter genes under the control of upstream LexA operators. The LexA-SIR4C construct was previously described (2) and includes the C-terminal region of Sir4p (aa 951 to 1358). Control strains expressed LexA from plasmid pBTM116 (4). GAD-INC fusions were constructed by inserting an XmaI-PstI fragment from pXW140 into pGAD (24); the GAD-inc construct is identical, except that it carries the S1094L mutation. The control expressed GAD from pGAD. Strains with the relevant plasmids were inoculated into 2 ml of selective medium and shaken at 30°C for 24 h. Tenfold serial dilutions of the cultures were made, and 5 μl of each dilution was plated onto synthetic complete medium (SC)-Trp-Leu or SC-Trp-Leu plus 5 mM 3-amino-1,2,4-triazole medium; plates were incubated at 30°C for 3 days.
Protein analyses.
Immunoblot analyses of Ty5 and GBD proteins were conducted as previously described using antibodies specific to the RGS-H6 tag (Qiagen) or GBD (Santa Cruz Biotechnology) (22). Two of the epitope-tagged Ty5 elements were described in that previous report (pWW32 and pIP19). The element with the TD replaced by the RGS-H6 epitope (pWW59) was constructed by PCR mutagenesis (3). The Ty5 fragment within pWW37 (see above) was amplified using the universal primer and the reverse primers DVO1180 (5′-TG A-TGG-TGA-TGC-GAT-CCT-CTC-GAT-GGA-GGA-GAT-TCG-ATT-G) and DVO1181 (5′-CGC-ATC-ACC-ATC-ACC-ATC-ACA-ATA-CCT-CAT-TTA-ACG-CGG-C). The BspEI-PflMI fragment contained within the amplification product was used to replace the corresponding fragment in the wild-type Ty5 element carried on pSZ152 (54).
To measure in vitro interactions between the TD and Sir4p, we first constructed a plasmid expressing the C terminus of SIR4 (aa 950 to 1358) by PCR amplifying pSZ269 (see above) with primers DVO1137 (5′-GAA-GGA-TCC-AGA - GGA - TCG - CAT - CAC - CAT - CAC - CAT - CAC - AGA - AGA - GTG - TCG - CAT-AGT-G) and DVO1085 (5′-TGA-TCT-CGA-GTC-AAT-ACG-GTT-TTA-TCT-CC). The amplification product was digested with BamHI and XhoI and inserted into pCITE-2a(+) (Novagen) to generate pWW56. pWW56 DNA (0.5 μg) was used in a 50-μl coupled transcription-translation reaction mixture (Promega) containing 20 μCi of [35S]methionine. GBD fusion proteins were immunoaffinity purified as previously described (41) from 250-ml yeast cultures (optical density at 600 nm, 0.8 to 1.2) with either pXW205, pXW213, or pGBDU. Cells were harvested, washed with ice-cold water, and resuspended in 1.8 ml of lysis buffer B (50 mM HEPES [pH 7.5], 0.5 M NaCl, 10% glycerol, 0.1% IGPEL CA-630 [Sigma], 2 mM dithiothreitol, 2.5 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin per ml, 2 μg of bestatin per ml, and 2 μg of pepstatin per ml). Cells were disrupted by the glass bead method (3), and the lysate was centrifuged at 12,000 rpm (JA-20 rotor; Beckman) at 4°C for 30 min. Levels of fusion protein in the supernatant were assessed by immunoblot analysis using anti-GBD antibodies (Santa Cruz Biotechnology). The GBD fusion proteins were immunoaffinity purified from 300 μl of supernatant using 10 μl of an anti-GBD agarose bead slurry (Santa Cruz Biotechnology). After a 2-h incubation, the beads were collected by centrifugation (500 × g, 2 min), washed twice with 300 μl of wash buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM Mg diacetate, protease inhibitors) and once with 1× PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.4 mM KH2PO4). The 50-μl in vitro transcription-translation reaction mixture containing the labeled Sir4p was then added to the washed anti-GBD agarose beads, and the mixture was incubated at room temperature for 30 min. The beads were collected by centrifugation (500 × g, 2 min) and washed three times with PBS. Ten microliters of 2× sodium dodecyl sulfate (SDS) sample buffer (3) was added to each tube; samples were heated (95°C for 10 min) and separated by SDS-polyacrylamide gel electrophoresis (PAGE). The gel was dried and exposed to X-ray film overnight.
RESULTS
Defining the Ty5 targeting domain.
We previously found that a single amino acid substitution at position 1094 in the Ty5 polyprotein (S1094L) dramatically decreased targeting to the telomeres and silent mating loci (14). This indicated that Ty5 plays an active role in selecting targets and pointed to a possible targeting domain around S1094. To further define Ty5-encoded targeting determinants, a 758-bp BspEI-PflMI restriction fragment encompassing S1094 was mutagenized by PCR (Fig. 1A). The PCR products were used to replace the corresponding wild-type fragment, and sequencing of several recombinants revealed that each carried two to eight base pair changes. Over 2,100 mutagenized elements were screened for targeting defects using our plasmid-based targeting assay, which we have previously shown is an effective measure of chromosomal integration patterns (14). In this assay, targeting is quantified as the percentage of integration events that occur at a plasmid-borne HMR locus. We identified 11 elements that were impaired in targeting to various degrees (Table 1). DNA sequencing revealed multiple nucleotide changes in the BspEI-PflMI fragments of these elements. Whereas some mutations were silent, the number of amino acid substitutions ranged from two (mutants ut41 and ut38) to eight (mutant ut46) (Table 1).
TABLE 1.
PCR mutagenesis defines the Ty5 TD
| Element | Transposition efficiency (10−5)a | Transposition fold decrease | % of targeted transpositionb | Targeting fold decrease | No. of base changesc | Amino acid sequence near S1094d |
|---|---|---|---|---|---|---|
| Wild type | 13.3 ± 2.8 | 1.0 | 7.9 | 1.0 | 0 | LDSSPP |
| Mutants identified in the screen | ||||||
| ut3 | 3.43 ± 0.92 | 3.9 | 1.3 | 6.1 | 4 | SDSSPP |
| ut29 | 0.97 ± 0.11 | 13.7 | 1.4 | 5.6 | 5 | VDSSPP |
| ut5 | 1.93 ± 0.42 | 6.9 | 0.4 | 19.8 | 5 | LDLSPP |
| ut35 | 0.52 ± 0.08 | 25.6 | 0 | NAe | 7 | LDPSPP |
| ut41 | 1.49 ± 0.27 | 8.9 | 0.2 | 39.5 | 2 | LDPSPP |
| ut38 | 1.14 ± 0.22 | 11.7 | 1.1 | 7.2 | 2 | LDSLPP |
| ut46 | 2.46 ± 0.68 | 5.4 | 0.5 | 15.8 | 8 | LDSPPP |
| ut15 | 0.44 ± 0.19 | 30.2 | 0 | NA | 4 | LDSSPL |
| ut23 | 1.42 ± 0.46 | 9.4 | 2.5 | 3.2 | 5 | LDSSPL |
| ut31 | 1.55 ± 0.02 | 8.6 | 1.8 | 4.4 | 6 | LDSSPQ |
| ut33 | 1.40 ± 0.06 | 9.5 | 2.1 | 3.8 | 4 | LDSSPQ |
| Mutants with single amino acid substitutions in the vicinity of S1094 | ||||||
| rut3 | 2.40 ± 0.10 | 5.5 | 0.7 | 11.3 | 1 | SDSSPP |
| pWW39 | NDf | 2.1 | 2.1 | 3.8 | 1 | VDSSPP |
| rut41 | 2.12 ± 0.16 | 6.3 | 0.7 | 11.3 | 1 | LDPSPP |
| rut38 | 2.25 ± 0.66 | 5.9 | 1.1 | 7.2 | 1 | LDSLPP |
| rut46 | 2.11 ± 0.98 | 6.3 | 0.6 | 13.2 | 1 | LDSPPP |
| rut15 | 2.64 ± 0.23 | 5.0 | 3.0 | 2.6 | 1 | LDSSPL |
| rut31 | 3.09 ± 0.89 | 4.3 | 2.0 | 4.0 | 1 | LDSSPQ |
Data compiled from three independent experiments.
Number of transposition events to the target plasmid divided by total number of transposition events. For the mutant whose value is 0%, transposition was too low to meaningfully determine the percentage of targeted transposition.
Base changes were identified by DNA sequencing of the BspEI-PflMI fragment from each of the mutants.
S1094 is in bold; missense mutations are underlined.
NA, not applicable.
ND, not determined.
We made three observations regarding the targeting mutants: (i) one mutant (ut5) had an S1094L substitution identical to the mutation described in our previous study; (ii) all of the remaining mutants had an amino acid substitution in the vicinity of S1094 (encompassing a span of 6 aa residues); and (iii) independent mutations in the same residue near S1094 had similar targeting defects (e.g., mutants ut23, ut31, and ut33). These observations suggested that the mutations near S1094 were primarily responsible for the loss of target specificity. To test this hypothesis, we constructed seven new mutants that carried only one amino acid substitution near S1094 (Table 1). The S1094L mutation was not included because it was previously characterized (14). Targeting assays indicated that in all seven cases, the single mutations conferred a targeting defect nearly identical to that of the original mutants. This was the case even for conservative substitutions; for example, the L1092V mutation dramatically decreased targeting. Based on these data, we concluded that Ty5 encodes a TD and that it may be limited to a short stretch of 6 aa (LDSSPP).
Because our mutagenesis recovered multiple substitutions in the same amino acid, this suggested that the mutagenesis of the PCR fragment was saturated. To confirm the boundaries of the TD and to ensure that all critical amino acid residues near the LDSSPP motif had been identified, directed PCR mutagenesis was used to change residues near S1094 to alanine that were not identified as important for targeting. This included two residues (Asp1093 and Pro1096) that are located between other amino acids critical for targeting as well as residues upstream and downstream of the LDSSPP motif (Table 2). These substitutions had at most a modest effect on targeting (e.g., P1096, pWW41) or no effect at all (e.g., N1098, pWW42). It is interesting that each of the targeting mutants identified in this study had transposition frequencies from two- to sixfold lower than that of the wild type (Table 1). In contrast, the mutants without altered target specificity transposed at near wild-type levels (data not shown). This observation was also made in our original study, wherein the S1094L mutation caused a fourfold decrease in transposition (14). This indicates that TD mutations also affect transposition efficiency.
TABLE 2.
Site-directed mutagenesis indicates that only 4 of the 6 aa in the Ty5 TD are required for integration specificity
| Element | % of targeted transpositiona | Amino acid sequence near the targeting domainb |
|---|---|---|
| Wild type | 7.9 | SPPSLDSSPPNTS |
| pWW40 | 7.0 | SPPSLASSPPNTS |
| pWW41 | 5.9 | SPPSLDSSAPNTS |
| pXW198 | 8.6 | APPALDSSPPNTS |
| pXW199 | 8.0 | SAPSLDSSPPNTS |
| pXW201 | 8.2 | SAASLDSSPPNTS |
| pXW200 | 8.6 | SPASLDSSPPNTS |
| pXW202 | 7.9 | SPPSLDSSPPNAA |
| pWW42 | 7.8 | SPPSLDSSPPATS |
Data compiled from three independent experiments.
The TD is in bold; missense mutations are underlined.
The TD is located in the integrase C terminus.
Ty5 encodes a single open reading frame that is processed by a Ty5 gene-encoded protease into several proteins, including IN (80 kDa) and reverse transcriptase (RT; 59 kDa) (22). Extrapolating molecular weights from the Ty5 amino acid sequence, we predict that the protease cleavage site separating IN from RT is within the vicinity of the TD and therefore that the TD may reside either within the C terminus of IN or within the N terminus of RT. To distinguish between these possibilities, the TD was replaced with an epitope tag (RGS-H6). Like the other TD mutants, the epitope-tagged element transposed at a level more than fivefold lower than that of the wild type (data not shown). Proteins were prepared from strains expressing the TD-tagged element (pWW59) as well as from control strains expressing elements with the same epitope at either the C terminus of RT (pIP19) or within the middle of the IN coding region (pWW32) (Fig. 1A). Immunoblot analysis indicated that the element with the tag located at the TD expressed a protein with the same mobility on SDS-PAGE gels as that of IN (Fig. 1B). Other tagged proteins revealed by immunoblotting represent various processing products or intermediates (22). All lanes contained equivalent amounts of total protein, yet levels of the TD-tagged IN were severalfold lower than those of the other tagged IN, suggesting that mutations in the TD may affect protein stability. Nonetheless, we could conclude from this experiment that the TD resides within IN, consistent with its role in target site selection.
Tethering the TD to DNA nucleates silent chromatin.
Our model for target specificity predicts that the TD interacts with silent chromatin to tether the integration apparatus to its target sites. If this is the case, then the converse may also be true: a TD tethered to DNA may recruit silencing factors and establish silent chromatin. To test this idea, we used an assay that evaluates a protein's ability to establish transcriptional silencing (10). The test protein was first fused to the GBD, and the fusion protein was expressed in a yeast strain with a weakened HMR-E silencer that contains Gal4p binding sites (UASg). The effectiveness of the tethered proteins in nucleating silent chromatin was measured by the transcriptional status of a reporter gene at HMR (e.g., TRP1). We constructed four Ty5-GBD fusion proteins (Fig. 1C), one of which has 258 aa of the Ty5 INC (GBD-INC) and another of which has only 9 Ty5 aa (GBD-TD), 6 of which constitute the TD. The remaining two fusion constructs differed only by the S1094L mutation (GBD-inc, GBD-td). Immunoblot analysis indicated that the wild-type and mutant forms of the fusion proteins were expressed equivalently in yeast (Fig. 1D).
The fusion constructs were introduced into yeast strains with two different HMR-E mutations: one lacking binding sites for ORC and Rap1p (aeB) and the other lacking binding sites for Rap1p and Abf1p (Aeb) (10). The ability of the fusion proteins to establish silencing was measured by spotting 10-fold serial dilutions of the various strains onto media lacking tryptophan. The GBD-INC fusion was found to repress transcription over 100-fold (Fig. 2). Surprisingly, even the 9-aa GBD-TD fusion protein silenced TRP1 more than 10-fold. In both cases, the transcriptional silencing required the presence of the UASg, and in agreement with previous work (1, 10), the fusion proteins were more effective at silencers with a wild-type ORC binding site (Aeb). If the TD interacts with silent chromatin as predicted, then mutations that disrupt targeting should decrease its effectiveness in recruiting silencing factors. Consistent with this hypothesis, both GBD-inc and GBD-td were unable to establish transcriptional silencing (Fig. 2C). All of the above observations were confirmed in a strain with a URA3 reporter gene at HMR, indicating that the silencing was not reporter gene dependent (data not shown).
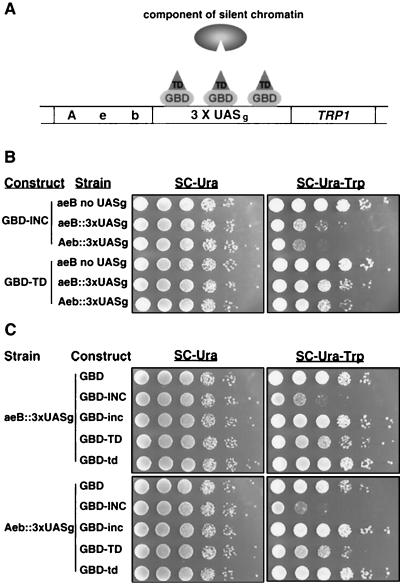
FIG. 2.
The Ty5 TD nucleates silent chromatin. (A) A cartoon depicting the tethered silencing assay. Yeast strains were used with deletions in two of the protein binding sites in HMR-E (A, E, and B) and three copies of UASG, binding sites for Gal4p (10). In the example depicted, the E and B binding sites are deleted (e, b), resulting in derepression of transcription at HMR. Expression of a fusion protein between GBD and the Ty5 TD (GBD-TD) was tested for its ability to recruit components of silent chromatin and restore silencing. Silencing is measured by expression of the adjacent TRP1 marker gene. (B) Silencing was established when GBD-INC and GBD-TD were tethered to the weakened HMR locus by the triple UASG. Serial, 10-fold dilutions of cells were plated onto control (SC-Ura) or test (SC-Ura-Trp) medium to measure silencing of the TRP1 reporter gene at HMR. (C) A point mutation that abolishes targeting fails to restore silencing for both fusion proteins (GBD-inc and GBD-td) in both test strains.
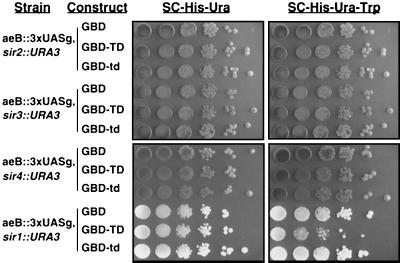
Silent chromatin, by definition, requires the actions of Sir2p, Sir3p, and Sir4p. To determine whether the TD fusions establish silent chromatin or rather act in some other way to occlude the transcriptional machinery from the TRP1 promoter, the TD fusions were introduced into various sirΔ strains (10) (Fig. 3). Sir1p was not required for TD-mediated silencing, consistent with its primary role in recruiting components of silent chromatin to the HM loci (48). However, the structural components of silent chromatin, Sir2p, Sir3p, and Sir4p, were all required, indicating that the Ty5 TD represses the reporter gene at HMR by establishing silent chromatin.
FIG. 3.
Silencing conferred by the TD is Sir dependent. The assay system is described in the legend to Fig. 2. In strains with deletions of SIR2, SIR3, or SIR4, GBD-TD fails to establish silencing at a weakened HMR locus. Silencing, however, does not require the SIR1 gene. The Ade− phenotype of the sir2Δ, sir3Δ, and sir4Δ strains confers their dark color; the sir1Δ stain is Ade+.
Overexpression of the TD disrupts telomeric silencing.
Transcriptional silencing is very sensitive to the expression level of some components of silent chromatin. Overexpression of Sir4p, for example, disrupts telomeric silencing, presumably by titrating away other components of silent chromatin or by disrupting complex formation (23, 39) (Fig. 4B). We tested whether overexpression of GBD-TD could disrupt telomeric silencing by monitoring the expression of telomeric (and therefore normally silenced) URA3 and ADE2 genes (16). Overexpression of GBD-TD (Fig. 1D) resulted in the loss of telomeric silencing, as evidenced by the ability of cells to grow on media lacking uracil (Fig. 4A). ADE2 expression was also evident by the white colony phenotype rather than the pinkish color characteristic of ADE2 repression and adenine precursor accumulation. As observed in the tethering experiments, expression of the fusion with the S1094L mutation did not affect telomeric silencing.
FIG. 4.
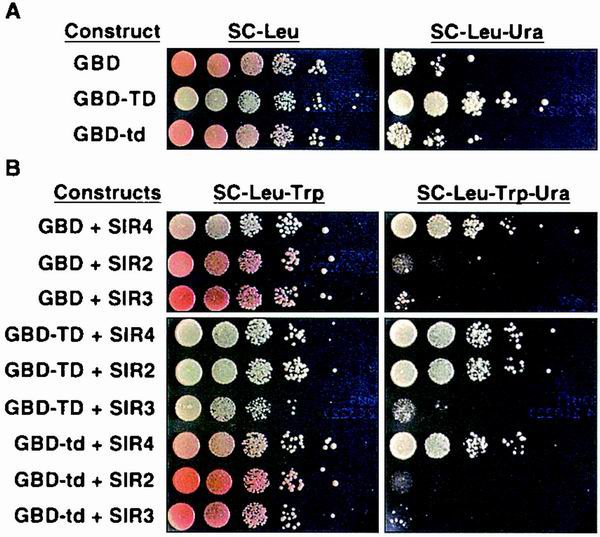
Overexpression of the Ty5 TD disrupts telomeric silencing and loss of silencing is complemented by overexpression of Sir3p. Two reporter genes, URA3 and ADE2, are located at telomeres VIIL and VR, respectively (16). URA3 expression was measured by growth of the yeast cells on selective media. ADE2 expression is indicated by colony color; when ADE2 is repressed, the colonies are red or pink. (A) Overexpression of GBD-TD disrupts telomeric silencing (i.e., causes URA3 expression), in contrast to what occurs in its mutant form (GBD-td) and with GBD alone. (B) Overexpression of Sir2p and Sir3p strengthen silencing, and Sir4p breaks silencing, as previously reported (23, 39). This is demonstrated in the GBD control strains and GBD-td strains. When the TD and SIR genes are overexpressed in the same strains, Sir3p, but not Sir2p or Sir4p, restores silencing.
To test whether GBD-TD disrupts telomeric silencing by titrating away components of silent chromatin, the SIR genes were ectopically expressed by introducing them on high-copy-number 2μm plasmids (Fig. 4B). Overexpression of Sir4p by this means has previously been shown to disrupt telomeric silencing (39), and we made the same observation regardless of whether GBD-TD was coexpressed. Overexpression of Sir2p did not restore telomeric silencing to GBD-TD-expressing strains; however, it did increase telomeric silencing in GBD or GBD-td strains. In contrast, overexpression of Sir3p overcame the GBD-TD-dependent loss of telomeric silencing. This suggests that Sir3p is titrated away (either directly or indirectly) by interacting with the TD. Alternatively, excess Sir3p could dominantly restore silencing by bypassing a factor being titrated away by TD. In addition, a growth defect was observed on nonselective media when Sir3p and GBD-TD were both overexpressed (Fig. 4B).
Sir4p interacts with Ty5 IN.
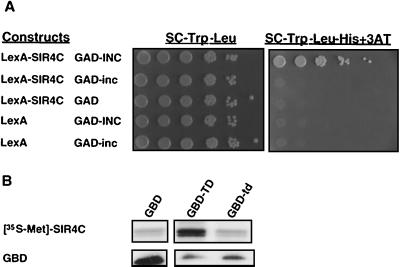
Several lines of evidence suggest that Sir3p and Sir4p are likely candidates for interacting with the Ty5 IN to mediate target specificity: (i) targeting is largely abolished in sir3Δ and sir4Δ strains (53); (ii) in sir4-42 strains, Ty5 integration specificity changes with the chromosomal localization of Sir3p and Sir4p; and (iii) as described above, loss of telomeric silencing due to overexpression of the TD can be complemented by overexpression of Sir3p. To test whether Ty5 IN interacts with Sir3p or Sir4p, two-hybrid assays were conducted. An interaction was detected between the C terminus of Sir4p (SIR4C, aa 951 to 1358 expressed as a LexA fusion protein) and the Ty5 INC (expressed as a GAD fusion protein). This interaction strongly activated the HIS3 reporter gene and enabled growth of yeast cells on selective media (Fig. 5A). Consistent with the role of the TD in silencing, this interaction required a wild-type TD: the S1094L mutation greatly weakened the two-hybrid interaction. These data indicate that INC binds to Sir4p (either directly or indirectly) and that the TD is required for this interaction.
FIG. 5.
The Ty5 INC interacts with the C terminus of Sir4p. (A) Yeast two-hybrid assays reveal an interaction between GAD-INC and LexA-SIR4C. Liquid cultures expressing the various LexA and GAD proteins were serially diluted 10-fold and spotted onto plates. A positive two-hybrid interaction was measured by transcriptional activation of the HIS3 reporter, which allowed for growth on selective media (SC-Trp-Leu-His with 5 mM 3-amino-1,2,4-triazole [3AT]). Activation of the HIS3 marker requires both SIR4C and the wild-type TD. (B) SIR4C interacts with GBD-TD in vitro. SIR4C was expressed and labeled with [35S]methionine by coupled transcription and translation. GBD-TD and GBD-td were expressed in yeast and immunoaffinity purified with anti-GBD agarose beads. The top lanes indicate the amount of labeled SIR4C bound by the various GBD proteins. The bottom lanes are from an immunoblot performed with anti-GBD antibodies, and they indicate the levels of GBD proteins in the extract used for immunoaffinity purification.
To confirm the two-hybrid data, we tested whether the Sir4p C terminus and the Ty5 TD could interact in vitro. GBD, GBD-TD, and GBD-td were expressed in yeast and immunoaffinity purified using anti-GBD agarose beads. Beads with the bound GBD proteins were incubated with SIR4C that had been labeled with [35S]methionine. The beads were washed, and the proteins were eluted and separated by SDS-PAGE. Sir4p bound to GBD-TD. Lower, background levels of binding were observed for GBD and GBD-td (Fig. 5B). These in vitro data support the results obtained in the two-hybrid assays and collectively suggest that the biological activity of the TD is mediated by interactions with Sir4p.
DISCUSSION
In a simple model to explain LTR retroelement target specificity, the interaction between the preintegration complex and DNA-bound proteins tethers the integration machinery to target sites and results in integration site biases (7). Support for this model comes from the study of the yeast Ty retrotransposons. These elements have different target preferences: Ty1 and Ty3 prefer sites of RNAP III transcription (9, 13), and Ty5 prefers silent chromatin (56). In both cases, DNA-bound protein complexes are required for targeting. Human immunodeficiency virus (HIV) IN interacts with a human homolog of the transcription factor SNF5 in two-hybrid assays (25); however, there is no evidence that this interaction mediates target site choice. In this study, we define a Ty5-encoded TD (LXSSXP) and show that it interacts with Sir4p, a component of silent chromatin. To our knowledge, this interaction between a retroelement-encoded protein and a chromatin factor provides the first direct evidence for the targeting model.
Integrase C terminus and the TD.
Retrotransposon and retroviral INs consist of three distinct domains (26): (i) an N-terminal region with a zinc-binding motif that is required for IN activity and likely binds cDNA, (ii) a catalytic domain that executes the integration reaction, and (iii) a C-terminal region which, for the retroviruses and some retrotransposons, is required for cDNA 3′-end processing. The retrotransposon integrase C termini are considerably larger than their retroviral counterparts; the Ty1 and Ty5 C termini constitute more than half of IN (e.g., HIV IN is 288 aa; Ty1 IN is 635 aa). Little is known about the function of the retrotransposon C-terminal extensions, with the exception that the very C terminus of Ty1 IN encodes a nuclear localization signal that is required for the preintegration complex to gain nuclear access (28, 42). For Ty1 and Ty5, the coding region of IN lies upstream of RT, and both are released from the polyprotein by proteolytic cleavage. In our earlier study of a Ty5 targeting mutant, we could not determine whether the targeting mutation was located in the C terminus of IN or the N terminus of RT (14). By replacing the TD with an epitope tag and comparing its electrophoretic mobility to those of other tagged forms of RT and IN, we demonstrate here that the TD resides within IN. This indicates that IN is responsible for target specificity and demonstrates a new function for the INC. For some members of the more distantly related Ty3/gypsy group retrotransposons (Metaviridae), the INC encodes a chromodomain, a motif implicated in targeting proteins to chromatin (37). The INC, therefore, may generally be used by retroelements for integration site selection.
Despite the large size of the INC, the Ty5 TD identified through our mutant screen spans only 6 aa residues. This short domain is biologically active: when as few as 9 aa encompassing the TD are expressed as part of a fusion protein (e.g., GBD-TD), they nucleate silent chromatin or disrupt telomeric silencing. Fusion proteins expressing larger fragments of the INC are at least 10-fold more effective in nucleating silent chromatin. This suggests that other regions of the INC play a role in targeting. These regions may not have been identified by our mutant screen if they are required for transposition. Transposition defects were observed during characterization of several targeting mutants. For example, the transposition efficiency of mutant ut15 was more than 30-fold lower than that of the wild type. This mutant had three missense mutations in the INC in addition to the mutation in the TD (Table 1). When the ut15 TD was evaluated in isolation (see rut15, Table 1), the level of transposition was only fivefold lower than that of the wild type. Additionally, we observed that most of the mutagenized elements (∼60%) were unable to transpose or showed greatly reduced levels of transposition (unpublished data). Some of these likely carry stop codons or frameshift mutations that prevent synthesis of the downstream RT; however, the frequency of nontransposing elements was too high based on the extent of mutagenesis (∼0.5%). This implies that regions of the C terminus outside of the TD are important for transposition.
In addition to loss of target specificity, a second phenotype shared by all of the TD mutants is an overall decrease in transposition (two- to sixfold). For wild-type Ty5, more than 94% of Ty5 integration events occur within regions of silent chromatin whereas the remainder appear to be randomly distributed throughout the genome (55, 57). If the TD is not required for integration, then TD mutations should not alter transposition frequencies. If the TD is required only for targeted integration, then the transposition frequencies would be predicted to drop ∼15-fold (94% divided by 6%). The intermediate effect of TD mutations on transposition suggests that the TD plays a role beyond tethering the integration apparatus to its preferred target sites. Possibilities include facilitating the integration reaction, facilitating reverse transcription (which is supported by recent data indicating that the INC is required to produce functional Ty1 RT in vitro [49]), or facilitating nuclear localization (the Ty1 nuclear localization signal is located in approximately the same region of the protein as the Ty5 TD [28, 42]). In strains expressing a modified Ty5 in which the TD was replaced by an epitope tag, we observed significantly lower levels of IN protein (Fig. 1). It is also possible that a wild-type TD is required for protein stability.
The TD and silent chromatin.
A large number of proteins are involved in the assembly and maintenance of silent chromatin (reviewed in references 35 and 36). These include proteins such as Sir3p and Sir4p that carry out structural or scaffolding roles and proteins like Sir1p and Rap1p that recruit and nucleate the structural proteins at the silencers. Tethering either class of these proteins to weakened HMR-E silencers effectively establishes silencing (10, 38). Likewise, the INC is very effective in nucleating silencing and causes a 100- to 1,000-fold decrease in the expression of reporter genes at HMR. The TD does not simply recruit a protein complex that occludes the RNAP II machinery from the reporter gene. Rather, it establishes silent chromatin as defined by the requirement for Sir2p, Sir3p, and Sir4p. TD-mediated silencing does not need Sir1p, which primarily acts at HM loci and is not required for silencing at the telomeres or rDNA. At the HM loci, Sir1p interacts with the ORC (48), and it is interesting that the TD is more effective when the ORC binding site is intact. This has also been observed with other nucleators of silent chromatin (1, 10). Although a number of S. cerevisiae proteins have motifs that match the TD consensus (LXSSXP), none are known components of silent chromatin.
We have previously shown that, in contrast to what occurs in sir3Δ and sir4Δ strains, targeting of Ty5 to the telomeres and HM loci is only partially impaired in sir2Δ strains and occurs at levels approximately 50% of those of the wild type (53). Furthermore, the loss of telomeric silencing caused by overexpressing the TD cannot be restored by overexpressing Sir2p. In light of recent findings that Sir2p is a histone deacetylase (21, 33, 47), this suggests that the Ty5 integration apparatus does not sense Sir2p-mediated acetylation patterns. The targeting defect in sir2Δ strains is likely a secondary consequence of perturbations in silent chromatin. In contrast, the loss of telomeric silencing caused by overexpressing the TD can be restored by additional SIR3 expression. The Sir4p C terminus interacts directly with Sir3p, and it may be that overexpression of the TD disrupts this interaction, which, in turn, is stabilized by additional Sir3p. In that regard, it has been observed that loss of telomeric silencing caused by overexpression of the Sir4p C terminus can be complemented by overexpressing Sir3p (15). Finally, it is important to note that the combined expression of Sir3p and a wild-type TD causes a growth defect, indicating that, although the TD may not interact directly with Sir3p, Sir3p does modulate the biological activity of the TD.
The TD and Sir4p.
Prior to the carrying out of our two-hybrid assays, no evidence distinguished the roles of Sir3p and Sir4p in Ty5 target specificity. The only notable difference was that Ty5 cDNA recombination increased more than 10-fold in sir4Δ strains and was only marginally affected in sir3Δ strains (53). In strains with the sir4-42 allele, which expresses a C-terminally truncated form of Sir4p (aa 1 to 1237), Ty5 integrates preferentially into the rDNA (53). Here we demonstrate that the Ty5 TD interacts with the Sir4p C terminus (aa 951 to 1358), suggesting that the relevant region of interaction is located between aa 951 and 1237. The Sir4p C terminus interacts with many proteins, including Sir2p and Sir3p (41), Rap1p (6, 43), Sif2p (12), and Dis1p (52). Two-hybrid interactions may result if one or more of these proteins serve as a bridge between the TD and Sir4p. However, we have not observed two-hybrid interactions between IN and other components of silent chromatin (data not shown). Furthermore, the observed in vitro binding between Sir4p and the TD argues that these molecules interact directly. Nonetheless, because we used fusion proteins purified from yeast cells for these experiments, we cannot exclude the possibility that other factors copurified or modified the TD to yield productive interactions.
Concluding remarks.
Previous studies demonstrated that DNA bound in silent chromatin is inaccessible to proteins like HO endonuclease, restriction enzymes, and transcription factors (34). However, the ability of Ty5 to integrate into silent chromatin suggests that this DNA is accessible to Ty5 IN. Our model suggesting that target specificity results from simply tethering the preintegration complex to chromatin may therefore require additional refinements. For example, the integration complex may induce changes in silent chromatin during integration, and the role of the TD in such processes is an important area of future research.
An increased understanding of targeting mechanisms may make it possible to manipulate retroelement target site choice. It may be possible to change the integration preference of retrotransposons by replacing TDs with peptide motifs that interact with specific chromosomal proteins. Such engineered retrotransposons may become useful tools for studying chromatin organization and may provide novel methods for genome manipulation. Retroviral vectors are widely used for DNA delivery in human gene therapy; however, uncontrolled, random integration into the host genome is one of their major drawbacks. It may now be possible to better control retroviral integration site choice to improve the efficacies of these vectors in gene delivery.
ACKNOWLEDGMENTS
W. Xie and X. Gai contributed equally to this work.
We thank Phil Irwin for his expert technical assistance. J. Rine and D. Gottschling graciously provided plasmids used in this study.
This work was supported by a grant from the American Cancer Society (RPG9510106MBC) to D.F.V. and by Hatch Act and State of Iowa funds.
Footnotes
This is Journal Paper J-19154 of the Iowa Agriculture and Home Economics Experiment Station, Ames, project 3383.
REFERENCES
- 1.Andrulis E D, Neiman A M, Zappulla D C, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 2.Ansari A, Gartenberg M R. The yeast silent information regulator Sir4p anchors and partitions plasmids. Mol Cell Biol. 1997;17:7061–7068. doi: 10.1128/mcb.17.12.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Greene/Wiley Interscience; 1987. [Google Scholar]
- 4.Bartel P L, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, Devine S E. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 6.Buck S W, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 7.Bushman F. Targeting retroviral integration. Science. 1995;267:1443–1444. doi: 10.1126/science.7878462. [DOI] [PubMed] [Google Scholar]
- 8.Chalker D L, Sandmeyer S B. Sites of RNA polymerase III transcription initiation and Ty3 integration at the U6 gene are positioned by the TATA box. Proc Natl Acad Sci USA. 1993;90:4927–4931. doi: 10.1073/pnas.90.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chalker D L, Sandmeyer S B. Ty3 integrates within the region of RNA polymerase III transcription initiation. Genes Dev. 1992;6:117–128. doi: 10.1101/gad.6.1.117. [DOI] [PubMed] [Google Scholar]
- 10.Chien C T, Buck S, Sternglanz R, Shore D. Targeting of SIR1 protein establishes transcriptional silencing at HM loci and telomeres in yeast. Cell. 1993;75:531–541. doi: 10.1016/0092-8674(93)90387-6. [DOI] [PubMed] [Google Scholar]
- 11.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 12.Cockell M, Renauld H, Watt P, Gasser S M. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr Biol. 1998;8:787–790. doi: 10.1016/s0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- 13.Devine S E, Boeke J D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996;10:620–633. doi: 10.1101/gad.10.5.620. [DOI] [PubMed] [Google Scholar]
- 14.Gai X, Voytas D F. A single amino acid change in the yeast retrotransposon Ty5 abolishes targeting to silent chromatin. Mol Cell. 1998;1:1051–1055. doi: 10.1016/s1097-2765(00)80105-7. [DOI] [PubMed] [Google Scholar]
- 15.Gotta M, Palladino F, Gasser S M. Functional characterization of the N terminus of Sir3p. Mol Cell Biol. 1998;18:6110–6120. doi: 10.1128/mcb.18.10.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschling D E, Aparicio O M, Billington B L, Zakian V A. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 17.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 18.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Heslop-Harrison J S, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F. Polymorphisms and genomic organization of repetitive DNA from centromeric regions of Arabidopsis chromosomes. Plant Cell. 1999;11:31–42. doi: 10.1105/tpc.11.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 22.Irwin P A, Voytas D F. Expression and processing of proteins encoded by the Saccharomyces retrotransposon Ty5. J Virol. 2001;75:1790–1797. doi: 10.1128/JVI.75.4.1790-1797.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivy J M, Klar A J, Hicks J B. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 26.Katz R A, Skalka A M. The retroviral enzymes. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 27.Ke N, Voytas D F. High frequency cDNA recombination of the Saccharomyces retrotransposon Ty5: the LTR mediates formation of tandem elements. Genetics. 1997;147:545–556. doi: 10.1093/genetics/147.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy B K, Austriaco N R, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- 30.Kennedy B K, Gotta M, Sinclair D A, Mills K, McNabb D S, Murthy M, Pak S M, Laroche T, Gasser S M, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell. 1997;89:381–391. doi: 10.1016/s0092-8674(00)80219-6. [DOI] [PubMed] [Google Scholar]
- 31.Kim J M, Vanguri S, Boeke J D, Gabriel A, Voytas D F. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 32.Kirchner J, Connolly C M, Sandmeyer S B. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science. 1995;267:1488–1491. doi: 10.1126/science.7878467. [DOI] [PubMed] [Google Scholar]
- 33.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laurenson P, Rine J. Silencers, silencing, and heritable transcriptional states. Microbiol Rev. 1992;56:543–560. doi: 10.1128/mr.56.4.543-560.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 36.Lustig A J. Mechanisms of silencing in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1998;8:233–239. doi: 10.1016/s0959-437x(98)80146-9. [DOI] [PubMed] [Google Scholar]
- 37.Malik H S, Eickbush T H. Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcand S, Buck S W, Moretti P, Gilson E, Shore D. Silencing of genes at nontelomeric sites in yeast is controlled by sequestration of silencing factors at telomeres by Rap1 protein. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- 39.Marshall M, Mahoney D, Rose A, Hicks J B, Broach J R. Functional domains of SIR4, a gene required for position effect regulation in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4441–4452. doi: 10.1128/mcb.7.12.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moazed D, Johnson D. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- 41.Moazed D, Kistler A, Axelrod A, Rine J, Johnson A D. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore S P, Rinckel L A, Garfinkel D J. A Ty1 integrase nuclear localization signal required for retrotransposition. Mol Cell Biol. 1998;18:1105–1114. doi: 10.1128/mcb.18.2.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 44.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.SanMiguel P, Tikhonov A, Jin Y K, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer P S, Edwards K J, Lee M, Avramova Z, Bennetzen J L. Nested retrotransposons in the intergenic regions of the maize genome. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 46.Shafikhani S, Siegel R A, Ferrari E, Schellenberger V. Generation of large libraries of random mutants in Bacillus subtilis by PCR-based plasmid multimerization. BioTechniques. 1997;23:304–310. doi: 10.2144/97232rr01. [DOI] [PubMed] [Google Scholar]
- 47.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triolo T, Sternglanz R. Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing. Nature. 1996;381:251–253. doi: 10.1038/381251a0. [DOI] [PubMed] [Google Scholar]
- 49.Wilhelm M, Boutabout M, Wilhelm F X. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: a fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem J. 2000;2:337–342. [PMC free article] [PubMed] [Google Scholar]
- 50.Yieh L, Kassavetis G, Geiduschek E P, Sandmeyer S B. The brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J Biol Chem. 2000;275:29800–29807. doi: 10.1074/jbc.M003149200. [DOI] [PubMed] [Google Scholar]
- 51.Zaccolo M, Williams D M, Brown D M, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. J Mol Biol. 1996;255:589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Z, Buchman A R. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Zou S, Wright D, Voytas D. Tagging chromatin with retrotransposons: target specificity of the Saccharomyces Ty5 retrotransposon changes with the chromosomal localization of Sir3p and Sir4p. Genes Dev. 1999;13:2738–2749. doi: 10.1101/gad.13.20.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zou S, Ke N, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 1996;10:634–645. doi: 10.1101/gad.10.5.634. [DOI] [PubMed] [Google Scholar]
- 55.Zou S, Kim J M, Voytas D F. The Saccharomyces retrotransposon Ty5 influences the organization of chromosome ends. Nucleic Acids Res. 1996;24:4825–4831. doi: 10.1093/nar/24.23.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou S, Voytas D F. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc Natl Acad Sci USA. 1997;94:7412–7416. doi: 10.1073/pnas.94.14.7412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zou S, Wright D A, Voytas D F. The Saccharomyces Ty5 retrotransposon family is associated with origins of DNA replication at the telomeres and the silent mating locus HMR. Proc Natl Acad Sci USA. 1995;92:920–924. doi: 10.1073/pnas.92.3.920. [DOI] [PMC free article] [PubMed] [Google Scholar]