ABSTRACT
Proteases are an evolutionarily conserved family of enzymes that degrade peptide bonds and have been implicated in several common gastrointestinal (GI) diseases. Although luminal proteolytic activity is important for maintenance of homeostasis and health, the current review describes recent advances in our understanding of how overactivity of luminal proteases contributes to the pathophysiology of celiac disease, irritable bowel syndrome, inflammatory bowel disease and GI infections. Luminal proteases, many of which are produced by the microbiota, can modulate the immunogenicity of dietary antigens, reduce mucosal barrier function and activate pro-inflammatory and pro-nociceptive host signaling. Increased proteolytic activity has been ascribed to both increases in protease production and decreases in inhibitors of luminal proteases. With the identification of strains of bacteria that are important sources of proteases and their inhibitors, the stage is set to develop drug or microbial therapies to restore protease balance and alleviate disease.
KEYWORDS: Protease, microbiota, pain, mucosal barrier, gluten, gut-brain axis, gastroenteritis
Introduction
Proteases are a diverse group of enzymes that cleave peptide bonds.1 They are classified according to their catalytic mechanism into serine, cysteine, aspartic, glutamic, threonine and metalloproteases.2 These enzymes play multifunctional roles in essential physiological processes, including digestion of dietary proteins, apoptosis, cell differentiation, inflammation and nociception, to name a few.3–6 Proteases are tightly regulated to prevent excessive degradation of host proteins or inappropriate immune activation, and imbalances between proteolytic and anti-proteolytic activity have been described in patients with different gastrointestinal (GI) disorders. For example, colonic tissues from patients with inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) have increased serine proteolytic activity (PA),7,8 suggesting a role in the pathophysiology of the disease.
In the past, most investigations in IBD and IBS have focused mainly on studying proteases released by the host. However, the gut hosts a vast and diverse microbial ecosystem, the microbiota, that has important impacts on human homeostasis and disease. The intestinal microbiota is a rich source of proteases, as microbes release different proteases for metabolism, defense, and host invasion. Gut microbes also produce protease inhibitors and protease-degrading enzymes, reflecting the importance of tightly regulating proteolytic activity.9 In the gut, microbial proteases were first identified as virulence mechanisms of pathogens.10,11 Elevated PA was also observed in stool supernatants from IBD and IBS patients compared to samples from healthy subjects, suggesting that enhanced PA in IBD and IBS may be due to elevations in PA from both host and microbial origin.12,13
With the advent of omics-based technologies, including DNA sequencing14 and mass spectrometry,15 the identification of a large number of bacteria living in the gut and their metabolic products have opened new routes for studying their contributions to GI diseases.16,17 The recent insights into the interactions of the microbiota and their products with the host have strengthened the hypothesis that the proteolytic capacity of bacteria plays an important role in gut homeostasis and disease. For example, elastase-like activity of the pathogen Pseudomonas aeruginosa can lead to the production of peptides following gluten metabolism with increased immunogenicity in celiac disease (CeD) patients,18 and serine proteases from a consortium of gut bacteria modulate the excitability of nociceptors via activation of protease-activated receptor 4 (PAR-4).19 However, the mechanistic characterization of specific bacterial products such as proteases in gut disorders still represent an enormous challenge. With the accumulating evidence suggesting that bacterial proteases play a key role in GI diseases, in this review we aim to highlight some of the key findings about their involvement in the development of IBD, IBS,CeD and GI infections, and consider their physiological implications.
Proteases: functions and classification based on their catalytic mechanism
Proteases are found in all forms of life and are vital for the survival of all organisms.
Based on their catalytic mechanism, they are classified into serine, threonine, cysteine, asparagine, glutamic, aspartic or metalloproteases (MEROPs database).20 These enzymes use an amino acid residue (serine, threonine, cysteine, asparagine, glutamic acid or aspartic acid, respectively) located in the active site to carry out the catalytic reaction, with the exception of metalloproteases, which use metals for catalysis, with Zn2+ being the most common (Figure 1a). In humans, proteases are one of the largest enzyme families, representing up to 2% of the human genome with over 500 different proteases described. Serine (35.1%) and metalloproteases (29.5%) are the most densely populated classes (Figure 1b).21 In addition to their well known functions in meal digestion, host proteases play very important roles in the gut, including cell proliferation and differentiation, tissue morphogenesis and remodeling, angiogenesis, wound repair, stem cell mobilization, inflammation, immunity, autophagy and apoptosis.21,22
Figure 1.
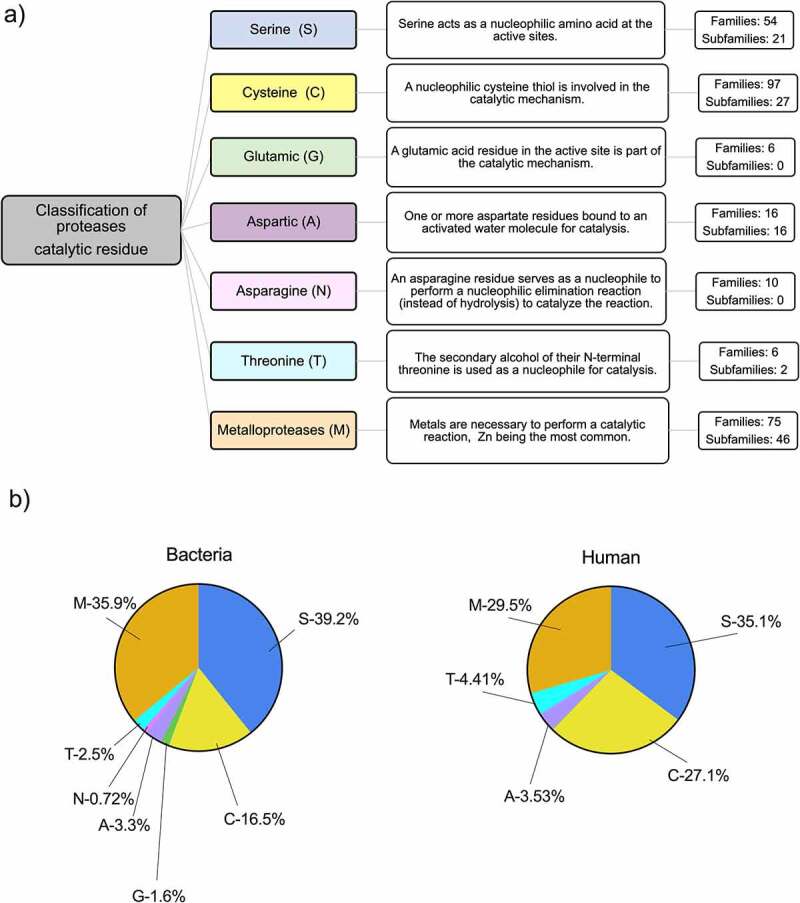
Classification and distribution of proteases based on their catalytic site. a) The MEROPs database classifies proteases based on the amino acid at the catalytic site used to carry out the catalytic process. Each group of proteases is identified by the letter of the amino acid that represents the catalytic type. All members are identified and classified based on structural similarities. b) Relative abundance of protease families in bacterial and human genomes, including putative proteases. Serine (S), cysteine (C), glutamic (G), aspartic (A), asparagine (N), threonine (T) and metalloproteases (M).
Bacterial proteases are mechanistically, structurally and functionally highly diverse compared to mammalian proteases. Microorganisms produce a vast array of proteases belonging to the same classes described for humans in addition to glutamic-proteases, which have not been found in mammals so far. The most abundant bacterial proteolytic enzymes are serine (39.2%), metalloproteases (35.9%) and cysteine (16.5%) proteases (Figure 1b). Although belonging to similar classes, bacterial proteases have different functional activity or substrate specificity and not always are well-characterized. Indeed, domain architectures of many protease classes identified in bacteria are very different from those observed in eukaryotes, suggesting distinct roles for proteases in prokaryotes.21,23 In addition, the type and proportion of the different proteases vary between taxonomic groups and strains. From a functional point of view, proteases play important roles in intercellular communication, cell viability, stress response and pathogenicity in bacteria.
Bacterial proteases are better classified based on functional location as 1) cell-associated protease complexes and 2) extracellular enzymes.24 The first group includes intracellular, conserved and highly regulated proteases which are in multimeric complexes and essential for cell viability. These proteases are ubiquitous within the eubacteria kingdom and include the serine proteases Clp, Lon and high-temperature requirement serine protease A (HtrA), Zn2+ metalloproteinase FtsH and threonine protease HslUV. With the exception of the HtrA, these proteins fall into the broad class of AAA+ enzymes (ATPases associated with diverse cellular activities). 24–26 On the other hand, extracellular enzymes are generally monomeric with high substrate specificity. They are often synthesized as inactive zymogens, protecting the cell from unregulated activity before secretion. Many of these secreted proteases are considered virulence factors and are unique to certain taxon or strains.11,24,25 Many of them have been linked to different GI disorders as we will discuss in the next section.
Involvement of bacterial proteases in gastrointestinal diseases
Inflammatory bowel disease
IBD is an umbrella term that encompasses several diseases associated with chronic relapsing and remitting inflammation of the GI tract. The two major subtypes of IBD are Crohn’s disease (CD) and ulcerative colitis (UC). Although there are differences in the nature and location of gut inflammation between these two diseases, they share several debilitating symptoms that include pain, altered bowel habits, weight loss and anemia. Despite the availability of drugs and monoclonal antibodies that target key inflammatory processes to induce remission, many IBD patients are unresponsive or lose responsiveness over time. Therefore, novel therapies for IBD are needed, and one promising candidate approach is to target luminal proteolytic activity.
A dysregulation of host proteolytic has been well characterized previously in IBD patients.8 In both CD and UC patients, increased host serine protease production from colonic tissue have been reported.27,28 Proteases such as cathepsin G and thrombin are overactive in supernatants of colonic tissues from IBD patients compared to healthy controls.8 Elastase-like activity has also garnered considerable attention in IBD. Motta et al.29 showed that colonic epithelial cells are a major source of elastase-like activity and this activity is markedly increased in IBD patients. The same study specifically identified elastase 2A (ELA2A) as enhanced in IBD patients. Protease activity within fecal samples from patients has been also tested and revealed an increase in serine protease activity during IBD.12 Unlike colonic tissue, fecal samples contain proteases from both the host and bacteria, and deciphering the origin of the proteases found in fecal samples remains a challenge. Consistent with a role of proteolytic overactivity in disease pathogenesis, inhibitors of serine proteases have beneficial effects in mouse models of IBD.27,30 Elafin, a Lactobacillus-derived inhibitor of serine proteases reduced the severity of colitis in mice exposed to dextran sulfate sodium (DSS) or trinitro benzene sulphonic acid.27,30,31
In 2021, Galipeau and colleagues proposed the use of bacterial proteases as a marker of disease in UC patients.32 In a longitudinal cohort of UC patients, it was found that increased fecal proteolytic activity was seen in UC patients, even prior to disease onset. This suggests that increased fecal proteolytic activity could be used as an early biomarker of disease. More importantly, enhanced fecal proteolytic activity may be an early step in UC pathogenesis, due to proteolytic effects on mucosal barrier function and immunoregulation. A bacterial source of this fecal proteolytic activity was suggested due to detection of an increase in bacterial protease gene expression by shotgun metagenomics. Authors also found an increase in fecal elastase-like activity in UC patients. Elastase-like activity was positively correlated with the relative abundance of Bacteroides vulgatus, a taxon known to have high proteolytic activity. Importantly, transfer of fecal microbiota from UC patients prior to disease onset into mice increased fecal proteolytic activity in the colon and activation of host inflammatory responses. These results suggest a bacterial contribution to fecal proteolytic activity and provides proof of concept that the proteolytic activity of UC patient microbiota is sufficient to induce gut inflammation. Findings from Galipeau et al., 2021 were recently confirmed by Mills and colleagues33 using proteomics and metabolomics, which can detect whether the proteases are of eukaryotic or prokaryotic origin. A subset of the clinically active UC patients had an overabundance of proteases derived from the bacterium Bacteroides vulgatus. Some of the correlated proteases included serine and metalloproteases that largely function in the extracellular space and may exacerbate disease activity. Taken together, there is evidence to suggest that an increase in bacterial proteases is associated with UC pathogenesis but the extent to which these proteases drive disease severity and whether a similar phenomenon occurs in CD remain to be determined.
Celiac disease
CeD is a chronic autoimmune inflammatory enteropathy that occurs in genetically susceptible individuals in response to the ingestion of gluten proteins. It has a worldwide prevalence of 1.4%.34,35 CeD primarily affects the small intestine, with the development of a mucosal immune response characterized by an increase of intraepithelial lymphocytes, villous atrophy and crypt hyperplasia.36 Currently, the only accepted treatment for CeD is a strict, life-long gluten-free diet.37 The role of microbes in CeD has gained considerable attention recently, based on reported alterations of the intestinal microbiome of CeD patients and associations between enteric infections with CeD onset in longitudinal studies.38–40
Along with changes in microbial composition, several studies have suggested a pivotal role of proteases in CeD pathogenesis. Contrary to a beneficial role of proteases in CeD, an increase in proteolytic activity towards gluten proteins has been observed in the duodenum and feces of CeD patients.41,42 Although the nature of these proteases is not well understood, recent reports have suggested a microbial origin. Duodenal biopsies from patients with active CeD have increased proteolytic activity against gluten that correlated with increased abundance of Pseudomonas, a well-known proteolytic taxon. Indeed, Pseudomonas aeruginosa producing the metalloprotease LasB induced food sensitivity in preclinical mouse models by different mechanisms, as described below.18 These studies suggest that proteases expressed by pathobionts impact gluten metabolism and immune activation in the small intestine of CeD patients.43 Consistent with a pathogenic role of proteolytic activity in CeD, a serine protease inhibitor produced by Bifidobacterium longum reduces gluten-induced immunopathology in a preclinical mouse model.44
For decades, it was suspected that missing digestive proteases may be the cause of CeD.45,46 This theory posited that defective digestion of gluten in CeD due to the lack of unknown host digestive proteases in susceptible individuals. Indeed, oral enzymatic therapy, a widely investigated therapeutic approach in CeD, focuses on the digestion of immunogenic gluten peptides in the human GI tract through peptidase supplementation.37 In this regard, microorganisms from the oral cavity have emerged as candidates that have the potential to produce enzymes that degrade luminal gluten. In vitro studies have shown that proteases released by Rothia strains (R. mucilaginosa and R. aeria) are potential sources of gluten-degrading enzymes that specifically target immunodominan gluten peptides.47 Subsequently, the enzyme produced by R. aeria was isolated and identified as belonging to the S8 subtilisin protease family with high capacity to effectively degrade gluten.48 Other researchers have shown that Prolyl endopeptidases from Flavobacterium meningosepticum, Sphingomonas capsulate and Myxococcus xanthus have promising applications in the treatment of CeD. The recombinant proteins from these microbes have the capacity to break down gluten peptides with different subsite specificities.49 These findings suggest the promising potential of bacterial proteases in the treatment of CeD and some formulations have already entered phase II clinical trials.37,50 Taken together, the evidence to date suggests that some proteases protect against CeD and other exacerbate the disease, depending on the substrate specificity of the protease in question and its ability to reduce or increase the immunogenicity of gluten catabolic products. The next decade is likely to witness a significant increase in studies of proteases of bacterial origin that aid in the metabolism of gluten, since it is only partially digested by human digestive enzymes, regardless of the disease.51
Irritable bowel syndrome
IBS is a common digestive disorder associated with chronic abdominal pain and altered bowel habit.52 Because IBS presents without the frank inflammatory damage that accompanies IBD, it is commonly viewed as a disorder of gut-brain communication. In addition to alterations in central nervous system processing of signaling from the gut, several changes within the bowel of IBS patients have been implicated in altered gut-brain communication, including altered serotonin release by enterochromaffin cells, altered mast cell-neuronal communication, and microbial dysbiosis.53–56 Similar to IBD, studies in mice and humans have provided evidence that host- and bacterial-derived proteases may contribute to pathogenesis and symptom generation.7,13,57,58 For example, Trypsin-like activity and tryptase release are increased in colonic biopsies from IBS patients.7,58 In a recent study, metagenomic analysis of faecal samples from patients with post-infectious IBS revealed an altered gut microbiota composition and high gut proteolytic activity driven by specific host serine proteases compared with controls. The authors also showed that β-glucuronidases released by commensal microbes suppressed host PA, thereby protecting the intestinal epithelium and suggest that a decrease in microbial β-glucuronidase activity may contribute to IBS pathogenesis.57 In agreement with a role for enhanced protease activity in IBS, nafamostat, an inhibitor of serine proteases, reduced visceral hyperalgesia in a rodent model of post-inflammatory IBS.31
Gastrointestinal infections
GI infections account for a large burden of acute and chronic disease, and proteases are essential to the ability of many microorganisms to infect the host. Bacterial pathogens rely on proteolysis for variety of purposes during the infection process. Intracellular and membrane proteases such as Clp, Lon or HtrA contribute to virulence through the timely degradation of virulence regulators and indirectly by providing tolerance to adverse conditions in the host. In contrast, pathogen-dependent extracellular proteases facilitate host invasion by degrading host extracellular matrix components or by interfering with host cell and immune signaling, as we discuss in the next section.11 A good example is Helicobacter pylori, a bacterium that infects approximately half of the world’s population and is the leading risk factor for peptic ulcer disease and gastric cancer.59 Although different virulence factors have been described in H. pylori,60 the zinc-protease PqqE and the serine-protease HtrA disrupt gastric mucosal integrity61–63 and thereby facilitate bacterial invasion.
Proteases are also pivotal virulence factors of infectious agents associated with gastroenteritis. Gastroenteritis is a diarrheal disease characterized by an increase in bowel movement frequency and stool water content, with or without fever, vomiting, and abdominal pain.64 Diarrhea caused by enteric infections is a major factor in morbidity and mortality worldwide. Although more than twenty microbial pathogens are known to cause acute gastroenteritis, several Escherichia coli strains are the most common, constituting a significant risk to human health and remaining an important cause of infant mortality in developing countries.65,66 This group of bacteria includes different pathotypes such as enterotoxigenic (ETEC), enteropathogenic (EPEC), enteroinvasive (EIEC), enterohemorrhagic (EHEC) or enteroaggregative E. coli (EAEC).66 Other clinically relevant microorganisms causing diarrhea are Shigella spp., Salmonella spp, Campylobacter jejuni/coli and Vibrio cholera.65 Enteric pathogens utilize a variety of sophisticated strategies to colonize the intestinal tract, evade the immune system, proliferate, and damage the host. Virulence factors related to these bacteria have a wide range of activities including adhesins, toxins, iron acquisition factors, lipopolysaccharides, polysaccharide capsules, invasins and proteases.
The serine protease autotransporters from enterobacteriacea (SPATE) constitute a superfamily of virulence factors. These are high molecular weight serine proteases generally secreted into the external milieu via the autotransport pathway and are highly prevalent among enteropathogens, including Shigella, Salmonella, Citrobacter and all Escherichia coli pathotypes. Several findings suggest that SPATEs degrade host intracellular or extracellular substrates, which trigger a variety of adverse effects on host cells.67 SPATEs can be classified in 2 types classes. Class-1 SPATEs target intracellular substrates, eliciting cytotoxic and endotoxin effects on the host.67 On the other hand, class-2 SPATEs seem to disrupt mucosal barriers and modulate the immune response by targeting host glycoproteins. In this class, the serine protease Pic produced by E. coli (EAEC), Citrobacter and Shigella flexneri is a virulence factor associated with adherence, colonization and evasion of the innate immune system.68–71 Class-2 SepA produced by Shigella flexneri and EAEC is also indispensable for barrier disruption. 72 Finally, the zinc metalloproteases StcE and SslE secreted by E. coli EHEC and EPEC/ETEC respectively, contribute to intimate adherence of these bacteria to host cells, a process that is essential for colonization.73–78
Other proteases have been described as virulence factors in gastroenteritis. As H. pylori, Salmonella typhimurium and Campylobacter jejuni, a bacterium responsible for foodborne infections, interact with the host cell epithelium and establish infection by HtrA.79–81 The extracellular Zn-dependent metalloprotease hemagglutinin (HA) also called vibriolysin, has been implicated in the pathogenicity of Vibrio cholerae. V. cholerae can cause cholera, a severe diarrheal disease that can be quickly fatal if untreated and is typically transmitted via contaminated water and person-to-person contact.82 Although the cholera toxin is the primary driver of the infection, vibriolysin presents a broad range of potentially pathogenic activities including degradation of the mucus barrier or disruption of epithelial tight junctions.10,83 Proteases could also mediate infections indirectly. This is the case of Clostridioides difficile, one of the leading causes of health-care-associated infections and diarrhoea in many countries. C. difficile causes mild to severe diarrhoea and can lead to life-threatening conditions such as colonic perforation, pseudomembranous colitis and toxic megacolon. The toxins A and B (TcdA and TcdB respectively) released by the pathogenic C. difficile are decisive for the infection. An internal Cys protease domain activates the toxin resulting in downstream effects on host cells.84 As proteases are essential to the ability of many bacteria to infect the host and cause disease, it has been proposed to block specific proteases to prevent common gastrointestinal infections; however, there are still no approved drugs with this mode of action.25
In the following sections, the pathophysiological consequences of proteolytic activity in GI diseases is discussed in the context of luminal actions, effects on mucosal barrier function, cell and immune signaling and impact on visceral sensation.
Effects of bacterial proteases in the gastrointestinal tract
Luminal actions of bacterial proteases: diet-microbiota interactions
Diet is a major driver of microbial composition and function.85,86 Our intestinal microbiota is capable of using different dietary components to generate microbial metabolites with bioactive properties.87 As happens in mammals, microbes use proteases to meet their nutritional amino acid requirements by hydrolyzing available proteins from the host or the diet. Consequently, microbial protease activity can be influenced by dietary choices. Patients with chronic inflammatory or functional GI conditions recognize diet as a driving factor in symptom onset/severity.88–90 Thus, diet should be considered when investigating the role of microbial proteases in inflammation or dysfunction. Indeed, there are multiple mechanisms by which microbial proteases could influence homeostasis through diet.
First, western diets are characterized by their high protein content, and many improperly digested dietary proteins are capable of inducing aberrant immune responses in the gut. 51,91 The functional diversity of the human gut microbiota implies a vast catalog of metabolic pathways that participate in the digestion of dietary components, even proteins that are difficult to digest by human enzymes.92,93 Thus, dietary proteins not used by the host become substrates for microbial proteases. This is particularly important in food sensitivities such as CeD.94 The main environmental trigger of CeD, gluten, is not fully digested by host digestive enzymes.51 It has been shown that the human gastrointestinal tract harbours bacteria with the capacity to metabolize gluten. These include commensal bacteria such as Actinomyces, Bacillus, Rothia, Staphylococcus, Streptococcus, Lactobacillus or Clostridium but also opportunistic pathogens such as Pseudomonas aeruginosa.95–98 Indeed, microbial proteases efficiently participate in gluten metabolism in vivo by modifying gluten’s mucosal absorption and immunogenicity, ultimately affecting the adaptive immune responses to gluten associated with CeD.43 In a recent study, microbial glutamate carboxypeptidase genes were associated with efficient gluten degradation.38 On the other hand, Pseudomonas aeruginosa, an opportunistic pathogen isolated from the duodenum of CeD patients, increased gluten immunogenicity by producing peptides that better translocate across the intestinal barrier and activate gluten-specific T-cells from CeD patients.18 P. aeruginosa degrades gluten through LasB, and this metalloprotease also leads to a gluten-independent upregulation of inflammatory pathways via protease-activated receptor (PAR)-2 activation. In mice expressing CeD risk genes, P. aeruginosa LasB synergizes with gluten to induce more severe inflammation that is associated with moderate villus blunting. Thus, the human intestine represents a rich source of microbial proteases helping in the digestion of common dietary proteins, which can increase or decrease their final immunogenicity.
A similar phenomenon has been demonstrated with other recalcitrant dietary proteins such as wheat amylase trypsin inhibitors (ATI). ATI are able to induce innate immune activation in the gut via toll-like receptor 4 activation, with downstream effects on intestinal inflammation and antigen sensitization.91,99,100 Gut microbial proteases are able to digest ATI, thereby reducing the intestinal dysfunction associated with wheat proteins.101 For example, Lactobacillus strains degrade both gluten and ATI peptides, reducing their immunogenic properties.18,101
In addition to the direct effects of microbial proteases on the host due to the catabolism of proteins, microbes release a plethora of metabolites impacting host homeostasis such as branched-chain fatty acids, amino acids, ammonia, phenols, hydrogen sulfide.102,103 Interestingly, tyrosine metabolites such as p-cresol and 4-ethylphenyl sulfate, which may contribute to gut-brain communication,are altered in IBS.104–108 Another bacterial metabolite, hydrogen sulfide, has multiple roles in physiological processes in the gut and has been linked to intestinal inflammation and colorectal cancer.109–111 Finally, tryptophan is a precursor for the synthesis of several important bioactive molecules, such as serotonin, melatonin, nicotinamide and vitamin B3, in addition to many other physiologically important intermediates.112 Tryptophan is an essential aromatic amino acid found in different dietary sources such as poultry, fish, oats, and dairy products. This unique amino acid can be metabolized by the gut microbiota into a range of indolic compounds, some of which can activate key homeostatic receptors such as the aryl hydrocarbon receptor (AhR) or the Pregnane X receptor (PXR).87 Indeed, these receptors have been implicated in intestinal inflammation and microbial tryptophan metabolism is altered in patients with IBD and CeD.113–116 Thus, microbial proteases could indirectly modulate different intestinal conditions by modifying common dietary antigens or facilitating the release of bioactive metabolites in the gut.
Impact of bacterial proteases on mucosal barrier function
The first line of host defence against both commensal bacteria and invading enteric pathogens is the intestinal mucosal barrier, which is a physical barrier that includes both biochemical and immunological components. The physical barrier consists of epithelial cells connected by tight junctions and protected by an overlying host-secreted mucus layer.117 The mucus layer in the gut forms a physical barrier between host epithelial cells and the gut microbiota. There is a continuous turnover of the mucous layer and deficiencies in this dynamic system have been linked to gastrointestinal diseases and colonic cancer. The primary component of mucus is the gel-forming mucin 2 (MUC2) protein, which is synthesized by goblet cells. MUC2 deficient mice are more susceptible to developing spontaneous colitis118 and MUC2 gene levels were found to be altered in both UC and CD compared to healthy controls.119,120 Mucus organization in the colon is vastly different from that of the small intestine. The mucus in the small intestine forms a single and penetrable layer but the bacteria are kept away from the epithelium by antibacterial mediators. The mucus in the colon forms a double layer. The inner mucus layer is firmly attached to the epithelium, impenetrable to bacteria and essential for inhibiting the interaction of microbes with host receptors on the epithelium. The outer mucus layer in the colon (secreted) is expanded and serves as a habitat for the microbiota.121
Although a major function of secreted mucus is to protect the host epithelium from both commensals and pathogens, the glycoproteins in this barrier also create a nutrient source for some colonic microorganisms. Mucin provides carbon and nitrogen sources to bacteria and the exposed O-glycan chains serve as attachment sites for bacterial colonization. The commensal microbiota adhering to mucins protect the host through colonization resistance. For decades the idea that contents found in fecal samples can degrade colonic mucus has been discussed.122,123 The microbial carbohydrate-active enzymes (CAZymes) required for mucus degradation have been intensively studied in recent years, especially among members of Bacteroides and Ruminococcus.117 Proteases from different microorganisms also exhibit strong proteolytic mucinase activity.124 Zinc metalloprotease ZmpB from Clostridium perfringens cleaves adjacent to glycosylated Serine and/or Threonine residues. Proteases from different infectious agents such as Pic in diarrheagenic E. coli and Shigella, or vibriolysin in V. cholera, degrade the colonic mucus, which is a key step facilitating epithelium invasion.10,68,69,71 StcE and SslE, metalloproteases from diarrheagenic E. coli strains, also cleave mucin glycoproteins, which may help the pathogen to reach the epithelium.74–76 In addition, the M60-like protease family cleaves the mucin glycoprotein backbone in a manner that is dependent on the presence of specific glycan sidechain structures.125,126 Many pathogens express this family of proteases for host invasion. Different mucin-degrading proteases have been also described in Bacteroides, a common commensal of the human intestine. These include proteases in B. thetaiotaomicron (BT4244) or Bacteroides caccae.126,127 Although mucin degradative capacity is considered a virulence factor of many GI pathogens, the implications in specific chronic intestinal diseases are not yet well understood.
In addition to effects on the mucus layer, proteases have been shown to disrupt the epithelial component of the mucosal barrier. The epithelial barrier function requires a contiguous layer of cells as well as the tight and adheren junctions that seal the paracellular spaces between them. Compromised intestinal barrier function has been associated with a number of disease states, both intestinal and systemic.128 Oral administration of the bacterial serine protease SP-1, produced by Clostridium spp resulted in impaired epithelial barrier, altered microbiota community compostion, and exacerbated DSS-induced colitis.129 Host proteases such as chymase or ELA2A are able to cleave tight and adherens junction proteins including zonula occludens-1 or E-cadherin.22,29,130 Microbial proteases also cleave inter-enterocyte junctions. The metalloprotease GelE, produced by Enterococcus faecalis, degrades E-cadherin leading to loss of barrier function that is evident before inflammation in a mouse model of spontaneous colitis.131 Tight junctions are also targets of infectious agents such as Pseudomonas aeruginosa (through LasB),132 H. pylori (PqqE and HtrA),61–63 C. jejuni (HtrA), 79,80 V. cholerae (vibrolysin),10,83 Shigella, Salmonella or pathogenic Escherichia coli (SepA).72,133 Thus, microbes use proteases for host invasion with important implications in the gut. Alteration in tight and adherens junctions lead to increased paracellular permeability of the epithelial barrier which is a pathophysiological hallmark of IBD and IBS. As mucin- and junction-degrading bacteria can cause damage, these enzymes may provide targets for protease inhibitors to treat or prevent intestinal diseases.25,117
Another mechanism whereby luminal proteases can modulate the function of the epithelial barrier is via the activation of protease activated receptors (PARs) expressed on the membrane of enterocytes. PARs are a family of receptors with pleiotropic effects which have been implicated in different gastrointestinal conditions and is covered in detail in the sections below. Regarding the role of PARs in intestinal function, apical administration of PAR-2 activating ligands led to prostanoid and interferon release as well as an increase in paracellular permeability due to ZO-1 degradation.6,134 PAR-4 activation by cathepsin G, a protease that is elevated in fecal samples from UC patients, led to increased mucosal permeability and inflammation in vivo in mice.28 Mucosal damage and inflammation caused by toxin A from the enteric pathogen C. difficile is markedly reduced when PAR-2 activation is prevented.135 Although it is clear that luminal proteases have pronounced PAR-dependent effects on enterocyte function, additional work is required to determine the importance of the contribution of microbial proteases to these effects.
Bacterial protease effects on cell and immune signaling
Proteases are signaling enzymes that can specifically regulate cell and immune signaling by different mechanistic pathways, including those mediated by PAR activation. PARs are G-protein coupled receptors that have seven transmembrane domains, an extracellular N-terminal and an intracellular C terminal. Proteolytic cleavage of the N-terminal initiates intracellular signaling by revealing a tethered ligand. The activation of the different members of the PAR family (PAR-1, 2,3 and 4) is protease specific, tightly regulated and influences a number of physiological functions in the gut such as motility, permeability and nociception.22 The functional consequence of a particular protease activating PARs depends on which PAR is activated and what its downstream signaling pathways are, including whether canonical or biased signaling pathways are initiated.136,137
PARs are ubiquitously expressed in the gastrointestinal tract (epithelial cells, neurons, mast cells, fibroblasts, etc) and mediate a wide array of pro-inflammatory, pronociceptive and proliferative effects after activation by proteases. PARs have been implicated in the pathogenesis of colorectal cancer138 and inflammatory and functional intestinal disorders.139–141 Moreover, elevated expression of par2 in the colon tissue of patients with UC has been described. 142 Different microbial proteases have been proposed as activators PARs. Of relevance, LasB from Pseudomonas aeruginosa,18,143 and GelE from Enterococcus faecalis144 degrade the N-terminal of PAR-2, contributing to food sensitivities and intestinal inflammation in preclinical mouse models. However, the full implications of activating PARs by microbial proteases in the context of GI diseases are still being resolved.
Proteases are also able to stimulate or diminish the production of key host immune mediators such as cytokines or immunoglobulins (Ig). Cytokine production is a dynamic event that is tightly regulated. Disturbances in its dynamics can provoke exacerbated responses in the host as they are involved in multiple cascades of intracellular signaling.
For example, cleavage and activation of PAR-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis induces the release of proinflammatory cytokines such as interleukin (IL)-6, IL-8 and tumour necrosis factor (TNF)-α.145 Interestingly, cytokines can also be degraded by bacterial proteases. Previous reports have shown that alkaline proteases and elastase from Pseudomonas aeruginosa can degrade IL-2 and interferon (INF)-γ.146,147 A Zn-metalloproteinase from Legionella pneumophila also has the ability to degrade IL-2.148 Likewise, gingipains, a trypsin-like cysteine proteinases produced by Porphyromonas gingivalis, can cleave IL-1β, IL-6, and IL-1ra.149 The same phenomenon can be observed with other protein-based mediators such as Ig. Ig are glycoproteins produced by plasma cells, which play an important role in adaptive immune responses by specifically recognizing particular antigens. In addition to the capacity of the microbiota to stimulate different Ig subtypes in the host, microbes can also degrade Ig helping immune system evasion. Many pathogens that infect mucosal surfaces encode proteases that cleave immunoglobulin such as Neisseria meningitides, N. gonorrhoeae or Streptococcus pneumoniae.150,151 It has been shown that the intestinal microbiota can degrade IgA, which plays a key role in mucosal immunology, and mice with documented degradation of secretory IgA are more susceptible to chemically-induced colitis.152 Host proteases such as trypsin are capable of degrading IgA, and microbial commensals such as Paraprevotella can prevent its degradation in the gut.9 Most studies have focused on understanding the interplay of bacterial exotoxins and the immune system to prevent disease. However, much remains to be done to understand whether proteases released by commensals promote or neutralize the production of anti- and pro- inflammatory mediators, respectively.
Bacterial proteases and abdominal pain
Activation of PARs by host-derived proteases has been implicated in abdominal pain for the past two decades. Agonists of PARs excite spinal afferent neurons that innervate the GI tract.153,154 Biopsy supernatants from IBS patients have also been shown to excite spinal afferent neurons via PAR-2 activation.7,58,155 These excitatory effects have been ascribed to sensitization of transient receptor potential (TRP) channels, including TRPV1, TRPV4 and TRPA1 as well as suppression of voltage-gated K+ channels. There is also evidence from models of colitis that PAR-2 activation contributes to nociceptor hyperexcitability.156 Importantly, activation of PAR-4 has opposite effects on nociceptor activation to PAR-2 activation in rodents. PAR-4 activation suppresses the excitability of colonic nociceptors in vitro,157 and in vivo.158
A role for bacterial proteases in the modulation of abdominal pain was first suggested by in vivo experiments using fecal supernatants from patients with IBD or IBS by Bueno and colleagues. Abdominal pain sensitivity was measured in rats and mice by quantifying the visceromotor response to colorectal distension. Intracolonic administration of fecal supernatants from IBS-D and IBS-C patients increased the visceromotor response to distension, with evidence of both allodynia and hyperalgesia.159,160 In contrast, fecal supernatants from UC patients had the opposite effect, decreasing the visceromotor response in a PAR-4-dependent manner.159 Thus, it appears that fecal proteases can either exascerbate or suppress abdominal pain in rodents, depending on the relative amount of PAR-2 or PAR-4 activation that occurs.
Another important consideration is where the sites of action of fecal proteases are relevant to pain modulation. PARs are expressed on many cells within the wall of the gut, including spinal afferent neurons and enterocytes. Based on the study of fecal proteases from IBS-C patients, enhancement of abdominal pain did not appear to be due to direct excitatory actions of proteases on neuronal PARs.160 Instead, intracolonic administration of cysteine proteases within IBS-C fecal supernatants to mice increased colonic permeability and led to degradation of occluding, which in turn led to increased visceral pain. Mucosal biopsies from IBS-C patients also displayed evidence of epithelial occludin degradation compared to biopsies from healthy controls.160 However, because actions of fecal proteases on neuronal activation were not assessed in this study, it remains possible that direct effects on nociceptor nerve terminals in the gut neu also contributes to visceral pain, as the reduction of mucosal barrier integrity would facilitate access of luminal proteases to spinal afferent nerve terminals.
Subsequent studies have identified Faecalibacterium prausnitzii as a potential source of anti-nociceptive mediators, including a PAR-4 activating serine protease. Using two well-established rodent models of IBS that lead to visceral hyperalgesia in vivo, it was found that the enhanced visceromotor response to colorectal distension was reversed following administration of F. prausnitzii. .161–164These antinociceptive effects were due to a reversal of the increase in mucosal permeability that is a feature of these IBS models. In vitro experiments on dorsal root ganglion neurons also support an anti-nociceptive role of F. prausnitzii.19 Media supernatant from cultures of F. prausnitzii acted directly on DRG neurons to suppress their excitability due to an increase in voltage-gated K+ conductance. This was the result of a cathepsin G-like serine protease that activated neuronal PAR-4.
In summary, PAR activation is able to suppress or augment abdominal pain, depending on which proteases predominate and which receptors they activate. Studies on samples from patients with abdominal pain indicate that both host and bacterial proteases could potentially contribute to pain. Given the evidence of luminal proteolytic imbalance in diseases associated with abdominal pain, including IBD and IBS, future studies directed towards further delineating the bacterial sources and cellular targets of these proteases will be valuable. The insights may lead to the development of next-generation probiotics that supress abdominal pain by shifting the balance of PAR activation to barrier-restoring and nociceptor-suppressing effects.
Conclusions
The impacts of proteases released by commensal bacteria on GI disease has received increasing attention in recent years. It has become evident that a complex balance between proteases, their host targets and protease inhibitors maintains the functionality and integrity of the gut. Dysregulation of this balance has a direct impact on intestinal health with serious consequences that lead to pathophysiological conditions (Figure 2). Furthermore, many pathogenic bacteria utilize proteases to colonise host tissues and cause disease (Table 1). Although recent findings have shown the importance of proteases from commensal gut bacteria in gut homeostasis, the study of these proteases and their contribution to disease is still in its infancy. However, as causal relationships between protease activity and disease are identified, along with mechanistic insights into how bacterial proteases promote or protect against disease, new opportunities to treat common GI diseases and infections may result.
Figure 2.
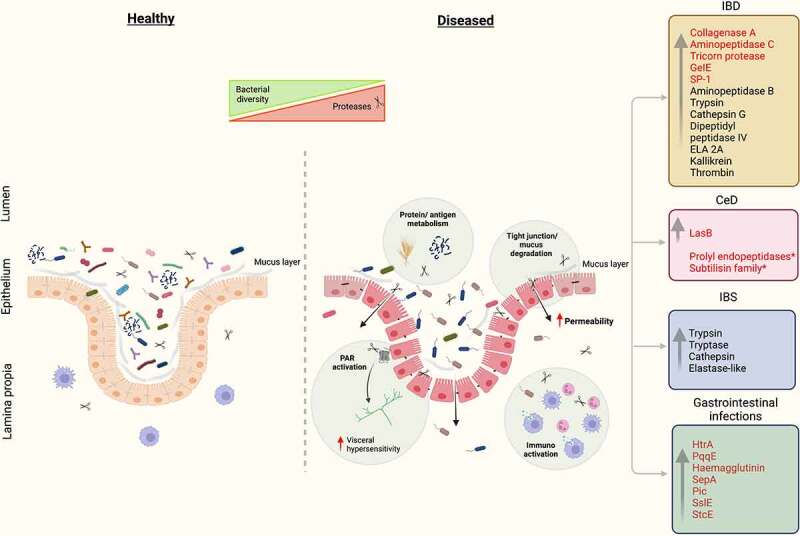
Proteases implicated in IBD, IBS, CeD and GI infections. In disease-related conditions, proteases induce structural and functional changes in the gut through multiple mechanisms of action, including effects on dietary protein metabolism, mucosal barrier function, neuronal excitability and immunoregulation. Luminal proteases impact GI function by a combination of PAR-dependent and independent effects. Proteases of microbial origin are highlighted in red. *Proteases with therapeutic potential. Figure was created with BioRender.com.
Table 1.
Summary of proteases of bacterial and host origin implicated in gastrointestinal diseases.
| Protease | Source | Classification | Activity/mechanism | Disease | Ref |
|---|---|---|---|---|---|
| Proteases released by bacteria | |||||
| Collagenase A | Clostridium perfringens | Metalloprotease | Intestinal barrier alteration/mucus | IBD | 124 |
| Dipeptidyl peptidase IV Dipeptidase Amino peptidase C Tricorn protease homolog |
Bacteroides/ B. vulgatus | Serine protease Metalloprotease Cysteine protease |
Contribute to UC disease activity, barrier dysfunction | IBD | 32,33 |
| ND | R. mucilaginosa | ND | Gluten degradation | CeD | 47 |
| ND | Lactobacillus | ND | ATI peptides/gluten degradation | CeD | 101 |
| LasB | P. aeruginosa | Zn metalloprotease | Gluten degradation, PAR-2 mediated signaling Intestinal barrier alteration Degrade IL-2 and IFN-γ |
CeD and others | 18,146,162 |
| Prolyl endopeptidases |
F. meningosepticum S. capsulate M. xanthus |
Serine peptidases | Gluten degradation | CeD | 49 |
|
BAV86562.1 Subtilisin family |
R. aeria | Serine protease | Gluten degradation | CeD | 48 |
| GeIE | E. faecalis | Metalloprotease | Compromise epithelial barrier | Colitis mouse model | 131 |
| SP-1 | C. ramosum | Serine protease | Epithelial barrier dysfunction | Colitis mouse model | 129 |
| HtrA |
H. pylori C. jejuni S. Typhimurium |
Serine protease | Epithelial barrier alteration Growth and infection |
Gastrointestinal infection | 63,80,81 |
| PqqE | H. pylori | Zn metalloprotease | Gastrointestinal infection | ||
| Haemagglutinin or vibriolysin | V. cholerae | Metalloprotease | Barrier integrity alteration/actin and tight junction rearrangement | Gastrointestinal infection | 83 |
| SepA |
S. flexneri E. coli |
Serine protease | Barrier dysfunction/invasion | Gastrointestinal infection | 72 |
| Pic |
E. coli Citrobacter S. flexneri |
Serine protease | Disrupt mucosal barriers/modulate immune response/ mucinase activity |
Gastrointestinal infection | 68–71 |
| SslE | E. coli | Zn metalloprotease | Mucin degradation | Gastrointestinal infection | 76 |
| StcE | E. coli | Metalloproteases | Mucus degradation | Gastrointestinal infection | 74,75 |
| ND | L. pneumophila | Zn Metalloproteases | Degrade IL-2 | Gastrointestinal infections | 148 |
| PROKKA_00509 | Paraprevotella | ND | Trypsin degradation | Others | 9 |
| M60-like proteases | B. caccae | Metalloproteases | Mucin degradation | Other | 127 |
| M60-like/PF13402 domain | B. thetaiotaomicron | Metalloprotease | Mucin degradation | Others | 125 |
| ZmpB | C. perfringens | Zn metalloproteases | Mucus degradation | Others | 117 |
| Proteases released by the host | |||||
| Aminopeptidase B | Human colonic tissues | Metalloprotease | Lysine-cleaving protease identified as active in CD | IBD | 8 |
| Cathepsin G | Human colonic tissue | Serine protease | Overactive | IBD | 8 |
| Human stool | Epithelial barrier alteration | IBD | 28 | ||
| Dipeptidyl Peptidase-4 | Human stool/intestinal tissue | Serine protease | Increased levels and expression | IBD | 163 |
| ELA 2A (Elastase 2A) |
Human colonic cells | Serine protease | Epithelial barrier dysfunction | IBD | 29 |
| Thrombin | Human colonic tissue | Serine protease | Overactive | IBD | 8 |
| Trypsin-like, Elastase-like Cathepsin G-like Proteinase 3-like |
Human stool | Serine proteases | Elevated activity | IBD | 12 |
| Kallikrein | Human intestinal tissue/stool | Serine protease | Inflammatory process in Crohn’s disease. | IBD/IBS | 13,164 |
| Trypsin-like Chymotrypsin-like Elastase-like |
Human stool | Serine protease | Elevated in patients with high proteolytic activity | IBS | 13 |
| Trypsin-3/Trypsin-like activity | Human colonic tissue | Serine protease | Upregulated in intestinal epithelial, signal to enteric neurons and to induce visceral hypersensitivity/somatic and visceral hyperalgesia and allodynia in colon mice | IBS | 7,58 |
| Tryptase-alphabeta-1/Tryptase | Rat colonic tissue | Serine protease | Expression increased in colon of rats pots-colitis with visceral hypersensitivity/somatic and visceral hyperalgesia and allodynia in colon mice | IBS | 31, 58 |
ND = Not determined
Funding Statement
This work was supported by the Canadian Institutes of Health Research
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–21. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 2.Di Cera E. Serine proteases. IUBMB Life. 2009;61:510–515. doi: 10.1002/iub.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukharev SA, Pleshakova OV, Sadovnikov VB. Role of proteases in activation of apoptosis. Cell Death Differ. 1997;4:457–462. doi: 10.1038/sj.cdd.4400263. [DOI] [PubMed] [Google Scholar]
- 4.Suresh B, Lee J, Kim H, Ramakrishna S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016;23:1257–1264. doi: 10.1038/cdd.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perisic Nanut M, Pecar Fonovic U, Jakos T, Kos J. The role of cysteine peptidases in hematopoietic stem cell differentiation and modulation of immune system function. Front Immunol. 2021;12:680279. doi: 10.3389/fimmu.2021.680279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong W, McConalogue K, Khitin LM, Hollenberg MD, Payan DG, Bohm SK, Bunnett NW. Luminal trypsin may regulate enterocytes through proteinase-activated receptor 2. Proc Natl Acad Sci U S A. 1997;94(16):8884–8889. doi: 10.1073/pnas.94.16.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rolland-Fourcade C, Denadai-Souza A, Cirillo C, Lopez C, Jaramillo JO, Desormeaux C, Cenac N, Motta J-P, Larauche M, Taché Y, et al. Epithelial expression and function of trypsin-3 in irritable bowel syndrome. Gut. 2017;66:1767–1778. doi: 10.1136/gutjnl-2016-312094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denadai-Souza A, Bonnart C, Tapias NS, Marcellin M, Gilmore B, Alric L, Bonnet D, Burlet-Schiltz O, Hollenberg MD, Vergnolle N, et al. Functional proteomic profiling of secreted serine proteases in health and inflammatory bowel disease. Sci Rep. 2018;8:7834. doi: 10.1038/s41598-018-26282-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Watanabe E, Kawashima Y, Plichta DR, Wang Z, Ujike M, et al. Identification of trypsin-degrading commensals in the large intestine. Nature. 2022;609(7927):582-589. doi: 10.1038/s41586-022-05181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benitez JA, Silva AJ. Vibrio cholerae hemagglutinin(HA)/protease: an extracellular metalloprotease with multiple pathogenic activities. Toxicon. 2016;115:55–62. doi: 10.1016/j.toxicon.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frees D, Brondsted L, Ingmer H. Bacterial proteases and virulence. Subcell Biochem. 2013;66:161–192. [DOI] [PubMed] [Google Scholar]
- 12.Jablaoui A, Kriaa A, Mkaouar H, Akermi N, Soussou S, Wysocka M, Wołoszyn D, Amouri A, Gargouri A, Maguin E, et al. Fecal Serine Protease Profiling in Inflammatory Bowel Diseases. Front Cell Infect Microbiol. 2020;10:21. doi: 10.3389/fcimb.2020.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edogawa S, Edwinson AL, Peters SA, Chikkamenahalli LL, Sundt W, Graves S, Gurunathan SV, Breen-Lyles M, Johnson S, Dyer R, et al. Serine proteases as luminal mediators of intestinal barrier dysfunction and symptom severity in IBS. Gut. 2020;69:62–73. doi: 10.1136/gutjnl-2018-317416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol. 2022;20:143–160. doi: 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 17.Metwaly A, Dunkel A, Waldschmitt N, Raj ACD, Lagkouvardos I, Corraliza AM, Mayorgas A, Martinez-Medina M, Reiter S, Schloter M, et al. Integrated microbiota and metabolite profiles link Crohn’s disease to sulfur metabolism. Nat Commun. 2020;11(1):4322. doi: 10.1038/s41467-020-17956-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caminero A, McCarville JL, Galipeau HJ, Deraison C, Bernier SP, Constante M, Rolland C, Meisel M, Murray JA, Yu XB, et al. Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2. Nat Commun. 2019;10(1):1198. doi: 10.1038/s41467-019-09037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sessenwein JL, Baker CC, Pradhananga S, Maitland ME, Petrof EO, Allen-Vercoe E, Noordhof C, Reed DE, Vanner SJ, Lomax AE, et al. Protease-Mediated suppression of DRG neuron excitability by commensal bacteria. J Neurosci. 2017;37(48):11758–11768. doi: 10.1523/JNEUROSCI.1672-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Otin C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vergnolle N. Protease inhibition as new therapeutic strategy for GI diseases. Gut. 2016;65:1215–1224. doi: 10.1136/gutjnl-2015-309147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tripathi LP, Sowdhamini R. Genome-wide survey of prokaryotic serine proteases: analysis of distribution and domain architectures of five serine protease families in prokaryotes. BMC Genomics. 2008;9:549. doi: 10.1186/1471-2164-9-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raju RM, Goldberg AL, Rubin EJ. Bacterial proteolytic complexes as therapeutic targets. Nat Rev Drug Discov. 2012;11:777–789. doi: 10.1038/nrd3846. [DOI] [PubMed] [Google Scholar]
- 25.Culp E, Wright GD. Bacterial proteases, untapped antimicrobial drug targets. J Antibiot (Tokyo). 2017;70:366–377. doi: 10.1038/ja.2016.138. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt M, Lupas AN, Finley D. Structure and mechanism of ATP-dependent proteases. Curr Opin Chem Biol. 1999;3:584–591. doi: 10.1016/S1367-5931(99)00013-7. [DOI] [PubMed] [Google Scholar]
- 27.Motta JP, Bermudez-Humaran LG, Deraison C, Martin L, Rolland C, Rousset P, Boue J, Dietrich G, Chapman K, Kharrat P, et al. Food-grade bacteria expressing elafin protect against inflammation and restore colon homeostasis. Sci Transl Med. 2012;4:158ra44. doi: 10.1126/scitranslmed.3004212. [DOI] [PubMed] [Google Scholar]
- 28.Dabek M, Ferrier L, Roka R, Gecse K, Annahazi A, Moreau J, Escourrou J, Cartier C, Chaumaz G, Leveque M, et al. Luminal cathepsin g and protease-activated receptor 4: a duet involved in alterations of the colonic epithelial barrier in ulcerative colitis. Am J Pathol. 2009;175:207–214. doi: 10.2353/ajpath.2009.080986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Motta JP, Rolland C, Edir A, Florence AC, Sagnat D, Bonnart C, Rousset P, Guiraud L, Quaranta-Nicaise M, Mas E, et al. Epithelial production of elastase is increased in inflammatory bowel disease and causes mucosal inflammation. Mucosal Immunol. 2021;14:667–678. doi: 10.1038/s41385-021-00375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Motta JP, Magne L, Descamps D, Rolland C, Squarzoni-Dale C, Rousset P, Martin L, Cenac N, Balloy V, Huerre M, et al. Modifying the protease, antiprotease pattern by elafin overexpression protects mice from colitis. Gastroenterology. 2011;140:1272–1282. doi: 10.1053/j.gastro.2010.12.050. [DOI] [PubMed] [Google Scholar]
- 31.Ceuleers H, Hanning N, Heirbaut J, Van Remoortel S, Joossens J, Van Der Veken P, Francque SM, De Bruyn M, Lambeir A-M, De Man JG, et al. Newly developed serine protease inhibitors decrease visceral hypersensitivity in a post-inflammatory rat model for irritable bowel syndrome. Br J Pharmacol. 2018;175:3516–3533. doi: 10.1111/bph.14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galipeau HJ, Caminero A, Turpin W, Bermudez-Brito M, Santiago A, Libertucci J, Constante M, Raygoza Garay JA, Rueda G, Armstrong S, et al. Novel fecal biomarkers that precede clinical diagnosis of ulcerative colitis. Gastroenterology. 2021;160:1532–1545. doi: 10.1053/j.gastro.2020.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Mills RH, Dulai PS, Vazquez-Baeza Y, Sauceda C, Daniel N, Gerner RR, Batachari LE, Malfavon M, Zhu Q, Weldon K, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. 2022;7(2):262–276. doi: 10.1038/s41564-021-01050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh P, Arora A, Strand TA, Leffler DA, Catassi C, Green PH, Kelly CP, Ahuja V, Makharia GK. Global prevalence of celiac disease: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2018;16(6):823–36 e2. doi: 10.1016/j.cgh.2017.06.037. [DOI] [PubMed] [Google Scholar]
- 35.Galipeau HJ, Verdu EF. The double-edged sword of gut bacteria in celiac disease and implications for therapeutic potential. Mucosal Immunol. 2022;15:235–243. doi: 10.1038/s41385-021-00479-3. [DOI] [PubMed] [Google Scholar]
- 36.Lindfors K, Ciacci C, Kurppa K, Lundin KEA, Makharia GK, Mearin ML, Murray JA, Verdu EF, Kaukinen K. Coeliac disease. Nat Rev Dis Primers. 2019;5(1):3. doi: 10.1038/s41572-018-0054-z. [DOI] [PubMed] [Google Scholar]
- 37.Kivela L, Caminero A, Leffler DA, Pinto-Sanchez MI, Tye-Din JA, Lindfors K. Current and emerging therapies for coeliac disease. Nat Rev Gastroenterol Hepatol. 2021;18:181–195. doi: 10.1038/s41575-020-00378-1. [DOI] [PubMed] [Google Scholar]
- 38.Constante M, Libertucci J, Galipeau HJ, Szamosi JC, Rueda G, Miranda PM, Pinto-Sanchez MI, Southward CM, Rossi L, Fontes ME, et al. Biogeographic variation and functional pathways of the gut microbiota in celiac disease. Gastroenterology. 2022. doi: 10.1053/j.gastro.2022.06.088. [DOI] [PubMed] [Google Scholar]
- 39.Caminero A, Verdu EF. Celiac disease: should we care about microbes? Am J Physiol Gastrointest Liver Physiol. 2019;317:G161–G70. doi: 10.1152/ajpgi.00099.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemppainen KM, Lynch KF, Liu E, Lonnrot M, Simell V, Briese T, Koletzko S, Hagopian W, Rewers M, She J-X, et al. Factors that increase risk of celiac disease autoimmunity after a gastrointestinal infection in early life. Clin Gastroenterol Hepatol. 2017;15:694–702 e5. doi: 10.1016/j.cgh.2016.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernardo D, Garrote JA, Nadal I, Leon AJ, Calvo C, Fernandez-Salazar L, Blanco-Quiros A, Sanz Y, Arranz E. Is it true that coeliacs do not digest gliadin? Degradation pattern of gliadin in coeliac disease small intestinal mucosa. Gut. 2009;58:886–887. doi: 10.1136/gut.2008.167296. [DOI] [PubMed] [Google Scholar]
- 42.Caminero A, Nistal E, Herran AR, Perez-Andres J, Ferrero MA, Vaquero Ayala L, Vivas S, Ruiz de Morales JMG, Albillos SM, Casqueiro FJ, et al. Differences in gluten metabolism among healthy volunteers, coeliac disease patients and first-degree relatives. Br J Nutr. 2015;114:1157–1167. doi: 10.1017/S0007114515002767. [DOI] [PubMed] [Google Scholar]
- 43.Caminero A, Galipeau HJ, McCarville JL, Johnston CW, Bernier SP, Russell AK, Jury J, Herran AR, Casqueiro J, Tye-Din JA, et al. Duodenal bacteria from patients with celiac disease and healthy subjects distinctly affect gluten breakdown and immunogenicity. Gastroenterology. 2016;151:670–683. doi: 10.1053/j.gastro.2016.06.041. [DOI] [PubMed] [Google Scholar]
- 44.McCarville JL, Dong J, Caminero A, Bermudez-Brito M, Jury J, Murray JA, et al. A commensal bifidobacterium longum strain prevents gluten-related immunopathology in mice through expression of a serine protease inhibitor. Appl Environ Microbiol. 2017;83(19):e01323-17. doi: 10.1128/AEM.01323-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frazer AC, Fletcher RF, Ross CA, Shaw B, Sammons HG, Schneider R. Gluten-induced enteropathy: the effect of partially digested gluten. Lancet. 1959;2:252–255. doi: 10.1016/S0140-6736(59)92051-3. [DOI] [PubMed] [Google Scholar]
- 46.Douglas AP, Peters TJ. Peptide hydrolase activity of human intestinal mucosa in adult coeliac disease. Gut. 1970;11:15–17. doi: 10.1136/gut.11.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamakhchari M, Wei G, Dewhirst F, Lee J, Schuppan D, Oppenheim FG, Helmerhorst EJ. Identification of Rothia bacteria as gluten-degrading natural colonizers of the upper gastro-intestinal tract. PLoS One. 2011;6:e24455. doi: 10.1371/journal.pone.0024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei G, Darwish G, Oppenheim FG, Schuppan D, Helmerhorst EJ. Commensal bacterium rothia aeria degrades and detoxifies gluten via a highly effective subtilisin enzyme. Nutrients. 2020;12:3724. doi: 10.3390/nu12123724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004;383:311–318. doi: 10.1042/BJ20040907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pultz IS, Hill M, Vitanza JM, Wolf C, Saaby L, Liu T, Winkle P, Leffler DA. Gluten Degradation, Pharmacokinetics, Safety, and Tolerability of TAK-062, an Engineered Enzyme to Treat Celiac Disease. Gastroenterology. 2021;161:81–93 e3. doi: 10.1053/j.gastro.2021.03.019. [DOI] [PubMed] [Google Scholar]
- 51.Shan L, Molberg O, Parrot I, Hausch F, Filiz F, Gray GM, Sollid LM, Khosla C. Structural basis for gluten intolerance in celiac sprue. Science. 2002;297:2275–2279. doi: 10.1126/science.1074129. [DOI] [PubMed] [Google Scholar]
- 52.Mearin F, Lacy BE, Chang L, Chey WD, Lembo AJ, Simren M, et al. Bowel disorders. Gastroenterology. 2016;S0016-5085(16): 00222-5. doi: 10.1053/j.gastro.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 53.Coates MD, Mahoney CR, Linden DR, Sampson JE, Chen J, Blaszyk H, Crowell MD, Sharkey KA, Gershon MD, Mawe GM, et al. Molecular defects in mucosal serotonin content and decreased serotonin reuptake transporter in ulcerative colitis and irritable bowel syndrom1. Gastroenterology. 2004;126(7):1657–1664. doi: 10.1053/j.gastro.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 54.Cremon C, Carini G, Wang B, Vasina V, Cogliandro RF, De Giorgio R, Stanghellini V, Grundy D, Tonini M, De Ponti F, et al. Intestinal serotonin release, sensory neuron activation, and abdominal pain in irritable bowel syndrome. Am J Gastroenterol. 2011;106(7):1290–1298. doi: 10.1038/ajg.2011.86. [DOI] [PubMed] [Google Scholar]
- 55.Bashashati M, Moossavi S, Cremon C, Barbaro MR, Moraveji S, Talmon G, et al. Colonic immune cells in irritable bowel syndrome: a systematic review and meta-analysis. Neurogastroenterol Motil. 2018. p. 30. [DOI] [PubMed] [Google Scholar]
- 56.Bhattarai Y, Muniz Pedrogo DA, Kashyap PC. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol. 2017;312:G52–G62. doi: 10.1152/ajpgi.00338.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edwinson AL, Yang L, Peters S, Hanning N, Jeraldo P, Jagtap P, Simpson JB, Yang T-Y, Kumar P, Mehta S, et al. Gut microbial beta-glucuronidases regulate host luminal proteases and are depleted in irritable bowel syndrome. Nat Microbiol. 2022;7:680–694. doi: 10.1038/s41564-022-01103-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cenac N, Andrews CN, Holzhausen M, Chapman K, Cottrell G, Andrade-Gordon P, Steinhoff M, Barbara G, Beck P, Bunnett NW, et al. Role for protease activity in visceral pain in irritable bowel syndrome. J Clin Invest. 2007;117(3):636–647. doi: 10.1172/JCI29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palrasu M, Zaika E, Paulrasu K, Caspa Gokulan R, Suarez G, Que J, El-Rifai W, Peek RM, Garcia-Buitrago M, Zaika AI, et al. Helicobacter pylori pathogen inhibits cellular responses to oncogenic stress and apoptosis. PLoS Pathog. 2022;18(6):e1010628. doi: 10.1371/journal.ppat.1010628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamaoka Y. Mechanisms of disease: helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marques MS, Costa AC, Osorio H, Pinto ML, Relvas S, Dinis-Ribeiro M, Carneiro F, Leite M, Figueiredo C. Helicobacter pylori PqqE is a new virulence factor that cleaves junctional adhesion molecule A and disrupts gastric epithelial integrity. Gut Microbes. 2021;13:1–21. doi: 10.1080/19490976.2021.1921928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tegtmeyer N, Wessler S, Necchi V, Rohde M, Harrer A, Rau TT, Asche CI, Boehm M, Loessner H, Figueiredo C, et al. Helicobacter pylori employs a unique basolateral type IV secretion mechanism for cagA delivery. Cell Host Microbe. 2017;22(4):552–60 e5. doi: 10.1016/j.chom.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 63.Ho Löwer M, Weydig C, Tegtmeyer N, Geppert T, Schröder P, Sewald N, Backert S, Schneider G, Hoy B, et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11(10):798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sattar SBA, Bacterial Gastroenteritis SS. StatPearls. Treasure Island (FL). 2022 [Google Scholar]
- 65.O’Ryan M, Prado V, Pickering LK. A millennium update on pediatric diarrheal illness in the developing world. Semin Pediatr Infect Dis. 2005;16:125–136. doi: 10.1053/j.spid.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 66.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, Choroszy-Krol I. Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 2019;11:10. doi: 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ruiz-Perez F, Nataro JP. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol Life Sci. 2014;71:745–770. doi: 10.1007/s00018-013-1355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–5596. doi: 10.1128/IAI.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abreu AG, Abe CM, Nunes KO, Moraes CT, Chavez-Duenas L, Navarro-Garcia F, Barbosa AS, Piazza RMF, Elias WP. The serine protease Pic as a virulence factor of atypical enteropathogenic Escherichia coli. Gut Microbes. 2016;7:115–125. doi: 10.1080/19490976.2015.1136775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bhullar K, Zarepour M, Yu H, Yang H, Croxen M, Stahl M, et al. The serine protease autotransporter pic modulates citrobacter rodentium pathogenesis and its innate recognition by the host. Infect Immun. 2015;83:2636–2650. doi: 10.1128/IAI.00025-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun. 2009;77:2465–2473. doi: 10.1128/IAI.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maldonado-Contreras A, Birtley JR, Boll E, Zhao Y, Mumy KL, Toscano J, Ayehunie S, Reinecker H-C, Stern LJ, McCormick BA, et al. Shigella depends on SepA to destabilize the intestinal epithelial integrity via cofilin activation. Gut Microbes. 2017;8:544–560. doi: 10.1080/19490976.2017.1339006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grys TE, Walters LL, Welch RA. Characterization of the StcE protease activity of Escherichia coli O157:H7. J Bacteriol. 2006;188:4646–4653. doi: 10.1128/JB.01806-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hews CL, Tran SL, Wegmann U, Brett B, Walsham ADS, Kavanaugh D, et al. The StcE metalloprotease of enterohaemorrhagic Escherichia coli reduces the inner mucus layer and promotes adherence to human colonic epithelium ex vivo. Cell Microbiol. 2017;19(6):e12717. doi: 10.1111/cmi.12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grys TE, Siegel MB, Lathem WW, Welch RA. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157: h7 to host cells. Infect Immun. 2005;73:1295–1303. doi: 10.1128/IAI.73.3.1295-1303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Valeri M, Rossi Paccani S, Kasendra M, Nesta B, Serino L, Pizza M, et al. Pathogenic E. Coli Exploits SslE Mucinase Activity to Translocate through the Mucosal Barrier and Get Access to Host Cells. PLoS One. 2015;10:e0117486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Luo Q, Kumar P, Vickers TJ, Sheikh A, Lewis WG, Rasko DA, Sistrunk J, Fleckenstein JM. Enterotoxigenic Escherichia coli secretes a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun. 2014;82:509–521. doi: 10.1128/IAI.01106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nesta B, Valeri M, Spagnuolo A, Rosini R, Mora M, Donato P, Alteri CJ, Del Vecchio M, Buccato S, Pezzicoli A, et al. SslE elicits functional antibodies that impair in vitro mucinase activity and in vivo colonization by both intestinal and extraintestinal Escherichia coli strains. PLoS Pathog. 2014;10:e1004124. doi: 10.1371/journal.ppat.1004124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharafutdinov I, Tegtmeyer N, Musken M, Backert S. Campylobacter jejuni serine protease HtrA Induces paracellular transmigration of microbiota across polarized intestinal epithelial cells. Biomolecules. 2022;12(4):521. doi: 10.3390/biom12040521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brondsted L, Andersen MT, Parker M, Jorgensen K, Ingmer H. The HtrA protease of Campylobacter jejuni is required for heat and oxygen tolerance and for optimal interaction with human epithelial cells. Appl Environ Microbiol. 2005;71:3205–3212. doi: 10.1128/AEM.71.6.3205-3212.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis C, Skovierova H, Rowley G, Rezuchova B, Homerova D, Stevenson A, Spencer J, Farn J, Kormanec J, Roberts M, et al. Salmonella enterica Serovar Typhimurium HtrA: regulation of expression and role of the chaperone and protease activities during infection. Microbiology (Reading). 2009;155:873–881. doi: 10.1099/mic.0.023754-0. [DOI] [PubMed] [Google Scholar]
- 82.Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, et al. Vibrio spp. Infections. Nat Rev Dis Primers. 2018;4:8. [DOI] [PubMed] [Google Scholar]
- 83.Wu Z, Nybom P, Magnusson KE. Distinct effects of Vibrio cholerae haemagglutinin/protease on the structure and localization of the tight junction-associated proteins occludin and ZO-1. Cell Microbiol. 2000;2:11–17. doi: 10.1046/j.1462-5822.2000.00025.x. [DOI] [PubMed] [Google Scholar]
- 84.Kordus SL, Thomas AK, Lacy DB. Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat Rev Microbiol. 2022;20:285–298. doi: 10.1038/s41579-021-00660-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 88.Moayyedi P, Simren M, Bercik P. Evidence-based and mechanistic insights into exclusion diets for IBS. Nat Rev Gastroenterol Hepatol. 2020;17:406–413. doi: 10.1038/s41575-020-0270-3. [DOI] [PubMed] [Google Scholar]
- 89.McIntosh K, Reed DE, Schneider T, Dang F, Keshteli AH, De Palma G, Madsen K, Bercik P, Vanner S. FODMAPs alter symptoms and the metabolome of patients with IBS: a randomised controlled trial. Gut. 2017;66:1241–1251. doi: 10.1136/gutjnl-2015-311339. [DOI] [PubMed] [Google Scholar]
- 90.Sasson AN, Ananthakrishnan AN, Raman M. Diet in treatment of inflammatory bowel diseases. Clin Gastroenterol Hepatol. 2021;19:425–35 e3. doi: 10.1016/j.cgh.2019.11.054. [DOI] [PubMed] [Google Scholar]
- 91.Junker Y, Zeissig S, Kim SJ, Barisani D, Wieser H, Leffler DA, et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J Exp Med. 2012;209:2395–2408. doi: 10.1084/jem.20102660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koppel N, Maini Rekdal V, Balskus EP. Chemical transformation of xenobiotics by the human gut microbiota. Science. 2017;356(6344):eaag2770. doi: 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Caminero A, Meisel M, Jabri B, Verdu EF. Mechanisms by which gut microorganisms influence food sensitivities. Nat Rev Gastroenterol Hepatol. 2019;16:7–18. doi: 10.1038/s41575-018-0064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caminero A, Verdu EF. Metabolism of wheat proteins by intestinal microbes: implications for wheat related disorders. Gastroenterol Hepatol. 2019;42:449–457. doi: 10.1016/j.gastrohep.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 95.Herran AR, Perez-Andres J, Caminero A, Nistal E, Vivas S. Ruiz de Morales JM, et al. Gluten-degrading Bacteria are Present in the Human Small Intestine of Healthy Volunteers and Celiac Patients. Res Microbiol. 2017;168:673–684. [DOI] [PubMed] [Google Scholar]
- 96.Caminero A, Herran AR, Nistal E, Perez-Andres J, Vaquero L, Vivas S, Ruiz de Morales JMG, Albillos SM, Casqueiro J. Diversity of the cultivable human gut microbiome involved in gluten metabolism: isolation of microorganisms with potential interest for coeliac disease. FEMS Microbiol Ecol. 2014;88:309–319. doi: 10.1111/1574-6941.12295. [DOI] [PubMed] [Google Scholar]
- 97.Tian N, Faller L, Leffler DA, Kelly CP, Hansen J, Bosch JA, et al. Salivary gluten degradation and oral microbial profiles in healthy individuals and celiac disease patients. Appl Environ Microbiol. 2017;83(6):e03330-16. doi: 10.1128/AEM.03330-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tian N, Wei G, Schuppan D, Helmerhorst EJ. Effect of Rothia mucilaginosa enzymes on gliadin (gluten) structure, deamidation, and immunogenic epitopes relevant to celiac disease. Am J Physiol Gastrointest Liver Physiol. 2014;307:G769–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bellinghausen I, Weigmann B, Zevallos V, Maxeiner J, Reissig S, Waisman A, Schuppan D, Saloga J. Wheat amylase-trypsin inhibitors exacerbate intestinal and airway allergic immune responses in humanized mice. J Allergy Clin Immunol. 2019;143(1):201–12 e4. doi: 10.1016/j.jaci.2018.02.041. [DOI] [PubMed] [Google Scholar]
- 100.Zevallos VF, Raker V, Tenzer S, Jimenez-Calvente C, Ashfaq-Khan M, Russel N, Pickert G, Schild H, Steinbrink K, Schuppan D, et al. Nutritional wheat amylase-trypsin inhibitors promote intestinal inflammation via activation of myeloid cells. Gastroenterology. 2017;152(5):1100–13 e12. doi: 10.1053/j.gastro.2016.12.006. [DOI] [PubMed] [Google Scholar]
- 101.Caminero A, McCarville JL, Zevallos VF, Pigrau M, Yu XB, Jury J, Galipeau HJ, Clarizio AV, Casqueiro J, Murray JA, et al. Lactobacilli degrade wheat amylase trypsin inhibitors to reduce intestinal dysfunction induced by immunogenic wheat proteins. Gastroenterology. 2019;156(8):2266–2280. doi: 10.1053/j.gastro.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 102.Hughes R, Kurth MJ, McGilligan V, McGlynn H, Rowland I. Effect of colonic bacterial metabolites on Caco-2 cell paracellular permeability in vitro. Nutr Cancer. 2008;60:259–266. doi: 10.1080/01635580701649644. [DOI] [PubMed] [Google Scholar]
- 103.Gill PA, Inniss S, Kumagai T, Rahman FZ, Smith AM. The role of diet and gut microbiota in regulating gastrointestinal and inflammatory disease. Front Immunol. 2022;13:866059. doi: 10.3389/fimmu.2022.866059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bermudez-Martin P, Becker JAJ, Caramello N, Fernandez SP, Costa-Campos R, Canaguier J, Barbosa S, Martinez-Gili L, Myridakis A, Dumas M-E, et al. The microbial metabolite p-Cresol induces autistic-like behaviors in mice by remodeling the gut microbiota. Microbiome. 2021;9(1):157. doi: 10.1186/s40168-021-01103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jeffery IB, Das A, O’Herlihy E, Coughlan S, Cisek K, Moore M, Bradley F, Carty T, Pradhan M, Dwibedi C, et al. Differences in fecal microbiomes and metabolomes of people with vs without irritable bowel syndrome and bile acid malabsorption. Gastroenterology. 2020;158(4):1016–28 e8. doi: 10.1053/j.gastro.2019.11.301. [DOI] [PubMed] [Google Scholar]
- 106.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli J, Chow J, Reisman S, Petrosino J, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Needham BD, Funabashi M, Adame MD, Wang Z, Boktor JC, Haney J, Wu W-L, Rabut C, Ladinsky MS, Hwang S-J, et al. A gut-derived metabolite alters brain activity and anxiety behaviour in mice. Nature. 2022;602(7898):647–653. doi: 10.1038/s41586-022-04396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pinto-Sanchez MI, Hall GB, Ghajar K, Nardelli A, Bolino C, Lau JT, Martin F-P, Cominetti O, Welsh C, Rieder A, et al. Probiotic Bifidobacterium longum NCC3001 reduces depression scores and alters brain activity: a pilot study in patients with irritable bowel syndrome. Gastroenterology. 2017;153(2):448–59 e8. doi: 10.1053/j.gastro.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 109.Miller TW, Wang EA, Gould S, Stein EV, Kaur S, Lim L, Amarnath S, Fowler DH, Roberts DD. Hydrogen sulfide is an endogenous potentiator of T cell activation. J Biol Chem. 2012;287(6):4211–4221. doi: 10.1074/jbc.M111.307819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo FF, Yu TC, Hong J, Fang JY. Emerging roles of hydrogen sulfide in inflammatory and neoplastic colonic diseases. Front Physiol. 2016;7:156. doi: 10.3389/fphys.2016.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wallace JL, Wang R. Hydrogen sulfide-based therapeutics: exploiting a unique but ubiquitous gasotransmitter. Nat Rev Drug Discov. 2015;14:329–345. doi: 10.1038/nrd4433. [DOI] [PubMed] [Google Scholar]
- 112.Barik S, Vanderschueren D, Antonio L. The uniqueness of tryptophan in biology: properties, metabolism, interactions and localization in proteins. Int J Mol Sci. 2020;22:21. doi: 10.3390/ijms22010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheng J, Shah YM, Gonzalez FJ. Pregnane X receptor as a target for treatment of inflammatory bowel disorders. Trends Pharmacol Sci. 2012;33:323–330. doi: 10.1016/j.tips.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Monteleone I, Rizzo A, Sarra M, Sica G, Sileri P, Biancone L, MacDonald TT, Pallone F, Monteleone G. Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology. 2011;141(237–48):48 e1. doi: 10.1053/j.gastro.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 115.Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G, Bridonneau C, Jegou S, Hoffmann TW, Natividad,J M., et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016; 22:598–605. 10.1038/nm.4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lamas B, Hernandez-Galan L, Galipeau HJ, Constante M, Clarizio A, Jury J, et al. Aryl hydrocarbon receptor ligand production by the gut microbiota is decreased in celiac disease leading to intestinal inflammation. Sci Transl Med. 2020;12(566):eaba0624. doi: 10.1126/scitranslmed.aba0624. [DOI] [PubMed] [Google Scholar]
- 117.Martens EC, Neumann M, Desai MS. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. 2018;16:457–470. doi: 10.1038/s41579-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 118.Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Büller HA, Dekker J, Van Seuningen I, Renes IB, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131:117–129. doi: 10.1053/j.gastro.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 119.Tytgat KM, Opdam FJ, Einerhand AW, Buller HA, Dekker J. MUC2 is the prominent colonic mucin expressed in ulcerative colitis. Gut. 1996;38:554–563. doi: 10.1136/gut.38.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Buisine MP, Desreumaux P, Leteurtre E, Copin MC, Colombel JF, Porchet N, et al. Mucin gene expression in intestinal epithelial cells in Crohn’s disease. Gut. 2001;49:544–551. doi: 10.1136/gut.49.4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Johansson ME, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016;16:639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Variyam EP, Hoskins LC. Mucin degradation in human colon ecosystems. Degradation of Hog Gastric Mucin by Fecal Extracts and Fecal Cultures. Gastroenterology. 1981;81:751–758. [PubMed] [Google Scholar]
- 123.Macfarlane GT, Hay S, Gibson GR. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989;66:407–417. doi: 10.1111/j.1365-2672.1989.tb05110.x. [DOI] [PubMed] [Google Scholar]
- 124.Pruteanu M, Hyland NP, Clarke DJ, Kiely B, Shanahan F. Degradation of the extracellular matrix components by bacterial-derived metalloproteases: implications for inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:1189–1200. doi: 10.1002/ibd.21475. [DOI] [PubMed] [Google Scholar]
- 125.Nakjang S, Ndeh DA, Wipat A, Bolam DN, Hirt RP, Permyakov EA. A novel extracellular metallopeptidase domain shared by animal host-associated mutualistic and pathogenic microbes. PLoS One. 2012;7:e30287. doi: 10.1371/journal.pone.0030287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Noach I, Ficko-Blean E, Pluvinage B, Stuart C, Jenkins ML, Brochu D, Buenbrazo N, Wakarchuk W, Burke JE, Gilbert M, et al. Recognition of protein-linked glycans as a determinant of peptidase activity. Proc Natl Acad Sci U S A. 2017;114(5):E679–E88. doi: 10.1073/pnas.1615141114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–53 e21. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14:9–21. doi: 10.1038/nrgastro.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kriaa A, Jablaoui A, Rhimi S, Soussou S, Mkaouar H, Mariaule V, et al. SP-1, a serine protease from the gut microbiota, influences colitis and drives intestinal dysbiosis in mice. In: Cells. 2021;10:2658. doi: 10.3390/cells10102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scudamore CL, Jepson MA, Hirst BH, Miller HR, Miller P. The rat mucosal mast cell chymase, RMCP-II, alters epithelial cell monolayer permeability in association with altered distribution of the tight junction proteins ZO-1 and occludin. Eur J Cell Biol. 1998;75:321–330. doi: 10.1016/S0171-9335(98)80065-4. [DOI] [PubMed] [Google Scholar]
- 131.Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F, et al. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. 2011;141(3):959–971. doi: 10.1053/j.gastro.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 132.Golovkine G, Faudry E, Bouillot S, Voulhoux R, Attree I, Huber P, Kazmierczak BI. VE-cadherin cleavage by LasB protease from Pseudomonas aeruginosa facilitates type III secretion system toxicity in endothelial cells. PLoS Pathog. 2014;10(3):e1003939. doi: 10.1371/journal.ppat.1003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Guttman JA, Finlay BB. Tight junctions as targets of infectious agents. Biochim Biophys Acta. 2009;1788:832–841. doi: 10.1016/j.bbamem.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 134.Cenac N, Chin AC, Garcia-Villar R, Salvador-Cartier C, Ferrier L, Vergnolle N, Buret AG, Fioramonti J, Bueno L. PART 2 activation alters colonic paracellular permeability in mice via IFN-γ-dependent and -independent pathways. J Physiol. 2004;558:913–925. doi: 10.1113/jphysiol.2004.061721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cottrell GS, Amadesi S, Pikios S, Camerer E, Willardsen JA, Murphy BR, Caughey GH, Wolters PJ, Coughlin SR, Peterson A, et al. Protease-activated receptor 2, dipeptidyl peptidase I, and proteases mediate Clostridium difficile toxin A enteritis. Gastroenterology. 2007;132(7):2422–2437. doi: 10.1053/j.gastro.2007.03.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Peach CJ, Edgington-Mitchell LE, Bunnett NW, Schmidt BL. Protease-Activated receptors in health and disease. Physiol Rev. 2022. doi: 10.1152/physrev.00044.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Adams MN, Ramachandran R, Yau MK, Suen JY, Fairlie DP, Hollenberg MD, Hooper JD. Structure, function and pathophysiology of protease activated receptors. Pharmacol Ther. 2011;130:248–282. doi: 10.1016/j.pharmthera.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 138.Darmoul D, Gratio V, Devaud H, Peiretti F, Laburthe M. Activation of proteinase-activated receptor 1 promotes human colon cancer cell proliferation through epidermal growth factor receptor transactivation. Mol Cancer Res. 2004;2:514–522. doi: 10.1158/1541-7786.514.2.9. [DOI] [PubMed] [Google Scholar]
- 139.Cenac N, Garcia-Villar R, Ferrier L, Larauche M, Vergnolle N, Bunnett NW, Coelho A-M, Fioramonti J, Bueno L. Proteinase-activated receptor-2-induced colonic inflammation in mice: possible involvement of afferent neurons, nitric oxide, and paracellular permeability. J Immunol. 2003;170(8):4296–4300. doi: 10.4049/jimmunol.170.8.4296. [DOI] [PubMed] [Google Scholar]
- 140.Chin AC, Vergnolle N, MacNaughton WK, Wallace JL, Hollenberg MD, Buret AG. Proteinase-activated receptor 1 activation induces epithelial apoptosis and increases intestinal permeability. Proc Natl Acad Sci U S A. 2003;100:11104–11109. doi: 10.1073/pnas.1831452100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jimenez-Vargas NN, Pattison LA, Zhao P, Lieu T, Latorre R, Jensen DD, Castro J, Aurelio L, Le GT, Flynn B, et al. Protease-activated receptor-2 in endosomes signals persistent pain of irritable bowel syndrome. Proc Natl Acad Sci U S A. 2018;115:E7438–E47. doi: 10.1073/pnas.1721891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim JA, Choi SC, Yun KJ, Kim DK, Han MK, Seo GS, Yeom -J-J, Kim T-H, Nah Y-H, Lee Y-M, et al. Expression of protease-activated receptor 2 in ulcerative colitis. Inflamm Bowel Dis. 2003;9:224–229. doi: 10.1097/00054725-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 143.Dulon S, Leduc D, Cottrell GS, D’Alayer J, Hansen KK, Bunnett NW, Hollenberg MD, Pidard D, Chignard M. Pseudomonas aeruginosa elastase disables proteinase-activated receptor 2 in respiratory epithelial cells. Am J Respir Cell Mol Biol. 2005;32(5):411–419. doi: 10.1165/rcmb.2004-0274OC. [DOI] [PubMed] [Google Scholar]
- 144.Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J, Djukic Z, Orlando R, Pawlinski R, Ellermann M, et al. Enterococcus faecalis Gelatinase Mediates Intestinal Permeability via Protease-Activated Receptor 2. Infect Immun. 2015;83(7):2762–2770. doi: 10.1128/IAI.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lourbakos A, Chinni C, Thompson P, Potempa J, Travis J, Mackie EJ, Pike RN. Cleavage and activation of proteinase-activated receptor-2 on human neutrophils by gingipain-R from Porphyromonas gingivalis. FEBS Lett. 1998;435:45–48. doi: 10.1016/S0014-5793(98)01036-9. [DOI] [PubMed] [Google Scholar]
- 146.Parmely M, Gale A, Clabaugh M, Horvat R, Zhou WW. Proteolytic inactivation of cytokines by Pseudomonas aeruginosa. Infect Immun. 1990;58:3009–3014. doi: 10.1128/iai.58.9.3009-3014.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Theander TG, Kharazmi A, Pedersen BK, Christensen LD, Tvede N, Poulsen LK, Odum N, Svenson M, Bendtzen K. Inhibition of human lymphocyte proliferation and cleavage of interleukin-2 by Pseudomonas aeruginosa proteases. Infect Immun. 1988;56:1673–1677. doi: 10.1128/iai.56.7.1673-1677.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mintz CS, Miller RD, Gutgsell NS, Malek T. Legionella pneumophila protease inactivates interleukin-2 and cleaves CD4 on human T cells. Infect Immun. 1993;61:3416–3421. doi: 10.1128/iai.61.8.3416-3421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fletcher J, Nair S, Poole S, Henderson B, Wilson M. Cytokine degradation by biofilms of Porphyromonas gingivalis. Curr Microbiol. 1998;36:216–219. doi: 10.1007/s002849900297. [DOI] [PubMed] [Google Scholar]
- 150.Janoff EN, Rubins JB, Fasching C, Charboneau D, Rahkola JT, Plaut AG, Weiser JN. Pneumococcal IgA1 protease subverts specific protection by human IgA1. Mucosal Immunol. 2014;7(2):249–256. doi: 10.1038/mi.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Woof JM, Russell MW. Structure and function relationships in IgA. Mucosal Immunol. 2011;4:590–597. doi: 10.1038/mi.2011.39. [DOI] [PubMed] [Google Scholar]
- 152.Moon C, Baldridge MT, Wallace MA, Burnham DCA, Virgin HW, Stappenbeck TS. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature. 2015;521:90–93. doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kayssi A, Amadesi S, Bautista F, Bunnett NW, Vanner S. Mechanisms of protease-activated receptor 2-evoked hyperexcitability of nociceptive neurons innervating the mouse colon. J Physiol. 2007;580:977–991. doi: 10.1113/jphysiol.2006.126599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Reed DE, Barajas-Lopez C, Cottrell G, Velazquez-Rocha S, Dery O, Grady EF, Bunnett NW, Vanner SJ. Mast cell tryptase and proteinase-activated receptor 2 induce hyperexcitability of Guinea-pig submucosal neurons. J Physiol. 2003;547:531–542. doi: 10.1113/jphysiol.2002.032011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valdez-Morales EE, Overington J, Guerrero-Alba R, Ochoa-Cortes F, Ibeakanma CO, Spreadbury I, Bunnett NW, Beyak M, Vanner SJ. Sensitization of peripheral sensory nerves by mediators from colonic biopsies of diarrhea-predominant irritable bowel syndrome patients: a role for PAR2. Am J Gastroenterol. 2013;108(10):1634–1643. doi: 10.1038/ajg.2013.241. [DOI] [PubMed] [Google Scholar]
- 156.Cattaruzza F, Lyo V, Jones E, Pham D, Hawkins J, Kirkwood K, Valdez–Morales E, Ibeakanma C, Vanner SJ, Bogyo M, et al. Cathepsin S is activated during colitis and causes visceral hyperalgesia by a PAR2-dependent mechanism in mice. Gastroenterology. 2011;141(1864–745):e1–3. doi: 10.1053/j.gastro.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Karanjia R, Spreadbury I, Bautista-Cruz F, Tsang ME, Vanner S. Activation of protease-activated receptor-4 inhibits the intrinsic excitability of colonic dorsal root ganglia neurons. Neurogastroenterol Motil. 2009;21:1218–1221. doi: 10.1111/j.1365-2982.2009.01353.x. [DOI] [PubMed] [Google Scholar]
- 158.Auge C, Balz-Hara D, Steinhoff M, Vergnolle N, Cenac N. Protease-activated receptor-4 (PAR 4): a role as inhibitor of visceral pain and hypersensitivity. Neurogastroenterol Motil. 2009;21:1189-e107. doi: 10.1111/j.1365-2982.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 159.Annahazi A, Gecse K, Dabek M, Ait-Belgnaoui A, Rosztoczy A, Roka R, Molnár T, Theodorou V, Wittmann T, Bueno L, et al. Fecal proteases from diarrheic-IBS and ulcerative colitis patients exert opposite effect on visceral sensitivity in mice. Pain. 2009;144(1):209–217. doi: 10.1016/j.pain.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 160.Annahazi A, Ferrier L, Bezirard V, Leveque M, Eutamene H, Ait-Belgnaoui A, Coëffier M, Ducrotté P, Róka R, Inczefi O, et al. Luminal cysteine-proteases degrade colonic tight junction structure and are responsible for abdominal pain in constipation-predominant IBS. Am J Gastroenterol. 2013;108(8):1322–1331. doi: 10.1038/ajg.2013.152. [DOI] [PubMed] [Google Scholar]
- 161.Miquel S, Martin R, Lashermes A, Gillet M, Meleine M, Gelot A, Eschalier A, Ardid D, Bermúdez-Humarán LG, Sokol H, et al. Anti-nociceptive effect of Faecalibacterium prausnitzii in non-inflammatory IBS-like models. Sci Rep. 2016;6(1):19399. doi: 10.1038/srep19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Azghani AO, Gray LD, Johnson AR. A bacterial protease perturbs the paracellular barrier function of transporting epithelial monolayers in culture. Infect Immun. 1993;61:2681–2686. doi: 10.1128/iai.61.6.2681-2686.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pinto-Lopes P, Melo F, Afonso J, Pinto-Lopes R, Rocha C, Melo D, Macedo G, Dias CC, Carneiro F, Magro F, et al. Fecal dipeptidyl peptidase-4: an emergent biomarker in inflammatory bowel disease. Clin Transl Gastroenterol. 2021;12(3):e00320. doi: 10.14309/ctg.0000000000000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Devani M, Cugno M, Vecchi M, Ferrero S, Di Berardino F, Avesani EC, Franchis R, Colman RW. Kallikrein-kinin system activation in Crohn’s disease: differences in intestinal and systemic markers. Am J Gastroenterol. 2002;97:2026–2032. doi: 10.1111/j.1572-0241.2002.05919.x. [DOI] [PubMed] [Google Scholar]


