Abstract
The presence of Streptococcus mutans expressing Cnm protein encoded by cnm (cnm-positive S. mutans) in the oral cavity is associated with immunoglobulin A (IgA) nephropathy (IgAN). However, the precise mechanism by which cnm-positive S. mutans is involved in the pathogenesis of IgAN remains unclear. The present study evaluated glomerular galactose-deficient IgA1 (Gd-IgA1) to clarify the association between the presence of cnm-positive S. mutans and glomerular Gd-IgA1 in patients with IgAN. The presence of S. mutans and cnm-positive S. mutans was evaluated by polymerase chain reaction in saliva specimens from 74 patients with IgAN or IgA vasculitis. Immunofluorescent staining of IgA and Gd-IgA1 using KM55 antibody in clinical glomerular tissues was then performed. There was no significant association between the glomerular staining intensity of IgA and the positive rate of S. mutans. However, there was a significant association between the glomerular staining intensity of IgA and the positive rate of cnm-positive S. mutans (P < 0.05). There was also a significant association between the glomerular staining intensity of Gd-IgA1 (KM55) and the positive rate of cnm-positive S. mutans (P < 0.05). The glomerular staining intensity of Gd-IgA1 (KM55) was not associated with the positive rate of S. mutans. These results suggest that cnm-positive S. mutans in the oral cavity is associated with the pathogenesis of Gd-IgA1 in patients with IgAN.
Introduction
Immunoglobulin A (IgA) nephropathy (IgAN) is the most prevalent type of primary glomerulonephritis worldwide [1, 2]. Approximately 30% of renal biopsy cases involve IgAN [3] and 30% to 40% of patients with IgAN progress to end-stage kidney disease within 20 years [1, 2]. In renal biopsy specimens, IgA1, but not IgA2, is predominantly deposited in the mesangial and peripheral capillary regions [4].
Although the precise mechanism is still unknown [5], several studies have suggested that galactose-deficient IgA1 (Gd-IgA1) is a key effector molecule in the pathogenesis of IgAN [6–9]. A novel lectin-independent enzyme-linked immunosorbent assay (ELISA) using an anti-Gd-IgA1 monoclonal antibody (KM55) was recently developed [10]. Glomerular Gd-IgA1 deposition has been shown by immunofluorescence with KM55 antibody, which provides new insights into the possibility that Gd-IgA1 functions as a key effector molecule of IgAN [10, 11]. Gd-IgA1 has been specifically detected in IgAN and IgA vasculitis, but not in other renal diseases [11].
Patients with IgAN sometimes present with macroscopic hematuria if they develop an upper respiratory tract infection, such as tonsillitis [12]. Several bacterial species have been reported to be potential contributors to the pathogenesis of IgAN [13–17], including periodontitis-related [18, 19] and dental caries-related [20–27] bacteria. Streptococcus mutans, which is a Gram-positive oral streptococcal species, is a major pathogen causative of dental caries [28]. S. mutans with the cnm gene (cnm-positive S. mutans), which encodes Cnm (collagen-binding cell surface protein) [29], can bind to the extracellular matrix [30]. Therefore, Cnm protein is considered as a virulence factor, such as in infective endocarditis [31], inflammatory bowel disease [32], cerebral hemorrhage [33–35], and non-alcoholic steatohepatitis [36, 37]. Recent clinical studies have suggested that cnm-positive S. mutans is associated with IgAN [20–22, 25, 26]. The results of recent studies in animal models also support this conclusion [23, 24]. Overall, the findings to date suggest that cnm-positive S. mutans is an important pathogen in IgAN.
Whether cnm-positive S. mutans is related to the production of Gd-IgA1 when IgAN is induced is unknown. If cnm-positive S. mutans proves to be associated with Gd-IgA1, this is a major discovery and could be a breakthrough in determining the pathogenic mechanism of IgAN. Although an association between bacterial infection and IgAN is assumed [13–17], there is no evidence that Gd-IgA1 production is involved in the mechanism of IgAN induced by infection.
In the present study, we analyzed Gd-IgA1 using KM55 antibody to clarify the association between the presence of cnm-positive S. mutans in the oral cavity and glomerular Gd-IgA1.
Materials and methods
Patients and clinical characteristics
Seventy-eight patients who underwent a renal biopsy at Seirei Hamamatsu General Hospital, Hamamatsu, Japan in 2017–2021 were initially included. All patients were over 18 years of age. These patients were diagnosed with IgAN (n = 68) or IgA vasculitis (Henoch–Schonlein purpura nephritis) (n = 10) by renal biopsies. Their diagnoses were made on the basis of light microscopy and immunohistochemical findings. Four of the 78 patients were excluded because there were no glomeruli in prepared sections for Gd-IgA1 (KM55) staining. Therefore, analysis was performed on 74 patients. Clinical data (age, sex, height, body weight, body mass index, blood pressure, serum creatinine, estimated glomerular filtration rate [eGFR], serum C3, serum IgA, urinary protein [g/g creatinine], and urinary sediment of red blood cells [RBCs] [>100/high-power field]) were obtained at the time of renal biopsy after patients had provided informed consent to participate in this study.
Analysis of cnm-positive S. mutans
Saliva specimens obtained from the patients and frozen at –20°C were used to test for the presence of S. mutans DNA and whether it was cnm-positive or cnm-negative by polymerase chain reaction, as described previously [21]. Campylobacter rectus and Porphyromonas gingivalis DNA was also evaluated, as described elsewhere [38].
Histological studies
Renal biopsy specimens were collected via percutaneous needle biopsy. Paraffin-embedded 3-μm thick sections of renal specimens were stained with periodic acid Schiff, silver methenamine, and Masson trichrome. For the immunofluorescence analysis, frozen sections were subjected to fluorescence by fluorescein-conjugated goat IgG fraction to human IgG (F110FC, American Qualex, San Clemente, CA, USA), fluorescein-conjugated goat IgG fraction to human IgA (55077, MP Biomedicals, Solon, OH, USA), fluorescein-conjugated goat IgG fraction to human IgM (55153, MP Biomedicals), fluorescein-conjugated goat IgG fraction to human C3 (55167, MP Biomedicals), fluorescein-conjugated rabbit anti-human C1q (F0254, DAKO Japan Inc., Kyoto, Japan), and fluorescein-conjugated goat IgG fraction to human fibrinogen (55169, MP Biomedicals). IgAN or IgA vasculitis was diagnosed on the basis of mesangial cell proliferation in light microscopic findings, mesangial IgA deposition in immunofluorescence findings, and mesangial electron dense deposits in electron microscopic findings. Histological findings were evaluated according to the Oxford classification [39–41]. The Oxford classification of IgAN includes the following five histological variables: mesangial hypercellularity (M0/M1 lesion), segmental glomerulosclerosis (S0/S1 lesion), endocapillary hypercellularity (E0/E1 lesion), tubular atrophy/interstitial fibrosis (T0/T1/T2 lesion), and crescents (C0/C1/C2 lesion) [41]. We compared the renal histology in the cnm-positive S. mutans group with that in the cmn-negative S. mutans group using the Oxford classification in a blind test.
Immunofluorescent staining of Gd-IgA1
Immunofluorescent staining of Gd-IgA1 and IgA in glomerular tissues obtained by renal biopsy was evaluated as described previously [11]. Paraffin-embedded 3 μm thick sections of the renal specimens were used for staining. Anti-human Gd-IgA1 antibody (KM55, Immuno-Biological Laboratories Co., Ltd, Gunma, Japan; 100 μg/ml) was used to evaluate Gd-IgA1, Alexa Fluor 555-conjugated goat anti-rat IgG antibody (1:1000; Life Technologies, Carlsbad, CA, USA) for the secondary antibody, and fluorescein-conjugated polyclonal rabbit anti-human IgA antibody (DAKO Japan; 100 μg/ml) for IgA [11]. The intensity of glomerular Gd-IgA1 and IgA was scored semiquantitatively (0–3 intensity) in a blind test [11].
Measurement of Gd-IgA1
Plasma was collected at the time of the renal biopsy. Plasma Gd-IgA1 concentrations were evaluated using the KM55 ELISA kit (Immuno-Biological Laboratories Co., Ltd). Briefly, the ELISA plates were incubated with plasma specimens (1:200 dilution in enzyme immunoassay buffer) and standard specimens for 1 hour at room temperature, washed 4 times with wash buffer, incubated with prepared-labeled antibodies, and then treated with 50 ml of tetramethylbenzidine solution for 30 minutes in the dark. Absorbance was evaluated at 450/650 nm by the Versamax Microplate Reader (Molecular Devices, Tokyo, Japan). The Gd-IgA1 concentrations was evaluated according to the standard curve.
Statistical analysis
All the results are expressed as the mean ± standard deviation (SD). When there was a significant difference, a further statistical analysis was conducted using Fisher’s PLSD test or Fisher’s exact test between 2 groups. The Cochran–Armitage trend test was used for trend analysis. A simple regression analysis was used for correlation analysis. In these analyses, P < 0.05 was considered statistically significant. The statistical analyses were conducted using Statview software (SAS Institute Inc., Cary, NC, USA) and GraphPad Prism 8 software (San Diego, CA, USA).
Results
Glomerular staining intensity of IgA and Gd-IgA1
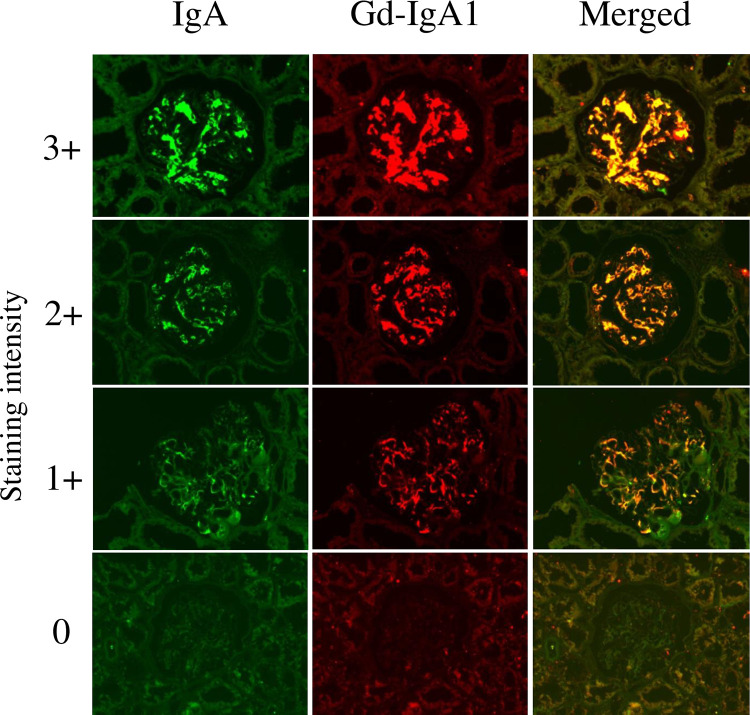
The glomerular staining intensity of IgA and Gd-IgA1 was defined by the intensity of 0 to +3 as shown in Fig 1. IgA and Gd-IgA1 were mainly positive in the mesangial region, and IgA and Gd-IgA1 had the same distribution. There was a significant association between the glomerular staining intensity of IgA and Gd-IgA1 using regression analysis (R = 0.787, P < 0.001).
Fig 1. Glomerular staining intensity of IgA and galactose-deficient IgA1.
Double staining with anti-IgA polyclonal antibody and Gd-IgA1 (KM55) monoclonal antibody was performed on biopsy specimens. Representative images of glomerular IgA and Gd-IgA1 (KM55) in patients with 0, 1+, 2+, and 3+ staining intensity. Seventy-four patients were divided into four groups according to the staining intensity of IgA: group 0 (n = 4), group 1+ (n = 46), group 2+ (n = 12), and group 3+ (n = 12). These patients were also divided into four groups according to the staining intensity of Gd-IgA1: group 0 (n = 4), group 1+ (n = 35), group 2+ (n = 23), and group 3+ (n = 12). Fluorescein-conjugated polyclonal rabbit anti-human IgA antibody (DAKO Japan Inc., Kyoto, Japan), anti-human Gd-IgA1 antibody (KM55) (Immuno-Biological Laboratories Co., Ltd, Gunma, Japan), and Alexa Fluor 555-conjugated goat anti-rat IgG antibody (Life Technologies) was used for immunofluorescence staining. Original magnification: ×200. Gd-IgA1, galactose-deficient IgA1.
Plasma Gd-IgA1 concentrations were measured in 33 of 74 patients. There was no significant association between the glomerular staining intensity of Gd-IgA1 and plasma Gd-IgA1 concentrations (n = 33) (S1 Fig).
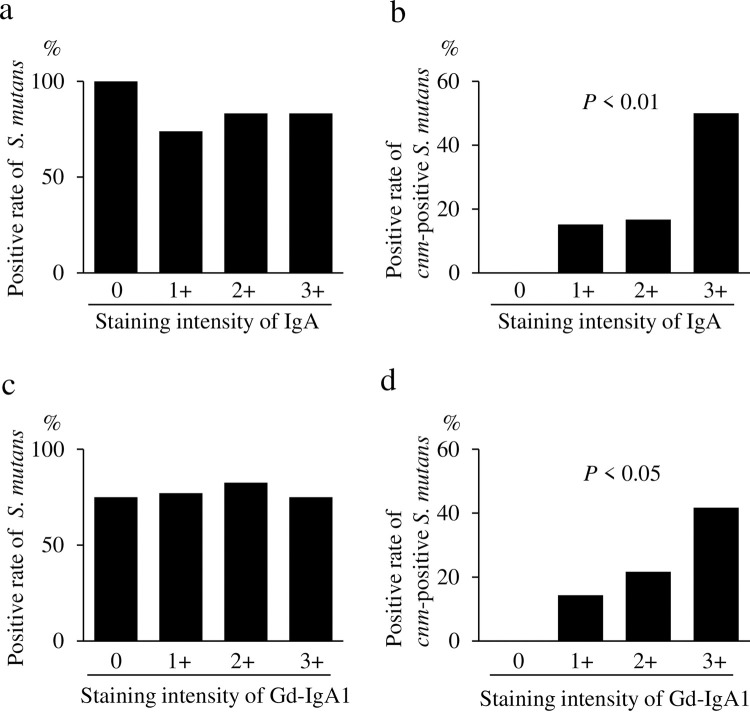
Association between glomerular staining intensity of IgA and Gd-IgA1, and the rate of cnm-positive S. mutans in the oral cavity
There was no significant association between the glomerular staining intensity of IgA and the positive rate of S. mutans (Fig 2A). However, there was a significant association between the glomerular staining intensity of IgA and the positive rate of cnm-positive S. mutans (Fig 2B). There was also no significant association between the glomerular staining intensity of Gd-IgA1 and positive rate of S. mutans (Fig 2C). However, there was a significant association between the glomerular staining intensity of Gd-IgA1 and the positive rate of cnm-positive S. mutans (Fig 2D).
Fig 2. Association between glomerular staining intensity of IgA and galactose-deficient IgA1 (Gd-IgA1) and the rate of cnm-positive S. mutans in the oral cavity.
Comparison of the glomerular staining intensity of IgA and the positive rate of S. mutans in the oral cavity (a). Comparison of the glomerular staining intensity of IgA and the positive rate of cnm-positive S. mutans in the oral cavity (b). Comparison of the glomerular staining intensity of Gd-IgA1 (KM55) and the positive rate of S. mutans in the oral cavity (c). Comparison of the glomerular staining intensity of Gd-IgA1 (KM55) and the positive rate of cnm-positive S. mutans in the oral cavity (d). Data were analyzed using the Cochran–Armitage trend test. P < 0.05 was considered statistically significant.
There was no significant association between the glomerular staining intensity of IgA and the positive rate of C. rectus (S2A Fig) or between the glomerular staining intensity of Gd-IgA1 and positive rate of C. rectus (S2B Fig). Furthermore, there was no significant association between the glomerular staining intensity of IgA and the positive rate of P. gingivalis (S2C Fig) or between the glomerular staining intensity of Gd-IgA1 and the positive rate of P. gingivalis (S2D Fig).
Comparison of the cnm-positive S. mutans and cnm-negative S. mutans groups
The percentage of the staining intensity of IgA: 3+ was significantly higher in the cnm-positive S. mutans group than in the cnm-negative S. mutans group (Table 1). The percentage of the staining intensity of Gd-IgA1: 3+ was also significantly higher in the cnm-positive S. mutans group than in the cnm-negative S. mutans group (Table 1). No significant differences were found in age, sex, height, body weight, body mass index, systolic blood pressure, diastolic blood pressure, serum creatinine concentrations, eGFR, serum C3 concentrations, serum IgA concentrations, or urinary protein concentrations between the groups (Table 1). The percentage of urinary sediment ≥100 RBCs/high-power field was higher in the cnm-positive S. mutans group than in the cnm-negative S. mutans group, but this was not significant (Table 1). No significant differences were found in the mesangial hypercellularity score, endocapillary hypercellularity score, segmental glomerulosclerosis score, tubular atrophy/interstitial fibrosis score, or the crescent score of the Oxford classification between the groups (Table 2).
Table 1. Comparison between the cnm-positive and cnm-negative S. mutans groups of patients.
| Characteristics | cnm-negative S. mutans (n = 59) | cnm-positive S. mutans (n = 15) | P value |
|---|---|---|---|
| Percentage of staining intensity of IgA: 3+ (%) | 10.2 | 40 | 0.0051 |
| Percentage of staining intensity of Gd-IgA1: 3+ (%) | 11.9 | 33.3 | 0.0440 |
| Age (years) | 44.9 ± 15.3 | 45.1 ± 13.3 | 0.9662 |
| Sex (M/F) | 27/32 | 9/6 | 0.3246 |
| Height (cm) | 162.6 ± 8.6 | 164.3 ± 7.5 | 0.4834 |
| Body weight (kg) | 62.5 ± 13.4 | 64.0 ± 11.3 | 0.6966 |
| BMI (kg/m2) | 23.6 ± 4.5 | 23.7 ± 3.7 | 0.9293 |
| Systolic blood pressure (mmHg) | 125.0 ± 21.3 | 121.5 ± 17.6 | 0.5582 |
| Diastolic blood pressure (mmHg) | 69.2 ± 11.6 | 68.4 ± 8.0 | 0.8046 |
| Serum creatinine (mg/dl) | 1.0 ± 0.7 | 1.0 ± 0.5 | 0.932 |
| eGFR (ml/minute/1.73 m2) | 67.3 ± 26.4 | 67.4 ± 22.6 | 0.9926 |
| Serum C3 (mg/dl) | 108.2 ± 16.1 | 107.0 ± 18.7 | 0.8029 |
| Serum IgA (mg/dl) | 348.3 ± 158.3 | 358.5 ± 164.6 | 0.8255 |
| Urinary protein (g/gCr) | 1.4 ± 0.8 | 0.8 ± 0.7 | 0.1857 |
| Percentage of urinary sediment ≥ 100 RBCs/HPF | 8.5 | 26.7 | 0.0543 |
Values are %, number, or mean ± standard deviation. Bold values indicate statistical significance at P < 0.05. BMI: body mass index, eGFR: estimated glomerular filtration rate.
Table 2. Analysis of renal biopsy specimens using the Oxford classification.
| Characteristic | cnm-negative S. mutans | cnm-positive S. mutans | P value |
|---|---|---|---|
| (n = 59) | (n = 15) | ||
| Positive mesangial hypercellularity score (%) | 72.9 | 86.7 | 0.2665 |
| Positive endocapillary hypercellularity score (%) | 50.8 | 46.7 | 0.7725 |
| Positive segmental glomerulosclerosis score (%) | 66.1 | 73.3 | 0.5932 |
| Tubular atrophy/interstitial fibrosis score ≥1+ (%) | 43.9 | 46.7 | 0.8565 |
| Crescent score ≥1+ (%) | 40.7 | 33.3 | 0.6029 |
Discussion
As far as we know, this is the first study to demonstrate an association between cnm-positive S. mutans in the oral cavity and glomerular Gd-IgA1. We discovered that there was a significant association between the positive rate of cnm-positive S. mutans in the oral cavity and glomerular staining intensity of IgA or Gd-IgA1. Although various factors are considered to be associated with Gd-IgA1, these data indicate that cnm-positive S. mutans in the oral cavity may be one of the factors associated with the development of IgA nephropathy via Gd-IgA1.
Recent clinical studies have demonstrated that cnm-positive S. mutans in the oral cavity is associated with IgAN [20–22, 26]. One study showed that the positive rate of cnm-positive S. mutans in the oral cavity was significantly higher in the IgAN group than in the healthy control group (32.1% vs. 14.0%) [20]. However, the positive rate of S. mutans was similar between the IgAN and healthy control groups [20]. Another study suggested that cnm-positive S. mutans in the oral cavity and the dental caries status were associated with urinary protein in patients with IgAN [21]. The Cnm protein in the tonsils may be associated with the severity of IgAN [22]. The report demonstrated the following: (1) the positive Cnm protein area/total tonsillar area rate was significantly higher in patients with IgAN than in the control (chronic tonsillitis) group; (2) Cnm protein existed in the tonsils, not in the glomerulus, in patients with IgAN; and (3) Cnm protein in the tonsils was associated with urinary protein in patients with IgAN [22]. These results suggest that IgAN is aggravated by immune reactions in the tonsils via an unknown mechanism induced by Cnm protein [22]. In rodent models, intravenous administration of cnm-positive S. mutans induced transient IgAN-like lesions [23] and severe dental caries induced by cnm-positive S. mutans caused IgAN-like glomerulonephritis [24]. As mentioned above, we found an association between cnm-positive S. mutans and IgAN. However, we could not identify how cnm-positive S. mutans induced IgAN.
Gd-IgA1 has been demonstrated as a key effector molecule in the pathogenesis of IgAN [6–9]. We showed an association between the presence of cnm-positive S. mutans and glomerular Gd-IgA1 in this study. This study indicates that cnm-positive S. mutans induced Gd-IgA1 production in the process of causing IgAN, suggesting that the most upstream factor for Gd-IgA1 production is oral bacterial infection, such as cnm-positive S. mutans. We hypothesize that repeated immune reactions involving IgA induced by infection such as cnm-positive S. mutans in the mucosal immune tissues of the oral cavity (e.g., the tonsils) may cause Gd-IgA1 [21]. We believe that this result is important in considering the pathogenic mechanism of IgAN. We do not have enough evidence to explain how cnm-positive S. mutans affects the induction of Gd-IgA1 at this stage and need to elucidate these mechanisms in a further study.
We also investigated the positive rate of C. rectus and P. gingivalis in the oral cavity because these bacteria are reported to be potentially associated with IgA nephropathy [19, 26, 27]. We could not confirm the association between these bacteria and Gd-IgA1 in this research.
Our study showed that the percentage of urinary sediment ≥100 RBCs/high-power field was higher in the cnm-positive S. mutans group than in the cnm-negative S. mutans group. Although this finding did not reach statistical significance, it suggests that cnm-positive S. mutans is associated with the hematuria that is the cardinal symptom of IgAN.
Unfortunately, because the exact mechanism of IgAN is not yet understood, there are no specific disease-targeted therapies for IgAN [5]. Our present findings are consistent with the new concept of an oral–kidney association and could establish a new treatment for IgAN [21]. Considering that S. mutans is a pathogen that causes dental caries, reducing it in the oral cavity to prevent dental caries may improve the prognosis of IgAN [27]. Further studies are needed to elucidate the effects of oral care as a treatment of IgAN.
There are some limitations in this study. First, although we demonstrated that cnm-positive S. mutans in the oral cavity was associated with Gd-IgA1 in patients with IgAN, how cnm-positive S. mutans contributes to Gd-IgA1 needs to be elucidated. Second, given that the only S. mutans protein we examined was Cnm, the possibility that other S. mutans proteins are associated with Gd-IgA1 cannot be excluded. Third, the sample size was small, and all patients were of the same ethnicity and from a single facility. Further studies in larger numbers of ethnically diverse patients from multiple facilities are needed to confirm our present findings.
In conclusion, cnm-positive S. mutans in the oral cavity is associated with Gd-IgA1 in glomeruli in patients with IgAN. This finding suggests that cnm-positive S. mutans is involved in the pathogenicity of IgAN through induction of Gd-IgA1.
Supporting information
There was no significant association between the glomerular staining intensity of Gd-IgA1 and the plasma Gd-IgA1 concentration. The data were examined for statistical significance using a simple regression analysis. Gd-IgA1: galactose-deficient IgA1.
(TIF)
Comparison of the glomerular staining intensity of IgA and the positive rate of Campylobacter rectus in the oral cavity (a). Comparison of the glomerular staining intensity of Gd-IgA1 (KM55) and the positive rate of C. rectus in the oral cavity (b). Comparison of the glomerular staining intensity of IgA and the positive rate of Porphyromonas gingivalis in the oral cavity (c). Comparison of the glomerular staining intensity of Gd-IgA1 (KM55) and the positive rate of P. gingivalis in the oral cavity (d). The data were examined for statistical significance using the Cochran–Armitage trend test. P < 0.05 was considered statistically significant.
(TIF)
Acknowledgments
Ethical statement
This study protocol fully adhered to the Declaration of Helsinki (64th WMA General Assembly, Fortaleza, Brazil, 2013). The protocol was approved by the Ethics Committee of Seirei Hamamatsu General Hospital (approval no. 3646), Osaka University Graduate School of Dentistry (approval no. H29-E9), and Okayama University Graduate School of Medicine (approval no. 1704–036). All patients were informed of the study protocol, and provided written informed consent prior to participating in the study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by Japan Society for the Promotion of Science (grant numbers 19K10098, Dr. Taro Misaki, 21H03149, Dr Kazuhiko Nakano and 21K08242, Dr Yasuyuki Nagasawa). The sponsors or funders did not play any role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.D’Amico G. The commonest glomerulonephritis in the world: IgA nephropathy. Q J Med. 1987;64(245):709–27. Epub 1987/09/01. . [PubMed] [Google Scholar]
- 2.Julian BA, Waldo FB, Rifai A, Mestecky J. IgA nephropathy, the most common glomerulonephritis worldwide. A neglected disease in the United States? Am J Med. 1988;84(1):129–32. Epub 1988/01/01. doi: 10.1016/0002-9343(88)90019-8 . [DOI] [PubMed] [Google Scholar]
- 3.Sugiyama H, Yokoyama H, Sato H, Saito T, Kohda Y, Nishi S, et al. Japan Renal Biopsy Registry and Japan Kidney Disease Registry: Committee Report for 2009 and 2010. Clin Exp Nephrol. 2013;17(2):155–73. Epub 2013/02/07. doi: 10.1007/s10157-012-0746-8 . [DOI] [PubMed] [Google Scholar]
- 4.Conley ME, Cooper MD, Michael AF. Selective deposition of immunoglobulin A1 in immunoglobulin A nephropathy, anaphylactoid purpura nephritis, and systemic lupus erythematosus. J Clin Invest. 1980;66(6):1432–6. Epub 1980/12/01. doi: 10.1172/JCI109998 ; PubMed Central PMCID: PMC371631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med. 2013;368(25):2402–14. Epub 2013/06/21. doi: 10.1056/NEJMra1206793 . [DOI] [PubMed] [Google Scholar]
- 6.Barratt J, Feehally J. IgA nephropathy. J Am Soc Nephrol. 2005;16(7):2088–97. Epub 2005/06/03. doi: 10.1681/ASN.2005020134 . [DOI] [PubMed] [Google Scholar]
- 7.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, et al. Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int. 2007;71(11):1148–54. Epub 2007/03/08. doi: 10.1038/sj.ki.5002185 . [DOI] [PubMed] [Google Scholar]
- 8.Suzuki H, Moldoveanu Z, Hall S, Brown R, Vu HL, Novak L, et al. IgA1-secreting cell lines from patients with IgA nephropathy produce aberrantly glycosylated IgA1. J Clin Invest. 2008;118(2):629–39. Epub 2008/01/04. doi: 10.1172/JCI33189 ; PubMed Central PMCID: PMC2157566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berthoux F, Suzuki H, Thibaudin L, Yanagawa H, Maillard N, Mariat C, et al. Autoantibodies targeting galactose-deficient IgA1 associate with progression of IgA nephropathy. J Am Soc Nephrol. 2012;23(9):1579–87. Epub 2012/08/21. doi: 10.1681/ASN.2012010053 ; PubMed Central PMCID: PMC3431415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yasutake J, Suzuki Y, Suzuki H, Hiura N, Yanagawa H, Makita Y, et al. Novel lectin-independent approach to detect galactose-deficient IgA1 in IgA nephropathy. Nephrol Dial Transplant. 2015;30(8):1315–21. Epub 2015/06/26. doi: 10.1093/ndt/gfv221 ; PubMed Central PMCID: PMC4513896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, Yasutake J, Makita Y, Tanbo Y, Yamasaki K, Sofue T, et al. IgA nephropathy and IgA vasculitis with nephritis have a shared feature involving galactose-deficient IgA1-oriented pathogenesis. Kidney Int. 2018;93(3):700–5. Epub 2018/01/14. doi: 10.1016/j.kint.2017.10.019 . [DOI] [PubMed] [Google Scholar]
- 12.Donadio JV, Grande JP. IgA nephropathy. N Engl J Med. 2002;347(10):738–48. Epub 2002/09/06. doi: 10.1056/NEJMra020109 . [DOI] [PubMed] [Google Scholar]
- 13.Suzuki S, Nakatomi Y, Sato H, Tsukada H, Arakawa M. Haemophilus parainfluenzae antigen and antibody in renal biopsy samples and serum of patients with IgA nephropathy. Lancet. 1994;343(8888):12–6. Epub 1994/01/01. doi: 10.1016/s0140-6736(94)90875-3 . [DOI] [PubMed] [Google Scholar]
- 14.Kusano K, Tokunaga O, Ando T, Inokuchi A. Helicobacter pylori in the palatine tonsils of patients with IgA nephropathy compared with those of patients with recurrent pharyngotonsillitis. Hum Pathol. 2007;38(12):1788–97. Epub 2007/08/24. doi: 10.1016/j.humpath.2007.04.012 . [DOI] [PubMed] [Google Scholar]
- 15.Iwama H, Horikoshi S, Shirato I, Tomino Y. Epstein-Barr virus detection in kidney biopsy specimens correlates with glomerular mesangial injury. Am J Kidney Dis. 1998;32(5):785–93. Epub 1998/11/20. doi: 10.1016/s0272-6386(98)70134-9 . [DOI] [PubMed] [Google Scholar]
- 16.Koyama A, Sharmin S, Sakurai H, Shimizu Y, Hirayama K, Usui J, et al. Staphylococcus aureus cell envelope antigen is a new candidate for the induction of IgA nephropathy. Kidney Int. 2004;66(1):121–32. Epub 2004/06/18. doi: 10.1111/j.1523-1755.2004.00714.x . [DOI] [PubMed] [Google Scholar]
- 17.Rollino C, Vischini G, Coppo R. IgA nephropathy and infections. J Nephrol. 2016. Epub 2016/01/24. doi: 10.1007/s40620-016-0265-x . [DOI] [PubMed] [Google Scholar]
- 18.Nagasawa Y, Iio K, Fukuda S, Date Y, Iwatani H, Yamamoto R, et al. Periodontal disease bacteria specific to tonsil in IgA nephropathy patients predicts the remission by the treatment. PLoS One. 2014;9(1):e81636. Epub 2014/02/04. doi: 10.1371/journal.pone.0081636 ; PubMed Central PMCID: PMC3904818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagasawa Y, Nomura R, Misaki T, Ito S, Naka S, Wato K, et al. Relationship between IgA Nephropathy and Porphyromonas gingivalis; Red Complex of Periodontopathic Bacterial Species. Int J Mol Sci. 2021;22(23). Epub 2021/12/11. doi: 10.3390/ijms222313022 ; PubMed Central PMCID: PMC8657970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misaki T, Naka S, Kuroda K, Nomura R, Shiooka T, Naito Y, et al. Distribution of Streptococcus mutans strains with collagen-binding proteins in the oral cavity of IgA nephropathy patients. Clin Exp Nephrol. 2015;19(5):844–50. Epub 2014/12/11. doi: 10.1007/s10157-014-1072-0 . [DOI] [PubMed] [Google Scholar]
- 21.Misaki T, Naka S, Hatakeyama R, Fukunaga A, Nomura R, Isozaki T, et al. Presence of Streptococcus mutans strains harbouring the cnm gene correlates with dental caries status and IgA nephropathy conditions. Sci Rep. 2016;6:36455. Epub 2016/11/05. doi: 10.1038/srep36455 ; PubMed Central PMCID: PMC5095553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ito S, Misaki T, Naka S, Wato K, Nagasawa Y, Nomura R, et al. Specific strains of Streptococcus mutans, a pathogen of dental caries, in the tonsils, are associated with IgA nephropathy. Sci Rep. 2019;9(1):20130. Epub 2019/12/29. doi: 10.1038/s41598-019-56679-2 ; PubMed Central PMCID: PMC6934739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naka S, Wato K, Misaki T, Ito S, Nagasawa Y, Nomura R, et al. Intravenous administration of Streptococcus mutans induces IgA nephropathy-like lesions. Clin Exp Nephrol. 2020;24(12):1122–31. Epub 2020/09/11. doi: 10.1007/s10157-020-01961-1 ; PubMed Central PMCID: PMC7599197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naka S, Wato K, Misaki T, Ito S, Matsuoka D, Nagasawa Y, et al. Streptococcus mutans induces IgA nephropathy-like glomerulonephritis in rats with severe dental caries. Sci Rep. 2021;11(1):5784. Epub 2021/03/13. doi: 10.1038/s41598-021-85196-4 ; PubMed Central PMCID: PMC7952735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagasawa Y, Misaki T, Ito S, Naka S, Wato K, Nomura R, et al. Title IgA Nephropathy and Oral Bacterial Species Related to Dental Caries and Periodontitis. Int J Mol Sci. 2022;23(2). Epub 2022/01/22. doi: 10.3390/ijms23020725 ; PubMed Central PMCID: PMC8775524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misaki T, Naka S, Wato K, Hatakeyama R, Nagasawa Y, Ito S, et al. Campylobacter rectus in the Oral Cavity Correlates with Proteinuria in Immunoglobulin A Nephropathy Patients. Nephron. 2018;139(2):143–9. Epub 2018/02/13. doi: 10.1159/000487103 . [DOI] [PubMed] [Google Scholar]
- 27.Misaki T, Naka S, Nagasawa Y, Matsuoka D, Ito S, Nomura R, et al. Simultaneous Presence of Campylobacter rectus and Cnm-Positive Streptococcus mutans in the Oral Cavity Is Associated with Renal Dysfunction in IgA Nephropathy Patients: 5-Year Follow-Up Analysis. Nephron. 2022:1–10. Epub 2022/08/24. doi: 10.1159/000525511 . [DOI] [PubMed] [Google Scholar]
- 28.Selwitz RH, Ismail AI, Pitts NB. Dental caries. Lancet. 2007;369(9555):51–9. Epub 2007/01/09. doi: 10.1016/S0140-6736(07)60031-2 . [DOI] [PubMed] [Google Scholar]
- 29.Naka S, Matsuoka D, Goto K, Misaki T, Nagasawa Y, Ito S, et al. Cnm of Streptococcus mutans is important for cell surface structure and membrane permeability. Front Cell Infect Microbiol. 2022;12:994014. Epub 2022/10/01. doi: 10.3389/fcimb.2022.994014 ; PubMed Central PMCID: PMC9513430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H. Streptococcus mutans strains harboring collagen-binding adhesin. J Dent Res. 2004;83(7):534–9. Epub 2004/06/26. doi: 10.1177/154405910408300705 . [DOI] [PubMed] [Google Scholar]
- 31.Nomura R, Naka S, Nemoto H, Otsugu M, Nakamura S, Ooshima T, et al. Potential high virulence for infective endocarditis in Streptococcus mutans strains with collagen-binding proteins but lacking PA expression. Arch Oral Biol. 2013;58(11):1627–34. Epub 2013/10/12. doi: 10.1016/j.archoralbio.2013.06.008 . [DOI] [PubMed] [Google Scholar]
- 32.Kojima A, Nakano K, Wada K, Takahashi H, Katayama K, Yoneda M, et al. Infection of specific strains of Streptococcus mutans, oral bacteria, confers a risk of ulcerative colitis. Sci Rep. 2012;2:332. Epub 2012/03/28. doi: 10.1038/srep00332 ; PubMed Central PMCID: PMC3312205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakano K, Hokamura K, Taniguchi N, Wada K, Kudo C, Nomura R, et al. The collagen-binding protein of Streptococcus mutans is involved in haemorrhagic stroke. Nat Commun. 2011;2:485. Epub 2011/09/29. doi: 10.1038/ncomms1491 ; PubMed Central PMCID: PMC3220351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tonomura S, Ihara M, Kawano T, Tanaka T, Okuno Y, Saito S, et al. Intracerebral hemorrhage and deep microbleeds associated with cnm-positive Streptococcus mutans; a hospital cohort study. Sci Rep. 2016;6:20074. Epub 2016/02/06. doi: 10.1038/srep20074 ; PubMed Central PMCID: PMC4742798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe I, Kuriyama N, Miyatani F, Nomura R, Naka S, Nakano K, et al. Oral Cnm-positive Streptococcus Mutans Expressing Collagen Binding Activity is a Risk Factor for Cerebral Microbleeds and Cognitive Impairment. Sci Rep. 2016;6:38561. Epub 2016/12/10. doi: 10.1038/srep38561 ; PubMed Central PMCID: PMC5146923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naka S, Nomura R, Takashima Y, Okawa R, Ooshima T, Nakano K. A specific Streptococcus mutans strain aggravates non-alcoholic fatty liver disease. Oral Dis. 2014;20(7):700–6. Epub 2014/11/02. doi: 10.1111/odi.12191 . [DOI] [PubMed] [Google Scholar]
- 37.Naka S, Hatakeyama R, Takashima Y, Matsumoto-Nakano M, Nomura R, Nakano K. Contributions of Streptococcus mutans Cnm and PA antigens to aggravation of non-alcoholic steatohepatitis in mice. Sci Rep. 2016;6:36886. Epub 2016/11/12. doi: 10.1038/srep36886 ; PubMed Central PMCID: PMC5105074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura R, Nagasawa Y, Misaki T, Ito S, Naka S, Okunaka M, et al. Distribution of periodontopathic bacterial species between saliva and tonsils. Odontology. 2022. Epub 2022/12/17. doi: 10.1007/s10266-022-00776-8 . [DOI] [PubMed] [Google Scholar]
- 39.Working Group of the International Ig ANN, the Renal Pathology S, Cattran DC, Coppo R, Cook HT, Feehally J, et al. The Oxford classification of IgA nephropathy: rationale, clinicopathological correlations, and classification. Kidney Int. 2009;76(5):534–45. Epub 2009/07/03. doi: 10.1038/ki.2009.243 . [DOI] [PubMed] [Google Scholar]
- 40.Working Group of the International Ig ANN, the Renal Pathology S, Roberts IS, Cook HT, Troyanov S, Alpers CE, et al. The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int. 2009;76(5):546–56. Epub 2009/07/03. doi: 10.1038/ki.2009.168 . [DOI] [PubMed] [Google Scholar]
- 41.Trimarchi H, Barratt J, Cattran DC, Cook HT, Coppo R, Haas M, et al. Oxford Classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group. Kidney Int. 2017;91(5):1014–21. Epub 2017/03/28. doi: 10.1016/j.kint.2017.02.003 . [DOI] [PubMed] [Google Scholar]