Abstract
Mammalian PERIOD2 protein (PER2) is the product of a clock gene that controls circadian rhythms, because PER2-deficient mice have an arrhythmic phenotype. The nuclear entry regulation of clock gene products is a key step in proper circadian rhythm formation in both Drosophila and mammals, because the periodic transcription of clock genes is controlled by an intracellular, oscillating, negative feedback loop. The present study used deletion mutants of rat PER2 (rPER2) to identify the functional nuclear localization signal (NLS) in rPER2. The elimination of putative NLS (residues 778 to 794) from the rPER2 fragment resulted in the loss of nuclear entry activity. Adding the NLS to the cytosolic protein (bacterial alkaline phosphatase) translocates the fusion protein to the nuclei. The data indicate the presence of a functional NLS in rPER2. Furthermore, intact rPER2 was preferentially translocated from the cytoplasm to the nucleus when coexpressed with human CRY1 (hCRY1). However, rPER2 mutants lacking a carboxyl-terminal domain could not enter the nucleus even in the presence of hCRY1. In addition, coexpression of the nuclear localization domain (residues 512 to 794) lacking rPER2 and CRY1 changed the subcellular localization of CRY1 from the nucleus to the cytoplasm. In vitro protein interaction studies demonstrated that the carboxyl-terminal domain of rPER2 is essential for binding to CRY1. The data suggested that both the rPER2 NLS and carboxyl-terminal CRY binding domain are essential for nuclear entry of the rPER2-CRY1 complex.
Circadian rhythms in organisms ranging from bacteria to humans are driven by endogenous biological clocks that regulate many biochemical, physiological, and behavioral processes with approximate 24-h periodicity (3). Three mammalian period genes (per1, per2, and per3) that resemble the clock-regulating gene of Drosophila melanogaster, period (per) (1, 19, 21–23, 29), have been cloned. Structural similarity indicated that the mammalian genes are components of a central oscillating mechanism. Transcript levels of all three per genes oscillate in the hypothalamic suprachiasmatic nucleus (SCN), where the principal mammalian circadian oscillator lies (8), as well as in peripheral tissues (2, 6, 15, 29). Among the three putative mammalian homologues, the per2 gene encodes a functional component of the mammalian clock, because mPER2brdm1 mutant mice lose circadian rhythms 2 weeks after release into constant darkness (28).
Circadian clocks are also apparently located in the peripheral tissues of mammals that are synchronized by the SCN (2, 6, 15, 22, 23). In both SCN and peripheral tissues, clock-related gene products including PER proteins precisely oscillate, and this oscillation is required to generate circadian rhythms (4, 10, 13, 17). Even in cultured cells, serum shock can induce the rhythmic expression of clock genes and clock-controlled genes (2), indicating that individual cells contain a set of molecules that drive their own oscillator.
Molecular dissection has revealed that circadian oscillation is driven by a transcription- and translation-based negative feedback loop, wherein positive elements induce the expression of negative regulators that in turn inhibit the transactivation of positive regulators (3). Thus, the timing of nuclear entry is critical to maintain the correct time of the biological clock in both Drosophila and mammals. In Drosophila, Drosophila PER (dPER) and dTIMELESS (dTIM) are located in the nuclei of pacemaker cells at night but not during the day. These two molecules accumulate in the cytoplasm until heterodimerization, which is a trigger for the nuclear entry of this complex that inhibits CLOCK/BMAL transactivation after dPER/dTIM expression decreases (14).
Mammalian PER proteins localize in the nucleus of SCN cells (5) and partially inhibit CLOCK-BMAL-dependent transactivation (7), which may proceed in the nucleus. Mammalian CRY proteins (CRY1 and CRY2), which were originally considered blue-light photoreceptors (cryptochromes), are essential components of the central clock mechanism, because mice lacking both murine CRY1 (mCRY1) and mCRY2 completely lose circadian rhythms (25). Kume et al. found that mCRY1 and mCRY2 form a complex with mouse PERs (mPERs) and appeared to facilitate the nuclear entry of mPERs (10). However, the nuclear entry mechanism and the binding domain of this complex have not been clarified. Another report suggested that nuclear entry can be accomplished by serum shock-induced heterodimerization of mPER1 and mPER2 with mPER3 (27). On the other hand, the phosphorylation of PER proteins by casein kinase Iɛ can modulate the nuclear localization of mPER1 and mPER3 but not of mPER2 (20, 26). These findings indicate that the pathway by which mPERs enter the nucleus is more complicated than that of Drosophila (14).
The domain structures of PER2 protein are predicted from the primary sequence. Based on the similarity of sequences to dPER, mammalian PER2 has PAS (PER-ARNT-SIM) domains near the amino terminus, which can interact with and regulate the activity of PAS domain transcriptional factors, such as CLOCK and BMAL (1, 15, 21). The region similar to the dPER cytoplasmic localization domain is located at the carboxyl region of the PAS domain (21). The nuclear localization signals (NLS) in mPER1 (26) and mPER3 (27) have been identified; the functional NLS in mPER2 remains unknown. Computer-aided motif prediction programs show a bipartite-type NLS in mPER2 and rat PER2 (rPER2) (15, 18).
Therefore, the nuclear entry mechanism of rPER2 was studied intensively by making deletion constructs. First, we developed systems with which to express truncated rPER2 protein in COS-1 cells and identified the sequence required for its nuclear localization. The nuclear import of cytosolic protein tagged with rPER2 NLS was observed. Coexpression of rPER2 with human CRY1 (hCRY1) induced the nuclear localization of both proteins. Deletion at the nuclear localization domain (NLD), which includes the NLS at the C terminus or carboxyl-terminal domain inhibited rPER2 nuclear localization facilitated by hCRY1. Subcellular localization analysis of hCRY1 and binding assays of each protein showed that the carboxyl-terminal domain of rPER2 is necessary for binding CRY1 and that the NLD of rPER2 is important in establishing the nuclear accumulation of both rPER2 and hCRY1.
MATERIALS AND METHODS
Construction of plasmids for overexpression.
To generate the rPER2 expression plasmid, cDNA encoding rPER2 (GenBank accession no. AB016532 [15]) was subcloned into the CAG-promoter-driven expression vector pCXN2 (a gift from J. Miyazaki [12]). The precise deletions of rPER2 were achieved using conventional restriction enzyme digestion and ligation. The FLAG-tagged carboxyl-terminal fragment of rPER2 was generated by excising the MroI-to-BglII (acids 1638 to 3878) or MroI-to-SmaI (acids 1638 to 3576) fragments of pCXN2-rPER2 and blunting and re-ligating them into the SmaI site of pFLAG-CMV2 (Sigma). The NLD and CRY1 binding domain fragments were amplified using oligonucleotides with EcoRI or BamHI sites and were sequenced and ligated into pFLAG-CMV2. rPER2 NLS insertion into the carboxyl-terminal end of bacterial alkaline phosphatase (BAP) was carried out by PCR amplification with oligonucleotides containing rPER2 nucleotide sequences coding the NLS. All constructs were verified by sequence analysis. hCRY1 was expressed by the plasmid pcDNA3.1-His (Invitrogen) containing the coding region for hCRY1 (GenBank accession no. NM004075). This expression plasmid was a gift from T. Todo.
Cell culture and transfection.
COS-1 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics in 5% CO2. Cells were seeded in 6- or 12-well plates, were cultured overnight to 30 to 50% confluence, and were then transfected with 1 or 0.5 μg of total plasmid DNA, respectively, using Fugene6 (Roche) according to the manufacturer's instructions. After 48 h, transfected cells were assayed as described below.
To determine the molecular sizes of overexpressed proteins, transfected cells were rinsed with phosphate-buffered saline (PBS) twice, lysed with sodium dodecyl sulfate (SDS) sample buffer, resolved by SDS-polyacrylamide gel electrophoresis, and transferred onto ProBlots (Applied Biosystems). The blots were blocked with 3% nonfat milk and were incubated with antibodies for 1 h at room temperature. After three rinses with 0.05% Tween 20 in PBS (T-PBS), proteins were detected using goat anti-rabbit or anti-mouse immunoglobulin G (IgG) antibody conjugated with horseradish peroxidase combined with enhanced chemiluminescence (NEN).
Immunocytochemistry.
Cells were cultured on glass coverslips in 12-well plates and were then transfected as indicated above. The cells were then rinsed once with PBS, fixed with 4% paraformaldehyde for 30 min, and permeabilized with 0.05% Triton X-100 in PBS including 300 nM 4′,6′-diamidino-2-phenylindole (DAPI) (Sigma) for 15 min. After two washes with PBS, cells were blocked with 5% bovine serum albumin for 30 min at room temperature. Anti-rPER2 antiserum (a gift from T. Nagase), anti-FLAG antibody (M2; Sigma), and/or anti-Xpress antibody (Invitrogen) were diluted in blocking solution and were incubated with the cells for 1 h at room temperature. The cells were then washed three times with T-PBS and incubated with fluorescein isothiocyanate- and/or rhodamine-conjugated secondary antibody. Coverslips containing the stained cells were washed with T-PBS and were mounted for fluorescence microscopy.
Coimmunoprecipitation.
Coimmunoprecipitation proceeded as described by Kume and colleagues (10) with some modifications. Forty-eight hours after transfection, cells in six-well plates were washed twice with PBS, were lysed with lysis buffer (150 mM NaCl, 5 mM EDTA, 0.5% NP-40, and 50 mM Tris-HCl [pH 7.5]) including protease inhibitor cocktail tablets (Complete; Roche) on ice for 30 min, and were then clarified by centrifugation for 10 min at 15,000 × g.
Protein A/G agarose beads (Santa Cruz Biotechnology, Inc.) were incubated with anti-rPER2 antibodies for 1 h at 4°C; then antibody-conjugated beads were collected by centrifugation, rinsed twice with PBS, and resuspended in lysis buffer. Protein A/G agarose-antibody complexes were incubated with the clarified cell lysate overnight at 4°C. Subsequently, beads were washed for 10 min at 4°C three times and boiled in 2× SDS sample buffer. Bound proteins were then eluted.
The supernatant was separated by SDS-polyacrylamide gel electrophoresis and immunoblotted against anti-rPER2 antiserum (1:2,000), anti-FLAG M2 antibodies (1:5,000), or anti-Xpress antibody (1:5,000) as described above.
RESULTS
Identification of region essential for nuclear localization of rPER2.
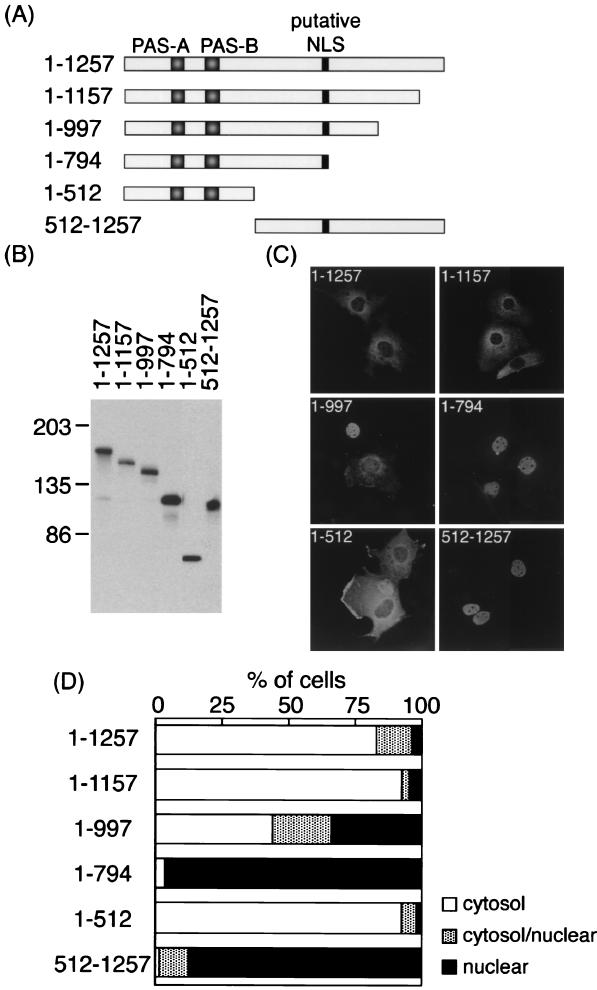
Although the primary structure of rPER2 shows a bipartite NLS in the central domain of rPER2 (residues 778 to 795), whether or not this putative sequence is functional remains unknown. To identify the precise molecular feature required for the nuclear entry system of rPER2, we expressed a series of deleted rPER2 proteins (Fig. 1A) and determined their subcellular localization in COS-1 cells.
FIG. 1.
Subcellular localization of truncated rPER2 mutants in COS-1 cells. (A) Diagrammatic representation of constructs used to identify the rPER2 NLD. PAS-A, PAS-B, and the putative bipartite NLS are shown as shaded and solid boxes. (B) Confirmation of the size of proteins expressed in COS-1 cells determined by immunoblotting. Sizes are indicated in kilodaltons. Anti-rPER2 antiserum was the first antibody. (C) Representative micrographs show the subcellular localization of rPER2. COS-1 cells were transiently transfected with truncated constructs 1–1257, 1–1157, 1–997, 1–794, 1–512, and 512–1257. Forty-eight hours later, cells were fixed and expressed proteins were visualized using anti-rPER2 antiserum and fluorescein isothiocyanate-conjugated secondary antibody. (D) Quantitative analysis of the above. Subcellular localization was categorized as cytoplasm, cytoplasm and nucleus, and nucleus. The ratio of cells with predominant localization to the total transfected cells was determined by counting 50 to 100 cells three to five times in each experiment under light microscopy.
Immunoblotting the overexpressed deletion proteins revealed that the predicted sizes of proteins were expressed in COS-1 cells (Fig. 1B). Figure 1C shows that full-length rPER2 predominated in the cytoplasm as reported in mPERs (10, 26, 27). Two deletions that included a mutant lacking a carboxyl-terminal domain (residues 1 to 1157) and an amino-terminal half-clone (residues 1 to 512) of rPER2 were also localized in the cytoplasm (Fig. 1C and D). In contrast, 1–794 and 512–1257 were located in the nuclei of 96.3 and 87.5% of transfected cells, respectively (Fig. 1C and D). These findings indicated that a region important for nuclear translocation, the NLD, is located between acids 512 and 794. The deletion of amino acids 998 to 1257 (1–997) intermediately affected localization (Fig. 1C and D), suggesting that the C-terminal domain affects the function of nuclear entry. Lacking the amino-terminal portion (residues 1 to 511) or carboxyl-terminal region (residues 795 to 1257) of NLD led to the facilitation of rPER2 mutants' nuclear entry, indicating that both portions are masking the NLD function of rPER2.
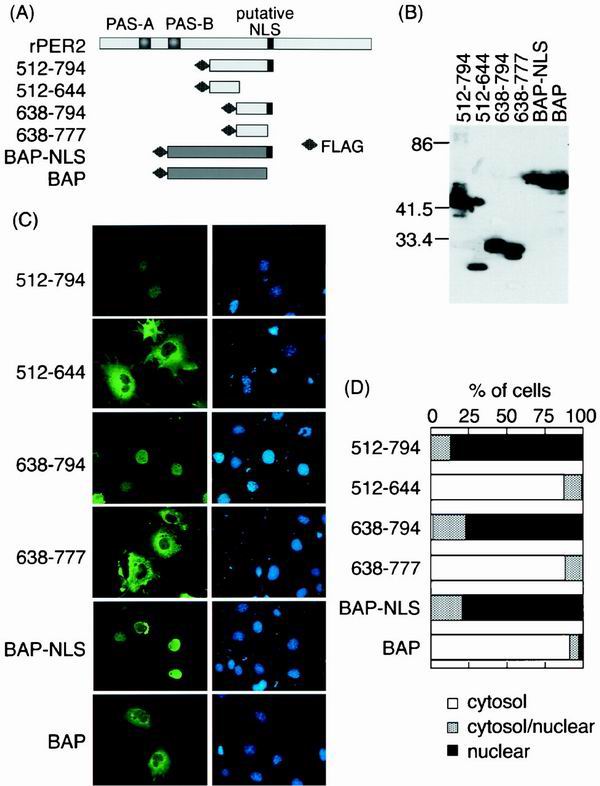
To narrow down the NLS region, we then expressed several FLAG-tagged constructs around the NLD region (Fig. 2A). The expressed proteins from two constructs (512–794 and 638–794) were localized in the nucleus, but 512–644 was found in the cytoplasm (Fig. 2C and D). In contrast to the nuclear localization of 638–794, the fragment of 638–777, lacking the predicted rPER2 NLS (778–794), was enriched in the cytoplasmic compartment.
FIG. 2.
Expression of NLDs of rPER2 and rPER2 NLS-tagged BAP (NLS-BAP). (A) Schematic diagrams of four constructs covering the NLD of rPER2. All fragments were tagged with FLAG at the amino terminus. rPER2 NLS was inserted at the carboxyl terminus of BAP. (B) The molecular size in kilodaltons of FLAG-tagged NLD fragments and BAP fragments was confirmed by immunoblotting using anti-FLAG M2 monoclonal antibody. (C) Subcellular localization of rPER2 NLD mutants (512–794, 512–644, 638–794, and 638–777) expressed in COS-1 cells and examined by immunofluorescence microscopy. Fragments of rPER2 NLD mutants were stained with a combination of anti-FLAG monoclonal antibody M2 and fluorescein isothiocyanate-conjugated anti-mouse IgG (green, left panels), and nuclei were visualized with DAPI (blue, right panels). The distribution of NLS-BAP and BAP was also confirmed as described above. (D) Quantitative analysis of above as described in the legend for Fig. 1D.
In order to show the ability of active nuclear transport of the predicted NLS of PER2, we inserted the NLS (778–794) into the C-terminal end of FLAG-tagged BAP (BAP-NLS, Fig. 2A). BAP-NLS was translocated in the nucleus, while BAP was retained in the cytoplasmic region as we expected (Fig. 2C and D). Thus, we concluded that the latent rPER2 nuclear localization feature is derived from the sequence of acids 778 to 794 rather than through coordination with another domain and that the predicted rPER2 NLS is functional.
Role of two rPER2 domains in nuclear localization of rPER2 by CRY1 coexpression.
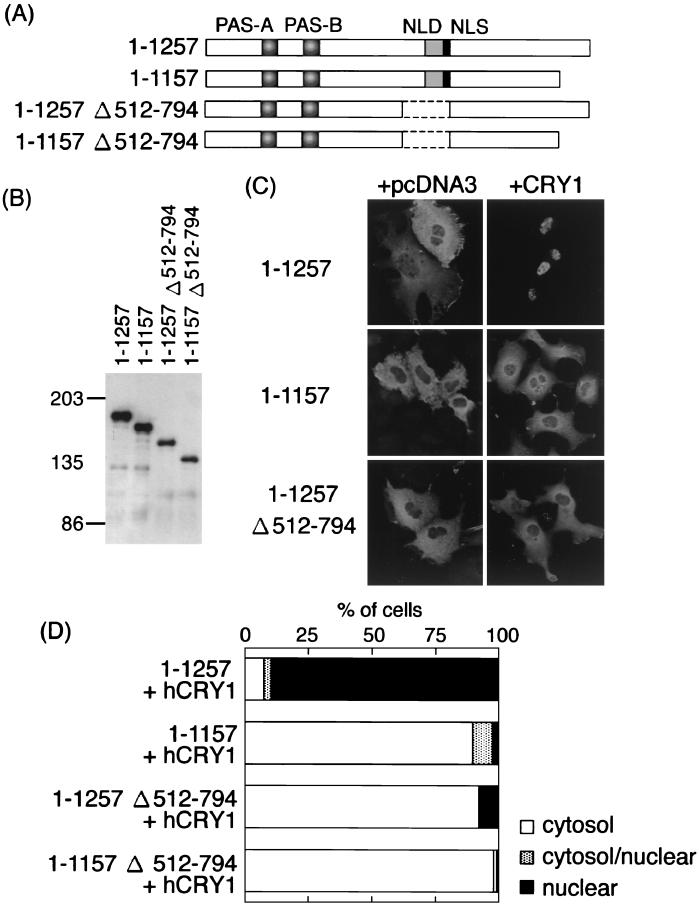
We surmised that the masking of rPER2 NLD would be disrupted by either binding or interaction with a partner molecule that modulates rPER2 nuclear entry like mCRY1 and mCRY2 (10). Moreover, whether or not the nuclear entry of mPERs with mCRYs is regulated by the NLS in PERs remained unknown. Thus, we coexpressed deletion mutants of rPER2 with hCRY1 and examined their subcellular localization.
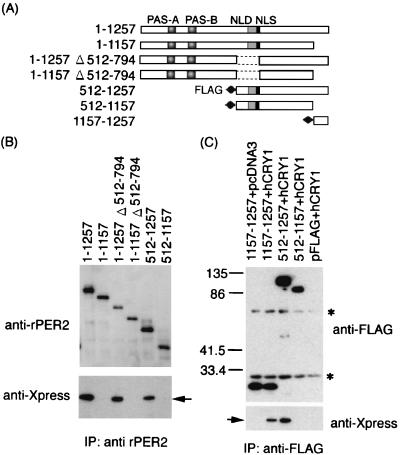
Intact rPER2 proteins coexpressed with hCRY1, rPER2 predominantly localized in the nucleus (Fig. 3A; 1–1257; 89.3% of transfected cells). Deletion of the carboxyl-terminal domain (1157–1257) changed the subcellular localization of rPER2 from the nucleus to the cytoplasm (Fig. 3C; 1–1157). Subcellular distribution of the other deletion mutants (Fig. 1A) was not affected by coexpression with hCRY1 (data not shown). These data suggested that the carboxyl-terminal domain is important for the nuclear localization of rPER2 in the presence of hCRY1. Furthermore, rPER2 lacking the NLD (Fig. 3A; 1–1257; Δ512–794) also caused the cytoplasmic retention of rPER2 (Fig. 3C; 92.1% of transfected cells). These findings suggested that the NLD of rPER2 is also important in deciding the nuclear entry of rPER2.
FIG. 3.
Role of two rPER2 domains in nuclear localization by CRY1 coexpression. (A) Schematic representation of truncated rPER2 constructs. The position of the rPER2 NLD is shown by shaded boxes as PAS-A and PAS-B. The putative NLS is shown as a solid box. Internal portion of constructs 1–1257 Δ512–794 and 1–1157 Δ512–794, containing NLD, was deleted (amino acids 512 to 794, dotted line). The carboxyl-terminal portion of clones 1–1157 Δ512–794 was also deleted to form clone 1–1157. (B) Molecular size in kilodaltons of clones with a truncation of the NLD was confirmed by immunoblotting using anti-rPER2 polyclonal antibody. (C) Full-length (1–1257) or truncated (1–1157 and 1–1257 Δ512–794) clones with a deletion of rPER2 were cotransfected with hCRY1-pcDNA3.1-His (right panels, + hCRY1) in COS-1 cells. Control used the mock vector, pcDNA3, instead of hCRY1-pcDNA3.1-His (left panels, + pcDNA3). Localization was examined by immunocytochemistry using anti-rPER2 antibody. (D) Quantitation of above as described in the legend for Fig. 1D.
Role of carboxyl-terminal domain of rPER2 in nuclear localization of rPER2-CRY1 complex.
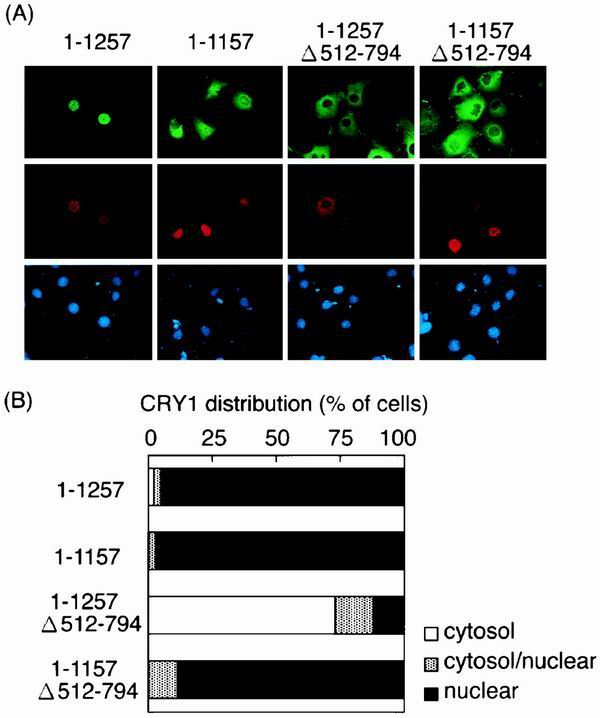
When singly expressed in COS-1 cells, hCRY1 was distributed in the nucleus (data not shown). We examined the localization of hCRY1 in COS-1 cells cotransfected with rPER2 deletion mutants. When hCRY1 was transfected with full-length rPER2 (1–1257) or carboxyl-terminal domain lacking rPER2 (1–1157), the nucleus became predominantly enriched with hCRY1 (Fig. 4). On the other hand, rPER2 localization differed between 1–1257 and 1–1157 (Fig. 4), which were enriched in the nucleus and cytosol, respectively, suggesting that the carboxyl-terminal domain is the binding domain of rPER2 for CRY1. hCRY1 coexpressed with 1–1257 Δ512–794, which lacks rPER2 NLD, accumulated in the cytoplasm (72.91% of transfected cells). Furthermore, 1–1157 Δ512–794, which lacks both NLD and the carboxyl-terminal domain, did not affect the nuclear entry of hCRY1 (Fig. 4). These data also indicated that the carboxyl-terminal domain of rPER2 is the hCRY1 binding site and suggested that after hCRY1 binds to rPER2, the NLD of rPER2 is essential for the nuclear translocation of the rPER2-hCRY1 complex.
FIG. 4.
Subcellular distribution of hCRY1 is affected by cotransfection with rPER2 mutants in COS-1 cells. (A) Full-length (1–1257) and deletion clones truncated at NLDs and/or the carboxyl-terminal portion were coexpressed with Xpress-tagged hCRY1. Transfected cells were triply stained with anti-rPER2 antibodies (green), anti-Xpress antibodies (red), and DAPI (blue). Anti-Xpress antibodies recognized recombinant hCRY and DAPI-stained nuclei. (B) Quantitative analysis of experiment shown in Fig. 4A as described in the legend for Fig. 1D.
Binding domain of rPER2 to hCRY1.
To determine whether or not the effect of rPER2 mutants on hCRY1 localization is due to the ability of each molecule to bind CRY1, we analyzed the binding activity of rPER2 mutants with hCRY1 using coimmunoprecipitation. The lysates from cells of cooverexpressed rPER2 mutants and Xpress-tagged hCRY1 was incubated with anti-rPER2 antibody-protein A/G agarose complex. The precipitated immunocomplexes were analyzed by immunoblotting with anti-rPER2 antibody and anti-Xpress antibody. Intact rPER2 and all mutants containing a carboxyl-terminal domain bound to hCRY1 (1–1257, 1–1257 Δ512–794, and 512–1257), while all deletions lacking the carboxyl-terminal domain of rPER2 lost the ability to bind hCRY1 (1–1157, 1–1157 Δ512–794, and 512–1157) (Fig. 5A and B).
FIG. 5.
Coimmunoprecipitation shows that hCRY1 interacts with rPER2 carboxyl-terminal domain. (A) Schematic representation of rPER2 mutant constructs for coimmunoprecipitation. The positions of rPER2 NLD, PAS-A, and PAS-B are shown by shaded boxes. The rPRE2 NLS is shown as a solid box. The mutants with a deletion at the amino-terminal half (512–1257 and 512–1157) and at the carboxyl-terminal fragment of rPER2 (1157–1257) were tagged with FLAG at their amino-terminal ends. (B) Total lysates from cells coexpressing hCRY1 and truncation mutants of rPER2 were immunoprecipitated (IP) and blotted with anti-rPER2 antibodies (top panel) and were detected using anti-Xpress antibodies (lower panel). Results were similar in replicate experiments. Arrow indicates position of hCRY1. (C) Identification of the CRY1 binding domain of rPER2. The FLAG-tagged truncation mutants of rPER2 proteins (1157–1257, 512–1257, and 512–1157) were expressed with hCRY1. Control was vector pcDNA3 or pFLAG instead of hCRY1 or rPER2 mutants. Lysates from transfected cells were immunoprecipitated (IP) and blotted with anti-FLAG antibodies (top panel) and were visualized using anti-Xpress antibodies (lower panel). The arrow indicates the position of hCRY1. Asterisks indicate nonspecific band corresponding to Ig derived from M2 monoclonal antibodies. Numerals indicate molecular weights in thousands.
We then constructed and overexpressed the three FLAG-tagged clones (1157–1257, 512–1257, and 512–1157) in COS-1 cells. Binding to hCRY1 was tested by coimmunoprecipitation with anti-FLAG antibody. The results showed that the minimal region of rPER2 required for binding hCRY1 was located in clone 1157–1257 (Fig. 5C). The data indicate that the carboxyl-terminal region (1157–1257) of rPER2 is the binding domain to hCRY1. These data easily explain the cytoplasmic colocalization of hCRY1 and rPER2 in Fig. 4A.
DISCUSSION
The present study focused on the nuclear entry mechanism of rPER2 and identified its latent functional NLS at acids 778 to 794; that NLS has been identified as the putative bipartite-type NLS. The full length of rPER2 overexpressed in COS-1 cells was predominantly distributed in the cytoplasm, like mPERs in COS-7 and NIH 3T3 cells (10, 27). However, the truncated mutagenesis of rPER2 (1–794 and 512–1257) promoted nuclear entry. Furthermore, the putative NLS deletion inactivated the nuclear localization of the rPER2 fragments, and BAP tagged with rPER2 NLS promoted the nuclear translocation of the molecule. The bipartite-type NLS in rPER2 is well conserved in mPER2 and in hPER2 at similar positions (1, 6, 11, 15, 18, 21). In the PER1 sequence, no obvious NLS was found, but the functional NLS was identified at the corresponding region to rPER2 NLS (26). In contrast, PER3 contains a simian virus 40 large-T-antigen type of NLS at the corresponding region, which is functional (26, 27).
The NLS appeared to be masked by two other regions (1–511 and 795–1257), and the deletion of masking regions may activate the nuclear translocating function (Fig. 1). Shearman et al. showed that mPER2 435–1257 could not enter into the nucleus (17), while our N-terminal deletion clone (512–1257) showed nuclear localization. We speculate that the domain for masking the NLS function or the portion for cytosolic retaining activity like NES may locate between residues 435 and 512. Furthermore, mPER2brdm1 (missing residues 348 to 434) could not be translocated in the nucleus, irrespective of the presence of mCRY1 (17). Because our results suggest that mPER2brdm1 can bind with mCRY1, the arrhythmic phenotype of mPER2brdm1 (28) may be caused by the inhibition of proper nuclear translocation of CRY proteins.
Kume et al. reported that mCRY1 and mCRY2 are nuclear proteins that interact with all three mPER proteins and translocate them from the cytoplasm to the nucleus (10). Constitutively overexpressed hCRY1 also localized in the nucleus (data not shown) like mCRY proteins (9, 10, 24) and promoted the nuclear entry of full-length rPER2 but failed to translocate rPER2 with a truncated carboxyl-terminal domain (Fig. 3). In fact, other reports also suggest that the amino-terminal half-region is not important for the nuclear translocation of mPER2 and mCRY proteins (17). Furthermore, our binding assays showed that the carboxyl-terminal domain was essential for rPER2 to bind hCRY1 followed by nuclear translocation. Recombinant rPER2 protein lacking the NLD did not enter the nucleus, even when coexpressing hCRY1 in COS-1 cells. Moreover, hCRY1 was also retained in the cytoplasm like rPER2 with a deletion of the NLD. This indicated that the rPER2 NLD, which becomes functional after binding with hCRY1, is essential for nuclear localization of the rPER2-hCRY1 complex. We speculate that the binding of hCRY1 to rPER2 changes the conformation of rPER2 and exposes the NLD to render it functional in translocating the complex to the nucleus.
The rat TIM-like protein (rTLP) homologous to dTIM (a partner in dPER nuclear translocation) was a candidate to expose rPER2 NLS like dTIM (14). However, rTLP did not influence the subcellular localization of rPER2 (16), in agreement with the results of other studies using mPER and mTIM (10). It is likely that CRYs play more important roles in the nuclear translocation of rPER2 than does rTLP.
When singly overexpressed in COS-1, hCRY1 was distributed in the nucleus (data not shown). However, hCRY1 located in the cytosol after coexpressing rPER2 with a deletion of the NLD. The sequence in rPER2 appeared to be abler to perform nuclear translocation than that in hCRY1.
Several pathways for the nuclear entry of mammalian PERs have been reported. Kume et al. found that mCRY proteins act as dimerization partners for the nuclear localization of mPER1, mPER2, and mPER3, with activity that is more effective on mPER1 and mPER2 than on mPER3 (10). The present study revealed details of the interaction mechanism between rPER2 and hCRY1 and suggests that hCRY1 translocates rPER2 to the nucleus by binding and inhibiting its ability to mask the rPER2 NLD. On the other hand, Yagita et al. demonstrated that the nuclear translocation of mPER1 and mPER2 is accelerated by serum shock after binding mPER3 (27), although the effect of mPER3 on the nuclear entry of mPER2 was weaker than that of mPER1. Casein kinase Iɛ modulates the nuclear translocation of mPER1 and mPER3 but not of mPER2 (20, 26). Thus, CRY appears to be the main partner involved in the nuclear translocation of PER2.
In the SCN of mCry1-mCry2 double-knockout mice, mPER1 weakly localizes in the nucleus (17, 27), whereas mPER2 does not accumulate in the nuclei of SCN neurons (17). Our results are consistent with these findings in that at least the nuclear translocation of PER2 will be mainly regulated by the CRY protein through direct binding.
The CRY1 binding domain of rPER2 shows higher homology with that of mPER2 but less with that of hPER2 and the PER subfamily, including PER1 and PER3. All mammalian PER proteins bind CRY proteins (10), suggesting that the 1156-to-1175 region, which is highly homologous to other PER proteins, is important for binding CRY proteins.
mCRY proteins are temporally expressed in the nucleus of SCN neurons, and the expression profile is synchronized with the accumulation of PER1 and PER2 in the nucleus (5, 10). The CRY proteins inhibit per1-promoter-driven reporter transactivation by the CLOCK-BMAL complex in NIH 3T3 cells (10). This inhibition might proceed in the nuclei of SCN neurons in vivo. Our conclusion that the NLS of rPER2 is essential for the nuclear entry of CRY1 indicates that PER2 plays an important role in the nuclear translocation of CRY, which may regulate the negative-feedback loop of the core circadian clock mechanism.
ACKNOWLEDGMENTS
We thank Takahiro Nagase for supplying us with anti-rPER2 antibody; Takeshi Todo for providing human CRY1 expression plasmid; Junichi Miyazaki for the constitutive expression vector pCXN2; Lino Saez for useful discussion; and Kazuko Suzuki and Yukako Soya for technical assistance.
This study was supported by a project grant for the Competition Research Program, AIST, MITI, Tokyo, Japan.
REFERENCES
- 1.Albrecht U, Sun Z S, Eichele G, Lee C C. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 2.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap J C. Molecular bases for circadian clocks. Cell. 1999;96:271–290. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- 4.Field M D, Maywood E S, O'Brien J A, Weaver D R, Reppert S M, Hastings M H. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 5.Hastings M H, Field M D, Maywood E S, Weaver D R, Reppert S M. Differential regulation of mPER1 and mTIM proteins in the mouse suprachiasmatic nuclei: new insights into a core clock mechanism. J Neurosci. 1999;0:RC11. doi: 10.1523/JNEUROSCI.19-12-j0001.1999. (1–7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishida N, Kaneko M, Allada R. Biological clocks. Proc Natl Acad Sci USA. 1999;96:8819–8820. doi: 10.1073/pnas.96.16.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin X, Shearman L P, Weaver D R, Zylka M J, de Vries G J, Reppert S M. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 8.Klein D C, Moore R Y, Reppert S M, editors. Suprachiasmatic nucleus: the mind's clock. New York, N.Y: Oxford University Press; 1991. [Google Scholar]
- 9.Kobayashi K, Kanno S, Smit B, van der Horst G T, Takao M, Yasui A. Characterization of photolyase/blue-light receptor homologs in mouse and human cells. Nucleic Acids Res. 1998;26:5086–5092. doi: 10.1093/nar/26.22.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kume K, Zylka M J, Sriram S, Shearman L P, Weaver D R, Jin X, Maywood E S, Hastings M H, Reppert S M. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 11.Nagase T, Ishikawa K, Nakajima D, Ohira M, Seki N, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;4:141–150. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 12.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 13.Oishi K, Fukui H, Ishida N. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem Biophys Res Commun. 2000;268:164–171. doi: 10.1006/bbrc.1999.2054. [DOI] [PubMed] [Google Scholar]
- 14.Saez L, Young M W. Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 15.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–27042. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 16.Sakamoto S, Miyazaki K, Fukui H, Oishi K, Hayasaka N, Okada M, Kamakura M, Taniguchi T, Nagai K, Ishida N. Molecular characterization and nuclear localization of rat timeless-like gene product. Biochem Biophys Res Commun. 2000;279:131–138. doi: 10.1006/bbrc.2000.3927. [DOI] [PubMed] [Google Scholar]
- 17.Shearman L P, Sriram S, Weaver D R, Maywood E S, Chaves I, Zheng B, Kume K, Lee C C, van der Horst G T, Hastings M H, Reppert S M. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 18.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F, Jr, Reppert S M. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 19.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 20.Takano A, Shimizu K, Kani S, Buijs R M, Okada M, Nagai K. Cloning and characterization of rat casein kinase 1epsilon. FEBS Lett. 2000;477:106–112. doi: 10.1016/s0014-5793(00)01755-5. [DOI] [PubMed] [Google Scholar]
- 21.Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, Okamura H. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells. 1998;3:167–176. doi: 10.1046/j.1365-2443.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 22.Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, Maebayashi Y, Okumura K, Takekida S, Yamamoto S, Yagita K, Yan L, Young M W, Okamura H. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 1998;17:4753–4759. doi: 10.1093/emboj/17.16.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 24.Thresher R J, Vitaterna M H, Miyamoto Y, Kazantsev A, Hsu D S, Petit C, Selby C P, Dawut L, Smithies O, Takahashi J S, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 25.van der Horst G T, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker A P, van Leenen D, Buijs R, Bootsma D, Hoeijmakers J H, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 26.Vielhaber E, Eide E, Rivers A, Gao Z-H, Virshup D M. Nuclear entry of the circadian regulator mPER1 is controlled by mammalian casein kinase I ɛ. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yagita K, Yamaguchi S, Tamanini F, van Der Horst G T, Hoeijmakers J H, Yasui A, Loros J J, Dunlap J C, Okamura H. Dimerization and nuclear entry of mPER proteins in mammalian cells. Genes Dev. 2000;14:1353–1363. [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng B, Larkin D W, Albrecht U, Sun Z S, Sage M, Eichele G, Lee C C, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 29.Zylka M J, Shearman L P, Weaver D R, Reppert S M. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–1110. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]