Abstract
Background
The COVID-19 pandemic led to severe health systems collapse, as well as logistics and supply delivery shortages across sectors. Delivery of PCR related healthcare supplies continue to be hindered. There is the need for a rapid and accessible SARS-CoV-2 molecular detection method in low resource settings.
Objectives
To validate a novel isothermal amplification method for rapid detection of SARS-CoV-2 across seven sub-Sharan African countries.
Study design
In this multi-country phase 2 diagnostic study, 3,231 clinical samples in seven African sites were tested with two reverse transcription Recombinase-Aided Amplification (RT-RAA) assays (based on SARS-CoV-2 Nucleocapsid (N) gene and RNA-dependent RNA polymerase (RdRP) gene). The test was performed in a mobile suitcase laboratory within 15 min. All results were compared to a real-time RT-PCR assay. Extraction kits based on silica gel or magnetic beads were applied.
Results
Four sites demonstrated good to excellent agreement, while three sites showed fair to moderate results. The RdRP gene assay exhibited an overall PPV of 0.92 and a NPV of 0.88. The N gene assay exhibited an overall PPV of 0.93 and a NPV 0.88. The sensitivity of both RT-RAA assays varied depending on the sample Ct values. When comparing sensitivity between sites, values differed considerably. For high viral load samples, the RT-RAA assay sensitivity ranges were between 60.5 and 100% (RdRP assay) and 25 and 98.6 (N assay).
Conclusion
Overall, the RdRP based RT-RAA test showed the best assay accuracy. This study highlights the challenges of implementing rapid molecular assays in field conditions. Factors that are important for successful deployment across countries include the implementation of standardized operation procedures, in-person continuous training for staff, and enhanced quality control measures.
Keywords: SARS-CoV-2, Diagnostics-in-a-suitcase, Recombinase polymerase amplification assay
1. Background
In March 2020, the World Health Organization (WHO) declared COVID-19 caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) as a global pandemic [1]. Early detection of infected cases is still regarded as essential to reduce the disease burden [2].
Real-time reverse transcription polymerase chain reaction (RT-PCR) is the standard approach in terms of sensitivity and accuracy. However, this technique requires well-established laboratory. Additionally, supply and delivery shortages have been reported across various sectors [3,4]. Antigen lateral flow tests were quickly developed and deployed, allowing carriers with high viral load to be diagnosed more easily. However, these tests often do not reach the WHO recommended minimum of >80% sensitivity and >97% specificity for new diagnostics tests [5]. Deployment of a simple, rapid molecular test method could offer marked advantages.
Recombinase-Polymerase/aided Amplification (RPA/RAA) assays have been described as a rapid and effective nucleic acid amplification technique, due to its simplicity and fast sample-to-result test time [6]. RPA/RAA is an isothermal probe-based nucleic acid detection method that neither requires template denaturation nor primer annealing steps [7]. The proper selection of a polymerase working at low temperature (39–42 °C), the robustness of the assay, and compactness of the fluorescence detection device make RPA/RAA an optimal technique for molecular diagnosis at the point of need. To enable the widespread use of the technology, mobile suitcase laboratories were deployed to many sub-Saharan African countries [8].
2. Objectives
To further clinically evaluate the system in real-life settings, a multi-country single blinded phase 2 study was conducted in seven sub-Saharan African countries. The aim was not only to determine the accuracy of RAA assay for detection of SARS-CoV-2 in local African settings, but also to assess performance differences between research institutions.
3. Material and methods
3.1. Sample size calculation
The sample size was calculated using the formula for comparing two independent proportions, which is used to estimate sample size for studies comparing sensitivity and/or specificity of two tests of unpaired design [9]. A minimum of 300 samples per site were required to achieve 95% confidence.
3.2. Study site and population
A multi-country, single blinded, phase 2 diagnostic evaluation study was conducted in seven sites: Institut Pasteur de Dakar (IPD), Senegal; Institut Pasteur de Madagascar (IPM), Madagascar; Kumasi Centre for Collaborative Research (KCCR), Ghana; University of Ibadan (UI), Nigeria; Institut National de Recherche Biomédicale (INRB), Democratic Republic of the Congo; University of Khartoum (UofK), Sudan; and Makerere University (MAK), Uganda. For the purpose of this study, a total of 3231 archived samples were used to evaluate the assay. Archived samples included in this study were from patients who tested positive for SARS-CoV-2 by the reference laboratory and patients who were suspected COVID-19 and tested negative by the reference laboratory. All samples were tested irrespective of age, sex, and race. During the study, all samples were handled anonymously. Samples included nasal, mid-turbinate swab or saliva in viral transport media (VTM), PBS or stored dry and maintained in −80 °C. Additional information regarding the samples was included in supplementary file #1.
3.3. Study design
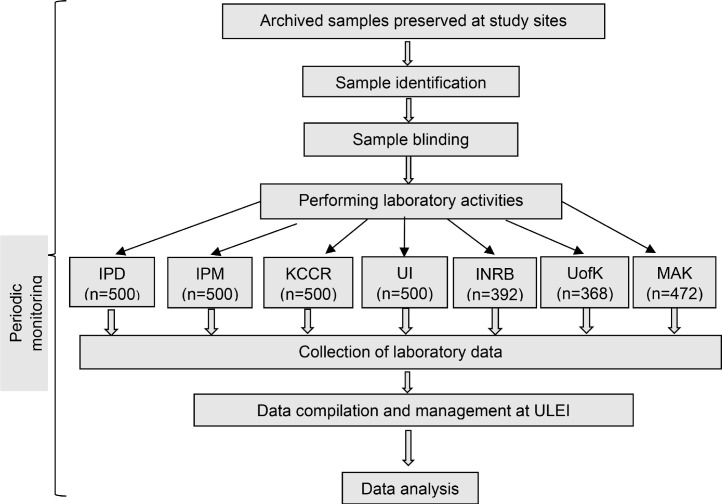
This study was conducted according to the guidelines for diagnostic kit evaluation (Fig. 1 ) [9].
Fig. 1.
Schematic representation of the multi-country, single blinded, phase 2 study to test archived samples with the RT-RAA assay for SARS-CoV-2. Institut Pasteur de Dakar (IPD); Institut Pasteur de Madagascar (IPM); Kumasi Centre for Collaborative Research (KCCR); University of Ibadan (UI); Institut National de Recherche Biomédicale (INRB); University of Khartoum (UofK); and Makerere University (MAK).
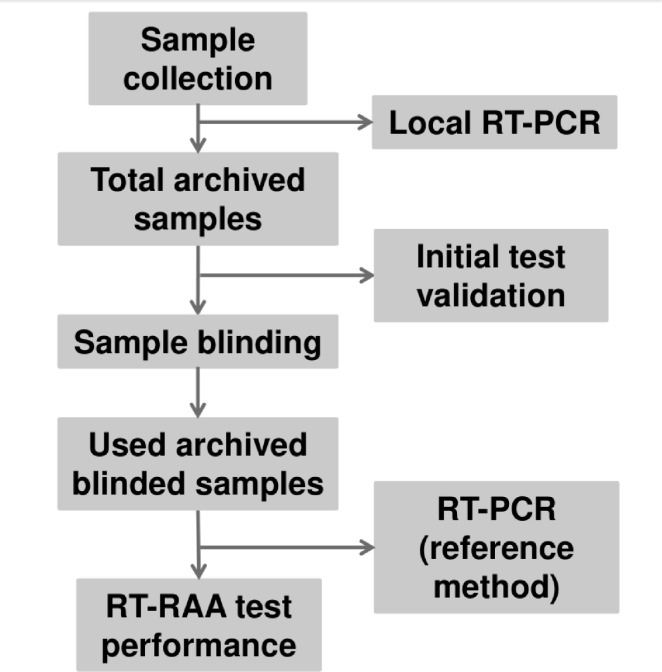
Each country initially tested an inactivated test panel of 20 samples using RT-RAA and real-time RT-PCR reagents to assure their preparedness to perform the tests (Fig. 2 ). Once successful (at least 90% accurate detection rate), each country started clinical sample testing as follows: all samples were labelled with random numbers by the site principal investigator, and the laboratory personnel were divided into two teams: one team performed the real-time RT-PCR and the other performed the RT-RAA. Both teams were blinded and did not know whether the sample was from a positive or negative subject. Simultaneously, the three parts carbonless copy paper-based laboratory report forms (LRFs) were made available in two modes, one by the unblinded staff and the other by the blinded staff (supplementary file #2). All laboratory data were reported in LRFs by both teams separately. The data was then decoded by the data management team, where it was merged and statistically analysed. Patient information was not shared between study sites, only positivity and negativity rates were recorded. To ensure the quality of the study activities, periodic monitoring was performed.
Fig. 2.
Workflow of the performed tests at the study sites. The new real-time RT-PCR performed was used as the reference method to validate the RT-RAA assays.
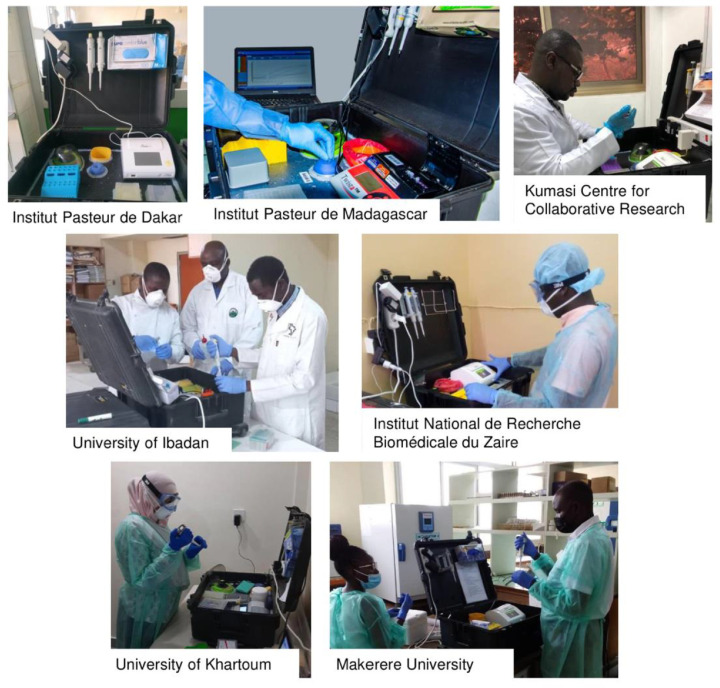
3.4. Mobile suitcase laboratory set up
The total set up consisted of a Glove Box (Bodo Koennecke, Berlin, Germany) and a mobile suitcase laboratory (Fig. 3 ). The Glove Box protects the technician while handling and inactivating the sample before nucleic acid extraction. The isothermal amplification test was performed in the Mobile Suitcase Lab (Fig. 4 ).
Fig. 3.
Example of the suitcase lab, which is fully equipped to perform molecular tests in the field.
Fig. 4.
The suitcase lab at various study sites.
3.5. Laboratory analysis
3.5.1. Real-time RT-PCR assay for detection of SARS-CoV-2 RNA
RNA was isolated from clinical samples of subjects, who were recently suspected of contracting COVID-19. At KCCR, UofK, UI and INRB, the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) was used. At MAK, Liferiver Viral DNA/RNA isolation kit (Shanghai ZJ Biotech co. let, Shanghai, China) was used. At IPD, the Veri-Q PREP M16 Automatic Nucleic Acid Extraction System (MiCo BioMed, South Korea) was used. IPM used the NucleoSpin Dx Virus Mini kit (Macherey-Nagel GmbH, Düren, Germany).
All samples were originally tested as part of routine diagnostics and divided into positive and negative according to the locally used real-time RT-PCR results (supplementary file #2). To assure integrity of RNA on the day of testing, an additional real-time RT-PCR was performed. This result was considered as reference method (Fig. 2). The archived patient material was tested at each site with a commercially available real-time RT-PCR assay combining oligonucleotide Lightmix Modular SARS-CoV-2 RNA-dependent RNA polymerase (RdRP) and the lyophilized one-step RT-PCR Polymerase Mix kit from TIB MOLBIOL (Berlin, Germany) according to the manufacturer's instructions. The real-time RT-PCR reaction comprised 15 μL, including 0.5 μL oligonucleotide mix, 4 μL PCR-grade water, 10 μL qPCR Master mix and 5 μL template or control. The following real-time PCR cycler was used: BioRad CFX96 Touch (Bio-Rad Laboratories, Hercules, United States) at IPD, KCCR and UI, Rotorgene Q (Qiagen, Hilden, Germany) at IPM, Applied Biosystems 7500 Fast (Thermo Fisher Scientific, Waltham, United States) at INRB, qTower (Biometra, Analytik Jena, Jena, Germany) at UofK and Applied Biosystems Quantstudio 7flex (Thermo Fisher Scientific, Waltham, United States) at MAK.
3.5.2. RT-RAA assay for detection of SARS-COV-2 RNA
Two RT-RAA assays were evaluated: one based on the RdRP gene and one on the Nucleocapsid (N) gene. The primers, probe and reaction conditions for SARS-CoV-2 genes N and RdRP RT-RAA assays were based on a previous study [8] Primer and probe were synthesized by TIB MOLBIOL (Berlin, Germany). The RT-RAA nucleic acid amplification kit (Fluorescent RT-RAA) from Jiangsu Qitian Gene Biotechnology Co. (Wuxi, China) was used. The kit comprises lyophilized enzymes, including the reverse transcriptase necessary for RNA amplification. The RT-RAA reaction total volume was 50 μL including 21.5 μL of the oligonucleotide mix (21 pMol for forward primer, 42 pMol for reverse primer and 6 pMol for exo-probe), 25 μL rehydration buffer, 2.5 μL Magnesium Acetate and 1 μL template or control. The mix was added into the lid of the reaction tube containing the freeze-dried pellet. The tube was closed, centrifuged, mixed, centrifuged, and placed immediately into the isothermal device: UofK and IPM used the TwistDx TS1 device (Cambridge, UK). UI used the Qiagen ESEquant TS2 model (Hilden, Germany), while all other countries used the Axxin T8 Isothermal instrument (Fairfield, Australia). The reaction was incubated at 42 °C for 15 min. A mixing step was conducted after 320 s for the N gene assay and after 230 s for the RdRP gene assay. For signal interpretation, a combined threshold time (TT) and first derivative analysis was used with the corresponding software of each device.
3.6. Data analysis
Standard formulas were used with MedCalc [10] to determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) [11]. Cohen's kappa coefficient (k) and McNemar's test were performed to determine the concordance and discordance between the RT-RAA assays and real-time RT-PCR-based method. The values of Cohen's kappa coefficients were interpreted according to Landis and Koch [12] and calculated using the online Graphpad version. The McNemar test was calculated via https://epitools.ausvet.com.au/mcnemar. A P-value <0.05 was considered to be statistically significant.
3.7. Mutational analysis of the RdRP amplicon
Over 10,000 SARS-CoV-2 RNA sequences from local strains in the involved countries were screened for potential mutations in the targeted RdRP region. A previously published pipeline was used [13].
4. Results
4.1. Performance of the deployed isothermal amplification assays
In total, 3231 samples were identified at study sites (Fig. 1) to validate the assays. After data curation, the N gene assay results were evaluated with a total of 1890 negative samples and 580 positive samples and the RdRP gene assay with a total of 2326 negative samples and 868 positive samples (supplementary file #3 displaying sample flowchart and raw data). The RdRP gene assay showed an overall PPV of 0.92 and a NPV of 0.88. The N gene assay showed an overall PPV of 0.93 and a NPV 0.88.
The sensitivity of both RT-RAA assays varied depending on the sample Ct values (Table 1 ). Real-time RT-PCR positive samples with high viral load (Ct <30) showed the best results with 90.8% and 81.8% overall sensitivities for the RdRP and N gene RT-RAA assays, respectively. When comparing sensitivity between sites, values differed considerably. For high viral load samples, the RdRP assay sensitivity ranged between 60.5 and 100%. The N gene did not perform as well as the RdRP gene, with a sensitivity between 25 and 98.6%. The specificity among all study sites ranged from 91.1 to 100% for the RdRP gene RT-RAA assay and from 94.3 to 100% for the N gene RT-RAA assay (Table 2 ). The overall sensitivity and specificity with both targets combined was 58.2% and 99.2%, respectively. The agreement between test methods varied across sites (Table 3 ). Sites IPD, KCCR, UI and INRB showed good to excellent agreement, while sites IPM, UofK and MAK showed fair to moderate results.
Table 1.
Overall clinical sensitivity of all samples across sites categorized according to the Ct values of real-time RT-PCR.
| RT-PCR Ct range | Target gene for RT-RAA | Overall Sensitivity (%) | RT-PCR positive | RT-RAA positive |
|---|---|---|---|---|
| 0–30 | RdRP | 90.8 (82.6 – 99.7) |
493 | 448 |
| N | 81.8 (72.5 – 92) |
341 | 279 | |
| ≥30–35 | RdRP | 45.6 (36.3 – 56.5) |
182 | 83 |
| N | 41.8 (30.98 – 55.4) |
117 | 49 | |
| ≥35–40 | RdRP | 13.5 (9 – 19.7) |
193 | 26 |
| N | 12.3 (6.9 – 20.2) |
122 | 15 | |
| Total | RdRP | 64.2 (58.9 – 69.7) |
868 | 557 |
| N | 59.1 (53 – 65.7) |
580 | 343 |
Table 2.
Clinical sensitivity and specificity at each study site. NA is not applicable as the site did not perform the assay. The 95% confidence interval is showed in parenthesis.
| Study site | RdRP sensitivity (%) | RdRP specificity (%) | N sensitivity (%) | N specificity (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| <30 Ct | 30–35 Ct | >35 Ct | Overall | <30 Ct | 30–35 Ct | >35 Ct | Overall | |||
| IPD | 100 (95 – 99.9) |
75 (47.6 –92.7 |
21.4 (4.7 – 50.8) |
85.3 (76.9 – 91.5) |
99 (97.4 – 99.7) |
98.6 (92.5 – 99.9) |
81.2 (54.3 – 95.9) |
14.3 (1.8 –42.8) |
84.3 (75.8 – 90.8) |
99 (97.8 – 99.8) |
| IPM | 94.4 (89.3 – 97.5) |
23.9 (12.6 –38.7) |
4.2 (0.5 – 14.2) |
62.4 (55.9 – 68.6) |
93.5 (89.8 – 96.2) |
82.5 (75.3 – 88.3) |
10.8 (3.6 –23.6) |
12.5 (4.7 –25.2) |
54,4 (47.9 –60.9) |
94.3 (90.8 – 96.8) |
| KCCR | 95.6 (78 – 99.9) |
84 (63.9 – 95.5) |
47.6 (25.7 – 70.2) |
76.8 (65 –86.1) |
99.7 (98.7 – 100) |
91.3 (72 – 98.9) |
68 (46.5 – 85) |
19 (5.4 – 41.9) |
60.8 (48.4 – 72.4) |
99 (97.6 –99.7) |
| UI | 95.5 (84.8 –99.5) |
90 (55.5 –99.7) |
42.9 (9.9 – 81.6) |
88.7 (78.1 – 95.3) |
100 (99.1–100) |
95.5 (84.8 –99.5) |
90 (55.5 –99.7) |
42.9 (9.9 – 81.6) |
88.7 (78.1 – 95.3) |
100 (99.1 – 100) |
| INRB | 89.2 (79.8 – 95.2) |
50 (28.2 –71.8) |
3.6 (0.1 –18.3) |
62.9 (53.8 – 71.4) |
99.6 (97.9 – 100) |
25 (8.7 – 49.1) |
16.6 (0.4 – 64.1) |
0 (0 –70.7) |
20.4 (8 – 39.7) |
100 (96 – 100) |
| UofK | 60.5 (40.4 – 76) |
23 (5 – 53.8) |
14.3 (4 – 32.7) |
38 (27.3 – 49.6) |
91.1 (87.2 – 94.3) |
55.3 (38.3 –71.4) |
28.6 (8.4 – 58.1) |
0 (0 – 30.9) |
30.8 (21.1 – 42.1) |
98.5 (96.4 – 99.6) |
| MAK | 87.9 (79.8 – 95.6) |
32 (19.5 – 46.7) |
6.5 (1.4 – 18) |
54.3 (47.09 – 61.5) |
99.3 (97.4 – 99.9) |
NA | NA | NA | NA | NA |
Table 3.
Agreement between real-time RT-PCR and RT-RPA assays at different sites. NA is not applicable as the site did not perform the assay. Fair = 0.21 – 0.40; moderate = 0.41 – 0.60; good = 0.61 – 0.80; excellent: 0.81 – 1.
| Study site | Kappa value RdRp | Agreement | p-value RdRP | Kappa value N | Agreement | p-value N |
|---|---|---|---|---|---|---|
| IPD | 0.878 | Excellent | 0.022 | 0.854 | Excellent | <0.001 |
| IPM | 0.568 | Moderate | <0.001 | 0.497 | Moderate | <0.001 |
| KCCR | 0.843 | Excellent | <0.001 | 0.697 | Good | <0.001 |
| UI | 0.901 | Excellent | 0.03 | 0.901 | Excellent | 0.03 |
| INRB | 0.696 | Good | <0.001 | 0.283 | Fair | <0.001 |
| UofK | 0.328 | Fair | 0.05 | 0.382 | Fair | <0.001 |
| MAK | 0.570 | Moderate | <0.001 | NA | NA | NA |
4.2. Mutational analysis of the amplicon
No significant mutations were found in the RdRP amplicon across countries (supplementary files #4 and #5).
5. Discussion
The worldwide spread of SARS-CoV-2 has taken viral diagnostics to a new level of importance and publicity. Alternative methods to real-time RT-PCR with equal sensitivity and specificity are urgently needed to overcome shortage in supply chains [14]. The WHO recommended the ASSURED criteria (affordable, sensitive, specific, user-friendly, robust, deliverable to end-users) for future diagnostics [15]. Isothermal amplification assays address most of these criteria as shown in many outbreak situations [16], [17], [18]. Although clinical research is rapidly progressing in the field of new diagnostic tests, multi-country approaches for SARS-CoV-2 are lacking, especially in sub-Saharan Africa [40]. Only one multicenter study was reported using RT-loop mediated amplification in four African countries in both east and west Africa with very promising sensitivity of 87% [19].
Using the RT-RAA technology helped to circumvent the worldwide supply shortages of real-time RT-PCR test kits [20,21]. The specificity for the two SARS-CoV-2 genome targets across sites ranged from 91.1 to 100% (RdRP gene RT-RAA) and 94.3 – 100% (N gene RT-RAA). In contrast the SARS-CoV-2 an E gene target RT-RAA, showed a high number of false positive results in a previous study and was not included in the current screening [22]. Detecting true negative samples accurately at high specificity avoids unnecessary clinical implications and social upset [23], [24], [25]. SARS-CoV-2 Rapid antigen tests have shown a higher false positive rate (96–99.7%) [26], and especially in low prevalence settings a molecular confirmatory test is needed [27].
When deploying both RT-RAA assays in this study, sensitivity showed a large range of intercountry variations. For high viral load samples, promising overall 90.8% (RdRP RT-RAA) and 81.8% (N RT-RAA) sensitivities were determined. For samples with Ct 31 – 40, values were inconsistent between sites. While higher assay accuracy was identified at three sites, two sites did not produce the expected outcomes despite the success during the preparatory phase. Compared to the RdRP gene, the N-gene RT-RAA assay demonstrated lower sensitivity. The insufficient performance of N gene RT-RAA assay led to its exclusion for further testing to maximize usage of laboratory materials and resources, further underlining the importance of adaptation to unanticipated events during a large diagnostic study. Overall, the performance of the RT-RAA assays is much better than the commercially available rapid antigen tests, whose sensitivity values differed considerably with sensitivities ranging from only 28–86% (Ct 17 – 36) [28,29]. Rapid antigen tests are suited best for detection of symptomatic carriers with high viral load [30,31]. Low viral load samples are often undetected as well as certain SARS-CoV-2 mutations [32,33].
The RdRP gene RT-RAA assay showed promising but very variable sensitivity values across sites. Some sites showed more than 90% sensitivity and others under 50% even with high viral load samples. A potential mutation in the target region of RdRP primers, was excluded by screening over 10,000 SARS-CoV-2 RNA sequences from local strains in the involved countries (supplementary file #4 and #5) using a recently published screening method [13], identifying no significant changes. Reagents deterioration during transportation was unlikely, since RAA reagents are lyophilized, cold-chain independent, robust, and stable over long periods of time [6]. In addition, a quality control check was conducted upon delivery of the kits to exclude this possibility. Clinical sample integrity is one of the factors for decreasing assay sensitivity especially since RNA is unstable [8]. Degraded RNA or samples contaminated with RNases can lead to poor assay performance [20]. In our study, all samples were tested with real-time RT-PCR and RT-RAA in a very short time window to assure sample integrity. Thus, it can be assumed that each sample had a similar viral load when tested with both methods. A limitation represents the use of different extraction kits and amplification devices across sites. This adds on the variability of the clinical settings of the study. It is difficult to standardize protocols and equipment across healthcare laboratories. Nonetheless, all devices and equipment used were approved to be used for in vitro diagnostics. Nucleic acid extraction remains the bottleneck of molecular diagnostics. In this study, standardized kits were used to ensure RNA quality to validate the isothermal amplification assays. However, to implement point-of-need molecular tests, extraction protocols need to be simplified and user-friendly. Different rapid methods have been described to extract SARS-CoV-2 RNA. Combinations of detergent, heat and magnetic beads can be used for quick extraction [34,35]. RPA/RAA has been shown to be more tolerant against inhibitors in clinical samples [36], [37], [38]. Thus, rapid extraction methods are feasible with this technology, as shown in various studies [39,40]. However, further refinement is needed to enhance RNA purity and yield. In contrast, the RT-PCR is more intolerant to inhibitors from different matrices and requires highly purified RNA [41], [42], [43]. Further large clinical studies are needed to combine both rapid extraction and SARS-CoV-2 molecular assays on site.
Surprisingly, one test site discovered an unusual cluster of discordance on certain days. After retesting those samples, better results were achieved. Deviations in performance of diagnostic tests have been attributed to sample quality/quantity, settings, and operators [22]. The latter could hinder the homogeneity of sensitivity values between sites. Thus, correct sample handling is an essential factor to be considered. Although the influence of individual operators on the results of a diagnostic test cannot be fully avoided, certain actions could help to reduce such events. In this study, continuous and in-person training was not possible due to the COVID-19 pandemic and travel restrictions. Thus, the variability of the assay's results could be partially explained due to this difference in quality of training. Furthermore, attention to details regarding workflow should not be underestimated. For example, after careful troubleshooting, inaccuracies while transcribing records and test results, or the influence of reduced concentration while working after a certain hour of the day were reported. As a consequence, standardized operations and in-person training for staff are of utmost importance before operating diagnostic samples, in addition to quality control checks [44].
The findings of this study provide evidence for the importance of the suitcase lab as a deployable and feasible setup for accurate, sensitive and specific pathogen detection, particularly in low-resource settings. Furthermore, the RT-RAA method, especially based on the RdRP gene, is a promising on-site detection method for SARS-CoV-2 infection, overall showing higher accuracy than commercially available rapid antigen test and bypassing supply shortages. However, variations in assay sensitivity between sites revealed the importance of quality control and face to face training for the staff. The influence of global and regional disruptions should not be underestimated in large multi-country diagnostic trials. Additionally, sample handling by staff was regarded as a bottleneck for test performance. Continuous in-person training is an essential tool for successful diagnostic testing, in case of excellent quality and the quantity of kits and devices. These lessons learned should be considered when planning and performing large multi-country diagnostic clinical trials in poor resource settings.
Funding
This project is part of the European and Developing Countries Clinical Trials Partnership (EDCTP) programme supported by the European Union (grant number: RIA2020EF-2937-Africa_Suitcaselab). The funding sources were not involved in any way regarding the views expressed in this manuscript. The authors alone are responsible for the study design, data collection, analysis and interpretation, as well as the realization and submission of this manuscript.
Ethical approval
Ethical approval was granted from each study site before performing the testing: 00000879MSAS/DPRS/DR (National Committee for Research and Ethics in Public Health, Senegal); CERBM: 023-MSANP/SG/AMM/CERBM (Ethics Committee of Biomedical Research, Madagascar); CHRPE/AP/078/21 (Committee on Human Research, Publication and Ethics, School of Medical Sciences, Ghana); UI/EC/21/0010 (UI/ UCH Ethics Committee, Nigeria); ESP/CE/60/2021(School of Public Health Ethics Committee, Democratic Republic of the Congo); 2–12–20 (National Research Ethics Review Committee, Sudan); SBS-REC-883 (School of Biomedical Sciences Research and Ethics Committee, Uganda); 2021.05.13_eb_92 (Ethics Committee of Leipzig University, Germany).
Supplementary material
Supplementary file #1: additional information of the samples collected at each study site; Supplementary file #2: laboratory report forms; Supplementary file #3: sample flowcharts, results of each study site; Supplementary file #4: raw data of the mutational analysis; Supplementary file #5: summary of the mutational analysis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We are grateful to all involved laboratory technicians, students, and administrative staff for their participation and support throughout this study in Africa.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2023.105422.
Appendix. Supplementary materials
References
- 1.Organization, W.H. WHO announces COVID-19 outbreak a pandemic. 2020.
- 2.Rai P., Kumar B.K., Deekshit V.K., Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021;105:441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu G., Chou M.C., Tsai C.W. Lessons learned from the COVID-19 pandemic exposing the shortcomings of current supply chain operations: a long-term prescriptive offering. Sustainability. 2020;12:5858. [Google Scholar]
- 4.Iyengar K.P., Vaishya R., Bahl S., Vaish A. Impact of the coronavirus pandemic on the supply chain in healthcare. British J. Healthcare Manag. 2020;26:1–4. [Google Scholar]
- 5.Organization W.H. World Health Organization; 2021. Antigen-Detection in the Diagnosis of SARS-CoV-2 Infection: Interim Guidance, 6 October 2021. [Google Scholar]
- 6.Li J., Macdonald J., von Stetten F. Review: a comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 7.Li J., Macdonald J., von Stetten F. A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst. 2018;144:31–67. doi: 10.1039/c8an01621f. [DOI] [PubMed] [Google Scholar]
- 8.El Wahed A.A., Patel P., Maier M., Pietsch C., Rüster D., Böhlken-Fascher S., Kissenkötter J., Behrmann O., Frimpong M., Diagne M.M. Suitcase Lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal. Chem. 2021;93:2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Products C.f.P.M. Guideline on clinical evaluation of diagnostic agents. EMEA. 2009 CPMP/EWP/1119/98/Rev 1: 2009. [Google Scholar]
- 10.Ltd, M.S. Diagnostic test evaluation calculator, 2020. Available online: https://www.medcalc.org/calc/diagnostic_test.php (accessed on December).
- 11.Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front. Public Health. 2017;5:307. doi: 10.3389/fpubh.2017.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landis J.R., Koch G.G. The measurement of observer agreement for categorical data. Biometrics. 1977:159–174. [PubMed] [Google Scholar]
- 13.Weidmann M., Graf E., Lichterfeld D., Abd El Wahed A., Bekaert M. Efficient screening of long oligonucleotides against hundred thousands of SARS-CoV-2 genome sequences. Front. Virol. 2022;2 [Google Scholar]
- 14.Matthews Q., da Silva S.J.R., Norouzi M., Pena L.J., Pardee K. Adaptive, diverse and de-centralized diagnostics are key to the future of outbreak response. BMC Biol. 2020;18:1–5. doi: 10.1186/s12915-020-00891-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 16.Abd El Wahed A., Patel P., Faye O., Thaloengsok S., Heidenreich D., Matangkasombut P., Manopwisedjaroen K., Sakuntabhai A., Sall A.A., Hufert F.T., et al. Recombinase polymerase amplification assay for rapid diagnostics of dengue infection. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0129682. e0129682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abd El Wahed A., Sanabani S.S., Faye O., Pessôa R., Patriota J.V., Giorgi R.R., Patel P., Böhlken-Fascher S., Landt O., Niedrig M. Rapid molecular detection of Zika virus in acute-phase urine samples using the recombinase polymerase amplification assay. PLoS Curr. 2017:9. doi: 10.1371/currents.outbreaks.a7f1db2c7d66c3fc0ea0a774305d319e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faye O., Faye O., Soropogui B., Patel P., Abd El Wahed A., Loucoubar C., Fall G., Kiory D., Magassouba N.F., Keita S. Development and deployment of a rapid recombinase polymerase amplification Ebola virus detection assay in Guinea in 2015. Eurosurveillance. 2015;20:30053. doi: 10.2807/1560-7917.ES.2015.20.44.30053. [DOI] [PubMed] [Google Scholar]
- 19.Baba M.M., Bitew M., Fokam J., Lelo E.A., Ahidjo A., Asmamaw K., Beloumou G.A., Bulimo W.D., Buratti E., Chenwi C. Diagnostic performance of a colorimetric RT-LAMP for the identification of SARS-CoV-2: a multicenter prospective clinical evaluation in sub-Saharan Africa. EClinicalMedicine. 2021;40 doi: 10.1016/j.eclinm.2021.101101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagen, A. Laboratory supply shortages are impacting COVID-19 and non-COVID diagnostic testing. 2020.
- 21.Microbiology, A.S.f. Supply shortages impacting COVID-19 and non-COVID testing. 2021.
- 22.Ghosh P., Chowdhury R., Hossain M.E., Hossain F., Miah M., Rashid M.U., Baker J., Rahman M.Z., Rahman M., Ma X. Evaluation of recombinase-based isothermal amplification assays for point-of-need detection of SARS-CoV-2 in resource-limited settings. Int. J. Infect. Dis. 2022;114:105–111. doi: 10.1016/j.ijid.2021.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kretschmer A., Kossow A., Grüne B., Schildgen O., Mathes T., Schildgen V. False positive rapid antigen tests for SARS-CoV-2 in the real-world and their economic burden. J. Infect. 2022;84:248–288. doi: 10.1016/j.jinf.2021.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skittrall J.P., Wilson M., Smielewska A.A., Parmar S., Fortune M.D., Sparkes D., Curran M.D., Zhang H., Jalal H. Specificity and positive predictive value of SARS-CoV-2 nucleic acid amplification testing in a low-prevalence setting. Clin. Microbiol. Infect. 2021;27(469) doi: 10.1016/j.cmi.2020.10.003. e469-469e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wikramaratna P.S., Paton R.S., Ghafari M., Lourenço J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.50.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imöhl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star Sars-CoV-2 RT PCR kit. J. Virol. Methods. 2021;288 doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinnes J., Deeks J.J., Berhane S., Taylor M., Adriano A., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021 doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheiblauer H., Filomena A., Nitsche A., Puyskens A., Corman V.M., Drosten C., Zwirglmaier K., Lange C., Emmerich P., Müller M. Comparative sensitivity evaluation for 122 CE-marked rapid diagnostic tests for SARS-CoV-2 antigen, Germany, September 2020 to April 2021. Eurosurveillance. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.44.2100441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denzler, A., Jacobs, M.L., Witte, V., Schnitzler, P., Denkinger, C.M., Knop, M. Rapid comparative evaluation of SARS-CoV-2 rapid point-of-care antigen tests. medRxiv 2021. [DOI] [PMC free article] [PubMed]
- 30.Ciotti M., Maurici M., Pieri M., Andreoni M., Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. J. Med. Virol. 2021;93:2988–2991. doi: 10.1002/jmv.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Routsias J.G., Mavrouli M., Tsoplou P., Dioikitopoulou K., Tsakris A. Diagnostic performance of rapid antigen tests (RATs) for SARS-CoV-2 and their efficacy in monitoring the infectiousness of COVID-19 patients. Sci. Rep. 2021;11:1–9. doi: 10.1038/s41598-021-02197-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osterman A., Badell I., Basara E., Stern M., Kriesel F., Eletreby M., Öztan G.N., Huber M., Autenrieth H., Knabe R. Impaired detection of omicron by SARS-CoV-2 rapid antigen tests. Med. Microbiol. Immunol. 2022;211:105–117. doi: 10.1007/s00430-022-00730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toptan T., Eckermann L., Pfeiffer A.E., Hoehl S., Ciesek S., Drosten C., Corman V.M. Evaluation of a SARS-CoV-2 rapid antigen test: potential to help reduce community spread? J. Clin. Virol. 2021;135 doi: 10.1016/j.jcv.2020.104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fomsgaard A.S., Rosenstierne M.W. Copenhagen; Denmark: 2020. An Alternative Workflow For Molecular Detection of SARS-CoV-2 – Escape from the NA Extraction Kit-Shortage. MarchEurosurveillance 2020, 25, 2000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao Z., Cui H., Song W., Ru X., Zhou W., Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. BioRxiv. 2020 doi: 10.1016/j.talanta.2023.124479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ceruti A., Kobialka R.M., Ssekitoleko J., Okuni J.B., Blome S., Abd El Wahed A., Truyen U. Rapid extraction and detection of African swine fever virus DNA based on isothermal recombinase polymerase amplification assay. Viruses. 2021;13:1731. doi: 10.3390/v13091731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kersting S., Rausch V., Bier F.F., von Nickisch-Rosenegk M. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar. J. 2014;13:1–9. doi: 10.1186/1475-2875-13-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krõlov K., Frolova J., Tudoran O., Suhorutsenko J., Lehto T., Sibul H., Mäger I., Laanpere M., Tulp I., Langel Ü. Sensitive and rapid detection of Chlamydia trachomatis by recombinase polymerase amplification directly from urine samples. J. Mol. Diagn. 2014;16:127–135. doi: 10.1016/j.jmoldx.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 39.Chowdhury R., Ghosh P., Khan M.A.A., Hossain F., Faisal K., Nath R., Baker J., Wahed A.A.E., Maruf S., Nath P. Evaluation of rapid extraction methods coupled with a recombinase polymerase amplification assay for point-of-need diagnosis of post-kala-azar dermal leishmaniasis. Trop. Med. Infect. Dis. 2020;5:95. doi: 10.3390/tropicalmed5020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu J., Shen D., Dai T., Lu X., Xu H., Dou D. Rapid and equipment-free detection of Phytophthora capsici using lateral flow strip-based recombinase polymerase amplification assay. Lett. Appl. Microbiol. 2019;69:64–70. doi: 10.1111/lam.13166. [DOI] [PubMed] [Google Scholar]
- 41.Bessetti J. An introduction to PCR inhibitors. J. Microbiol. Methods. 2007;28:159–167. [Google Scholar]
- 42.Dalecka, B., Mezule, L. Study of potential PCR inhibitors in drinking water for Escherichia coli identification. 2018.
- 43.Sidstedt M., Hedman J., Romsos E.L., Waitara L., Wadsö L., Steffen C.R., Vallone P.M., Rådström P. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 2018;410:2569–2583. doi: 10.1007/s00216-018-0931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H., Li G., Zhao J., Li Y., Ai Y. An overview of nucleic acid testing for the novel coronavirus SARS-CoV-2. Front. Med. (Lausanne) 2021;7 doi: 10.3389/fmed.2020.571709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.