Abstract
In response to phosphate limitation, Saccharomyces cerevisiae induces transcription of a set of genes important for survival. A phosphate-responsive signal transduction pathway mediates this response by controlling the activity of the transcription factor Pho4. Three components of this signal transduction pathway resemble those used to regulate the eukaryotic cell cycle: a cyclin-dependent kinase (CDK), Pho85; a cyclin, Pho80; and a CDK inhibitor (CKI), Pho81. Pho81 forms a stable complex with Pho80-Pho85 under both high- and low-phosphate conditions, but it only inhibits the kinase when cells are starved for phosphate. Pho81 contains six tandem repeats of the ankyrin consensus domain homologous to the INK4 family of mammalian CKIs. INK4 proteins inhibit kinase activity through an interaction of the ankyrin repeats and the CDK subunits. Surprisingly, we find that a region of Pho81 containing 80 amino acids C terminal to the ankyrin repeats is necessary and sufficient for Pho81's CKI function. The ankyrin repeats of Pho81 appear to have no significant role in Pho81 inhibition. Our results suggest that Pho81 inhibits Pho80-Pho85 with a novel motif.
Cyclin-dependent kinases (CDKs) were originally identified as critical regulatory components of the eukaryotic cell division cycle (24, 25). They have also been implicated in the regulation of gene transcription, signal transduction, and other important cellular processes. To function as active kinases, CDKs require association with specific cyclin subunits (24). The activity of many CDK-cyclin complexes is controlled by CDK inhibitors (CKIs). There are two classes of CKIs in mammalian cells, both of which are important regulators of the cell cycle. The Cip/Kip family inhibits CDK4-, CDK6-, and CDK2-containing cyclin-CDK complexes involved in G1 and G1/S control (9), and the INK4 family inhibits cyclin D-CDK4 and cyclin D-CDK6 complexes involved in G1 control (30). There are three CKIs that have been identified in Saccharomyces cerevisiae (22): Sic1, which inhibits Clb5-Cdc28 and Clb6-Cdc28 complexes during G1 to control the timing of S phase (23); Far1, which inactivates Cln-Cdc28 complexes in the presence of mating pheromone (32); and Pho81, which inhibits the Pho80-Pho85 complex involved in a phosphate-responsive signal transduction pathway (34).
Studies of CKI inhibition and regulation have focused on mammalian CKIs. Members of the Cip/Kip family make extensive contacts with both the cyclin and the CDK subunits. Binding of Cip/Kip proteins to the cyclin may block the binding of substrates, while binding to the CDK subunit inhibits catalysis (30). Regulation of these CKIs occurs through transcriptional induction in response to intracellular and extracellular signals (3, 9). These CKIs are also negatively regulated through phosphorylation-induced, ubiquitin-mediated proteolysis (3, 9). The INK4 CKIs bind exclusively to CDK4 and CDK6, altering CDK structure so that the CDK is unable to bind to and be activated by its partner, cyclin D (3, 30). The INK4 CKIs are also capable of inhibiting intact cyclin D-CDK4 and cyclin D-CDK6 complexes, although there do not seem to be any significant contacts between the CKIs and cyclin D (33, 37). All members of the INK4 family consist of four tandem repeats of the ankyrin consensus domain, which make contacts with and inhibit the CDK subunits. The ankyrin consensus domain is a ubiquitous protein-protein interaction domain present in a number of proteins with different functions (36).
Although the structures and inhibitory mechanisms of mammalian CKIs have been well studied, less is known about these aspects of the CKIs in S. cerevisiae. We have chosen to study how the CKI Pho81 functions in the phosphate-responsive signal transduction pathway in S. cerevisiae. In response to phosphate limitation, budding yeast induces transcription of a set of genes important for its survival (29). The phosphate-responsive signal transduction pathway (the PHO pathway) mediates this response by controlling the activity of a transcription factor, Pho4 (19, 28). Three components of this signal transduction pathway resemble those used to regulate the eukaryotic cell cycle: a CDK, Pho 85 (43, 45); a cyclin, Pho80 (21, 42, 45); and a CKI, Pho81 (4, 27, 34). Under high-phosphate conditions, the Pho80-Pho85 cyclin-CDK complex phosphorylates and inactivates the transcription factor Pho4 (12). When yeast is starved for phosphate, the CDK inhibitor Pho81 inhibits Pho80-Pho85, resulting in accumulation of the unphosphorylated form of Pho4 and transcription of phosphate-responsive genes including PHO5, an acid phosphatase gene (12, 15). Because most of the PHO genes are not essential and the activity of the PHO pathway can be controlled by the level of phosphate in the medium, this pathway provides a useful system to investigate the function of these types of cell cycle regulatory proteins.
Pho81 contains six tandem repeats of the ankyrin consensus domain that are homologous to the INK4 family of mammalian CDK inhibitors, which includes an inhibitor of cyclin D-CDK4 and cyclin D-CDK6, p16 (27, 34, 37). A region of Pho81 containing six ankyrin repeats plus some neighboring sequence is sufficient to inhibit Pho80-Pho85 in vitro and partially complements the pho81Δ phenotype when expressed in vivo (27), suggesting that Pho81 might inhibit Pho80-Pho85 via the ankyrin repeats by a mechanism similar to that of p16. However, there are important differences between Pho81 and p16 with respect to regulation of the cyclin-CDK complex. p16 interacts directly and exclusively with CDK4 and CDK6. The activity of p16 is regulated primarily by transcriptional induction, and whenever p16 is present, it binds to and inhibits CDK4 and CDK6 (30, 33). In contrast, Pho81 forms a stable complex with Pho80-Pho85 under both high- and low-phosphate conditions, but it only inhibits the kinase under low-phosphate conditions, suggesting that the complex is regulated posttranslationally (34). Additionally, Pho81 can associate with the cyclin Pho80 and the Pho80-Pho85 complex, but not with Pho85 alone (34). It is not known how Pho81 inhibits Pho80-Pho85 in response to phosphate conditions.
In this report, our functional analysis of Pho81 has revealed a minimum domain of Pho81 containing 80 amino acids (aa) (aa 645 to 724) that is necessary and sufficient for Pho81 inhibition of Pho80-Pho85 in response to phosphate conditions. This Pho81 minimum domain resides C terminal to the six ankyrin repeats that appear to have no significant role in the inhibition of Pho80-Pho85. In contrast to p16, which inhibits the kinase through interactions of the ankyrin repeats with the CDK subunit, our findings suggest a novel inhibitory mechanism of Pho81 as a CKI.
MATERIALS AND METHODS
Strains, plasmids, media, and general methods.
Strains of S. cerevisiae used in this study are listed in Table 1, and plasmids are listed in Table 2. Standard rich (YEPD) and synthetic (SD) media were used as described (38). No-phosphate medium is SD medium consisting of yeast nitrogen base lacking inorganic phosphate. Yeast nitrogen base lacking inorganic phosphate was made with components described in the Difco manual, except that potassium phosphate was replaced by the same amount of potassium chloride. Low-phosphate medium was made from no-phosphate medium with the addition of 10 mg of potassium phosphate/liter. High-phosphate medium was made from no-phosphate medium with the addition of 1.5 g of potassium phosphate/liter. Yeast cultures were grown at 30°C for all experiments. Yeast transformations were performed by the lithium acetate method (7).
TABLE 1.
Strains of S. cerevisiae used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| K699 | mataade2-1 trp1-1 can1-100 leu2-3,112 his3-11,15 ura3 GAL+ | 24a |
| EY0313 | K699 PHO81-GFP integrated at the PHO81 locus | This work |
| EY0518 | K699 pho81Δ::HIS3 | This work |
| EY0520 | K699 pho81Δ::HIS3 pho80Δ::H1S3 | This work |
| EY0607 | K699 pho81Δ::HIS3 pho80Δ::HIS3 pho85Δ::HIS3 | This work |
| EY0622 | K699 pho81Δ::HIS3 pho3Δ::LEU2 | This work |
| EY0134 | K699 pho80Δ::HIS3 | This work |
| EY0619 | K699 pho3Δ::LEU2 | This work |
| EY0323 | K699 pho80Δ::HIS3 PHO81c integrated at the PHO81 locus | This work |
| EY0439 | K699 PHO81c integrated at the PHO81 locus and PHO5pr-CAN1 integrated at the CAN1 locus | This work |
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant genotype | Source |
|---|---|---|
| EB1339 | pPHO4pr-PHO81-GFP in pRS314 | This work |
| EB0049 | pGPD1pr-PHO80 in pRS426 | This work |
| EB0082 | pGPD1pr-PHO85-HA in pRS426 | This work |
| EB1352 | pPHO80pr-PHO80 in pRS316 with a BglII site introduced at the start codon | This work |
| EB1086 | Same as EB1352 except with another BglII site introduced at the stop codon | This work |
| EB1353 | pPHO80pr-pho80-59 in pRS316 (containing pho80 allele 59) | This work |
| EB1354 | pPHO80pr-pho80-60 in pRS316 (containing pho80 allele 60) | This work |
| EB1355 | pPHO80pr-pho80-126 in pRS316 (containing pho80 allele 126) | This work |
| EB1356 | pho80-126 in pSBETA | This work |
| EB0036 | GST-PHO85 in pGEX-2T | This work |
| EB1255 | PHO81 in pSBETA | This work |
| EB1357 | PHO81 (aa 1–376)-zz in pET16b | This work |
| EB1358 | PHO81 (aa 725–1179) in pSBETA | This work |
| EB0121 | His6-PHO81 (aa 400–724) in pET16b | This work |
| EB1359 | His6-PHO81 (aa 400–660) in pET16b | This work |
| EB1360 | His6-PHO81 (aa 645–724) in pET16b | This work |
| EB1361 | pADH1pr-PHO81-Py2 in pRS316 | This work |
| EB0730 | pADH1pr-Py2 in pRS316 | This work |
| EB1362 | pADH1pr-PHO81(aa 400–724)-Py2 in pRS316 | This work |
| EB1363 | pADH1pr-PHO81(aa 400–660)-Py2 in pRS316 | This work |
| EB1364 | pADH1pr-PHO81(aa 645–724)-Py2 in pRS316 | This work |
Plasmid construction.
Plasmids containing pho80 mutants were obtained as described below (see “PCR mutagenesis and isolation of pho80 mutants”). PHO81 deletion constructs were made by PCR amplification of selected regions of the PHO81 open reading frame. Vectors for expression in yeast are generally based on the pRS316 series (40); a low-copy-number CEN-ARS Escherichia coli-S. cerevisiae shuttle vector. Each PHO81 construct for in vitro transcription-translation was made using a 5′ primer containing an NdeI restriction endonuclease site and a 3′ primer containing a stop codon immediately following a BamHI restriction endonuclease site. Each PHO81 construct for in vivo complementation was made using a 5′ primer containing a ClaI restriction endonuclease site and a 3′ primer containing a stop codon immediately following a KpnI restriction endonuclease site. Plasmids containing pho81 mutants were constructed as described below (see “Site-directed mutagenesis and generation of pho81 mutants”). Plasmids based on pSBETA (10) were constructed by cloning coding sequences into the NdeI and BamHI sites of the vector. Plasmids containing PCR products were confirmed by sequencing analysis (ABI Prism). Plasmid-expressed, tagged genes were tested for their ability to complement the null phenotype of the appropriate deletion strains. The Py2 (34) and zz (14) tags are as previously described.
Fluorescence microscopy.
For all microscopy experiments, cells were freshly transformed with appropriate plasmids. Cells were first grown overnight to an optical density at 600 nm (OD600) of 0.6 to 1.0 in SD medium supplemented with amino acids. The overnight cultures were pelleted and washed with sterile water. Each pellet was then diluted in high-phosphate medium or no-phosphate medium and grown for 2 to 3 h to an OD600 of 0.1 to 0.3. When indicated, cycloheximide was added to a final concentration of 0.1 mg/ml. To stain cells with DAPI (4′,6′-diamidino-2-phenylindole), each washed pellet of overnight cultures was diluted into high-phosphate medium with 0.5 mg of DAPI per liter and grown for 2 h to an OD600 of 0.4 to 0.6. The cultures were then washed, rediluted, and grown for 2 to 3 h to an OD600 of 0.1 to 0.3 in high-phosphate medium or no-phosphate medium, each with 0.5 mg of DAPI per liter. Two microliters of the culture was placed on a microscope slide and examined directly by fluorescence microscopy. Liquid acid phosphatase assays were performed as described (15) for the control cultures to confirm that cells were starved for phosphate. All fluorescence images were collected with an Olympus BX-60 microscope with a charge-coupled device camera (Photometrics) using identical exposures and settings.
PCR mutagenesis and isolation of pho80 mutants.
PCR mutagenesis was performed with 1.38 mM Mg2+ and 0.12 mM Mn2+ as described (20). The plasmid EB1086 was used as the template for PCR mutagenesis. EB1086 is a pRS316 (40) derivative containing a copy of the PHO80 gene under the control of the PHO80 promoter, with BglII restriction endonuclease sites introduced at the start and stop codons. The primers used were 5′-GCCCCAAGCCATCATAAATAGCC and 5′-ATTAACCCTCACTAAAGGGA. The primer pair was designed to amplify the PHO80 open reading frame plus 285 bases of sequence upstream of the start codon and 187 bases downstream of the stop codon. The gapped vector was the larger fragment from the BglII digest of EB1086. The gel-purified PCR products and the gapped vector were cotransformed into a PHO81c pPHO5-CAN1 strain (EY0439), which carries the CAN1 gene (7) under the control of the PHO5 promoter. PHO81c is a dominant point mutation in the PHO81 gene that results in constitutive activation of the CDK inhibitor Pho81 (due to the mutation S161F) (41). The transformed yeast cells were plated onto SD plates lacking uracil and arginine containing 10 mg of canavanine per ml to select for cells containing Pho80 mutants. Acid phosphatase plate assays were used to identify Pho80 mutants that suppress the Pho81c Pho phenotype (44). The pho80 mutants were sequenced to determine the nature of mutations. Since the introduced BglII site at the stop codon of Pho80 affected the interaction of Pho80 and an anti-Pho80 peptide antibody, the SacII/SalI fragment of each pho80 mutant was replaced with the wild-type sequence from EB1352.
Site-directed mutagenesis and generation of pho81 mutants.
All pho81 mutants were generated using site-directed mutagenesis as previously described (16). The mutagenesis was performed with single-stranded pRS316 constructs carrying the coding sequences for the Pho81 minimum region (amino acids 645 to 724) or full-length Pho81 using the same set of oligonucleotides. Each oligonucleotide was designed to replace three adjacent amino acids in the Pho81 minimum region by alanine in a nonoverlapping manner. The DNA sequence coding for three alanines was designed to introduce a NotI restriction endonuclease site for the primary analysis of potential mutants. Mutations were confirmed by sequencing analysis.
Recombinant protein expression and purification.
GST-Pho85 (EB0036) was expressed alone or coexpressed with Pho80 (EB1076) (10) or Pho80-126 (EB1356) in BL21(DE3). Cells were grown in Luria-Bertani medium with 50 μg of carbenecillin per ml and 70 μg of kanamycin per ml (Pho80 coexpression only) to an OD600 of 0.4 to 0.6 and induced for 18 h at 24°C with 40 μM IPTG (isopropyl-β-d-thiogalactopyanoside). Cells were harvested at 4°C, frozen at −20°C, and thawed at 4°C. Cells were resuspended in 30 ml of lysis buffer (10% [vol/vol] glycerol, 50 mM Tris-HCl [pH 7.5], 0.3 M NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 mM benzamidine) per liter of culture. The cell suspension was sonicated as described for the purification of Pho81-His6, and the lysate was then spun for 15 min at 16,000 rpm with in an SS34 rotor at 4°C. DTT was added to a final concentration of 10 mM, and the lysate was incubated in batch with 1 ml of glutathione (GSH)-agarose (Sigma) per liter of cells for 3 to 4 h at 4°C. The resin was spun down in a clinical centrifuge for 5 min, the flowthrough was removed, and the resin was poured into a 10-ml plastic disposable column (Pierce). The resin was washed with 45 ml of wash buffer (10% glycerol, 50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM DTT, 1 mM EDTA, 1 mM PMSF, 2 mM benzamidine), and bound protein was eluted with 1-ml aliquots of wash buffer containing 5 mM GSH. Fractions containing purified glutathione S-transferase (GST)-Pho85, GST-Pho85-Pho80, or GST–Pho85–Pho80-126 were pooled and dialyzed into storage buffer (10% glycerol, 50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF, 2 mM benzamidine). Yield was >5 mg of purified protein per liter. GST was also purified by the same method. The purity of all preparations was estimated to be >95% by Coomassie staining.
Binding assays using in vitro transcription-translation products.
All binding reactions were performed with siliconized tubes. Plasmids used for in vitro transcription-translation contain sequences expressing different regions of Pho81 under the control of the T7 promoter (Table 2). These plasmids were transcribed and translated in vitro in the presence of [35S]methionine using rabbit reticulocyte lysate as described by the manufacturer (Promega TNT kit). The transcription-translation reaction mixtures were incubated at 30°C for 90 min and used immediately for the binding assay. The average yield for full-length Pho81 was 2.5 × 10−16 mol/μl.
For each binding reaction mixture, 10 μl of TNT reaction mixture was added to 40 μl of binding buffer (10% [vol/vol] glycerol, 50 mM Tris-HCl [pH 7.5], 0.15 M NaCl, 0.01% [vol/vol] NP-40, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine, 0.1 mg of bovine serum albumin [Boehringer Mannheim] per ml) containing GST-Pho85-Pho80, GST-Pho85-Pho80-126, or GST. For each deletion construct of Pho81, three binding reactions were carried out in which 200, 50, or 10 nM kinase complex or control GST was added. All binding reaction mixtures were incubated at room temperature (23 to 25°C) for 1 h and were then transferred to a new tube containing 10 μl of GSH-agarose (Sigma) equilibrated in the binding buffer. This reaction mixture was incubated at room temperature with rotation for 1 h and washed quickly three times with 0.8 ml of binding buffer. Samples were transferred to new tubes after the last wash, and all supernatant was removed. Pellets were resuspended in 20 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and boiled, and the suspensions were loaded on gels. The quantitation of 35S-labeled Pho81 was performed with a PhosphorImager (Molecular Dynamics).
Preparation of antibodies and antibody beads.
Polyclonal serum recognizing Pho85 was obtained (34) and was then affinity purified from immunoblots (8). Peptide antibodies were made that recognize the last 11 amino acids of Pho80 (AHIYNKRSKPD) and Pho81 (ELLFENNIDM). Peptides were synthesized with an N-terminal cysteine, coupled to keyhole limpet hemocyanin and injected into rabbits (BAbCo). Antibodies were affinity purified from rabbit serum by being bound to a peptide-agarose column (Affigel) and eluted with 0.1 M glycine, pH 2.0. Monoclonal anti-Py2 was purified with protein G columns (8).
Anti-Pho85 beads and anti-Pho80 beads were prepared by adding 5 μg of each antibody per 10 μl of protein A-Sepharose beads (Pharmacia) and incubating the mixture at room temperature with rotation for 1 h. The mixture was washed three times with phosphate-buffered saline (PBS) (10× volume) and was then equilibrated in the appropriate binding buffer.
Preparation of cell extracts and coimmunoprecipitation experiments.
Cells were grown in high- or low-phosphate medium as indicated, and extracts were prepared as described (37), except with a low-salt buffer (10% [vol/vol] glycerol, 250 mM Hepes [pH 7.5], 0.1 M NaCl, 0.01% [vol/vol] NP-40, 1 mM β-mercaptoethanol, 1 mM EDTA, 1 mM PMSF, 2 mM benzamidine, 10 mM NaF, 30 mM β-glycerophosphate, 1-μg/ml pepstatin A, 1-μg/ml leupeptin, and 10 nM calyculin A).
For examining levels of protein expression, 50 to 100 μg of each cell extract was analyzed by SDS-PAGE, followed by Western blotting. In the coimmunoprecipitation experiments, 3 mg of each cell extract was mixed with approximately 1.25 μg of anti-Pho80 antibody in 300 μl of low-salt buffer. The mixture was incubated at 4°C with rotation for 1 h and was then spun for 10 min at 3,000 rpm in a Microfuge at 4°C. The supernatant was transferred to a new tube containing 15 μl of protein A-Sepharose beads equilibrated in PBS and incubated at 4°C with rotation for 1 h. The mixture was washed four times at 4°C with 0.5 ml of PBS supplemented with 0.1% NP-40, 1 mM PMSF, 2 mM benzamidine, 10 mM NaF, and 30 mM β-glycerophosphate. After the first wash, all material was transferred to a new tube, and the third wash was done with rotation for 5 min. Pellets were resuspended in 20 μl of SDS-PAGE sample buffer and boiled, and the suspensions were loaded on gels. All gels were analyzed by Western blotting using anti-Pho80 or anti-Py2 antibodies.
RESULTS
Nuclear localization of Pho81 is not regulated by phosphate conditions and is dependent on Pho80.
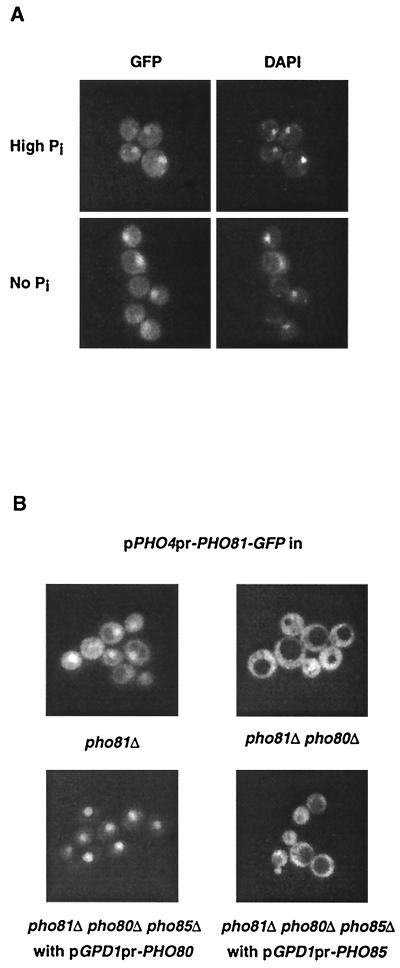
Many proteins are regulated through control of their subcellular localization (13), including the CKI Far1. To investigate the subcellular localization of Pho81, we tagged Pho81 with the green fluorescent protein (GFP) and followed its localization in live cells grown under high- and low-phosphate conditions. This Pho81-GFP fusion complemented the uninducible Pho5 phenotype of a pho81Δ mutant (data not shown). Pho81 transcription is under the control of the PHO pathway and predominantly localized to the nucleus under both high- and low-phosphate conditions (Fig. 1A), suggesting that the activity of Pho81 is not controlled through regulation of its nuclear localization. We also observed Pho81-GFP in the cytoplasm and at the plasma membrane under low-phosphate conditions (data not shown). The fact that Pho81-GFP localizes to the nucleus under high- and low-phosphate conditions is expected, given that Pho80-Pho85 is localized to the nucleus under these conditions (14).
FIG. 1.
Nuclear localization of Pho81. (A) Pho81 is localized to the nucleus independent of phosphate conditions. Yeast strain EY0313 (integrated Pho81-GFP) was grown in high- or no-phosphate medium, and the live cells were examined by fluorescence microscopy. The nuclear disposition of GFP-based fluorescence was confirmed by its colocalization with DAPI-stained nuclear DNA. (B) Nuclear localization of Pho81 is dependent on Pho80 but not Pho85. pho81Δ (EY0518) cells were transformed with pPHO4pr-PHO81-GFP, pho81Δ pho80Δ (EY0520) cells were transformed with pPHO4pr-PHO81-GFP, and pho81 Δpho80Δ pho85Δ (EY0607) cells were cotransformed with pPHO4pr-PHO81-GFP and pGPD1pr-PHO80 or pGPD1pr-PHO85. All strains were grown in high-phosphate medium and were examined by fluorescence microscopy. Similar results were obtained when the strains were grown in low-phosphate medium (data not shown).
We wanted to test whether the localization of Pho81 to the nucleus depended on Pho80 or Pho85. This analysis was complicated by the fact that Pho81-GFP is strongly induced in pho80Δ and pho85Δ mutants because Pho4 is constitutively active (43, 44). When Pho81-GFP was expressed under the control of the PHO81 promoter, pho80Δ and pho85Δ mutant cells fluoresced bright green, and Pho81-GFP appeared completely cytoplasmic under high- and low-phosphate conditions (data not shown). Since the high level of expression of Pho81-GFP under these conditions could obscure our ability to detect Pho81-GFP in different parts of the cell, we expressed Pho81-GFP from the constitutive PHO4 promoter. Expression of Pho81-GFP from this promoter results in levels of fluorescence midway between the levels seen with high- and low-phosphate concentrations when Pho81-GFP was expressed from the PHO81 promoter, but expression was high enough to complement the pho81Δ mutant phenotype (data not shown). Interestingly, Pho81-GFP expressed under the control of the PHO4 promoter in a pho80Δ strain was cytoplasmic (Fig. 1B), suggesting that Pho80 is necessary for nuclear localization of Pho81-GFP. Additionally, when we overexpressed Pho80 using the GPD1 promoter in a pho80Δ pho85Δ strain, all of the Pho81-GFP was nuclear (Fig. 1B), indicating that overexpression of Pho80 is sufficient to transport Pho81-GFP into the nucleus. In contrast, Pho81-GFP was still cytoplasmic when Pho85 was overexpressed using the GPD1 promoter in a pho80Δ pho85Δ strain. These findings demonstrate that localization of Pho81 to the nucleus is dependent on Pho80 but not on Pho85. This is consistent with previous findings that Pho81 can be coimmunoprecipitated with Pho80 in a yeast strain lacking Pho85 but failed to be coimmunoprecipitated with Pho85 in a yeast strain lacking Pho80 (34).
PCR mutagenesis and isolation of pho80 mutants.
Since the interaction of Pho81 with the Pho80-Pho85 complex involves significant contacts with Pho80 (S. Huang and E. K. O'Shea, unpublished observations; 34), we wished to identify the domains of Pho80 required for the interaction with Pho81 in vivo. We performed a genetic selection for Pho80 mutants that cannot be inhibited by a Pho81 mutant but that are able to form functional kinase complexes with Pho85 and phosphorylate Pho4. PHO81c is a dominant point mutation in the PHO81 gene that results in constitutive activation of the CKI Pho81 (41). Whereas wild-type Pho81 only inhibits Pho80-Pho85 under low-phosphate conditions, Pho81c inhibits Pho80-Pho85 under both conditions, resulting in constitutive production of the acid phosphatase Pho5. There might be two types of Pho80 mutation that could suppress Pho81c: a mutation that prevents Pho81c binding and therefore affects inhibition of Pho80-Pho85 and a mutation that does not affect Pho81c binding but prevents Pho81c from inhibiting the kinase.
In this selection, PCR-mutagenized PHO80 was introduced into a PHO81c yeast strain carrying the CAN1 gene under the control of the promoter of the phosphate-responsive gene PHO5. The CAN1 gene encodes an arginine permease that will uptake the toxic arginine analog, canavanine (7). Under high-phosphate conditions, wild-type expression of Pho80-Pho85 is inhibited by Pho81c, resulting in the expression of PHO5 and CAN1 and in sensitivity to canavanine. Strains carrying loss-of-function mutations in PHO80 are also sensitive to canavanine. In contrast, Pho80 mutants that cannot be inhibited by Pho81c but still have full kinase activity will be able to repress transcription of PHO5 and CAN1, resulting in survival on medium containing canavanine.
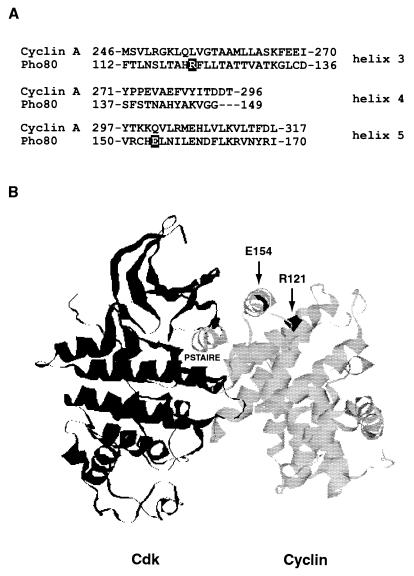
This selection yielded several interesting pho80 mutants. The two most frequent mutations result in an arginine-to-lysine change at residue 121 (R121K) and a glutamic acid-to-valine substitution at residue 154 (E154V) (Fig. 2A). Pho80-59 (E154V), Pho80-60 (R121K), and Pho80-126 (R121, S69T, and F81L) are three representative mutants used in the following studies. Interestingly, residues R121 and E154 are conserved among Pho80 and all other cyclins in the Pcl family (1). Based on sequence alignment with cyclin A (Fig. 2B) (2, 11), these two residues are predicted to be located on a solvent-exposed region of helices 3 and 5. Helices 3 and 5 of the cyclin box interact with conserved residues in the PSTAIRE helix of the CDK (Fig. 2B) (11).
FIG. 2.
Sequence and structural analysis of pho80 mutants isolated from PCR mutagenesis. (A) Protein sequence alignment of cyclin A and Pho80 (2). From the sequencing analysis, the two most frequent Pho80 mutations are an arginine (R)-to-lysine (K) change at residue 121 and a glutamic acid (E)-to-valine (V) substitution at residue 154. R121 and E154 (highlighted in black) are localized to helices 3 and 5 of the cyclin box (2), respectively. (B) Possible physical locations of R121 and E154 of Pho80, based on the crystal structure of Cdk2-cyclin A and the protein sequence alignment of cyclin A and Pho80 (2, 11). The residues of Cdk2 are in black, except that the PSTAIRE helix is colored gray; and the residues of cyclin A are in gray, except that the two equivalent residues of R121 and E154 from Pho80 are colored black.
Characterization of the pho80 mutants.
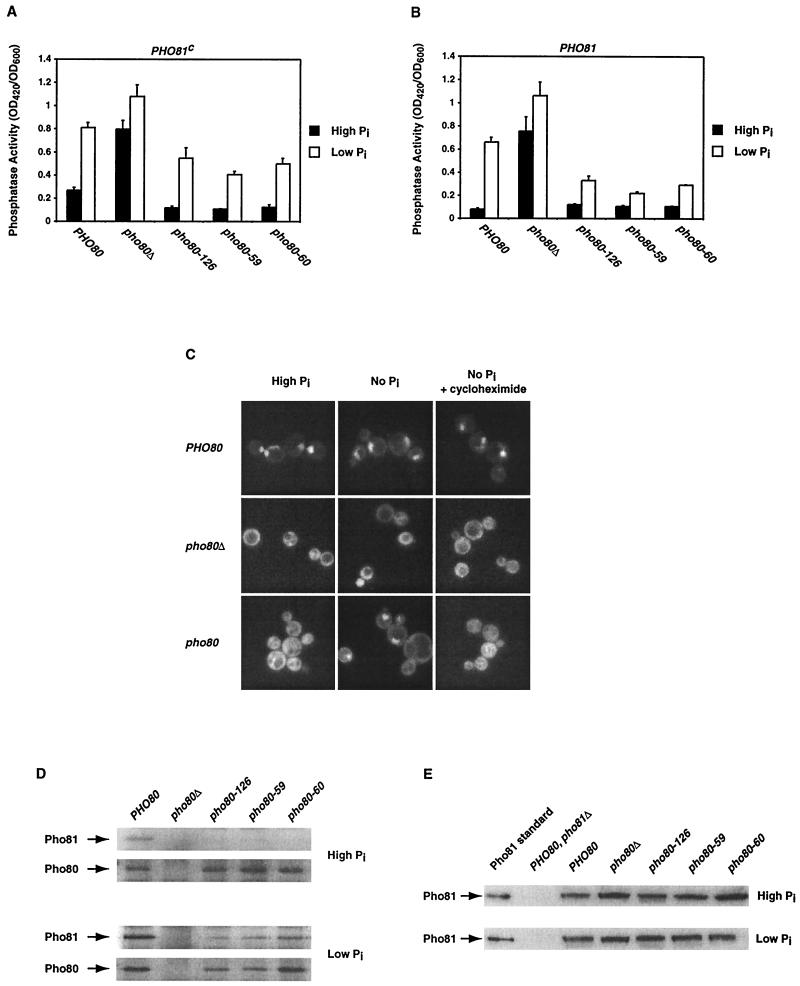
To further characterize the effect of the PHO80 mutations, we quantitatively examined the PHO phenotype of yeast strains expressing the Pho80 mutants in a PHO81c or PHO81 background, when grown in high- and low-phosphate medium. In the PHO81c yeast strain background, cells containing the Pho80 mutants had less Pho5 activity than cells containing wild-type Pho80 when grown under either high- or low-phosphate conditions (Fig. 3A). In the PHO81 strain background, cells containing the Pho80 mutants had less Pho5 activity than cells containing wild-type Pho80, only when grown in low-phosphate medium (Fig. 3B). These results indicate that the Pho80 mutants are not as effectively inhibited by Pho81.
FIG. 3.
Characterization of the pho80 mutants isolated from the PCR mutagenesis. (A) In the PHO81c background (a dominant point mutation in the PHO81 gene that results in constitutive activation of the CKI Pho81), pho80 mutant strains have reduced phosphatase activity compared to a PHO81c strain containing wild-type PHO80 grown in either high- or low-phosphate medium. pho80Δ PHO81c (EY0323) cells were transformed with pPHO80pr-PHO80, pRS316, pPHO80pr-pho80-126, pPHO80pr-pho80-50, pPHO80pr-pho80-60. The phosphatase activity was then quan- titated from cells grown in high-phosphate (solid column) and low-phosphate (open column) media. (B) In a wild-type PHO81 background, pho80 mutant strains have reduced phosphatase activity under low- but not high-phosphate conditions. pho80Δ PHO81 (EY0134) cells were transformed with pPHO80pr-PHO80, pRS316, pPHO80pr-pho80-126, pPHO80pr-pho80-50, or pPHO80pr-pho80-60. The phosphatase activity was then quantitated from cells grown in high-phosphate (solid column) and low-phosphate (open column) media. (C) Pho81 is localized to the cytoplasm in the pho80 yeast strain grown in high-phosphate medium but relocalized within the nucleus when grown in low-phosphate medium, and this relocalization is blocked in the presence of cycloheximide. pho81Δ pho80Δ (EY0520) cells were cotransformed with pPHO4pr-PHO81-GFP and pPHO80pr-PHO80, pRS316, or pPHO80pr-pho80-126. Strains were grown in high- no-, or low-phosphate medium with cycloheximide, followed by examination by fluorescence microscopy. (D) Pho81 fails to coimmunoprecipitate with mutant Pho80 from yeast grown in high- but not in low-phosphate medium. pho80Δ (EY0134) cells were transformed with pPHO80pr-PHO80, pRS316, pPHO80pr-pho80-126, pPHO80pr-pho80-59, or pPHO80pr-pho80-60. Yeast extracts were made from cells containing wild-type or mutant Pho80 grown in high- or low-phosphate medium. An anti-Pho80 antibody was then used to immunoprecipitate Pho80 from yeast extracts, and the immunoprecipitated proteins were analyzed by SDS-PAGE, followed by Western blotting with anti-Pho81 or anti-Pho80 antibodies. (E) Pho81 protein levels are similar in yeast strains expressing wild-type or mutant Pho80. A total of 50 μg of the same yeast extracts shown in Fig. 3D were analyzed by SDS-PAGE, followed by Western blotting with anti-Pho81 antibody. As controls, a recombinant Pho81 standard and yeast extracts from cells lacking Pho81 (EY0518) grown in high- and low-phosphate media were included in each blot.
There might be two ways to impair Pho81 inhibition: by preventing Pho81 binding or by affecting the ability of Pho81 to inhibit the kinase without interfacing with binding. If the Pho80 mutants are defective in binding to Pho81, the subcellular localization of Pho81 might be altered in yeast cells containing these Pho80 mutants. We found that Pho81-GFP was cytoplasmic in the pho80 mutant cells when grown in high-phosphate medium (Fig. 3C, High Pi). However, when the same strains were shifted from high- to no-phosphate medium, Pho81-GFP was relocalized to the nucleus in the majority of the cells (Fig. 3C, No Pi). These results suggest that Pho81 does not efficiently bind to the Pho80 mutants in high-phosphate medium, but under no-phosphate conditions Pho81 regains an interaction with Pho80 that is sufficient to relocalize it to the nucleus. Since Pho80 localizes to the nucleus independent of Pho81 and Pho85 (data not shown), relocalization of Pho81 in the pho80 mutant background in low-phosphate medium might be the result of binding to newly synthesized cytoplasmic Pho80. If this is true, inhibition of new protein synthesis should prevent Pho81 in the pho80 background from being relocalized in response to low-phosphate conditions. We found that Pho81-GFP no longer relocalized to the nucleus when Pho80 mutants were shifted from high- to no-phosphate medium in the presence of cycloheximide (Fig. 3C, No Pi+ cycloheximide).
To further examine the hypothesis that the Pho80 mutants are defective in binding to Pho81 in high-phosphate medium but regain the interaction when starved for phosphate, we directly investigated the interactions of the Pho80 mutants with Pho81 in vivo. Yeast extracts were made from cells containing wild-type or mutant Pho80 grown in high- or low-phosphate medium. Pho80 was immunoprecipitated from yeast extracts, and the immunoprecipitated proteins were analyzed by SDS-PAGE, followed by Western blotting (Fig. 3D). Pho81 was not efficiently coimmunoprecipitated with Pho80 mutants from the high-phosphate extracts but was coimmunoprecipitated from the low-phosphate extracts. These differences in the ability of Pho81 to be coimmunoprecipitated with wild-type and mutant Pho80 were not due to the Pho81 protein levels, which were similar in both conditions (Fig. 3E). These results indicate that the Pho80 mutants are partially defective in binding to Pho81 and that the interaction between Pho81 and the Pho80 mutants is significantly increased when cells are starved for phosphate.
Deletion analysis of Pho81 using an in vitro transcription-translation system.
Since we had identified Pho80 residues important for interaction with Pho81, we also wished to identify domains of Pho81 required for binding to the Pho80-Pho85 complex. We established a system to study binding that used in vitro-transcribed and -translated Pho81 and GST-Pho85-Pho80 expressed and purified from E. coli. Radioactively labeled Pho81 was incubated either with wild-type GST-Pho85-Pho80 or with mutant GST-Pho85-Pho80 containing a Pho80 binding mutant (Pho80-126 [R121] isolated from the selection described above. Both wild-type and mutant GST-Pho85-Pho80 complexes have full kinase activity in vitro (data not shown). The incubated reactions were bound to GSH-agarose beads, and bound Pho81 was analyzed on SDS-PAGE gels. We found that full-length Pho81 has significantly higher affinity for wild-type kinase than for the mutant kinase (Fig. 4A). This result suggests that this in vitro system mimics the in vivo state of high-phosphate conditions, since Pho81 does not bind to the mutant kinase under high-phosphate conditions but binds under low-phosphate conditions in vivo.
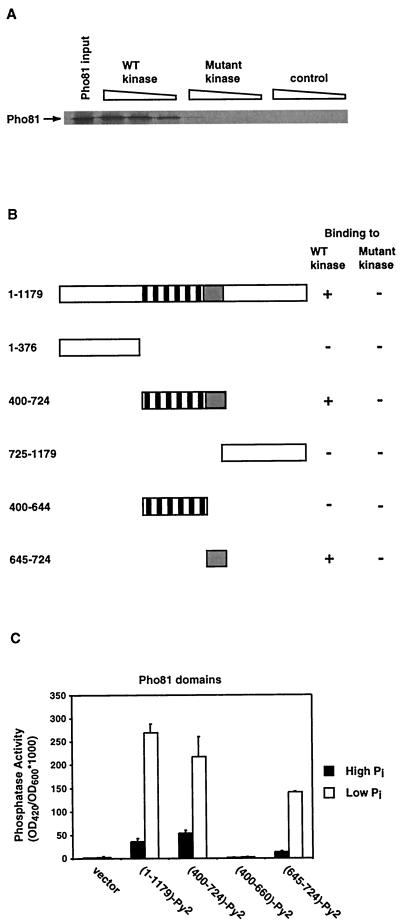
FIG. 4.
Identification of the Pho81 minimum domain. (A) In vitro-translated full-length Pho81 has significantly higher affinity for wild-type kinase than for the mutant kinase. Radioactively labeled Pho81 was made in a eukaryotic in vitro transcription-translation system (Promega TNT kit) and was then incubated either with wild-type GST-Pho85-Pho80 or with mutant GST-Pho85-Pho80 (Pho80-126) expressed and purified from E. coli. The incubated reaction mixtures were bound to GSH-agarose beads, and bound Pho81 was analyzed with SDS-PAGE gels. (B) Summary of the binding of different Pho81 domains to Pho80-Pho85. Deletion constructs containing different domains of Pho81 were in vitro transcribed and translated and then tested for their ability to bind GST-Pho85-Pho80 and GST-Pho85-Pho80 (Pho80-126). (C) The Pho81 minimum domain aa (645 to 724) is sufficient for Pho81 function in vivo. pho81Δpho3Δ (EY0622) cells were transformed with pADH1pr-Py2 (vector), pADH1pr-PHO81(1–1179 a.a.)-Py2 (full-length Pho81), pADH1pr-PHO81(400–724 a.a.)-Py2, pADH1pr-PHO81(400–660 a.a.)-Py2, or pADH1pr-PHO81(645–724 a.a.)-Py2. The phosphatase activity was then quantitated from cells grown in high-phosphate (solid column) and low-phosphate (open column) media.
To identify the domains of Pho81 involved in binding to the kinase complex, we made deletion constructs containing different domains of Pho81 and tested their ability to bind Pho80-Pho85 with the in vitro system (Fig. 4B). We found that neither the N-terminal nor the C-terminal domain of Pho81 was able to bind the kinase complex. The domain containing the six ankyrin repeats and 80 additional aa C terminal to the repeats appeared to have the same binding characteristics as full-length Pho81. These findings are not surprising, because this domain (aa 400 to 724), but not the N-terminal and C-terminal domains, was able to partially complement the uninducible Pho5 phenotype of a pho81Δ mutant in low-phosphate medium (27). Surprisingly, the domain containing only the six ankyrin repeats was not able to bind the kinase complexes. In contrast, we found that the small domain containing the 80 aa C terminal to the ankyrin repeats was able to bind. These results suggest that these 80 aa, but not the ankyrin repeats, are involved in binding to the kinase.
The Pho81 minimum domain is sufficient for Pho81 function in vivo.
It has been shown that ankyrin repeats of other CKIs are required to inhibit CDK function (33). We hypothesized that the Pho81 ankyrin repeats are required for Pho81 inhibition, whereas the 80 aa C terminal to the ankyrin repeats are involved in stable binding to the kinase. To test this hypothesis, we examined the PHO phenotype of yeast strains expressing different domains of Pho81 when grown in high- and low-phosphate media (Fig. 4C). As expected, yeast cells expressing full-length Pho81 or the domain (aa 400 to 724) containing the ankyrin repeats and the additional 80 aa induce Pho5 expression in response to phosphate starvation. Yeast cells expressing the domain (aa 400 to 660) containing only the ankyrin repeats have no detectable Pho5 activity under either high- or low-phosphate conditions. Surprisingly, yeast cells expressing the 80 aa (aa 645 to 724) C- terminal to the ankyrin repeats induce Pho5 expression in response to phosphate starvation. The inability of the ankyrin repeat domain to function is not due to a lack of expression, because this domain is present in cells at a level similar to full-length Pho81 and the other domains (data not shown). We now refer to this 80-aa region as the Pho81 minimum domain, since it is sufficient to bind to Pho80-Pho85 in vitro and to induce Pho5 expression in vivo in response to phosphate starvation.
The Pho81 minimum domain is necessary for Pho81 function in vivo.
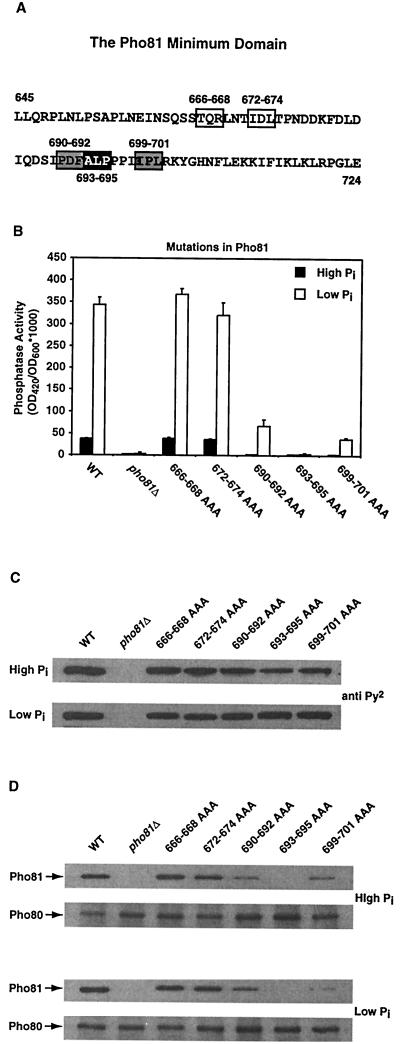
Although we had shown that the Pho81 minimum domain is sufficient for Pho81 function in vivo, it was not clear whether this domain was necessary in the context of the full-length protein. We investigated this question with a mutagenesis strategy in which we replaced three adjacent aa in the Pho81 minimum domain with alanine in a nonoverlapping manner. Site-directed mutagenesis was first performed on templates containing the Pho81 minimum domain (data not shown). We found five mutants that were defective in inducing Pho5 expression in response to phosphate limitation in the context of the Pho81 minimum domain (Fig. 5A).
FIG. 5.
The Pho81 minimum domain (aa 645 to 724) is necessary for Pho81 function in vivo. (A) Alanine scan of the Pho81 minimum domain. Site-directed mutagenesis was designed to replace three adjacent aa in the Pho81 minimum domain with alanine in a nonoverlapping manner. All mutants were tested for complementation of the pho81Δ phenotype. Only five mutants were found to be defective in inducing Pho5 expression in response to phosphate limitation: mutations in aa 666 to 668 and 672 to 674 (open boxes) appeared to have a less severe defect than mutations in aa 690 to 692, 699 to 701 (gray boxes), and 693 to 695 (black box), which resulted a complete loss of function. (B) The effects of the five alanine scan mutations when introduced into full-length Pho81. pho81Δ pho3Δ (EY0622) cells were transformed with pADH1pr-PHO81-Py2(wild-type Pho81), pADH1pr-Py2 (vector), pADH1pr-PHO81(666–668AAA)-Py2 (alanine substitution of residues 666 to 668), pADH1pr-PHO81(672–674AAA)-Py2, pADH1pr-PHO81(690–692AAA)-Py2, pADH1pr-PHO81(693–695AAA)-Py2, pADH1pr-PHO81(699–701AAA)-Py2. Phosphatase activity was then quantitated from cells grown in high-phosphate (solid column) and low-phosphate (open column) media. (C) Protein expression levels of Pho81 alanine substitution mutants are similar to the levels of wild-type Pho81. Yeast extracts were made from cells containing wild-type or mutant Pho81 grown in high- and low-phosphate media. A total of 100 μg of each extract was then analyzed by SDS-PAGE and Western blotting with an anti-Py2 antibody. (D) Pho81 alanine substitution mutants detective in inducing Pho5 expression are unable to be coimmunoprecipitated with Pho80. Yeast extracts were made from cells containing wild-type or mutant Pho81 grown in high- and low-phosphate media. An anti-Pho80 antibody was then used to immunoprecipitate Pho80 from yeast extract, and the immunoprecipitated proteins were analyzed by SDS-PAGE, followed by Western blotting with an anti-Pho81 or anti-Pho80 antibody.
To determine if these residues are necessary for Pho81 function, we examined the phenotype resulting from the same mutations in the context of full-length Pho81 (Fig. 5B). Yeast cells expressing full-length Pho81 with mutations in aa 666 to 668 and 672 to 674 appeared to have the same ability to induce Pho5 expression in response to phosphate starvation as those expressing wild-type Pho81. However, yeast cells expressing full-length Pho81 with mutations in aa 690 to 695 and 699 to 701 had significant defects in inducing Pho5 expression. Mutations in residues 693 to 695 completely abolished Pho81 function. These differences in the ability to induce Pho5 were not due to differences in protein levels, because wild-type Pho81 and the Pho81 mutants had similar expression levels (Fig. 5C). We then investigated the ability of the Pho81 mutants to bind the Pho80-Pho85 complex in vivo. Yeast extracts were made from cells containing wild-type or mutant Pho81 grown in high- and low-phosphate medium, and Pho81-Pho80-Pho85 complexes were immunoprecipitated with an anti-Pho80 antibody (Fig. 5D). The Pho81 mutant containing the alanine substitutions in residues 693 to 695 was not efficiently coimmunoprecipitated with Pho80 under either low- or high-phosphate conditions. The Pho81 mutants containing the alanine substitutions in residues 690 to 692 and 699 to 701 also appeared to have defects in binding to the kinase complex. These results indicate that the Pho81 minimum domain is required for Pho81 function in vivo, and that residues 690 to 695 and 699 to 701 are particularly important for Pho81 binding to Pho80-Pho85.
DISCUSSION
Previous studies have shown that mammalian CKIs interact with cyclin-CDK complexes in one of two ways. They can make extensive contacts with both the cyclin and CDK subunits (Cip/Kip family), interfering with substrate binding and catalysis (9, 30). Alternatively, they interact exclusively through the CDK subunit (INK4 family), altering the ability of the CDK to bind to and be activated by its cyclin partner (3, 30). In contrast to the INK4 family, Pho81 can bind to the cyclin subunit in the absence of the CDK but not to the CDK subunit in the absence of the cyclin (Huang and O'Shea, unpublished; 34). Consistent with this model, we found that nuclear localization of Pho81 is dependent on Pho80 but not on Pho85 (Fig. 1).
To determine how Pho81 inhibits the Pho80 Pho85 complex, we wished to first understand how Pho81 binds to this kinase. Pho80 mutants isolated from a genetic selection are defective in binding to Pho81 under high-phosphate conditions but regain interaction with Pho81 when cells are starved for phosphate. Though the Pho80 mutants are partially defective in binding to Pho81, the affinity of Pho81 for the mutant kinase is increased in response to low-phosphate conditions (Fig. 3C and D). Due to binding defects, the mutant Pho80-Pho85 complexes are less inhibited by Pho81 than wild-type Pho80-Pho85, and therefore induction of Pho5 expression is reduced compared to induction observed in a PHO80 background (Fig. 3B). These observations strongly suggest that the interaction between Pho81 and the Pho80-Pho85 complex is regulated in response to phosphate levels. One simple model is the following: the interaction of Pho81 and the kinase complex in high-phosphate medium allows the inhibitor to associate with Pho80-Pho85 without inhibition; when the cells are starved for phosphate, the affinity of the interaction is increased or altered so that Pho81 now inhibits the kinase.
We were surprised to discover that the ankyrin repeats are dispensable for Pho81 inhibition of Pho80-Pho85. It has been shown previously that a Pho81 domain containing six ankyrin repeats and 80 amino acids C -terminal to the ankyrin repeats was able to partially complement the uninducible Pho5 phenotype of a pho81Δ mutant in low-phosphate medium (27). From this observation and because of the homology of Pho81 to the INK4 CKIs, it was originally hypothesized that Pho81 might inhibit Pho80-Pho85 through the ankyrin repeats (27, 34). We have now determined that the region of Pho81 containing 80 aa (aa 645 to 724) C terminal to the ankyrin repeats is necessary and sufficient for regulated inhibition of Pho80-Pho85 (Fig. 4C and 5). Alanine scanning of the minimum functional region identified nine residues which are required for binding to Pho80-Pho85 and inhibition of the kinase in vivo (Fig. 5). Mutations of these nine residues impair interaction of Pho81 with Pho80-Pho85 and therefore impair its ability to inhibit the kinase. These results strongly suggest that the way in which Pho81 binds to and inhibits Pho80-Pho85 is different from the way the INK4 CKIs bind and inhibit CDK4-CDK6. Interestingly, this region of nine residues is highly conserved between Pho81 and Nuc-2, a Neurospora crassa CKI that has sequence homology to Pho81 (31). The unique sequence of this Pho81 minimum domain identifies a new type of CKI motif.
It is remarkable that 80 residues of the Pho81 contain all the information needed for stable binding and regulated inhibition in response to phosphate conditions. How does Pho81 inhibit the kinase? Two of the Pho80 mutations obtained repeatedly in the selection are in residues R121 and E154, which are predicted to be localized on a solvent-exposed region of helices 3 and 5 of the highly conserved cyclin box (Fig. 2). It has been shown that the helices 3 and 5 of the cyclin box interact with the PSTAIRE helix of the CDK (Fig. 2) (11). These observations suggest that Pho81 inhibits the kinase by acting through the cyclin to perturb the interactions of cyclin helices 3 and 5 and the PSTAIRE helix of the CDK. The simplest model is that Pho81 binds to the region of R121 and E154 on helices 3 and 5 of the cyclin Pho80. Interestingly, this region is adjacent to a hydrophobic patch on cyclin A, which contacts the RXL motif of p27 (35). This hydrophobic patch is also important for tight physical interaction and phosphorylation of RXL-containing substrates by cyclin A-CDK2. Although it is known that Pho80 interacts tightly with its substrate Pho4, the region of Pho80 responsible for this interaction has not been identified. It will be interesting to determine if Pho80 binds to Pho4 using a hydrophobic patch and if Pho81 binding to Pho80 affects the interaction with Pho4.
If these 80 aa are all that are required for inhibition of Pho80-Pho85, what do the other 1,099 aa in Pho81 do? Although the Pho81 minimum domain is necessary and sufficient for Pho81 inhibition of Pho80-Pho85, some point mutations in the N-terminal region result in constitutive activation of the CDK inhibitor Pho81 (5). These observations, coupled with the fact that cells containing full-length Pho81 are more inducible for Pho5 activity than cells containing the Pho81 minimum domain (Fig. 4C), suggest that the N-terminal region might also contribute to Pho81 inhibition of Pho80-Pho85. Several observations suggest that Pho81 may have a function in addition to its role as a CKI. Pho81 appears to be involved in regulation of Pcl7-Pho85 (Huang and O'Shea, unpublished; 17). Also, overexpression of Pho81 can suppress the temperature-sensitive phenotype of a phospholipase C mutant (6), suggesting that Pho81 may play a role in lipid metabolism. Pho81 appears to localize to the cytoplasm and to the plasma membrane in addition to its predominant nuclear localization (D. Jeffery and E. K. O'Shea, unpublished observations). Localization to the nucleus requires binding to Pho80, while localization to the plasma membrane is mediated by the first 200 aa of Pho81 localization (Jeffery and O'Shea, unpublished). Although the only identified phenotype exhibited by a pho81Δ mutant is the PHO5 uninducible expression phenotype, it is possible that Pho81 performs another function(s) in the cell and that the parts of Pho81 that are not required for inhibiting Pho80-Pho85 are important for those functions. Another S. cerevisiae CKI, Far1, also has more than one function. Far1 was originally identified as a CKI which inactivates Cln-Cdc28 complexes and contributes to G1 arrest in the presence of mating pheromone (32). It has also been shown that Far1 plays an important role in controlling morphogenesis through its interaction with the guanine-nucleotide exchange factor Cdc24 (39).
An important remaining question is how Pho81 activity is regulated in response to phosphate conditions. Inhibition of Pho80-Pho85 by Pho81 under low-phosphate conditions leads to nuclear localization of Pho4 and transcriptional induction of PHO5, both of which occur even when cells are shifted to low-phosphate medium in the presence of cycloheximide (18, 26). Although Pho81 is transcriptionally induced under low-phosphate conditions, overexpression of Pho81 from the GAL1 promoter or on a high-copy-number plasmid does not lead to significant induction of PHO5 in high-phosphate medium (5, 27). These data point strongly to a posttranslational mechanism of regulation of Pho81 in response to phosphate starvation. In contrast, the activity of INK4 CKIs is regulated primarily by transcriptional induction (3). Several possibilities exist for the way in which Pho81 is regulated. Pho81 could be bound by another protein or a metabolite of phosphate that affects its activity in response to phosphate conditions. Although we have been unable to detect any proteins bound to Pho81 or to the Pho81-Pho80-Pho85 complex by using silver staining or metabolic labeling of proteins with [35S]-methionine (Jeffery and O'Shea, unpublished), we cannot exclude the possibility that a regulator of this complex dissociated during the isolation procedure. At this time, we favor a model in which Pho80, Pho85, or Pho81 is covalently modified in response to phosphate levels. The identification of a small 80-aa region that responds to phosphate levels should greatly facilitate analysis of such modifications.
In conclusion, our study demonstrates that Pho81 associates with the Pho80-Pho85 kinase complex through binding to the cyclin Pho80 and that a minimum domain of Pho81 containing 80 aa (aa 645 to 724) is necessary and sufficient for Pho81 inhibition of Pho80-Pho85 in response to phosphate conditions. The unique sequence of this Pho81 minimum domain identifies a new type of CKI motif. Interesting questions still remain to fully understand the novel inhibitory mechanism and the regulation of Pho81 as a CKI, as well as other possible functions of Pho81.
ACKNOWLEDGMENTS
We thank J. Weissman and members of the O'Shea lab for critical reading of the manuscript.
This work was supported by grants from the N.I.H. (GM51377) and the Howard Hughes Medical Institute (to E.K.O.). M.D.A. was supported by a summer fellowship awarded by the UCSF Student Research Committee.
REFERENCES
- 1.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 2.Bazan J F. Helical fold prediction for the cyclin box. Proteins. 1996;24:1–17. doi: 10.1002/(SICI)1097-0134(199601)24:1<1::AID-PROT1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Carnero A, Hannon G J. The INK4 family of CDK inhibitors. Curr Top Microbiol Immunol. 1998;227:43–55. doi: 10.1007/978-3-642-71941-7_3. [DOI] [PubMed] [Google Scholar]
- 4.Coche T, Prozzi D, Legrain M, Hilger F, Vandenhaute J. Nucleotide sequence of the PHO81 gene involved in the regulation of the repressible acid phosphatase gene in Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:2176. doi: 10.1093/nar/18.8.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creasy C L, Madden S L, Bergman L W. Molecular analysis of the PHO81 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:1975–1982. doi: 10.1093/nar/21.8.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flick J S, Thorner J. An essential function of a phosphoinositide-specific phospholipase C is relieved by inhibition of a cyclin-dependent protein kinase in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:33–47. doi: 10.1093/genetics/148.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guthrie C, Fink G R, editors. Methods in enzymology. 194. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [PubMed] [Google Scholar]
- 8.Harlow E, Lane D. Using antibodies. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 9.Hengst L, Reed S I. Inhibitors of the Cip/Kip family. Curr Top Microbiol Immunol. 1998;227:25–41. doi: 10.1007/978-3-642-71941-7_2. [DOI] [PubMed] [Google Scholar]
- 10.Jeffery D A, Springer M, King D S, O'Shea E K. Multi-site phosphorylation of Pho4 by the cyclin-CDK Pho80-Pho85 is semi-processive with site preference. J Mol Biol. 2001;306:997–1010. doi: 10.1006/jmbi.2000.4417. [DOI] [PubMed] [Google Scholar]
- 11.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 12.Kaffman A, Herskowitz I, Tjian R, O'Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 13.Kaffman A, O'Shea E K. Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol. 1999;15:291–339. doi: 10.1146/annurev.cellbio.15.1.291. [DOI] [PubMed] [Google Scholar]
- 14.Kaffman A, Rank N M, O'Neill E M, Huang L S, O'Shea E K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- 15.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 17.Lee M, O'Regan S, Moreau J-L, Johnson A L, Johnston L H, Goding C R. Regulation of the Pcl7-Pho85 cyclin-cdk complex by Pho81. Mol Microbiol. 2000;38:411–422. doi: 10.1046/j.1365-2958.2000.02140.x. [DOI] [PubMed] [Google Scholar]
- 18.Lemire J M, Willcocks T, Halvorson H O, Bostian K A. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1985;5:2131–2141. doi: 10.1128/mcb.5.8.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenburg M E, O'Shea E K. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 20.Leung D W, Chen E Y, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- 21.Madden S L, Creasy C L, Srinivas V, Fawcett W, Bergman L W. Structure and expression of the PHO80 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1988;16:2625–2637. doi: 10.1093/nar/16.6.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendenhall M D. Cyclin-dependent kinase inhibitors of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Curr Top Microbiol Immunol. 1998;227:1–24. doi: 10.1007/978-3-642-71941-7_1. [DOI] [PubMed] [Google Scholar]
- 23.Mendenhall M D. An inhibitor of p34CDC28 protein kinase activity from Saccharomyces cerevisiae. Science. 1993;259:216–219. doi: 10.1126/science.8421781. [DOI] [PubMed] [Google Scholar]
- 24.Morgan D O. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 24a.Nasmyth K, Adolf G, Lydall D, Seddon A. The identification of a second cell cycle control on the HO promoter in yeast: cell cycle regulation of SWI5 nuclear entry. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- 25.Nurse P, Thuriaux P. Regulatory genes controlling mitosis in the fission yeast Schizosaccharomyces pombe. Genetics. 1980;96:627–637. doi: 10.1093/genetics/96.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill E M, Kaffman A, Jolly E R, O'Shea E K. Regulation of PHO4 nuclear localization by the PHO80–PHO85 cyclin-CDK complex. Science. 1996;271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa N, Noguchi K-I, Sawai H, Yamashita Y, Yompakdee C, Oshima Y. Functional domains of Pho81p, an inhibitor of Pho85p protein kinase, in the transduction pathway of Pi signals in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:997–1004. doi: 10.1128/mcb.15.2.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 29.Oshima Y. Regulatory circuits for gene expression: the metabolism of galactose and phosphate. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 159–180. [Google Scholar]
- 30.Pavletich N P. Mechanisms of cyclin-dependent kinase regulation: structures of Cdks, their cyclin activators, and Cip and 1NK4 inhibitors. J Mol Biol. 1999;287:821–829. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 31.Peleg Y, Aramayo R, Kang S, Hall J G, Metzenberg R L. NUC-2, a component of the phosphate-regulated signal transduction pathway in Neurospora crassa, is an ankyrin repeat protein. Mol Gen Genet. 1996;252:709–716. doi: 10.1007/BF02173977. [DOI] [PubMed] [Google Scholar]
- 32.Peter M, Herskowitz I. Direct inhibition of the yeast cyclin-dependent kinase Cdc28-Cln by Far1. Science. 1994;265:1228–1231. doi: 10.1126/science.8066461. [DOI] [PubMed] [Google Scholar]
- 33.Russo A A, Tong L, Lee J O, Jeffrey P D, Pavletich N P. Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a. Nature. 1998;395:237–243. doi: 10.1038/26155. [DOI] [PubMed] [Google Scholar]
- 34.Schneider K R, Smith R L, O'Shea E K. Phosphate-regulated inactivation of the kinase PHO80–PHO85 by the CDK inhibitor PHO81. Science. 1994;266:122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- 35.Schulman B A, Lindstrom D L, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sedgwick S G, Smerdon S J. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- 37.Serrano M, Hannon G J, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 38.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 39.Shimada Y, Gulli M P, Peter M. Nuclear sequestration of the exchange factor Cdc24 by Far1 regulates cell polarity during yeast mating. Nat Cell Biol. 2000;2:117–124. doi: 10.1038/35000073. [DOI] [PubMed] [Google Scholar]
- 40.Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toh-e A, Oshima Y. Characterization of a dominant, constitutive mutation, PHOO, for the repressible acid phosphatase synthesis in Saccharomyces cerevisiae. J Bacteriol. 1974;120:608–617. doi: 10.1128/jb.120.2.608-617.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toh-e A, Shimauchi T. Cloning and sequencing of the PHO80 gene and CEN15 of Saccharomyces cerevisiae. Yeast. 1986;2:129–139. doi: 10.1002/yea.320020209. [DOI] [PubMed] [Google Scholar]
- 43.Toh-e A, Tanaka K, Uesono Y, Wickner R B. PHO85, a negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 44.Toh-e A, Ueda Y, Kakimoto S-I, Oshima Y. Isolation and characterization of acid phosphatase mutants in Saccharomyces cerevisiae. J Bacteriol. 1973;113:727–738. doi: 10.1128/jb.113.2.727-738.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uesono Y, Tanaka K, Toh-e A. Negative regulators of the PHO system in Saccharomyces cerevisiae: isolation and structural characterization of PHO85. Nucleic Acids Res. 1987;15:10299–10309. doi: 10.1093/nar/15.24.10299. [DOI] [PMC free article] [PubMed] [Google Scholar]