Abstract
Pyrophosphorolysis-activated polymerization (PAP) was initially developed to enhance the specificity of allele-specific PCR for detection of known mutations in the presence of a great excess of wild-type allele. The high specificity of PAP derives from the serial coupling of pyrophosphorolysis-mediated activation of a pyrophosphorolysis-activatable oligonucleotide (P*) followed by extension of the activated oligonucleotide. Herein, we demonstrate that genetically engineered DNA polymerases greatly improve the efficiency of PAP, making it a practical technique for detection of rare mutations. We also show that P* oligonucleotides have the novel and unexpected property of high sensitivity to mismatches throughout at least the 16 3′-terminal nucleotides. Thus, PAP constitutes a technology platform of potential utility whenever high specificity is required along the length of an oligonucleotide.
INTRODUCTION
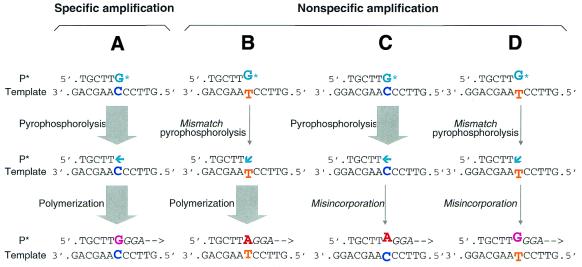
Pyrophosphorolysis is the reverse reaction of DNA polymerization. In the presence of pyrophosphate, the 3′ nucleotide is removed from duplex DNA to generate the triphosphate and the 3′-terminal shortened duplex DNA: [dNMP]n + PPi → [dNMP]n – 1 + dNTP (1). Pyrophosphorolysis-activatable oligonucleotides (P*) underlie PAP, which was developed to detect rare mutations by increasing the specificity of PCR amplification of specific alleles (Fig. 1) (2). The specificity results from serial coupling of pyrophosphorolysis and polymerization, since significant non-specific amplification requires both mismatch pyrophosphorolysis and misincorporation by the DNA polymerase, an extremely rare event the probability (frequency) of which is estimated to be 3.3 × 10–11.
Figure 1.
Pyrophosphorolysis-activated polymerization (PAP) for high specificity amplification. The high specificity of PAP derives from the serial coupling of two reactions: activation of a dideoxy 3′-terminated oligonucleotide by pyrophosphorolysis followed by polymerization. In this example, a P* matches the G variant (GV) template but mismatches the A variant (AV) template at the 3′-terminus. Column A shows the primary process of specific amplification indicated by thick arrows. When the P* hybridizes to the GV template, activation by pyrophosphorolytic removal of the 3′-terminal ddGMP* occurs, which then permits extension by polymerization. Non-specific amplification generating the AV product occurs rarely when either mismatch pyrophosphorolysis (indicated by thin arrows) (column B, estimated frequency 10–5) or misincorporation (column C, estimated frequency 3.3 × 10–6) occurs. Non-specific amplification (column D) to generate the GV product occurs only if both errors occur (estimated frequency 3.3 × 10–11). In an exponential PAP with two oligonucleotides, only the non-specific product in column D, once it occurs, can be amplified exponentially in subsequent cycles.
In the initial report, PAP was performed with native Tfl and Taq DNA polymerases (2). The practical application of PAP was limited by very low efficiency. Herein, we have overcome this limitation using genetically engineered DNA polymerases with greatly reduced discrimination against ddNMP pyrophosphorolysis. We now substantially extend the range of potential applications of PAP by the surprising observation that PAP is highly sensitive to mismatches throughout the length of the P* oligonucleotide.
MATERIALS AND METHODS
Preparation of template by PCR
A 640 bp region of the human D1 dopamine receptor gene was amplified by PCR with two primers (T, 5′-GAC CTG CAG CAA GGG AGT CAG AAG-3′; U, 5′-TCA TAC CGG AAA GGG CTG GAG ATA-3′). The TU:UT duplexed product spans nucleotides 33–672 in GenBank accession no. X55760 and the G+C content of the product is 55% (Fig. 2A). Common G or A variants at nucleotide 229, denoted GV and AV, produce the three genotypes GV/GV, AV/AV and GV/AV (3). The PCR volume was 50 µl, containing 50 mM KCl, 10 mM Tris–HCl, pH 8.3, 1.5 mM MgCl2, 200 µM each of the four dNTPs, 0.1 µM each primer, 2% DMSO, 1 U Taq DNA polymerase (Boehringer Mannheim) and 250 ng genomic DNA from the GV/GV homozygote, AV/AV homozygote or GV/AV heterozygote. Cycling conditions were 94°C for 15 s, 55°C for 30 s and 72°C for 1 min, for a total of 35 cycles, with a GeneAmp PCR System 9600 (Perkin Elmer Applied Biosytems). The PCR product used as the model template was purified from primers and other small molecules ∼10 000-fold by three rounds of retention on a Centricon 100 microconcentrator (Amicon). The amount of recovered PCR product was determined by UV absorbance at 260 nm.
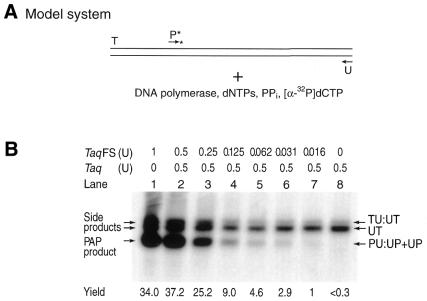
Figure 2.
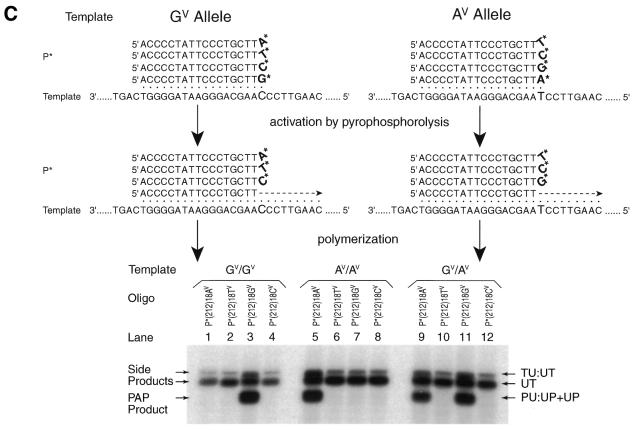
AmpliTaqFS DNA polymerase dramatically increases PAP efficiency and decreases discrimination among ddNMP termini of P*s. (A) Model system. The duplexed GV or AV templates, denoted TU:UT, were amplified with two oligonucleotides P* and U, DNA polymerase, dNTPs, pyrophosphate and [α-32P]dCTP. TU:UT, a 640 bp segment of the dopamine D1 receptor gene; P*, pyrophosphorolysis-activatable oligonucleotide. (B) AmpliTaqFS DNA polymerase dramatically increases PAP efficiency. The duplexed GV template was amplified with P*(210)20GV and U (Table 1, P* oligonucleotide no. 3). Different amounts of AmpliTaqFS and native Taq DNA polymerase were added to each 25 µl reaction. The radioactively labeled specific products of 463 nt (duplex PU:UP and excess antisense strand UP) are produced. Other side products UT and UT:TU are indicated. The effect of AmpliTaqFS amount is indicated as the yield in arbitrary units, which was quantitated by scanning the autoradiograph with a densitometer. Lane 8, which contains only Taq polymerase, has a yield below the level of detection (<0.3 U). (C) Efficiency and specificity of P*s with different 3′-terminal dideoxynucleotides. Each of the four P*s has a ddAMP, ddTMP, ddGMP or ddCMP at the 3′-terminus (Table 1, P* oligonucleotide no. 4 and three additional P*s that have ddA, ddC and ddT at the 3′-terminus). The 3′-terminal nucleotide is either specific to the complementary strand of the GV or AV template or not matched. An autoradiogram of PAP from the GV/GV, AV/AV and GV/AV DNA templates of the human dopamine receptor gene is shown. Radioactively labeled specific products of 461 nt (duplex PU:UP and excess antisense strand UP) are produced. PU:UP and UP are separated when they are electrophoresed longer. Other side products, UT and UT:TU, are indicated. Note that TU:UT derives from annealing excess radioactively labeled UT with non-radioactively labeled TU original template.
Synthesis of P* by adding a 3′-dideoxynucleotide
The informative name for P* is described in footnote a of Table 1. The deoxynucleotide oligonucleotide was synthesized in a Perseptive Biosystems 8909 Synthesizer (Framingham) and purified on oligopure cartridges (Hamilton) in the City of Hope DNA/RNA Chemistry Laboratory. The 3′-terminal dideoxynucleotide was added with terminal transferase. The mixture contained, in a total volume of 30 µl, 100 mM potassium cacodylate, pH 7.2, 2.0 mM CoCl2, 0.2 mM DTT, 2500 pM oligonucleotide, 2 mM 2′,3′-ddNTP (the molar ratio of the 3′-OH terminus to ddNTP was 1:24) (Boehringer Mannheim), 100 U terminal transferase (Gibco BRL). The reaction was incubated at 37°C for 4 h and then stopped by adding EDTA to a 5 mM final concentration. After desalting using a Centri-spin column (Princeton Separations), P* was purified by preparative 7 M urea–20% polyacrylamide gel electrophoresis in TBE buffer (90 mM Tris–borate, 1 mM EDTA, pH 8.3) (4). The amount of recovered P* was determined by UV absorbance at 260 nm.
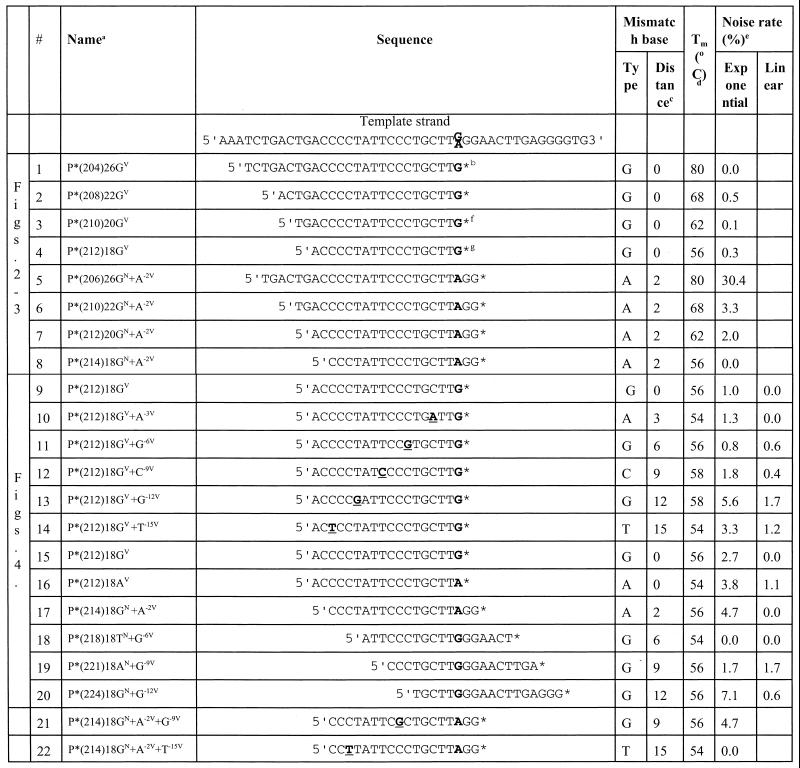
Table 1. List of P*s.
aMost P*s match either the G or A polymorphism at position 229 of the human dopamine D1 gene. A few P*s mismatch the GV template by 1 nt. The informative nomenclature provides critical information about the P* including: (i) allele specificity, if any; (ii) location of the allele-specific base within the oligonucleotide; and (iii) any mismatches from the GV or AV templates. Each PAP oligonucleotide is indicated by P*, followed in parentheses by the base number of the 5′-nucleotide, by the length of the oligonucleotide and by the identity of the 3′ dideoxy nucleotide and the internal identity. Specificity for a known variant (a polymorphism or a mutation) at the 3′-terminus is indicated by superscripted V. If the 3′-terminus does not confer specificity for any known variant, the superscript is N, for non-variant. If there is allele specificity or a mismatch internal to the oligonucleotide, the allele-specific or mismatched nucleotide is indicated with the distance from the 3′-terminus and the superscript V. For example, P*(204)26GV is a P* with a G dideoxynucleotide at the 3′-terminus. ddG is the allele-specific nucleotide. The first base at the 5′-terminus corresponds to nucleotide 204 in GenBank accession no. X55760. Its length is 26 nt. P*(206)26GN+A–2V is a P* that begins at base 206. It is 26 nt in length and the 3′-terminus is a dideoxy G that is not known to recognize any variance. In addition there is an A 2 nt from the 3′-terminus that is specific to the A allele. Thus, this oligonucleotide perfectly matches the AV template. A few oligonucleotides were synthesized to mismatch the G and A templates. These oligonucleotides mismatch both the G and the A alleles by at least 1 nt. The informative names are useful in day-to-day communication about experiments. The table shows the complete nucleotide sequences.
bThe bold G or A is at the GV or AV variant and the underlined nucleotide in five P*s used in Figure 3A have a designed mismatch.
cThe distance from the 3′-terminus to the GV or AV variant or to a mismatched nucleotide, e.g. 0, at the 3′-terminus; 3, 3 nt from the 3′-terminus.
dThe Tm for each oligonucleotide was estimated to be 4°C × (G + C) + 2°C × (T + A) at 1 M NaCl.
eAmplification from the GV and AV templates by exponential PAP with two oligonucleotides or by linear PAP with one P*. The noise rate of PAP (%) is the relative yield of the mismatched product to the matched product. A specific signal is defined as <10% noise rate.
fP* used in Figure 2B.
gThe core P* used in Figures 2–4. Figure 2C contains three additional P*s that have ddA, ddC and ddT at the 3′-terminus.
Since small amounts of unterminated oligonucleotide would result in unexpected PCR amplification, each P* was 32P-labeled at the 5′-terminus with T4 polynucleotide kinase and then was electrophoresed through a 7 M urea–20% polyacrylamide gel. Only P* products were visible, even when the gel was overexposed (data not shown). It is estimated that >99.99% of P* contained a dideoxynucleotide at the 3′-terminus. Purity of P* was supported by the absence of PCR product or PAP product at pH 8.3, at which pyrophosphorolysis is inhibited.
PAP amplification
Regions from 445 to 469 bp within the TU:UT duplexed template were amplified by PAP with oligonucleotides P* and U or with P* only. The PU:UP duplexed product corresponds to nucleotides 204–228 to 672 in GenBank accession no. X55760 (G+C content 56%). The PAP reaction mixture contained, in a total volume of 25 µl, 50 mM KCl, 10 mM Tris–HCl, pH 7.6, 1.5 mM MgCl2, 100 µM each of the four dNTPs (dATP, dTTP, dGTP and dCTP), 0.1 µM P*, 0.1 µM U, 300 µM Na4PPi, 2% DMSO, 1 µCi [α-32P]dCTP (3000 Ci/mmol; Amersham), 1 U AmpliTaqFS DNA polymerase (PE Applied Biosystems) or 0.5 U each of AmpliTaqFS and Taq DNA polymerases and 10 ng TU:UT. ThermoSequenase (Amersham Pharmacia) was also tested under the same conditions except that 8 U ThermoSequenase or 4 U ThermoSequenase/0.5 U Taq and 2.5 mM MgCl2 were used. The cycling conditions were 94°C for 10 s, 60°C for 1 min (at 55°C for ThermoSequenase) and 72°C for 2 min, for a total of 15 cycles.
The product was electrophoresed through a standard 2% agarose gel. The gel was stained with ethidium bromide for UV photography with a CCD camera (Bio-Rad Gel Doc 1000) using Multi-Analyst software, dried and subjected to Kodak X-OMAT AR film for autoradiography. The PAP yield was quantitated with a PhosphorImager with ImageQuant software (Molecular Dynamics) as the total amount of signal in the PAP band minus the background (arbitrary unit).
RESULTS
Mutated polymerase makes PAP a practical method
AmpliTaqFS and ThermoSequenase contain a F667Y mutation in the active site, which results in the incorporation of dNTPs and ddNTPs at similar rates (5–7). AmpliTaqFS increased the yield of PAP >100-fold relative to that obtained with native Taq (Fig. 2B, compare lane 1 with 8), allowing a single copy human genomic gene to be amplified by PAP. The effect of AmpliTaqFS on PAP efficiency was examined using different amounts of this enzyme. The level of PAP-specific signal is proportional to the amount of AmpliTaqFS over a broad dynamic range. Similar results were obtained with ThermoSequenase (data not shown).
When using native Tfl or Taq DNA polymerase, P* blocked at the 3′-terminus with ddGMP could be activated, but not that terminated with ddAMP (2). AmpliTaqFS DNA polymerase produced a much higher efficiency of PAP with much less discrimination against the four different terminal dideoxynucleotides. Figure 2C shows an experiment in which the G variant (GV) and A variant (AV ) templates of the dopamine D1 gene (3) were amplified by PAP only when they matched their specific P* oligonucleotides. The heterozygote was amplified with a yield ratio estimated to be 1.4 (compare lane 11 with 9), indicating comparable efficiency of PAP with ddGMP and ddAMP terminated P*s. In other experiments P*s with either ddCMP or ddTMP at the 3′-terminus were also activated and extended efficiently.
Inhibition of PAP by mismatches along P*
A series of P*s that precisely match either the GV or AV template were synthesized (Table 1). In addition, a few P*s were synthesized in which at least one mismatch occurred with the GV and AV templates (Table 1). Multiple P*s of different lengths and with different mismatches were examined using AmpliTaqFS.
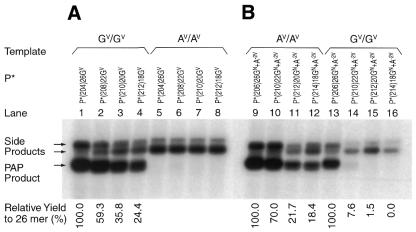
PAP was highly specific for the GV template when the position of the GV variant was at the 3′-terminus of P* (Fig. 3 and Table 1, P* oligonucleotides 1–4). The specificity was unaffected by size with 3′-co-terminal P*s ranging from 18 to 26 nt. When the position of the AV variant was 2 nt from the 3′-terminus of P*, the specificity was retained for P*s 18, 20 and 22 nt long (Fig. 3 and Table 1, P* oligonucleotides 5–8).
Figure 3.
Effect of P* length. PAP was performed with P*s of different lengths and U (Table 1, P* oligonucleotides 1–8). (A) A PAP product is seen when the 3′-co-terminal P*s of length 18–26 nt match the GV template at the 3′-terminus (lanes 1–4) (Table 1, P* oligonucleotides 1–4). A PAP product is not seen with the AV template mismatched at the 3′-terminus (lanes 5–8). (B) A PAP product is seen when P*s of length 18–26 nt match the AV template 2 nt from the 3′-terminus (lanes 9–12) (Table 1, P* oligonucleotides 5–8). When the GV template was used (lanes 13–16) substantial non-specific PAP product is seen with the 26mer. In both situations the yield of PAP product increased somewhat with length. The length effect is indicated as the yield ratio (%) for the 26mer from the same template, which varied from 0.0 to 70.0% with each 2 or 4 nt shortening in length. The yield ratio is not shown in lanes 5–8 because the signals are at or close to the background.
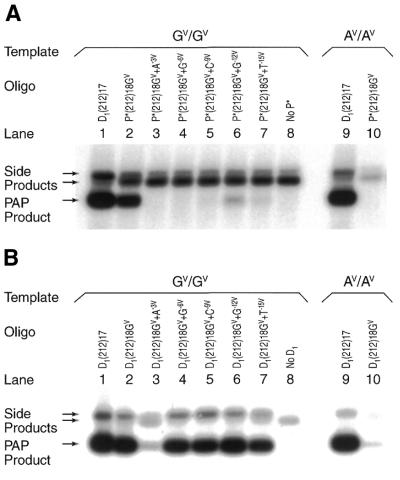
The 18mers were examined further with a series of P*s that measured the effect of a single mismatch at varying distances from the 3′-terminus (Fig. 4A and Table 1, P* oligonucleotides 9–14). P*s were specific to the GV template at the 3′-terminus. When P* matched the GV template along the length, the PAP product was amplified (lane 2). If a mismatch occurred at either 3, 6, 9, 12 or 15 nt from the 3′-terminus, little or no amplification occurred (lanes 3–7). The noise rate varied from 0.8 to 5.6% (Table 1). Thus, a mismatch at 3–15 nt from the 3′-terminus inhibited PAP amplification.
Figure 4.
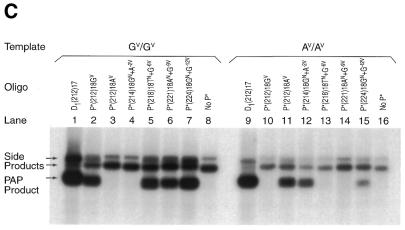
Inhibition of PAP by mismatches along P*. (A) PAP specificity with differently mismatched P*s. PAP was performed with a series of P*s and U (Table 1, P* oligonucleotides 9–14). P*s are specific to the GV variant at the 3′-terminus. P* matches the GV template throughout the length in lane 2, but a mismatch was introduced in each of lanes 3–7 at different positions from the 3′-terminus. A negative control with only U is shown in lane 8 and PCR positive controls with D1(212)17 and U [D1(212)17 is an unblocked primer whose 3′-terminus ends 1 nt before the GV or AV variant] in lanes 1 and 9. Another PAP control indicates that the P* has a mismatch at the 3′-terminus and does not amplify the AV template (lane 10). (B) Comparison of PASA specificity. PASA specificity with unblocked primers of identical sequences. (C) PAP specificity with differently localized P*s. PAP was performed with a ‘staggered’ series of P*s and U (Table 1, oligonucleotides 15–20). P*s were constructed in which the GV or AV variant was located 0, 3, 6, 9 and 12 nt from the 3′-terminus so that each P* matches either the GV or AV template. Four P*s matched the GV template (lanes 2 and 5–7) but not the AV template (lanes 10 and 13–15). Two P*s were specific to the AV template but not the GV template (lanes 11 and 12 versus 3 and 4). Positive or negative controls are as in (A) (lanes 1, 8, 9 and 16).
The specificity of PCR amplification of specific alleles (PASA) was compared with that of PAP by using an identical set of unblocked 18mer primers (Fig. 4B). The PCR primer was specific for the GV template at its 3′-terminus (lanes 2–7). When the GV template was utilized, the primer with a mismatch 3 nt from the 3′-terminus had a reduced yield with PASA. However, the primers with mismatches 6, 9, 12 and 15 nt from the 3′-terminus did not inhibit PASA with the GV template.
The stringency of PAP to mismatches was also shown with a ‘staggered’ series of P*s (Fig. 4C and Table 1, P* oligonucleotides 15–20). The P*s were constructed so that the position of the GV or AV variant was 0, 3, 6, 9 or 12 nt from the 3′-terminus. Thus, each P* matches either the GV or AV template. Four P*s matched and amplified the GV template (lanes 2 and 5–7), but little or no amplification occurred from the AV template (lanes 10 and 13–15). Two P*s were specific for and only amplified the AV template (lanes 11 and 12 versus 3 and 4). The noise rate ranged from 0.0 to 7.1% (Table 1, P* oligonucleotides 15–20). Thus, all the mismatches examined in both experiments inhibited PAP amplification. Two additional P*s, P*(214)18GN+A–2V+G–9V and P*(214)18GN+A–2V+T–15V, with mismatches to the AV template at 9 and 15 nt, respectively, from the 3′-terminus also inhibited PAP amplification (Table 1, P* oligonucleotides 21–22).
As an alternative to exponential PAP with two oligonucleotides, linear PAP with only one P* was performed using the 18mer P*s (Table 1, footnote e, P* oligonucleotides 9–20). Higher specificity was observed, as judged by a lower noise rate (see Discussion).
DISCUSSION
PAP is a practical method
We show that two genetically engineered polymerases that tolerate ddNTP incorporation are highly efficient at pyrophosphorolysis of ddNMP (Fig. 2). The enhanced efficiency of AmpliTaqFS and ThermoSequenase presumably derives from a F667Y mutation in the active site (5–7). The commercially available AmpliTaqFS and ThermoSequenase are formulated with pyrophosphatase, which can hydrolyze PPi. It is anticipated that unformulated enzymes will generate even stronger signals.
PAP allele-specific amplification (PAP-A) is now a practical technique and has the potential for detecting rare mutations at 1 per 1011 nt (Fig. 1) (2). However, the actual specificity of PAP-A may be limited by the error rates associated with other components of the reaction, such as sequence context, mutation type, purity of P* oligonucleotides and DNA polymerase. PAP-A can be used to measure minimal residual disease. If a specific point mutation has been defined in a tumor, such as a mutation in the p53 gene, the presence of rare tumor cells can be assayed at the margins or within lymph nodes. In head and neck tumors, where surgery is disfiguring, minimizing the margins is helpful. In breast cancer, the presence of micrometastases to the lymph node may determine whether chemotherapy is appropriate. Finally, early recurrence of tumor can be monitored. The current methods for detecting point mutations have a routine specificity of 10–3 (8). The specificity of PAP-A should be orders of magnitude higher.
Inhibition of PAP by mismatches along P*
Inhibition of PAP by single base mismatches was shown in one series of 3′-co-terminal P*s in which mismatches occurred up to 15 nt from the 3′-terminus (Fig. 4A). Similar inhibition was also observed with a ‘staggered’ series of P*s with different sequences (Fig. 4C).
It is unclear why mismatches along the length of P* inhibit PAP. Perhaps pyrophosphorolysis requires more cooperativity of DNA melting due to mismatches, as has been reported when oligonucleotides are attached to gold particles (9). PAP requires annealing of P*, activation by pyrophosphorolysis and extension by polymerization. If a single base mismatch inhibits each of the steps by a factor of 10, the total inhibitory effect would be 1000-fold. Thus, the higher sensitivity to mismatches along P* may reflect the serial coupling of pyrophosphorolysis and polymerization.
Although the experiments demonstrate the above points in a model system, further experiments are needed to explore the robustness of the inhibition of PAP by single base mismatches to sequence context, size and type of 3′-terminal dideoxynucleotide of P*s, the template sequence and the DNA polymerase.
Scanning for unknown sequence variants using PAP
P*s should be advantageous whenever high specificity is desired along the length of an oligonucleotide. Two potential applications of PAP illustrate the diversity of possible utility of this observation.
For many molecular epidemiological analyses and for molecular diagnosis, the overwhelming majority of DNA sequences analyzed are negative for a mutation, making scanning for unknown mutations particularly cost effective. Sequencing by hybridization (SBH) can be applied for mutation scanning by analyzing positively hybridized oligonucleotides (10–17). While one oligonucleotide can be optimized to distinguish single base mismatches (18), it has proved very difficult to find a generic set of conditions that will distinguish single base mismatches in all sequence contexts. The oligonucleotides in an array are 11–20 nt in length and have a 7–9 nt-specific region. However, non-specific signal can be generated by mismatched hybridization. The duplex stability between match and mismatch is also affected by the terminal mismatch and the flanking sequence (14,15,17). To date SBH cannot detect most microdeletions and microinsertions and the signal-to-noise ratio is such that single base changes can be missed in routine practice (16).
The inhibition of PAP by mismatches along the length of P*s suggests that PAP can form the basis of microarray-based scanning or a resequencing method to detect virtually all mutations. A particularly attractive approach would combine PAP with maskless photochemical synthesis of oligonucleotides on digital mirror-based combinatorial parallel synthesis addressable oligonucleotide microarrays (19,20). This flexible alternative to a large number of photolithographic masks utilizes virtual masks generated on a computer and relayed to a digital micromirror. By repeating cycles of programmed chemical coupling with virtual masks, it is possible to synthesize an oligonucleotide array with any desired sequence. Photochemical oligonucleotide synthesis in the 5′→3′ direction has been achieved (Beier et al., submitted for publication). These oligonucleotides can be blocked by utilizing available dideoxy substrates for the 3′-terminal reactions.
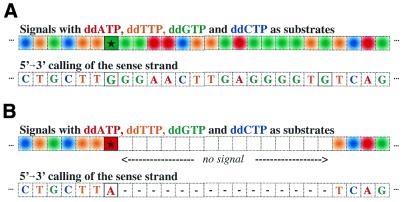
One of a number of possible strategies is depicted in Figure 5A and B. For wild-type samples, the color of every spot would correspond to the wild-type sequence. A single base substitution would change the color at the mutation site and result in the absence of signal for the subsequent 15 nt.
Figure 5.
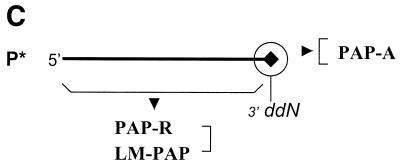
Microarray-based resequencing to detect a G→A mutation. (A) The wild-type template. (B) The G→A mutant template. Each spot contains four P*s with ddAMP*, ddTMP*, ddGMP* and ddCMP*. Linear PAP was performed with the four different dye-labeled dideoxy nucleoside triphosphates as substrates for single base extensions. P*s are 18mers and the specific region at the 3′-terminus is assigned to be only 16 nt. For a sequence of n nucleotides, a total of 4n P*s are synthesized such that on each nucleotide in the sense strand of the wild-type sequence a set of four P*s will have identical sequence except for the four dideoxynucleotides at the 3′-terminus. Homozygous or hemizygous DNA templates are assigned. Furthermore, the four P*s can be immobilized in one single spot when the four dye-labeled ddNTPs are used as substrates for single base extensions in linear PAP. In the example shown, the P* with a ddAMP at the 3′-terminus is activated at the position of the G→A mutation. The mutation also creates a 15 base ‘gap’ of little or no PAP signal for the subsequent overlapping 15 sets of P*s. Additional redundancy may be achieved by synthesis of P*s for the antisense strand. An unknown single base substitution can be determined by combination of the two sets of P*s . For heterozygous changes, the G and A signal will be seen at the site of the sequence change and a signal of half intensity will be seen in the subsequent 15 nt. An unknown small deletion and insertion can be detected and localized. (C) PAP as a technology platform. The ultra-high specificity at the 3′-terminus is applied to PAP-A. The inhibition of PAP by mismatches along the length of the P*s are the basis for PAP-RE and LM-PAP.
Ligation-mediated PAP (LM-PAP)
The specificity of PAP can be helpful for analyzing in vivo chromatin structure, which affects mammalian gene regulation and development. Somatically heritable changes in chromatin structure are an important contributing factor to the development of cancer. One of the few methods for assaying in vivo chromatin structure, and the only method with resolution at the single nucleotide level, is ligation-mediated PCR (LM-PCR) (21,22).
LM-PCR is completely dependent on polymerase priming by sequence-specific synthetic oligonucleotides. For LM-PCR, total genomic mammalian DNA is used, so extreme specificity is required. Approximately 80% of primer pairs work adequately enough to obtain some useful information, even though some general background caused by non-specific priming is often visible. However, LM-PCR remains a difficult technique which many investigators are reluctant to use. Some primer sets work much better than others.
When LM-PCR was compared directly with LM-PAP utilizing identical oligonucleotides for the GV and AV alleles of the dopamine D1 receptor gene, except that the 3′-terminus for LM-PAP was blocked with a dideoxy nucleotide, the signal-to-noise ratio was dramatically higher with LM-PAP (manuscript in preparation). When the 3′-terminal nucleotide of P* is specific for a heterozygote, allele-specific LM-PAP may be performed to distinguish chromatin patterns in imprinted genes or active and inactive X-chromosomal genes in females.
Conclusions
We have demonstrated that PAP can occur efficiently and that the reaction is inhibited by single base mismatches along the length of P*. These experiments suggest that PAP is a technology platform in which at least three novel methods can be envisioned (Fig. 5C).
REFERENCES
- 1.Duetcher M.P. and Kornberg,A. (1969) Enzymatic synthesis of deoxyribonucleic acid. 28. The pyrophosphate exchange and pyrophosphorolysis reactions of deoxyribonucleic acid polymerase. J. Biol. Chem., 244, 3019–3028. [PubMed] [Google Scholar]
- 2.Liu Q. and Sommer,S.S. (2000) Pyrophosphorolysis activated polymerization (PAP): application to allele-specific amplification. Biotechniques, 29, 1072–1083. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q., Sobell,J.L. and Sommer,S.S. (1995) Screening the dopamine D1 receptor gene in 131 schizophrenics and eight alcoholics: identification of polymorphisms but lack of functionally significant sequence changes. Am. J. Med. Genet. Neuropsych. Genet., 60, 165–171. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 5.Innis M.A. and Gelfand,D.H. (1999) Optimization of PCR: conversations between Michael and David. In Innis,M.A., Gelfand,D.H. and Sninsky,J.J. (eds), PCR Applications: Protocols for Functional Genomics. Academic Press, pp. 3–22.
- 6.Tabor S. and Richardson,C.C. (1995) A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA, 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vander Horn P.B., Davis,M.C., Cunniff,J.J., Ruan,C., McArdle,B.F., Samols,S.B., Szasz,J., Hu,G., Hujer,K.M., Domke,S.T., Brummet,S.R., Moffett,R.B. and Fuller,C.W. (1997) Thermo Sequenase DNA polymerase and T.acidophilum pyrophosphatase: new thermostable enzymes for DNA sequencing. Biotechniques, 22, 758–762. [DOI] [PubMed] [Google Scholar]
- 8.Parsons B.L. and Heflich,R.H. (1997) Genotypic selection methods for the direct analysis of point mutations. Mutat. Res., 387, 97–121. [DOI] [PubMed] [Google Scholar]
- 9.Taton T.A., Mirkin,C.A. and Letsinger,R.L. (2000) Scanometric DNA array detection with nanoparticle probes. Science, 289, 1757–1760. [DOI] [PubMed] [Google Scholar]
- 10.Southern E.M., Maskos,U. and Elder,J.K. (1992) Analyzing and comparing nucleic acid sequences by hybridization to arrays of oligonucleotides: evaluation using experimental models. Genomics, 13, 1008–1017. [DOI] [PubMed] [Google Scholar]
- 11.Lysov I., Florent’ev,V.L., Khorlin,A.A., Khrapko,K.R. and Shik,V.V. (1988) Determination of the nucleotide sequence of DNA using hybridization with oligonucleotides. A new method. Dokl. Akad. Nauk. SSSR, 303, 1508–1511. [PubMed] [Google Scholar]
- 12.Bains W. and Smith,G.C. (1988) A novel method for nucleic acid sequence determination. J. Theor. Biol., 135, 303–307. [DOI] [PubMed] [Google Scholar]
- 13.Drmanac R., Labat,I., Brukner,I. and Crkvenjakov,R. (1989) Sequencing of megabase plus DNA by hybridization: theory of the method. Genomics, 4, 114–128. [DOI] [PubMed] [Google Scholar]
- 14.Khrapko K.R., Lysov,Y., Khorlyn,A.A., Shick,V.V., Florentiev,V.L. and Mirzabekov,A.D. (1989) An oligonucleotide hybridization approach to DNA sequencing. FEBS Lett., 256, 118–122. [DOI] [PubMed] [Google Scholar]
- 15.Pevzner P.A. (1989) 1-Tuple DNA sequencing: computer analysis. J. Biomol. Struct. Dyn., 7, 63–73. [DOI] [PubMed] [Google Scholar]
- 16.Hacia J.G. and Collins,F.S. (1999) Mutational analysis using oligonucleotide microarrays. J. Med. Genet., 36, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ginot F. (1997) Oligonucleotide micro-arrays for identification of unknown mutations: how far from reality? Hum. Mutat., 10, 1–10. [DOI] [PubMed] [Google Scholar]
- 18.Wallace R.B., Shaffer,J., Murphy,R.F., Bonner,J., Hirose,T. and Itakura,K. (1979) Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res., 6, 3543–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh-Gasson S., Green,R.D., Yue,Y., Nelson,C., Blattner,F., Sussman,M.R. and Cerrina,F. (1999) Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nature Biotechnol., 17, 974–978. [DOI] [PubMed] [Google Scholar]
- 20.LeProust E., Pellois,J.P., Yu,P., Zhang,H., Gao,X., Srivannavit,O., Gulari,E. and Zhou,X. (2000) Digital light-directed synthesis. A microarray platform that permits rapid reaction optimization on a combinatorial basis. J. Comb. Chem., 2, 349–354. [DOI] [PubMed] [Google Scholar]
- 21.Mueller P.R. and Wold,B. (1989) In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science, 246, 780–786. [Erratum, 1990, 248, 802]. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer G.P., Steigerwald,S.D., Mueller,P.R., Wold,B. and Riggs,A.D. (1989) Genomic sequencing and methylation analysis by ligation mediated PCR. Science, 246, 810–813. [DOI] [PubMed] [Google Scholar]