Abstract
The nucleotide-binding site in a variety of DNA polymerases was probed by analyzing incorporation of dideoxy and acyclic chain terminators. Family B archaeon DNA polymerases Vent, Deep Vent, 9°N and Pfu incorporated acyclic in preference to dideoxy terminators, while the Family A DNA polymerases Taq and Klenow preferred dideoxy terminators. These divergent biases suggest that significant differences exist in sugar recognition in these two classes of polymerases. Mutants of Vent (A488L) and Taq (F667Y) that increase incorporation of dideoxy terminators maintained the acyclic/dideoxy bias of the parent enzyme, while more efficiently incorporating both dideoxy and acyclic terminators. The preference of archaeon DNA polymerases for acyclic analogs was exploited in chain terminator DNA sequence and genotype analysis. This technology was additionally aided by identification of specific dye-modified bases that improve terminator incorporation over that of the unmodified terminator. These three enhancing effects, (i) acyclic analogs, (ii) archaeon variants and (iii) specific dyes, appear to act additively and independently to increase terminator incorporation efficiency, collectively enhancing incorporation ∼8000-fold over the wild-type incorporation of dideoxynucleotides. Fluorescent chain terminator DNA sequence traces demonstrate the applicability of these advances in improving terminator incorporation, as required in DNA sequence and genotype determinations.
INTRODUCTION
All organisms depend on DNA polymerases to replicate and maintain their genomes. These polymerases are thought to be among the earliest evolutionary forms of enzymatic activity (1,2). DNA polymerases also play a central role in molecular biology. Their use is at the core of a wide range of laboratory protocols, including cloning, PCR (3), DNA sequencing (4) and single nucleotide polymorphism (SNP) detection (5–8). These applications rely on the ability of polymerases to faithfully replicate DNA, yielding a detectable product that accurately represents the initial template. In many techniques, this process requires base-specific insertion of nucleotides with modified sugars or bases, either as a detection probe or chain terminator. Insofar as these modifications interfere with specificity contacts by the polymerase, incorporation can be compromised. Accordingly, a great deal of effort has been expended screening analogs, polymerases and polymerase variants for combinations that yield complete, uniform incorporation of nucleotides and nucleotide analogs. Such screening can also expand our understanding of normal nucleotide addition.
Primary amino acid sequence similarities allow the classification of many DNA polymerases into one of three families, A, B or C, according to similarities with Escherichia coli polymerases I, II and III, respectively (9,10). Kinetic and crystallographic studies of Family A and B DNA polymerases reveal a common structural core for the nucleotide-binding site (11), suggesting analogous mechanisms for nucleotide recognition and addition (1). DNA polymerases of Family A (e.g. E.coli pol I, T7 and Taq DNA polymerases) are the predominant enzymes used in DNA sequence determination and thus have been most extensively studied with regard to their ability to incorporate nucleotides and nucleotide analogs, including chain terminators and dye-labeled nucleotides.
DNA polymerases of other families have also been tested for the efficiency of incorporation of modified nucleotides. Since a number of these applications involve a heat treatment step for DNA strand denaturation, thermostable enzymes of Family B have been examined, including a number derived from hyperthermophilic archaea (e.g. Vent DNA polymerase). Early experiments with these polymerases noted difficulties incorporating modified nucleotides such as ddNTPs (12), particularly those labeled with dyes (B. Slatko, personal communication). (While recognizing that it is the NMP form of nucleotides and analogs that is incorporated into DNA products, we have chosen to refer to NTP as both the substrate and incorporated product to improve readability of the manuscript.) These observations discouraged use of these enzymes in sequencing applications and suggested that nucleotide binding determinants differed from those of Family A DNA polymerases.
More recently, the development of new generations of dyes prompted a re-examination of the use of archaeon DNA polymerases for dye–nucleotide insertion. Tests of the extent of incorporation of nine indocyanine and rhodamine dCTP analogs by Vent and Taq DNA polymerases identified two dye-substituted dCTP analogs whose incorporation was preferred over dCTP: the indocyanine analog IC3-dCTP and the rhodamine analog R6G-dCTP (13). These studies did not, however, address incorporation of the corresponding dye-labeled chain terminators.
Although ddNTPs are the dominant chain terminators utilized in DNA sequence analysis, other analogs could potentially be used as chain terminators. For example, cordycepin (3′-deoxyadenosine) triphosphate incorporation by Vent DNA polymerase is similar to that of ddNTPs (12). Recently, dye-labeled AcycloTerminators have been commercialized by PerkinElmer Life Sciences for use in DNA sequencing and SNP detection. Such acyclic derivatives substitute a 2-hydroxyethoxymethyl group for the 2′-deoxyribofuranosyl sugar normally present in dNTPs and thus are not substrates for polymerase extension (Scheme 1).
Scheme 1.
The use of unlabeled acyclo terminators (acyNTPs) in DNA sequencing reportedly yields sequencing ladders virtually identical to those formed with ddNTPs, although 10 times higher terminator concentrations are required when used with Klenow fragment or AMV reverse transcriptase (14).
DNA sequencing technology has been enhanced by the availability of polymerase variants that more readily incorporate terminators. A prime example has been the use of Family A Taq DNA polymerase variants such as Thermo Sequenase and AmpliTaq FS. Family B DNA polymerase variants with altered incorporation properties have also been described, although the magnitude of the effect is smaller than that observed with the Family A DNA polymerases (12,15).
We have compared the influence of dyes and terminator type on terminator incorporation using a variety of polymerases and polymerase variants. These findings provide information on the structural basis for nucleotide recognition by polymerases and suggest methods for improving DNA sequencing technologies.
MATERIALS AND METHODS
DNA polymerases
All DNA polymerases described here lack 3′→5′ exonuclease activity. Taq DNA polymerase (Amersham Pharmacia Biotech, Piscataway, NJ) and its F667Y derivative Thermo Sequenase (Amersham Pharmacia Biotech) naturally lack this activity, while the archaeon DNA polymerases have been genetically modified within the conserved exonuclease motif DIE to AIA to eliminate this activity (16–18). Vent and Deep Vent DNA polymerases were from New England Biolabs and Pfu DNA polymerase was from Stratagene (La Jolla, CA). 9°N DNA polymerase and the A485L variant of this polymerase were purified according to Southworth et al. (17). The Vent A488L and Y499L variants were isolated as previously described (12). Vent A488L and 9°N A485L activities were measured as previously described (13) using 15 nM primed M13 as template. Polymerase units were those defined by the supplier, in all cases reflecting conversion of 10 nmol dNTP into an acid-insoluble form in 30 min under defined reaction conditions.
The A485L variant of 9°N DNA polymerase was created using two successive rounds of PCR (19). In the first, primers 216–153 (5′-CAGGCAGAGGCTTATAAAAATCCTCGCCAACAGCTT-3′) and 175–70 (5′-GGTGGCAGCAGCCAACTCAGCTTCCT-3′) were used together with pNEB917 (M. Southworth, New England Biolabs) to generate a PCR product containing the desired DNA sequence changes within the polymerase gene. The PCR product was diluted and amplified using primers 175–70 and 216–155 (5′-GATTCTCATGATAAGCTACGCCGA-3′). The resulting product was cut with BamHI and BsiWI and the resulting fragment was used to replace the similarly sized fragment in pNEB917. DNA sequence analysis of the resulting construct confirmed that only the desired changes had occurred. The polymerase produced by this altered coding sequence (Therminator DNA polymerase) was purified using minor modifications of the method of Southworth et al. (17).
Nucleotides and analogs
Dye terminators were either purchased from PerkinElmer Life Sciences (Boston, MA) or were the kind gift of Phil Buzby (PerkinElmer Life Sciences). AcyCTP was prepared by Roger Knott (New England Biolabs Organic Synthesis Division, Beverly, MA).
Titration assay for incorporation of chain terminators
The propensity of a series of DNA polymerases to incorporate CTP analogs (Table 1) was evaluated using titration assays. Incorporation of modified bases was assayed by mixing (A) 2.5 µl of 0.04 µM 32P-5′-end-labeled S1224-primed M13mp18 (materials from New England Biolabs, substrate prepared as described in 16), 0.04 U/µl thermostable pyrophosphatase (New England Biolabs), 0.1 mM each dNTP, 0.15 U/µl DNA polymerase in 2× reaction buffer with (B) 2.5 µl of nucleotide analog solution to yield the final ratios of (analog:dNTP) indicated in the figures. The 2× reaction buffers contained: Family A DNA polymerase buffer (Amersham Pharmacia Biotech), 52 mM Tris–HCl pH 9.5, and 13 mM MgCl2; Family B DNA polymerase buffer, 20 mM KCl, 40 mM Tris–HCl, pH 8.8 at 25°C, 20 mM (NH4)2SO4, 4 mM MgSO4, 0.2% Triton X-100. Reactions with Pfu DNA polymerase additionally included 0.1 mg/ml BSA (final concentration). After incubating at 72°C for 20 min, the reactions were halted by addition of 4 µl stop/dye solution (deionized formamide containing 0.3% xylene cyanole FF, 0.3% bromophenol blue, 0.37% EDTA, pH 7.0). Samples were then heated at 72°C for 3 min and separated on a QuickPoint DNA sequencing gel (Invitrogen, Carlsbad, CA) run at 1200 V. The gel was fixed by soaking in 10% acetic acid/10% ethanol, dried and polymerization products visualized by autoradiography. The extent of terminator incorporation was determined by visual inspection of the autoradiographs.
Table 1. Dye–terminator incorporation efficiency by Vent DNA polymerase.
| Equivalent incorporation | BODIPY Fl–ddCTP |
| BODIPY R6G–ddCTP | |
| IRD40–ddCTP | |
| IRD700–ddCTP | |
| Cyanine 5–acyCTP | |
| Decreased incorporation | Cyanine 3–ddCTP |
| Cyanine 5–ddCTP | |
| FAM–ddCTP | |
| JOE–ddCTP | |
| R110–ddCTP | |
| Cyanine 3–acyCTP | |
| Enhanced incorporation | BODIPY TMR–ddCTP |
| BODIPY TR–ddCTP | |
| R6G–ddCTP | |
| ROX–ddCTP | |
| TAMRA–ddCTP | |
| BODIPY FL–acyGTP | |
| FAM–acyGTP | |
| Fl-12–acyCTP | |
| IRD40–acyNTP | |
| IRD700–acyNTP | |
| R110–acyGTP | |
| R6G–acyATP | |
| ROX–acyCTP | |
| TAMRA–acyCTP | |
| TAMRA–acyUTP |
Incorporation efficiencies are relative to that of the analogous ddNTP and were determined by the titration assay described in the text. The designation NTP is given for cases in which analogs of all four nucleotide bases were tested.
DNA sequence analysis using 9°N A485L DNA polymerase
Four separate reactions (one for each dye–acyNTP terminator) generated DNA sequencing termination ladders for analysis. Reactions contained (in 20 µl) 37.5 ng/µl pUC19, 0.16 µM S1224 M13/pUC sequencing primer, 0.2 mM dNTP, 50 mM Tris–HCl pH 8 at 25°C, 0.2 M KCl, 2 mM MgSO4, 0.02 U/µl thermostable inorganic pyrophosphatase, 0.04 U/µl 9°N A485L DNA polymerase and one IR700–acyNTP (2 µM IR700–acyATP, 0.3 µM IR700–acyCTP, 0.4 µM IR700–acyGTP or 0.8 µM IR700–acyUTP). Reactions were thermally cycled as follows: 94°C for 5 min; 30 cycles of 94°C for 30 s, 50°C for 30 s and 72°C for 30 s; 72°C for 7 min.
Unincorporated dye-labeled terminators were removed by gel filtration using CentriSep mini-columns (Princeton Separations, Adelphia, NJ), lyophilized and suspended in 5 µl of deionized formamide containing 0.3% xylene cyanole FF, 0.3% bromophenol blue, 0.37% EDTA pH 7.0. Reactions were loaded onto a 3.7% KBplus gel (Li-COR Biosciences, Lincoln, NE) and reaction products were separated and detected with an automated NEN Model 4200 Global IR2 DNA sequencer (distibuted by Li-COR). DNA sequencing traces were processed and displayed using the accompanying e-Seq software.
RESULTS
Titration assay to determine relative incorporation efficiency
The relative efficiency of terminator incorporation was assessed using a titration assay (12). A primed single-stranded DNA substrate was incubated in a series of reaction mixtures, each containing a fixed concentration of enzyme, template and dNTPs with increasing amounts of terminator. Following an extension reaction, termination products were separated by denaturing polyacrylamide gel electrophoresis and visualized using autoradiography. Parallel reactions containing no terminator confirmed sufficient polymerase had been added to complete replication in the absence of the terminator. Thus, the observed bands arose from terminator incorporation rather than incomplete replication by the DNA polymerase.
Once the spectrum of termination products was determined, a comparison of the length, uniformity and clarity of these patterns was used to evaluate incorporation of the terminator. Reaction conditions producing shorter products at a given terminator concentration were those in which the terminator was more efficiently incorporated by the DNA polymerase. Conversely, when comparisons revealed identical banding patterns at different terminator concentrations, the lower concentration identified conditions more favorable to terminator incorporation.
The size distribution of termination products is determined by the relative rates of dNTP and terminator incorporation. These competing reactions utilize the same pool of template and continue until replication halts, either by incorporation of a terminator or by extension to the end of the template. The incorporation efficiency of two terminators can be compared using parallel reactions differing only in the type and concentration of terminator. Under conditions where these parallel reactions yield identical products, the relative rates of terminator and dNTP incorporation must be identical in each reaction. In this situation, dNTP incorporation dynamics are expected to be identical in each of the parallel reactions. Accordingly, the relative incorporation efficiency of the two terminators will be reflected in the concentrations of terminators in the two reactions. For example, if a first reaction contains 10-fold more terminator than a second reaction, insertion of the first terminator is 10-fold less efficient than that of the second. We note that all reactions described here have at least a 50-fold excess of terminator over template, and thus the terminator concentration would not be expected to change significantly during the reaction.
AcyNTP incorporation by Vent DNA polymerase
AcyNTPs, lacking the ribose moiety and more particularly the 3′-OH needed for chain extension, are potential alternatives to ddNTPs in chain terminator DNA sequence analysis. The propensity of DNA polymerases to incorporate acyNTPs was evaluated by generating DNA sequencing ladders in the presence of increasing concentrations of terminator and comparing the range of reaction products.
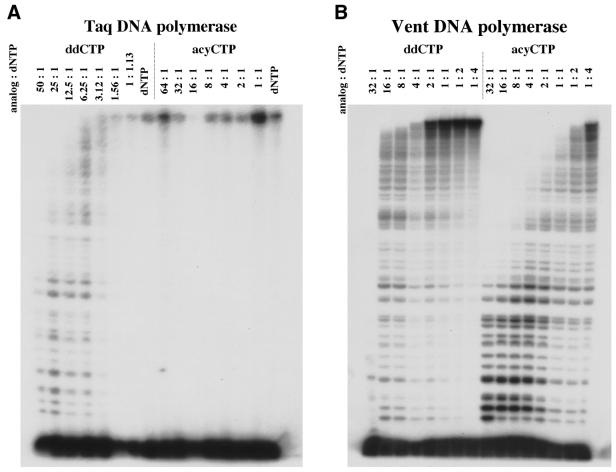
Taq DNA polymerase showed a large discrimination against ddNTPs and an even larger discrimination against acyNTPs, as evidenced by the higher concentration of acyNTP required to give equivalent sequencing patterns (Fig. 1A). In contrast, Vent DNA polymerase incorporated ddNTPs ∼16-fold less efficiently than the corresponding acyNTPs in the experiment shown in Figure 1B. Specifically, a 16:1 ratio of ddCTP:dNTP generated a similar banding pattern to a 1:1 ratio of acyCTP:dNTP.
Figure 1.
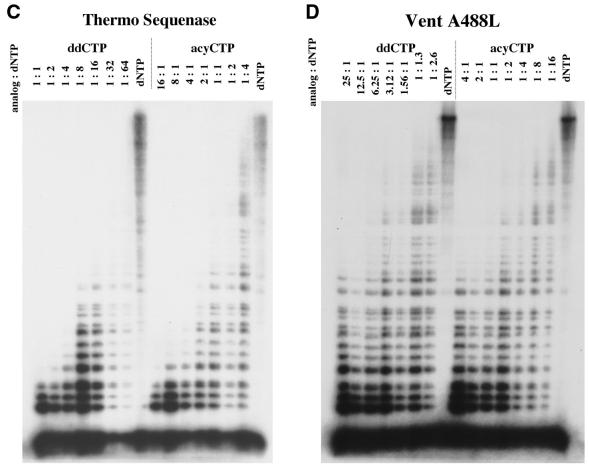
Incorporation of ddCTP and acyCTP. Titration assay for the indicated terminators using (A) Taq (B) Vent, (C) ThermoSequenase or (D) Vent A488L DNA polymerase. Extension of a 32P-labeled primer on a M13mp18 single-stranded substrate was examined in the presence of the indicated ratios of terminator to dNTP. The lane marked dNTP is a control reaction performed in the absence of terminators.
Thermo Sequenase and Vent A488L are variants of Taq and Vent DNA polymerases, respectively, with increased capacity to incorporate ddNTPs (12,20). As anticipated, each variant more effectively incorporated ddNTPs than the parental enzyme (Fig. 1). In addition, each variant had an increased capacity to incorporate acyNTP terminators and, in fact, the concentration range in which acyCTP was maximally incorporated is similar for both enzymes (Fig. 1C and D). In these and other experiments, Thermo Sequenase displayed a 10–20-fold preference for ddCTP over acyCTP, while Vent A488L reversed that bias, favoring acyCTP over ddCTP by 10–20-fold (Fig. 1C and D). We note that similar biases were observed in the parental polymerases and thus appear to be fundamental, distinguishing properties of these two classes of polymerases.
Dye substitutions affect the efficiency of ddNTP incorporation
Dye substitutions are typically attached to the nucleoside base. A reasonable concern is whether these modifications impede incorporation. Indeed, this was the case with Vent DNA polymerase using the first generation of dye–ddNTPs developed for fluorescence sequencing (B. Slatko, personal communication). Subsequently, Ilsey and Buzby identified dye-labeled dCTP analogs whose incorporation by Vent DNA polymerase is enhanced relative to unmodified dCTP (13), leaving open the question as to whether this new generation of dyes might also enhance incorporation when coupled to chain terminators. This hypothesis was tested by comparative titrations using ddCTP and a variety of dye–ddCTP analogs (Table 1). Three classes of dye terminators were identified based on the patterns of termination products produced with Vent DNA polymerase. In the first class, banding patterns for ddCTP and the analog had similar concentration dependencies, indicating that the modified base was incorporated no better than the corresponding ddCTP. In the second class, incorporation of the dye-substituted base was less than that observed with the normal ddCTP, as indicated by a dominance of higher molecular weight bands at comparable terminator concentrations. In the final class of analogs the distribution of terminated products was shifted to lower molecular weights, indicating an increased incorporation relative to the parent ddCTP. Importantly, similar banding patterns were produced by this latter group of analogs, signaling retention of base specificity by these analogs. Assays with Thermo Sequenase yielded similar groupings as had been generally observed for dye–dCTP analogs (13; data not shown).
Additive effects of dye substitutions and acyNTP chain terminators
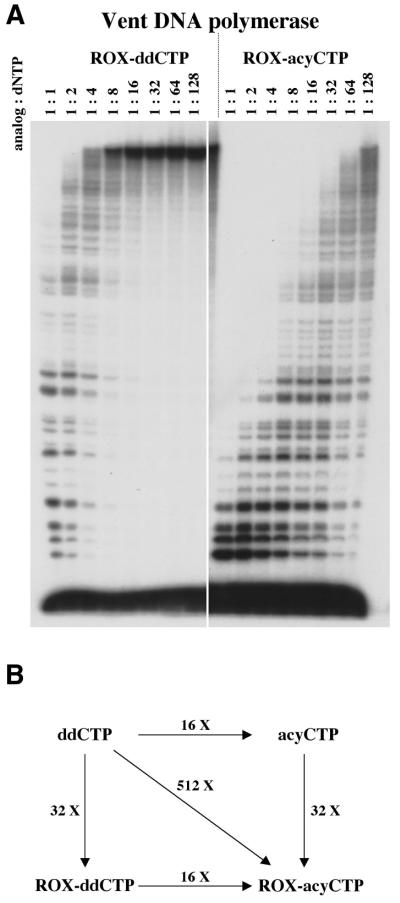
We next asked whether attached dyes could also enhance incorporation of acyNTPs by Vent DNA polymerase. Again using the titration assay, we found that dye–acyCTPs were incorporated ∼32-fold more efficiently than the corresponding dye–ddCTPs (Fig. 2). This value was similar to that observed for acyCTP versus ddCTP incorporation, suggesting that dye and terminator structures had largely independent effects upon enhanced incorporation. Consistent with this prediction, the enhancement for ddCTP versus ROX–acyCTP incorporation is ∼500-fold, the product of acyclo versus dideoxy (16-fold) and ROX dye (32-fold) enhancements. The hierarchy of dye effects on incorporation observed with dideoxy derivatives was preserved with the acyclic derivatives. This hierarchy was maintained with Thermo Sequenase as well, although incorporation of the ddNTP analog was greater than the corresponding acyclic derivative (data not shown).
Figure 2.
ROX dye attachment enhances both dideoxy and acyclic terminator addition by Vent DNA polymerase. (A) Titration assays were used to compare incorporation of the indicated terminators by Vent DNA polymerase. Numbers refer to the ratio of terminator analog to dNTP. (B) Relative incorporation of ddCTP, acyCTP, ROX–ddCTP and ROX–acyCTP by Vent DNA polymerase is diagrammed to emphasize the elements that enhance terminator incorporation.
Hyperthermophilic archaeon DNA polymerases have similar terminator preferences
The similarity in amino acid sequence of Family B archaeon DNA polymerases suggests that these enzymes might share similar dye–terminator incorporation properties. To test this proposition, terminator titration assays were repeated using an expanded set of archaeon DNA polymerases, specifically Vent, Deep Vent, Pfu and 9°N. The pattern of ROX–acyCTP incorporation was identical for these four enzymes (Fig. 3). Each polymerase also incorporated dye–acyNTPs more efficiently than the corresponding dye–ddNTPs (data not shown). Thus, we anticipate that the dye–terminator incorporation properties of other hyperthermophilic archaeon DNA polymerases will be similar.
Figure 3.
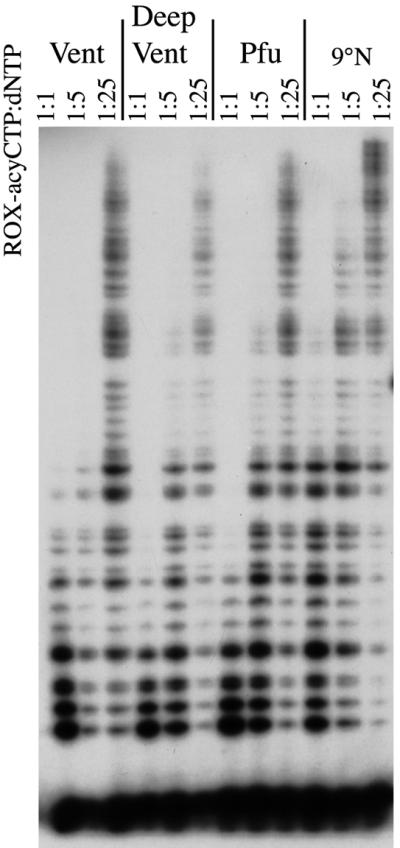
Hyperthermophilic Family B archaeon DNA polymerases share terminator incorporation properties. Incorporation efficiencies of ROX-acyCTP by Family B DNA polymerases Vent, Deep Vent, Pfu and 9°N DNA polymerases were compared using the titration assay. Numbers refer to the ratio ROX–acyCTP:dNTP in the reaction mixture.
Archaeon DNA polymerase variants that enhance dye–acyNTP terminator incorporation
Previous reports have demonstrated the increased ability of Vent DNA polymerase variants to incorporate ddNTPs and cordycepin 5′-triphosphate (3′-dATP) (12). We extended these results to additional compounds using the most promiscuous variant, Vent A488L DNA polymerase. Incorporation of TAMRA–acyCTP was enhanced ∼16-fold with this variant when compared to the wild-type polymerase (Fig. 4). In separate experiments, the preference for acyNTPs as opposed to ddNTPs was also displayed by this polymerase, as were the dye incorporation preferences noted for the wild-type polymerases. Consequently, the increased incorporation reflects effects above and beyond dye and terminator influences.
Figure 4.
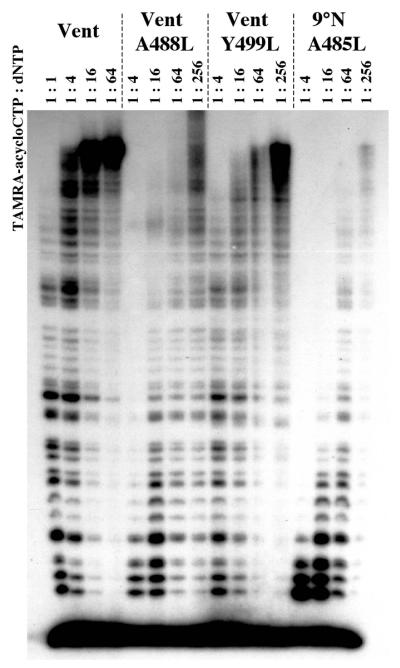
Polymerase variants can improve dye–acyNTP terminator incorporation. The incorporation efficiencies of TAMRA–acyCTP by Vent, Vent A488L, Vent Y499L and 9°N A485L DNA polymerases were compared using the titration assay. Numbers refer to the ratio TAMRA–acyCTP:dNTP in the reaction mixture.
To broaden the applicability of this observation, the analogous 9°N DNA polymerase variant A485L was created and employed in the same assays. Relative incorporation of dye–terminators was similar to those observed with the Vent A488L DNA polymerase with regard to both terminator type (i.e. ddNTP or acyNTP) and dye label (Fig. 4), establishing the common effects of mutants at this locus among archaeon DNA polymerases.
Additional experiments with a second Vent DNA polymerase variant, Y499L, displayed increased incorporation similar in nature to the A488L variant, demonstrating that increased incorporation was not limited to the A488L variant (Fig. 4).
Use of dye–acyNTP terminators with archaeon DNA polymerase variants for DNA sequencing
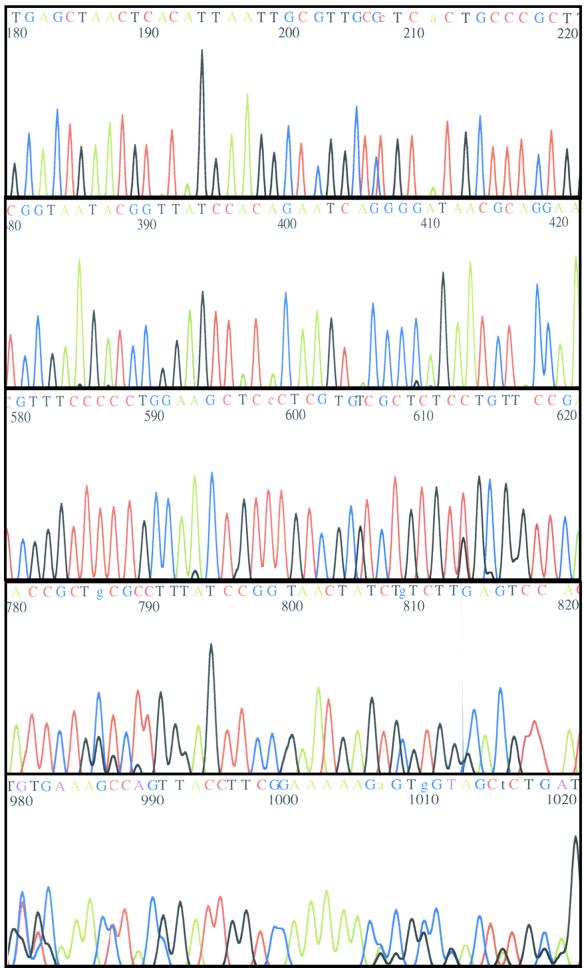
The enhanced incorporation of modified terminators noted in the examples enables a new system of polymerases and reagents for use in automated DNA sequencing. These reactions rely upon incorporation of four chain terminators, each corresponding to one of the four bases normally present in DNA, and each labeled with a detectable fluorescent dye. The feasibility of such a reaction was tested using dye-labeled acyNTPs. Such reactions tested not only the ability to incorporate the terminator, but also gave a basis for assessing the specificity of the reaction. Reaction products were separated by denaturing polyacrylamide gel electrophoresis and detected via fluorescence upon laser excitation (Fig. 5). Sequencing reactions were productive, providing useful sequence beyond 1000 bp (Table 2). Sequencing reactions using labeled primers and unlabeled acyNTPs yielded comparable data (data not shown).
Figure 5.
DNA sequence determination with dye–acyNTP terminators. The output of an NEN Model 4200 Global IR2 automated DNA sequencer is illustrated. Reactions were generated with 9°N A485L DNA polymerase. pUC19 was sequenced with primer S1224 using IR700–acyNTPs. Confidence levels for this sequencing reaction are given in Table 2.
Table 2. 9°N A485L DNA polymerase sequencing accuracy.
| Region | Quality value | No. of errors | Accuracy (%) |
|---|---|---|---|
| 1–250 | 35 | 2 | 99.2 |
| 251–500 | 34 | 2 | 99.2 |
| 501–750 | 30 | 5 | 98.0 |
| 751–1000 | 25 | 11 | 95.6 |
Regions were numbered beginning with the first called base nearest the primer. Quality values, as calculated by eSeq Phred algorithms (25), were averaged over the indicated regions. A quality value of 30 corresponds to a 0.001 probability of being incorrect. Values listed in No. of errors include ambiguous calls. The percent accuracy is calculated as (no. of errors/no. of bases in the region) × 100.
DISCUSSION
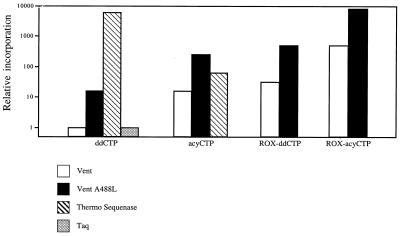
The quest for optimal DNA sequencing reagents presents a formidable challenge given the variety of polymerases, nucleotide analogs and reaction conditions available. These studies identify three elements that enhance terminator incorporation, thus facilitating DNA sequence determination. These elements are (i) acyNTPs, a preferred substrate for archaeon DNA polymerases, (ii) dyes that enhance incorporation when conjugated to nucleoside bases and (iii) archaeon DNA polymerase variants with enhanced ability to incorporate chain terminators such as ddNTPs and acyNTPs. To a first approximation, these elements act in an additive way, with each element making a distinct contribution (Fig. 6). Thus, the 8000-fold enhancement of ROX–acyCTP incorporation by Vent A488L DNA polymerase, compared with ddCTP incorporation by Vent DNA polymerase, can be broken down into its component parts: a 32-fold enhancement due to the ROX dye, 16-fold due to the acyCTP and 16-fold due to the A488L mutation.
Figure 6.
Incorporation enhancement attributable to terminator and polymerase variations. Incorporation is represented on a logarithmic scale relative to ddCTP incorporation by Vent DNA polymerase. Values for ThermoSequenase incorporation of ddCTP are inferred from Tabor and Richardson (20). Taq DNA polymerase failed to incorporate detectable amounts of acyCTP and thus is not represented. ROX analog incorporation was only examined for Vent and Vent A488L DNA polymerases.
Incorporation of acyNTPs by Family B DNA polymerases has previously been studied in the context of the antiviral activity of analogs such as acyclovir (i.e. acycloguanine). The Family B DNA polymerases from Herpes virus type 2 and human cytomegalovirus insert acyGTP more readily than ddGTP (21), a characteristic shared with the hyperthermophilic archaeon DNA polymerases tested here. In contrast, human DNA polymerase α incorporates acyGTP less efficiently than ddGTP (21). This brief survey of polymerases demonstrates that classification in Family B is insufficient to predict whether ddNTPs or acyNTPs will be more efficiently incorporated. Nonetheless, we anticipate that, given the high degree of sequence similarity, most, if not all, hyperthermophilic archaeon Family B DNA polymerases will share the terminator incorporation patterns we have observed. Sequence similarities suggest that other archaeon non-thermophilic DNA polymerases may also follow this pattern.
Taq DNA polymerase discriminates against ddNTPs and even more strongly against acyNTPs (Fig. 1A). In the case of ddNTP, discrimination is manifest in a decreased rate of phosphodiester bond formation, as opposed to a decrease in the affinity of substrate binding (22). The decreased rate has been ascribed to loss of hydrogen bonding between the 3′-OH of the substrate dNTP and the pro-Sp oxygen in the same dNTP (23), possibly affecting alignment of the reactive groups. In contrast, the F667Y variant of Taq DNA polymerase (Thermo Sequenase; 20) modestly discriminates against ddNTPs and only slightly more against acyNTPs (Fig. 1C), presumably reflecting the ability of the substituted tyrosine hydroxyl group to substitute for the missing 3′-OH. Indeed, T7 DNA polymerase containing tyrosine at the analogous position does not distinguish between dNTP and ddNTP incorporation, a fact reflected in virtually identical kinetic parameters involving nucleotide binding and phosphodiester bond formation (22).
While the activity of the F667Y Taq variant can be rationalized in direct terms, the effects of DNA polymerase variants 9°N A485L, Vent A488L and Pfu A486Y (15) are less apparent. The crystal structures of the apo 9°N DNA polymerase (24) and of the Family B RB69 DNA polymerase replication complex (11) predict that this residue will face away from the active site, with little likelihood of directly interacting with the nucleotide substrate. Thus, this mutation appears to operate indirectly, perhaps affecting the conformational shift noted prior to polymerization.
The RB69 replication complex exhibits stacking interactions between the ribose of the substrate TTP and Y416, an amino acid conserved in Family B archaeon DNA polymerases and implicated in ribonucleotide discrimination (12). Local sequence and crystallographic similarities suggest that this stacking will also be available in the archaeon DNA polymerases. Even though such stacking cannot occur with acyclic analogs, such analogs are preferred substrates over dideoxynucleotides, perhaps due to the increased conformational flexibility inherent in the acyclic structure. It seems somewhat paradoxical that in the case of the T7 DNA replication complex no stacking interactions with the ribose ring are predicted and yet dideoxy are favored over acyclic analogs. Indeed, given the differing acyNTP and ddNTP sensitivities, it seems probable that alternate sugar contacts are involved in Family A and B polymerases, contacts that ultimately affect positioning of active site residues. Thus, despite global similarities, significant differences appear to exist in the active sites of these two families of DNA polymerases.
At present it is difficult to predict the structural or chemical basis by which dye substituents enhance nucleotide incorporation. Ilsey and Buzby noted in the series of compounds tested that the most hydrophobic indocyanine or rhodamine dyes were preferred substrates for polymerization (13). However, a limited number of dyes were tested, making it difficult to predict which elements are most important in the observed incorporation bias. Presumably, the dye effect arises from enhanced binding of the base, although effects on the transition into an active polymerization complex cannot be excluded.
The apparent additivity of dye substitution, terminator type and mutations in facilitating terminator incorporation suggests that these effects act independently. In the absence of further kinetic studies it is not possible to state whether they act on the same rate limiting step or even if the same rate limiting step is encountered in terminator incorporation as opposed to dNTP incorporation. Nor have the effects of binding and the chemistry of bond formation been separately identified. More detailed kinetic and structural studies hold the promise of dissecting the components involved in the observed discrimination. Hopefully such studies will illuminate not only the basis of the observed effects, but also lend insight into the operation of the active site in DNA polymerization.
One outcome of these studies is an alternative environment for DNA sequencing, applicable in both long range primary sequence determination (Fig. 5) and in the short extensions required for SNP analysis. Incorporation dynamics are comparable to dye–ddNTP incorporation by Taq F667Y DNA polymerase variants, allowing the use of low concentrations of dye–terminators while retaining sequence specificity. As such, these reagents are poised to complement and extend currently available methods of DNA sequence analysis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Phil Buzby and Jim DiMeo of PerkinElmer Life Sciences for providing many of the dye-labeled compounds used in these studies and Roger Knott for acyCTP synthesis. We gratefully acknowledge the technical assistance of Donna Novi of PerkinElmer Life Sciences in performing DNA sequence analyses. Insightful discussions with Barton Slatko, Tom Evans, Sriharsha Pradhan and Phil Buzby aided greatly in the preparation of this manuscript. We are also indebted to Don Comb for fostering a supportive research environment at New England Biolabs.
REFERENCES
- 1.Steitz T.A. (1999) DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem., 274, 17395–17398. [DOI] [PubMed] [Google Scholar]
- 2.Joyce C.M. and Steitz,T.A. (1994) Function and structure relationships in DNA polymerases. Annu. Rev. Biochem., 63, 777–822. [DOI] [PubMed] [Google Scholar]
- 3.Saiki R.K., Scharf,S., Faloona,F., Mullis,K.B., Horn,G.T., Erlich,H.A. and Arnheim,N. (1985) Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 230, 1350–1354. [DOI] [PubMed] [Google Scholar]
- 4.Sanger F., Nicklen,S. and Coulson,A.R. (1977) DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkar G., Yoon,H.S. and Sommer,S.S. (1992) Dideoxy fingerprinting (ddE): a rapid and efficient screen for the presence of mutations. Genomics, 13, 441–443. [DOI] [PubMed] [Google Scholar]
- 6.Nikiforov T.T., Rendle,R.B., Goelet,P., Rogers,Y.H., Kotewicz,M.L., Anderson,S., Trainor,G.L. and Knapp,M.R. (1994) Genetic Bit Analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res., 22, 4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X. and Kwok,P.Y. (1997) Template-directed dye-terminator incorporation (TDI) assay: a homogeneous DNA diagnostic method based on fluorescence resonance energy transfer. Nucleic Acids Res., 25, 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Levine,L. and Kwok,P.Y. (1999) Fluorescence polarization in homogeneous nucleic acid analysis. Genome Res., 9, 492–498. [PMC free article] [PubMed] [Google Scholar]
- 9.Ito J. and Braithwaite,D.K. (1991) Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res., 19, 4045–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heringa J. and Argos,P. (1994) Evolution of viruses as recorded in their polymerase sequences. In Morse,S.S. (ed.), The Evolutionary Biology of Viruses. Raven Press, New York, NY, pp. 87–103.
- 11.Franklin M.C., Wang,J. and Steitz,T.A. (2001) Structure of the replicating complex of a pol α family DNA polymerase. Cell, 105, 657–667. [DOI] [PubMed] [Google Scholar]
- 12.Gardner A.F. and Jack,W.E. (1999) Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res., 27, 2545–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilsey D.D. and Buzby,P.R. (1999) Incorporation of fluorescent nucleotides by DNA polymerases. FASEB J., 13, A144. [Google Scholar]
- 14.Trainor G.L. (1996) DNA sequencing method using acyclonucleoside triphosphates. US patent 5,558,991.
- 15.Evans S.J., Fogg,M.J., Mamone,A., Davis,M., Pearl,L.H. and Connolly,B.A. (2000) Improving dideoxynucleotide-triphosphate utilisation by the hyper-thermophilic DNA polymerase from the archaeon Pyrococcus furiosus. Nucleic Acids Res., 28, 1059–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong H., Kucera,R.B. and Jack,W.E. (1993) Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement and exonuclease activities. J. Biol. Chem., 268, 1965–1975. [PubMed] [Google Scholar]
- 17.Southworth M.W., Kong,H., Kucera,R.B., Ware,J., Jannasch,H.W. and Perler,F.B. (1996) Cloning of thermostable DNA polymerases from hyperthermophilic marine Archaea with emphasis on Thermococcus sp. 9 degrees N-7 and mutations affecting 3′-5′ exonuclease activity. Proc. Natl Acad. Sci. USA, 93, 5281–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathur E.J. (1996) Exonuclease-deficient thermostable Pyrococcus furiosus DNA polymerase I. US patent 5,558,991.
- 19.Colosimo A., Xu,Z., Novelli,G., Dallapiccola,B. and Gruenert,D.C. (1999) Simple version of “megaprimer” PCR for site-directed mutagenesis. Biotechniques, 26, 870–873. [DOI] [PubMed] [Google Scholar]
- 20.Tabor S. and Richardson,C.C. (1995) A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA, 92, 6339–6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid R., Mar,E.C., Huang,E.S. and Topal,M.D. (1988) Insertion and extension of acyclic, dideoxy and ara nucleotides by herpesviridae, human alpha and human beta polymerases. A unique inhibition mechanism for 9-(1,3-dihydroxy-2-propoxymethyl)guanine triphosphate. J. Biol. Chem., 263, 3898–3904. [PubMed] [Google Scholar]
- 22.Brandis J.W., Edwards,S.G. and Johnson,K.A. (1996) Slow rate of phosphodiester bond formation accounts for the strong bias that Taq DNA polymerase shows against 2′,3′-dideoxynucleotide terminators. Biochemistry, 35, 2189–2200. [DOI] [PubMed] [Google Scholar]
- 23.Doublie S., Tabor,S., Long,A.M., Richardson,C.C. and Ellenberger,T. (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature, 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez A.C., Park,H.-W., Mao,C. and Besse,L.S. (2000) Crystal structure of a pol alpha family DNA polymerase from the hyperthermophile archaeon Thermococcus sp. 9°N7. J. Mol. Biol., 299, 447–462. [DOI] [PubMed] [Google Scholar]
- 25.Ewing B. and Green,P. (1998) Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res., 8, 186–198. [PubMed] [Google Scholar]