Abstract
The Brain Imaging Data Structure (BIDS) is a specification accompanied by a software ecosystem that was designed to create reproducible and automated workflows for processing neuroimaging data. BIDS Apps flexibly build workflows based on the metadata detected in a dataset. However, even BIDS valid metadata can include incorrect values or omissions that result in inconsistent processing across sessions. Additionally, in large-scale, heterogeneous neuroimaging datasets, hidden variability in metadata is difficult to detect and classify. To address these challenges, we created a Python-based software package titled “Curation of BIDS” (CuBIDS), which provides an intuitive workflow that helps users validate and manage the curation of their neuroimaging datasets. CuBIDS includes a robust implementation of BIDS validation that scales to large samples and incorporates DataLad—a version control software package for data—as an optional dependency to ensure reproducibility and provenance tracking throughout the entire curation process. CuBIDS provides tools to help users perform quality control on their images’ metadata and identify unique combinations of imaging parameters. Users can then execute BIDS Apps on a subset of participants that represent the full range of acquisition parameters that are present, accelerating pipeline testing on large datasets.
Keywords: BIDS, MRI, Brain, Neuroimaging, Software, Curation, Validation, Metadata, Version control, Heterogeneity
1. Introduction
The Brain Imaging Data Structure (BIDS) specification provides a standardized format for organizing and describing neuroimaging data (Gorgolewski et al., 2016). BIDS relies on specific nested directory structures and filename conventions and requires that each MR image file (e.g. Neuroimaging Informatics Technology Initiative or NIfTI) be accompanied by a JavaScript Object Notation (JSON) sidecar—a data dictionary detailing its corresponding image’s metadata. BIDS is especially helpful when dealing with large, multimodal studies; as the number of subjects and runs increases, generalizable structures and standards become not only beneficial but essential. Pipelines that ingest BIDS datasets—commonly referred to in the BIDS software ecosystem as “BIDS Apps ” (Gorgolewski et al., 2017)—such as fMRIPrep and QSIPrep—rely heavily on correct specification of BIDS, as they build workflows based on the metadata encountered (Esteban et al., 2019; Cieslak et al., 2021). While generally an important and useful feature, this workflow construction structure can also be a vulnerability: if the BIDS metadata is inaccurate, a BIDS app may build an inappropriate (but technically “correct”) preprocessing pipeline. For example, a fieldmap with no IntendedFor field specified in its JSON sidecar is not technically incorrect but rather incomplete. When a participant containing a fieldmap missing an IntendedFor field is run through a MRI image type processing pipeline such as fMRIPrep and QSIPrep, the pipeline will execute with neither errors nor warnings but will skip distortion correction. Curating the dataset with CuBIDS before running it through such pipelines will allow users to easily identify all instances of fieldmaps missing IntendedFor references—CuBIDS places those scans in separate groups. Users must understand and verify that the metadata present in BIDS is correct. This usually requires meticulous curation—the process of checking and fixing filename or metadata issues present in a dataset. In the context of the lifecycle of a neuroimaging dataset, the CuBIDS curation workflow begins directly after the data has been organized into a BIDS directory structure with BIDS-like filenames.
While large, multi-modal neuroimaging datasets constitute extremely valuable data resources, they also frequently possess substantial heterogeneity in their image acquisition parameters. BIDS provides an ideal structure for organizing neuroimaging data, but the size and complexity of large-scale datasets can render curation both tedious and difficult. Data curation can be an ad-hoc process that involves substantial manual intervention; such manual curation is usually neither well tracked nor reproducible. Thus, curation constitutes a major vulnerability in the field-wide effort to create reproducible, analytic workflows for neuroimaging data. Finally, many current BIDS tools, including the BIDS Validator, MatlabBIDS, and PyBIDS (Yarkoni et al., 2019), that parse and interact with BIDS datasets were optimized for small fMRI studies and may behave erratically when given large quantities of heterogeneous data.
With these challenges in mind, we developed “Curation of BIDS” (CuBIDS): a software package that provides easy-to-use workflows that help users curate large BIDS datasets. CuBIDS provides users with customizable features to visualize heterogeneity in complex BIDS datasets and includes a robust, scalable implementation of BIDS validation that can be applied to arbitrarily-sized datasets. Critically, CuBIDS renders curation reproducible via an easy-to-use, wrapped implementation of DataLad (Halchenko et al., 2021) as an optional feature the user can access at will. Finally, CuBIDS provides tools to identify unique combinations of imaging parameters in a dataset so that users can test BIDS Apps on a subset of participants that represents the parameter space of the entire dataset. This option dramatically speeds up pipeline testing, as users can be assured that they have tested a BIDS App on the full range of acquisition parameters present in a dataset. As described below, CuBIDS facilitates an understanding of what is present in an MRI BIDS dataset, allows for reproducible BIDS curation, and accelerates successful data processing at scale.
2. Materials and methods
The standard lifecycle of a neuroimaging study begins with acquisition and ends with image analysis and hypothesis testing. CuBIDS’ role in this process begins directly after the data has been organized into a BIDS directory structure with BIDS-like filenames. The CuBIDS workflow ends with curated data in a repository, allowing for further exploration, pooling, meta-analysis, and runs of preprocessing pipelines. As curation occurs quite early in this timeline of preparing neuroimaging data for analysis, decisions made during curation will affect every subsequent stage.
2.1. Data and code availability statement
A copy of the small, example dataset whose curation we walk through in the Results section is compressed into a ZipFile and submitted with this paper under “Supplementary Material.” Additionally, the Philadelphia Neurodevelopmental Cohort (PNC), the dataset whose curation we summarize in the second portion of the Results, is publicly available in the Database of Genotypes and Phenotypes (dbGaP accession phs000607.v3.p2). The source code for CuBIDS is publicly available at https://github.com/PennLINC/CuBIDS, the documentation for our software is available at https://cubids.readthedocs.io/en/latest/, and our package is available for download on the Python Package Manager (pypi) https://pypi.org/project/cubids/.
2.2. Ethics statement
No new data was collected specifically for this paper. The Philadelphia Neurodevelopmental Cohort (PNC) (Satterthwaite et al., 2014) was approved by IRBs of The University of Pennsylvania and Children’s Hospital of Philadelphia. All adult participants in the PNC provided informed consent to participate; minors provided assent alongside the informed consent of their parents or guardian.
2.3. Overview
CuBIDS provides a workflow that aids users in curating large, heterogeneous BIDS datasets. CuBIDS summarizes a dataset’s metadata, enabling users to visualize and understand the variability in critical scanning parameters and fix errors when they are present. To do this, CuBIDS features several command line interface (CLI) programs (Table 1). Notably, all CuBIDS CLI programs wrap DataLad as an optional dependency so that the user can implement reproducible tracking at any stage of curation or revert to a prior state of their data. If the user wants to apply DataLad version control while using CuBIDS, they can run the CLI programs with the ---use-datalad optional flag set.
Table 1. CuBIDS command line interface programs.
CuBIDS features several command line interface (CLI) programs that help users curate and process BIDS datasets. We use the color-coded backgrounds to map each program to a stage in the curation workflow seen in Fig. 1. These programs were built for the steps of a study’s curation process. Some programs—such as print-metadata-fields, group, validate, and copy-exemplars--require only “read” access to the data and aid the user in visualizing a dataset’s heterogeneity. Others—such as apply, purge, undo, and remove-metadata-fields—require "write" access, as they involve modifying metadata, changing filenames, or removing entire subjects altogether.
| COMMAND | POSITIONAL ARGUMENTS | OPTIONAL ARGUMENTS | OUTPUT FILES | DESCRIPTION |
|---|---|---|---|---|
cubids-print-metadata-fields |
bids_dir the root of a BIDS dataset. It should contain sub-X directories and dataset_description.json |
Prints out all sidecar field names present in the dataset | ||
cubids-remove-metadata-fields |
bids_dir |
--fields FIELDS [FIELDS …] space-separated list of metadata fields to remove (default: []) |
Removes a desired list of metadata fields and can be used to delete Patient Health Information (PHI) from datasets | |
cubids-add-nifti-info |
bids_dir |
--use-datalad ensure that there are no untracked changes before finding groups and save dataset after NifTI info is added to sidecars (default: False) --force-unlock unlock dataset before adding NifTI information (default: False) |
Adds the following information from the headers of images in NifTI format: Obliquity, NumVolumes, Dim*Size, ImageOrientation, VoxelSizeDim* to its corresponding sidecar | |
cubids-validate |
bids_dir the root of a BIDS dataset. It should contain sub-X directories and dataset_description.json output_prefix file prefix to which tabulated output file(s) are written. If users pass in just a filename prefix (e.g. V1), then CuBIDS will put the four grouping outputs in bids_dir/code/CuBIDS. If the user a path (e.g. /Users/scovitz/BIDS/V1), then output files will go to the specified location. |
--sequential run the BIDS Validator sequentially on each subject (default: False) --ignore_nifti_headers disregard NlfTI header content during validation (default: False) --ignore_subject_consistency skip checking that any given file for one subject is present for all other subjects (default: False) --sequential-subjects SEQUENTIAL_SUBJECTS List: Filter the sequential run to only include the listed subjects, e.g. --sequential-subjects sub-01 sub-02 sub-03 (default: None) |
output_prefix_validation.tsTSV TSV file with one row per file containing a BIDS validation error. Columns: files, type, severity, description, code, url, subject |
Stable, robust version of the BIDS-validator |
cubids-group |
bids_dir output_prefix |
--acq-group-level ACQ GROUP LEVEL Levelat which acquisition groups are created options: "subject" or "session" (default: subject) --config CONFIG path to a config file for grouping (default: None) |
output_prefix_summary.tsv output_prefix_files.tsv output_prefix_AcqGrouping.tsv output_prefix_AcqGroupInfo.txt |
Produces four files that display the heterogenelt y present in the dataset in a user-friendly format |
cubids-purge |
bids_dir the root of a BIDS dataset. It should contain sub-X directories and dataset_description.json scans path to the txt file of runs whose associations should be purged |
--use-datalad save deletions after purge runs (default: False) |
Takes in a list of NlfTI images and removes them and all their association files and IntendedFor references from the dataset | |
cubids-apply |
bids_dir edited_summary_tsv the *_summary.csv that has been edited in the Mergelnto and RenameKeyGroup columns. files_tsv the *_files.csv that the *_summary.csv corresponds to. **Note: If the edited summary and files tsvs are located at bids_dir/code/CuBIDS, users may pass just the filename, if not, they mut pass the path to the file. output_prefix file prefix to which tabulated validator output is written. If users pass in just a filename prefix (e.g. V1), then CuBIDS will put the four grouping outputs in bids_dir/code/CuBIDS. If the user a path (e.g. /Users/scovitz/BIDS/V1), then output files will go to the specified location. |
--use-datalad save changes applied to the dataset (default: False) --acq-group-level ACQ_GROUP_LEVEL level at which acquisition groups are created options: "subject" or "session" (default: subject) --config CONFIG path to a config file for grouping (default: None) |
output_prefix_summary.tsv output_prefix_files.tsv output_prefix_AcqGrouping.tsv output_prefix_AcqGroupInfo.txt |
Applies the user’s edits to the *_summary.tsv file to the BIDS dataset |
cubids-copy-exemplars |
bids_dir path to the root of a BIDS dataset. It should contain sub-X directories and dataset_description.json exemplars_dir path to the root of a BIDS dataset containing one subject from each Acquisition Group. It should contain sub-X directories and dataset_description.json exemplars_tsv path to the .csv file that lists one subject from each Acquisition Group (*_AcqGrouping.csv from the cubids-group output) |
--use-datalad ensure that there are no untracked changes before finding groups and save exemplar dataset post creation (default: False) --min-group-size MIN_GROUP_SIZE minimum number of subjects an Acquisition Group must have in order to be included in the exemplar dataset (default: 1) |
copies one subject from each exemplar group into a separate directory | |
cubids-undo |
bids_dir |
reverts dataset to its state prior to the most recent saved modifications (only available to users using the DataLad-based version control option) |
2.4. Software development practices
We applied test-driven development while building CuBIDS, prioritizing writing tests for each new feature concurrent with its construction. We integrated CircleCI—a web-based continuous integration testing platform—into our GitHub repository so that each new commit is run through the full suite of tests. We apply a standardized approach to fixing bugs and adding features: first creating an issue on our GitHub page and then creating a new branch of our code base named specifically for fixing that issue. Once the issue is fixed on the new branch, a pull request merges the new branch into the main branch with the issue tagged. If all continuous integration tests pass and the merge is successful, the issue gets automatically closed. Centering our development process around both tests and issues has ensured the integrity of the code and facilitated both organization and documentation.
2.5. Installation, setup, and version control
We recommend users install CuBIDS inside an Anaconda-based Python environment. Users can install Anaconda/Miniconda/Miniforge, create and activate an environment, and then obtain CuBIDS locally by either installing from the Python Package Manager (Pypi) using pip or cloning directly from the CuBIDS GitHub repository. Documentation regarding use of CuBIDS is publicly available on our Read the Docs page. Notably, CuBIDS commands incorporate version control using DataLad as an optional dependency. Checking their BIDS dataset into DataLad and operationalizing command line programs with the ---use-datalad flag set allows users to access several extra version-control based functionalities. These include tracking changes they make to their dataset, reverting their dataset back to earlier versions, and automatically saving changes CuBIDS makes to the data with detailed commit messages. If users would like to access this functionality, they must separately install both DataLad and Git Annex (a dependency of Data-Lad). Although users can run CuBIDS programs without DataLad, opting to leverage the version control capabilities is recommended, as it renders the CuBIDS workflow portion of curation fully reproducible.
2.6. Definitions
The CuBIDS workflow relies upon five main concepts, all delineating different ways to categorize and catalog data: Key, Parameter, Acquisition, Dominant, and Variant Groups. The first is a “Key Group” -- the set of runs whose filenames share all BIDS filename key-value pairs, except for subject and session. For example, CuBIDS would place a T1w NIfTI file named sub-X_ses-A_acq-refaced_T1w.nii.gz, which contains the BIDS key-value pair “acq-refaced”—in the following Key Group: acquisition-refaced_datatype-anat_suffix-T1w. Notably, Key Groups only consider the scan’s BIDS filename; they do not account for the variance in metadata fields that might be present in the JSON sidecars.
For this reason, within each Key Group, we define a “Parameter Group” as the set of runs with identical metadata parameters contained in their sidecars. Parameter Groups exist within Key Groups and are denoted numerically—each Key Group will have n Parameter Groups, where n is the number of unique sets of scanning parameters present in that Key Group. For example, a T1w can belong to Key Group acquisition-refaced_datatype-anat_suffix-T1w and Parameter Group 1. CuBIDS defines Parameter Groups within Key Groups because differences in parameters can affect how BIDS Apps will configure their pipelines (e.g. Fieldmap availability, multiband factor, etc.).
Next, we define a “Dominant Group” as the Parameter Group that contains the most runs in its Key Group. Analogously, we define a “Variant Group” as any Parameter Group that is non-dominant. This is an important term because (as described below) CuBIDS can optionally rename all Variant Groups in an automated and reproducible fashion.
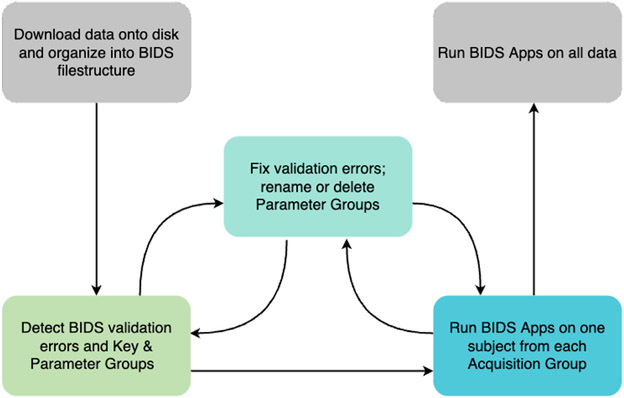
Finally, we define an “Acquisition Group” as a collection of sessions across participants that contain the exact same set of Key and Parameter Groups. Since Key Groups are based on the BIDS filenames—and therefore both MRI image type and acquisition specific—each BIDS session directory contains images that belong to a set of Parameter Groups. CuBIDS assigns each session—or set of Parameter Groups—to an Acquisition Group such that all sessions in an Acquisition Group possesses an identical set of acquisitions and metadata parameters across all MRI image types present in the dataset. We find Acquisition Groups to be a particularly useful categorization of BIDS data, as they identify homogeneous sets of sessions (not individual scans) in a large dataset. They are also useful for expediting the testing of pipelines; if a BIDS App runs successfully on a single subject from each Acquisition Group, one can be confident that it will handle all combinations of scanning parameters in the entire dataset. These various sets of methods by which one can group a BIDS dataset are critical to the CuBIDS workflow (see Fig. 1).
Fig. 1. CuBIDS workflow.
The CuBIDS workflow begins after the generation of NIfTI files and JSON sidecars and ends directly before the execution of pre-processing pipelines. We start with a BIDS dataset, which can be validated using CuBIDS’ robust version of the BIDS-validator. After purging the dataset of any sensitive fields, users can move to the next workflow stage: detecting Parameter Groups. Users can then rename or delete Parameter Groups. At any point in the workflow, users can implement version control to track changes made to the data using an easy-to-use, wrapped version of DataLad. Finally, users test one Exemplar Subject from each Acquisition Group on BIDS Apps to ensure each set of scanning parameters can run through pipelines error-free. CuBIDS includes command line programs for each step of the workflow (see Table 1).
2.7. Accounting for NIfTI header information
Information from NIfTI headers—including number of volumes, voxel size, image dimensions, and image obliquity—is often important but is usually absent from JSON sidecars. We created a program, cubids-add-nifti-info, that reads information from the NIfTI header and adds it to the JSON sidecar. For example, knowing the number of volumes in a run may be particularly useful when performing an initial quality assessment—i.e., identifying and removing runs with unexpectedly short durations (i.e., 20 vol in an fMRI timeseries). Similarly, runs with vastly different voxel sizes or fields of view may be easily identified and removed if desired.
2.8. BIDS validation
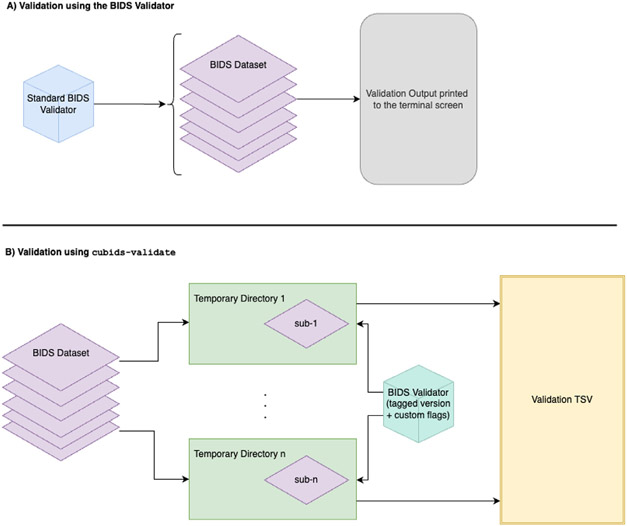
An essential first stage of curation is validation: finding the errors present in a BIDS dataset. This step is usually accomplished using the BIDS Validator. However, while BIDS validation is essential to the curation process, the standalone BIDS Validator can exhibit unstable file I/O behavior when validating large datasets (n>100). As a result, it sometimes fails unpredictably. To combat this issue, cubids-validate checks the BIDS layout using a wrapped, stable, scalable version of the standard BIDS Validator. To ensure scalability, cubids-validate parallelizes validation across participants, validating each subject directory on its own and deferring the detection of parameters that may vary across subjects. Thereafter, cubids-validate aggregates all validation errors found across participants in an easy-to-read TSV (see Fig. 2), which is accompanied by a data dictionary JSON sidecar. This table includes one row for each file that contains a BIDS validation error and displays that filename along with a description of the error (see Fig. 2)).
Fig 2. Stable, scalable BIDS validation.
CuBIDS wraps a stable version of the BIDS Validator and adds a few additional features including the ability to reorganize the validator output into an easy to read, tabular structure and save it as a TSV. A) The standard BIDS Validator’s default option (Gorgolewski et al., 2016) validates an entire BIDS dataset and outputs a summary of the errors and warnings it discovers to the terminal screen. B) In addition to visualizing the output in a scalable and easy-to-read format, cubids-validate includes the standard BIDS Validator’s ability to ignore cross-session comparisons or metadata from NIfTI headers while also adding an option for sequential participant-by-participant validation. This feature, which we recommend users leverage, parallelizes validation and validates each subject directory as its own BIDS dataset.
In designing cubids-validate, we also intended to separate metadata heterogeneity detection from BIDS error detection. By default, the validator does both—providing large amounts of unactionable information concerning the metadata variance in the terminal output. For example, if a sample includes participants with different sets of scans, the standalone BIDS Validator will print warnings alerting the user to the presence of incongruencies across subjects, often producing copious output that can obscure critical issues. If there are errors or forms of inconsistency users would prefer excluded from the CuBIDS validation TSV, they can run cubids-validate with optional BIDS Validator flags such as ---ignore_nifti_headers, which disregards NIfTI header content during validation and --ignore_subject_consistency, which we set as the default and skips checking that any given file for one subject is present for all other subjects. Furthermore, we implemented --sequential, which parallelizes validation by running the BIDS Validator sequentially on each subject (i.e. treating each participant as a standalone BIDS dataset and performing validation inside a temporary filesystem directory) (see Fig. 2B), and ---sequential-subjects, which filters the sequential run to only include the listed subjects, e.g. --sequential-subjects sub-01 sub-05 sub-09. These flags allow users to focus the validation process exclusively on the issues and subjects they would like to evaluate, and the sequential option, which parallelizes validation, addresses the standalone BIDS Validator’s scalability issue.
2.9. Grouping: heterogeneity detection and classification
While cubids-validate will find and display BIDS validation errors present in a dataset, it does not identify metadata parameters that might be inconsistent or omitted. For this reason, we developed cubids-group: a grouping function that classifies the heterogeneity present in a BIDS dataset and displays it in readable TSVs. Each grouping output is accompanied by a data dictionary JSON sidecar. The input to cubids-group is the path to the root of a BIDS Dataset, and the program produces four outputs, each of which gives a different view of the underlying data. The first (and most important) is summary.tsv, which contains one row per Parameter Group, and one column per metadata parameter present in the dataset. To understand the relative prevalence of each group, the program also counts, and includes in summary.tsv, the number of files in each Key and Parameter Group; this documentation is very useful for visualizing metadata heterogeneity across the entire dataset.
The next output of cubids-group is files.tsv, which contains one row per NIfTI file in the BIDS directory. This table keeps track of every scan’s assignment to Key and Parameter Groups and includes a field that allows users to easily identify the Key and Parameter Groups to which each image belongs. The next two grouping outputs organize the dataset by Acquisition Group. AcqGrouping.tsv organizes the dataset by session and tags each one with its Acquisition Group number. Finally, AcqGroupInfo.txt lists all Key Groups that belong to a given Acquisition Group along with the number of sessions each group possesses.
When applied to large datasets, cubids-group will often reveal issues within a BIDS dataset, some of which validation alone does not always catch. Such issues include missing metadata parameters and runs with low numbers of volumes or unusual image and voxel dimensions. For this reason, cubids-group can aid users in performing first pass quality assurance on their BIDS dataset. Since summary.tsv breaks down the dataset by Parameter Group with one column per scanning parameter, users can then search that TSV by desired parameters. Next, users can set a threshold or requirement for a certain parameter (e.g. number of volumes or dimension/voxel size) and use cubids-purge to remove runs that do not possess the desired values for those parameters. For example, a user may want to remove all fMRI runs with a low number of volumes before data processing with a BIDS App such as fMRIPrep.
2.10. Applying changes
The cubids-apply program provides an easy way for users to manipulate their datasets. Specifically, cubids-apply can rename files according to the users’ specification in a tracked and organized way. Here, the summary.tsv functions as an interface modifications; users can mark Parameter Groups they want to rename (or delete) in a dedicated column of the summary.tsv and pass that edited TSV as an argument to cubids-apply.
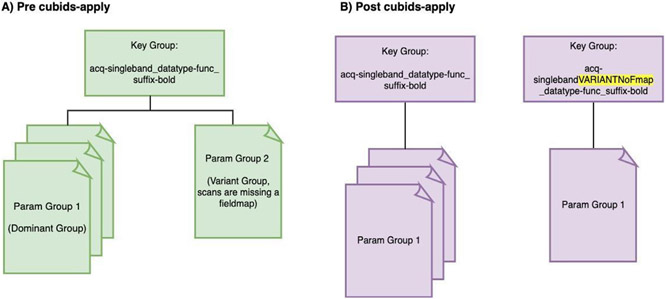
Additionally, cubids-apply can automatically rename files in Variant Groups based on their scanning parameters that vary from those in their Key Groups’ Dominant Parameter Groups. Renaming is automatically suggested when the summary.tsv is generated from a cubids-group run, with the suggested new name listed in the TSV’s “Rename Key Group” column. CuBIDS populates this column for all Variant Groups—e.g., every Parameter Group except the Dominant one. Specifically, CuBIDS will suggest renaming all Non-Dominant Parameter Groups to include VARIANT* in their acquisition field where * is the reason the Parameter Group varies from the Dominant Group. For example, when CuBIDS encounters a Parameter Group with a repetition time that varies from the one present in the Dominant Group, it will automatically suggest renaming all runs in that Variant Group to include acquisition-VARIANTRepetitionTime in their filenames. When the user runs cubids-apply, filenames will get renamed according to the auto-generated names in the “Rename Key Group” column in the summary.tsv (see Fig. 3)).
Fig. 3. Parsing the dataset by Parameter Group.
The summary.tsv file is a cubids-group output that contains one row per Parameter Group and one column per scanning parameter. Thus, this TSV summarizes all metadata present within a dataset. A) Before cubids-apply is run, a given Key Group may have multiple Parameter Groups, each containing a different set of scanning parameters. This summary table includes a “Rename Key Group” column that auto-configures when cubids-apply is run and labels each non-dominant Parameter Group as a Variant Group based on the scanning parameters that differentiate that group from the Dominant Group. Specifically, CuBIDS represents this variance by adding “VARIANT*”—where * indicates the metadata parameters that cause those files to vary from the Dominant Group—to the “acq” field of those files in non-dominant Parameter Groups. For example, in A), the metadata in the Param Group 2 image differs from that of the Dominant Group (Param Group 1) image because that run is missing a fieldmap. The result of running cubids-apply can be seen in B) where the Param Group 2 image ends up in a new Key group because CuBIDS added “VARIANTNoFmap” to the acquisition field of its filename when cubids-apply was run.
2.11. Customizable configuration
CuBIDS also features an optional, customizable, MRI image type-specific configuration file. This file can be passed as an argument to cubids-group and cubids-apply using the ---config flag and allows users to customize grouping settings based on image type and parameter. Each Key Group is associated with one (and only one) MRI image type, as BIDS filenames include MRI image type-specific values as their suffixes. This easy-to-modify configuration file provides several benefits to curation. First, it allows users to add and remove metadata parameters from the set that determines groupings. This can be very useful if a user deems a specific metadata parameter irrelevant and wishes to collapse variation based on that parameter into a single Parameter Group. Second, the configuration file allows users to apply tolerances for parameters with numerical values. This functionality allows users to avoid very small differences in scanning parameters (i.e., a TR of 3.0 s vs 3.0001 s) being split into different Parameter Groups. Third, the configuration file allows users to determine which scanning parameters are listed in the acquisition field when auto-renaming is applied to Variant Groups.
2.12. Exemplar testing
In addition to facilitating curation of large, heterogeneous BIDS datasets, CuBIDS also prepares datasets for testing BIDS Apps. This portion of the CuBIDS workflow relies on the concept of the Acquisition Group: a set of sessions that have identical scan types and metadata across all MRI image types present in the session set. Specifically, cubids-copy-exemplars copies one subject from each Acquisition Group into a separate directory, which we call an Exemplar Dataset. Since the Exemplar Dataset contains one randomly selected subject from each unique Acquisition Group in the dataset, it will be a valid BIDS dataset that spans the entire metadata parameter space of the full study. If users run copy-exemplars with the ---use-datalad flag, the program will ensure that the Exemplar Dataset is tracked and saved in DataLad. If the user chooses to forgo this flag, the Exemplar Dataset will be a standard directory located on the filesystem. Once the Exemplar Dataset has been created, a user can test it with a BIDS App (e.g., fMRIPrep or QSIPrep) to ensure that each unique set of scanning parameters will pass through the pipelines successfully. Because BIDS Apps auto-configure workflows based on the metadata encountered, they will process all runs in each Acquisition Group in the same way. By first verifying that BIDS Apps perform as intended on the small sub-sample of participants present in the Exemplar Dataset (that spans the full variation of the metadata), users can confidently move forward processing the data of the complete BIDS dataset.
3. Results
The CuBIDS workflow is currently being used in neuroimaging labs at a number of institutions including the University of Pennsylvania, the Children’s Hospital of Philadelphia, the Child Mind Institute, and the University of Minnesota’s Masonic Institute for the Developing Brain. To demonstrate the utility of CuBIDS, here we apply the software to two datasets. First, we curate a small example dataset that is included in the software’s GitHub repository and can be downloaded here. Second, we apply CuBIDS to the large-scale data of the Philadelphia Neurodevelopmental Cohort.
3.1. The CuBIDS workflow for curating a BIDS dataset (example dataset)
The following walkthrough displays the process of curating a dataset using CuBIDS on a Linux machine. This example walkthrough is also documented on the CuBIDS Read the Docs page. To do so, we use an example dataset that is bundled with the software. For this demonstration, we install CuBIDS inside a conda environment. Note that if you are using an Apple M1 chip machine, you will need to install Miniforge instead of Miniconda. Once we have conda installed we create and activate a new environment using the following commands:
conda create -n test-env python=3.8 conda activate test-env
To obtain CuBIDS locally, we can use pip to download our software from the Python Package Manager (Pypi) using the following command:
pip install CuBIDS
Alternatively, we can clone from the CuBIDS GitHub repository using the following command:
git clone https://github.com/PennLINC/CuBIDS.git
Now that we have a copy of the source code, we can install it by running
cd CuBIDS pip install -e.
We will now need to install some dependencies of CuBIDS. To do this, we first must install nodejs. We can accomplish this using the following command:
conda install nodejs
Now that we have npm installed, we can install the bids-validator using the following command:
npm install -g bids-validator@1.7.2
In this example, we use the bids-validator v1.7.2. using a different version of the validator may result in slightly different validation TSV printouts, but CuBIDS is compatible with all versions of the validator at or above v1.6.2. Throughout this example walkthrough, we use DataLad for version control, so we will also need to install both DataLad and git-annex, the large file storage software DataLad runs under the hood. Installation instructions for DataLad and git-annex can be found here.
Now that we have installed CuBIDS and all necessary dependencies, we are ready to begin the curation process on our example dataset. We create a CuBIDS_Test directory to function as the working directory and navigate to it as follows:
mkdir $PWD/CuBIDS_Test cd CuBIDS_Test
Throughout this walkthrough, we will run all commands from the CuBIDS_Test directory. Next, we download BIDS_Dataset.zip (a ZipFile containing the example dataset) and unzip as follows:
curl -sSLO https://github.com/PennLINC/CuBIDS/raw/main/cubids/testdata/BIDS_Dataset.zip unzip BIDS_Dataset.zip rm BIDS_Dataset.zip
As a first step, we use CuBIDS to identify the metadata fields present in the dataset. This is accomplished with the following command:
cubids-print-metadata-fields BIDS_Dataset
This command returns a total of 66 fields, including acquisition parameters and other metadata fields present in the dataset’s JSON sidecars. Some of these fields contain simulated protected health information (PHI) such as PatientName that we wish to remove. Completing this step prior to checking the BIDS dataset into DataLad is critical, as we must ensure PHI is not tracked as part of version control. To remove the PatientName field from the sidecars, we can use the command:
cubids-remove-metadata-fields BIDS_Dataset --fields PatientName
If we were to run cubids-print-metadata-fields once more, we would see that PatientName is no longer present in the dataset. Now that all PHI has been removed from the metadata, we are ready to check our dataset into DataLad. To do this, we run the following command:
datalad create -c text2git BIDS_Dataset_DataLad
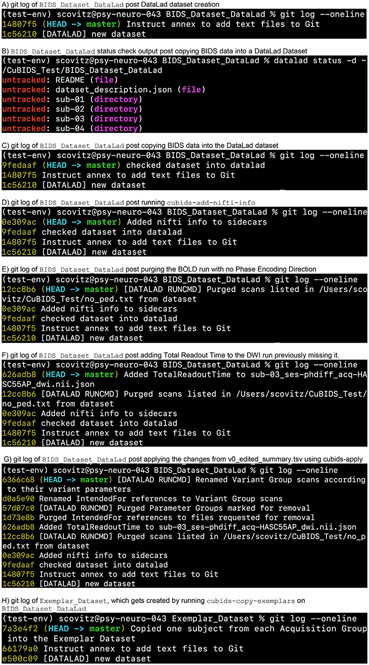
The creation of our DataLad dataset will be accordingly reflected in the dataset’s version control history, or “git log” (see example in Fig. 4A). At any point in the CuBIDS workflow, we can view a summary of our dataset’s version history by running the following commands:
Fig. 4.
Version history throughout the curation process.
cd BIDS_Dataset_DataLad git log --oneline cd ..
Next, we copy the contents of our BIDS dataset into the newly created and currently empty DataLad dataset:
cp -r BIDS_Dataset/* BIDS_Dataset_DataLad
In addition to being able to access the version history of our data, any point in this workflow, we can also check the status of untracked (not yet saved) changes using the datalad status command, as seen below:
datalad status -d BIDS_Dataset_DataLad
This command produces a description of the changes we have made to the data since the last commit (see Fig. 4B). The command above shows all files untracked, as we have copied the BIDS data into BIDS_Dataset_DataLad but have not yet saved those changes. Our next step is to run save. It is best practice to provide a detailed commit message, for example:
datalad save -d BIDS_Dataset_DataLad -m "checked dataset into datalad"
This commit is reflected in our git log (see Fig. 4C). Now that the dataset is checked into DataLad, at any point in the workflow going forward, we can run the following command to revert the dataset back to the previous commit:
cubids-undo BIDS_Datast_DataLad
At this stage, we also recommend removing the BIDS_Dataset directory — its contents are safely copied into and tracked in BIDS_Dataset_DataLad.
Next, we seek to add new fields regarding our image parameters that are only reflected in the NIfTI header to our metadata; these include important details such as image dimensions, number of volumes, image obliquity, and voxel sizes. To do this, we run:
cubids-add-nifti-info BIDS_Dataset_DataLad --use-datalad
This command adds the NIfTI header information to the JSON sidecars and saves those changes. In order to ensure that this command has been executed properly, we can run cubids-print-metadata-fields once more, which reveals that NIfTI header information has been successfully included in the metadata. Since we ran add-nifti-info with the --use-datalad flag set, CuBIDS will automatically save the changes made to the dataset to the git log (see Fig. 4D)).
The above panels display the version history of the small, example DataLad dataset we curated to display the effectiveness of the CuBIDS workflow. These panels are screenshots of the git history of the dataset taken after each change was made to the data. A shasum (yellow string of letters and numbers to the left of each commit message) is assigned to each commit, and each commit is recorded with a message (white text describing the changes made to the data). If users would like more information about each commit, they can run the git log command without the oneline flag to get a detailed summary of each commit. This summary will include files that were changed, exact changes that were made to each file, date and time of the commit, and information about the git user who made the changes. At any point in the workflow after checking the dataset into DataLad, we can use cubids-undo to revert the dataset back to the previous commit.
The next step in the CuBIDS workflow is to understand what BIDS validation errors may be present (using cubids-validate) as well as the structure, heterogeneity, and metadata errors present in the dataset (using cubids-group). Notably, neither of these two programs requires write access to the data, as each simply reads in the contents of the data and creates TSVs that parse the metadata and validation errors present. Validation can be accomplished by running the following command:
cubids-validate BIDS_Dataset_DataLad v0 --sequential
The use of the sequential flag forces the validator to treat each participant as its own BIDS dataset. This command produces v0_validation.tsv (see Supplementary Data 1A).
This initial validation run reveals that Phase Encoding Direction (PED) is not specified for one of the BOLD task-rest scans. We can clearly see that we either need to find the PED for this run elsewhere and edit that sidecar to include it or remove that run from the dataset, as this missing scanning parameter will render field map correction impossible. For the purpose of this demonstration, we elect to remove the scan. To do this, we run the following command:
cubids-purge BIDS_Dataset_DataLad no_ped.txt --use-datalad
Here, no_ped.txt (see Supplementary Data 1B) is a text file containing the full path to the dwi run flagged in v0_validation.txt for missing PED. The user must create this file before running cubids-purge (a command such as echo $PWD/BIDS_Dataset_DataLad/sub-02/ses-phdiff/func/sub-02_ses-phdiff_task-rest_bold.nii.gz > no_ped.txt will work).
We elect to use cubids-purge instead of simply removing the run because cubids-purge will ensure all associated files, including sidecars and IntendedFor references in the sidecars of fieldmaps, are also deleted. This change will be reflected in the git history (see Fig. 4E).
Returning again to v0_validation.tsv, we can also see that there is one DWI run missing TotalReadoutTime, a metadata field necessary for certain pipelines. In this case, we determine that TotalReadoutTime (TRT) was erroneously omitted from the DWI sidecars. For the purpose of this example, we assume we are able to obtain the TRT value for this run (perhaps by asking the scanner technician). Once we have this value, we manually add it to the sidecar for which it is missing by opening BIDS_Dataset_DataLad/sub-03/ses-phdiff/dwi/sub-03_ses-phdiff_acq-HASC55AP_dwi.json in an editor and adding the following line:
"TotalReadoutTime": 0.0717598, on a new line anywhere inside the curly braces between lines containing parameters and their values, save the changes, and close the JSON file. We then save the latest changes to the dataset with a detailed commit message as follows:
datalad save -d BIDS_Dataset_DataLad -m "Added TotalReadoutTime to sub-03_ses-phdiff_acq-HASC55AP_dwi.nii.json"
This change will be reflected in the git history (see Fig. 4F).
To verify that there are no remaining validation errors, we rerun validation with the following command:
cubids-validate BIDS_Dataset_DataLad v1 --sequential
This command will produce no TSV output and instead print “No issues/warnings parsed, your dataset is BIDS valid” to the terminal, which indicates that the dataset is now free from BIDS validation errors and warnings.
Along with parsing the BIDS validation errors in our dataset, it is important to understand the dataset’s structure, heterogeneity, and metadata errors. To accomplish these tasks, we use cubids-group. Large datasets almost inevitably contain multiple validation and metadata errors. As such, it is typically useful to run both cubids-validate and cubids-group in parallel, as validation errors are better understood within the context of a dataset’s heterogeneity. Additionally, being able to see both the metadata errors that grouping reveals alongside BIDS errors that the validator catches gives users a more comprehensive view of the issues they will need to fix during the curation process. Note that if users choose to provide just a pass in just a filename prefix (e.g. v0) for the second argument, then CuBIDS will put the four grouping outputs in bids_dir/code/CuBIDS. If users provide a path (e.g. $PWD/v0), then output files will go to the specified location. The command to run the grouping function is as follows:
cubids-group BIDS_Dataset_DataLad v0
As noted in Table 1, this command will produce four tables that display the dataset’s heterogeneity in different ways. First, v0_summary.tsv contains all detected Key and Parameter groups and provides a high-level overview of the heterogeneity in the entire dataset (see Supplementary Data 1C). Second, v0_files.tsv (see Supplementary Data 1D) maps each imaging file in the BIDS directory to a Key and Parameter group. Third, v0_AcqGrouping.tsv (see Supplementary Data 1E) maps each session in the dataset to an Acquisition Group. Finally, v0_AcqGroupInfo.txt (see Supplementary Data 1F) lists the set of scanning parameters present in each Acquisition Group.
The next step in the CuBIDS curation process is to examine v0_summary.tsv, which allows for automated metadata quality assurance (QA)—the identification of incomplete, incorrect, or unusable parameter groups based on acquisition fields such as dimension and voxel sizes, number of volumes, etc. While v0_validation.tsv identified all BIDS validation errors present in the dataset, it will not identify several issues that might be present with the sidecars. Such issues include instances of erroneous metadata and missing sidecar fields, which may impact successful execution of BIDS Apps.
Examining v0_summary.tsv (see Supplementary Data 1C) we can see that one DWI Parameter Group—acquisition-HASC55AP_datatype-dwi_suffix-dwi--2--contains only one image (see “Counts” column) with only 10 vol (see “NumVolumes” column). Since the majority of DWI runs in this dataset have 61 vol, CuBIDS assigns this single run to a “Non-Dominant”, or “Variant” Parameter Group and populates that Parameter Group’s “RenameKeyGroup” column in v0_summary.tsv with acquisition-HASC55APVARIANTNumVolumes_datatype-dwi_suffix-dwi. For the purpose of this demonstration, we elect to remove this run because it does not have enough volumes to be usable for most analyses. To do this, we can either use cubids-purge, or we can edit v0_summary.tsv by adding “0” to the “MergeInto” column in the row (Parameter Group) we want to remove. For this walkthrough, we chose the latter. To do this, we open v0_summary.tsv in an editor, navigate to row 4, which contains all information for Key Group acquisition-HASC55AP_datatype-dwi_suffix-dwi Parameter Group 2. If we scroll to the NumVolumes column (row 4, column S), we see this Parameter Group has only 10 vol, which explains why it received an auto-generated Rename Key Group value of acquisition-HASC55APVARIANTNumVolumes_datatype-dwi_suffix-dwi. Remaining in this same row, we navigate back to column C, which is labeled “MergeInto” and manually a “0” to the cell in row 4 column C. This will ensure all runs in that Parameter Group (in this example, just one scan) are removed when we run cubids-apply. We then export and save the TSV in our CuBIDS_Test working directory as v0_edited_summary.tsv (see Supplementary Data 1 G). We will then save this edited version of v0_summary.tsv as v0_edited_summary.tsv, which will be passed into cubids-apply in our next curation step.
Now that all metadata issues have been remedied--both the validation and summary outputs appear problem-free--we are ready to rename our files based on their Rename Key Group values and apply the requested deletion in v0_edited_summary.tsv. The cubids-apply function renames runs in each Variant Parameter Group according to the metadata parameters with a flag “VARIANT”, which is useful because the user will then be able to see, in each scan’s filename, which metadata parameters associated with that run vary from those in the acquisition’s Dominant Group. Note that like in cubids-group, cubids-apply requires full paths to the BIDS Dataset, summary and files TSVs, and output prefix. If the edited summary and files TSVs are located in the bids_dir/code/CuBIDS directory, the user may just pass in those filenames. Otherwise, specifying the path to those files is necessary. We execute cubids-apply with the following command:
cubids-apply BIDS_Dataset_DataLad v0_edited_summary.tsv v0_files.tsv v1 --use-datalad
Checking our git log, we can see that all changes from apply have been saved (see Fig. 4G). As a final step, we can check the four grouping TSVs cubids-apply produces (see Supplementary Data 1H-K) to ensure they look as expected--that all files with variant scanning parameters have been renamed to indicate the parameters that vary in the acquisition fields of their filenames (and therefore Key Group names).
At this stage, the curation of the dataset is complete; next is preprocessing. CuBIDS facilitates this subsequent step through the creation of an Exemplar Dataset: a subset of the full dataset that spans the full variation of acquisitions and parameters by including one subject from each Acquisition Group. By testing only one subject per Acquisition Group, users are able to pinpoint both the specific metadata values and runs that may be associated with pipeline failures; these acquisition groups could then be evaluated in more detail and flagged for remediation or exclusion. The Exemplar Dataset can easily be created with the cubids-copy-exemplars command, to which we pass in v1_AcqGrouping.tsv—the post-apply acquisition grouping TSV (see Supplementary Data 1 J).
cubids-copy-exemplars BIDS_Dataset_DataLad Exemplar_Dataset v1_AcqGrouping.tsv --use-datalad
Since we used the use-datalad flag, Exemplar_Dataset is a DataLad dataset with the version history tracked in its git log (see Fig. 4H). Once a preprocessing pipeline completes successfully on the Exemplar Dataset, the full dataset can be executed with confidence, as a pipeline’s behavior on the full range of metadata heterogeneity in the dataset will have already been discovered during exemplar testing.
3.2. Application to a large-scale study of brain development
In addition to applying the CuBIDS workflow to a toy dataset, here we describe the workflow applied to the Philadelphia Neurodevelopmental Cohort (PNC), a multimodal dataset of n = 1601 participants. The PNC data is publicly available (Satterthwaite et al., 2014) and is one of many datasets encompassing the forthcoming Reproducible Brain Chart (RBC)—a large, developmental neuroimaging aggregation initiative led jointly by the University of Pennsylvania and the Child Mind Institute. CuBIDS was developed, in part, to help manage the data for RBC. The curated version of the dataset will be publicly available as part of the forthcoming RBC data release. The PNC curation workflow involved iterative rounds of checking and fixing due to the heterogeneity and size of the dataset, so the following section will be a summary of how we used CuBIDS to curate this dataset (rather than a step-by-step walkthrough).
One of our early curation actions was to take inventory regarding the metadata heterogeneity of PNC by obtaining the initial summary table. To do this, we ran cubids-group, which requires approximately 15 min to finish on PNC. For smaller datasets including our toy dataset from the walkthrough described above, cubids-group completes in just seconds. Examining the initial summary table (see Supplementary Data 2B), we find that PNC contains 144 Parameter Groups—runs containing both identical BIDS filename key-value pairs and identical metadata parameters present in their sidecars. The summary table is organized by MRI image type, so we can easily see that some MRI image types and acquisitions in the dataset are much more heterogeneous with respect to their metadata parameters than are others. For example, from the table, we can see that PNC has only one Key Group for T1w runs and only three Parameter Groups. Furthermore, according to the “Counts” column of the tsv, the vast majority of T1w runs (n = 1597), are in the Dominant Parameter Group. By contrast, in this same summary table we can see that there are 62 different Parameter Groups in the dataset for task-frac2back BOLD fMRI scans. Examining the “RenameKeyGroup” column of those frac2back rows in the summary table, we can see that the primary source of variance is the number of volumes acquired.
Our team relied upon the summary table to make a number of curation decisions—especially inclusion/exclusion based on metadata. The summary table provided a platform for collaboration and discussion among the team that was curating, validating, and modifying the dataset. Since PNC was curated with DataLad and is saved as a DataLad Dataset, a detailed history of the curation decisions can be found in the commit history. Each change to the dataset was saved with a commit message, so all modifications we made to the dataset are tracked and tagged with a shasum. If we want to restore PNC to a previous curation stage, we can so using cubids-undo.
Modifications to PNC during the curation stage, documented in the dataset’s git log, include creating and adding previously missing events tsvs, adding NIfTI header information to all sidecars in the dataset (using the cubids-add-nifti-info command), removing DWI runs that do not have enough volumes to successfully run through diffusion preprocessing pipelines (e.g. QSIPrep), and adding Parallel Reduction Factor in Plane to two sidecars that were initially missing this field. Once the validation and grouping outputs revealed we had no more BIDS and metadata issues to fix, we ran cubids-apply on the dataset to produce a new set of TSVs (see Supplementary Data 2 G-J). As can be seen in the summary table (see Supplementary Data 2G) generated by cubids-apply, all runs in Variant (e.g., non-Dominant) Groups were renamed based on the scanning parameters that are variant (see Fig. 3).
We then executed the final step of the CuBIDS workflow, which entailed running cubids-copy-exemplars. This command creates the Exemplar Dataset—a DataLad-tracked BIDS dataset containing one subject from each Acquisition Group (see Supplementary Data 2I). The final, curated version of PNC contains 1601 participants, 15,077 scans, and 65 Acquisition Groups. Thus, the Exemplar Dataset contains only 65 participants but spans the entire dataset’s parameter space, reducing the scope of the pipeline testing by 96%. We then used this Exemplar Dataset to test MRI image type-specific preprocessing pipelines such as fMRIPrep and QSIPrep.
For large datasets especially, exemplar testing can be a necessary step; users will often need to go back and re-curate aspects of the BIDS data based on metadata errors that only become apparent during pipeline runs on the Exemplar Dataset. For example, after we ran the PNC Exemplar Dataset through QSIPrep, we noticed that one participant (whose Acquisition Group includes 34 participants), failed to complete the pipeline successfully. After examining the error log, we realized that for this participant, the number of bvals did not match the number of volumes in the scan’s sidecar (which was pulled directly from the NIfTI header and added to the sidecar during the cubids-add-nifti-info step of curation). Since all participants in the same Acquisition Group possess identical scanning parameters, when a pipeline encounters a metadata error in an Exemplar Subject, all participants in that Acquisition Group will have that same error and thus require the same fix. Accordingly, we used cubids-purge to remove all DWI runs from that Exemplar Group and reran cubids-group to obtain our final CuBIDS outputs (see Supplementary Data 2K-N). Since all other Exemplar Subjects passed through QSIPrep successfully, we were then able to run the pipeline through the entire dataset without concern that erroneous metadata would impact preprocessing.
4. Discussion
Ample recent evidence has emphasized the challenges to reproducibility in neuroimaging research (Kang et al., 2016). Although often overlooked, curation can be a critical part of the scientific workflow. Because curation is often the first step after data acquisition, errors in curation can ramify throughout each subsequent stage. BIDS apps adapt to metadata encountered in an automatic and flexible way, which can be a vulnerability in ensuring datasets are processed identically. If BIDS data are improperly curated, pre-processing pipelines may mis-configure, with the potential to impact eventual results. Curation challenges are particularly acute in large-scale data resources, which continue to proliferate (e.g., UK Biobank, ABCD, PNC, HBN, HCP, etc.) (Bycroft et al., 2018; Karcher and Barch, 2021; Satterthwaite et al., 2014; Alexander et al., 2017; Van Essen et al., 2013). In large datasets, curation is often an iterative, manual process that is neither well documented nor reproducible. To address these challenges, CuBIDS allows for reproducible data curation at scale. As discussed below, our software provides five main advantages: stability in validation, reproducibility in curation, the ability to identify and manage heterogeneity, transparency in naming, and accelerated pipeline testing.
4.1. Stable BIDS validation at scale
The BIDS Validator is the current standard tool for validation of all BIDS datasets. It is widely used and plays an essential role in the standard BIDS workflow; it effectively identifies the ways in which a dataset does not comply with BIDS standards. However, it does not scale well, at times failing unpredictably on larger samples. Furthermore, when run in a Linux shell, the validator prints (often a large volume) of text describing the errors and warnings to the terminal screen. For large datasets with many errors and warnings, such information is often quite difficult to visualize and comprehend. We wrapped the BIDS Validator in the cubids-validate CLI to address these challenges, creating a scalable implementation that yields a readable TSV. This allows users to easily identify the range of validation issues that may be present in a large-scale dataset.
4.2. Reproducible data curation
Curation of large, heterogeneous BIDS datasets is an iterative, multistep task. However, this process is often not reproducible, which, in turn, may compromise the reproducibility of subsequent workflows. Without version control, any decision made during the curation process—such as inclusion/exclusion decisions or editing metadata—will go unrecorded. Further, if the person curating the data makes a mistake, they will have no clear way to undo that mistake and revert the data to a prior state. In leveraging CuBIDS’ use of DataLad, users can save each change made to a dataset with a detailed commit message (e.g. “Removed all DWI runs with less than 30 vol ”). If a user erroneously changes the data and wants to undo those changes, cubids-undo reverts the most recent commit.
4.3. Parsing heterogeneity in large-scale data resources
While cubids-validate will catch instances where the data does not comply with BIDS format, it has important limitations. For example, validation does not always account for missing JSON sidecars or empty NIfTI headers. In addition, it will neither identify runs that have errant metadata values nor those with parameters that might render the runs unusable. This functionality is provided by cubids-group, which produces parameter-based summary tables that parse the dataset based on metadata, allowing for users to visualize and assess metadata quality in ways that validation cannot.
This functionality is especially critical in the curation of large datasets. Scaling up both the number of participants and the number of scanners within a single data resource has the potential to introduce a massive amount of heterogeneity to that study’s eventual BIDS dataset. Heterogeneity in scanning parameters can result in heterogeneity in preprocessing pipelines; if users are do not appreciate the metadata heterogeneity in their dataset, they may be surprised by inconsistencies in preprocessing settings and outcomes. Further, parameter groups could be explicitly modeled when accounting for batch effects rather than just using scanner or site. Thus, being able to identify and correct metadata errors in a heterogeneous dataset is a critical part of the data curation process, as such decisions may impact the derived images from preprocessing pipelines.
4.4. Enhancing transparency with dominant and variant groups
In order to provide transparent documentation of parameter heterogeneity, the cubids-apply function renames runs in each MRI image type according to their variant metadata parameters. For example, if the majority of BOLD task-rest runs in a dataset are Oblique but sub-X’s image is not oblique, CuBIDS users can choose to accept and apply the automatically suggested renaming of “acq-VARIANTObliquity” to that run’s filename. When performing sensitivity analyses on derivatives from datasets that have been curated using CuBIDS, researchers may choose to exclude any runs in Non-Dominant Groups to ensure that scanning parameters variance does not affect their results. Alternatively, researchers could use image harmonization tools (e.g., ComBat) to ensure that such variation does not impact analyses (Fortin et al., 2018).
4.5. Accelerating pipeline testing with exemplar datasets
Even after careful curation, the best way to verify successful image processing is empirical testing. In a highly heterogeneous dataset, pipeline testing often reveals errors that were not immediately apparent on initial curation, which usually require minor additional adjustments to the metadata or exclusion of specific scans. Finding such edge cases while processing a large dataset can slow down the workflow, so it is advantageous to conduct pipeline testing before full deployment on a large data set. CuBIDS facilitates this process through the creation of Exemplar Datasets that include data from each Acquisition Group and thus span the full variation of the metadata present. After successful testing on the Exemplar Dataset, the likelihood that unexpected outcomes occur when the full dataset is processed is dramatically reduced. Furthermore, resource usage can be monitored during the exemplar runs to estimate the runtime and storage demands for processing the entire dataset.
4.6. Limitations
CuBIDS possesses several limitations that should be acknowledged. First, at present, CuBIDS does not possess a GUI, so running the software requires basic knowledge of the terminal and Linux machines. However, such skills are likely to be a prerequisite for curating large-scale imaging datasets. Second, if users are curating BIDS Datasets with n>2500 participants and using the DataLad-enabled version control option, CuBIDS programs that rely on saving changes made to the dataset might experience runtimes that extend to over an hour—due to the need for DataLad to index such a large dataset. Third, our add-nifti-info program does pull Obliquity, Number of Volumes, Dimension Size, Image Orientation, and Voxel Sizes from the NIfTI header and adds those values to the sidecars. However, we do not currently cross-reference existing sidecar metadata with NIFTI header values. This additional functionality would be a good future direction for the software but is currently outside the scope of this paper. Finally, CuBIDS is currently only able to handle MRI BIDS, not other MRI image types, and can only be run on disk—either a local machine or a high performance computing cluster; users cannot currently run CuBIDS using either cloud-based computing (e.g., Amazon’s S3) or neuroimaging databases such as the eXtensible Neuroimaging Archive Toolkit (XNAT), Longitudinal Online Research and Imaging System (LORIS), Collaborative Informatics Neuroimaging Suite (COINS), and the commercial platform Flywheel (Herrick et al., 2016; Das et al., 2012; Landis et al., 2016). Such functionality may be added to the software in future releases.
5. Conclusions
Curating large, heterogeneous neuroimaging datasets can be a difficult and frustrating task. As the size and heterogeneity of data resources continues to expand, tools that allow for reproducible curation are not only helpful but necessary. CuBIDS facilitates efficient identification and correction of issues present in the metadata of heterogeneous BIDS datasets in a reproducible manner. Furthermore, CuBIDS Exemplar Datasets allow users to verify that BIDS Apps perform as intended on a small sub-sample of participants that spans the entire parameter space of the dataset, accelerating the processing of all data from the complete study. Together, CuBIDS allows users to simultaneously streamline curation and ameliorate metadata issues while maximizing reproducibility.
Supplementary Material
Acknowledgements
This study was supported by grants from the National Institutes of Health: R01MH120482, R37MH125829, R01MH113550, R01EB022573, RF1MH116920, R01MH112847, R01MH123550, R01NS112274, R01MH123563. Additional support was provided by the CHOP-Penn Lifespan Brain Institute, the Penn Brain Science Center, and the Center for Biomedical Image Computing and Analytics
Footnotes
APPENDIX A. SUPPLEMENTARY MATERIAL
Supplementary Data 1 (for toy dataset in Results)
Supplementary Data files 1A-O are zipped into Supplementary_Data_1.zip (submitted as supplementary material)
Declaration of Competing Interest
The authors of Curation of BIDS (CuBIDS) declare that they have no competing or conflicting interests.
Credit authorship contribution statement
Sydney Covitz: Software, Writing – original draft, Writing – review & editing, Conceptualization. Tinashe M. Tapera: Writing – review & editing. Azeez Adebimpe: Writing – review & editing. Aaron F. Alexander-Bloch: Writing – review & editing. Maxwell A. Bertolero: Writing – review & editing. Eric Feczko: Writing – review & editing. Alexandre R. Franco: Writing – review & editing, Data curation. Raquel E. Gur: Writing – review & editing. Ruben C. Gur: Writing – review & editing. Timothy Hendrickson: Software, Writing – review & editing, Data curation. Audrey Houghton: Writing – review & editing. Kahini Mehta: Writing – review & editing, Data curation. Kristin Murtha: Writing – review & editing. Anders J. Perrone: Software, Writing – review & editing. Tim Robert-Fitzgerald: Writing – review & editing. Jenna M. Schabdach: Writing – review & editing. Russell T Shinohara: Writing – review & editing. Jacob W. Vogel: Writing – review & editing. Chenying Zhao: Writing – review & editing. Damien A. Fair: Software, Writing – review & editing, Data curation. Michael P. Milham: Writing – review & editing, Data curation. Matthew Cieslak: Software, Writing – original draft, Writing – review & editing, Conceptualization, Supervision. Theodore D. Satterthwaite: Software, Writing – original draft, Writing – review & editing, Data curation, Conceptualization, Supervision.
Data and code availability statement
A copy of the small, example dataset whose curation we walk through in the Results section is compressed into a ZipFile and available for download here. Additionally, the Philadelphia Neurodevelopmental Cohort (PNC), the dataset whose curation we summarize in the second portion of the Results, is publicly available in the Database of Genotypes and Phenotypes (dbGaP accession phs000607.v3.p2).
The source code for CuBIDS is publicly available at https://github.com/PennLINC/CuBIDS, the documentation for our software is available at https://cubids.readthedocs.io/en/latest/, and our package is available for download on the Python Package Manager (pypi) https://pypi.org/project/cubids/.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2022.119609.
References
- Alexander L, Escalera J, Ai L, et al. , 2017. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data 4, 170181. doi: 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bycroft C, Freeman C, Petkova D, et al. , 2018. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslak M, Cook PA, He X, et al. , 2021. QSIPrep: an integrative platform for preprocessing and reconstructing diffusion MRI data. Nat. Methods 18, 775–778. doi: 10.1038/s41592-021-01185-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Zijdenbos AP, Harlap J, Vins D, Evans AC, 2012. LORIS: a web-based data management system for multi-center studies. Front. Neuroinformatics 5, 37. doi: 10.3389/fninf.2011.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban O, Markiewicz CJ, Blair RW, et al. , 2019. fMRIPrep: a robust preprocessing pipeline for functional MRI. Nat. Methods 16 (1), 111–116. doi: 10.1038/s41592-018-0235-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, Auer T, Calhoun V, et al. , 2016. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044. doi: 10.1038/sdata.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Cullen N, Sheline YI, Taylor WD, Aselcioglu I, Cook PA, Adams P, Cooper C, Fava M, McGrath PJ, McInnis M, Phillips ML, Trivedi MH, Weissman MM, Shinohara RT, 2018. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage 167, 104–120. doi: 10.1016/j.neuroimage.2017.11.024, 2018 Feb 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgolewski K, et al. , 2017. BIDS apps: improving ease of use, accessibility, and reproducibility of neuroimaging data analysis methods. PLoS Comput. Biol doi: 10.1371/journal.pcbi.1005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halchenko, et al. , 2021. DataLad: distributed system for joint management of code, data, and their relationship. J. Open Source Softw 6 (63), 3262. doi: 10.21105/joss.03262. [DOI] [Google Scholar]
- Herrick R, Horton W, Olsen T, McKay M, Archie KA, Marcus DS, 2016. XNAT central: open sourcing imaging research data. Neuroimage 124, 1093–1096. doi: 10.1016/j.neuroimage.2015.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, et al. , 2016. Editorial: recent advances and challenges on big data analysis in neuroimaging. Front. Neurosci doi: 10.3389/fnins.2016.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher NR, Barch DM, 2021. The ABCD study: understanding the development of risk for mental and physical health outcomes. Neuropsychopharmacology 46, 131–142. doi: 10.1038/s41386-020-0736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis D, Courtney W, Dieringer C, Kelly R, King M, Miller B, et al. , 2016. COINS data exchange: an open platform for compiling, curating, and disseminating neuroimaging data. Neuroimage 124, 1084–1088. doi: 10.1016/j.neuroimage.2015.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterthwaite T, Elliott M, Ruparel K, Prabhakaran K, Calkins M, Hopson R, Jackson C, Keefe J, Riley M, Mensh Frank, Sleiman Patrick, Verma Ragini, Davatzikos Christos, Gur Ruben, Gur Raquel., 2014. Neuroimaging of the philadelphia neurodevelopmental cohort. Neuroimage 86. doi: 10.1016/j.neuroimage.2013.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, et al. , 2013. The WU-Minn Human Connectome project: an overview. Neuroimage 80, 62–79. doi: 10.1016/j.neuroimage.2013.05.041, ISSN 1053-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni, et al. , 2019. PyBIDS: python tools for BIDS datasets. J. Open Source Softw 4 (40), 1294. doi: 10.21105/Joss.01294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A copy of the small, example dataset whose curation we walk through in the Results section is compressed into a ZipFile and submitted with this paper under “Supplementary Material.” Additionally, the Philadelphia Neurodevelopmental Cohort (PNC), the dataset whose curation we summarize in the second portion of the Results, is publicly available in the Database of Genotypes and Phenotypes (dbGaP accession phs000607.v3.p2). The source code for CuBIDS is publicly available at https://github.com/PennLINC/CuBIDS, the documentation for our software is available at https://cubids.readthedocs.io/en/latest/, and our package is available for download on the Python Package Manager (pypi) https://pypi.org/project/cubids/.