Abstract
The impact of water deficit on sucrose metabolism in sink organs like the fruit remains poorly known despite the need to improve fruit crops resilience to drought in the face of climate change. The present study investigated the effects of water deficit on sucrose metabolism and related gene expression in tomato fruits, aiming to identify candidate genes for improving fruit quality upon low water availability. Tomato plants were subjected to irrigated control and water deficit (−60% water supply compared to control) treatments, which were applied from the first fruit set to first fruit maturity stages. The results have shown that water deficit significantly reduced fruit dry biomass and number, among other plant physiological and growth variables, but substantially increased the total soluble solids content. The determination of soluble sugars on the basis of fruit dry weight revealed an active accumulation of sucrose and concomitant reduction in glucose and fructose levels in response to water deficit. The complete repertoire of genes encoding sucrose synthase (SUSY1-7), sucrose-phosphate synthase (SPS1-4), and cytosolic (CIN1-8), vacuolar (VIN1-2) and cell wall invertases (WIN1-4) was identified and characterized, of which SlSUSY4, SlSPS1, SlCIN3, SlVIN2, and SlCWIN2 were shown to be positively regulated by water deficit. Collectively, these results show that water deficit regulates positively the expression of certain genes from different gene families related to sucrose metabolism in fruits, favoring the active accumulation of sucrose in this organ under water-limiting conditions.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-023-01288-7.
Keywords: Drought, Fruit quality, Invertase, Solanum lycopersicum, Sucrose-phosphate synthase, Sucrose synthase
Introduction
In the majority of plant species, the assimilated carbon is transported from source—leaves to sink organs in the form of sucrose (Nguyen-Quoc and Foyer 2001; Dahiya et al. 2017). Sucrose is an essential element of plant life cycle, being the main product of photosynthesis. In addition, sucrose plays a crucial role in the plant development, storage, productivity, signal transduction, and osmotic homeostasis under abiotic stress conditions, generating a variety of sugars to stimulate growth, synthesize essential compounds, and act as signals to regulate transcription factors and other genes (Braun et al. 2014; Jiang et al. 2015). During its metabolism, the action of at least nine enzymes contribute to the sucrose synthesis and degradation, including fructokinase, invertase (INV), hexokinase, phosphoglucomutase, phosphoglucose isomerase, sucrose synthase (SUSY), sucrose-phosphate synthase (SPS), sucrose-phosphate phosphatase (SPP), and uracil-diphosphate (UDP)-glucose pyrophosphorylase (UGPase). Of these, the last four enzymes are more involved in the sucrose synthesis than degradation process, when compared to the other enzymes (Jiang et al. 2015).
The synthesis, transportation, storage, and degradation of sucrose are determining steps for biomass allocation and crop productivity (Braun et al. 2014). The sucrose use as a carbon and energy source depends on its hydrolysis to hexose. In plants, this reaction is catalyzed reversibly by SUSY (SUS; EC 2.4.1.13), producing UDP-glucose and fructose, and irreversibly by INVs (β-fructokinase; EC 3.2.1.26), producing glucose and fructose. SUSY is a cytosolic enzyme that plays an important role in the sucrose utilization during fruit development and there is a strong correlation between SUSY activity, growth rate, and starch content in tomato fruits (Nguyen-Quoc and Foyer 2001). SUSY has been identified and characterized in several plants species, including Arabidopsis thaliana (Baud et al. 2004), Oryza sativa (Hirose et al. 2008), Zea mays (An et al. 2014), and Malus domestica (Tong et al. 2018), and their expression levels have been demonstrated to be altered under salt, drought, cold, and light stresses (Xiao et al. 2014; Zhu et al. 2017).
Based on their optimum pH for activity, the INVs are classified into two main groups: acid and neutral (alkaline) INVs (CIN). Acid INVs are still classified in vacuolar (VIN) and cell wall (CWIN) INVs that belong to the glycosyl hydrolase 32 (GH32) family, while CINs are cytosolic and belong to the glycosyl hydrolase 100 (GH100) family (Roitsch and González 2004). Compared to CWINs and VINs, there is less information available in the literature on the functional characterization of plant CINs. However, evidences suggest the importance of CINs in the plant development and responses to biotic and abiotic stresses in several plant species, such as A. thaliana (Xiang et al. 2011), O. sativa (Jia et al. 2008), and Lotus japonicus (Welham et al. 2009). Vargas et al. (2007) proposed that the wheat alkaline invertase (Ta-A-Inv) activity would be associated with the cytosolic degradation of sucrose more efficiently during environmental stresses. The acidic INVs have an important role in the thermal stress tolerance in reproductive organs (Xiang et al. 2011) and heat stress tolerance in young fruits (Juárez-Colunga et al. 2018). Under water deficit conditions, the induction of VINs may occur in leaf tissues, resulting in the increase of hexoses (glucose and fructose). These results suggest the responsiveness of INVs when exposed to abiotic stresses (Juárez-Colunga et al. 2018).
SPS (SPS; EC 2.4.1.14) is the key regulatory enzyme of sucrose biosynthesis from UDP-glucose and has been associated to the control of crop growth and yield (Castleden et al. 2004). The SPS activity in corn (Z. mays) was correlated with growth in young plants and dry matter yield and associated with the quantitative trait locus (QTL) of grain yield (Prioul et al. 1999). In rice (O. sativa), the activity of OsSPS1 induced leaf expansion and greater plant height (Seneweera et al. 1995), while in sugarcane (Saccharum officinarum) the accumulation of sucrose in the stems was dependent on the SPS activity (Zhu et al. 1997). Additionally, SPS has been shown to play a crucial role in plants response to abiotic stresses, including drought and extreme temperatures (Almadanim et al. 2017; Bilska-Kos et al. 2020; Zhang et al. 2022), and associated to the activity of the apoplastic ascorbate oxidase to control carbon partitioning and yield improvement of tomato under water deficit (Garchery et al. 2013).
The soluble sugars content, including sucrose, glucose, and fructose, has been shown to be increased in various fleshy fruit under water deficit, depending on genotype, stress intensity, and fruit development stage (Ripoll et al. 2014, 2016), contributing to the fruit tolerance to dehydration and improvement of its organoleptic quality. Although well documented in the literature the important role that sucrose metabolizing enzymes perform in the metabolism of sugars (reviewed by Beckles et al. 2012), the effects of water deficit on these enzymes in sink organs like fruits remain poorly documented (Ripoll et al. 2014; Hou et al. 2020). For instance, an increase in SUSY activity was observed in sweet orange fruits (Citrus sinensis L. Osb.) under water deficit (Hockema and Etxeberria 2001). In another recent study, a SUSY isoform was reported to be up-regulated by water deficit in the transcriptome of tomato fruits (Bai et al. 2023). This knowledge may contribute to the development of breeding strategies to increase drought tolerance and fruit quality, in order to improve the adaptation of fleshy fruits plants to low water availability in face of climate change.
The objective of the present study was to investigate the water deficit effects on the sucrose metabolism in tomato fruits, as a model of fleshy fruit, and related gene families coding for the main sucrose metabolizing enzymes. This study focused on gene families coding for sucrose synthase, sucrose-phosphate synthase, and cytosolic, vacuolar and cell wall invertases, as these enzymes are more directly associated with the activity of synthesis and degradation of sucrose.
Materials and methods
Plant material and experimental conditions
The experiment was carried out under greenhouse conditions on the campus of the Universidade Estadual de Santa Cruz, located near the urban region of the city of Ilhéus, BA (14°47′00" S, 39°02′00" W). Tomato seeds (Solanum lycopersicum L.) cv. Santa Clara, a drought-susceptible commercial cultivar, were germinated and the seedlings, with 4–6 true leaves, were transplanted into 5-L plastic pots containing a mixture of soil and washed sand (ratio of 2:1) and cultivated under optimal conditions of water availability and nutrients in a greenhouse, before the application of the treatments. At the fruit set stage, which took place 38 days after transplanting, the pots were sealed with aluminum foil to prevent water loss by evaporation, and the plants (10 plants/treatment) were then subjected to the following treatments: (i) irrigated control, in which the plants were irrigated up to 90% of the substrate field capacity (CC; cm−3 cm−3) whenever the water content decreased to 75% of the CC, and (ii) water deficit, in that the plants received a deficit irrigation of 40% of the total volume of irrigation applied in the control treatment. Preliminary experiments demonstrated that this level of deficit irrigation induced a moderate drought stress based on several plant indicators (i.e. leaf water potential, stomatal conductance, specific leaf mass, relative growth rate, and fruit number, diameter and weight). Monitoring of substrate moisture was performed by gravimetry, and irrigation was applied to plants in the water deficit treatment whenever the plants in the control treatment were irrigated. The water treatments that started at the fruit set stage were maintained until the first fruit maturation stage.
Plant phenotyping
The second or third fully expanded and mature leaf from the apex of the plant was used to determine the pre-dawn leaf water potential (Ψw), between 1 to 3 am, using a Scholander pressure chamber (PMS Instrument Co., Albany, OR, USA), according to the methodology described by Scholander et al. (1965). Leaf gas exchange measurements were performed on fully expanded and mature leaves from 7 to 10 am, using a portable Li-COR photosynthesis measurement system (LI-6400 XT, Nebraska, USA). The net photosynthetic rate (A), stomatal conductance (gs), transpiration (E), and the ratio between internal and external CO2 concentrations (Ci/Ca) were measured under artificial saturating light of 1000 μmol photons m−2 s−1 and atmospheric concentration of CO2 (Ca) of 400 μmol mol−1. The measurements of Ψw and leaf gas exchange were carried out concomitantly, using the same plants, when their fruits reached the breaker (B), breaker plus 7 days (B + 7; red ripe stage) and breaker plus 14 days (B + 14) stages. Instantaneous water use efficiency (A/E) was obtained by the ratio between the net photosynthetic rate (A) and the transpiration rate (E), intrinsic water use efficiency (A/gs) was calculated by the ratio between the net photosynthetic rate (A) and stomatal conductance to water vapor (gs). The carboxylation efficiency (A/Ci) was obtained by the ratio between the net photosynthetic rate (A) and the intercellular concentration of CO2 (Ci).
Dry biomass of roots, stems, leaves, and fruits were determined at the beginning and end of water treatments application by drying in a forced air circulation oven at 70 °C until reaching constant weight. The productive efficiency of fruits was calculated by the ratio of average dry biomass of fruits per plant and average dry biomass of the fruits of their corresponding treatment.
Fruit phenotyping
Fruits were collected at the red ripe stage (B + 7), on average 35 days after the application of the treatments, and the equatorial, lower and upper polar diameters were measured using a manual caliper. pH was determined by direct reading in pulp solution from samples of at least three fruits randomly chosen, using a digital pH meter (PHS-3E-BI, Ion, Araucária, Brazil). The same samples of fruit pulp were used to measure the total soluble solids (SSC; oBrix), using an analog refractometer (0 to 32% Brix; Akso, RHB32, São Leopoldo, Brazil).
Soluble sugars were extracted from lyophilized tissue samples from the pericarp of the fruit which were macerated in a mortar with a pestle and transferred to micro-tubes with lid containing 1.5 ml of 80% ethanol per 20 mg of tissue, placed in a water bath at 80 °C for 20 min, and centrifuged to obtain the supernatant. Extraction was performed four times with the same volume of ethanol and the combined supernatants were concentrated in vacuo (ThermoScientific R Savant SC 250 EXP), resuspended in 1 ml of water and 1 ml of chloroform, and used for sugar analysis. Sucrose, fructose, and glucose concentrations were determined by High-Performance Anion Exchange Chromatography with Pulsed Amperometric Detection (HPAEC-PAD) on a Dionex R system (ICS 5000), using a CarboPac PA1 column and elution with 150 µM of sodium hydroxide in an isocratic run of 27 min, according to the methodology described by Pagliuso et al. (2018).
Identification and analysis of the sucrose metabolism gene families in tomato
Annotation information and BLAST searches using conserved domain sequences as input were employed to identify all putative genes encoding SUSY, SPS, CIN, VIN, and CWIN enzymes in the S. lycopersicum reference genome available on Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html) and Sol Genomics (https://solgenomics.net/) databases. Information on coding sequences (CDS), physical location, exon–intron structure, and predicted amino acid sequences were also obtained from Phytozome and Sol Genomics. The exon/intron structures were constructed using the GSDS—Gene Structure Display Server 2.0 (Guo et al. 2007) (http://gsds.cbi.pku.edu.cn/) and the MapChart software (2.30) (https:/ /www.wur.nl/en/show/Mapchart-2.30.htm) was used to plot the location of genes on the respective chromosomes. Analysis of cis-acting regulatory elements present in the 1500 bp promoter region before the start site of gene transcription was performed using the plantCARE tool (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The Grand Average of Hydropathy (GRAVY), molecular mass (MW), and isoelectric point (pI) of the deduced amino acid sequences were predicted with the PROTPARAM tool available in the Expert Analysis System (ExPASy) (http://web.expasy.org/protparam/). The prediction of subcellular location was performed using the PSORTII tool available on the GenScript database (https://www.genscript.com/tools/psort) and Deep-Loc tool 1.0 (http://www.cbs.dtu.dk/services/DeepLoc/). The amino acid sequences were aligned using the ClustalX 2.1 software (Thompson et al. 1994), and the similarity dendrogram was generated using the Neighbor-Joining (NJ) method (Saitou and Nei 1987), with a bootstrap of 1000 replications. The dendrograms were constructed using the MEGA7 program (Kumar et al. 2007). Potential protein–protein interactions (PPI) of the sucrose metabolism genes positively regulated by water deficit were analyzed using the STRING database (https://string-db.org), which collects, integrates, and scores all available PPI data from different sources, including experiments, databases, co-expression, and co-occurrence, to show comprehensive physical and functional PPI networks. A high confidence cut-off of 0.7 was applied.
Publicly available gene expression data
Publicly available data from the tomato transcriptome were used to investigate the expression profiles of SUSY, SPS, CIN, VIN, and CWIN genes at different stages of fruit maturation (The Tomato Genome Consortium 2012) or in response to treatments with abscisic acid (ABA) and its inhibitor NDGA (Nordihydroguaiaretic acid) during fruit maturation (Mou et al. 2015).
Quantitative real-time PCR (RT-qPCR) analysis
Total RNA was extracted from the pericarp of fruits in the red ripe (B + 7) stage using the TRIzol® reagent, following the manufacturer's instructions. The quality and integrity of the isolated RNA were evaluated by 1% agarose gel analysis and quantified with the aid of NANOdrop (Thermo Scientific™, ND2000USCAN, Wilmington, DE, USA). The RNA samples were then treated with RNAse-free DNAse I (Invitrogen, Carlsbad, CA, USA), and cDNA synthesis was performed using the RevertAid H Minus kit (Fermentas Life Science, Hanover, MD, USA), following the instructions of the manufacturer. All RT-qPCR procedures, including tests, validations, and experiments, were performed on the Mx 3005P device (Agilent Technologies, Santa Clara, CA, USA), using the Ampliqon Real QPlus 2 × Master Mix Green Low Rox™ kit (Ampliqon Company, Denmark), according to the manufacturer's instructions. The reference genes GAPDH (Solyc05g014470), RPL2 (Solyc10g006580), and ACT (Solyc03g078400) were amplified together with the target genes as endogenous controls to normalize the expression among different samples. To choose the best reference gene, the NormFinder program (Andersen et al. 2004) (https://moma.dk/normfinder-software) was used. The sequences of target genes and endogenous controls are described in Table S1. To quantify gene expression, the 2−ΔΔCt method was used (Livak et al. 2001), using data from at least three biological replicates that were validated individually. Control reactions, devoid of cDNA (NTC), were also used in all experiments.
Statistical analysis
The experimental design adopted in the water deficit experiment was performed in completely randomized blocks, with ten replicates of one plant per pot and two water regimes (control and water deficit). Data were initially tested for distribution using the Shapiro–Wilk test, with a significance level of 5%. Once the hypothesis was confirmed, the statistical differences were evaluated based on the analysis of variance (ANOVA), and the averages were separated by the Student's t-test, with a critical value of P ≤ 0.05. The heatmaps of gene expression and RPKM data were plotted using the “ComplexHeatmap” package. All statistical analyzes were performed in R software (R Development Core Team 2017).
Results
Identification of the tomato sucrose metabolism gene families
Although some genes of the SUSY, SPS, CIN, VIN, and CWIN families have been previously identified in tomato, their sequences, structure characteristics, phylogenetic relationships, and expression responses to environmental stresses have not yet been comprehensively analyzed. The recent advances in plant genome sequencing and analysis technologies and the release of updated versions of the S. lycopersicum reference genome allowed us to identify and characterize these gene families in a more comprehensive manner. The number of genes identified within each gene family, their chromosomal locations, CDS (coding sequence) length, and the predicted polypeptide sizes, molecular weights, isoelectric points (pI), and subcellular locations are shown in Table 1 and Fig. S1.
Table 1.
Characteristics of genes encoding sucrose metabolizing enzymes in Solanum lycopersicum L. GRAVY: Grand average of hydropathy; MW: molecular weight; pI: isoelectric point
| Enzyme | Transcript name | Chromosomal location | CDS (bp) | Polypeptide size (aa) | MW (KDa) | pI | GRAVY | Subcellular location prediction |
|---|---|---|---|---|---|---|---|---|
| SlSUSY1 | Solyc12g009300 | ch12:2,573,935..2,577,879 | 2418 | 805 | 92.5 | 5.94 | −0.251 | Cytoplasm |
| SlSUSY2 | Solyc12g040700 | ch12:43,028,228..43,031,386 | 771 | 256 | 29.5 | 7.53 | −0.062 | Cytoplasm |
| SlSUSY3 | Solyc07g042550 | ch07:55,976,889..55,982,494 | 2418 | 805 | 92.5 | 5.96 | −0.254 | Cytoplasm |
| SlSUSY4 | Solyc09g098590 | ch09:72,380,198..72,385,562 | 2439 | 812 | 92.9 | 5.91 | −0.292 | Cytoplasm |
| SlSUSY5 | Solyc07g042520 | ch07:55,816,657..55,820,439 | 2412 | 803 | 91.6 | 5.97 | −0.298 | Cytoplasm |
| SlSUSY6 | Solyc03g098290 | ch03:60,636,360..60,640,569 | 2676 | 891 | 100.7 | 5.87 | −0.296 | Cytoplasm |
| SlSUSY7 | Solyc02g081300 | ch02:45,315,741..45,320,028 | 2655 | 884 | 100.6 | 8.42 | −0.39 | Cytoplasm |
| SlSPS1 | Solyc08g042000 | ch08:24,345,525..24,352,756 | 3138 | 1045 | 117.5 | 6.21 | −0.383 | Cytoplasm |
| SlSPS2 | Solyc09g092130 | ch09:71,269,146..71,277,256 | 3195 | 1064 | 119.5 | 6.13 | −0.477 | Cytoplasm |
| SlSPS3 | Solyc11g045110 | ch11:31,768,096..31,775,364 | 3003 | 1000 | 113.1 | 6.59 | −0.404 | Cytoplasm |
| SlSPS4 | Solyc07g007790 | ch07:2,438,924..2,447,625 | 3165 | 1054 | 118.4 | 6.05 | −0.44 | Cytoplasm |
| SlCIN1 | Solyc01g100810 | ch01:90,737,614..90,743,352 | 1962 | 653 | 74.4 | 8.18 | −0.317 | Chloroplast |
| SlCIN2 | Solyc01g111100 | ch01:97,477,472..97,482,570 | 1818 | 605 | 69.0 | 6.95 | −0.3 | Chloroplast |
| SlCIN3 | Solyc04g081440 | ch04:65,414,803..65,419,467 | 1713 | 571 | 65.2 | 5.97 | −0.29 | Cytoplasm |
| SlCIN4 | Solyc06g065210 | ch06:40,659,646..40,663,785 | 1656 | 551 | 62.7 | 6.16 | −0.201 | Cytoplasm |
| SlCIN5 | Solyc11g007270 | ch11:1,652,454..1,656,955 | 1968 | 655 | 73.5 | 5.84 | −0.231 | Chloroplast |
| SlCIN6 | Solyc11g020610 | ch11:11,747,788..11,752,875 | 1608 | 535 | 60.8 | 6.11 | −0.221 | Cytoplasm |
| SlCIN7 | Solyc11g067050 | ch11:52,804,241..52,807,353 | 1926 | 641 | 72.4 | 6.35 | −0.223 | Chloroplast |
| SlCIN8 | Solyc01g058010 | ch01:64,953,319..64,958,618 | 1521 | 506 | 57.1 | 5.38 | −0.274 | Chloroplast |
| SlCWIN1 | Solyc09g010080 | ch09:3,475,480..3,479,343 | 1755 | 584 | 67.2 | 9.20 | −0.361 | Cell wall |
| SlCWIN2 | Solyc10g083290 | ch10:63,110,526..63,115,912 | 1749 | 582 | 65.8 | 9.23 | −0.393 | Cell wall |
| SlCWIN3 | Solyc09g010090 | ch09:3,480,545..3,484,159 | 1752 | 583 | 66.1 | 6.93 | −0.428 | Cell wall |
| SlCWIN4 | Solyc10g083300 | ch10:63,123,100..63,127,293 | 1770 | 589 | 66.8 | 8.94 | −0.347 | Cell wall |
| SlVIN1 | Solyc08g079080 | ch08:62,722,555..62,726,754 | 1959 | 652 | 72.7 | 6.21 | −0.325 | Vacuole |
| SlVIN2 | Solyc03g083910 | ch03:53,851,092..53,855,368 | 1947 | 648 | 71.3 | 5.54 | −0.219 | Vacuole |
Sucrose synthase (SUSY)
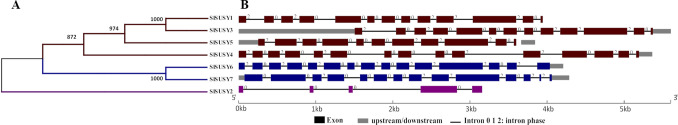
Analysis based on sequence searches in the tomato genomic databases resulted in the identification of seven genes encoding SUSY (Table 1). All seven SlSUSY amino acid sequences share two conserved sucrose synthase and glycosyl transferase domains, which are typical signatures of SUSY proteins (Fig. S2). The similarity dendrogram based on the amino acid sequences revealed three different clusters (Fig. 1A). The first group contains four enzymes, the second two enzymes, and the last group only one enzyme. These results are consistent with those of the exon–intron structure analysis (Fig. 1B). SlSUSYs contain five to 15 exons, with SlSUSY4 and -6 characterized by having an additional exon (exon 13), SlSUSY1 and -3 by having the longest exon 11, and SlSUSY7 by having the shortest exon 13. SlSUSY5 has only 11 exons due to the union of exons 2 and 3, exons 5 and 6, and exons 9 and 10, as compared to SlSUSY4 and -6. SlSUSY2 presented the lowest number (5) of exons (Fig. 1B).
Fig. 1.
Characteristics of the sucrose synthase (SUSY) gene family in Solanum lycopersicum. Dendrogram of amino acid sequence similarity (A) and exon–intron structure (B) of the SUSY gene family in S. lycopersicum
Sucrose-phosphate synthase (SPS)
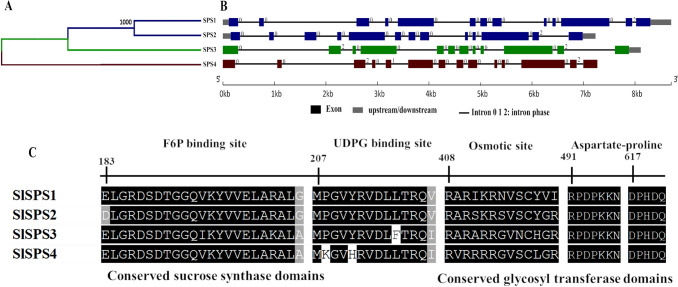
Four genes encoding SPS (SlSPS1-4) were identified in the tomato genome (Table 1). The similarity dendrogram based on amino acid sequences showed three groups, the first containing two members (SlSPS1 and -2) and both second and third with only one member each (SlSPS3 and SlSPS4) (Fig. 2A). SlSPS1 and -2 have similar structures and numbers of exons and introns, while SlSPS3 has 12 exons, due to the absence of exon 2 and exon 5, which were found in SlSPS1, -2, and -4, respectively (Fig. 2B). The SPS family has three conserved protein domains, sucrose synthase (N-terminal), glycosyl transferase and the sucrose-6F-phosphate phosphohydrolase domain (C-terminus), three conserved binding sites (F6P binding site, UDPG binding site, osmotic site), and presence of aspartate-proline (DP) (Figs. 2C and S3).
Fig. 2.
Characteristics of the sucrose-phosphate synthase (SPS) gene family in Solanum lycopersicum. Dendrogram of amino acid sequence similarity (A), exon–intron structure (B), and alignment of the conserved amino acid regions (C). Black and gray indicate conserved amino acids
Cytosolic invertase (CIN)
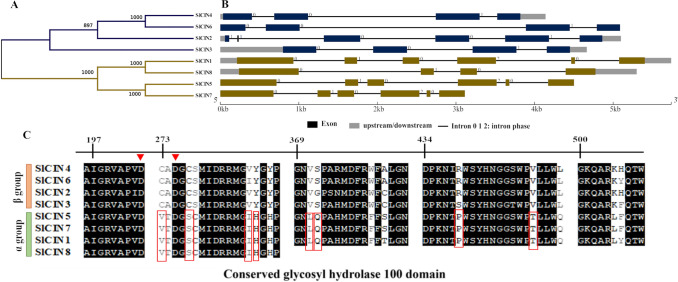
Eight genes of neutral/cytosolic INVs (SlCIN1-8) were identified in the tomato genome (Table 1). SlCINs contain a glycosyl hydrolase 100 protein domain covering approximately 80% of the protein (Fig. S4). The similarity dendrogram divided the SlCINs into two main groups (Fig. 3A). The α group contains SlCIN1, -5, -7, and -8, while the β group contains SlCIN2, −3, −4, and −6 (Fig. 3A). The exon–intron structure analysis evidenced that they usually consist of 3 or 6 exons alternated by extensive intronic regions (Fig. 3B). SlCIN1, −2, −5 and −7 have six exons and SlCIN1 shows a pattern of exonic structure distinct from the other genes of the family. SlCIN3, -4, and -8 have four exons and SlCIN8 show an increase in exon 1 and a decrease in exons 2 and 3 (Fig. 3B). The two groups consistently differ by eight amino acid residues in the conserved motifs (C273V, C277S, V286I, Y287H, V371L, S372Q, R439P, and V450T), based on the amino acid numbering of SlCIN4 (Fig. 3C).
Fig. 3.
Characteristics of the cytosolic invertase (CIN) gene family in Solanum lycopersicum. Similarity dendrogram (A), exon–intron structure (B), and alignment of conserved regions (C). Black indicates conserved amino acids. The red boxes indicate the amino acids present in only one group of the SlCIN family. Red triangles highlight conserved Aspartate residues
Vacuolar (VIN) and cell wall (CWIN) invertases
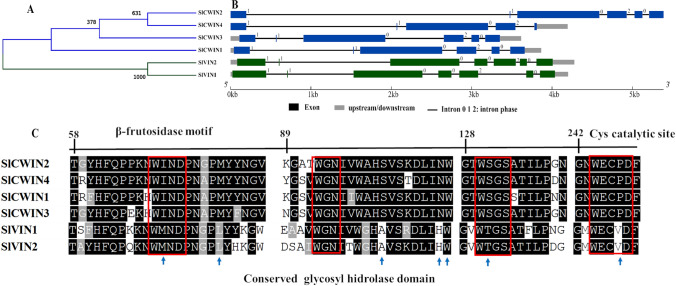
Six genes encoding acid INVs were identified in S. lycopersicum (Table 1). Among these, four of them are located in the cell wall (SlCWIN1-4) and two in the vacuole (SlVIN1, and -2). The similarity dendrogram divided the acid INVs into two clades, one of which is inferred to be directed to the cell wall and the other to the vacuole (Fig. 4A). The SlCWIN gene family has 6 exons, with a high standard of conservation. The SlCWIN1 and -3 and SlCWIN2 and -4 showed the same pattern of exon conservation, as they are duplicated in tandem on chromosomes 9 and 10, respectively (Fig. 4B; Fig. S1). The SlCVINs have seven well-conserved exons (Fig. 4B). Analysis of SlVIN enzymes revealed three domains, of which one in the N-terminal portion is of unknown function (DUF3357) and the other two in the C-terminal portion are glycosyl hydrolases domains (Fig. S5). On the other hand, SlCWINs contain the glycosyl hydrolase domain in the N and C-terminal portions of all enzymes (Figs. 4C and S6).
Fig. 4.
Characteristics of the acid invertases (SlCWIN and SlVIN) in Solanum lycopersicum. Dendrogram of amino acid sequence similarity (A), exon–intron structure (B), and alignment of conserved regions (C). Black and gray indicate conserved amino acids. Blue arrows indicate conserved amino acids characteristic of each gene family according to their subcellular location. The red boxes indicate the conserved motifs of the acid INVs
Stress-responsive cis-acting regulatory elements in the promoter regions of the tomato sucrose metabolism gene families
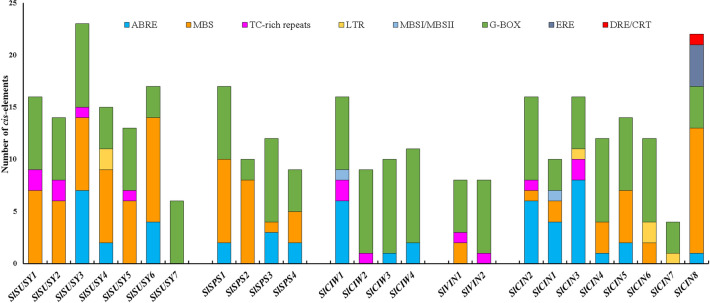
Analysis of the promoter regions of the sucrose metabolism genes identified the presence of several cis-acting regulatory elements responsive to stress, including ABRE (ABA-responsive element), MBS (MYB binding site), TC-rich repeats (involved in defense and stress responsiveness), G-box (light, ABA, methyl-jasmonate and anaerobic responses), DRE/CRT (dehydration-responsive element/C-repeat), LTR (low-temperature responsiveness), flavonoid biosynthesis (MBSI), and ERE (ethylene-responsive element (Fig. 5). Cis-acting regulatory elements involved in processes related to plant development were also found, including those involved in the regulation of metabolism (O2-site), anaerobic induction (ARE), expression in endosperm (GCN4_motif), mesophyll cell differentiation (HD-Zip 1), MeJA response element, TATC-box (gibberellin response), TCA-element (salicylic acid response), and TGA-element (auxin response) (Fig. S7).
Fig. 5.
Cis-acting regulatory elements responsive to stress present in the promoter regions of the sucrose metabolism genes from S. lycopersicum. The cis-acting regulatory elements were analyzed in the 1500 kb region upstream of the translation start site
Plant and fruit responses and transcriptional regulation of sucrose metabolism genes under water deficit
Publicly available RNA-Seq expression data show that the transcription of the sucrose metabolism genes is highly regulated during the normal process of fruit ripening and in response to ABA (Figs. S8A, B). For instance, SlSUSY2, -3 and -6, SlSPS1, SlCIN1 and -5, and SlVIN2 are down-regulated in the mature green stage of fruit ripening but up-regulated in the red ripe stage, while the other genes analyzed showed an opposite behavior of gene expression (Fig. S8A). In addition, SlSUSY1, -3, -4, and -6, SlSPS1 and -4, SlCIN1, SlVIN2, and SlCWIN2 are up-regulated by ABA and down-regulated by its inhibitor NDGA, while an opposite expression behavior was observed for SlSUSY7, SlSPS3, SlCIN3, -4, -5, and -6, SlVIN1, and SlCWIN1 (Fig. S8B).
A set of sucrose metabolism genes regulated by ABA were further selected for analysis of gene expression in response to water deficit in fruits as described in ‘Materials and Methods’. Plant and fruit phenotyping firstly showed that water deficit caused a decrease in the values of Ѱw, A, gs, and E (Fig. S9), Ci/Ca and A/Ci (Table S2), and dry biomass of leaves, stems, and fruits and fruit number (Table S3), an increase in the instantaneous (A/E) and intrinsic (A/gs) water use efficiencies (Table S2), root dry biomass (Table S3), and SSC (Fig. S10), and no significant changes in fruit diameters and productive efficiency (Table S3) and pH (Fig. S10). Determination of the concentration soluble sugars on a fruit dry weight basis showed that water deficit caused a significant reduction in the levels of glucose and fructose, but a significant increase in the sucrose content (Fig. S10). RT-qPCR analysis showed that SlSUSY4, SlSPS1, SlCIN3, SlVIN2, and SlCWIN2 were positively regulated by water deficit in fruits, while SlSUSY1 was down-regulated in the two water regimes analyzed (Fig. 6).
Fig. 6.
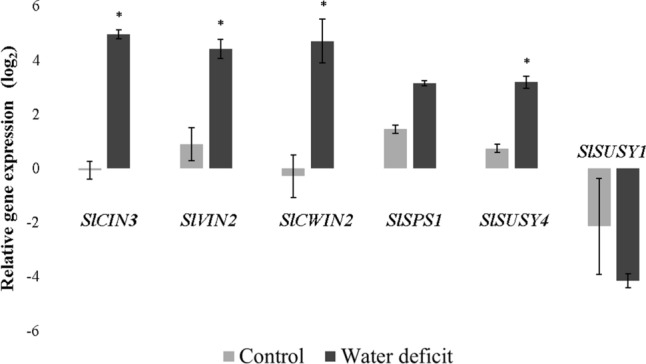
RT-qPCR expression analysis of sucrose metabolism genes in response to irrigated control or water deficit treatments in fruits of Solanum lycopersicum L. cv. Santa Clara at the red ripe stage (B + 7). The data are means ± SE of three biological replicates in which ACT transcripts were used as internal controls. *Significantly different from control treatment at P ≤ 0.05 by the Student's t-test
Protein interaction network analysis of the water deficit-induced sucrose metabolism genes
We further explored the protein–protein interaction network of the validated sucrose metabolism candidate genes which were induced by water deficit. The resulting interaction network indicated that all proteins analyzed have a high connectivity with other proteins of the carbohydrate metabolism (Fig. 7 and Table S4). SlSUSY1 and -4 are able to interact with each other, as well as SlSPS1 and -4. These proteins show, in turn, prominent interactions with diverse proteins of carbohydrate metabolism, including ectonucleotide pyrophosphatase/phosphodiesterase, UTP-glucose-1-phosphate uridylyltransferase, starch synthase, and sucrose-phosphatase. SlVIN2 and SlCWIN2 showed interaction with a number of fructokinase (FRK) and hexokinase (HXK) proteins, which have been involved in responses of plants to several abiotic stress.
Fig. 7.
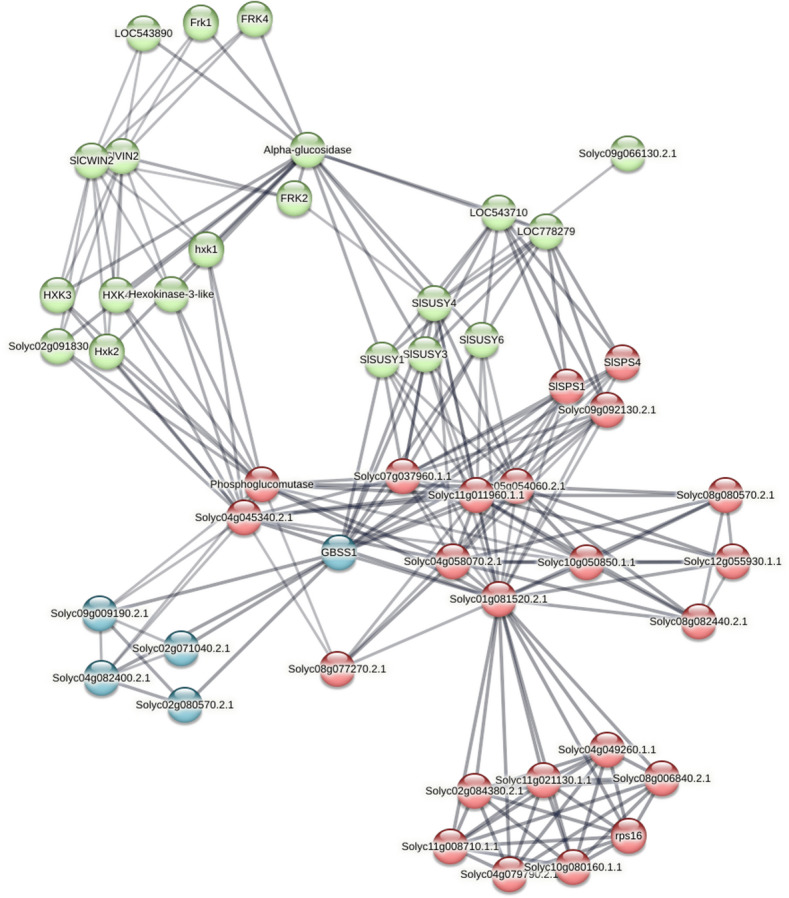
The protein interaction network of the sucrose metabolism genes positively regulated by water deficit. Graphical representation of the network was retrieved from the STRING database. In red, primary interaction proteins, in blue, secondary interaction proteins and in green, tertiary interaction proteins. Line thickness indicates the strength of data support
Discussion
The identification and molecular characterization of genes involved in sucrose metabolism provide an initial basis for understanding their functions during the maturation process of fleshy fruits under water deficit conditions. The S. lycopersicum genome encodes seven SlSUSY, four SlSPS, eight SlCIN, two SlVIN, and four SlCWIN genes (Table 1). This study identified an additional SlCIN gene to the seven previously described in tomato (Ruan 2014). This number of genes in the respective families is similar to those found in Arabidopsis thaliana, and the differences may be explained by the recent triplication of the S. lycopersicum genome (The Tomato Genome Consortium 2012).
The size and position of introns and exons provide important information about gene evolution. Structural analysis of the tomato genes related to sucrose metabolism indicates that SlSUSY has 10 to 14 introns (Fig. 1B), consistent with analyzes of SUSY in the apple tree (Tong et al. 2018) and cotton (Ruan et al. 2008), which contain 10 to 14 introns, and A. thaliana (Zhang et al. 2015) with 11 to 14 introns. The inferences indicate that these similar characteristics in SUSY family genes among plants (dicotyledon and monocotyledon) may contribute to their functional similarity within the same group (Zhang et al. 2015; Tong et al. 2018). Although there were differences in the number and size of exons-introns among the seven SlSUSY genes (Fig. 1B), a high level of conservation can be observed within the groups, as well as the high similarity observed by multiple alignments among the seven predicted amino acid sequences (Fig. 1C). The amino acid sequences of all seven enzymes showed typical characteristics of the SUSY family observed in previous studies (Ruan et al. 2008; Zhang et al. 2015; Tong et al. 2018), such as the presence of the closely connected domains sucrose synthase and glycosyl transferase.
The tomato SPS genes showed a conserved gene structure and their predicted amino acid sequences were separated into three groups (Figs. 2A, B), corroborating with information reported in other plant species such as A. thaliana (Sun et al. 2011), O. sativa (Castleden et al. 2004), and Z. mays (Lutfiyya et al. 2007). The phylogeny of the SPS family suggests divergence between monocotyledon and dicotyledon, showing successive gene duplication events. Structural and evolutionary differences can originate enzymes with distinct biological functions (Lutfiyya et al. 2007). Tomato SPS present several regulatory mechanisms, such as regulation by F6P, UDPG, and osmotic stress (Toroser and Huber 1997), commonly found in spinach and rice, in which the phosphorylation site of Ser-424 was identified. This site is reversibly phosphorylated in response to osmotic stress (Toroser and Huber 1997) and may be associated with the regulatory responses in plants (Taylor et al. 2000; Lunn and Macrae 2003).
Concerning the cytosolic INVs (Figs. 3A-C), the differences in the structure of the groups suggest that these genes may have arisen from distinct ancestors and their intronic regions suffered changes during evolution. Similar events were observed in M. domestica (Hyun et al. 2011), Brassica rapa (Eom et al. 2019), O. sativa, and A. thaliana (Ji et al. 2005). Analysis of their amino acid sequences resulted in the identification of eight amino acid residues in the conserved motifs, which have also been reported in O. sativa (Ji et al. 2005), B. rapa (Eom et al. 2019), A. thaliana (Qi et al. 2007), and M. domestica (Hyun et al. 2011).
The tomato acid INVs (Figs. 4A-C) also demonstrated a conserved exons-intron structure, regardless of their predicted subcellular location, which may correspond to the ancestral gene structure of acid INVs (Ji et al. 2005). The structure of exons-intron in the cell wall INVs (Figs. 4A-C) indicates that duplication of intron 1 occurred in the 5' region of the SlCWIN2 and -4 genes in comparison to the SlCWIN1 gene. The SlCWIN3 gene had the loss of intron 1 (3' region) and the duplication of intron 2, with all genes having the same number of exons and introns (Fig. 4B). Among the four CWIN isoforms in tomato, SlCWIN1 is closely related to SlCWIN3 expressed in reproductive organs, while SlCWIN2 is more closely related to SlCWIN4, expressed mainly in vegetative tissues (Godt and Roitsch 1997). Similar results were found in A. thaliana, where AtCWIN2 and AtCWIN4 are predominantly expressed in reproductive organs, whereas AtCWIN1 and AtCWIN5 were expressed constitutively (Wang and Ruan 2012). Analysis of their amino acid sequences showed that both SlCWIN and SlVIN have the β-fructokinase motif (NDPN) in the N-terminal portion and a catalytic site of WECPD containing a proline (P) in CWINs and a valine (V) WECVD in VINs (Le Roy et al. 2007; Wan et al. 2018) (Fig. 4C). The CWINs show a conserved hydrophobic region (WIN/WGN/WSGS), which stabilizes sucrose binding, and Asp-239/Lys-242 residues (D and R) (Le Roy et al. 2007). In in vitro mutagenesis studies, a single D to A amino acid substitution (Asp-239) transformed AtCWIN1 from A. thaliana (At3g13790) into a protein incapable of hydrolyzing sucrose. Thus, the presence or absence of Asp-239 (D) has been proposed as a reliable determinant for the identification of functional or defective INVs (Le Roy et al. 2007).
The gene expression data evidenced a functional specialization of the sucrose metabolism genes during the fruit ripening process and in response to ABA (Figs. S8A, B). The group of genes induced in the early stages of maturation (mature green and breaker), or by treatment with ABA, is distinct from that induced in the red ripe stage or by treatment with the ABA NDGA inhibitor (Figs. S8A, B). An ABA-induced expression of genes acting on sucrose metabolism has also been observed in other studies (Yoshida et al. 2019; Siebeneichler et al. 2020). The presence of cis-acting regulatory elements of the type ABRE in the promoter regions of some sucrose metabolism genes (Fig. 5) supports these findings, indicating that these genes are transcriptionally activated in the ABA signaling pathway (Yoshida et al. 2019). On the other hand, sucrose has also been suggested to function as a signal that acts upstream in the ABA signaling pathway, playing an important role in the regulation of strawberry ripening induced by ABA (Jia et al. 2013).
Water deficit caused, among other effects, a significant reduction in the fruit dry biomass and number (Table S3), but significantly increased SSC values (Fig. S10), as previously reported in tomato (Bertin et al. 2000; Bai et al. 2023), grape (Castellarin et al. 2007), strawberry (Terry et al. 2007), and apple (Wang et al. 2019). SSC are the main osmotic compounds that accumulate in fleshy fruits, in addition to being determinant components of their organoleptic quality (Beckles et al. 2012). In the present study, the higher SSC values in fruits under water deficit can be attributed to the significant increase in the sucrose content (Fig. S10). Such an increase in sucrose concentration and concomitant reduction of glucose and fructose levels in fruits under water deficit suggest that, under these conditions, sucrose synthesis predominates over its degradation, as a strategy for the fruit to tolerate water deficit. In fact, sucrose has been shown to influence the cellular water relations in fruits and hence fruit water uptake that determines fruit size and solute concentrations (Hou et al. 2020), besides to act as a potent osmoprotectant and osmoregulation agent, protecting cellular components from damage caused by stress (Bertin et al. 2000; Ma et al. 2017; Zhang et al. 2016). RT-qPCR analysis showed that SlSUSY4, SlSPS1, SlCIN3, SlCWIN2, and SlVIN2 genes were positively regulated by water deficit treatment (Fig. 6), suggesting their involvement in the sucrose accumulation in tomato fruits under water deficit (Fig. S10). The AtSUS1 (AT5G20830) and AtSUS4 (AT3G43190) genes of A. thaliana and the MeSUS2 gene (Manes.01G221900) of M. esculenta had also their transcriptional activity induced in conditions of water deficit, and MeSUS2 was considered a determining factor in the metabolism of sucrose under water deficit (Baud et al. 2004; Liao et al. 2017). SlSUSY4 is an ortholog of AtSUS4 and MeSUS2 (Baud et al. 2004; Liao et al. 2017) and may share similar functions. Similarly, the overexpression of SlSPS1 was shown to increase the growth and biomass, including fruit weight, SPS activity, sucrose content, and heat stress tolerance of transgenic tomato plants (Zhang et al. 2022), whereas the Arabidopsis ortholog of SlCIN3 was identified as a candidate gene responsible for the difference in frost tolerance between different genotypes of A. thaliana (Meissner et al. 2013). SlCWIN2 was also demonstrated to be highly expressed during cold stress (Wei et al. 2020), whereas the wheat ortholog of SlVIN2 was more expressed in the drought-tolerant than in the drought-susceptible genotype under water stress conditions (Li et al. 2012). Previous studies on Nicotiana tabacum (Wang et al. 2015), M. domestica (Tong et al. 2018), B. rapa (Eom et al. 2019), S. tuberosum (Geigenberger et al. 1999), and S. lycopersicum (Bai et al. 2023) also reported induction of transcripts for some enzymes that metabolize sucrose under water deficit conditions, but information regarding the activity of these enzymes in fruits subjected to water deficit is still limited.
Collectively, the results suggest that genes belonging to the SUSY, SPS, and INVs families of S. lycopersicum play an important role in sucrose metabolism under water deficit conditions. This is further supported by the presence in their promoter regions of other ABA and drought cis-acting responsive elements besides ABRE, including DRE/CRT, MBS, G-BOX, and TC-rich repeats (Fig. 5; Hernandez-Garcia and Finer 2014), and also by their protein interactions with other proteins implicated in abiotic stress response, such as FRKs and HXKs (Fig. 7). These are important enzymes that catalyze the key metabolic step of fructose (FRKs) and glucose (HXKs) phosphorylation required for these free hexoses enter metabolic pathways, after sucrose cleavage by SUSY and INVs. They have been shown to respond to different types of abiotic stress, including anoxia, salt, drought, and wounding, and implicated in conducting carbohydrate metabolism toward distinct metabolic pathways and regulating the amount of carbohydrate metabolized under varied environmental conditions, especially in sink tissues (Granot et al. 2014; Stein and Granot 2018).
Conclusion
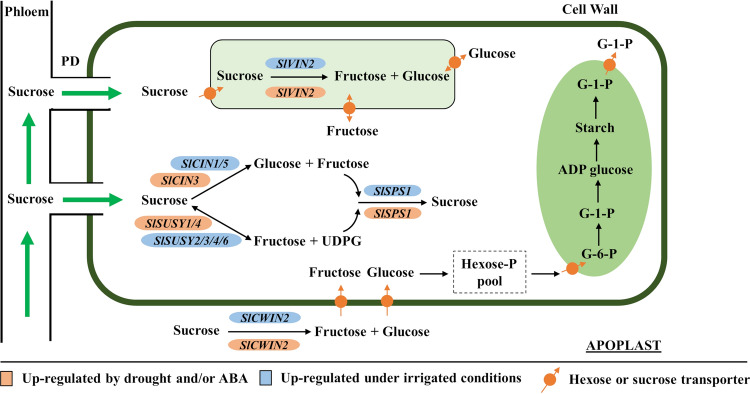
In the present study, 25 genes distributed over five families involved in the sucrose metabolism were identified and characterized at molecular level. The enzymes showed conserved domains similar to sequences previously characterized in other plant species, indicating that they are functional proteins. The expression of these genes can be regulated at the transcriptional level by endogenous and environmental signals, including ABA and drought, as demonstrated by the cis-acting regulatory elements present in their promoter regions. The increase in the soluble sugar content of the fruit in response to water deficit is caused by the significant increase in sucrose levels, which correlated with the significant induction in the expression levels of SlSUSY4, SlSPS1, SlCIN3, SlCWIN2, and SlVIN2. Finally, a model of the effects of water deficit on genes encoding sucrose metabolism enzymes in the fruit is proposed in Fig. 8. According to the proposed model, water deficit induces an up-regulation of SlCWIN2, whose enzymatic activity contributes to the increase of the hexose pool in the apoplast. Most of the hexoses generated in the apoplast are reconverted to sucrose in the cytosol by the action of SlSPS1, whose expression is induced by water deficit. Sucrose pools are inverted into hexoses in the cytosol and vacuole by the respective actions of SlSUSY4, SlCIN3, and SlVIN2, whose genes are also transcriptionally induced by water deficit. These genes are valuable targets for further functional studies to confirm their functions in the adaptation of fleshy fruits to low water availability and for potential applications in the improvement of drought tolerance and organoleptic quality in fruit crops.
Fig. 8.
A proposed model of the effects of water deficit on sucrose metabolism in tomato fruits. PD – plasmodesmata
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Marcos Silveira Buckeridge (IB-USP) for providing the HPAEC-PAD instrumental facilities.
Author contributions
ACOB, DSRJ and MGCC conceived and designed the experiments; ACOB, DSRJ, GCBS, MGMS, PHGAO, and AAC performed the experiments; ACOB, DSRJ, MGMS, LRC and AAC analyzed the experimental data; ACOB wrote the article; and LRC and MGCC revised the article. All the authors read and approved the final manuscript.
Funding
This work was supported by research grant from CNPq (Process # 304878/2018–9). ACOB was recipient of a master's degree scholarship provided by CAPES Foundation (Brasília, DF, Brazil). MGMS was recipient of a Scientific initiation program (IC) scholarship provided by UESC. MGCC is a CNPq Research Fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Almadanim MC, Alexandre BM, Rosa MTG, Sapeta H, Leitão AE, Ramalho JC, Lam TT, Negrão S, Abreu IA, Oliveira MM. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017;40:1197–1213. doi: 10.1111/pce.12916. [DOI] [PubMed] [Google Scholar]
- An X, Chen Z, Wang J, Ye M, Ji L, Wang J, Liao W, Ma H. Identification and characterization of the Populus sucrose synthase gene family. Gene. 2014;539:58–67. doi: 10.1016/j.gene.2014.01.062. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bai C, Zuo J, Watkins CB, Wang Q, Liang H, Zheng Y, Liu M, Ji Y. Sugar accumulation and fruit quality of tomatoes under water deficit irrigation. Postharvest Biol Technol. 2023;195:112112. doi: 10.1016/j.postharvbio.2022.112112. [DOI] [Google Scholar]
- Baud S, Vaultier MN, Rochat C. Structure and expression profile of the sucrose synthase multigene family in Arabidopsis. J Exp Bot. 2004;55:397–409. doi: 10.1093/jxb/erh047. [DOI] [PubMed] [Google Scholar]
- Beckles DMB, Ong NH, Tamova LS, Uengwilai KL. Biochemical factors contributing to tomato fruit sugar content: a review. Fruits. 2012;67:49–64. doi: 10.1051/fruits/2011066. [DOI] [Google Scholar]
- Bertin N, Guichard S, Leonardi C, Longuenesse JJ, Langlois D, Navez B. Seasonal evolution of the quality of fresh glasshouse tomatoes under Mediterranean conditions, as affected by air vapour pressure deficit and plant fruit load. Ann Bot. 2000;85:741–750. doi: 10.1006/anbo.2000.1123. [DOI] [Google Scholar]
- Bilska-Kos A, Mytych J, Suski S, Magoń J, Ochodzki P, Zebrowski J. Sucrose phosphate synthase (SPS), sucrose synthase (SUS) and their products in the leaves of Miscanthus × giganteus and Zea mays at low temperature. Planta. 2020;252:23. doi: 10.1007/s00425-020-03421-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DM, Wang L, Ruan YL. Understanding and manipulating sucrose phloem loading, unloading, metabolism, and signalling to enhance crop yield and food security. J Exp Bot. 2014;65:1713–1735. doi: 10.1093/jxb/ert416. [DOI] [PubMed] [Google Scholar]
- Castellarin SD, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G. Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ. 2007;30:1381–1399. doi: 10.1111/J.1365-3040.2007.01716.X. [DOI] [PubMed] [Google Scholar]
- Castleden CK, Aoki N, Gillespie VJ, Macrae EA, Quick WP, Buchne P, Foyer CH, Furbank RT, Lunn JE. Evolution and function of the sucrose-phosphate synthase gene families in wheat and other grasses. Plant Physiol. 2004;135:1753–1764. doi: 10.1104/pp.104.042457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya DA, Saini R, Saini HS, Devi A. Sucrose metabolism: controls the sugar sensing and generation of signalling molecules in plants. J Pharmacogn Phytochem. 2017;6:1563–1572. [Google Scholar]
- Eom SH, Rim Y, Hyun TK. Genome-wide identification and evolutionary analysis of neutral/alkaline invertases in Brassica rapa. Biotechnol Biotec Eq. 2019;33:1158–1163. doi: 10.1080/13102818.2019.1643784. [DOI] [Google Scholar]
- Garchery C, Gest N, Do PT, Alhagdow M, Baldet P, Menard G, Rothan C, Massot C, Gautier H, Aarrouf J, Fernie AR, Stevens R. A diminution in ascorbate oxidase activity affects carbonal location and improves yield in tomato under water deficit. Plant Cell Environ. 2013;36:159–175. doi: 10.1111/j.1365-3040.2012.02564.x. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Reimholz R, Deiting U, Sonnewald U, Stitt M. Decreased expression of sucrose phosphate synthase strongly inhibits the water stress-induced synthesis of sucrose in growing potato tubers. Plant J. 1999;19:119–129. doi: 10.1046/j.1365-313X.1999.00506.x. [DOI] [PubMed] [Google Scholar]
- Godt DE, Roitsch T. Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiol. 1997;115:273–282. doi: 10.1104/pp.115.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot D, Kelly G, Stein O, David-Schwartz R. Substantial roles of hexokinase and fructokinase in the effects of sugars on plant physiology and development. J Exp Bot. 2014;65:809–819. doi: 10.1093/jxb/ert400. [DOI] [PubMed] [Google Scholar]
- Guo AY, Zhu QH, Chen X, Luo JC. GSDS a gene structure display server. Hereditas. 2007;29:1023–1026. doi: 10.1360/yc-007-1023. [DOI] [PubMed] [Google Scholar]
- Hernandez-Garcia CM, Finer JJ. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014;217–218:109–119. doi: 10.1016/j.plantsci.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Hirose T, Scofield GN, Terao T. An expression analysis profile for the entire sucrose synthase gene family in rice. Plant Sci. 2008;174:534–543. doi: 10.1016/j.plantsci.2008.02.009. [DOI] [Google Scholar]
- Hockema BR, Etxeberria E. Metabolic contributors to drought-enhanced accumulation of sugars and acids in oranges. J Am Soc Hortic Sci. 2001;126:599–605. doi: 10.21273/jashs.126.5.599. [DOI] [Google Scholar]
- Hou X, ZhangDuKangDavies WTSWJ. Responses of water accumulation and solute metabolism in tomato fruit to water scarcity and implications for main fruit quality variables. J Exp Bot. 2020;71:1249–1264. doi: 10.1093/jxb/erz526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun TK, Eom SH, Kim J. Genomic analysis and gene structure of the two invertase families in the domesticated apple (Malus × domestica Borkh) Plant Omics. 2011;4:391–399. [Google Scholar]
- Ji X, Van Den Ende W, Van Laere A, Cheng S, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. J Mol Evol. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Jia H, Wang Y, Sun M, Li B, Han Y, Zhao Y, Li X, Ding N, Li C, Ji W, Jia W. Sucrose functions as a signal involved in the regulation of strawberry fruit development and ripening. New Phytol. 2013;198:453–465. doi: 10.1111/NPH.12176. [DOI] [PubMed] [Google Scholar]
- Jia L, Zhang B, Mao C, Li J, Wu Y, Wu P, Wu Z. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.) Planta. 2008;228:51–59. doi: 10.1007/s00425-008-0718-0. [DOI] [PubMed] [Google Scholar]
- Jiang S, Chi Y, Wang J, Zhou J, Cheng Y, Zhang L, Ma A, Vanitha J, Ramachandran S. Sucrose metabolism gene families and their biological functions. Sci Rep. 2015;5:17583. doi: 10.1038/srep17583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juárez-Colunga S, López-González C, Morales-Elías NC, Massange-Sánchez JA, Trachsel S, Tiessen A. Genome-wide analysis of the invertase gene family from maize. Plant Mol Biol. 2018;97:385–406. doi: 10.1007/s11103-018-0746-5. [DOI] [PubMed] [Google Scholar]
- Kumar P, Rouphael Y, Cardarelli M, Cola G. Vegetable grafting as a tool to improve drought resistance and water use efficiency. Front Plant Sci. 2007;8:1130. doi: 10.3389/fpls.2017.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy K, Lammens W, Verhaest M, De Coninck B, Rabijns A, Van Laere A, Van Den Ende W. Unraveling the difference between invertases and fructan exohydrolases: A single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiol. 2007;145:616–625. doi: 10.1104/pp.107.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Meng FR, Zhang CY, Zhang N, Sun MS, Ren JP, Niu HB, Wang X, Yin J. Comparative analysis of water stress-responsive transcriptomes in drought-susceptible and -tolerant wheat (Triticum aestivum L.) J Plant Biol. 2012;55:349–360. doi: 10.1007/s12374-011-0032-4. [DOI] [Google Scholar]
- Liao WB, Li YY, Lu C, Peng M. Expression of sucrose metabolism and transport genes in cassava petiole abscission zones in response to water stress. Biol Plant. 2017;61:219–226. doi: 10.1007/s10535-016-0658-7. [DOI] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lunn JE, Macrae E. New complexities in the synthesis of sucrose. Curr Opin Plant Biol. 2003;6:208–214. doi: 10.1016/S1369-5266(03)00033-5. [DOI] [PubMed] [Google Scholar]
- Lutfiyya LL, Xu N, Ordine RLD, Morrell JA, Miller PW, Duff SMG. Phylogenetic and expression analysis of sucrose phosphate synthase isozymes in plants. J Plant Physiol. 2007;164:923–933. doi: 10.1016/j.jplph.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Ma QJ, Sun MH, Lu J, Zhu XP, Gao WS, Hao YJ. Ectopic expression of apple MdSUT2 gene influences development and abiotic stress resistance in tomato. Sci Hortic. 2017;220:259–266. doi: 10.1016/j.scienta.2017.04.013. [DOI] [Google Scholar]
- Meissner M, Orsini E, Ruschhaupt M, Melchinger AE, Hincha DK, Heyer AG. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation. Plant Cell Environ. 2013;36:1256–1267. doi: 10.1111/pce.12054. [DOI] [PubMed] [Google Scholar]
- Mou W, Li D, Luo Z, Mao L, Ying T. Transcriptomic analysis reveals possible influences of ABA on secondary metabolism of pigments, flavonoids and antioxidants in tomato fruit during ripening. PLoS ONE. 2015;10:e0129598. doi: 10.1371/journal.pone.0129598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Quoc B, Foyer CH. A role for “futile cycles” involving invertase and sucrose synthase in sucrose metabolism of tomato fruit. J Exp Bot. 2001;52:881–889. doi: 10.1093/jexbot/52.358.881. [DOI] [PubMed] [Google Scholar]
- Pagliuso D, Grandis A, Igarashi ES, Lam E, Buckeridge MS. Correlation of apiose levels and growth rates in duckweeds. Front Chem. 2018;6:291. doi: 10.3389/FCHEM.2018.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prioul JL, Pelleschi S, Séne M, Thévenot C, Causse M, De Vienne D, Leonardi A. From QTLs for enzyme activity to candidate genes in maize. J Exp Bot. 1999;50:1281–1288. doi: 10.1093/jxb/50.337.1281. [DOI] [Google Scholar]
- Qi X, Wu Z, Li J, Mo X, Wu S, Chu J, Wu P. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Mol Biol. 2007;64:575–587. doi: 10.1007/s11103-007-9177-4. [DOI] [PubMed] [Google Scholar]
- Ripoll J, Urban L, Brunel B, Bertin N. Water deficit effects on tomato quality depend on fruit developmental stage and genotype. J Plant Physiol. 2016;190:26–35. doi: 10.1016/j.jplph.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Ripoll J, Urban L, Staudt M, Lopez-Lauri F, Bidel LPR, Bertin N. Water shortage and quality of fleshy fruits - making the most of the unavoidable. J Exp Bot. 2014;65:4097–4117. doi: 10.1093/jxb/eru197. [DOI] [PubMed] [Google Scholar]
- Roitsch T, González MC. Function and regulation of plant invertases: sweet sensations. Trend Plant Sci. 2004;9:606–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ruan Y. Sucrose metabolism : gateway to diverse carbon use and sugar signaling. Ann Rev Plant Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- Ruan YL, Llewellyn DJ, Liu Q, Xu SM, Wu LM, Wang L, Furbank RT. Expression of sucrose synthase in the developing endosperm is essential for early seed development in cotton. Funct Plant Biol. 2008;35:382–393. doi: 10.1071/FP08017. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstruction of phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scholander PF, Hammel HT, Bradstreet ED, Hemmingsen EA. Sap pressure in vascular plants. Science. 1965;148:339–346. doi: 10.1126/SCIENCE.148.3668.339. [DOI] [PubMed] [Google Scholar]
- Seneweera SP, Basra AS, Barlow EW, Conroy JP. Diurnal regulation of leaf blade elongation in rice by CO2. Is it related to sucrose-phosphate synthase activity. Plant Physiol. 1995;108:1471–1477. doi: 10.1104/pp.108.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebeneichler TJ, Crizel RL, Camozatto GH, Paim BT, da Silva MR, Rombaldi CV, Galli V. The postharvest ripening of strawberry fruits induced by abscisic acid and sucrose differs from their in vivo ripening. Food Chem. 2020;317:126407. doi: 10.1016/j.foodchem.2020.126407. [DOI] [PubMed] [Google Scholar]
- Stein O, Granot D. Plant fructokinases: evolutionary, developmental, and metabolic aspects in sink tissues. Front Plant Sci. 2018;9:339. doi: 10.3389/fpls.2018.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhang J, Larue CT, Huber SC. Decrease in leaf sucrose synthesis leads to increased leaf starch turnover and decreased RuBP regeneration-limited photosynthesis but not Rubisco-limited photosynthesis in Arabidopsis null mutants of SPSA1. Plant Cell Environ. 2011;34:592–604. doi: 10.1111/j.1365-3040.2010.02265.x. [DOI] [PubMed] [Google Scholar]
- Taylor P, Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Crit Rev Plant Sci. 2000;1:31–67. doi: 10.1080/07352680091139178. [DOI] [PubMed] [Google Scholar]
- Terry LA, Chope GA, Bordonaba JG. Effect of water deficit irrigation and inoculation with Botrytis cinerea on strawberry (Fragaria × ananassa) fruit quality. J Agr Food Chem. 2007;55:10812–10819. doi: 10.1021/jf072101n. [DOI] [PubMed] [Google Scholar]
- The Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X-l, Wang Z-y, Ma B-q, Zhang C-x, Zhu L-c, Ma F-w, Li M-j. Structure and expression analysis of the sucrose synthase gene family in apple. J Integr Agr. 2018;17:847–856. doi: 10.1016/S2095-3119(17)61755-6. [DOI] [Google Scholar]
- Toroser D, Huber SC. Protein phosphorylation as a mechanism for osmotic-stress activation of sucrose-phosphate synthase in spinach leaves. Plant Physiol. 1997;114:947–955. doi: 10.1104/pp.114.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL. Differential expression of alkaline and neutral invertases in response to environmental stresses: characterization of an alkaline isoform as a stress-response enzyme in wheat leaves. Planta. 2007;226:1535–1545. doi: 10.1007/s00425-007-0590-3. [DOI] [PubMed] [Google Scholar]
- Wan H, Wu L, Yang Y, Zhou G, Ruan Y. Evolution of sucrose metabolism: the dichotomy of invertases and beyond. Trend Plant Sci. 2018;23:163–177. doi: 10.1016/j.tplants.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Wang L, Ruan YL. New insights into roles of cell wall invertase in early seed development revealed by comprehensive spatial and temporal expression patterns of GhCWIN1 in cotton. Plant Physiol. 2012;160:777–787. doi: 10.1104/pp.112.203893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu L, Wang Y, Tao H, Fan J, Zhao Z, Guo Y. Effects of soil water stress on fruit yield, quality and their relationship with sugar metabolism in ‘Gala’apple. Sci Hortic. 2019;258:108753. doi: 10.1016/j.scienta.2019.108753. [DOI] [Google Scholar]
- Wang Z, Wei P, Wu M, Xu Y, Li F, Luo Z, Zhang J, Chen A, Xie X, Cao P. Analysis of the sucrose synthase gene family in tobacco: structure, phylogeny, and expression patterns. Planta. 2015;242:153–166. doi: 10.1007/s00425-015-2297-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Chai S, Ru L, Pan L, Cheng Y, Ruan M, Ye Q, Wang R, Yao Z, Zhou G, Chen Y, Wan H. New insights into the evolution and expression dynamics of invertase gene family in Solanum lycopersicum. Plant Growth Regul. 2020;92:205–217. doi: 10.1007/s10725-020-00631-2. [DOI] [Google Scholar]
- Welham T, Pike J, Horst I, Flemetakis E, Katinakis P, Kaneko T, Sato S, Tabata S, Perry J, Parniske M, Wang TL. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J Exp Bot. 2009;60:3353–3365. doi: 10.1093/jxb/erp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang L, Le Roy K, Bolouri-Moghaddam MR, Vanhaecke M, Lammens W, Rolland F, Van Den Ende W. Exploring the neutral invertase-oxidative stress defence connection in Arabidopsis thaliana. J Exp Bot. 2011;62:3849–3862. doi: 10.1093/jxb/err069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Tang C, Fang Y, Yang M, Zhou B, Qi J. Structure and expression profile of the sucrose synthase gene family in the rubber tree: indicative of roles in stress response and sucrose utilization in the laticifers. FEBS J. 2014;281:291–305. doi: 10.1111/febs.12595. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Anjos L, Medeiros DB, Araújo WL, Fernie AR, Daloso DM. Insights into ABA-mediated regulation of guard cell primary metabolism revealed by systems biology approaches. Prog Biophys Mol Biol. 2019;146:37–49. doi: 10.1016/j.pbiomolbio.2018.11.006. [DOI] [PubMed] [Google Scholar]
- Zhang B, Tieman DM, Jiao C, Xu Y, Chen K, Fe Z, Giovannoni JJ, Klee HJ. Chilling-induced tomato flavor loss is associated with altered volatile synthesis and transient changes in DNA methylation. P Natl Acad Sci USA. 2016;113:12580–12585. doi: 10.1073/pnas.1613910113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Yu M, Ma R, Shen Z, Zhang B, Korir NK. Structure, expression profile, and evolution of the sucrose synthase gene family in peach (Prunus persica) Acta Physiol Plant. 2015;37:81. doi: 10.1007/s11738-015-1829-4. [DOI] [Google Scholar]
- Zhang Y, Zeng D, Liu Y, Zhu W. SlSPS, a sucrose phosphate synthase gene, mediates plant growth and thermotolerance in tomato. Horticulturae. 2022;8:491. doi: 10.3390/horticulturae8060491. [DOI] [Google Scholar]
- Zhu X, Wang M, Li X, Jiu S, Wang C, Fang J. Genome-wide analysis of the sucrose synthase gene family in grape (Vitis vinifera): structure, evolution, and expression profiles. Genes. 2017;8:111. doi: 10.3390/genes8040111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YJ, Komor E, Moore PH. Sucrose accumulation in the sugarcane stem is regulated by the difference between the activities of soluble acid invertase and sucrose phosphate synthase. Plant Physiol. 1997;115:609–616. doi: 10.1104/pp.115.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.