Abstract
Salt stress is one of the most critical abiotic stresses having significant contribution in global agriculture production. Chickpea is sensitive to salt stress at various growth stages and a better knowledge of salt tolerance in chickpea would enable breeding of salt tolerant varieties. During present investigation, in vitro screening of desi chickpea by continuous exposure of seeds to NaCl-containing medium was performed. NaCl was applied in the MS medium at the rate of 6.25, 12.50, 25, 50, 75, 100, and 125 mM. Different germination indices and growth indices of roots and shoots were recorded. Mean germination (%) of roots and shoots ranged from 52.08 to 100%, and 41.67–100%, respectively. The mean germination time (MGT) of roots and shoots ranged from 2.40 to 4.78 d and 3.23–7.05 d. The coefficient of variation of the germination time (CVt) was recorded as 20.91–53.43% for roots, and 14.53–44.17% for shoots. The mean germination rate (MR) of roots was better than shoots. The uncertainty (U) values were tabulated as 0.43–1.59 (roots) and 0.92–2.33 (shoots). The synchronization index (Z) reflected the negative impact of elevated salinity levels on both root and shoot emergence. Application of NaCl exerted a negative impact on all growth indices compared to control and decreased gradually with elevated NaCl concentration. Results on salt tolerance index (STI) also revealed the reduced STI with elevated NaCl concentration and STI of roots was less than shoot. Elemental analysis revealed more Na and Cl accumulation with respective elevated NaCl concentrations. The In vitro growth parameters and STI values validated and predicted by multilayer perceptron (MLP) model revealed the relatively high R2 values of all growth indices and STI. Findings of this study will be helpful to broaden the understanding about the salinity tolerance level of desi chickpea seeds under in vitro conditions using various germination indices and seedling growth indices.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-023-01282-z.
Keywords: Artificial neural network, Chickpea, Mathematical expressions, NaCl stress, Salt tolerance
Introduction
Plant growth, development, and production are largely influenced by several abiotic stresses like salinity, drought, and high or low temperature. These abiotic stresses influence more than 50% of plant production (Munns and Tester 2008). Among these, salinity is considered one of the main limiting factors to plant production affecting nearly 80 million hectares of agricultural lands worldwide (Yamaguchi and Blumwald 2005). The salt present in the soil solution lowers the availability of water to the roots (osmotic stress) and salt taken up by the plant also accumulate in various plant parts exceeding toxicity limits and ultimately contributing to reduced crop production (Munns et al. 1995). Legumes are considered an important pillar of our agriculture production system serving as a cheap source of high quality nutrition (Nadeem et al. 2021). However, legumes have been found highly sensitive to salt stress, especially during vegetative and reproductive stages (Nadeem et al. 2019).
Chickpea (Cicer auritinum L.) is one of the leading edible legumes serving as a vital source of food for millions of people all over the world (Kalefetoğlu et al. 2017), and is generally divided into two different types; known as Kabuli and Desi (Wang et al. 2010) based on their characteristics and phenotype. Kabuli chickpea has a smooth seed coat with a relatively larger size as compared to desi chickpea, which is not only smaller in size but also has a rough seed coat (Purushothaman et al. 2014; Aasim and Khan 2019). The cultivation of desi chickpea is mainly confined to a specific part of the world like the Indian subcontinent (Aydemir and Yemenicioğlu 2013) along with Ethiopia, Iran, Mexico, and Turkey (Ghribi et al. 2015; Mohammadi 2015). In the Indian subcontinent, desi chickpea is used for different purposes ranging from edible legumes to medicinal purposes (Kalefetoğlu et al. 2017; Aasim and Khan 2019). In Turkey, desi chickpea is cultivated in the Southeastern region for both animal fodder and human consumption (Aasim and Khan 2019). It is also a habitually cultivated crop in various other parts of the world and is considered a highly significant medicinal plant mainly due to the presence of dietary fibers and certain biochemical contents in its seed (Aharon et al. 2011; Jukanti et al. 2012).
Germination is a complex process and regulates the plant's establishment and development under variable conditions ranging from open field to controlled conditions under greenhouse or in vitro conditions. However, the process is complex, and different biological and environmental factor governs it (Bewley et al. 2012) by regulating the molecular mechanism. Optimum temperature, moisture contents, gases, light, salt type, concentration, etc. are the significant abiotic factors that alter or hinder the germination process (Kumar et al. 2013). Salinity is among one of the most challenging factors for farmers to germinate plants on salt-containing soils all over the World. The problem is continuously increasing due to ill practices like excessive use of fertilizers, brackish water, and excessive irrigation, especially in semi-arid and arid regions of the world. Low yield with more input enforces the farmers to adopt new technologies to overcome this issue. In recent years, different priming techniques have been employed to overcome the salinity issue by exposing the seeds to salinity under different cultural conditions to check the response of seeds.
Germination of seeds is the first step to examine the impact of salinity on seeds followed by the impact on seedlings. There are a lot of established parameters for the screening based on qualitative or quantitative measures. The germination ability and seed behavior during the germination process along with quantitative aspects of germination is highly significant in this aspect (Ranal and Santana 2006; Bewley et al. 2012). Quantitative parameters like germination time, rate, homogeneity, and synchronization of the whole germination process are considered to be highly significant. To date, several variable germination indices have been established to check the germination and viability of seeds under variable conditions ranging from natural to induced artificial environments. These germination indices are helpful for seed physiologists, seed technologists, and ecologists to better understand the germination process to make appropriate field plans from germination to harvesting.
Different germination measurement indices with different names and abbreviations make it difficult to regulate and understand the germination process (Ranal and Santana 2006). Researchers from different research areas established and employed the mathematical-based germination indices to understand the germination process and measurement from different angles and aspects (de Santana and Ranal 2004; Ranal and Santana 2006). Following germination, seedling establishment is the next step for the completion of a successful plant growth cycle. To understand germination and seedling establishment under laboratory conditions, in vitro plant tissue culture techniques can be employed successfully. It offers to germinate recalcitrant crops, endangered or seeds with high dormancy. It is also possible to check the efficacy of different stimulants or stressors like light, temperature, or stress conditions on agronomic characteristics more easily and efficiently (Phat et al. 2017).
Artificial intelligence (AI) and its subfield machine learning (ML) is the application of data science to solve complex problems in all scientific fields. However, the use of AI/ML approaches in plant and agricultural sciences is somewhat limited as compared to other scientific fields. It is still successfully employed in different disciplines of plant science like plant breeding, gene function and networks, (Mahood et al. 2020; van Dijk et al. 2021), in vitro germination (Hesami et al. 2021; Pepe et al. 2021a; Aasim et al. 2022c), and regeneration studies (Hesami et al. 2019, 2020a; Hesami and Jones 2020; Aasim et al. 2022a, c; Kirtis et al. 2022), in vitro mutagenesis (Mirza et al. 2022), plant identification (Grinblat et al. 2016), plant system biology (Hesami et al. 2022), pathogen identification (Mishra et al. 2019), nutrient deficiency (Rico-Chávez et al. 2022), and agrochemical applications (Soltis et al. 2020). Application of ML modeling for salt stress response is very circumscribed and these studies focused mainly on the plant stress phenotyping (Singh et al. 2016; Gao et al. 2020) and stress physiology (Jafari and Shahsavar 2020).
In vitro induced stress provides an alternative and efficient way of screening of shoots/plants/lines/genotypes etc. against different types of stressors in a relatively short time with more accuracy. However, understanding the impact of stress followed by making decision on the basis of scientific results is rather a challenging task. The problem can be solved by employing different models and algorithms to make considerably more precise and accurate prediction. Application of mathematical expressions and artificial intelligence-based models under in vitro conditions to understand the germination process is very limited. However, successful application of AI-based models have been employed to data prediction and validation of in vitro germination (Hesami et al. 2021; Aasim et al. 2022c) and in vitro growth indices of C. sativa (Aasim et al. 2022c) recently. There is an imperative need to investigate the impact of different stressors on germination and plant growth of plants via artificial intelligence tools. Keeping in view, the present study was designed to investigate the salinity tolerance level of chickpea seeds under in vitro conditions using various germination indices and seedling growth indices, followed by data validation and prediction through multilayer perceptron model.
Materials and methods
Seed sterilization
The chickpea seeds (desi) were procured from the Department of Field Crops, Faculty of Agriculture, Dicle University, Diyarbakir. Before sterilization, all damaged and non-uniform seeds were removed. Seeds were sterilized by using 5% w/v NaOCl for 15 min (Kirtiş and Aasim 2019; Kirtis and Aasim 2020). Sterilized seeds were placed on agar gelled medium augmented with different levels of NaCl.
In vitro culture conditions and medium preparation
The culture medium was comprised of Murashige and Skoog (MS) basal medium (Murashige and Skoog 1962). The basal medium was prepared according to the standard by using MS (0.44%), sucrose (3.0%), and agar (0.65%). NaCl at variable concentration (0, 6.25, 12.50, 25.00, 50.00, 75.00, 100.00 and 150.00 mM) were added in the culture medium prior to autoclave. The pH of the medium was automated to around 5.8 with the aid of 1N NaOH or HCl after adding NaCl. The medium was autoclaved for 20 min. at 121 °C (1.5 kg cm− 2 pressure). The culture medium was poured into culture vials having approximately 50 ml. The seeds were placed on the medium for seed germination and Magentas were placed in the growth room equipped with white LEDs (~1500 Lux) for 16/8 h light/dark photoperiod and a temperature of 24 ± 1 °C. All chemicals used in this study were procured from Duchefa Biochemie, The Netherland and Sigma-Aldrich, United States.
Germination indices
The seeds inoculated on NaCl supplemented medium was observed after 12 h and 24 h followed by an interval of 24 h for the next 8 days for data collection. Radicle emergence (approximately 2–3 mm long) was used as a standard for germination count. After tabulation of data, different germination indices like germination percentage (G), mean germination time (MGT or MT), coefficient of variation of the germination time (CVt), mean germination rate (MR), uncertainty (U), and synchronization index (Z) were calculated by using respective formulas and methodology (Ranal et al. 2009). Notably, the germination indices were computed for both root and shoot separately.
Mean germination time (MGT)
Mean germination time or mean emergence time (MET) or mean length of incubation time (MLIT) or mean days for germination (M-days) is used to calculate the average time length for maximum germination of a given seed lot (Ranal et al. 2009) and is represented via Eq. (1).
| 1 |
ni: number of newly germinated seeds on the ith day or the daily germination percentage at time (ti) from sowing,
ti: time from the start of the experiment to the ith day,
k: last day of observation.
Germination percentage (G)
Germination percentage or germination rate is used to calculate the time course of seed germination. Alternatively, it also helps to estimate the seed viability of a given population (Ranal et al. 2009) and is defined as Eq. (2).
| 2 |
ni: number of seeds newly germinated on the ith day,
k: last day of observation used,
N: total number of seeds per replicate (Magenta).
The coefficient of variation of the germination time (CVt)
The coefficient of variation of the germination time (CVt) is the measurement of uniformity or variability of germination in relation to the mean germination time (Ranal et al. 2009) and it can be written as Eq. (3).
| 3 |
st: the standard deviation of the germination time,
the mean germination time
ti: time between the start of the experiment,
ith: day,
ni: number of seeds germinated on the ith day,
k: the last day of observation.
CVt representation in percentage is given in Eq. (4).
| 4 |
Mean germination rate (MR)
Mean germination rate (MR) is the reciprocal of the mean germination time and it is presented as Eq. (5), where is the mean germination time (Ranal et al. 2009).
| 5 |
Uncertainty (U)
Uncertainty (U) is used to assess the degree of uncertainty linked with the distribution of the relative germination frequency and is presented in Eq. (6). Here, lower values of U express highly synchronized germination (Ranal et al. 2009).
| 6 |
fi: relative frequency of germination,
ni: number of seeds germinated on the ith day,
k: the last day of observation.
Synchronization index (Z)
The synchronization index (Z) was originally developed to estimate the degree of overlapping of flowering among individuals in a population. Later, it was also used as germination indices and the value of Z = 1 expresses the germination of entire seeds at the same time and Z = 0 expresses the germination of at least two seeds with one at each time. More importantly, Z only produces a non-zero value if and only if two seeds complete the seed germination process at the same time (Ranal et al. 2009). Its formula is expressed via Eq. (7).
| 7 |
Combination of seeds germinated in the ith time, two by two,
ni: number of seeds germinated on the ith day.
In vitro seedling growth indices
After the successful establishment of in vitro seedlings on NaCl containing medium, different growth indices like shoot length, root length, fresh shoot wt, dry shoot wt, fresh root wt, and dry root wt were tabulated after three weeks of in vitro culture. Randomly selected 10 plants/replicate were selected for the estimation of variable growth indices. The shoot and root length of all selected samples were measured by using a measuring scale. Thereafter, they were weighed on a sensitive electric weighing balance for the estimation of fresh shoot or root weight. For the estimation of dry wt, both shoot and root samples (weighed) were placed and wrapped in aluminum foil followed by placing them in an oven at 65 °C (Anawar et al. 2011) for a period of 72 h and weighed thereafter. All the data presented as growth indices are based on a per plant by dividing the data with the number of shoots.
Element analysis
Element analysis (K+, Na+, Cl−) was performed at Agriculture Ankara, Turkey. Shoots and root samples were oven-dried at 65 °C for 5 days followed by grinding. 0.25 g oven-dried samples were subjected to the mixture of Nitric Acid (HNO3) and Throcoles (HCLO4). Standard solutions were prepared for both sodium and potassium and the flame photometer was calibrated accordingly. "Standard curve” was created based on the recorded readings (Kacar and Inal 2008). For Cl− ion analysis, 0.1 g oven-dried samples were used. Potassium chromate indicator and AgNO3 were used for taking the values. The Cl− contents were used by protocol and formula reported (Kacar and Inal 2008).
Salt tolerance index (STI)
Salt tolerance index (STI) is the estimation of the dry wt of salinity treated plants to control plants and can be calculated by the formula given below (Karakullukçu 2008).
| 8 |
The STI value of roots and shoots were estimated separately to check their respective tolerance level.
Multilayer perceptron analysis
NaCl concentrations were used as input variables, and growth parameters along with STI were used as output variables for ANN based MLP model (Silva et al. 2019). The data was splitted into training and testing sets by using leave-one-out cross-validation (LOO-CV) technique (Webb et al. 2011). For hyperparameter optimization, grid search method was employed. The open-source Python programming language (Van Rossum and Drake 2009) was used for coding with the aid of sklearn library (Pedregosa et al. 2011). The coefficient of determination (R2), mean square error (MSE), mean absolute error (MAE), and mean absolute percentage error (MAPE) were calculated to validate the performance of the models. The R2 estimates the strength of the relationship between the model and the dependent variables. The MSE exhibits that how close a regression line is to the measured data points. MAE specifies the average magnitude of the differences between the prediction of an observation and its actual value. Whereas MAPE is a prediction accuracy of forecasting system. The mathematical representation of the above-mentioned performance metrics is given in Eqs. 9–12.
| 9 |
| 10 |
| 11 |
| 12 |
where = actual value, = predicted value, = mean of the actual values and n = sample count. Moreover, all features (inputs) were standardized to scale the values to be centered on the mean with a unit standard deviation by using the below formula before training and testing the models.
| 13 |
standardized value. actual data. mean of the feature values. standard deviation of the featured values.
Statistical analysis
The experiment was designed with four replicates. The SPSS 26 statistical program for Windows (SPSS Inc. Chicago, IL, USA) was used for the univariate statistical analysis (ANOVA, standard error) for both germination indices and growth indices. Comparison of means was performed by applying Duncan’s Multiple Range Test (DMRT) at p < 0.05 or p < 0.01 level of significance. The arrangement of data before ANOVA and comparison of means were done according to arcsine (√X) transformation (Snedecor and Cochran 1967). The Pearson correlation coefficient was performed through XLSTAT statistical software (www.xlstat.com).
Results
In vitro germination indices
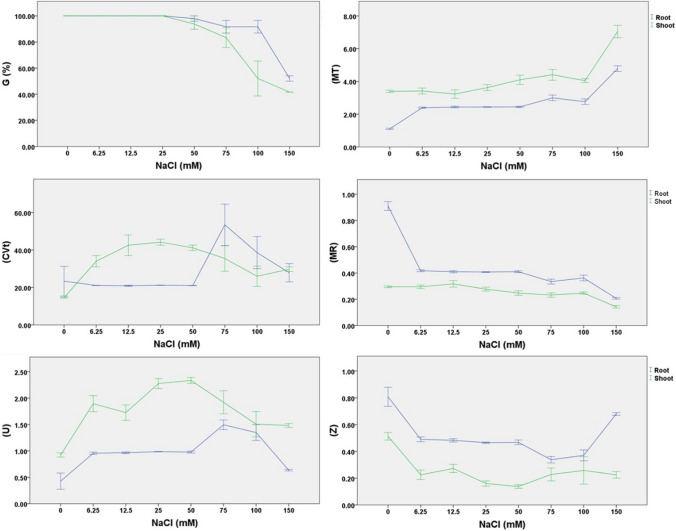
The impact of different NaCl concentrations on the germination of chickpea was studied by using different mathematical expressions to understand the whole germination process. Results revealed the more rapid germination from the control group compared to the medium enriched with NaCl. It was observed that exposing seeds to NaCl exerted stress on both root and shoot emergence followed by their respective growth. Therefore, mathematical expressions of germination indices of both shoot and root were performed individually and are presented in Fig. 1. Mean germination of roots and shoots ranged from 52.08 to 100% and 41.67–100%, respectively (Fig. 1, Table S1). Mean germination of chickpea seeds revealed a direct association with NaCl concentrations (p < 0.01). Almost similar germination patterns were attributed for both shoots and root emergence in response to NaCl concentration. In general, elevated NaCl concentration resulted in delayed germination, which in turn affected the other growth indices. However, the delayed emergence was more prominent for shoots as compared to roots especially at higher salinity levels of 100 and 150 mM NaCl. Exposing seeds to 100 mM NaCl resulted in mean germination of 52.08% (shoots) and 91.67% (root). Whereas the provision of 150 mM NaCl was detrimental for both root and shoot emergence and recorded 52.08% and 41.67%, respectively (Fig. 1, Table S1). The mean germination time of chickpea seeds exhibited a negative impact of NaCl concentration with delayed root and shoot emergence as compared to control. The MGT for roots exposed to different NaCl concentrations ranged from 2.40 to 4.78 d which was higher than the control (1.10 d). MGT for shoots ranged from 3.23 to 7.05 d when exposed to different NaCl concentrations. The MGT for shoots exposed to 6.25–25 mM NaCl concentration was statistically similar to control shoots (3.40 d). The highest MGT for both roots (4.78 d) and shoots (7.05 d) were recorded on a medium supplemented with 150 mM NaCl exhibited the negative impact of salinity level on the MGT of chickpea seeds (Fig. 1, Table S1).
Fig. 1.
An overview of the impact of different NaCl concentrations on in vitro germination indices of chickpea
Provision of NaCl concentrations exhibited variable CVt of roots and ranged from 20.91 to 53.43%. The highest CVt (roots) was recorded from a medium supplemented with 75 mM NaCl (53.43%) followed by 100 mM NaCl (38.60%). Supplementation of 6.25 to 50 and 150 mM NaCl resulted in the least and statistically similar CVt values. On the other hand, CVt of shoots ranged from 14.53 to 44.17% with the highest CVt of shoots (44.17%) being attributed to a 25 mM NaCl-containing medium. The CVt of roots and shoots were different from each other and response was associated with the salinity level (NaCl concentration) of the culture medium. The CVt (%) of roots was more than CVt of shoots on the medium supplemented with 0, 75, and 100 mM NaCl. Whereas CVt (%) of shoots was almost double than CVt (%) of roots on medium supplemented with 6.25 – 50 mM NaCl (Fig. 1, Table S1). Supplementation of NaCl in the culture medium significantly affected the MR values of both roots and shoot emergence. The highest MR values of roots were recorded in the control group (0.909) and exposure of seeds to different salinity levels resulted in 2 to fourfold less MR than in the control (0.210–0.418) and decreased elevated salinity level (Fig. 1, Table S1). The MR values of shoots were less than roots and ranged from 0.143 to 0.316. The highest MR value of shoots was recorded on a medium supplemented with 12.5 mM NaCl but statistically similar to control, 6.25, and 25 mM NaCl. Further increase in salinity level was more detrimental and resulted in reduced MR which showed the negative impact of salinity on mean germination rate. The least MR value of roots (0.210) and shoots (0.143) were recorded on a medium supplemented with 150 mM NaCl.
The uncertainty (U) values of roots and shoots ranged from 0.43 to 1.59 and 0.92–2.33. The lowest U values of both roots and shoots were attributed to the control group which shows more synchronized germination. The uncertainty (U) values of roots and shoots ranged from 0.43 to 1.59 and 0.92–2.33. Results further revealed that U values of roots were relatively less than shoots and it reflected more salt tolerance in roots as compared to shoots. The U values of roots on medium supplemented with 6.25–50 mM NaCl were statistically similar to each other and resulted in more synchronized root emergence and can be confirmed by synchronized MT and MR values. Further increase of NaCl resulted in elevated U values with a highest U value of 1.50 from medium supplemented with 75 mM NaCl. The U values of shoots increased with elevated NaCl concentration and highest U value (2.33) were recorded on a medium supplemented with 50 mM NaCl (Fig. 1, Table S1). Results on the synchronization index reflected the negative impact of elevated salinity levels on both root and shoot emergence of chickpea. The highest Z value of both roots and shoots were recorded for control which reflects the more synchronization during the germination process and can be confirmed by low U values. The Z values of roots declined with elevated salinity levels up to 100 mM NaCl followed by a significant increase of Z value on medium supplemented with 150 mM NaCl. This might be due to the release of initial shock caused by high salinity level and development of stress mechanism system which enables the radicle to tolerate and resulted in high Z value and can be confirmed by low U values of roots. On the other hand, Z values of shoots were 2–fourfold less than the control group, but these all were statistically significant with each other and ranged from 0.14 to 0.26 compared to control (0.51) (Fig. 1, Table S1).
In vitro seedling growth traits
Application of NaCl in the culture medium affected the various growth indices in a similar fashion of less than control and exhibited decreased growth with elevated NaCl concentration. Shoot numbers and root numbers ranged from 1.0 to 2.50 and 1.0–16.75, respectively. In the case of shoot length and root length, highest values were observed in control followed by low concentrations of NaCl-containing medium. The incorporation of NaCl up to 25 mM did not pose any negative impact on shoot length. However, further increase of NaCl concentration was detrimental and resulted in decreased shoot length. On the other hand, the application of 50 mM NaCl and more was highly detrimental and resulted in low root length. In the case of shoot length: root length ratio, lowest values were observed at 12.5 mM, while the highest was observed at 100 mM (Table 1).
Table 1.
Impact of different NaCl (mM) concentrations on plant growth of in vitro regenerated plantlets of desi chickpea
| NaCl (mM) | Shoot numbers | Root numbers | Shoot length (cm) | Root length (cm) |
|---|---|---|---|---|
| 0 | 2.50 ± 0.18a | 16.75 ± 0.95a | 9.24 ± 0.18a | 11.21 ± 1.46a |
| 6.25 | 2.21 ± 0.63ab | 15.69 ± 1.90a | 8.29 ± 0.55a | 8.50 ± 1.46ab |
| 12.5 | 2.31 ± 0.16a | 16.00 ± 2.01a | 8.23 ± 0.25a | 10.87 ± 1.38a |
| 25 | 2.00 ± 0.00ab | 10.63 ± 2.21bc | 8.48 ± 0.69a | 7.06 ± 1.33b |
| 50 | 1.88 ± 0.13ab | 12.40 ± 1.45ab | 6.27 ± 0.46b | 3.39 ± 0.27c |
| 75 | 1.50 ± 0.10bc | 7.75 ± 1.04c | 5.83 ± 0.62b | 2.36 ± 0.25c |
| 100 | 1.06 ± 0.06c | 7.21 ± 0.76c | 6.23 ± 0.79b | 2.25 ± 0.10c |
| 150 | 1.00 ± 0.00c | 1.00 ± 0.00d | 3.66 ± 0.52c | 1.96 ± 0.19c |
| NaCl (mM) | Fresh shoot wt (g) | Dry shoot wt (g) | Fresh root wt (g) | Dry root wt (g) |
|---|---|---|---|---|
| 0 | 0.296 ± 0.026a | 0.028 ± 0.001a | 0.329 ± 0.012a | 0.027 ± 0.001a |
| 6.25 | 0.233 ± 0.017b | 0.026 ± 0.002ab | 0.144 ± 0.018bc | 0.017 ± 0.002bc |
| 12.5 | 0.237 ± 0.019b | 0.026 ± 0.001ab | 0.179 ± 0.023b | 0.021 ± 0.002b |
| 25 | 0.211 ± 0.019bc | 0.024 ± 0.002ab | 0.157 ± 0.039b | 0.013 ± 0.003 cd |
| 50 | 0.179 ± 0.023 cd | 0.023 ± 0.001b | 0.074 ± 0.016de | 0.010 ± 0.001de |
| 75 | 0.151 ± 0.012de | 0.018 ± 0.001c | 0.115 ± 0.046cde | 0.008 ± 0.001ef |
| 100 | 0.113 ± 0.011ef | 0.015 ± 0.002c | 0.047 ± 0.008de | 0.005 ± 0.001ef |
| 150 | 0.063 ± 0.011f | 0.008 ± 0.001d | 0.018 ± 0.002e | 0.003 ± 0.00f |
Means followed by different small letters within columns are significantly different (p < 0.01)
Results on fresh wt and dry wt of shoot and root showed a similar pattern of high values on control. Application of different NaCl concentrations exerted a negative impact on all growth indices and gradually decreased with elevated NaCl concentration. The values were recorded as 0.063–0.296 g (fresh shoot wt), 0.018–0.0329 g (fresh root wt), 0.008–0.028 g (dry shoot wt), and 0.003–0.027 g (dry root wt). Results further revealed that NaCl exerted a more negative impact on root fresh wt and dry wt compared to fresh and dry wt of shoots. Overall, highest fresh shoot weight was observed in the control. The same pattern was observed for dry shoot weight, fresh root weight, and dry root weight as highest values for these traits were observed with an increase in NaCl dose.
Correlation analysis
During this study, a highly significant, positive, and very strong correlation was observed among the studied traits. Dry shoot wt reflected a highly significant, positive, and very strong correlation (r = 0.878*) with dry root weight (Table 2). Most of the traits reflected a highly significant and positive correlation except fresh root wt, which reflected a non-significant correlation with root numbers and dry shoot wt. A strong and positive correlation among studied traits showed that these traits can be used as a trustable criterion in the selection of salt stress-tolerant chickpea genotypes.
Table 2.
An overview of Correlation analysis in vitro seedling growth traits of desi chickpea
| Variables | Shoot numbers | Root numbers | Shoot length (cm) | Root length (cm) | Fresh shoot wt (g) | Dry shoot wt (g) | Fresh root wt (g) | Dry root wt (g) |
|---|---|---|---|---|---|---|---|---|
| Shoot numbers | 1 | 0.943 | 0.893 | 0.914 | 0.976 | 0.962 | 0.851 | 0.942 |
| Root numbers | 1 | 0.902 | 0.856 | 0.950 | 0.973 | 0.773 | 0.899 | |
| Shoot length (cm) | 1 | 0.881 | 0.947 | 0.936 | 0.844 | 0.880 | ||
| Root length (cm) | 1 | 0.911 | 0.835 | 0.863 | 0.962 | |||
| Fresh shoot wt (g) | 1 | 0.968 | 0.916 | 0.961 | ||||
| Dry shoot wt (g) | 1 | 0.795 | 0.878 | |||||
| Fresh root wt (g) | 1 | 0.943 | ||||||
| Dry root wt (g) | 1 |
Salt tolerance index (STI)
Results on STI illustrated the clear impact of NaCl concentration and plant part. Relatively higher STI values were attributed to roots compared to shoots. The decline in STI (shoots) was relatively slow on mediums supplemented with 6.25–50 mM NaCl followed by a sharp decline on mediums containing 100 mM and above NaCl. The lowest STI (shoots) was recorded (28.57) on a medium supplemented with 150 mM NaCl which was almost half of 100 mM NaCl (53.57). On the contrary, STI for roots behaved variably and the highest STI (roots) was recorded on a medium containing 12.5 mM NaCl. Thereafter, gradually decreased STI (roots) were attributed to elevated NaCl and the lowest (11.11) was recorded on a medium containing 150 mM NaCl (Table 3). On the other hand, decreased STI was attributed to elevated NaCl concentration for both plant parts. This section may be divided into subheadings.
Table 3.
Salt tolerance index (STI) analysis of in vitro seedling of desi chickpea
| NaCl (mM) | Shoot | Root | ||
|---|---|---|---|---|
| Actual | Predicted | Actual | Predicted | |
| 6.25 | 92.86 | 100.00 | 62.96 | 63.51 |
| 12.5 | 92.86 | 93.83 | 77.78 | 62.09 |
| 25.0 | 85.71 | 80.85 | 48.15 | 53.55 |
| 50.0 | 82.14 | 76.99 | 37.04 | 43.46 |
| 75.0 | 64.29 | 65.35 | 29.63 | 22.05 |
| 100.0 | 53.57 | 51.58 | 18.52 | 16.16 |
| 150.0 | 28.57 | 40.45 | 11.11 | 8.08 |
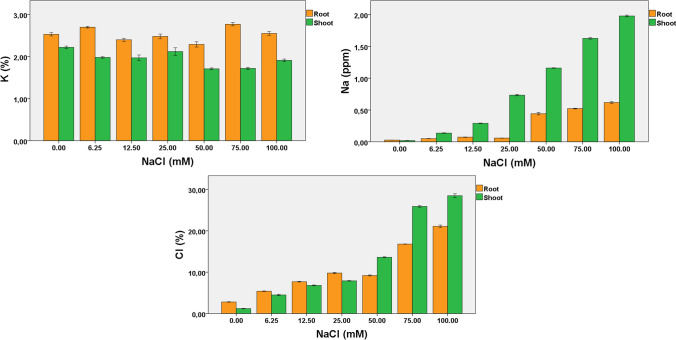
Estimation of Na+, K+, and Cl− in root and shoot
The results obtained revealed the variable concentration of K+ (%) in both shoots and roots and ranged from 2.29 to 2.77 (shoots) and 1.91–2.22 (roots). Highest K+ (%) in shoots (2.77%) and roots (2.22%) were recorded on a medium supplemented with 75 mM NaCl and control, respectively (Fig. 2, Table S2). Na+ contents in both shoots and roots were linked with their respective salinity concentration. Relatively low Na+ contents were recorded on medium supplemented with 6.25–50 mM followed by an exponential increase of Na+ contents in both shoots and roots on medium supplemented with 50–100 mM NaCl (Fig. 2, Table S2). A comparison of NaCl concentration revealed more Na+ contents in shoots compared to roots. The results revealed that both shoots and roots exhibited high K+ uptake with low Na+ contents on medium supplemented with 6.25–25 mM NaCl. Likewise, Na+, Cl− contents in both shoots and roots varied with NaCl concentration, and low Cl− contents in shoots were recorded on a medium supplemented with 0–12.5 mM NaCl (Fig. 2, Table S2). Thereafter, more Cl− contents were attributed to roots compared to shoots on all NaCl concentrations. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
Fig. 2.
Impact of different NaCl concentrations on element analysis of chickpea
Artificial neural network modeling
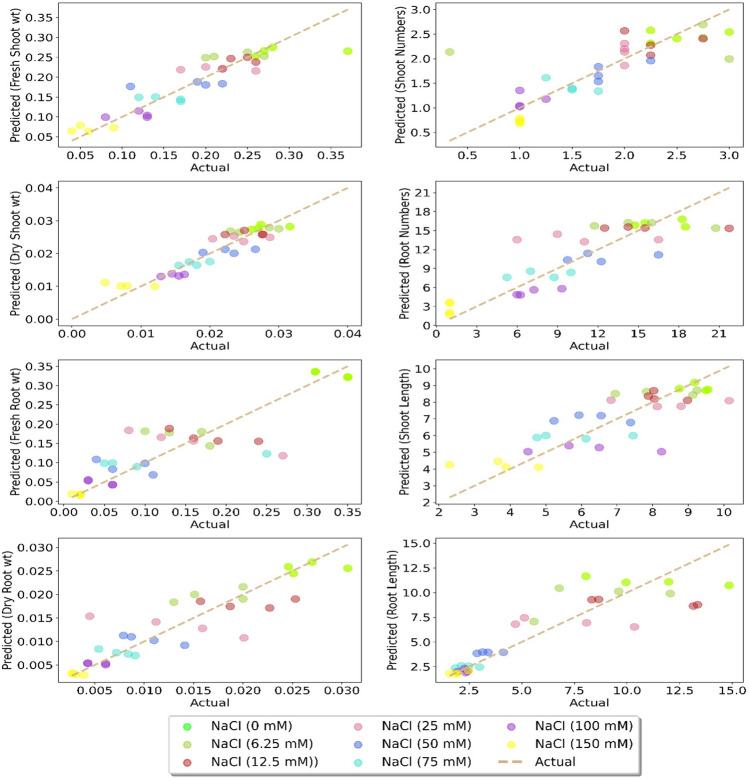
The results attained regarding in vitro growth parameters and STI were subjected to ANN analysis using MLP algorithm, and results were validated and predicted by employing four different performance metrics. Results regarding R2 were recorded as 0.5749 (shoot number), 0.718 (root number), 0.6742 (shoot length), 0.7662 (root length), 0.8193 (fresh shoot wt), 0.8530 (dry shoot wt), 0.7262 (root fresh wt) and 0.7886 (dry root wt). The MSE values were generally recorded low except root numbers (9.1091) and root length (3.8797). The similar trend was also observed for MAE values for all growth parameters. On the other hand, relatively high MAPE values were recorded for all growth parameters and ranged between 12.69–38.37%. The highest MAPE value was recorded for fresh root wt followed by shoot numbers. Whereas minimum MAPE values were attributed for dry shoot wt and shoot numbers, respectively (Table 4). The distribution of actual and predicted values of all growth parameters is presented in Fig. 3. The ANN models were also employed for the STI for both shoot and root (Table 4) followed by comparing actual and predicted values (Table 3). Results revealed the relatively high R2 values for STI of both shoot (0.9272) and root (0.8865). The MAE and MAPE values (Table 4) were also high but still fall in the satisfactory level. However, relatively high MSE values were recorded for both outputs (35.850 for shoot and 55.5808 for root) which reflected in the difference among actual and predicted values of STI. Results on STI of shoots revealed the variable difference among actual and predicted values at different NaCl concentration. The major difference between actual and predicted values were recorded at 150 mM NaCl followed by 6.25 mM NaCl and 50.0 mM NaCl, respectively. Whereas relatively minor difference among actual and predicted values were observed at other concentrations of NaCl. On the other hand, the minor difference between mM NaCl for STI of root were recorded for 6.25 mM NaCl followed by 100 mM NaCl and 150 mM NaCl, respectively (Table 3). All other NaCl concentrations (12.5–75.0 mM NaCl) yielded major difference between the actual and predicted values.
Table 4.
Performance metrics for the MLP model
| R2 | MSE | MAE | MAPE (%) | |
|---|---|---|---|---|
| Shoot numbers | 0.5749 | 0.1956 | 0.2470 | 28.65 |
| Root numbers | 0.7184 | 9.1091 | 2.2146 | 20.71 |
| Shoot length | 0.6742 | 1.2676 | 0.8698 | 13.56 |
| Root length | 0.7662 | 3.8797 | 1.3513 | 20.43 |
| Fresh shoot wt | 0.8193 | 0.0011 | 0.0243 | 16.29 |
| Dry shoot wt | 0.8530 | 1e−05 | 0.0022 | 12.69 |
| Fresh root wt | 0.7262 | 0.0028 | 0.0379 | 38.37 |
| Dry root wt | 0.7886 | 1e−05 | 0.0026 | 26.96 |
| STI (Shoot) | 0.9272 | 35.8150 | 4.6329 | 9.31 |
| STI (Root) | 0.8865 | 55.5808 | 5.4736 | 12.28 |
Fig. 3.
Distribution of predicted values of different growth parameters of chickpea
Discussion
Salinity caused by different salts in water or soil is one of the most threatening abiotic factors which affects the plant's metabolic processes, and enzymatic activities and ultimately affects the plant growth and productivity. The inhibitory impact varies with genotype and salinity level (natural or artificially induced) and the first and immediate impact is on plant germination followed by respective plant growth (Mungala et al. 2008). Screening of genotypes or cultivars are highly significant, and in vitro screening involves artificially induced salinity by incorporating different salts at variable concentration (Day et al. 2016; Day and Aasim 2017). Therefore, seeds were inoculated on the saline medium to investigate the in vitro germination indices, in vitro growth, indices salt tolerance index (STI), and mineral ion contents. Overall, NaCl exerted a significant impact on in vitro plant growth and development.
The most common method used for checking the seed vigor and viability is the estimation of the mean germination percentage in response to any external stimulant or exposing the seed to any stressor. NaCl is one of the most widely used stressors for checking the salinity tolerant level on germination under variable culture conditions ranging from field conditions to in vitro conditions (Day and Aasim 2017). However, the salinity tolerant level is dependent mainly on genotype (Atieno et al. 2017). The study carried out on cowpea cultivars revealed the reduced or total inhibition of germination on a medium enriched with a high NaCl level of 150 mM (Cokkizgin 2012). The results further revealed that further provision of NaCl concentration in the culture medium may be more lethal with less root and shoot emergence. In vitro study on common bean resulted in 100% germination on medium supplemented with 150 mM NaCl followed by the sharp decline of germination on medium supplemented with 200 mM NaCl (40%) and 250 mM (10%) and completed inhibition on 300 mM NaCl (Mena et al. 2015). High germination percentage of common beans on medium supplemented with 150 mM and 200 mM NaCl have also been registered (Thiam et al. 2013). The variable response of different plants to salinity conditions is dependent on other factors like cultivar and genetic variability (Kouam et al. 2017). Results showed that both roots and shoot emergence exhibited the same pattern of delayed emergence with elevated NaCl concentration. possibly due to low seed hydration at higher salinity levels (Asmare and Ambo 2013; Mena et al. 2015).
The weighted mean of germination depending on germination rate in response to time (Bewley et al. 2012) is known as mean germination time (MGT) and is another important mathematical expression for germination. It is a reciprocal of germination rate and is successfully used for different types of plants (Demir et al. 2008; Khajeh-Hosseini et al. 2009). Results obtained reflected the negative impact of elevated NaCl concentration on MGT values which resulted in delayed germination. The delayed GMT at a higher salinity level might be due to the delayed germination process and reduced speed possibly due to enzymes and hormones alterations in the seed in response to a high salinity level (Botía et al. 1998; Cokkizgin 2012). The other possible reason is the elevated osmotic potential in the culture medium, which affects the seed hydration followed by inhibition of the radicle emergence and subsequent seedling emergence (Gill et al. 2003).
Uniform germination is the prerequisite for the successful establishment of plants/crops for their life cycle. Different internal or external factors regulate the germination process, and any type of variation or delayed germination may lead to variable plant growth and ultimately low crop yield. The uniform or non-uniform germination of seeds in association with MGT (Cruz et al. 2001; Dorneles et al. 2005) can be checked by the coefficient of variation of the germination time (CVt). The low CVt value reflects the more uniform seed germination at a specific time and vice-versa. The results revealed the variable response of root and shoots on Cvt of chickpea seeds. The response of salinity in vitro and in vivo may differ due to variable culture conditions. Although a lot of studies reported the salinity impact of NaCl on in vitro germination indices, the impact of NaCl concentration on Cvt is very limited. However, work done under field conditions or greenhouse conditions highlights the impact of salinity level on Cvt values of a given seed lot. The highest Cvt values at 100 mM NaCl followed by a declining pattern on 200 and 300 mM NaCl of Elymus farctu seeds have been documented (Carvalho et al. 2005). Contrarily, the insignificant impact of NaCl concentrations on CVt of sea fennel seeds (El-Katony et al. 2015) has also been registered. Mean germination rate (MR) is the reciprocal of mean germination time and it generally expresses the seed vigor and germination speed of a given seed lot. The value of MR falls from 0 to 1 (0 < v ≤ 1 day-1) with MR values of 1 or near to 1 reflecting more vigorous seeds with high germination speed in a given time and vice- versa (Ranal and Santana 2006). Results illustrated the impact of NaCl concentration and plant part on MR values. Relatively more MR values were attracted to roots compared to shoots, which reflects the more salinity tolerance of roots compared to shoots.
The germination process in seeds is generally regulated by variable internal (seeds), external (environmental) factors (Nimac et al. 2018), and induced factor-like seeds scarification (Regnier 2020), which may result in non-uniform germination. This asynchronization of germination reflects the certain uncertainty of germination, and its quantification can be linked between uncertainty and relative germination frequency. The values of uncertainty (U) are 0 ≤ U ≤ , and a value of “0” or close to “0” presents more synchronized germination and vice-versa (Bewley et al. 2012). A single seed in a given seed lot is highly significant, and may lead to uncertainty (U). On the other hand, the overlapping of germination of two seeds simultaneously is known as the synchronization index (Z) and expresses as 0 ≤ Z ≤ 1 (Bewley et al. 2012). The Z values are contrary to the U values and Z value of 1 or near to 1 reflects more synchronized germination and Z value of “0” means no germination and values near to “0” mean asynchronized germination of a given seed lot. Results revealed the significant impact of induced stress on U values compared to the control group. A study on Cassia ferruginea also revealed the high U values due to mechanical scarification of seeds (Regnier 2020). Results further revealed that an increase in NaCl resulted in decreased U values. These results suggested the recovery of shoots from the initial shock of high salinity level followed by root and shoot development. However, the negative impact of high salinity levels was prominent on root and shoot growth and development. Interestingly, the U values on medium supplemented with 150 mM NaCl was less than all other NaCl concentration. Uncertainty of germination is a common feature that can be affected by variable induced factors (Regnier 2020) or uncontrolled factors like temperature (Nimac et al. 2018). On the other hand, the negative impact of salinity level on the synchronization index of shoots which was relatively less than roots on all salinity levels was also registered.
The impact of various NaCl concentrations was checked on various growth indices and a similar pattern was observed for the shoot and root numbers which were less than control and gradually decreased with elevated NaCl concentration in the culture medium. The incorporation of the highest NaCl in the culture medium was most detrimental and resulted in a single shoot and root. The negative impact of elevated NaCl concentrations on shoot numbers is a common feature of salinity in plants (Bahmani et al. 2012). Stunting shoots and roots of cowpea attained on various doses of NaCl confirmed the findings of this study (Cokkizgin 2012). Similarly, the negative impact of elevated NaCl concentration on different growth variables has been documented for different plants like lentils (Ashraf and Waheed 1993), wheat (Akbarimoghaddam et al. 2011), and Sorghum bicolor (El Naim et al. 2012). However, exposure time is also another significant factor, which controlled and affects the different seedling growth traits (indices) (Akbarimoghaddam et al. 2011; Yousef et al. 2020). Another important observation was the multiple shoot induction from the seed (Kendir et al. 2009; Kirtis et al. 2022) and shoot count depicted the negative impact and showed decreased pattern with elevated concentration of NaCl.
Correlation analysis is the most used analysis for the determination of the magnitude of association between two or more traits and can be used as a selection index. Therefore, breeders give much importance to correlation analysis while selecting genotypes. When two traits are correlated significantly, the selection of one trait will exert variations in its mean through additive gene effects and reflect an indirect effect in its correlated trait (Mudasir et al. 2012) Whereas the phenomenon of correlation was concluded as the correlation between traits and their genetic linkage or epistatic effects among various genes (Özer et al. 2010). Correlation analysis also showed a highly significant and positive correlation between the traits. Similarly, a highly significant, positive, and very strong correlation between dry shoot weight with dry root weight in chickpeas has also been documented (Zawude and Shanko 2017; Yousef et al. 2020). Moreover, these traits also showed significant association with each other. Therefore, breeders should use these traits as a selection criterion for the evaluation of salt-tolerant chickpea genotypes.
The salt tolerance index reflects the tolerance potential of a given species under natural or artificially induced salinity conditions. It is the ratio between NaCl-treated plants with control plants (Wu et al. 2019). Both in vitro germinated shoots and roots were considered for the estimation of STI of chickpeas (Karakullukçu 2008). According to the results, the STI of shoots was relatively higher than the roots on all mediums containing NaCl. The possible reason for lower STI values for roots is the direct contact of roots to the culture medium supplemented with NaCl concentration and relatively low root dry wt compared to shoots. Comparing NaCl concentration, STI decreased with elevated NaCl concentration for both shoots and roots. A similar impact has already been reported in tomatoes when exposed to different NaCl concentrations under in vitro conditions (Zaki and Yokoi 2016). These results reflected that STI values can vary with NaCl concentration and plant part. Previous studies also emphasized the role of different factors like explant type (Mungala et al. 2008), NaCl concentration (Day and Aasim 2017), and genotype (Wu et al. 2019) on STI for different crops.
Estimation of different elements like Na+, K+, and Cl− in plant parts are helpful for salinity tolerance of that given plant and both root and shoot can be used for estimating the salinity impact on nutrient uptake and plant growth (Wei-hua et al. 2013; Muchate et al. 2019). The K+ uptake at a higher salinity level induced by NaCl is the desired character for a given trait (Yeo and Flowers 1989) and generally, low K+ concentration at a higher salinity level is a common feature (Wu et al. 2019). Comparing plant parts, K (%) was relatively low in shoots compared to roots on all mediums, and previous studies also revealed the variable K+ contents in different plant parts (Zaki and Yokoi 2016; Wu et al. 2019). On the contrary, previous studies revealed elevated K+ contents in leaves with elevated NaCl contents (Atieno et al. 2017). Similarly, variable Na+ contents were attributed to plant part and NaCl concentration and increased Na+ contents with elevated NaCl concentration for both shoots and roots (Zaki and Yokoi 2016; Al-Khateeb et al. 2020). The relationship between Na+ and K+ is highly important and low Na+ contents and high K contents exhibit a positive impact on salinity (Liang et al. 2018). Although K+/Na+ ratio is highly significant for salinity tolerance (Almeida et al. 2017), the focus must be given to K uptake rather than K+/Na ratio for screening against salinity. However, it may vary with other factors like genotype (Škrubej et al. 2015). Results on Cl contents exhibited increased Cl− contents in response to elevated NaCl concentration (Zaki and Yokoi 2016).
In order to comprehend the impact of different stresses on growth parameters and germination indices one need to fully grasp the relationship between the input and output variables which can be accomplished with the aid of statistical or computer-aided algorithms. Lately, validation, prediction and optimization of data is done by AI/ML based models and performance of these models is tested by scores of different performance metrics. In vitro studies on plant revealed the reliability and accuracy of different ML in different disciplines ranging from in vitro germination to callus and shoot induction (Hesami et al. 2020b; Hesami and Jones 2021; Pepe et al. 2021a, b; Aasim et al. 2022b). Application of ML or ANN models for germination or growth indices has also been employed for different crops (Škrubej et al. 2015). These studies generally involved the deep learning models for detecting germination (Genze et al. 2020) and seedling establishment (Samiei et al. 2020). Whereas, in vitro germination and seedling growth parameters using ML/ANN models has been documented for industrial hemp (Aasim et al. 2022c). In this study, four different performance metrics were utilized for predicting the data for salinity impact on in vitro growth parameters by MLP model. The four performance indicators that were employed corroborated the findings and accurately anticipated the outcomes. However, each output exhibited variable R2 scores with relatively less R2 scores were registered for shoot length and shoot numbers. All remaining output variables exhibited relatively high R2 scores. Another important aspect of the current study was validation and prediction of STI of shoot and root at different NaCl concentration. The results revealed the clear impact of NaCl concentration and organ type, and highest R2 scores were documented for STI (shoot) and STI (root). The actual and predicted scores for both STI parameters were close to each other confirmed by the R2 scores of both output parameters. Application of ML/ANN modeling for estimating the salt stress is a key tool to screen the plants against different types of stresses. Limited number of reports have publicized the successful use of AI/ML models for identification, quantification, classification and prediction of salt stress in plants like rice (Moghimi et al. 2018), wheat (Das et al. 2020; Kecoglu et al. 2022), and Arabidopsis (Kang et al. 2018; Vakilian 2020). Whereas, phenotyping screening of plants against salt stress (Singh et al. 2016; Feng et al. 2020; Gao et al. 2020) have also been employed. The results of the study have successfully demonstrated that AI/ML can be employed for in vitro screening against different types of stresses.
Conclusion
This study presents the impact of different NaCl concentrations on germination indices and growth indices for desi chickpea seeds cultured under in vitro conditions. A negative impact of elevated NaCl concentration on all germination and growth indices was clearly observed when compared to the control group. Majority of the traits reflected a highly significant and positive correlation except fresh root wt, which reflected a non-significant correlation with root numbers and dry shoot wt. Elemental analysis revealed more Na and Cl accumulation with respective elevated NaCl concentrations. Application of MLP models to predict various outputs with high precision was achieved efficaciously and it was also validated through four different performance metrics—R2, MSE, MAE and MAPE. Moreover, STI of desi chickpea was precisely predicted via MLP with high precision for both shoot and root. This work clearly highlights a necessity to employ more AI models or hybrid models to predict and optimize the stress level under natural or controlled conditions. Above all, the unprecedented findings in this study may serve to start a new era to frequently employ AI based models for estimating the impact of different types of stresses under either in vitro conditions or in field studies in conjunction with traditonal statistical analysis.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
Conceptualization, MA and SAA; methodology, MA; software, SAA; validation, FA and MA; formal analysis, MA, SAA; investigation, FA, MBT and MSC; resources, MA and KMK; data curation, FA, MBT and MSC; writing—original draft preparation, MA; writing—review and editing, MA, SAA and KMK; visualization, MA; supervision, MA; project administration, MA, and SAA; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data availability and materials
All data has been submitted and cited.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Muhammad Aasim, Email: mshazim@gmail.com.
Fatma Akin, Email: akn.fatma@gmail.com.
Seyid Amjad Ali, Email: syedali@bilkent.edu.tr.
Mehmet Burak Taskin, Email: mbtaskin@ankara.edu.tr.
Muslume Sevba Colak, Email: smyilmaz@ankara.edu.tr.
Khalid Mahmood Khawar, Email: kmkhawar@gmail.com.
References
- Aasim M, Ali SA, Bekiş P, Nadeem MA. Light-emitting diodes induced in vitro regeneration of Alternanthera reineckii mini and validation via machine learning algorithms. Vitr Cell Dev Biol. 2022;58:1–10. [Google Scholar]
- Aasim M, Katirci R, Baloch F, et al. Innovation in the breeding of common bean through a combined approach of in vitro regeneration and machine learning algorithms. Front Genet. 2022;13:1–13. doi: 10.3389/fgene.2022.897696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aasim M, Katırcı R, Akgur O, et al. Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L) Ind Crops Prod. 2022;181:114801. doi: 10.1016/j.indcrop.2022.114801. [DOI] [Google Scholar]
- Aasim M, Khan AA (2019) Nutritional values, health benefits and multiple uses of desi chickpea. In: Lund AT, Schultz ND (eds) Handbook of chickpeas: nutritional value, health benefits and management. pp 57–73
- Aharon S, Hana B, Liel G, et al. Total phenolic content and antioxidant activity of chickpea (Cicer arietinum L.) as affected by soaking and cooking conditions. Food Nutr Sci. 2011;2011:1–7. [Google Scholar]
- Akbarimoghaddam H, Galavi M, Ghanbari A, Panjehkeh N. Salinity effects on seed germination and seedling growth of bread wheat cultivars. Trakia J Sci. 2011;9:43–50. [Google Scholar]
- Al-Khateeb SA, Al-Khateeb AA, Sattar MN, Mohmand AS. Induced in vitro adaptation for salt tolerance in date palm (Phoenix dactylifera L.) cultivar Khalas. Biol Res. 2020 doi: 10.1186/s40659-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida DM, Oliveira MM, Saibo NJM. Regulation of Na+ and K+ homeostasis in plants: towards improved salt stress tolerance in crop plants. Genet Mol Biol. 2017;40:326–345. doi: 10.1590/1678-4685-gmb-2016-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anawar H, Canha N, Freitas M, et al. Effects of different drying processes on the concentrations of metals and metalloids in plant materials. J Radioanal Nucl Chem. 2011;289:29–34. doi: 10.1007/s10967-011-1051-9. [DOI] [Google Scholar]
- Asefpour Vakilian K. Machine learning improves our knowledge about miRNA functions towards plant abiotic stresses. Sci Rep. 2020;10:1–10. doi: 10.1038/s41598-020-59981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf M, Waheed A. Responses of some local/exotic accessions of lentil (Lens culinaris Medic.) to salt stress. J Agron Crop Sci. 1993;170:103–112. doi: 10.1111/j.1439-037X.1993.tb01063.x. [DOI] [Google Scholar]
- Asmare HA, Ambo E. Impact of salinity on tolerance, vigor, and seedling relative water content of haricot bean (Phaseolus vulgaris L.) cultivars. J Plant Sci. 2013;1:22–27. [Google Scholar]
- Atieno J, Li Y, Langridge P, et al. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci Rep. 2017;7:1–11. doi: 10.1038/s41598-017-01211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydemir LY, Yemenicioğlu A. Potential of Turkish Kabuli type chickpea and green and red lentil cultivars as source of soy and animal origin functional protein alternatives. LWT-Food Sci Technol. 2013;50:686–694. doi: 10.1016/j.lwt.2012.07.023. [DOI] [Google Scholar]
- Bahmani R, Gholami M, Mozafari A-A, Alivaisi R. Effects of salinity on in vitro shoot proliferation and rooting of apple rootstock MM. 106. World Appl Sci J. 2012;17:292–295. [Google Scholar]
- Bewley JD, Bradford K, Hilhorst H. Seeds: physiology of development, germination and dormancy. Cham: Springer Science & Business Media; 2012. [Google Scholar]
- Botía P, Carvajal M, Cerdá A, Martínez V. Response of eight Cucumis melo cultivars to salinity during germination and early vegetative growth. Agronomie. 1998;18:503–513. doi: 10.1051/agro:19980801. [DOI] [Google Scholar]
- Carvalho MP, Santana DG, Ranal MA. Emergência de plântulas de Anacardium humile A. St.-Hil. (Anacardiaceae) avaliada por meio de amostras pequenas. Brazilian J Bot. 2005;28:627–633. doi: 10.1590/S0100-84042005000300018. [DOI] [Google Scholar]
- Cokkizgin A. Salinity stress in common bean (Phaseolus vulgaris L.) seed germination. Not Bot Horti Agrobot Cluj-Napoca. 2012;40:177–182. doi: 10.15835/nbha4017493. [DOI] [Google Scholar]
- Cruz ED, Martins FDEO, Jeude C. Biometria de frutos e sementes e germinação de jatobá-curuba (Hymenaea intermedia Ducke, Leguminosae-Caesalpinioideae) Brazilian J Bot. 2001;24:161–165. doi: 10.1590/S0100-84042001000200005. [DOI] [Google Scholar]
- Das B, Manohara KK, Mahajan GR, Sahoo RN. Spectroscopy based novel spectral indices, PCA-and PLSR-coupled machine learning models for salinity stress phenotyping of rice. Spectrochim Acta Part A Mol Biomol Spectrosc. 2020;229:117983. doi: 10.1016/j.saa.2019.117983. [DOI] [PubMed] [Google Scholar]
- Day S, Aasim M. In vitro screening of preconditioned plumular apices explants of peanut (Arachis Hypogeae) to different salts concentration. Fresenius Environ Bull. 2017;26:354–587. [Google Scholar]
- Day S, Aasim M, Bakhsh A. Effects of preconditioning, plant growth regulators and Kcl on shoot regeneration of peanut (Arachis hypogea) J Anim Plant Sci. 2016;26:294–300. [Google Scholar]
- Demir I, Ermis S, Mavi K, Matthews S. Mean germination time of pepper seed lots (Capsicum annuum L.) predicts size and uniformity of seedlings in germination tests and transplant modules. Seed Sci Technol. 2008;36:21–30. doi: 10.15258/sst.2008.36.1.02. [DOI] [Google Scholar]
- Dorneles MC, Ranal MA, Santana DG. Germinação de diásporos recém-colhidos de Myracrodruon urundeuva Allemão (Anacardiaceae) ocorrente no cerrado do Brasil Central. Brazilian J Bot. 2005;28:399–408. doi: 10.1590/S0100-84042005000200018. [DOI] [Google Scholar]
- El Naim AM, Mohammed KE, Ibrahim EA, Suleiman NN. Impact of salinity on seed germination and early seedling growth of three sorghum (Sorghum biolor L. Moench) cultivars. Sci Technol. 2012;2:16–20. doi: 10.5923/j.scit.20120202.03. [DOI] [Google Scholar]
- El-Katony TM, Khedr A-HA-F, Soliman NG. Nutrients alleviate the deleterious effect of salinity on germination and early seedling growth of the psammophytic grass Elymus farctus. Botany. 2015;93:559–571. doi: 10.1139/cjb-2015-0096. [DOI] [Google Scholar]
- Feng X, Zhan Y, Wang Q, et al. Hyperspectral imaging combined with machine learning as a tool to obtain high-throughput plant salt-stress phenotyping. Plant J. 2020;101:1448–1461. doi: 10.1111/tpj.14597. [DOI] [PubMed] [Google Scholar]
- Gao Z, Luo Z, Zhang W, et al. Deep learning application in plant stress imaging: a review. AgriEngineering. 2020;2:29. doi: 10.3390/agriengineering2030029. [DOI] [Google Scholar]
- Genze N, Bharti R, Grieb M, et al. Accurate machine learning-based germination detection, prediction and quality assessment of three grain crops. Plant Methods. 2020;16:1–11. doi: 10.1186/s13007-020-00699-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghribi A, Maklouf I, Blecker C, et al. Nutritional and com-positional study of desi and kabuli chickpea (Cicer arietinum L.) flours from Tunisian cultivars. Adv Food Technol Nutr Sci Open J. 2015;1:38–47. doi: 10.17140/AFTNSOJ-1-107. [DOI] [Google Scholar]
- Gill PK, Sharma AD, Singh P, Bhullar SS. Changes in germination, growth and soluble sugar contents of Sorghum bicolor (L.) Moench seeds under various abiotic stresses. Plant Growth Regul. 2003;40:157–162. doi: 10.1023/A:1024252222376. [DOI] [Google Scholar]
- Grinblat GL, Uzal LC, Larese MG, Granitto PM. Deep learning for plant identification using vein morphological patterns. Comput Electron Agric. 2016;127:418–424. doi: 10.1016/j.compag.2016.07.003. [DOI] [Google Scholar]
- Hesami M, Jones AMP. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol. 2020;2020:1–37. doi: 10.1007/s00253-020-10888-2. [DOI] [PubMed] [Google Scholar]
- Hesami M, Jones AMP. Modeling and optimizing callus growth and development in Cannabis sativa using random forest and support vector machine in combination with a genetic algorithm. Appl Microbiol Biotechnol. 2021;2021:1–12. doi: 10.1007/s00253-021-11375-y. [DOI] [PubMed] [Google Scholar]
- Hesami M, Naderi R, Tohidfar M. Modeling and optimizing In vitro sterilization of chrysanthemum via multilayer perceptron-non-dominated sorting genetic algorithm-II (MLP-NSGAII) Front Plant Sci. 2019;10:1–13. doi: 10.3389/fpls.2019.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesami M, Condori-Apfata JA, Valencia MV, Moham M. Application of artificial neural network for modeling and studying in vitro genotype-independent shoot regeneration in wheat. Appl Sci. 2020;10:1–10. doi: 10.3390/APP10155370. [DOI] [Google Scholar]
- Hesami M, Naderi R, Tohidfar M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: the establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl Microbiol Biotechnol. 2020;104:10249–10263. doi: 10.1007/s00253-020-10978-1. [DOI] [PubMed] [Google Scholar]
- Hesami M, Pepe M, Monthony AS, et al. Modeling and optimizing in vitro seed germination of industrial hemp (Cannabis sativa L.) Ind Crops Prod. 2021;170:113753. doi: 10.1016/j.indcrop.2021.113753. [DOI] [Google Scholar]
- Hesami M, Alizadeh M, Jones AMP, Torkamaneh D. Machine learning: its challenges and opportunities in plant system biology. Appl Microbiol Biotechnol. 2022;2022:1–24. doi: 10.1007/s00253-022-11963-6. [DOI] [PubMed] [Google Scholar]
- Jafari M, Shahsavar A. The application of artificial neural networks in modeling and predicting the effects of melatonin on morphological responses of citrus to drought stress. PLoS One. 2020;15:e0240427. doi: 10.1371/journal.pone.0240427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukanti AK, Gaur PM, Gowda CLL, Chibbar RN. Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br J Nutr. 2012;108:S11–S26. doi: 10.1017/S0007114512000797. [DOI] [PubMed] [Google Scholar]
- Kacar B, Inal A, editors. Bitki Analizleri. 1. Şti: Nobel Yayın Dağıtım Ltd.; 2008. [Google Scholar]
- Kalefetoğlu T, Macar O, Dürdane İ. Variability in some biochemical and nutritional characteristics in desi and Turkish kabuli chickpea (Cicer arietinum L.) types. Celal Bayar Univ J Sci. 2017;13:677–680. [Google Scholar]
- Kang D, Ahn H, Lee S, et al (2018) Identifying stress-related genes and predicting stress types in Arabidopsis using logical correlation layer and CMCL loss through time-series data. In: 2018 IEEE International conference on bioinformatics and biomedicine (BIBM). IEEE, pp 399–404
- Karakullukçu E. Bazı nohut (Cicer arietinum L.) çeşitlerinin tuza toleranslarının belirlenmesi. J Agric Sci. 2008;14:1–10. [Google Scholar]
- Kecoglu I, Sirkeci M, Unlu MB, et al. Quantification of salt stress in wheat leaves by Raman spectroscopy and machine learning. Sci Rep. 2022;12:1–10. doi: 10.1038/s41598-022-10767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendir H, Sahin-Demirbag N, Aasim M, Khawar KM. In vitro plant regeneration from Turkish Narbon Vetch (Vicia narbonensis L. var. narbonensis L.) African J Biotechnol. 2009;8:1–5. [Google Scholar]
- Khajeh-Hosseini M, Lomholt A, Matthews S. Mean germination time in the laboratory estimates the relative vigour and field performance of commercial seed lots of maize (Zea mays L.) Seed Sci Technol. 2009;37:446–456. doi: 10.15258/sst.2009.37.2.17. [DOI] [Google Scholar]
- Kirtis A, Aasim M. In vitro axillary shoot regeneration from shoot tip explant of desi chikpea (Cıcer arıetınum L.) J Glob Innov Agric Soc Sci. 2020;8:65–69. doi: 10.22194/jgiass/8.903. [DOI] [Google Scholar]
- Kirtis A, Aasim M, Katırcı R. Application of artificial neural network and machine learning algorithms for modeling the in vitro regeneration of chickpea (Cicer arietinum L.) Plant Cell, Tissue Organ Cult. 2022;2022:1–12. [Google Scholar]
- Kirtiş A, Aasim M. Thidiazuron (TDZ) induced in vitro axillary shoot regeneration of desi chikpea (Cicer arietinum L.) J Appl Biol Sci. 2019;13:17–20. [Google Scholar]
- Kouam EB, Ndo SM, Mandou MS, et al. Genotypic variation in tolerance to salinity of common beans cultivated in Western Cameroon as assessed at germination and during early seedling growth. Open Agric. 2017;2:600–610. doi: 10.1515/opag-2017-0064. [DOI] [Google Scholar]
- Kumar B, Gupta E, Mali H, et al. Constant and alternating temperature effects on seed germination potential in Artemisia annua L. J Crop Improv. 2013;27:636–642. doi: 10.1080/15427528.2013.832458. [DOI] [Google Scholar]
- Liang W, Ma X, Wan P, Liu L. Plant salt-tolerance mechanism: a review. Biochem Biophys Res Commun. 2018;495:286–291. doi: 10.1016/j.bbrc.2017.11.043. [DOI] [PubMed] [Google Scholar]
- Mahood EH, Kruse LH, Moghe GD. Machine learning: a powerful tool for gene function prediction in plants. Appl Plant Sci. 2020;8:e11376. doi: 10.1002/aps3.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena E, Leiva-Mora M, Jayawardana EKD, et al. Effect of salt stress on seed germination and seedlings growth of Phaseolus vulgaris L. Cultiv Trop. 2015;36:71–74. [Google Scholar]
- Mirza K, Aasim M, Katırcı R, et al. Machine learning and artificial neural networks-based approach to model and optimize ethyl methanesulfonate and sodium azide induced in vitro regeneration and morphogenic traits of water hyssops (Bacopa monnieri L.) J Plant Growth Regul. 2022;2022:1–15. [Google Scholar]
- Mishra B, Kumar N, Mukhtar MS. Systems biology and machine learning in plant–pathogen interactions. Mol Plant-Microbe Interact. 2019;32:45–55. doi: 10.1094/MPMI-08-18-0221-FI. [DOI] [PubMed] [Google Scholar]
- Moghimi A, Yang C, Marchetto PM. Ensemble feature selection for plant phenotyping: a journey from hyperspectral to multispectral imaging. IEEE Access. 2018;6:56870–56884. doi: 10.1109/ACCESS.2018.2872801. [DOI] [Google Scholar]
- Mohammadi K. Nutritional composition of Iranian desi and kabuli chickpea (Cicer arietinum L.) cultivars in autumn sowing. Int J Agric Biosyst Eng. 2015;9:550–553. [Google Scholar]
- Muchate NS, Rajurkar NS, Suprasanna P, Nikam TD. NaCl induced salt adaptive changes and enhanced accumulation of 20-hydroxyecdysone in the in vitro shoot cultures of Spinacia oleracea (L.) Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-48737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudasir S, Sofi PA, Khan MN, et al. Genetic diversity, variability and character association in local common Bean (Phaseolus vulgaris L.) germplasm of Kashmir. Electron J Plant Breed. 2012;3:883–891. [Google Scholar]
- Mungala AJ, Radhakrishnan T, Dobaria JR. In vitro screening of 123 Indian peanut cultivars for sodium chloride induced salinity tolerance. World J Agric Sci. 2008;4:74–582. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Munns R, Schachtman DP, Condon AG. The significance of a two-phase growth response to salinity in wheat and barley. Funct Plant Biol. 1995;22:561–569. doi: 10.1071/PP9950561. [DOI] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- Nadeem M, Li J, Yahya M, et al. Grain legumes and fear of salt stress: focus on mechanisms and management strategies. Int J Mol Sci. 2019;20:799. doi: 10.3390/ijms20040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeem MA, Yeken MZ, Shahid MQ, et al. Common bean as a potential crop for future food security: an overview of past, current and future contributions in genomics, transcriptomics, transgenics and proteomics. Biotechnol Biotechnol Equip. 2021;35:758–786. doi: 10.1080/13102818.2021.1920462. [DOI] [Google Scholar]
- Nimac A, Lazarević B, Petek M, et al. Effects of salinity and seed priming on germination of sea fennel (Crithmum maritimum L.) Agric Conspec Sci. 2018;83:181–185. [Google Scholar]
- Özer S, Karaköy T, Toklu F, et al. Nutritional and physicochemical variation in Turkish kabuli chickpea (Cicer arietinum L.) landraces. Euphytica. 2010;175:237–249. doi: 10.1007/s10681-010-0174-3. [DOI] [Google Scholar]
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- Pepe M, Hesami M, Jones AMP. Machine learning-mediated development and optimization of disinfection protocol and scarification method for improved in vitro germination of cannabis seeds. Plants. 2021;10:2397. doi: 10.3390/plants10112397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe M, Hesami M, Small F, Jones AMP. Comparative analysis of machine learning and evolutionary optimization algorithms for precision micropropagation of Cannabis sativa: prediction and validation of in vitro shoot growth and development based on the optimization of light and carbohydrate sou. Front Plant Sci. 2021;12:757869. doi: 10.3389/fpls.2021.757869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phat P, Ju H-J, Noh J, et al. Effects of hydropriming and explant origin on in vitro culture and frequency of tetraploids in small watermelons. Hortic Environ Biotechnol. 2017;58:495–502. doi: 10.1007/s13580-017-0242-x. [DOI] [Google Scholar]
- Purushothaman R, Upadhyaya HD, Gaur PM, et al. Kabuli and desi chickpeas differ in their requirement for reproductive duration. F Crop Res. 2014;163:24–31. doi: 10.1016/j.fcr.2014.04.006. [DOI] [Google Scholar]
- Ranal MA, de Santana DG. How and why to measure the germination process? Brazilian J Bot. 2006;29:1–11. doi: 10.1590/S0100-84042006000100002. [DOI] [Google Scholar]
- Ranal MA, de Santana DG, Ferreira WR, Mendes-Rodrigues C. Calculating germination measurements and organizing spreadsheets. Brazilian J Bot. 2009;32:849–855. doi: 10.1590/S0100-84042009000400022. [DOI] [Google Scholar]
- Regnier L. Influence of mechanical scarification and open-field sowing procedure over Cassia ferruginea germination process. J Hortic Plant Res. 2020;9:30–38. doi: 10.18052/www.scipress.com/JHPR.9.30. [DOI] [Google Scholar]
- Rico-Chávez AK, Franco JA, Fernandez-Jaramillo AA, et al. Machine learning for plant stress modeling: a perspective towards hormesis management. Plants. 2022;11:970. doi: 10.3390/plants11070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum G, Drake FL. Python 3 Reference Manual. Scotts Valley, CA: CreateSpace; 2009. [Google Scholar]
- Samiei S, Rasti P, Ly VuJ, et al. Deep learning-based detection of seedling development. Plant Methods. 2020;16:1–11. doi: 10.1186/s13007-020-00647-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Santana DG, Ranal M (2004) Análise da germinação: um enfoque estatístico. UnB
- Silva JCF, Teixeira RM, Silva FF, et al. Machine learning approaches and their current application in plant molecular biology: a systematic review. Plant Sci. 2019;284:37–47. doi: 10.1016/j.plantsci.2019.03.020. [DOI] [PubMed] [Google Scholar]
- Singh A, Ganapathysubramanian B, Singh AK, Sarkar S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016;21:110–124. doi: 10.1016/j.tplants.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Škrubej U, Rozman Č, Stajnko D. Assessment of germination rate of the tomato seeds using image processing and machine learning. Eur J Hortic Sci. 2015;80:68–75. doi: 10.17660/eJHS.2015/80.2.4. [DOI] [Google Scholar]
- Snedecor G, Cochran W (1967) Statistical methods. Ames, Lowa, the Lowa state University
- Soltis PS, Nelson G, Zare A, Meineke EK. Plants meet machines: prospects in machine learning for plant biology. Appl Plant Sci. 2020;8:e11371. doi: 10.1002/aps3.11371. [DOI] [Google Scholar]
- Thiam M, Champion A, Diouf D, Ourèye SYM. NaCl effects on in vitro germination and growth of some senegalese cowpea (Vigna unguiculata (L.) Walp.) cultivars. Int Sch Res Not. 2013;2013:1–12. doi: 10.5402/2013/382417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk ADJ, Kootstra G, Kruijer W, de Ridder D. Machine learning in plant science and plant breeding. Iscience. 2021;24:101890. doi: 10.1016/j.isci.2020.101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb GI, Sammut C, Perlich C, et al. Encyclopedia of machine learning. US, Boston: Springer; 2011. Leave-one-out cross-validation; pp. 600–601. [Google Scholar]
- Wei-hua L, Hui-ming PU, Jie-fu Z, et al. Screening of Brassica napus for salinity tolerance at germination stage. Chinese J Oil Crop Sci. 2013;35:271. [Google Scholar]
- Wu H, Guo J, Wang C, et al. An effective screening method and a reliable screening trait for salt tolerance of Brassica napus at the germination stage. Front Plant Sci. 2019;10:530. doi: 10.3389/fpls.2019.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Blumwald E. Developing salt-tolerant crop plants: challenges and opportunities. Trends Plant Sci. 2005;10:615–620. doi: 10.1016/j.tplants.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Yeo AR, Flowers TJ (1989) Selection for physiological characters--examples from breeding for salt tolerance. In: Seminar series-society for experimental biology
- Yousef F, Shafique F, Ali Q, Malik A. Effects of salt stress on the growth traits of chickpea (Cicer arietinum L.) and pea (Pisum sativum L.) seedlings. Biol Clin Sci Res J. 2020;10:1–10. doi: 10.54112/bcsrj.v2020i1.29. [DOI] [Google Scholar]
- Zaki HEM, Yokoi S (2016) A comparative in vitro study of salt tolerance in cultivated tomato and related wild species. Plant Biotechnol 16–1006 [DOI] [PMC free article] [PubMed]
- Zawude S, Shanko D. Effects of salinity stress on chickpea (Cicer arietinum L.) landraces during early growth stage. Int J Sci Reports. 2017;3:214–219. doi: 10.18203/issn.2454-2156.IntJSciRep20173093. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data has been submitted and cited.