Abstract
A functional homolog (rhp23) of human HHR23A and Saccharomyces cerevisiae RAD23 was cloned from the fission yeast Schizosaccharomyces pombe and characterized. Consistent with the role of Rad23 homologs in nucleotide excision repair, rhp23 mutant cells are moderately sensitive to UV light but demonstrate wild-type resistance to γ-rays and hydroxyurea. Expression of the rhp23, RAD23 or HHR23A cDNA restores UV resistance to the mutant, indicating that rhp23 is a functional homolog of the human and S.cerevisiae genes. The rhp23::ura4 mutation also causes a delay in the G2 phase of the cell cycle which is corrected when rhp23, RAD23 or HHR23A cDNA is expressed. Rhp23 is present throughout the cell but is located predominantly in the nucleus, and the nuclear levels of Rhp23 decrease around the time of S phase in the cell cycle. Rhp23 is ubiquitinated at low levels, but overexpression of the rhp23 cDNA induces a large increase in ubiquitination of other proteins. Consistent with a role in protein ubiquitination, Rhp23 binds ubiquitin, as determined by two-hybrid analysis. Thus, the rhp23 gene plays a role not only in nucleotide excision repair but also in cell cycle regulation and the ubiquitination pathways.
INTRODUCTION
Rad23 homologs are highly conserved eukaryotic proteins, thus far identified in more than 20 organisms spanning yeast, algae, fruit fly, plants and mammals (1–3). Rad23 homologs have an unusual structure in which the N-terminal end, referred to as UbL for ubiquitin-like, is similar to ubiquitin, the small 76 amino acid long protein that serves as a signal for degradation of proteins by the proteasome (4). The Rad23 homologs also have a central and a C-terminal ubiquitin-associated (UBA) domain. UBA domains are highly conserved structural motifs containing about 50 amino acids, originally identified in proteins that participate in the ubiquitin pathway and whose function has not been clearly established (5–8).
Studies of rad23 mutations in the budding yeast Saccharomyces cerevisiae first indicated a role for this gene in nucleotide excision repair (NER) of UV-damaged DNA since rad23 mutant cells are moderately sensitive to UV light due to a deficiency in this type of repair (3). The Rad23 protein binds to Rad4 and other NER factors. Furthermore, the Rad23–Rad4 complex binds preferentially to UV-damaged DNA (9,10) as part of the NER process. The UbL domain of Rad23 plays a role in NER since deletion of this region causes UV sensitivity intermediate between the levels demonstrated by a wild-type and a rad23 deletion strain (3). The interaction of this UbL domain with the proteasome suggested that proteolysis is important for NER (11). However, using both in vivo and in vitro approaches, Russell et al. (12) found that the interaction with the proteasome but not proteolysis per se was essential for the role of Rad23 in NER. These studies suggested that the UbL domain of Rad23 is important for the assembly of protein complexes required for efficient NER (12).
One of the human homologs of Rad23, HHR23B, was originally identified in a complex with XP-C. The latter protein is a human homolog of budding yeast Rad4 and represents one of the eight complementation groups for the genetic disorder xeroderma pigmentosum (XP) (13). Patients with XP are highly sensitive to UV and demonstrate a greatly increased frequency of skin cancer due to defective NER (13). While HHR23B and HHR23A, a second human homolog of RAD23, are not XP genes and no naturally occurring mutations have yet been identified in them, studies with in vitro systems indicate that NER takes place without the two encoded proteins. However, either HHR23A or HHR23B activates XP-C for its required function in NER (14). Thus, Rad23 homologs in both S.cerevisiae and humans appear to play a role in NER as part of complexes with Rad4 homologs and other NER factors (9).
In addition to their well-established role in NER, some evidence suggests involvement of Rad23 homologs in other cellular processes. For example, only a fraction of HHR23B is complexed with XP-C and HHR23A is not found in a complex with XP-C in the cell (2), raising the possibility that these free forms function in processes other than NER. Since the UbL domain of Rad23 and HHR23A/B interacts with the proteasome (11,15), Rad23 homologs are thought to participate in ubiquitin-dependent proteolysis by the proteasome (11,15,16). This possibility was supported by in vitro studies with the human homologs and by studies of budding yeast strains with mutations in both rad23 and rpn10 (15–17). Rad23 homologs may also be involved in cell cycle regulation since they are degraded during S phase (11,18), spindle pole body duplication is defective in a budding yeast strain with mutations in both rad23 and dsk2 (19) and HHR23A interacts with the HIV-1 Vpr protein known to induce G2 arrest in both human and fission yeast cells (8,20–22).
To help better define these putative multiple functions of Rad23, we isolated and characterized the Schizosaccharomyces pombe homolog. In addition to its expected role in NER, as indicated by the UV sensitivity of an rhp23 deletion mutant, we find that S.pombe Rhp23 also plays a role in the cell cycle and the ubiquitin–proteasome pathway. Moreover, the involvement of Rhp23 in cell cycle control seems to be a common feature of the Rad23 homologs, as both HHR23A and RAD23 complement the cell cycle defect.
MATERIALS AND METHODS
Yeast strains and media
Genotypes and sources of S.pombe strains and plasmids used in this study are summarized in Table 1. The S.pombe wild-type (SP223), mutant rhp23::ura4 and rad9-192 cells containing various plasmid constructs were maintained in standard Edinburgh minimal medium (EMM) (23) supplemented with uracil and adenine at 75 µg/ml. Thiamine was added to the medium at a final concentration of 20 µM to repress expression from the regulated nmt1 (no message in thiamine) promoter (24).
Table 1. Schizosaccharomyces pombe strains and plasmids.
| Name | Genotype | Source |
|---|---|---|
| Strains | ||
| SP223 | Wild-type, h– ade6 leu1-32 ura4-294 | David Beach |
| Rad9-192 | h– rad9-192 ade6 leu1-32 ura4-294, isogenic to SP223 | (36) |
| Rhp23::ura4 | h– rhp23::ura4+ ade6 leu1-32 ura4-294, isogenic to SP223 | This study |
| RE70 | A single copy integrant of a gfp–rhp23 fusion | This study |
| RE74 | A single copy integrant of gfp | This study |
| Plasmids | ||
| pYZ1N | Derivative of pREP1N | (25) |
| pYZ3N | pYZ1N with GFP | (25) |
| pYZ3N-rhp23 | S.pombe gfp–rhp23 fusion | This study |
| pYZ1N-rhp23 | S.pombe rhp23 in pYZ1N | This study |
| pYZ1N-RAD23 | S.cerevisiae RAD23 in pYZ1N | This study |
| pYZ1N-HHR23A | Human HHR23A in pYZ1N | This study |
| pSF173-rhp23 | HA–rhp23 fusion | This study |
Molecular cloning and nucleotide sequence determination of the S.pombe rhp23 gene
The rhp23 sequence reported in this paper was deposited in the GenBank database on July 29, 1999 (accession no. AF174293). The rhp23 cDNA was cloned from a cDNA library made from S.pombe wild-type strain SP972 (a gift from Greg Hannon). The 1286 bp cDNA was amplified by PCR using primers rhp23-NdeI d(GCAGGACATAACAAGGTAGGATCTCTAC) and rhp23-BamHI d(GGTACAAGCAGTTTTAGGCTGACTTC). The amplified DNA fragments were then digested with BamHI and NdeI and ligated to S.pombe expression vector pYZ1N (25) digested with the same enzymes. The resulting ligation mixtures were transformed into Escherichia coli DH5α for identification and amplification of plasmids containing inserted rhp23 cDNA by formation of white colonies on Xgal-containing agar plates. The complete nucleotide sequence of the rhp23 cDNA insert in pYZ1N was determined with an ABI automated DNA Sequencer Model 377 (Perkin-Elmer, CA). For Rhp23 nuclear localization, green fluorescent protein (GFP) was fused in-frame at the 5′-end of rhp23 by introduction of a SacII site during PCR with the rhp23-GFP primer d(ACAGACCGCGGATATGAATTTGACATTCAAAAATCTAC) and the rhp23-BamHI primer, followed by cloning at the SacII and BamHI sites of the pYZ3N-GFP vector (25). After removing the EcoRI fragment containing the ars1 sequence, plasmids were integrated at the ura4 locus as described previously (26) and a single copy integrant was identified by PCR analysis. Cellular localization of GFP–Rhp23 in S.pombe cells after staining of nuclei with Hoechst 33342 (27) was examined using fluorescent microscopy as described previously (25). The time series for GFP–Rhp23 was performed on a microscope slide with a well containing agarose medium as described (28). The HA-tagged rhp23 gene was cloned into expression vector pSF173 at the NotI and BglII sites as previously described (29). Plasmid DNAs were transformed into S.pombe cells by the lithium acetate procedure (30,31). The same strategies were used to clone and express the human HHR23A and S.cerevisiae RAD23 cDNAs in S.pombe.
Construction of the rhp23::ura4 deletion/disruption mutant
Insertion of S.pombe ura4 into the rhp23 locus was performed to generate a rhp23::ura4 disruption mutant using a one-step gene disruption procedure described previously (32,33). Briefly, the ura4 gene was PCR amplified using primers containing 40 bases of Rhp23 sequence at the 5′-end to generate a ura4 fragment flanked at each end with 40 bp of rhp23 sequence. The PCR product was purified with a Qiagen column (Qiagen, CA) and used to transform a S.pombe strain in which the ura4 gene had been deleted (h– mik1::leu2 ade6 leu1-32 ura4-D18). The ura+ transformants were screened by PCR for colonies in which the rhp23 gene had been disrupted and the rhp23::ura4 disruption was confirmed by DNA sequencing of the rhp23–ura4 junctions. The rhp23 sequences in the PCR product used for the transformation led to deletion of amino acids 86–238 from the Rhp23 protein. The original transformant strain with a rhp23::ura4 disruption was crossed with SP224 (h+ ade6-216 leu1-32 ura4-294) and sporulated to generate the rhp23::ura4 strain with a leu1 marker used for plasmid transformations.
UV light, γ-ray and hydroxyurea (HU) resistance
UV irradiation (254 nm) was performed at a dose rate of ∼3 J/m2/s. γ-Ray irradiation was performed using a GammaCell 220 (Nordion, Ontario, Canada) with a 60Co source at a dose rate of 1 Gy/s. To prepare cells for radiation exposure, S.pombe cultures containing the pYZ1N-cDNA constructs or vector control were first grown to stationary phase in the presence of 20 µM thiamine (primary culture). Cells were then washed three times with distilled water, diluted to a final concentration of ∼2 × 105 cells/ml in 20 ml of EMM medium with (low gene expression) or without (high gene expression) thiamine and grown at 30°C with shaking (200 r.p.m.) for 18 h to reach log phase (secondary culture, 1–5 × 107 cells/ml) prior to irradiation. The densities of the S.pombe cell cultures were ascertained using a hemocytometer and then the cells were either left unirradiated or exposed to graded doses of γ-rays. Samples were diluted for plating in a volume of 100 µl onto EMM plates supplemented with adenine, uracil and thiamine at predetermined concentrations. Data represent the averages of three independent trials. For UV radiation, cells were irradiated after plating.
For HU treatment, cell cultures were prepared as described above and the drug was added to a concentration of 12 mM. Cell density was diluted to 5 × 106 cells/ml at the start of the experiment. Samples of mock-treated and treated cells were collected at various times and seeded onto supplemented EMM plates at predetermined concentrations to assess cell survival.
To measure cell survival in response to UV, γ-rays and HU treatment, agar plates were incubated at 30°C for 3–4 days to obtain individual colonies. Percent survival was calculated as the number of colonies in treated versus mock-treated control cell populations. Data represent the averages of three independent trials.
Analyses of cell cycle-dependent stability and ubiquitination of Rhp23
The SP223 strain was grown in EMM to log phase as described above. The cell population was synchronized in S phase by treatment with 12 mM HU for 4 h at 30°C, with shaking prior to sample collection. Synchronized cells were washed three times with EMM to remove HU and then incubated at 30°C with shaking. Samples were collected at 30 min intervals, up to 7 h. Passage of the synchronized culture through the cell cycle was monitored by Calcofluor staining of the septum (34) that forms as cells divide shortly after mitosis.
Cell extracts were prepared as described previously (http://www.bio.uva.nl/pombe/handbook/section3/section3-3.html). Briefly, cells were washed once with water prior to adding cold stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3, pH 8.0). Cells were then pelleted and resuspended in 3 vol of lysis solution (25 mM MOPS, pH 7.2, 60 mM β-glycerophosphate, 15 mM p-nitrophenylphosphate, 15 mM MgCI2, 15 mM EGTA, 1% Triton X-100, 1 mM DTT). A mixture of protease inhibitors (1 mM PMSF, 20 µg/ml leupeptin, 40 µg/aprotinin and 0.1 mM sodium vanadate) was added immediately before lysis by glass-bead agitation. The cells were disrupted for 60 s using an homogenizer (Biospec Products, Bartlesville, OK). Cell breakage was checked under a microscope and disruption was repeated 3–5 times if necessary. Equal amounts of protein (20–50 µg) were resolved on a SDS–polyacrylamide gel and transferred to a nitrocellulose membrane. Rhp23 protein levels and potential ubiquitination of Rhp23 were assayed by immunoblotting procedures using an anti-Rad23 serum (a gift of K. Madura) and an anti-ubiquitin monoclonal antibody (mAb 1510; Chemicon Inc.), respectively. Immunoblots were developed using the enhanced chemiluminescence (ECL) system (Amersham). α-Tubulin was used as an internal protein control by immunoblotting with anti-α-tubulin antibodies (35).
The anti-Rad23 antibodies cross-reacted with Rhp23 but did not cross-react with HHR23A. This conclusion was supported by the observations that no band around 50 kDa appeared in extracts from the rhp23::ura4 strain, overexpression of the rhp23 cDNA from the nmt1 promoter in the rhp23::ura4 strain gave a very strong band around 50 kDa, overexpression of the RAD23 cDNA gave a very strong band of slightly larger size and overexpression of the HHR23A cDNA did not result in the appearance of a band (data not shown).
RESULTS
Molecular cloning of the rhp23 gene and evolutionary conservation of the protein
A S.pombe amino acid sequence, which shows 36% identity and 53% similarity to amino acid sequences of the human HHR23A gene product over the entire protein sequence, was initially identified from the S.pombe database. The cDNA for this putative HHR23A homolog was amplified from a wild-type S.pombe SP223 cDNA library using PCR and cloned into the fission yeast expression vector pYZ1N (25). Cloning of the cDNA was confirmed by nucleotide sequence analysis of the entire insert. As HHR23A is known to be a functional homolog of the budding yeast RAD23 gene, the newly identified fission yeast gene was named rhp23 (rad homolog of pombe 23; GenBank accession no. AF174293). Consistent with the sequence analysis found in the S.pombe database, the rhp23 gene has four introns and encodes a protein containing 368 amino acids, with a predicted molecular weight of ∼41 kDa. Comparison of the predicted S.pombe Rhp23 protein sequence with sequences of 12 other Rad23 homologs isolated from various eukaryotes indicated a high degree of similarity (32–45% identical, 50–63% similar). Rhp23 also has the structural characteristics of HHR23A and other Rad23 homologs since a UbL domain at the N-terminus and two UBA domains (7,8) in the C-terminal portion are present (see Supplementary Material).
An rhp23::ura4 mutation causes UV sensitivity
A rhp23 deletion was created in S.pombe by replacing part of the rhp23 gene with ura4 using a PCR-based method (32,33). Correct insertion of the ura4 gene at the rhp23 locus was confirmed by PCR and sequence analyses. The rhp23::ura4 mutant strain is viable, but UV resistance and cell cycle characteristics are aberrant. The rhp23::ura4 strain was tested for resistance to UV as well as ionizing radiation and HU, the latter two of which induce DNA strand breaks and block DNA synthesis, respectively. In comparison to a S.pombe rad9-192 strain, which is highly sensitive to UV light, ionizing radiation and HU (36), the rhp23::ura4 strain is moderately sensitive to UV light but demonstrates near wild-type levels of resistance to the other agents, except possibly at very high doses (Fig. 1A–C). This possible difference at high doses of ionizing radiation or HU may be due to strain differences between the wild-type and rhp23::ura4 deletion strains since introducing the rhp23 cDNA into mutant cells had no effect on sensitivity to ionizing radiation or HU. In contrast, the rhp23 cDNA fully complements the UV sensitivity of the mutant (Fig. 1A). This pattern of sensitivity is consistent with Rhp23 playing a role in NER, as is found for Rad23 and HHR23B in budding yeast and human cells, respectively (3,14). The moderate UV sensitivity of the fission yeast mutant is also similar to that observed for a rad23 mutant of S.cerevisiae (3).
Figure 1.
The rhp23::ura4 mutant is sensitive to UV light but not ionizing radiation or HU. Cell survival after UV irradiation (A), ionizing radiation exposure (B) or HU treatment (C) of wild-type SP223, rhp23::ura4, rhp23::ura4 (pYZ1N-rph23) and rad9-192 cells. The rad9-192 mutant was used as a control since it is highly sensitive to all three agents (36). The rhp23::ura4 disruption does not cause increased sensitivity to ionizing radiation or HU, but mutant cells are moderately sensitive to UV light in comparison with the resistant wild-type SP223 strain and the highly sensitive rad9-192 mutant strain. The pYZ1N-rhp23 plasmid containing the rhp23 cDNA restores UV resistance to wild-type levels in the rhp23::ura4 disruption strain at both high (no thiamine) and low expression (plus thiamine; data not shown) levels. Experiments in (A) and (B) were performed in medium without thiamine (high expression) and the experiments in (C) were performed in the presence of thiamine (low expression). Error bars represent the standard deviation of three measurements.
Complementation of the UV sensitivity of rhp23::ura4 cells by human HHR23A and S.cerevisiae RAD23
To test whether human HHR23A and budding yeast RAD23 can restore UV resistance to the rhp23::ura4 strain, both cDNAs were cloned into pYZ1N, containing a thiamine-repressible nmt1 promoter. The cloning was confirmed by DNA sequence analysis. Expression of the HHR23A or RAD23 cDNAs from this nmt1 promoter at high levels restored UV resistance to wild-type levels in the rhp23::ura4 strain (Fig. 2A and B). However, when the nmt1 promoter was repressed by thiamine, the fission yeast rhp23 cDNA still complemented the UV sensitivity of the rhp23::ura4 mutant, while the HHR23A cDNA showed little complementation (Fig. 2C). Thus, efficient complementation of the UV defect in the rhp23::ura4 mutant strain requires high levels of HHR23A.
Figure 2.
Human HHR23A and S.cerevisiae RAD23 restore UV resistance to S.pombe rhp23::ura4. High level expression of RAD23 (A) or HHR23A (B) cDNAs from the pYZ1N plasmid in the S.pombe rhp23::ura4 mutant restores UV resistance. cDNAs in (A) and (B) were expressed under inducing conditions (no thiamine) from the nmt1 promoter (24). In (C) thiamine is present to repress the nmt1 promoter, and at these low expression levels the HHR23A cDNA does not efficiently restore UV resistance to the rhp23::ura4 mutant. Error bars represent the standard deviation of three measurements.
The rhp23::ura4 mutation causes a cell cycle G2 delay
Increased cell length in fission yeast indicates a cell cycle delay (37) and rhp23::ura4 mutant cells are somewhat longer than the isogenic rhp23 cell population (Fig. 3A). The mean cell length of the rhp23::ura4 strain is 12.9 ± 0.38 µm (SEM) and is statistically significant at the P < 0.001 level when compared with the wild-type length of 11.1 ± 0.26 µm. In addition to an increased mean cell length in the rhp23::ura4 mutant population, some of the cells are longer than 18 µm, while no such cells are seen in the wild-type strain. Transforming the rhp23::ura4 mutant strain with a plasmid containing the rhp23 cDNA reduces the mean cell length to 10.6 ± 0.3 µm and eliminates the appearance of cells longer than 18 µm (Fig. 3A). Transformation with a plasmid carrying the HHR23A cDNA gives an identical mean cell length and also eliminates cells longer than 18 µm (data not shown), indicating that the human homolog complements the rhp23::ura4 phenotype of increased length. Transformation with a plasmid carrying the S.cerevisiae RAD23 cDNA gives a mean cell length of 11.8 ± 0.38 µm, intermediate between the wild-type and mutant values, indicating that the budding yeast homolog partially complements the length phenotype. These experiments on complementation of the cell cycle defect in the rhp23::ura4 mutant strain were performed at low levels of expression when the nmt1 promoter was repressed, indicating that, in contrast to UV sensitivity, low level expression of HHR23A is sufficient to fully complement the cell cycle defect.
Figure 3.
The S.pombe rhp23::ura4 mutation causes a cell cycle G2 delay. Cell length was measured for at least 70 cells during log phase growth in medium with thiamine to repress nmt1 promoter activity. (A) Cell length distribution shown for three strains. (B) The mean cell length shown along with the standard error of the mean.
Characterization of the long cells in the rhp23::ura4 cultures indicates that they are delayed in G2. Flow cytometry of cells stained with propidium iodide (38) found no difference between wild-type and rhp23::ura4 cells in the fraction of G1 cells (data not shown), indicating that the long rhp23::ura4 cells are not in G1. When 26 long cells (mean cell length 25 µm, range 19–40 µm) were examined by fluorescence microscopy after propidium iodide staining, it was observed that 65% had a single nucleus and 35% had two nuclei. Actin staining also indicates that the majority of the large cells are in G2. In late G2, actin is distributed as discrete foci near the growing ends of the cell, and approximately at the time of mitosis actin redistributes to form a ring at the center of the cell (39). Phalloidin staining of actin in rhp23::ura4 cells showed that the majority (71%) of the 34 long cells examined (mean cell length 26 µm, range 20–51 µm) have actin dots at both ends of the cell, indicative of a cell in late G2 phase of the cell cycle. The remaining 29% of the long cells have an actin ring at their center, indicating the onset of mitosis.
The long cells in the rhp23::ura4 culture appear to be delayed in G2 rather than terminally arrested in G2. The observations that 35% of the long cells are bi-nucleated and 29% have a medial actin ring support the conclusion that, after a period of delay in G2, the long cells eventually undergo mitosis and cell division. In addition, when a rhp23::ura4 log phase culture is shifted to medium without nitrogen, where fission yeast cells go into stationary phase as small cells (38), essentially all of the rhp23::ura4 cells become small, indicating that the long cells in the log phase culture remain capable of undergoing mitosis and division.
The level and subcellular localization of Rhp23 are cell cycle dependent
To determine the cellular distribution of Rhp23, the Rhp23 protein was fused at its N-terminus to GFP, encoded by pYZ3N (25). This gfp–rhp23 fusion gene is functional as it restores UV resistance to the rhp23 deletion strain even at low levels of expression (data not shown). As a control, gfp alone was also expressed from the pYZ3N vector in S.pombe cells. As shown in Figure 4A (top), GFP is evenly dispersed throughout the cell. In contrast, even though some of the GFP–Rhp23 fusion protein is present in the cytoplasm, most is located in the nucleus (Fig. 4A, bottom). This predominantly nuclear localization is similar to that reported for budding yeast Rad23 and human HHR23A (2,3).
Figure 4.
Rhp23 is located predominantly in the nucleus, and levels of Rhp23 protein are cell cycle dependent. (A) The top three panels show cells containing a single integrated copy of gfp expressed from the nmt1 promoter under inducing conditions. The left panel is a picture of GFP fluorescence; the middle panel illustrates DNA after staining with Hoechst 33342; the right panel shows a differential interference contrast picture of the cells. The bar in the right panel represents 10 µm. The bottom three panels are of cells containing a single integrated copy of the gfp–rhp23 fusion gene under inducing conditions. (B) Wild-type SP223 cells were synchronized in S phase by treatment with HU, an inhibitor of DNA synthesis. After removal of the drug, immunoblots of extracts prepared every 30 min were probed with antibodies against α-tubulin and Rad23. The anti-Rad23 antibodies cross-react with Rhp23 (see Materials and Methods). The lane on the left labeled – is for an extract prepared from the rhp23 deletion strain, and the two lanes on the right labeled + are two dilutions of an extract prepared from a strain overexpressing rhp23 cDNA. The graph represents the amount of Rhp23 relative to α-tubulin determined from scanning the immunoblot. (C) Levels of GFP–Rhp23 in the nucleus decrease at the time of nuclear division. Confocal images of GFP–Rhp23 were taken every 2 min, and the time of the image is shown in each panel. The arrow indicates the cell undergoing nuclear division during this time period.
The levels of both budding yeast Rad23 and human HHR23A are cell cycle dependent (11,18). To determine whether Rhp23 levels are also cell cycle dependent, wild-type fission yeast SP223 cells were synchronized in S phase by treatment with HU for 4 h as described (http://www.bio.uva.nl/pombe/handbook/section3/section3-3.html). After removing the drug, protein extracts were prepared every 30 min for up to 7 h to allow approximately two rounds of cell division. Progression through the cell cycle was monitored by the septation index, which measures the fraction of cells containing the septum that forms between dividing cells shortly after mitosis around the time of S phase in S.pombe (34). In the HU synchronized cultures, peaks of septation were found around 0, 2.5 and 5.5 h. The levels of Rhp23 protein, measured by immunoblot analysis and standardized to the levels of α-tubulin (Fig. 4B), oscillated with the cell cycle. Rhp23 abundance was lowest at 0, 2.5 and 5.5–6 h, corresponding to S phase, and highest at 2 and 5 h, corresponding to passage through mitosis. Thus, the levels for all three Rad23 protein homologs are lowest near S phase (11,18).
To determine whether the cellular distribution of Rhp23 protein is cell cycle dependent, a GFP–Rhp23 fusion was followed in live cells in a time series of fluorescent microscopy images. In the series taken at 2 min intervals, shown in Figure 4C, the intensity of GFP–Rhp23 in the single nucleus in the lower cell did not change during the duration of the experiment. In contrast, for the upper cell, in which the nucleus divided during the experiment, the amount of GFP–Rhp23 in the nucleus decreased around the time of nuclear division. Thus, at about the time the lowest levels of Rhp23 are seen in a synchronous culture (Fig. 4B), the amount of GFP–Rhp23 in the nucleus decreases relative to the amount in the cytoplasm (Fig. 4C).
Overexpression of rhp23 induces a large increase in ubiquitinated proteins
The correlation of cell cycle phase with Rhp23 levels, and the presence of a UbL and two UBA domains in Rhp23, raises the possibility that the abundance of Rhp23 is regulated by ubiquitination and degradation. Figure 5A shows that Rhp23 is ubiquitinated at low levels. Cell extracts from a wild-type (SP1208) and a mts2 mutant strain (SP1212), which has a temperature-sensitive mutation in a subunit of the proteasome (40,41), were probed with anti-Rad23 antibodies. For cell extracts from the wild-type strain incubated at 25°C or incubated for 4 h at 37°C, only Rhp23 without added ubiquitin was detected (Fig. 5A, lanes 1 and 2). In contrast, in the mts2 mutant strain, higher molecular weight ubiquitinated forms of Rhp23 were detectable at the permissive temperature of 25°C and increased amounts were seen at the restrictive temperature of 37°C (Fig. 5A, lanes 3 and 4). The mts2 mutation is in a subunit of the proteasome and inactivation of the proteasome at the restrictive temperature leads to accumulation of poly-ubiquitinated proteins (40,41). Thus, the detection of ubiquitinated forms of Rhp23 only in the mts2 strain indicates that Rhp23 is ubiquitinated at a low level.
Figure 5.
Rhp23 is ubiquitinated at low levels, and overexpression of rhp23 leads to an accumulation of ubiquitinated proteins. (A) Ubiquitinated forms of Rhp23 are found only in a strain with a mts2 mutation in the proteasome subunit. The molecular weights of size standards (kDa) are indicated to the left of the immunoblot. (B) Overexpression of HA–Rhp23 leads to accumulation of large amounts of ubiquitinated proteins. Cell extracts were prepared from the wild-type strain SP223 without (–) or with (+) a plasmid carrying HA-tagged Rhp23 (HA–Rhp23). Cells were grown in media without thiamine to overexpress the tagged rhp23. Extracts were immunoprecipitated with anti-HA antibodies and the immunoprecipitate (indicated in the IP row by P) and the supernatant (indicated by S) were immunoblotted and probed with anti-ubiquitin (top panel) and anti-Rad23 antibodies (bottom panel). The positions of HA–Rhp23 and the immunoglobin heavy chain (IgHc), which was used as a protein loading control, are indicated. (C) In the left panel, cell extracts from the rhp23::ura4 strain carrying a plasmid with the rhp23, RAD23 or HHR23 cDNA or a vector control were prepared under inducing (+) or repressing (–) conditions for the nmt1 promoter expressing the cDNAs. For all extracts, 120 µg protein per lane were loaded on the gel, except for the induced rhp23 cDNA condition, where dilutions of 120, 12 and 2.4 µg were loaded (lanes 1–3). The upper immunoblot was probed with anti-ubiquitin antibodies. The lower immunoblot was probed with anti-Rad23 antibodies, which cross-react with Rhp23 but do not cross-react with HHR23A. The positions of Rhp23 and the slightly larger Rad23 are indicated. In the right panels, extracts were prepared from the rhp23::ura4 strain carrying a plasmid with the rhp23 cDNA either without (lane 1) or with induction (lanes 2–6). For the induced samples, cycloheximide (Chx) was added to 100 µg/ml at time 0 and samples taken 30, 60, 90 and 120 min later. Immunoblots with 120 µg protein were probed with anti-ubiquitin antibodies (upper panel) and with anti-Rad23 antibodies (lower panel). The samples in the right panel were run on a higher percentage polyacrylamide gel than in the left panel, so the anti-ubiquitin bands appear more compressed in the right panel.
While ubiquitination of Rhp23 was being examined, we found that overexpression of rhp23 leads to a large increase in the abundance of other ubiquitinated proteins. One of these experiments is shown in Figure 5B, where HA-tagged Rhp23 protein was used in an immunoprecipitation experiment. Equal amounts of extracts from cells with or without a plasmid carrying the HA–rhp23 fusion were used in an anti-HA immunoprecipitation and immunoprecipitated proteins were probed with anti-ubiquitin antibody (Fig. 5B). Expression of HA–Rhp23 in the pull-down extracts was confirmed by immunoblot analysis using anti-Rad23 antibodies (bottom). No higher molecular weight forms of HA–Rhp23 were seen in the immunoprecipitate with the anti-ubiquitin antibody, indicating that the levels of ubiquitinated HA–Rhp23 in these extracts from wild-type cells are too low to be detected by this method. Interestingly, however, strong anti-ubiquitin signals were detected in the original extract from cells overproducing HA–Rhp23 (lane 4) compared with those not synthesizing large amounts of this protein (lane 3). Thus, overproduction of HA–Rhp23 leads to a large increase in ubiquitinated proteins even though ubiquitination of HA–Rhp23 itself remains undetectable.
To confirm that overexpression of rhp23 promotes ubiquitination of other cellular proteins, extracts from rhp23::ura4 mutant cells overexpressing S.pombe rhp23, S.cerevisiae RAD23, human HHR23A or cells containing a pYZ1N vector control with no insert were probed with anti-ubiquitin antibodies (Fig. 5C, left upper). Consistent with the results for HA–Rhp23, much stronger anti-ubiquitin signals were detected throughout the lane only when the rhp23 gene was induced [lane 1 compared to lanes 4 (uninduced) and 9 (induced vector control)]. A series of dilutions (lanes 1–3) indicate that ubiquitinated protein abundance increases ∼10-fold when rhp23 is overexpressed. This increased ubiquitination does not prevent colony formation on plates or growth in liquid media, but it may slow growth, since the doubling time after overexpression of rhp23 increases to 5.2 h, compared with 2.8 h for a strain carrying only the vector. In contrast to the results with rhp23, no significant increase in anti-ubiquitin signals was observed when human HHR23A (lanes 7 and 8) or budding yeast RAD23 was overexpressed (lanes 5 and 6), even though high levels of Rad23 were produced (bottom, lane 5). Thus, only overexpression of fission yeast rhp23 leads to a large accumulation of ubiquitinated proteins in S.pombe.
To determine whether the accumulation of ubiquitinated proteins is due to decreased rates of degradation, a cycloheximide (Chx) experiment was performed. This inhibitor prevents the synthesis of new proteins so that the stability of proteins can be observed. When Chx is added to cultures overexpressing rhp23, the amount of ubiquitinated proteins and of Rhp23 remains stable for 2 h (Fig. 5C, right), indicating that under these conditions they are not rapidly degraded. Since ubiquitinated proteins are ordinarily rapidly degraded through the proteasome (4), this stability of the ubiquitinated proteins indicates that overexpression of rhp23 most likely inhibits degradation of ubiquitinated proteins by the proteasome. Similar to the results from the immunoprecipitation experiment (Fig. 5B, upper), no larger forms of Rhp23 are seen after overexpression (Fig. 5C, lower right), indicating that there is no detectable accumulation of ubiquitinated Rhp23. Thus, even though ubiquitination of Rhp23 itself remains undetectable, overproduction of Rhp23 leads to a large increase in ubiquitinated proteins, probably because proteasome activity is inhibited.
Rhp23 binds ubiquitin
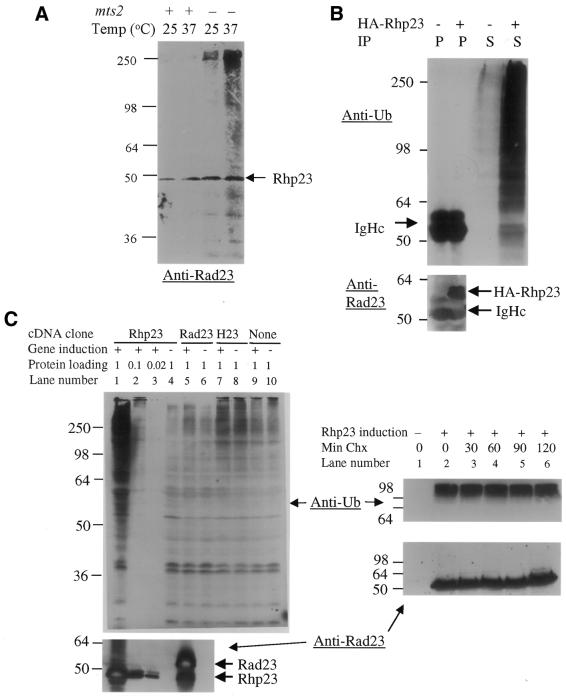
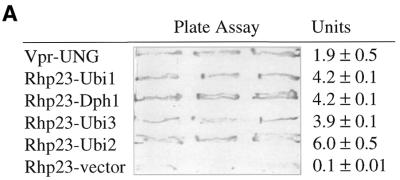
The yeast two-hybrid system (ClonTech, CA) (42) was used to screen for proteins capable of interacting with Rhp23, and ubiquitin was the most frequently identified protein in this screen (Table 2). Starting with 105 transformants from a S.pombe cDNA library, 480 colonies were found to be his+ and to have β-galactosidase activity, indicating that the cDNA encoded a protein interacting with Rhp23. The cDNA insert was identified in 241 of these colonies by DNA sequencing and PCR analysis, and 224 (92%) of these cDNA clones were ubiquitin-encoding genes, of which 205 were ubi3, 12 were ubi1, six were ubi2 and one was dph1. All four of these genes give approximately the same signal in a plate assay for the strength of interaction in the two-hybrid system (Fig. 6A). The ubi1, ubi2 and ubi3 genes have not been characterized previously in fission yeast. The dph1 gene, which is a fission yeast homolog of the budding yeast DSK2 gene, encodes a protein with a UbL at the N-terminal end and a UBA domain at the C-terminal end (43). Budding yeast DSK2 and RAD23 have been shown to interact genetically, with either gene being required for spindle pole body duplication (19), but this is the first report that Dsk2 and Rad23 homologs physically interact. Protein sequence alignment of these four Rhp23-binding proteins (Fig. 6B) indicated that the only common feature is the ubiquitin sequence in the first 76 amino acids, suggesting that Rhp23 binds specifically to ubiquitin.
Table 2. Interaction of Rhp23 with ubiquitin-encoding proteins.
| Clone | Gene designation | Protein accession no. | Putative function | Human/budding yeast homolog | Reference |
|---|---|---|---|---|---|
| Rbp1H9 | ubi1 | CAB16209 | Ub fusion protein | UbBa/Ubi1 | This study |
| Rbp4E9 | ubi2 | CAB55853 | Ub fusion protein | UbBa/Ubi2 | This study |
| Rbp2C7 | ubi3 | CAB11297 | Ub fusion protein | UbBa/Ubi3 | This study |
| Rbp5E3 | dph1 | CAA93239 | Ub-like protein | Chap1/Dsk2 | (43) |
The ubi1 and ubi2 genes encode identical proteins.
aThe first 76 amino acids are nearly identical (98% identity) to human ubiquitin B (see also Fig. 6B).
Figure 6.
Rhp23-binding proteins. (A) Interacting proteins identified in the yeast two-hybrid system. The β-galacatosidase activity of strains carrying the indicated cDNAs in the yeast two-hybrid system was determined in triplicate by a plate assay and a liquid assay (42). The units column shows the mean and standard deviation for three measurements in the liquid assay. The positive control on the top line is HIV-1 Vpr and human UNG, which interact in the two-hybrid system (49). The negative control on the bottom line is rhp23 with the pGAD GH vector (ClonTech, CA) not containing an insert. (B) Alignment of Rhp23-binding proteins. The amino acid sequences were aligned using a multiple sequence alignment Jellyfish program (Biowire). Identical amino acids are in dark color. The highly similar and moderately similar residues are indicated by light color. Consensus amino acids are listed under each residue. Note that the first 76 amino acids, which correspond to the ubiquitin sequence, are the only homologous sequences shared by all four proteins.
DISCUSSION
Fission yeast S.pombe Rhp23 plays a role in promoting resistance to UV light, most likely by participation in NER, since budding yeast Rad23 and human HHR23 function in this DNA repair pathway. A specific role for Rhp23 in NER is further supported by our observations that a rhp23 deletion causes moderate UV sensitivity but does not affect resistance to HU or ionizing radiation (Fig. 1). Indeed, an independent study (44) showed that fission yeast rhp23 partially complements the NER defect in a budding yeast rad23 deletion strain and that Rhp23 is required for both transcription-dependent and transcription-independent NER in fission yeast. We show here that RAD23 and HHR23A also complement the UV sensitivity of a S.pombe rhp23 mutant, although HHR23A needs to be expressed at high levels for full restoration of resistance to wild-type levels (Fig. 2). This complementation provides further support for functional conservation of the Rad23 homologs with respect to NER. There are other similarities among these three Rad23 homologs in that all three show a nuclear localization (Fig. 4A) (3), that the amounts of these proteins are cell cycle dependent (Fig. 4B) (11,18) and that they are ubiquitinated at low levels (Fig. 5A) (11,18). These similarities suggest a high degree of functional conservation for Rad23 homologs.
Besides these similar properties of the Rad23 homologs, there is evidence that they may also play a role in cell cycle G2/M control (Fig. 3). However, S.pombe Rhp23 clearly differs from budding yeast Rad23 in its role in the cell cycle. Schizosaccharomyces pombe with a rhp23 mutation shows a G2 delay (Fig. 3). In contrast, S.cerevisiae rad23 deletion mutants have a normal cell cycle (19), although combinations of the rad23 deletion with other mutations can affect the cell cycle in budding yeast (see below). A potential role for Rad23 in cell cycle control is suggested by our observation that RAD23 partially complements the cell cycle defect in a rhp23 mutant S.pombe strain (Fig. 3B). Cells with mutations in RAD23 homologs are available only in these two yeasts and no human cells with mutations in HHR23A or HHR23B have been reported, so the role of these genes in the human cell cycle cannot currently be determined directly. However, HHR23A fully complements the cell cycle defect of the S.pombe rhp23 mutant even at low levels of expression, suggesting that HHR23A plays an equivalent role in the human cell cycle. Thus, complementation of the cell cycle defect in the S.pombe mutant strain by all of the Rad23 homologs suggests that they play a role in controlling the cell cycle of their respective organisms.
Rhp23 also plays a role in the ubiquitination–proteasome pathway since overexpression leads to a large increase in the abundance of ubiquitinated proteins (Fig. 5B and C). This increase in ubiquitinated proteins is most likely due to inhibition of proteasome activity since the level of ubiquitinated proteins is not affected by inhibition of protein synthesis (Fig. 5C, right upper). There are two simple models based on the properties of Rad23 homologs that might explain the increase in ubiquitinated proteins. The first model is based on the binding of Rad23 homologs to ubiquitin (Fig. 6; 6,17,45). By interacting with ubiquitin, Rhp23 may inhibit degradation through the proteasome, by preventing either the assembly of multi-ubiquitin chains, as suggested by Ortolan et al. (17), or their recognition by the proteasome. While the ubiquitin-binding property of Rad23 homologs may be part of the reason for the increase in ubiquitinated proteins after rhp23 overexpression, it seems unlikely to be a complete explanation, since overexpression of RAD23 or HHR23A did not lead to an increase in ubiquitinated proteins (Fig. 5C, left upper). The second model is based on an interaction between the proteasome and the UbL domain of Rad23 homologs (11,12,15,46). If the interaction with Rhp23 inhibits proteasome activity and this interaction is species specific, this would explain why in fission yeast only overexpression of rhp23 leads to an increase in ubiquitinated proteins. Clearly, further work is required to resolve the roles of ubiquitin binding and the UbL interaction with the proteasome in the increased ubiquitination seen after rhp23 is overexpressed.
The relationships between the NER-, cell cycle- and ubiquitin–proteolysis-related roles of Rad23 homologs remain to be clearly established, but their function in NER and the cell cycle are likely to be independent. First, the G2 delay seen in the rhp23 deletion is unique to this gene and is not characteristic of mutations in NER genes (9). Specifically, the rad9 strain, which is much more sensitive to UV than the rhp23 mutant (Fig. 1), does not show any unusually long cells (data not shown). Second, expression of HHR23A at a low level fully complements the cell cycle defect of the rhp23 deletion strain (Fig. 3) but complements UV sensitivity weakly (Fig. 2).
There are a number of reasons, however, to think that the roles of Rhp23 in the cell cycle and in ubiquitin–proteolysis are related. One reason is that a double rad23 rpn10 mutation in S.cerevisiae (the latter gene encodes a subunit of the proteasome) caused a pronounced G2/M delay (16). Another double mutation combining alterations in rad23 and dsk2, a gene that like RAD23 homologs contains UbL and UBA domains, also causes a G2/M arrest (19). Both of these double mutation combinations suggest that Rad23 plays a role in the cell cycle of budding yeast, but that this cell cycle function overlaps with activities of other budding yeast genes. Also, since the other gene (rpn10 and dsk2) in both cases has some connection to the ubiquitination pathway (16,47,48), these results with budding yeast Rad23 suggest that the cell cycle function is related to the ubiquitination pathway. A second reason for thinking that the cell cycle and ubiquitin–proteasome functions of Rhp23 are related is that ubiquitination is known to have a number of roles in the cell cycle. For example, degradation of the cell cycle regulator Cdc25 through ubiquitin-mediated proteolysis affects the cell cycle in fission yeast (41). Thus, it seems likely that the cell cycle and ubiquitin–proteasome functions of Rad23 homologs will turn out to be related in some way.
In summary, S.pombe rhp23 has a role in NER but also functions in the cell cycle and the ubiquitination pathway. Saccharomyces cerevisiae RAD23 and human HHR23A complement the UV sensitivity and cell cycle defects of the rhp23 deletion mutant, suggesting that these are common features of Rad23 homologs. However, only overexpression of rhp23 leads to accumulation of ubiquitinated proteins in S.pombe, indicating that some aspect of this effect is species specific. As S.pombe Rhp23 plays roles in NER, the cell cycle and ubiquitination, this organism serves as an excellent model system for defining the relationships and mechanisms underlying these multiple roles of Rad23 homologs.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Karen Chiu, Cori Green, Kelly Burger, Erin Zhang and Patrick Reed for technical assistance, Susan Forsburg for the pSF173 vector and K. Madura for the anti-Rad23 antibodies. This research was supported in part by grants from the Medical Research Institute Council, Medical Research Junior Board Foundation, John Lloyd Foundation, Illinois Department of Public Aid and National Institutes of Health grants AI40891 (Y.Z.) and GM52493 (H.B.L.).
DDBJ/EMBL/GenBank accession no. AF174293
REFERENCES
- 1.Sturm A. and Lienhard,S. (1998) Two isoforms of plant RAD23 complement a UV-sensitive rad23 mutant in yeast. Plant J., 13, 815–821. [DOI] [PubMed] [Google Scholar]
- 2.van der Spek P.J., Eker,A., Rademakers,S., Visser,C., Sugasawa,K., Masutani,C., Hanaoka,F., Bootsma,D. and Hoeijmakers,J.H. (1996) XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res., 24, 2551–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins J.F., Sung,P., Prakash,L. and Prakash,S. (1993) The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol. Cell. Biol., 13, 7757–7765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka K., Suzuki,T. and Chiba,T. (1998) The ligation systems for ubiquitin and ubiquitin-like proteins. Mol. Cell, 8, 503–512. [PubMed] [Google Scholar]
- 5.Hofmann K. and Bucher,P. (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci., 21, 172–173. [PubMed] [Google Scholar]
- 6.Bertolaet B.L., Clarke,D.J., Wolff,M., Watson,M.H., Henze,M., Divita,G. and Reed,S.I. (2001) UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nature Struct. Biol., 8, 417–422. [DOI] [PubMed] [Google Scholar]
- 7.Withers-Ward E.S., Mueller,T.D., Chen,I.S. and Feigon,J. (2000) Biochemical and structural analysis of the interaction between the UBA(2) domain of the DNA repair protein HHR23A and HIV-1 Vpr. Biochemistry, 39, 14103–14112. [DOI] [PubMed] [Google Scholar]
- 8.Dieckmann T., Withers-Ward,E.S., Jarosinski,M.A., Liu,C.F., Chen,I.S. and Feigon,J. (1998) Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nature Struct. Biol., 5, 1042–1047. [DOI] [PubMed]
- 9.Prakash S. and Prakash,L. (2000) Nucleotide excision repair in yeast. Mutat. Res., 451, 13–24. [DOI] [PubMed] [Google Scholar]
- 10.Jansen L.E., Verhage,R.A. and Brouwer,J. (1998) Preferential binding of yeast Rad4.Rad23 complex to damaged DNA. J. Biol. Chem., 273, 33111–33114. [DOI] [PubMed] [Google Scholar]
- 11.Schauber C., Chen,L., Tongaonkar,P., Vega,I., Lambertson,D., Potts,W. and Madura,K. (1998) Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature, 391, 715–718. [DOI] [PubMed] [Google Scholar]
- 12.Russell S.J., Reed,S.H., Huang,W., Friedberg,E.C. and Johnston,S.A. (1999) The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol. Cell, 3, 687–695. [DOI] [PubMed] [Google Scholar]
- 13.Berneburg M. and Lehmann,A.R. (2001) Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet., 43, 71–102. [DOI] [PubMed]
- 14.Masutani C., Araki,M., Sugasawa,K., van der Spek,P.J., Yamada,A., Uchida,A., Maekawa,T., Bootsma,D., Hoeijmakers,J.H. and Hanaoka,F. (1997) Identification and characterization of XPC-binding domain of hHR23B. Mol. Cell. Biol., 17, 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiyama H., Yokoi,M., Masutani,C., Sugasawa,K., Maekawa,T., Tanaka,K., Hoeijmakers,J.H. and Hanaoka,F. (1999) Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J. Biol. Chem., 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- 16.Lambertson D., Chen,L. and Madura,K. (1999) Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics, 153, 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortolan T.G., Tongaonkar,P., Lambertson,D., Chen,L., Schauber,C. and Madura,K. (2000) The DNA repair protein Rad23 is a negative regulator of multi-ubiquitin chain assembly. Nature Cell Biol., 2, 601–608. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S., Talis,A.L. and Howley,P.M. (1999) Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem., 274, 18785–18792. [DOI] [PubMed] [Google Scholar]
- 19.Biggins S., Ivanovska,I. and Rose,M.D. (1996) Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol., 133, 1331–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gragerov A., Kino,T., Ilyina-Gragerova,G., Chrousos,G.P. and Pavlakis,G.N. (1998) HHR23A, the human homologue of the yeast repair protein RAD23, interacts specifically with Vpr protein and prevents cell cycle arrest but not the transcriptional effects of Vpr. Virology, 245, 323–330. [DOI] [PubMed] [Google Scholar]
- 21.Withers-Ward E.S., Jowett,J.B., Stewart,S.A., Xie,Y.M., Garfinkel,A., Shibagaki,Y., Chow,S.A., Shah,N., Hanaoka,F., Sawitz,D.G., Armstrong,R.W., Souza,L.M. and Chen,I.S. (1997) Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J. Virol., 71, 9732–9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Cao,J., O’Gorman,M.R.G., Yu,M. and Yogev,R. (1996) Effect of human immunodeficiency virus Type 1 protein R (vpr) gene expression on basic cellular functions of fission yeast Schizosaccharomyces pombe. J. Virol., 70, 5821–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fantes P. (1977) Control of cell size and cycle time in Schizosaccharomyces pombe. J. Cell Biol., 24, 51–67. [DOI] [PubMed] [Google Scholar]
- 24.Maundrell K. (1993) Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene, 123, 127–130. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y., Elder,R.T., Chen,M. and Cao,J. (1998) Fission yeast expression vectors adapted for large scale cloning and GFP fusion with positive screening. Biotechniques, 25, 438–444. [DOI] [PubMed] [Google Scholar]
- 26.Keeney J.B. and Boeke,J.D. (1994) Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics, 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding D.Q., Chikashige,Y., Haraguchi,T. and Hiraoka,Y. (1998) Oscillatory nuclear movement in fission yeast meiotic prophase is driven by astral microtubules, as revealed by continuous observation of chromosomes and microtubules in living cells. J. Cell Sci., 111, 701–712. [DOI] [PubMed] [Google Scholar]
- 28.Chang F. (2000) Microtubule and actin-dependent movement of the formin Cdc12p in fission yeast. Microsc. Res. Tech., 49, 161–167. [DOI] [PubMed]
- 29.Forsburg S.L. and Sherman,D.A. (1997) General purpose tagging vectors for fission yeast. Gene, 191, 191–195. [DOI] [PubMed] [Google Scholar]
- 30.Okazaki K., Okazaki,N., Kume,K., Jinno,S., Tanaka,K. and Okayama,H. (1990) High-frequency transformation method and library transducing vectors for cloning mammalian cDNAs by trans-complementation of Schizosaccharomyces pombe. Nucleic Acids Res., 18, 6485–6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y. and Lu,Y. (1996) Yeast DNA transformation. In Li,Y.M. and Zhao,Y. (eds), Practical Protocols in Molecular Biology. Science Press, New York, NY, pp. 61–63.
- 32.Rothstein R. (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol., 194, 281–301. [DOI] [PubMed] [Google Scholar]
- 33.Kaur R., Ingavale,S.S. and Bachhawat,A.K. (1997) PCR-mediated direct gene disruption in Schizosaccharomyces pombe. Nucleic Acids Res., 25, 1080–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Localization of cell organelles with fluorescent probes: Calcofluor. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 19–22.
- 35.Alfa C.E. and Hyams,J.S. (1990) Distribution of tubulin and actin through the cell division cycle of the fission yeast Schizosaccharomyces japonicus var. versatilis: a comparison with Schizosaccharomyces pombe. J. Cell Sci., 96, 71–77. [DOI] [PubMed] [Google Scholar]
- 36.Lieberman H.B., Hopkins,K.M., Laverty,M. and Chu,H.M. (1992) Molecular cloning and analysis of Schizosaccharomyces pombe rad9, a gene involved in DNA repair and mutagenesis. Mol. Gen. Genet., 232, 367–376. [DOI] [PubMed] [Google Scholar]
- 37.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) General introduction to S. pombe. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–11.
- 38.Alfa C., Fantes,P., Hyams,J., McLeod,M. and Warbrick,E. (1993) Nutritional control of entry into stationary phase determined by flow fluorocytometry. Experiments with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 52–56.
- 39.Marks J., Hagan,I.M. and Hyams,J.S. (1986) Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. Suppl., 5, 229–241. [DOI] [PubMed] [Google Scholar]
- 40.Gordon C., McGurk,G., Dillon,P., Rosen,C. and Hastie,N.D. (1993) Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature, 366, 355–357. [DOI] [PubMed] [Google Scholar]
- 41.Nefsky B. and Beach,D. (1996) Pub1 acts as an E6-AP-like protein ubiquitiin ligase in the degradation of Cdc25. EMBO J., 15, 1301–1312. [PMC free article] [PubMed] [Google Scholar]
- 42.Fields S. and Song,O. (1989) A novel genetic system to detect protein-protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- 43.He X., Jones,M.H., Winey,M. and Sazer,S. (1998) Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell Sci., 111, 1635–1647. [DOI] [PubMed] [Google Scholar]
- 44.Lombaerts M., Goeloe,J.I., den Dulk,H., Brandsma,J.A. and Brouwer,J. (2000) Identification and characterization of the rhp23(+) DNA repair gene in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun., 268, 210–215. [DOI] [PubMed] [Google Scholar]
- 45.Clarke D.J., Mondesert,G., Segal,M., Bertolaet,B.L., Jensen,S., Wolff,M., Henze,M. and Reed,S.I. (2001) Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Mol. Cell. Biol., 21, 1997–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T., Park,H., Kwofie,M.A. and Lennarz,W.J. (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26 S proteasome in yeast. J. Biol. Chem., 276, 21601–21607. [DOI] [PubMed] [Google Scholar]
- 47.Kleijnen M.F., Shih,A.H., Zhou,P., Kumar,S., Soccio,R.E., Kedersha,N.L., Gill,G. and Howley,P.M. (2000) The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell, 6, 409–419. [DOI] [PubMed] [Google Scholar]
- 48.Funakoshi M., Geley,S., Hunt,T., Nishimoto,T. and Kobayashi,H. (1999) Identification of XDRP1; a Xenopus protein related to yeast Dsk2p binds to the N-terminus of cyclin A and inhibits its degradation. EMBO J., 18, 5009–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bouhamdan M., Benichou,S., Rey,F., Navarro,J.M., Agostini,I., Spire,B., Camonis,J., Slupphaug,G., Vigne,R., Benarous,R. and Sire,J. (1996) Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J. Virol., 70, 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.