Abstract
Exosomes are small extracellular vesicles with a lipid bilayer structure secreted from different cell types which can be found in various body fluids including blood, pleural fluid, saliva and urine. They carry different biomolecules including proteins, metabolites, and amino acids such as microRNAs which are small non-coding RNAs that regulate gene expression and promote cell-to-cell communication. One main function of the exosomal miRNAs (exomiRs) is their role in cancer pathogenesis. Alternation in exomiRs expression could indicate disease progression and can regulate cancer growth and facilitate drug response/resistance. It can also influence the tumour microenvironment by controlling important signaling that regulating immune checkpoint molecules leading to activation of T cell anti-tumour immunity. Therefore, they can be used as potential novel cancer biomarkers and innovative immunotherapeutic agents. This review highlights the use of exomiRs as potential reliable biomarkers for cancer diagnosis, treatment response and metastasis. Finally, discuses their potential as immunotherapeutic agents to regulate immune checkpoint molecules and promote T cell anti-tumour immunity.
Keywords: exosomal miRNA, extracellular vesicles (EVs), cancer, small non-coding RNAs (sncRNAs), biomarker, clinical implication, liquid bioposy, immune checkpoint molecules
1 Introduction
Extracellular vesicles (EVs) were initially described by Harding in 1983 (Harding et al., 1983) and later confirmed by Pan in 1985 (Pan et al., 1985). At first, they were known as vehicles for clearance of cellular “waste” which results from cell metabolism with no influence on neighboring cells. This concept, however, switched after the finding of other biomolecules, e.g., amino acids, fatty acid, and nucleic acids including small RNAs particularly microRNA in 2007 (Théry et al., 2002; Valadi et al., 2007). Upon release from cells, they can circulate to the neighboring cells and internalized via endocytosis (Larrea et al., 2016) and ultimately result in cell-to-cell communication and contribute to reprogram the recipient cells (Meldolesi, 2018). Thus, exosomal microRNAs (exomiRs) play a major role in intercellular communication to regulate gene expression (Théry, 2011; Kozomara and Griffiths-Jones, 2014). All type of cells including cancer cells can naturally secrete exosomes (Théry et al., 2002) in which can be isolated from different bio-fluids including urine and serum. This secretion is a result of cells undergoing difference condition such as apoptosis/necrosis or chronic inflammation which suggest a possible source of less-invasive method of the so-called “liquid biopsy”.
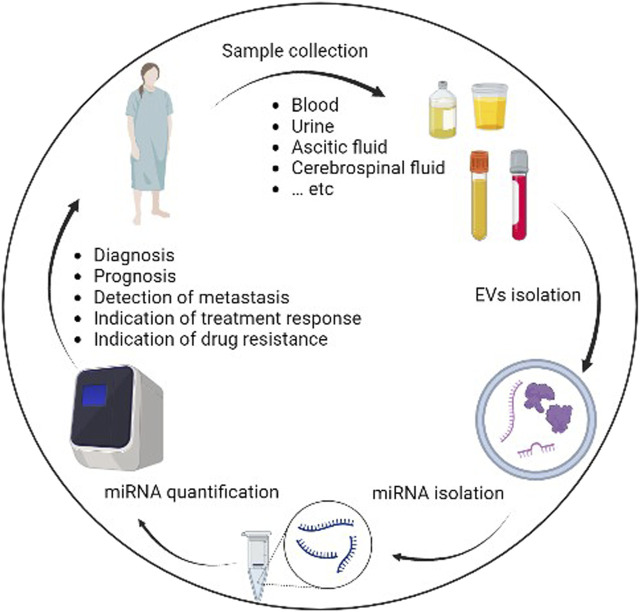
ExomiRs can be isolated from variety of body fluid including blood, saliva, urine, and breast milk (Figure 1) by differential ultracentrifugation (Amorim et al., 2017) which results in separation of exomiRs from contaminated cells, cellular debris and other EVs subtype such as apoptotic bodies and microparticles (Théry et al., 2006). Further techniques based on size exclusion such as chromatography and Optiprep™ density gradient have been used to increase the purity of exomiRs isolation (Boing et al., 2014; Lobb et al., 2015). In 2014, the International Society for EVs has published recommendations for EVs definition and their functions (Lotvall et al., 2014). Protein expression for at least three EV markers (e.g., CD63, CD81 and CD9) is typically used to identify the purity of the sample (Ekström et al., 2022). Furthermore, the use of two techniques (e.g., electron microscopy and nanoparticle tracking analysis) are recommended to illustrate the degree of heterogeneity in the sample (Dragovic et al., 2011; Van der Pol et al., 2014). To help standardize the protocols, EV-TRACK knowledgebase project (Can be access at http://evtrack.org) was developed and conducted by international collaboration from several countries (Van Deun et al., 2017). This helps researchers to add methodological parameters into central repository to quantifies their protocol. It is recommended to follow protocol based on the biofluid resources (Ramirez et al., 2018). For example, when working with blood as a source for the exomiRs, challenges like hemolysis has been shown to decrease the expression of some microRNAs and agitation used to stimulate blood transportation can lead to release of platelets (Ramirez et al., 2018). Another important factor is the storage time which can affect exomiRs yield (Ramirez et al., 2018). Comparison study shows increase amount of exomiRs isolated from the blood after storage for 3 h compared to freshly isolated blood which can be due to platetets-drived EVs (Ramirez et al., 2018).
FIGURE 1.
Workflow of processing and utilization of exomiRs for cancer clinical implication.
In cancer, alternation in exomiRs expression is a well-known feature in cancer (Alotaibi, 2021). It can bind to messenger RNA which results in posttranscriptional gene regulation that promote cancer cell processes including progression and metastasis (Lin and Gregory, 2015). This process is critical for carcinogenesis, which help cancer cells thrive in the tumour microenvironment. For example, crosstalk between cancer cell and surrounding cells including immune cells and stromal cells leading to pre-metastatic niche development. Therefore, investigating exomiRs secreted from cancer cells is important to reveal cancer behavior and metastasis and understand their potential role as cancer biomarkers and to develop novel immunotherapeutic agents for cancer. This review describes the use of exomiRs as potential biomarkers for cancer diagnosis/prognosis, treatment response, and cancer metastasis and explore their role as innovative immunotherapeutic agents for cancer.
2 ExomiRs as novel biomarkers for cancer
Early treatment intervention for cancer can positively impact overall survival and result in desirable treatment outcome (Hiom, 2015). The standard method to confirm diagnosis is usually invasive biopsy of suspected tissue. Although is reliable, such a method can be difficult to perform due to tissue inaccessibility and possible damage to the normal tissue with the risk to stimulate metastasis (Shyamala et al., 2014). Therefore, using less invasive and effective method is important to improve early cancer diagnosis and predict treatment response and metastasis. Changes in levels of exomiRs can be noticed before the patients can develop clear symptoms for cancer and during cancer development (Chen et al., 2008a). Screening variant expression of tissue-specific exomiRs isolated from various body fluids are shown to prove the diagnosis and prognosis of different type of cancer (Table 1). Predict treatment response (Table 2) and metastasis (Table 3).
TABLE 1.
List of exomiRs used for cancer diagnosis.
| Centralize the miRNAs ID | Pattern of expression | Cancer type | Tumor stage | Body fluid source | References |
|---|---|---|---|---|---|
| ExomiR-92 | Increased | Colorectal cancer | All stages | Plasma and tissue samples | Ng et al. (2009) |
| ExomiR-17-5p and exomiR-92a-3p | Increased | Colorectal cancer | Higher clinical stages | Serum | Fu et al. (2018) |
| ExomiR-17-5p, exomiR-21, exomiR-106a and exomiR-106b | Increased | Gastric cancer | Stages I-IV | Plasma | Tsujiura et al. (2010) |
| ExomiR-21, exomiR-155, exomiR-210 and exomiR-196a | Increased | Pancreatic adenocarcinoma patients | All stages | Plasma | Wang et al. (2009) |
| ExomiR-500 | Increased | Hepatocellular carcinoma | Not mentioned | Serum | Yamamoto et al. (2009) |
| ExomiR-184 | Increased | Squamous-cell carcinoma of the tongue | Not mentioned | Plasma | Wong et al. (2008) |
| ExomiR-125a and exomiR-200a | Decreased | Oral squamous-cell carcinoma | Stages I-IV | Saliva | Park et al. (2009) |
| 63 exomiRs | Increased | Non-small-cell lung cancer | Stages I-IV | Serum | (Chen et al., 2008a) |
| 34 exomiRs | Increased | Asymptomatic NSCLC | Early-stage nodule (Ia or Ib) | Serum | Bianchi et al. (2011) |
| 21 exomiRs | Increased | Lung cancer | 12–28 before and at the time of diagnosis | Plasma | Boeri et al. (2011) |
| hsa-miR-212, -214, −205, −210, −203, −191, −192, −146, −155, −21, −106a and -17-3p | Increased | Lung adenocarcinoma | Stages I-IV | Tissues biopsy | Rabinowits et al. (2009) |
| ExomiR-200-5p, exomiR-379, exomiR-139-5p and exomiR-378a | Increased | Lung adenocarcinoma | Early-stage nodule (Ia or Ib) | Tissue biopsy and plasma | Cazzoli et al. (2013) |
| ExomiR-141 and other 15 exomiRs | Increased | Prostate cancer | Stage 3 and 4 | Serum | Lodes et al. (2009) |
| ExomiR-21-5p, exomiR-574-3p, and exomiR-141-5p | Increased | Prostate cancer | Not mentioned | Urine | Samsonov et al. (2016) |
| ExomiR-92, exomiR-93 and exomiR-126 | Increased | Epithelial ovarian cancer | Stages I-IV | Serum | Resnick et al. (2009) |
| ExomiR-21, exomiR-141, exomiR-200a, exomiR-200c, exomiR-200b, exomiR-203, exomiR-205 and exomiR-214) | Increased | Ovarian cancer | Various stages | Serum | Taylor and Gercel-Taylor (2008) |
| ExomiR-195 | Increased | Breast cancer | Stage IV | Serum, plasma, or whole blood | Heneghan et al. (2010) |
| ExomiR-101 and exomiR-372 | Increased | Breast cancer and triple-negative breast cancer | pT1 pT2-4 |
Serum | Eichelser et al. (2014) |
| ExomiR-199a-3p | Increased | Pediatric neuroblastoma | Not mentioned | Plasma | Ma et al. (2019) |
| ExomiR-16 | Increased | Pediatric acute lymphoblastic leukemia | Not mentioned | Blood | Kaddar et al. (2009) |
| ExomiR-21 | Increased | Pediatric Hepatoblastoma | Not mentioned | Plasma | Liu et al. (2016b) |
| ExomiR-7112-5p, exomiR-885-3p and exomiR-1245a | Increased | Pediatric acute myeloid leukaemia | Risk of disease recurrence | Plasma | Zampini et al. (2017) |
| ExomiR-25-3p | Increased | Osteosarcoma | Not mentioned | Serum | Fujiwara et al. (2017) |
| ExomiR-125b | Decreased | Ewing’s sarcoma | Metastasis and non-metastasis | Serum | Nie et al. (2015) |
| ExomiR-21 | Increased | Diffuse large B cell lymphoma | Stages I-IV | Serum | Lawrie et al. (2008) |
| ExomiR-92a | Decreased | Acute leukemias | Not mentioned | Plasma | Tanaka et al. (2009) |
| ExomiR-148a, exomiR-181a, exomiR-20a, exomiR-221, exomiR-625, and exomiR-9 | Increased | Multiple myeloma | Not mentioned | Plasma | Huang et al. (2012a) |
| ExomiR-32, exomiR-98 and exomiR-374 | Decreased | Chronic lymphocytic leukemia | Not mentioned | Blood | Rahimi et al. (2021) |
| ExomiR-451 | Increased | Chronic myelogenous leukemia | Chronic stage | Plasma | Keramati et al. (2021) |
TABLE 2.
List of exomiRs associated with drug-response.
| Centralize the miRNAs ID | Pattern of expression | Cancer type | References |
|---|---|---|---|
| ExomiR-181b | Increased | Better response to 5-fluorouracil in Colorectal cancer | Nakajima et al. (2006) |
| ExomiR-140 and ExomiR-215 | Increased | Resistance to methotrexate, 5-fluorouacil, and Tomudex in human osteosarcoma and colon cancer cells | Song et al. (2009), Song et al. (2010) |
| ExomiR-19a | Increased | FOLFOX resistance in Advanced Colorectal Cancer Cases | Chen et al. (2013) |
| ExomiR-155 | Increased | Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma | Mikamori et al. (2017) |
| ExomiR‐34a | Decreased Increased |
Docetaxel resistance in castration-resistant prostate cancer Increased sensitivity to sorafenib in hepatocellular carcinoma cell lines |
Corcoran et al. (2014)
(Yang et al., 2014) |
| ExomiR-21 | Increased | Cisplatin resistance in gastric cancer (In mice) | Zheng et al. (2017) |
| ExomiR-122 | Decreased | Resistance to taxol in liver cancer | Sun et al. (2016) |
| ExomiR-128b | Loss of expression | Better response to gefitinib in non-small cell lung cancer | Weiss et al. (2008) |
| ExomiR-21 | Increased | Resistant to docetaxel-based chemotherapy in prostate cancer | Zhang et al. (2011a) |
| ExomiR-9 | Increased | Increase sensitivity of ovarian cancer to DNA damaging chemotherapy | Sun et al. (2013) |
| ExomiR‐125 b | Deletion | Better respond to chemotherapy with anthracycline in breast cancer | Climent et al. (2007) |
| ExomiR-195, ExomiR-455-3p, and ExomiR-10a miR-221 |
Increased Increased |
Temozolomide resistance in glioma |
Ujifuku et al. (2010)
Yang et al. (2017a) |
| ExomiR-29a and exomiR-100 | Increased | Drug resistance in pediatric acute promyelocytic leukemia | Zhang et al. (2011b) |
| ExomiR-142-3p and exomiR-17-92 | Increased | Glucocorticoid-resistant B cell precursor acute lymphoblastic leukemia | Sakurai et al. (2019) |
| ExomiR-99a, exomiR-100, and exomiR-125b | Increased | Resistance to daunorubicin and vincristine in pediatric acute lymphoblastic leukemia | Schotte et al. (2011) |
| ExomiR-142-5p, exomiR-199b, exomiR-217, exomiR-221, and exomiR-365a-3p | Decreased | Tyrosine kinase inhibitors resistance in chronic myelogenous leukemia | Yeh et al. (2016), Jiang et al. (2018), Klümper et al. (2020) |
| ExomiR-145-3p and exomiR-155 | Decreased | Bortezomib resistance in multiple myeloma | Amodio et al. (2019), Wu et al. (2020) |
| ExomiR-217 | Increased | Sensitizes AML to doxorubicin | Xiao et al. (2017) |
| ExomiR-143 | Increased | Enhances chemosensitivity of acute myeloid leukaemia to cytosine arabinoside | Zhang et al. (2020a) |
| ExomiR-181a/b | Increased | Fludarabine response in chronic lymphocytic leukemia | Zhu et al. (2012) |
TABLE 3.
List of exomiRs associated with tumour recurrence.
| Centralize the miRNAs ID | Pattern of expression | Tumour type | References |
|---|---|---|---|
| ExomiR-106a-5p | Increased | Gastric cancer metastasis | Yuan et al. (2016) |
| ExomiR-4772-3p | Increased | Recurrence in stage II and III colon cancer | Liu et al. (2016a) |
| ExomiR-19a | Increased | Recurrence in Colorectal cancer | Matsumura et al. (2015) |
| ExomiR-3653 | Increased | Pancreatic neuroendocrine tumour | Gill et al. (2019) |
| ExomiR-1307-5p and exomiR-103 | Increased | Hepatocellular carcinoma | Fang et al. (2018a), Eun et al. (2020) |
| ExomiR-1247-3p | Increased | Liver cancer | Fang et al. (2018b) |
| ExomiR-718 | Decreased | Recurrence in hepatocellular carcinoma | Sugimachi et al. (2015) |
| ExomiR-21 | Increased | Esophageal squamous cell cancer | Tanaka et al. (2013) |
| ExomiR-497-5p | Decreased | Non-small cell lung cancer | Huang et al. (2019) |
| ExomiR-141, exomiR-146b-3p and exomiR-194 | Increased | Recurrence in prostate cancer | Selth et al. (2013) |
| ExomiR-275 | Increased | Recurrence in bone marrow In prostate cancer |
Jiang et al. (2022) |
| ExomiR-148a-3p | Decreased | Lymph node metastasis in ovarian cancer | Gong et al. (2016) |
| ExomiR-21 | Increased | Recurrence in glioma | Shi et al. (2015) |
| ExomiR‐375 | Increased | Bone marrow metastases in patients with neuroblastoma | Colletti et al. (2020) |
2.1 Gastrointestinal tumors
In a study of colorectal cancer (CRC), a set of exomiRs including exomiR-92 was significantly overexpressed in plasma and tissue samples (Ng et al., 2009). This study has also suggested that exomiR-92 can be used as potential biomarker to detect colorectal cancer since it was differentially expressed in patients with colorectal cancer compared to gastric cancer (Ng et al., 2009). In another study of CRC patients, there were significant increased in exomiR-92a-3p and exomiR-17-5p levels in serum samples and this increase correlates with the stage and grade of the cancer (Fu et al., 2018) which suggest the value of exomiR not only as diagnostic biomarker but as prognostic biomarker. A study on gastric cancer reported significant levels of exomiR-17-5p, exomiR-21, exomiR-106a and exomiR-106b in plasma from patients with gastric cancer compared to healthy individuals (Tsujiura et al., 2010). Expression analysis of exomiR profile including exomiR-21, exomiR-155, exomiR-210 and exomiR-196a isolated from plasma is shown to be associated with pancreatic adenocarcinoma patients (Wang et al., 2009). ExomiR-210 from plasma was also altered in two independent cohorts with pancreatic cancer (Ho et al., 2010). Increase expression of exomiR-500 was observed in serum of patients with hepatocellular carcinoma (HCC) (Yamamoto et al., 2009). Expression levels of exomiR-184 in plasma were significantly higher in patients with squamous-cell carcinoma of the tongue compared to healthy individuals and its levels decreased significantly after tumour removal (Wong et al., 2008). Although the majority of the studies focused on circulating exomiRs in serum and plasma, further studies have investigated the potential use of exomiRs as diagnostic/prognostic biomarker for cancer in other body fluids. For example, expression levels of exomiR-125a and exomiR-200a in saliva from patients with oral squamous-cell carcinoma were significantly reduced compared to healthy individuals (Park et al., 2009).
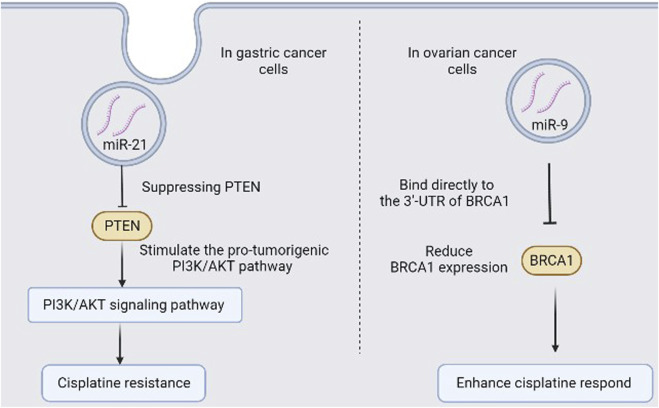
Chemotherapy is one of the most common therapeutic approaches to treat cancer. Successful response to initial therapy is often dependent on type of treatment and tumour type and can be identified by disease progression while resistance to drugs often accompanies with recurrence of tumour. The mechanisms of how cancer can resist treatment have been previously identified (Zahreddine and Borden, 2013) and exomiRs mediating intercellular communication have been identified as one of the mechanisms (Maacha et al., 2019). Tumour cells in their microenvironment can exchange genetic materials and mediate intracellular communications through secreted exomiRs which can promote tumour progression (Lin and Gregory, 2015). In CRC study, exomiR-181b was over-expressed in tumour biopsy compared to normal tissues and was associated with response to 5-fluorouracil treatment (Nakajima et al., 2006). Colon cancer cells and human osteosarcoma cells with elevated levels of exomiR-140 and exomiR-215 have been shown resistance to methotrexate, 5-fluorouacil, and Tomudex (Song et al., 2009; Song et al., 2010). FOLFOX chemotherapy is usually giving to patients with advanced CRC as first-line treatment, and half of the patients acquire resistance with no reliable approach to predict resistance. ExomiR-19a was noticed to be upregulated in patients serum with FOLFOX-resistance and further analysis showed that exomiR-19a can predict acquired drug resistance (Chen et al., 2013) which suggest the use of serum exomiR-19a as a potential biomarker to predict resistance in FOLFOX for advanced CRC patients. Gemcitabine (GEM), a common chemotherapy drug used to treat cancer patients, with promising results, but cancer patients often develop resistance after going long-term treatment. Alternation in specific exomiRs level may play a role since long-term administration with GEM has been shown to associate with increase exomiR-155 level which mediate anti-apoptotic activity that led to chemoresistance in pancreatic ductal adenocarcinoma (Mikamori et al., 2017). Therefore, exomiR-155 could predict GEM-resistance and could be used as novel therapeutic target for GEM treatment in pancreatic ductal adenocarcinoma, based on its function as a driver of resistance. Docetaxel can induce tumour cell apoptosis through Bcl-2 which can be regulated by exomiR-34a (Corcoran et al., 2014). Increasing exomiR-34a level can reduce cell viability and enhance hepatocellular carcinoma cell lines sensitivity to sorafenib through reduced Bcl-2 expression (Yang et al., 2014). When exomiRs internalized through endocytosis in tumour cells, exomiRs can regulate their response to cell signals (Hannafon and Ding, 2013) and inhibit cell apoptosis by suppressing PTEN and stimulate the pro-tumorigenic PI3K/AKT pathway (Zheng et al., 2017). Such a process can promote cisplatin resistance (Radisavljevic, 2013) (Figure 2). In addition, elevated level of exomiR-122 in patients could predict better response to taxol as suppressing exomiR-122 can lead to increase septin-9 in liver cancer which correlates with resistance to taxol (Sun et al., 2016). Hence, exomiRs can mediate communication to tumour cells, tumour progression and resistance to drugs (Table 2).
FIGURE 2.
Schematic for two examples of exomiRs in drug response/drug resistance in cancer.
Metastasis can result in tumour recurrence and plays a key role in reducing survival rate (Gandaglia et al., 2015). Currently, there are no approaches to predict recurrence of tumour at any stage of the disease. Several exomiRs shave been shown to associate with different stages of cancer (Calin and Croce, 2006), therefore, they could be used as potential biomarkers to predict recurrence of tumour (Table 3). Increase levels of exomiR-106a-5p in gastric cancer patients is correlated with the potential to promote metastases (Yuan et al., 2016). In colon cancer patients diagnose at stage II and III, exomiR-4772-3p levels have been associated with elevated risk of tumour recurrence and reduced overall survival (Liu et al., 2016a), and a increased expression of exomiR-19a has been associated with tumour recurrence in colorectal cancer (Matsumura et al., 2015). In pancreatic neuroendocrine tumour study, upregulation of exomiR-3653 was associated with high risk of tumour recurrence through interaction with ATRX (Gill et al., 2019). Circulating exomiR-1307-5p was reported to promote tumour metastasis in HCC through promoting epithelial–mesenchymal transition (EMT) (Eun et al., 2020). ExomiR-103 can reduce the integrity of endothelial cell junction and promote the permeability of vessels through targeting of VE-Cadherin and p120-catenin and ZO-1 which results in transendothelial infiltration of HCC cells and promote metastasis (Fang et al., 2018a). Furthermore, increased expression levels of exomiR-1247-3p is associated with lung metastasis in patients with liver cancer (Fang et al., 2018b). HCC recurrence is important factor of therapy such as liver transplantation. Screening exomiR-718 levels in HCC can predict poor prognosis after liver transplantation and HCC recurrence (Sugimachi et al., 2015). Furthermore, in esophageal squamous cell cancer patients, elevated exomiR-21 level is correlated with metastasis (Tanaka et al., 2013).
2.2 Lung cancer
A study examined exomiR-expression profile from serum and found 63 exomiRs to be associated with non-small-cell lung cancer (NSCLC) (Chen et al., 2008a). This study showed that the level of these exomiRs profile differed from serum and blood cells from NSCLC patients while it was the same in healthy individual which suggests that tumour-specific exomiRs in serum were derived from cancer cells. In addition, another board of 34 exomiRs were found in asymptomatic NSCLC patients serum (Bianchi et al., 2011). These reports suggest potential use of exomiRs as non-invasive surrogate diagnostic markers for cancer, potentially of value in screening of asymptomatic populations. A study generated exomiR profile consist of 21 exomiRs analysed from plasma samples collected 12–28 months before lung cancer diagnosis and at the time of detection which suggest a potential diagnosis and prognosis biomarkers for lung cancer (Boeri et al., 2011). Further exomiRs analysis was tested on tissues biopsy took from lung adenocarcinoma patients shows increased levels in panel containing 12 exomiRs including hsa-miR-212, -214, −205, −210, −203, −191, −192, −146, −155, −21, −106a and -17-3p (Rabinowits et al., 2009). A following study used the same approach in screening exomiRs and found similar exomiRs profile in both the tissue biopsy and plasma-derived exosomes from lung adenocarcinoma patients with elevated level of exomiR-200-5p, exomiR-379, exomiR-139-5p and exomiR-378a which suggest a useful biomarker for lung adenocarcinoma (Cazzoli et al., 2013). Loss of exomiR-128b, an EGFR regulator, was associated with better response to gefitinib, an EGFR inhibitor, in patients with relapsed NSCLC (Weiss et al., 2008) and high expression levels of exomiR-21 were found in patients with vertebral column metastasis through increased expression of COX-19 (Guo et al., 2015). NSCLC patients with reduced level of exomiR-497-5p have high chance of metastases (Huang et al., 2019). ExomiR-497-5p can regulate multiple mRNAs including FGF2-encoding mRNAs which can result in migration and invasion (Huang et al., 2019).
2.3 Prostate cancer
A study shows the expression levels of exomiR-141 detected in serum were used to distinguish prostate cancer patients from heathy individuals (Lodes et al., 2009), with other 15 exomiRs including exomiR-16 and exomiR-92a/b were highly expressed in prostate cancer patients (Lodes et al., 2009). In addition, increase levels of exomiR-21-5p, exomiR-141-5p and exomiR-574-3p were also observed in urine samples from patients with prostate cancer (Samsonov et al., 2016). Furthermore, serum exomiR-21 expression levels were associated with resistant to docetaxel-based chemotherapy compared to patients with chemosensitive response in prostate cancer (Zhang et al., 2011a). Prostate cancer patients with increase levels of exomiR-194, exomiR-146b-3p and exomiR-141 have high chance of poor prognosis and recurrence (Selth et al., 2013). Bone metastasis is common in patients with prostate cancer and mediate disease complication (Kfoury et al., 2021). ExomiR-275 derived from prostate cancer has been reported to mediate bone metastasis in prostate cancer patients (Jiang et al., 2022).
2.4 Ovarian and breast cancer
A study on epithelial ovarian cancer found eight exomiRs from serum including exomiR-92, exomiR-93 and exomiR-126 were highly expressed in 19 patients compared to 11 healthy individuals (Resnick et al., 2009). A panel of exomiRs containing exomiR-205, exomiR-214, exomiR-200b, exomiR-203, exomiR-200a, exomiR-200c, exomiR-21 and exomiR-141 were noticed to be escalated in exosomes isolated from patients serum with ovarian cancer (Taylor and Gercel-Taylor, 2008). A prospective analysis showed significant alternation in exomiR-195 in serum, plasma, or whole blood collected from patients with breast cancer (BC) (Heneghan et al., 2010). The serum levels of exomiR-195 was remarkably decreased after tumour removal (Heneghan et al., 2010). Furthermore, BC patents have elevated level of serum exomiR-101 and exomiR-372 compared to healthy controls (Eichelser et al., 2014) and patients with triple-negative BC have increased level of exomiR-373 increased level of exomiR-373 (Eichelser et al., 2014). This suggests, the use of these exomiRs as potential diagnostic markers for BC.
Increase level of certain exomiRs can enhance sensitivity of many chemotherapy drugs. For instance, exomiR-9 downregulates BRCA1 protein through direct binding to the 3′-UTR of BRCA1 mRNA and reduce the ability of the BRCA complex to repair damage in DNA. Therefore, increase exomiR-9 can suppress DNA damage repair in ovarian cancer and enhance ovarian cancer respond to chemotherapy, such as cisplatin (Figure 2) (Sun et al., 2013). BC patients with genetically deleted chromosome 11q which containing the miR‐125 b gene often show better respond to chemotherapy with anthracycline (Climent et al., 2007) which suggests a potential association between exomiR-125b dysregulation and response to drugs containing anthracycline in BC patients. Reduced levels of exomiR-148a-3p have been shown to increase chance of tumour metastasis in ovarian cancer (Gong et al., 2016) which suggest the use of exomiR-148a as a potential marker for tumour recurrence and, possibly, tumour invasion and migration in ovarian cancer.
2.5 Neuroblastoma
Patients with recurrent glioma have higher cerebrospinal fluid exomiR-21 levels compared to non-tumour control group, however, no differences were observed in exomiR-21 isolated from serum (Shi et al., 2015). ExomiR‐375 has been reported to promote bone marrow metastases in patients with neuroblastoma (NB) (Colletti et al., 2020) by downregulating YAP1 levels which enhance osteogenic differentiation of mesenchymal stromal cells (Colletti et al., 2020). Additional screening of exomiRs in metastatic sites and primary tumour sites is important to enhance prediction of recurrence and metastasis.
First-line treatment for glioma patients is usually temozolomide (TMZ) (Chibbaro et al., 2004), however, there is no reliable approach to predict which patients will be resistance to TMZ. Interestingly, downregulation of exomiR-195, exomiR-455-3p, and exomiR-10a has been associated with acquired TMZ-resistance (Ujifuku et al., 2010). Furthermore, TMZ resistance in glioma cells have been reported to be associated with dysregulation with exomiR-221 level (Yang et al., 2017a). These studies, recommend screening for exomiR-10a, exomiR-122, exomiR-455-3p and exomiR-195 levels before and during TMZ therapy to identify better treatment approach for the patient.
2.6 Pediatric tumours
The non-invasive diagnosis, limited-risk and availability of the exomiRs in the body fluids make them attractive method to diagnose cancer in pediatric patients (Galardi et al., 2019). NB is one of the most common tumour in children with heterogeneous clinical characteristics (Ma et al., 2019). Profile for exomiRs isolated from 17 NB patients plasma have been shown different expression compared to healthy controls (Ma et al., 2019). The significant expression of exomiR-199a-3p was correlated with severity of NB patients (Ma et al., 2019), as it increases proliferation and migration of NB cells in vitro (Ma et al., 2019). In another study, high expression levels of exomiR-16 were linked to poor prognosis in childhood acute lymphoblastic leukemia (ALL) (Kaddar et al., 2009).
Another pediatric tumour accounts for 80% of primary tumour liver in young children infant is Hepatoblastoma (HB) (Ranganathan et al., 2020). The expression of exomiR-21 in plasma was higher in HB patients compared to healthy control (Liu et al., 2016b), which makes it a good diagnosis and prognosis biomarker for HB. Further analysis studies of exomiRs profile have shown increased expression of three-miRNA-based expression signature (exomiR-7112-5p, exomiR-885-3p and exomiR-1245a) in plasma of acute myeloid leukaemia patients (AML) (Zampini et al., 2017), increased expression of exomiR-25-3p in serum of osteosarcoma patients (Fujiwara et al., 2017) and decreased expression of exomiR-125b in serum of ewing’s sarcoma patients (Nie et al., 2015). These results showed evidence of using exomiRs as a potential diagnostic and prognostic biomarkers for pediatric tumours. In pediatric AML, both exomiR-29a and exomiR-100 were indicator for better response to chemotherapy (Said et al., 2022). Increased expression levels of exomiR-125b were associated with drug resistance in pediatric acute promyelocytic leukemia (Zhang et al., 2011b).
2.7 Hematologic tumours
Alternation in exomiRs expression is potential biomarker not only in solid tumours, but also in non-solid hematologic tumours. For example, increase expression of exomiR-21, exomiR-155, and exomiR-210 was discovered in the serum of diffuse large B cell lymphoma (DLBCL) patients (Lawrie et al., 2008). Moreover, there was a significant decrease of exomiR-92a in plasma collected from patients with acute leukemias compared to healthy individuals (Tanaka et al., 2009). In addition, plasma samples collected from multiple myeloma (MM) patients have elevated levels of exomiR-148a, exomiR-181a, exomiR-20a, exomiR-221, exomiR-625, and exomiR-99b compared to healthy individuals (Huang et al., 2012a). The increased levels of both exomiR-20a and exomiR148a were associated with short relapse-free survival rate (Huang et al., 2012a). The expression profile of exomiR-128a, exomiR-128b, let-7b, exomiR-223 can help distinguish ALL from AML with over 95% accuracy rate (Mi et al., 2007). Alternation expression of exomiR-32, exomiR-98 and exomiR-374 in blood samples is observed in chronic lymphocytic leukemia (CLL) patients (Rahimi et al., 2021). In a chronic myelogenous leukemia (CML) studies, increased levels of exomiR-451 was observed in plasma samples from CML patients in the chronic stage (Keramati et al., 2021) while increased levels of exomiR-126, exomiR-155, and exomiR-222 were observed during blast crisis (Machová Poláková et al., 2011).
Increased expression levels of exomiR-142-3p and the exomiR-17-92 were associated with glucocorticoid-resistant B cell precursor acute lymphoblastic leukemia (Sakurai et al., 2019). Furthermore, resistance to daunorubicin and vincristine in pediatric ALL was associated with increased expression levels of exomiR-99a, exomiR-100, and exomiR-125b (Schotte et al., 2011).
In CML, tyrosine kinase inhibitors (TKIs) resistance was associated with decreased expression levels of exomiR-142-5p, exomiR-199b, exomiR-217, exomiR-221, and exomiR-365a-3p (Yeh et al., 2016; Jiang et al., 2018; Klümper et al., 2020). In DLBCL studies, upregulation of exomiR-155 and reduced expression levels of exomiR-193b-5p and exomiR-1244 were linked to treatment failure with rituximab plus doxorubicin, cyclophosphamide, prednisone, and vincristine (Iqbal et al., 2015; Bento et al., 2022). Reduced expression levels of both exomiR-145-3p and exomiR-155 were associated with bortezomib resistance in MM (Amodio et al., 2019; Wu et al., 2020). ExomiR-217 sensitizes AML to doxorubicin via targeting KRAS (Xiao et al., 2017) while exomiR-143 enhances chemosensitivity of AML to cytosine arabinoside by targeting ATG7-and ATG2B-dependent autophagy (Zhang et al., 2020a). In CLL, fludarabine response was associated with increased expression levels of exomiR-181a/b (Zhu et al., 2012). Another study has shown a role of exomiR-181a in GC resistance in MM cells (Huang et al., 2012b). Elevated expression of exomiR-485-3p increased sensitivity to Top2 inhibitors in CEM/VM-1-5 cells through regulating the NF-YB expression level (Chen et al., 2011). Downregulation of exomiR-451 was associated with Imatinib resistance in CML (Scholl et al., 2012). In patients with refractory AML, downregulation of exomiR-let-7f was associated with Adriamycin resistance (Dai et al., 2014). Plasma levels of let-7a and exomiR-16 were significantly decreased in patients with myelodysplastic syndrome which predict with both progression-free survival and overall survival (Zuo et al., 2011).
3 ExomiRs as immunotherapeutic targets to regulate immune checkpoint molecules
The therapeutic potential of Immune-checkpoint blockade in cancer has improved the overall survival in the last years (Postow et al., 2015). The first checkpoint blockade to receive FDA approval was ipilimumab, which is an antibody that target cytotoxic T-lymphocyte-associated protein 4 (CTLA4) (Hodi et al., 2010). FDA has also approved additional two immune checkpoint blockade antibodies that target programmed cell death protein 1 (PD-1) known as pembrolizumab and nivolumab to treat stage IV melanoma (Robert et al., 2015a; Robert et al., 2015b) and NSCLC (Brahmer et al., 2015; Garon et al., 2015). Although these targeting antibodies have shown promising results in cancer treatment, the systematic administration of these protein-format and murine-origin blocking antibodies can result in undesirable immune-related adverse events (irAEs) (Van Hoecke and Roose, 2019). ExomiRs secreted from cancer cells can regulate stromal cells and promote cancer angiogenesis (Wu et al., 2019). In addition, they play a key role in intercellular transmission of signals that regulate immune checkpoint molecules and influence the function of several immune cells such as dendritic cells and T cells which are important cells in cancer immunotherapy (Huemer et al., 2021). Several exomiRs can regulate the expressions of immune checkpoint molecules, mimicking the therapeutic impact of immune checkpoint blocking antibodies and controlling the irAEs associated with the administration of the blocking antibodies (Van Hoecke and Roose, 2019). Therefore, they have a potential role as immunotherapeutic agents to regulate immune checkpoint molecules expression either as exomiR mimics or exomiR antagonists. The exomiR mimics function to restore the tumour suppressor capabilities of exomiR while the exomiR antagonists serve as inhibitors (Bader et al., 2011).
The first exomiR mimic to enter the clinical trial phase was the use of MRX34, an exomiR-34a mimic (Bader, 2012; Austin, 2013). In AML, exomiR-34a was found to target PD-L1 mRNA and downregulate PD-L1 expression (Wang et al., 2015). In addition, combination therapy of radiotherapy (XRT) and administration of MRX34 result in increased CD8+ T cell infiltration and reduced Treg in NSCLC (Cortez et al., 2016). These results suggest a potential role of exomiR-34a mimic to enhance anti-tumour immunity and reduce tumour growth. Further studies have examined new potential targets for exomiR mimics. For example, exomiR-424 has the ability to supress PD-L1 and CD80 expression, and restores cytotoxic CD8+T cells effector function and improves the survival in ovarian carcinoma mouse model (Xu et al., 2016). The exomiR inhibitors have also shown immunotherapeutic potential. Cobomarsen (MRG105) is an anti-exomiR-155 agent has shown reduce tumour growth when administrated systematically (Van Roosbroeck et al., 2017). Another study of melanoma, adoptive transfer of CD8+ cytotoxic T lymphocytes treated with exomiR-23a inhibitor has shown decreased in tumour growth and increased effector function of CD8+ cytotoxic T lymphocytes (Lin et al., 2014). Furthermore, exomiR-149-3p reduces inhibitory receptors and revised CD8+ T cell exhaustion in breast cancer cells (Zhang et al., 2019), and exomiR-5119 enhance BC immunotherapy through regulation of immune checkpoints in dendritic cells (Zhang et al., 2020b). In addition, exomiR-34a-5p can regulate the expression of PDL-1 in AML (Wang et al., 2015). ExomiR-138-5p can regulate PDL-1 expression in colorectal cancer (Zhao et al., 2016) and regulate PD-1 expression in glioma (Wei et al., 2016). Low expression levels of exomiR-200 in NSCLC cells are associated with increased expression levels of PD-L1 since exomiR-200 have been shown to target 3′UTR of PD-L1 and decreases its expression (Chen et al., 2014). Therefore, NSCLC patients with exomiR-200 low pattern expression may benefit from the use of miR-200 mimics. Furthermore, decrease expression levels of exomiR-197 are associated with increased expression levels of PD-L1 and promote drug resistance and reduced overall survival in patients with NSCLC (Fujita et al., 2015). Thus, treatment with exomiR-197 mimics may benefit patients with PD-L1-positive NSCLC.
CTLA-4 is another important immune checkpoint molecule which can bind to either CD80 or CD86 on antigen presenting cells and result in suppression the effector function of T cell (Teft et al., 2006). CTLA-4 can be regulated by exomiR-138-5p (Wei et al., 2016). In vivo treatment of exomiR-138 mimic results in downregulation of CTLA-4, PD-1 and Foxp3 on tumor infiltrating CD4+T cells leading to significant decrease of T reg in glioma mouse model (Wei et al., 2016). In addition, exomiR-424 reduced CD80 expression in dendritic cells and results in CD80/CTLA-4 blockade and increased T cell activity (Xu et al., 2016). Furthermore, T cell immunoglobulin and mucin-domain containing-3 (TIM-3) is expressed on activated T cell and reduces T cell activity by inducing T cell exhaustion and tolerance (Ferris et al., 2014). In glioma, exomiR-15a/16 knock-out Mice have decreased TIM-3 and PD-1 expression and increased cytokines secretion in tumor-infiltrating CD8+ T cells which result in better overall mice survival (Yang et al., 2017b). These examples show a potential role of targeting exomiRs as immunotherapeutic agents to regulate immune checkpoint molecules and enhance anti-tumour immunity.
4 Conclusion remarks
Treatment to cancer has improved significantly over the past few decades. However, many patients do not benefit from the treatment due to late-stage diagnosis, tumour recurrence and sometime treatment resistance. Therefore, early diagnosis of cancer and reliable biomarkers for cancer recurrence and treatment resistance are critical to determine the best therapeutic approach and to improve overall survival. To develop and identify a sensitive, and less invasive biomarkers for cancer implication, exomiRs should not be ignored. These circulating exomiRs have the potential for new cancer biomarkers due to several characteristic factors. First, oncogenic pathways can be regulated by exomiRs (Sumazin et al., 2011) which can result in tumour development, suppression and can mediate treatment response which makes it good candidate for cancer progression biomarker. The unique expression profiles of exomiRs in tumours helps in providing wide range of information for tumour stage, recurrence and treatment resistance which makes it a good candidate for liquid biopsy without the need for tissue biopsy. The lipid biolayer structure in the exosomal membrane protect exomiRs from degranulation in the biofluid; this protection makes it more desirable compared to other molecules which can be degraded in harsh conditions like extreme temperature and prolong storage (Chen et al., 2008b). The easy accessibility is another reason for their desirable use, exomiRs can be obtained in less-invasive way from liquid biopsy including blood, urine and saliva. The change in exomiRs expression profile observed in these bio-fluid can be seen as early as early stage of tumour (Jiménez-Avalos et al., 2021).
Based on the advantages of exomiRs derived from tumour, it can be used as a potential biomarker for cancer implication as it can predict the growth, spread, recurrence and treatment-resistance of tumours. However, several studies reported unsuccessful validation in using exomiRs as biomarkers for cancer (Sapre et al., 2014; Wan et al., 2014). This could be due to several factors including limited methodologies to access and obtain exomiRs and time of collection and the cancer stage (Mateescu et al., 2017). These studies highlight the significance of incorporating methodological approaches to obtain exomiRs and to ensure the reproducibility of the result in future studies.
The discovery of immune checkpoint blockade as cancer immunotherapy has improved the overall survival significantly, however, new strategies on how to regulate their expression using something other than protein-format and murine-origin antibodies are crucial to avoid undesirable irAE. ExomiRs play a key role in posttranscriptional control of protein expression and therefore, may be of better target since some exomiRs expression levels are associated with expression of immune checkpoint molecules. Further investigations are important to deeply show the effect of exomiRs on cancer biology.
5 Future direction
The variability of cancer lends itself in the growing field of personalized medicine which provide great patient benefit (Verma, 2012). Artificial intelligence (AI) technique is emerging in personalized medicine and biomedical research which include cancer clinical implications including cancer diagnosis, treatment, and the discovery of new potential therapy (Schork, 2019; Elemento et al., 2021). The big data obtained from thousands of studies related to exomiRs in cancer can leverage an opportunity to implement AI in cancer clinical implication and improve cancer diagnosis (Paolini et al., 2022). Together, these computational platforms can provide a new technique in cancer clinical implication and provide a modern approach on the validity of exomiRs signature as biomarker in cancer and immunotherapeutic agents.
Acknowledgments
The author would like to thank Prof. James Koropatnick from the departments of Oncology and Pathology, Schulich School of Medicine and Dentistry, University of Western Ontario, London, Ontario, Canada, for all the years of guidance and support.
Author contributions
FA reviewed the literature, wrote the manuscript and designed the figures and tables.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Alotaibi F. (2021). Exosomal microRNAs: Potential biomarkers for cancer diagnosis, treatment response and prognosis, in Role of exosomes in biological communication systems, 321–336. [Google Scholar]
- Amodio N., Gallo Cantafio M. E., Botta C., Agosti V., Federico C., Caracciolo D., et al. (2019). Replacement of miR-155 elicits tumor suppressive activity and antagonizes bortezomib resistance in multiple myeloma. Cancers 11 (2), 236. 10.3390/cancers11020236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim M. G., Valieris R., Drummond R. D., Pizzi M. P., Freitas V. M., Sinigaglia-Coimbra R., et al. (2017). A total transcriptome profiling method for plasma-derived extracellular vesicles: Applications for liquid biopsies. Sci. Rep. 7 (1), 1–11. 10.1038/s41598-017-14264-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin T. b. M. (2013). First microRNA mimic enters clinic. Nat. Biotechnol. 31 (7), 577. 10.1038/nbt0713-577 [DOI] [PubMed] [Google Scholar]
- Bader A., Brown D., Stoudemire J., Lammers P. (2011). Developing therapeutic microRNAs for cancer. Gene Ther. 18 (12), 1121–1126. 10.1038/gt.2011.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader A. G. (2012). miR-34–a microRNA replacement therapy is headed to the clinic. Front. Genet. 3, 120. 10.3389/fgene.2012.00120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bento L., Vogler O., Sas-Barbeito A., Muncunill J., Ros T., Martinez J., et al. (2022). Screening for prognostic microRNAs associated with treatment failure in diffuse large B cell lymphoma. Cancers 14 (4), 1065. 10.3390/cancers14041065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F., Nicassio F., Marzi M., Belloni E., Dall'olio V., Bernard L., et al. (2011). A serum circulating miRNA diagnostic test to identify asymptomatic high‐risk individuals with early stage lung cancer. EMBO Mol. Med. 3 (8), 495–503. 10.1002/emmm.201100154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., et al. (2011). MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. 108 (9), 3713–3718. 10.1073/pnas.1100048108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boing A., Edwin V, Anita E G, Frank A W C, Auguste S, Rienk N, et al. (2014), Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles. J Extracell Vesicles 8, 3, 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmer J., Reckamp K. L., Baas P., Crino L., Eberhardt W. E. E., Poddubskaya E., et al. (2015). Nivolumab versus docetaxel in advanced squamous-cell non–small-cell lung cancer. N. Engl. J. Med. 373 (2), 123–135. 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G. A., Croce C. M. (2006). MicroRNA signatures in human cancers. Nat. Rev. cancer 6 (11), 857–866. 10.1038/nrc1997 [DOI] [PubMed] [Google Scholar]
- Cazzoli R., Buttitta F., Di Nicola M., Malatesta S., Marchetti A., Rom W. N., et al. (2013). microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 8 (9), 1156–1162. 10.1097/JTO.0b013e318299ac32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-F., He X., Arslan A. D., Mo Y. Y., Reinhold W. C., Pommier Y., et al. (2011). Novel regulation of nuclear factor-YB by miR-485-3p affects the expression of DNA topoisomerase IIα and drug responsiveness. Mol. Pharmacol. 79 (4), 735–741. 10.1124/mol.110.069633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Gibbons D. L., Goswami S., Cortez M. A., Ahn Y. H., Byers L. A., et al. (2014). Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 5 (1), 5241. 10.1038/ncomms6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Xia H. W., Ge X. J., Zhang Y. C., Tang Q. L., Bi F. (2013). Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac. J. Cancer Prev. 14 (12), 7421–7426. 10.7314/apjcp.2013.14.12.7421 [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18 (10), 997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., et al. (2008). Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18 (10), 997–1006. 10.1038/cr.2008.282 [DOI] [PubMed] [Google Scholar]
- Chibbaro S., Benvenuti L., Caprio A., Carnesecchi S., Pulera F., Faggionato F., et al. (2004). Temozolomide as first-line agent in treating high-grade gliomas: Phase II study. J. neuro-oncology 67 (1-2), 77–81. 10.1023/b:neon.0000021728.36747.93 [DOI] [PubMed] [Google Scholar]
- Climent J., Dimitrow P., Fridlyand J., Palacios J., Siebert R., Albertson D. G., et al. (2007). Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 67 (2), 818–826. 10.1158/0008-5472.CAN-06-3307 [DOI] [PubMed] [Google Scholar]
- Colletti M., Tomao L., Galardi A., Paolini A., Di Paolo V., De Stefanis C., et al. (2020). Neuroblastoma-secreted exosomes carrying miR‐375 promote osteogenic differentiation of bone-marrow mesenchymal stromal cells. J. Extracell. vesicles 9 (1), 1774144. 10.1080/20013078.2020.1774144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran C., Rani S., O'Driscoll L. (2014). miR‐34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate 74 (13), 1320–1334. 10.1002/pros.22848 [DOI] [PubMed] [Google Scholar]
- Cortez M. A., Ivan C., Valdecanas D., Wang X., Peltier H. J., Ye Y., et al. (2016). PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 108 (1), djv303. 10.1093/jnci/djv303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C.-W., Bai Q. W., Zhang G. S., Cao Y. X., Shen J. K., Pei M. F., et al. (2014). MicroRNA let-7f is down-regulated in patients with refractory acute myeloid leukemia and is involved in chemotherapy resistance of adriamycin-resistant leukemic cells. Leukemia Lymphoma 55 (7), 1645–1648. 10.3109/10428194.2013.847936 [DOI] [PubMed] [Google Scholar]
- Dragovic R. A., Gardiner C., Brooks A. S., Tannetta D. S., Ferguson D. J. P., Hole P., et al. (2011). Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine Nanotechnol. Biol. Med. 7 (6), 780–788. 10.1016/j.nano.2011.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelser C., Stuckrath I., Muller V., Milde-Langosch K., Wikman H., Pantel K., et al. (2014). Increased serum levels of circulating exosomal microRNA-373 in receptor-negative breast cancer patients. Oncotarget 5 (20), 9650–9663. 10.18632/oncotarget.2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström K., Crescitelli R., Petursson H. I., Johansson J., Lasser C., Olofsson Bagge R. (2022). Characterization of surface markers on extracellular vesicles isolated from lymphatic exudate from patients with breast cancer. BMC cancer 22 (1), 50–17. 10.1186/s12885-021-08870-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elemento O., Leslie C., Lundin J., Tourassi G. (2021). Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 21 (12), 747–752. 10.1038/s41568-021-00399-1 [DOI] [PubMed] [Google Scholar]
- Eun J. W., Seo C. W., Baek G. O., Yoon M. G., Ahn H. R., Son J. A., et al. (2020). Circulating exosomal MicroRNA-1307-5p as a predictor for metastasis in patients with hepatocellular carcinoma. Cancers 12 (12), 3819. 10.3390/cancers12123819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J. H., Zhang Z. J., Shang L. R., Luo Y. W., Lin Y. F., Yuan Y., et al. (2018). Hepatoma cell‐secreted exosomal microRNA‐103 increases vascular permeability and promotes metastasis by targeting junction proteins. Hepatology 68 (4), 1459–1475. 10.1002/hep.29920 [DOI] [PubMed] [Google Scholar]
- Fang T., Lv H., Lv G., Li T., Wang C., Han Q., et al. (2018). Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 9 (1), 191. 10.1038/s41467-017-02583-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris R. L., Lu B., Kane L. P. (2014). Too much of a good thing? Tim-3 and TCR signaling in T cell exhaustion. J. Immunol. 193 (4), 1525–1530. 10.4049/jimmunol.1400557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu F., Jiang W., Zhou L., Chen Z. (2018). Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Transl. Oncol. 11 (2), 221–232. 10.1016/j.tranon.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., et al. (2015). The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 23 (4), 717–727. 10.1038/mt.2015.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Uotani K., Yoshida A., Morita T., Nezu Y., Kobayashi E., et al. (2017). Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 8 (20), 33375–33392. 10.18632/oncotarget.16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galardi A., Colletti M., Di Paolo V., Vitullo P., Antonetti L., Russo I., et al. (2019). Exosomal MiRNAs in pediatric cancers. Int. J. Mol. Sci. 20 (18), 4600. 10.3390/ijms20184600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandaglia G., Karakiewicz P. I., Briganti A., Passoni N. M., Schiffmann J., Trudeau V., et al. (2015). Impact of the site of metastases on survival in patients with metastatic prostate cancer. Eur. Urol. 68 (2), 325–334. 10.1016/j.eururo.2014.07.020 [DOI] [PubMed] [Google Scholar]
- Garon E. B., Rizvi N. A., Hui R., Leighl N., Balmanoukian A. S., Eder J. P., et al. (2015). Pembrolizumab for the treatment of non–small-cell lung cancer. N. Engl. J. Med. 372 (21), 2018–2028. 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- Gill P., Kim E., Chua T. C., Clifton-Bligh R. J., Nahm C. B., Mittal A., et al. (2019). MiRNA-3653 is a potential tissue biomarker for increased metastatic risk in pancreatic neuroendocrine tumours. Endocr. Pathol. 30, 128–133. 10.1007/s12022-019-9570-y [DOI] [PubMed] [Google Scholar]
- Gong L., Wang C., Gao Y., Wang J. (2016). Decreased expression of microRNA-148a predicts poor prognosis in ovarian cancer and associates with tumor growth and metastasis. Biomed. Pharmacother. 83, 58–63. 10.1016/j.biopha.2016.05.049 [DOI] [PubMed] [Google Scholar]
- Guo Q., Zhang H., Zhang L., He Y., Weng S., Dong Z., et al. (2015). MicroRNA-21 regulates non-small cell lung cancer cell proliferation by affecting cell apoptosis via COX-19. Int. J. Clin. Exp. Med. 8 (6), 8835–8841. [PMC free article] [PubMed] [Google Scholar]
- Hannafon B. N., Ding W.-Q. (2013). Intercellular communication by exosome-derived microRNAs in cancer. Int. J. Mol. Sci. 14 (7), 14240–14269. 10.3390/ijms140714240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C., Heuser J., Stahl P. (1983). Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 97 (2), 329–339. 10.1083/jcb.97.2.329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneghan H. M., Miller N., Lowery A. J., Sweeney K. J., Newell J., Kerin M. J. (2010). Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann. Surg. 251 (3), 499–505. 10.1097/SLA.0b013e3181cc939f [DOI] [PubMed] [Google Scholar]
- Hiom S. (2015). Diagnosing cancer earlier: Reviewing the evidence for improving cancer survival. Br. J. Cancer 112, S1–S5. Nature Publishing Group. 10.1038/bjc.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A. S., Huang X., Cao H., Christman-Skieller C., Bennewith K., Le Q. T., et al. (2010). Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl. Oncol. 3 (2), 109–113. 10.1593/tlo.09256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi F. S., O'Day S. J., McDermott D. F., Weber R. W., Sosman J. A., Haanen J. B., et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363 (8), 711–723. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.-j., Yu J., Li J. y., Liu Y. t., Zhong R. q. (2012). Circulating microRNA expression is associated with genetic subtype and survival of multiple myeloma. Med. Oncol. 29, 2402–2408. 10.1007/s12032-012-0210-3 [DOI] [PubMed] [Google Scholar]
- Huang X., Wang L., Liu W. (2019). MicroRNA-497-5p inhibits proliferation and invasion of non-small cell lung cancer by regulating FGF2. Oncol. Lett. 17 (3), 3425–3431. 10.3892/ol.2019.9954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Yang M., Jin J. (2012). Triptolide enhances the sensitivity of multiple myeloma cells to dexamethasone via microRNAs. Leukemia lymphoma 53 (6), 1188–1195. 10.3109/10428194.2011.638069 [DOI] [PubMed] [Google Scholar]
- Huemer F., Leisch M., Geisberger R., Zaborsky N., Greil R. (2021). miRNA-based therapeutics in the era of immune-checkpoint inhibitors. Pharmaceuticals 14 (2), 89. 10.3390/ph14020089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J., Shen Y., Huang X., Liu Y., Wake L., Liu C., et al. (2015). Global microRNA expression profiling uncovers molecular markers for classification and prognosis in aggressive B-cell lymphoma. J. Am. Soc. Hematol. 125 (7), 1137–1145. 10.1182/blood-2014-04-566778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q., Tan X. P., Zhang C. H., Li Z. Y., Xu Y., et al. (2022). Non-coding RNAs of extracellular vesicles: Key players in organ-specific metastasis and clinical implications. Cancers 14 (22), 5693. 10.3390/cancers14225693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Cheng Y., Hu C., Zhang A., Ren Y., Xu X. (2018). MicroRNA-221 sensitizes chronic myeloid leukemia cells to imatinib by targeting STAT5. Leukemia lymphoma 60, 1709–1720. 10.1080/10428194.2018.1543875 [DOI] [PubMed] [Google Scholar]
- Jiménez-Avalos J. A., Fernández-Macías J. C., González-Palomo A. K. (2021). Circulating exosomal MicroRNAs: New non-invasive biomarkers of non-communicable disease. Mol. Biol. Rep. 48 (1), 961–967. 10.1007/s11033-020-06050-w [DOI] [PubMed] [Google Scholar]
- Kaddar T., Chien W. W., Bertrand Y., Pages M. P., Rouault J. P., Salles G., et al. (2009). Prognostic value of miR-16 expression in childhood acute lymphoblastic leukemia relationships to normal and malignant lymphocyte proliferation. Leukemia Res. 33 (9), 1217–1223. 10.1016/j.leukres.2008.12.015 [DOI] [PubMed] [Google Scholar]
- Keramati F., Jafarian A., Soltani A., Javandoost E., Mollaei M., Fallah P. (2021). Circulating miRNAs can serve as potential diagnostic biomarkers in chronic myelogenous leukemia patients. Leukemia Res. Rep. 16, 100257. 10.1016/j.lrr.2021.100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kfoury Y., Baryawno N., Severe N., Mei S., Gustafsson K., Hirz T., et al. (2021). Human prostate cancer bone metastases have an actionable immunosuppressive microenvironment. Cancer Cell 39 (11), 1464–1478. e8. 10.1016/j.ccell.2021.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klümper T., Bruckmueller H., Diewock T., Kaehler M., Haenisch S., Pott C., et al. (2020). Expression differences of miR-142-5p between treatment-naïve chronic myeloid leukemia patients responding and non-responding to imatinib therapy suggest a link to oncogenic ABL2, SRI, cKIT and MCL1 signaling pathways critical for development of therapy resistance. Exp. Hematol. Oncol. 9 (1), 26–15. 10.1186/s40164-020-00183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A., Griffiths-Jones S. (2014). miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic acids Res. 42 (D1), D68–D73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrea E., Sole C., Manterola L., Goicoechea I., Armesto M., Arestin M., et al. (2016). New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 17 (5), 627. 10.3390/ijms17050627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrie C. H., Gal S., Dunlop H. M., Pushkaran B., Liggins A. P., Pulford K., et al. (2008). Detection of elevated levels of tumour‐associated microRNAs in serum of patients with diffuse large B‐cell lymphoma. Br. J. Haematol. 141 (5), 672–675. 10.1111/j.1365-2141.2008.07077.x [DOI] [PubMed] [Google Scholar]
- Lin R., Chen L., Chen G., Hu C., Jiang S., Sevilla J., et al. (2014). Targeting miR-23a in CD8+ cytotoxic T lymphocytes prevents tumor-dependent immunosuppression. J. Clin. investigation 124 (12), 5352–5367. 10.1172/JCI76561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Gregory R. I. (2015). MicroRNA biogenesis pathways in cancer. Nat. Rev. cancer 15 (6), 321–333. 10.1038/nrc3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Eng C., Shen J., Lu Y., Takata Y., Mehdizadeh A., et al. (2016). Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 7 (46), 76250–76260. 10.18632/oncotarget.12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Chen S., Liu B. (2016). Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: A Chinese population-based study. Pediatr. Surg. Int. 32 (11), 1059–1065. 10.1007/s00383-016-3960-8 [DOI] [PubMed] [Google Scholar]
- Lobb R. J., Becker M., Wen S. W., Wong C. S. F., Wiegmans A. P., Leimgruber A., et al. (2015). Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. vesicles 4 (1), 27031. 10.3402/jev.v4.27031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodes M. J., Caraballo M., Suciu D., Munro S., Kumar A., Anderson B. (2009). Detection of cancer with serum miRNAs on an oligonucleotide microarray. PloS one 4 (7), e6229. 10.1371/journal.pone.0006229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotvall J., Hill A. F., Hochberg F., Buzas E. I., Di Vizio D., Gardiner C., et al. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the international society for extracellular vesicles. J. Extracell. Vesicles 3 (26913), 26913. 10.3402/jev.v3.26913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Xu M., Yin M., Hong J., Chen H., Gao Y., et al. (2019). Exosomal hsa-miR199a-3p promotes proliferation and migration in neuroblastoma. Front. Oncol. 9, 459. 10.3389/fonc.2019.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maacha S., Bhat A. A., Jimenez L., Raza A., Haris M., Uddin S., et al. (2019). Extracellular vesicles-mediated intercellular communication: Roles in the tumor microenvironment and anti-cancer drug resistance. Mol. cancer 18 (1), 55. 10.1186/s12943-019-0965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machová Poláková K., Lopotova T., Klamova H., Burda P., Trněny M., Stopka T., et al. (2011). Expression patterns of microRNAs associated with CML phases and their disease related targets. Mol. cancer 10 (1), 41–13. 10.1186/1476-4598-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Kowal E. J. K., van Balkom B. W. M., Bartel S., Bhattacharyya S. N., Buzas E. I., et al. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA–an ISEV position paper. J. Extracell. vesicles 6 (1), 1286095. 10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Sugimachi K., Iinuma H., Takahashi Y., Kurashige J., Sawada G., et al. (2015). Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. cancer 113 (2), 275–281. 10.1038/bjc.2015.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J. (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol. 28 (8), R435–R444. 10.1016/j.cub.2018.01.059 [DOI] [PubMed] [Google Scholar]
- Mi S., Lu J., Sun M., Li Z., Zhang H., Neilly M. B., et al. (2007). MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc. Natl. Acad. Sci. 104 (50), 19971–19976. 10.1073/pnas.0709313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikamori M., Yamada D., Eguchi H., Hasegawa S., Kishimoto T., Tomimaru Y., et al. (2017). MicroRNA-155 controls exosome synthesis and promotes gemcitabine resistance in pancreatic ductal adenocarcinoma. Sci. Rep. 7, 42339. 10.1038/srep42339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima G., Hayashi K., Xi Y., Kudo K., Uchida K., Takasaki K., et al. (2006). Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer genomics & proteomics 3 (5), 317–324. [PMC free article] [PubMed] [Google Scholar]
- Ng E. K., Chong W. W. S., Jin H., Lam E. K. Y., Shin V. Y., Yu J., et al. (2009). Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 58 (10), 1375–1381. 10.1136/gut.2008.167817 [DOI] [PubMed] [Google Scholar]
- Nie C., Ren W. H., Ma Y., Xi J. S., Han B. (2015). Circulating miR-125b as a biomarker of Ewing’s sarcoma in Chinese children. Genet. Mol. Res. 14, 1904919049–1904919056. 10.4238/2015.December.29.12 [DOI] [PubMed] [Google Scholar]
- Pan B.-T., Teng K., Wu C., Adam M., Johnstone R. M. (1985). Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 101 (3), 942–948. 10.1083/jcb.101.3.942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini A., Baldassarre A., Bruno S. P., Felli C., Muzi C., Ahmadi Badi S., et al. (2022). Improving the diagnostic potential of extracellular miRNAs coupled to multiomics data by exploiting the power of artificial intelligence. Front. Microbiol. 13, 888414. 10.3389/fmicb.2022.888414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N. J., Zhou H., Elashoff D., Henson B. S., Kastratovic D. A., Abemayor E., et al. (2009). Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 15 (17), 5473–5477. 10.1158/1078-0432.CCR-09-0736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow M. A., Callahan M. K., Wolchok J. D. (2015). Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 33 (17), 1974–1982. 10.1200/JCO.2014.59.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowits G., Gercel-Taylor C., Day J. M., Taylor D. D., Kloecker G. H. (2009). Exosomal microRNA: A diagnostic marker for lung cancer. Clin. lung cancer 10 (1), 42–46. 10.3816/CLC.2009.n.006 [DOI] [PubMed] [Google Scholar]
- Radisavljevic Z. (2013). AKT as locus of cancer multidrug resistance and fragility. J. Cell. physiology 228 (4), 671–674. 10.1002/jcp.24176 [DOI] [PubMed] [Google Scholar]
- Rahimi Z., Ghorbani Z., Motamed H., Jalilian N. (2021). Aberrant expression profile of miR-32, miR-98 and miR-374 in chronic lymphocytic leukemia. Leukemia Res. 111, 106691. 10.1016/j.leukres.2021.106691 [DOI] [PubMed] [Google Scholar]
- Ramirez M. I., Amorim M. G., Gadelha C., Milic I., Welsh J. A., Freitas V. M., et al. (2018). Technical challenges of working with extracellular vesicles. Nanoscale 10 (3), 881–906. 10.1039/c7nr08360b [DOI] [PubMed] [Google Scholar]
- Ranganathan S., Lopez-Terrada D., Alaggio R. (2020). Hepatoblastoma and pediatric hepatocellular carcinoma: An update. Pediatr. Dev. Pathology 23 (2), 79–95. 10.1177/1093526619875228 [DOI] [PubMed] [Google Scholar]
- Resnick K. E., Alder H., Hagan J. P., Richardson D. L., Croce C. M., Cohn D. E. (2009). The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 112 (1), 55–59. 10.1016/j.ygyno.2008.08.036 [DOI] [PubMed] [Google Scholar]
- Robert C., Long G. V., Brady B., Dutriaux C., Maio M., Mortier L., et al. (2015). Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med. 372 (4), 320–330. 10.1056/NEJMoa1412082 [DOI] [PubMed] [Google Scholar]
- Robert C., Schachter J., Long G. V., Arance A., Grob J. J., Mortier L., et al. (2015). Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372 (26), 2521–2532. 10.1056/NEJMoa1503093 [DOI] [PubMed] [Google Scholar]
- Said F., Tantawy M., Sayed A., Ahmed S. (2022). Clinical significance of MicroRNA-29a and MicroRNA-100 gene expression in pediatric acute myeloid leukemia. J. Pediatr. Hematology/Oncology 44 (2), e391–e395. 10.1097/MPH.0000000000002168 [DOI] [PubMed] [Google Scholar]
- Sakurai N., Komada Y., Hanaki R., Morimoto M., Ito T., Nakato D., et al. (2019). Role of microRNAs in glucocorticoid-resistant B-cell precursor acute lymphoblastic leukemia. Oncol. Rep. 42 (2), 708–716. 10.3892/or.2019.7191 [DOI] [PubMed] [Google Scholar]
- Samsonov R., Shtam T., Burdakov V., Glotov A., Tsyrlina E., Berstein L., et al. (2016). Lectin‐induced agglutination method of urinary exosomes isolation followed by mi‐RNA analysis: Application for prostate cancer diagnostic. Prostate 76 (1), 68–79. 10.1002/pros.23101 [DOI] [PubMed] [Google Scholar]
- Sapre N., Hong M. K. H., Macintyre G., Lewis H., Kowalczyk A., Costello A. J., et al. (2014). Curated microRNAs in urine and blood fail to validate as predictive biomarkers for high-risk prostate cancer. PLoS One 9 (4), e91729. 10.1371/journal.pone.0091729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl V., Hassan R., Zalcberg I. R. (2012). miRNA-451: A putative predictor marker of Imatinib therapy response in chronic myeloid leukemia. Leukemia Res. 36 (1), 119–121. 10.1016/j.leukres.2011.08.023 [DOI] [PubMed] [Google Scholar]
- Schork N. J. (2019). “Artificial intelligence and personalized medicine,” in Precision medicine in cancer therapy (Springer; ), 265–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte D., De Menezes R. X., Akbari Moqadam F., Khankahdani L. M., Lange-Turenhout E., Chen C., et al. (2011). MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. haematologica 96 (5), 703–711. 10.3324/haematol.2010.026138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth L., Townley S. L., Bert A. G., Stricker P. D., Sutherland P. D., Horvath L. G., et al. (2013). Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. cancer 109 (3), 641–650. 10.1038/bjc.2013.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Wang P. Y., Li X. Y., Chen J. X., Li Y., Zhang X. Z., et al. (2015). Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget 6 (29), 26971–26981. 10.18632/oncotarget.4699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala K., Girish H., Murgod S. (2014). Risk of tumor cell seeding through biopsy and aspiration cytology. J. Int. Soc. Prev. Community Dent. 4 (1), 5–11. 10.4103/2231-0762.129446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Titmus M. A., Botchkina G., Formentini A., Kornmann M., et al. (2010). Molecular mechanism of chemoresistance by miR-215 in osteosarcoma and colon cancer cells. Mol. cancer 9 (1), 96. 10.1186/1476-4598-9-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Xi Y., Kudo K., Bruheim S., Botchkina G. I., et al. (2009). Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 28 (46), 4065–4074. 10.1038/onc.2009.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimachi K., Matsumura T., Hirata H., Uchi R., Ueda M., Ueo H., et al. (2015). Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br. J. cancer 112 (3), 532–538. 10.1038/bjc.2014.621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumazin P., Yang X., Chiu H. S., Chung W. J., Iyer A., Llobet-Navas D., et al. (2011). An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147 (2), 370–381. 10.1016/j.cell.2011.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Li N., Yang Z., Zhou B., He Y., Weng D., et al. (2013). miR-9 regulation of BRCA1 and ovarian cancer sensitivity to cisplatin and PARP inhibition. J. Natl. Cancer Inst. 105 (22), 1750–1758. 10.1093/jnci/djt302 [DOI] [PubMed] [Google Scholar]
- Sun H.-L., Cui R., Zhou J., Teng K. Y., Hsiao Y. H., Nakanishi K., et al. (2016). ERK activation globally downregulates miRNAs through phosphorylating exportin-5. Cancer Cell 30 (5), 723–736. 10.1016/j.ccell.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Oikawa K., Takanashi M., Kudo M., Ohyashiki J., Ohyashiki K., et al. (2009). Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS one 4 (5), e5532. 10.1371/journal.pone.0005532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Kamohara H., Kinoshita K., Kurashige J., Ishimoto T., Iwatsuki M., et al. (2013). Clinical impact of serum exosomal microRNA‐21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 119 (6), 1159–1167. 10.1002/cncr.27895 [DOI] [PubMed] [Google Scholar]
- Taylor D. D., Gercel-Taylor C. (2008). MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110 (1), 13–21. 10.1016/j.ygyno.2008.04.033 [DOI] [PubMed] [Google Scholar]
- Teft W. A., Kirchhof M. G., Madrenas J. (2006). A molecular perspective of CTLA-4 function. Annu. Rev. Immunol. 24, 65–97. 10.1146/annurev.immunol.24.021605.090535 [DOI] [PubMed] [Google Scholar]
- Théry C., Amigorena S., Raposo G., Clayton A. (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 30 (1), Unit 3.22–3.22. 29. 10.1002/0471143030.cb0322s30 [DOI] [PubMed] [Google Scholar]
- Théry C. (2011). Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 3, 15. 10.3410/B3-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Zitvogel L., Amigorena S. (2002). Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2 (8), 569–579. 10.1038/nri855 [DOI] [PubMed] [Google Scholar]
- Tsujiura M., Ichikawa D., Komatsu S., Shiozaki A., Takeshita H., Kosuga T., et al. (2010). Circulating microRNAs in plasma of patients with gastric cancers. Br. J. cancer 102 (7), 1174–1179. 10.1038/sj.bjc.6605608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujifuku K., Mitsutake N., Takakura S., Matsuse M., Saenko V., Suzuki K., et al. (2010). MiR-195, miR-455-3p and miR-10a∗ are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer Lett. 296 (2), 241–248. 10.1016/j.canlet.2010.04.013 [DOI] [PubMed] [Google Scholar]
- Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J. J., Lotvall J. O. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9 (6), 654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- Van der Pol E., Coumans F. A. W., Grootemaat A. E., Gardiner C., Sargent I. L., Harrison P., et al. (2014). Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thrombosis Haemostasis 12 (7), 1182–1192. 10.1111/jth.12602 [DOI] [PubMed] [Google Scholar]
- Van Deun J., Mestdagh P., Agostinis P., Akay O., Anand S., et al. (2017). EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. methods 14 (3), 228–232. 10.1038/nmeth.4185 [DOI] [PubMed] [Google Scholar]
- Van Hoecke L., Roose K. (2019). How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 17 (1), 54. 10.1186/s12967-019-1804-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roosbroeck K., Fanini F., Setoyama T., Ivan C., Rodriguez-Aguayo C., Fuentes-Mattei E., et al. (2017). Combining anti-miR-155 with chemotherapy for the treatment of lung cancers. Clin. Cancer Res. 23 (11), 2891–2904. 10.1158/1078-0432.CCR-16-1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M. (2012). Personalized medicine and cancer. J. personalized Med. 2 (1), 1–14. 10.3390/jpm2010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.-W., Mach C. M., Allen G. I., Anderson M. L., Liu Z. (2014). On the reproducibility of TCGA ovarian cancer microRNA profiles. PloS one 9 (1), e87782. 10.1371/journal.pone.0087782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen J., Chang P., LeBlanc A., Li D., Abbruzzesse J. L., et al. (2009). MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. 2 (9), 807–813. 10.1158/1940-6207.CAPR-09-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li J., Dong K., Lin F., Long M., Ouyang Y., et al. (2015). Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 27 (3), 443–452. 10.1016/j.cellsig.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Wei J., Nduom E. K., Kong L. Y., Hashimoto Y., Xu S., Gabrusiewicz K., et al. (2016). MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro-oncology 18 (5), 639–648. 10.1093/neuonc/nov292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss G., Bemis L. T., Nakajima E., Sugita M., Birks D. K., Robinson W. A., et al. (2008). EGFR regulation by microRNA in lung cancer: Correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann. Oncol. 19 (6), 1053–1059. 10.1093/annonc/mdn006 [DOI] [PubMed] [Google Scholar]
- Wong T.-S., Liu X. B., Wong B. Y. H., Ng R. W. M., Yuen A. P. W., Wei W. I. (2008). Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 14 (9), 2588–2592. 10.1158/1078-0432.CCR-07-0666 [DOI] [PubMed] [Google Scholar]
- Wu F., Li F., Lin X., Xu F., Cui R. R., Zhong J. Y., et al. (2019). Exosomes increased angiogenesis in papillary thyroid cancer microenvironment. Endocrine-Related Cancer 26 (5), 525–538. 10.1530/ERC-19-0008 [DOI] [PubMed] [Google Scholar]
- Wu H., Liu C., Yang Q., Xin C., Du J., Sun F., et al. (2020). MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy 16 (4), 683–697. 10.1080/15548627.2019.1635380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Deng T., Su C., Shang Z. (2017). MicroRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting KRAS. Oncol. Lett. 13 (6), 4986–4994. 10.3892/ol.2017.6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Tao Z., Hai B., Liang H., Shi Y., Wang T., et al. (2016). miR-424 (322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 7 (1), 11406. 10.1038/ncomms11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Kosaka N., Tanaka M., Koizumi F., Kanai Y., Mizutani T., et al. (2009). MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers 14 (7), 529–538. 10.3109/13547500903150771 [DOI] [PubMed] [Google Scholar]
- Yang F., Li Q. j., Gong Z. b., Zhou L., You N., Wang S., et al. (2014). MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol. cancer Res. Treat. 13 (1), 77–86. 10.7785/tcrt.2012.500364 [DOI] [PubMed] [Google Scholar]
- Yang J.-K., Tong J., Jing S. Y., Fan B., Wang F., et al. (2017). Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J. neuro-oncology 131 (2), 255–265. 10.1007/s11060-016-2308-5 [DOI] [PubMed] [Google Scholar]
- Yang J., Liu R., Deng Y., Qian J., Lu Z., Wang Y., et al. (2017). MiR‐15a/16 deficiency enhances anti‐tumor immunity of glioma‐infiltrating CD8+ T cells through targeting mTOR. Int. J. cancer 141 (10), 2082–2092. 10.1002/ijc.30912 [DOI] [PubMed] [Google Scholar]
- Yeh C.-H., Moles R., Nicot C. (2016). Clinical significance of microRNAs in chronic and acute human leukemia. Mol. cancer 15 (1), 37–16. 10.1186/s12943-016-0518-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan R., Wang G., Xu Z., Zhao H., Chen H., Han Y., et al. (2016). Up-regulated circulating miR-106a by DNA methylation promised a potential diagnostic and prognostic marker for gastric cancer. Anti-Cancer Agents Med. Chem. Former. Curr. Med. Chemistry-Anti-Cancer Agents) 16 (9), 1093–1100. 10.2174/1871520615666150716110657 [DOI] [PubMed] [Google Scholar]
- Zahreddine H., Borden K. (2013). Mechanisms and insights into drug resistance in cancer. Front. Pharmacol. 4, 28. 10.3389/fphar.2013.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampini M., Bisio V., Leszl A., Putti M. C., Menna G., Rizzari C., et al. (2017). A three-miRNA-based expression signature at diagnosis can predict occurrence of relapse in children with t (8; 21) RUNX1-RUNX1T1 acute myeloid leukaemia. Br. J. Haematol. 183 (2), 298–301. 10.1111/bjh.14950 [DOI] [PubMed] [Google Scholar]
- Zhang H., Kang J., Liu L., Chen L., Ren S., Tao Y. (2020). MicroRNA-143 sensitizes acute myeloid leukemia cells to cytarabine via targeting ATG7-and ATG2B-dependent autophagy. Aging (Albany NY) 12 (20), 20111–20126. 10.18632/aging.103614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Luo X. Q., Feng D. D., Zhang X. J., Wu J., Zheng Y. S., et al. (2011). Upregulation of microRNA-125b contributes to leukemogenesis and increases drug resistance in pediatric acute promyelocytic leukemia. Mol. cancer 10, 108–113. 10.1186/1476-4598-10-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. L., Yang L. F., Zhu Y., Yao X. D., Zhang S. L., Dai B., et al. (2011). Serum miRNA‐21: Elevated levels in patients with metastatic hormone‐refractory prostate cancer and potential predictive factor for the efficacy of docetaxel‐based chemotherapy. Prostate 71 (3), 326–331. 10.1002/pros.21246 [DOI] [PubMed] [Google Scholar]
- Zhang M., Gao D., Shi Y., Wang Y., Joshi R., Yu Q., et al. (2019). miR-149-3p reverses CD8+ T-cell exhaustion by reducing inhibitory receptors and promoting cytokine secretion in breast cancer cells. Open Biol. 9 (10), 190061. 10.1098/rsob.190061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Shi Y., Zhang Y., Wang Y., Alotaibi F., Qiu L., et al. (2020). miRNA-5119 regulates immune checkpoints in dendritic cells to enhance breast cancer immunotherapy. Cancer Immunol. Immunother. 69, 951–967. 10.1007/s00262-020-02507-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Yu H., Yi S., Peng X., Xiao Z., et al. (2016). The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget 7 (29), 45370–45384. 10.18632/oncotarget.9659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P., Chen L., Yuan X., Luo Q., Liu Y., Xie G., et al. (2017). Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 36 (1), 53. 10.1186/s13046-017-0528-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.-X., Zhu W., Fang C., Fan L., Zou Z. J., Wang Y. H., et al. (2012). miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis 33 (7), 1294–1301. 10.1093/carcin/bgs179 [DOI] [PubMed] [Google Scholar]
- Zuo Z., Calin G. A., de Paula H. M., Medeiros L. J., Fernandez M. H., Shimizu M., et al. (2011). Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood, J. Am. Soc. Hematol. 118 (2), 413–415. 10.1182/blood-2011-01-330704 [DOI] [PMC free article] [PubMed] [Google Scholar]