Abstract
The cellular homeostasis of proteins (proteostasis) and RNA metabolism (ribostasis) are essential for maintaining both the structure and function of the brain. However, aging, cellular stress conditions, and genetic contributions cause disturbances in proteostasis and ribostasis that lead to protein misfolding, insoluble aggregate deposition, and abnormal ribonucleoprotein granule dynamics. In addition to neurons being primarily postmitotic, nondividing cells, they are more susceptible to the persistent accumulation of abnormal aggregates. Indeed, defects associated with the failure to maintain proteostasis and ribostasis are common pathogenic components of age-related neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. Furthermore, the neuronal deposition of misfolded and aggregated proteins can cause both increased toxicity and impaired physiological function, which lead to neuronal dysfunction and cell death. There is recent evidence that irreversible liquid–liquid phase separation (LLPS) is responsible for the pathogenic aggregate formation of disease-related proteins, including tau, α-synuclein, and RNA-binding proteins, including transactive response DNA-binding protein 43, fused in sarcoma, and heterogeneous nuclear ribonucleoprotein A1. Investigations of LLPS and its control therefore suggest that chaperone/disaggregase, which reverse protein aggregation, are valuable therapeutic targets for effective treatments for neurological diseases. Here we review and discuss recent studies to highlight the importance of understanding the common cell death mechanisms of proteostasis and ribostasis in neurodegenerative diseases.
Keywords: proteostasis, ribostasis, neurodegenerative disease, liquid–liquid phase separation, cell death mechanism
INTRODUCTION
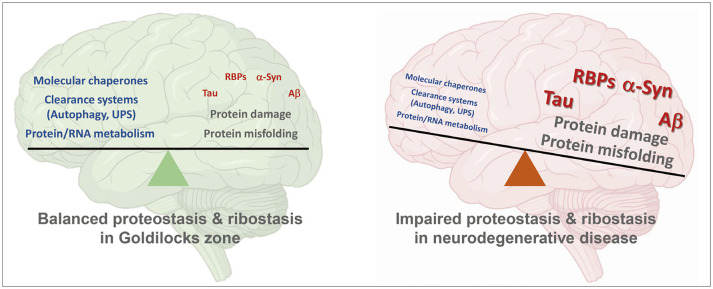
The most common neurodegenerative diseases, such as Alzheimer’s disease (AD), Parkinson’s disease (PD), and amyotrophic lateral sclerosis (ALS), are characterized by the progressive and selective loss of vulnerable neurons in the affected brain regions.1 Although the types and regional deposition of disease-associated proteins vary between diseases, there is accumulating evidence that neurodegenerative diseases share a common pathogenic mechanism that involves aberrant deposition of misfolded and aggregated proteins, which leads to impaired protein homeostasis (proteostasis) and ribostasis in the central nervous system.1 The ‘Goldilocks zone’ concept in astronomy states that planets that could support life need to revolve around stars within habitable regions that are suitable for life, neither too close to (too hot) nor too far from (too cold) the star. In biology, this Goldilocks zone can be applied to the balance or homeostasis of proteins and/or RNA metabolism that maintains normal cellular function (Fig. 1). Specifically, the molecular mechanism responsible for effective proteostasis and ribostasis is essential to remain within the Goldilocks zone, which improves the probability of cell survival. However, spatial and temporal disturbances in cellular homeostasis are closely associated with normal aging. Because differentiated neurons are postmitotic and cannot dilute the defects like in dividing cells, they progressively accumulate deleterious defects as age increases. Numerous molecular mechanisms that contribute to cellular homeostasis are also disrupted in age-related neurodegenerative diseases,2 which results in the deposition of aberrant aggregates in affected neurons, such as tau, α-synuclein (α-syn), and some RNA-binding proteins (RBPs) in AD, PD, and ALS, respectively.
Fig. 1. Balance or homeostasis of protein and RNA metabolism in the Goldilocks zone. The ideal Goldilocks zone facilitates molecular chaperones, clearance systems, and protein/RNA to perform their function in the most effective way. Molecular chaperones are components of the cellular protein quality control mechanism that suppresses irreversible dysfunctional protein aggregation. Autophagy and the UPS are clearance systems that regulate protein damage and misfolding caused by dysfunctional proteins, which contribute to defective protein and RNA metabolism in neurodegeneration. Damaged and misfolded proteins that contribute to neurodegeneration (tau, RBPs, α-syn, and Aβ) cause imbalance in the proteostasis and ribostasis of the brain, which leads to cell death. Figure was created using BioRender (https://biorender.com/). Aβ, amyloid-beta; RBPs, RNA-binding proteins; UPS, ubiquitin proteasome system; α-syn, α-synuclein.
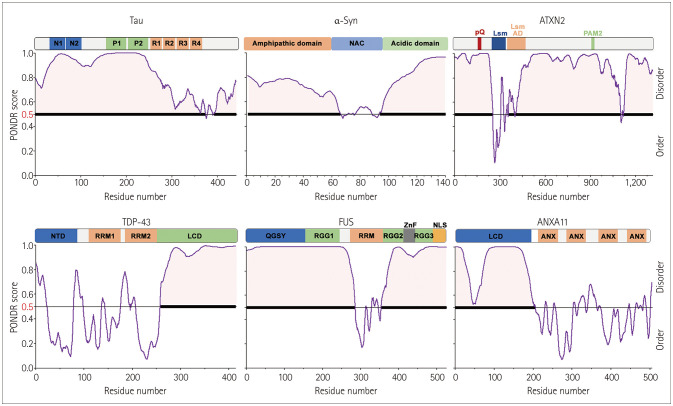
There is recent evidence that liquid–liquid phase separation (LLPS) is a key biophysical mechanism that drives disease-related protein aggregation.3 LLPS is a reversible thermodynamic process that occurs when biomolecules (proteins and RNAs) self-assemble, which produces distinct cytosolic or nuclear compartments known as biomolecular condensates.4 Furthermore, intrinsically disordered regions (IDRs) or low-complexity domains (LCDs) are polypeptide segments of amino acid sequences that do not mediate folding into specific secondary or tertiary structures. The nature of proteins that contain IDRs mean that they can enhance LLPS driven by protein–protein interactions. Moreover, tau, α-syn, and some ALS-linked RBPs also possess predicted IDRs and undergo reversible dynamic LLPS (Fig. 2). It is particularly interesting that disease-linked mutations and posttranslational modifications (e.g., phosphorylation) in these IDR-containing proteins can cause a phase transition from the reversible to the irreversible state, which leads to pathogenic aggregation.3 For example, many annexin A11 mutations in ALS are found in the N-terminal LCD, which alters their propensity for LLPS and the dynamics of stress granules (SGs).5,6 Transactive response DNA-binding protein 43 (TDP-43) mutations linked to ALS are frequently found in the C-terminal LCD; however, mutations of fused in sarcoma (FUS) are located in the N-terminal LCD. These ALS-linked mutations in LCDs play central roles in the ALS-related pathology of aberrant phase separation in RBPs that results in irreversible condensates forming in neurons and glia.7 Numerous proteins and mRNAs are formed in healthy cells during the assembly of SGs in the cytoplasm and can be continually disassembled by cellular mechanisms. However, continuously altered mutant RBP and RNA assembly generates irreversible ribonucleoprotein (RNP) granules and perturbs RNA homeostasis or ribostasis, which eventually leads to cell death. Therefore, effective control of ribostasis, which means proper cellular transcriptome regulation, and proteostasis, which is a suitable protein folding regulatory mechanism, is critical to cell function and survival.
Fig. 2. Intrinsic disorder predisposition of neurodegenerative-disease-causing proteins evaluated using the Predictor of Natural Disordered Regions (www.pondr.com/). Domain architecture of tau, α-syn, ATXN2, TDP-43, FUS, and ANXA11. The following domains are indicated in tau: N1 and N2 (N-terminal domains); P1 and P2 (proline-rich region); R1, R2, R3, and R4 (repeated sequences at the microtubule-binding domain). The six proteins share similar domain architectures and protein disorders. ANX, annexin domain; ANXA11, annexin A11; ATXN2, ataxin-2; Aβ, amyloid-beta; FUS, fused in sarcoma; LCD, low-complexity domain; Lsm, Like-Sm; Lsm AD, Lsm-associated domain; NAC, non-Aβ component; NLS, nuclear localization signal; NTD, N-terminal domain; PAM2, PABP-interacting motif 2; polyQ, polyglutamine domain; QGSY, QGSY-rich prion-like domain; RBPs, RNA-binding proteins; RGG, RGG-rich domain; RRM, RNA recognition motif; RRM1, RNA-recognition motif 1; RRM2, RNA-recognition motif 2; TDP-43, TAR DNA-binding protein 43; UPS, ubiquitin proteasome system; ZnF, zinc finger; α-syn, α-synuclein.
This review discusses the significance of understanding the common cell death mechanisms in proteostasis and ribostasis in neurodegenerative diseases.
Proteostasis network
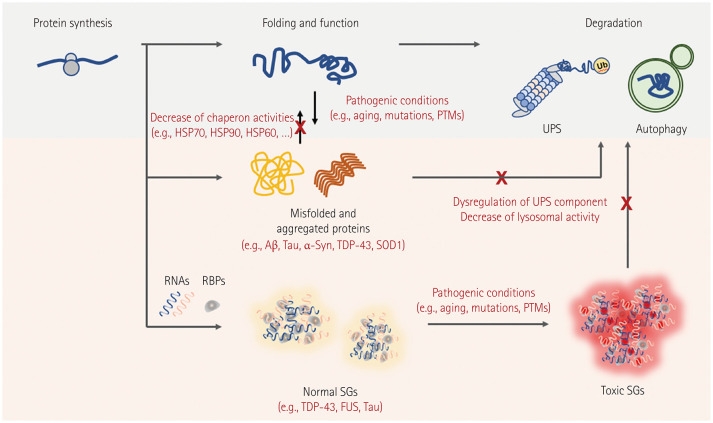
Proteostasis networks are highly interactive pathways that maintain the integrity and balance of proteins in order to maintain their functionality.8 Several cellular proteostasis networks have been developed in eukaryotic cells to ensure the proper folding of newly synthesized polypeptides, degradation of misfolded proteins, and control of protein aggregates, thus maintaining cellular proteostasis (Fig. 3). Importantly, the main components of the complex proteostasis network are molecular chaperones, the ubiquitin-proteasome system (UPS), and the autophagy-lysosomal pathway (ALP).9 These three types of modules delicately control proteome balance through interconnectivity centered on molecular chaperones, and precise regulation of these networks is essential to reducing the proteotoxicity caused by misfolded and aggregated proteins. Molecular chaperones play major roles in numerous cellular processes, including protein folding, disaggregation, degradation, trafficking, and signal transduction within cells.10 They are categorized into different functional families, including small heat-shock proteins (sHSPs), HSP90, HSP70, HSP60, and HSP40. Specifically, these factors enable the de novo folding of newly synthesized proteins and maintain their soluble and native conformations. Molecular chaperone expression is mostly regulated by heat-shock transcription factor 1 (HSF1), which adjusts cytoplasmic proteostasis during stress conditions.11,12 HSF1 is mostly monomeric and transcriptionally suppressed under normal circumstances by binding to its target HSPs, including HSP70 and HSP90. It therefore lacks the capacity to bind the heat-shock elements in the promoter regions of the HSP genes.13,14,15 The chaperones are dissociated from HSF1 and bound to denatured substrates during heat-shock and proteotoxic stresses. The released monomeric HSF1 subsequently translocates to the nucleus, where it undergoes posttranslational modification (i.e., phosphorylation) and induces the transcription of target HSPs by binding to heat-shock elements.13,14,15
Fig. 3. Altered proteostasis and ribostasis are common pathogenic mechanisms in neurodegenerative diseases. Under physiological conditions, newly synthesized proteins fold correctly to ensure their normal functions, and misfolded proteins are targeted to maintain proteostasis in degradation systems. Two protein degradation systems, the UPS and autophagy, are essential to maintaining cellular homeostasis. However, mutations and/or PTMs of disease-associated proteins (Aβ and tau in Alzheimer’s disease, α-syn in Parkinson’s disease, and SOD1, TDP-43, and FUS in ALS) form pathogenic aggregates and induce degradation system dysregulation. During stress conditions, ALS-associated RBPs (TDP-43 and FUS) are often reversibly assembled into SGs with target RNAs, and prolonged SGs are removed through the autophagy system. Chronic stress and/or RBP mutations lead to the irreversible accumulation of SGs and inhibit the normal functions of RBPs and target RNAs in maintaining RNA metabolism. Aβ, amyloid-beta; ALS, amyotrophic lateral sclerosis; HSP, heat-shock protein; PTMs, posttranslational modifications; SGs, stress granules; SOD1, superoxide dismutase-1; UPS, ubiquitin proteasome system; α-syn, α-synuclein.
To prevent the toxic effects caused by aberrant aggregates, misfolded and aggregated proteins must to be correctly eliminated through proteolysis. The UPS and ALP are the two primary proteolytic degradation pathways in eukaryotic cells. The UPS is a highly regulated mechanism of precisely coordinated enzymatic processes that lead to target protein ubiquitination through an enzymatic cascade that involves the ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes, and proteolytic degradation by the proteasome.16,17,18 Ubiquitin is a 76-amino acid protein that is covalently bounded to target proteins and initiates ubiquitination. Most proteins labeled by ubiquitination are also degraded by proteasomes. The balance between free- and ubiquitin-tagged proteins is regulated by the competing functions of E3 ubiquitin ligases and deubiquitinating enzymes. The UPS cooperates closely with molecular chaperones during protein quality control to recognize substrates and misfolded toxic aggregate proteins for in proteasomal degradation. For example, the carboxyl terminus of HSP70-interacting protein (CHIP) is an E3 ubiquitin ligase that selectively ubiquitylates misfolded proteins by collaborating with the molecular chaperones HSP70 and HSP90.19,20
An autophagy system is a critical mechanism that balances proteostasis via protein degradation with the UPS. However, the UPS is primarily responsible for removing individual proteins from the proteasome, while the autophagy system degrades larger aggregates in the lysosome. Autophagy is a cellular system for the lysosome-mediated degradation of toxic aggregated proteins and damaged organelles (e.g., mitochondria) that are engulfed by double-membrane vesicles known as autophagosomes.21 Notably, the following three autophagy types are commonly mentioned in eukaryotic cells depending on the mechanism of cargo delivery to the lysosome: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA).22,23,24 Furthermore, autophagy enhances survival as an adaptive response to stress in the presence of disease, whereas it may also promote apoptosis and disease in other cases.25,26
Mammalian cells use macroautophagy and CMA to selectively degrade misfolded and aggregated proteins. During macroautophagy, autophagy receptors or adaptors such as SQSTM1/p62 and NBR1 selectively recognize their cargos that contain ubiquitin-tagged proteins, and receptor–cargo complexes are loaded into the autophagosome via receptor–LC3 interaction.27 The resulting autophagosome is subsequently fused with a lysosome to produce an autolysosome, the contents of which are degraded by lysosomal hydrolases.28 Compared with macroautophagy, CMA is a non-vesicle-mediated autophagic mechanism that enhances the translocation of cytosolic cargos containing consensus sequences linked to the KFERQ motif from the cytosol to the lysosomal lumen by crossing its membrane.29,30 The constitutively expressed molecular chaperone, heat-shock cognate protein 70 (HSC70), and the lysosomal-associated membrane protein-2a subunit that functions as a lysosomal receptor are two critical factors for the multistep CMA process, which includes target recognition, unfolding, translocation into the lysosome, and degradation via lysosomal hydrolases.30 Collectively, misfolded and aggregated proteins that are resistant to removal via the UPS can be cleared by these lysosomal degradation processes, macroautophagy, and CMA.
The activities of the UPS and autophagy in various cell types gradually decrease with aging.31,32 This results in the accumulation of several toxic protein aggregates, particularly in postmitotic cells such as muscles and neurons that cannot dilute toxic aggregates via cell division. Proteostasis deficits are therefore closely linked to aging and neurodegenerative-disease pathogenesis.
Ribostasis and RNA-binding protein
RBPs regulate gene expression by controlling every stage of the RNA life cycle in the nucleus and cytoplasm.33,34 More than 50% of all known RBPs are expressed in the brain, where they are crucial for neuronal gene expression and play several roles in both normal and pathological brain function.35,36 Dysfunctional mutant RBPs in patients with neurodegenerative and neuromuscular diseases alter translational activity and dysregulate RNA metabolism in neurons. Dysregulation of critical steps in RNA metabolism can alter the phase transitions and granule-formation equilibrium of RBPs, which impairs their target RNAs and increases cellular stress and neurodegeneration.
RBPs are the main components of SGs that interact with each other, and these granules rapidly consolidate under stress conditions. Specifically, SGs are RNP granules without membranes that are responsible for the transient recruitment of mRNA transcripts in eukaryotic cells, which is one of the first responses to stress. SGs comprise mRNAs that are stalled in preinitiation translation complexes and can be triggered by different stimuli such as oxidative stress, osmotic stress, or heat shock, and are disassembled upon recovery.37 As a potent driver of RBP nucleation, SGs recruit proteins and cause the aggregates to form a core-protein–RNA interaction network.
The accumulation of SGs with pathological protein aggregates can lead to neurodegenerative disorders (Fig. 3). Specific RBPs self-assemble and drive the pathological RNP aggregate formation, which disrupts the normal ribostasis of cells and leads to their death. The pathological interplay between RBPs and SGs has emerged as a major pathogenic process causing neurotoxicity-related aggregation formation and contributing to the pathology of neurological disease processes. Furthermore, the disease-causing mutations in RBPs increase RNA granule formation, which leads to greater SG formation and disrupted axonal granule trafficking, which could cause more-widespread cellular toxicity.38 Pathological protein aggregates comprise different RBPs and SG components and are associated with distinct neuropathologies such as those in ALS, frontotemporal dementia (FTD), AD, or PD.
Proteostasis and ribostasis in neurodegenerative diseases
Alzheimer’s disease
AD is a common neurodegenerative disease characterized by progressive declines in cognition and memory. Its pathogenesis involves the deposition of extracellular amyloid-beta (Aβ) plaques and the production of intracellular neurofibrillary tangles (NFTs) composed of hyperphosphorylated tau/microtubule-associated protein tau aggregates.39 These two characteristics imply that impaired molecular chaperone function and/or protein quality control may contribute to AD pathology, since they are responsible for the degradation of misfolded and aggregated proteins. Molecular chaperone HSPs limit Aβ and NFT aggregation and enhance the binding of ubiquitin to misfolded proteins for degradation. Proteomic analyses of transgenic mice with amyloid precursor protein (APP) also revealed changes in the expression of cellular chaperones such as HSP60 and HSP70, indicating that the Aβ peptide may influence chaperone expression.40 HSP70 overexpression exerts a neuroprotective effect against the toxic effects of intracellularly expressed Aβ in neurons, suppresses AD-related phenotypes in mice, and inhibits the early stage of Aβ aggregation in vitro.41,42,43 Similarly, numerous studies have found that HSP90 prevents Aβ aggregation.42,44 Furthermore, the UPS plays a crucial role in regulating Aβ accumulation in neurons by either reducing Aβ synthesis or increasing its proteolytic destruction. Macroautophagy has been proposed as a pathway that generates Aβ.45 The autophagosome is believed to be the principal location of Aβ processing because Aβ-processing enzymes and Aβ itself colocalize with autophagosomes.45,46 In the brains of patients and mouse models with AD, abnormal accumulation of autophagic compartments containing both APP and Aβ was observed.45,47,48 Reversing autophagy malfunction by enhancing lysosomal activities in an AD mouse model had therapeutic effects, such as reduced amyloid pathology and alleviation of memory impairment.49 Moreover, tau aggregates are also degraded through the UPS and ALPs. HSP70 and HSP90 can decrease the production of NFTs by promoting their dephosphorylation and degradation. The CHIP–HSC70 complex cooperates with the E2-conjugating enzyme UbcH5B to ubiquitinate phosphorylated tau (P-tau) for proteasomal degradation and inhibits P-tau-induced cell death.50 HSP90 and the cochaperone CDC37 coordinate tau phosphorylation by regulating the activation of some kinases, including cyclin-dependent kinase 5 (CDK5) and AKT.51,52,53 Consistent with these findings, CDK5 activity is also higher in the prefrontal cortex of the brains of patients with AD.54
In addition to AD, tau mutations that cause tau accumulation, increase tau phosphorylation, and increase neuron loss have been discovered in patients with FTD.55,56 Although tau is not an RBP, it engages RNA sequestered by tau inclusions and promotes tau phase separation and fibrillization in tauopathies.57 The specific RNAs in tau aggregates that contribute to disease progression are still unclear. However, a recent study identified RNAs, small nucleolar RNAs, small nuclear RNAs, and some tRNAs that were enriched in tau aggregates and revealed related perturbations in splicing speckles. Nuclear speckles are mislocalized to cytoplasmic tau aggregates in cell models, transgenic mouse brains based on an FTD-linked tau model, and the brains of patients with AD and FTD.58 Considering the discovery of specific RNAs sequestered by nuclear and cytoplasmic tau aggregates, further investigations are still required to determine how tau aggregation alters cellular RNA and which therapeutic strategies disrupt abnormal tau–RNA interactions to mitigate tauopathies. A recent study demonstrated that human tau forms dynamic liquid droplets in vitro at the physiological protein level.59 The disease-associated modifications, including the AT8 phosphoepitope and the P301L tau mutation that is linked to inherited tauopathy, enhanced LLPS in vitro and the P301L mutant presented the greatest oligomer formation following extended phase separation. That study suggested that tau phase separation facilitates the formation of neurotoxic pathogenic tau conformations in the neurons of patients with the tau P301L mutation or pathological phospho-tau.
Several SG components have been linked to tau pathogenesis. HuD/ELAVL4, a splicing regulator that encodes a neuronal RBP needed for brain development, stabilizes tau translation by transporting tau mRNA from the neuron cell body to the axon in the form of granules. ELAVL4 shows RNA-dependent accumulation within G3BP1-positive SGs under oxidative stress and colocalization of ELAVL4 and tau in a 2-month tau mutant organoids, supporting the interactions among tau, ELAVL4, and SG function.60 Besides G3BP1, tau also interacts with RBP TIA1 in SGs, which regulates tau misfolding and aggregation and promotes tau-mediated primary neuron degeneration.61 A study found that TIA1 knockdown and knockout inhibited tau misfolding and tau-mediated toxicity in cultured hippocampal neurons.62 These processes also prevented tau granule formation in neurons via the translational inhibitor cycloheximide, which prevents elongation and inhibits SG formation, suggesting that pharmacological interventions that synergistically modulate both SG and tau can inhibit tauopathies.
Parkinson’s disease
PD is clinically characterized by bradykinesia, muscular rigidity, rest tremor, and postural and gait impairments caused by the progressive loss of dopaminergic neurons in the substantia nigra.63 A major pathogenic feature of PD is the deposition of misfolded α-syn into Lewy bodies, indicating dysfunction in the quality control of proteins.64,65 Numerous characterizations of Lewy bodies have indicated that molecular chaperones (HSP27, HSP60, HSP70, HSP90, HSP110, and CHIP) and UPS components (proteasome subunits, ubiquitination, deubiquitination enzymes, and proteasome activators) are prevalent in these aggregates and have highly ubiquitinated proteins.66,67,68 Furthermore, human genetics studies to identify PD-related genes have identified mutations in genes that encode Parkin/PARK2 (E3 ubiquitin ligase) and UCH-L1/PARK5 (ubiquitin C-terminal hydrolase), which are closely associated with UPS function.69,70,71 Dysregulation of ALP components has also been reported in PD. A significant decrease in the number of lysosomes in dopaminergic neurons preceded autophagosome accumulation and dopaminergic cell death in patients and mouse models with PD.72 Several other studies related to PD and dementia with Lewy bodies (DLB) also found an increase in the autophagosome marker LC3-II and a decrease in the lysosomal proteins LAMP1 and LAMP2A, suggesting the presence of ALP impairments in α-synucleinopathies.73,74,75,76,77,78 Moreover, several genetics findings also support the role of ALP in PD pathogenesis.
LRRK2 mutations are the most common genetic risk factors for PD. LRRK2 is degraded by macroautophagy and CMA, but G2019S (a common mutation) impairs this degradation.79 LRRK2-G2019S also acts on the lysosomal receptor LAMP2A and impairs CMA, resulting in the accumulation of CMA substrates such as α-syn. LRRK2-G2019S expression also increased autophagic vacuoles and neurite reorganization in neuroblastoma cells.80 Besides these findings, identifying mutations in the genes that encode the lysosomal protein ATP13A2/PARK9, lysosomal enzyme glucocerebrosidase, and vacuolar protein sorting-associated protein 35 support the idea that ALP malfunction contributes to the pathogenesis of PD.81,82,83,84,85 The autophagy system can also be involved in the selective degradation of specific organelles. Parkin/PARK2 and PINK1/PARK6 promote the selective autophagic clearance of damaged mitochondria (mitophagy), and their inability to remove defective mitochondria (for example due to disease-associated mutations) has been linked to PD pathology.86 In cultured cell and animal models, the modulation of molecular chaperones, UPS, and ALP has shown protective efficacy against α-syn aggregation. Overall, these findings indicate that defects in protein quality control may be responsible for the α-syn accumulation and Lewy-body production in PD.
α-syn is a DNA- and lipid-binding protein that is typically localized to the nucleus and plasma membrane, but it has also been found to form abnormal cytoplasmic aggregates in patients with PD. α-syn also forms toxic soluble oligomers and fibrillar proteins in PD and related Lewy-body disorders (PD with dementia and DLB).87 Nuclear α-syn modulates DNA repair, in which the protein can be found in foci that colocalize with DNA damage markers in the cortical neurons of mice.88 α-syn removal from mouse neurons was associated with increased levels of double-strand breaks in DNA and a reduced ability to repair them, which could be rescued by α-syn re-expression, indicating that α-syn regulates cellular repair responses. A recent study suggested that α-syn LLPS and subsequent liquid-to-solid phase transition could be pathological, which can only be triggered by the critical concentration of phase separation and LLPS-mediated aggregation or the amyloid formation of α-syn.89 Overall, many studies have proposed that α-syn aggregation is neurotoxic and that LLPS is the crucial mechanism causing abnormal α-syn accumulation.
Amyotrophic lateral sclerosis
ALS is a devastating neurodegenerative disease that is characterized by a progressive loss of upper and lower motor neurons in the motor cortex, brain stem, and spinal cord, which leads to muscle weakness, atrophy, and spasticity.90 There is abundant evidence that protein misfolding and aggregation play roles in ALS development and progression, similar to in other neurodegenerative diseases.91 Three major types of pathogenic aggregates are deposited independently within the motor neurons of patients with ALS. TDP-43-positive inclusions were the most common inclusions in patients with ALS (–97%), followed by superoxide dismutase-1 (SOD1, –2%) and FUS (–1%).91 These inclusions are closely linked to proteostasis and/or ribostasis impairment as major contributors to ALS pathogenesis.
Many ALS-related proteins, including SQSTM1/p62, optineurin (OPTN), ubiquilin2 (UBQLN2), valosin-containing protein, and TANK-binding kinase 1 (TBK1), are involved in the UPS and ALP for proteostasis.92,93,94,95,96,97 This suggests that disruption of protein quality control is important in ALS pathology. SQSTM1/p62 and OPTN also function as autophagic receptors that recognize ubiquitinated cargo and promote the autophagy-mediated degradation of misfolded and aggregated proteins. A neuropathological investigation of patients with ALS and mutations in SQSTM1/p62 and OPTN revealed p62- and OPTN-immunoreactive cytoplasmic inclusions, respectively, accompanied by Ub- and TDP43-positive inclusions, implying proteostasis disturbance in the motor neurons.98,99 However, how these ALS-associated mutations contribute to ALS etiology remains unknown. TBK1, which is a serine/threonine protein kinase, binds and phosphorylates SQSTM1/p62 and OPTN, which improves their linkage ability to LC3 and ubiquitinated cargo as well as promoting autophagic degradation.100,101 However, TBK1 haploinsufficiency causes familial ALS and FTD, and may cause impaired autophagic degradation and lead to the accumulation of protein aggregates.96,97 In contrast, UBQLN2 is a member of the ubiquilin family that may function in the UPS and ALP for degradation.102 ALS-associated UBQLN2 mutations lead to ubiquitin-mediated proteasomal degradation impairment in cultured cells and the accumulation of skein-like inclusions, which are immunoreactive for antibodies against UBQLN2, ubiquitin, TDP43, and FUS in the spinal motor neurons of patients with ALS.94,103
SOD1 was the first gene discovered to cause ALS.104 ALS-associated mutant SOD1 is highly susceptible to structural alterations that cause protein aggregation.105,106 Consistent with this, misfolded SOD1 inclusions were found in the motor neurons of patients with ALS who carried SOD1 mutations and in those who carried mutations in ALS-causing genes other than SOD1.107 Moreover, SOD1 inclusions also sequester ubiquitin, molecular chaperones (e.g., HSP70 and HSP27), and the proteasome.108,109 Wild-type and mutant SOD1 can be removed by proteasomes and by macroautophagy.110 Studies involving cultured cells and mouse models have found that misfolded SOD1 mutations can inhibit proteasomal and autophagic processes, resulting in the accumulation of large mutant SOD1 aggregates.111,112
Toxic RBP aggregation and RNA-protein granule accumulation are found in the brains and spinal cords of both patients with ALS and with FTD.113 Alterations in RBPs cause the malfunction of essential RNA metabolism pathways that leads to motor neuron degeneration. SG components colocalize with neuropathology in the brain tissue of patients and in animal models with ALS and with FTD.114 Several disease-causative or disease-associated RBP mutations involved in SG formation, such as TDP-43, ataxin-2 (ATXN2), FUS, heterogeneous nuclear RNP (hnRNP) A1, hnRNP A2B1, and TIA1, have been found to increase the risks of ALS and FTD.115
RBP TDP-43 regulates transcription and forms transient cytoplasmic aggregates that colocalize with SGs under stress conditions. Specifically, TDP-43 is the main component that accumulates in the cytoplasmic, ubiquitin-positive protein inclusions in the neurons of patients with ALS and with FTD.114 Disease mutations in the prion-like domain in the C-terminus and fragment of TDP-43 cause TDP-43 mislocalization and consequently sequestration within SGs, disturbing the assembly and disassembly dynamics of SGs.116 Studies have found that increased aggregation propensities accelerate the disease onset in patients with familial ALS who carry the D169G, G298S, M337V, and N352S TDP-43 mutations. G298S and M337V were in particular found to decrease motility, increase TDP-43 granule viscosity, and disrupt axonal transport functions.117 The ALS-linked D169G mutant is the only TDP-43 mutation within the RNA-recognition motif 1 (RRM1) domain that abnormally accumulates in SGs, enhances the formation of TDP-43 inclusions, and prevents the binding capacity of the RRM1 domain to ATP, suggesting that altered charge or hydrophobicity trigger dynamic changes and enhance aggregation.118
TDP-43 is posttranslationally modified by ubiquitination, SUMOylation, and phosphorylation. Abnormal TDP-43 hyperphosphorylation of S379, S403/404, and S409/410 has been implicated in protein aggregation in the tissues of patients with ALS and with FTD-TDP.119 Among the various kinases suggested to be involved in TDP-43 phosphorylation, the constitutively hyperactive form of CSNK1E and coexpression of TDP-43 in SH-SY5Y cells caused insoluble phosphorylation of TDP-43 at S393/395. The insoluble phosphorylated TDP-43, in turn, functions as a seed for the accumulation of cytoplasmic mislocalized TDP-43, indicating that TDP-43 phosphorylation by an abnormal kinase form induces both aggregation and cytotoxicity in TDP-43.120
Many studies have investigated the potential therapeutic targets for pathological aggregation in TDP-43. For example, drug screening in primary mouse cortical neurons identified novel agents that decrease SG formation, which further reduced the formation of TDP-43 inclusions, making this a potential therapy for ALS.121 Given the interplay between TDP-43 phosphorylation and aggregation, pharmacological inhibition of cell division cycle 7 kinase122 and protein casein kinase-1d can prevent TDP-43 phosphorylation123 and aggregation,124 while maintaining or restoring the nuclear localization of TDP-43.
Mutations in other RBPs, in addition to TDP-43 mutations, have been found in patients with ALS. For example, mutant expansion in the CAG polyglutamine tract of ATXN2 causes spinocerebellar ataxia 2, and intermediate-length expansion in the same protein is associated with an increased risk of ALS. ATXN2 is abnormally aggregated in the spinal cord neurons of patients with ALS and is a potent modifier of TDP-43 toxicity in an RNA- and dose-dependent in yeast and drosophila.125 Recent studies have similarly found that reducing ATXN2 using antisense oligonucleotides is effective in preventing TDP-43 aggregation and increasing the survival rates of transgenic mice.126 FUS is a predominantly nuclear RBP that is involved in nuclear mRNA metabolism and DNA damage repair. Consequently, abnormal accumulation of insoluble FUS protein in ALS is caused by mutations mostly in the C-terminus of the protein that disrupt a nuclear localization signal (NLS); however, in FTD cases, wild-type FUS inclusions are found in the absence of tau or TDP-43 pathological inclusions, suggesting that the two diseases have distinct pathogenic mechanisms.127 hnRNP proteins are the most abundant RBPs that regulate alternative pre-mRNA splicing in cells; specifically hnRNPA1 function in mRNA transcription and splicing. Pathogenic mutations in the prion-like domains of hnRNPA1 induce exacerbated assembly into self-seeding fibrils, altering the dynamics of RNA granule assembly and causing ALS.128 Loss of RBP hnRNPA2B1 results in alternative splicing of ALS-associated D-amino acid oxidase, which reduces D-serine metabolism. Furthermore, the fibroblasts and induced pluripotent stem-cell-derived motor neurons of patients with the hnRNPA2B1 D290V mutations displayed widespread splicing changes. Mutant motor neurons also cause abnormal cytoplasmic aggregates and decreased cell survival.129 RBPs in ALS therefore have multiple functional roles in RNA metabolism that lead to ALS pathogenesis. However, additional investigations of RBP/RNA alterations in ALS are needed to address the complexity and multifactorial nature of the disease.
A therapeutic strategy to combat neurodegenerative diseases
The diversity in the genetic associations, pathologies, and clinical features of neurodegenerative diseases such as AD, PD, and ALS indicate that they are complex and heterogeneous. Such traits represent obstacles to researchers developing effective treatments. Over the past few decades, the development of disease-preventing or disease-modifying drugs for the treatment of neurodegenerative diseases has accelerated, but none of the identified agents are currently curative. Most of these drugs have been developed to target single molecules and/or mechanisms, and some of the FDA-approved drugs have neuroprotective and beneficial effects on patients. For example, riluzole, an FDA-approved drug for ALS that downregulates the glutamatergic neurotransmission pathway, slows disease progression and extends the lifespan by several months.130,131 Donepezil acts as a cholinesterase inhibitor and is most commonly used to treat AD.132 However, there is still currently no curative therapeutic strategy available for AD, PD, or ALS. Addressing this requires new concepts to understand the complex mechanisms underlying neurodegenerative diseases and the develop of more effective treatments. Given the complex and multifactorial nature of neurodegenerative diseases, treatment strategies that can initially target multiple risk factors and disease mechanisms may be the most effective in slowing or stopping disease progression. To develop effective drugs, we also need to apply emerging concepts of stratification (i.e., subgrouping of patients according to their clinical profiles, genetic backgrounds, or other risk factors) as well as personalized medicine.133
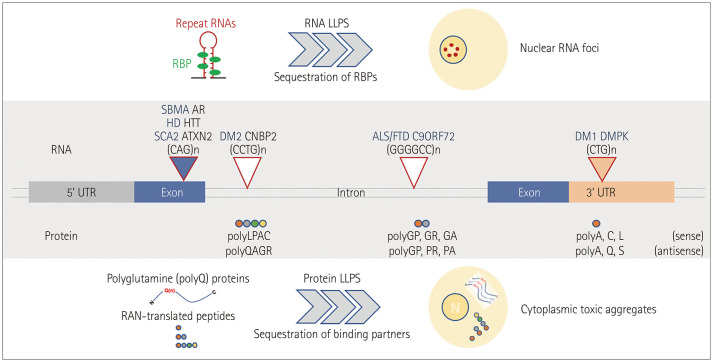
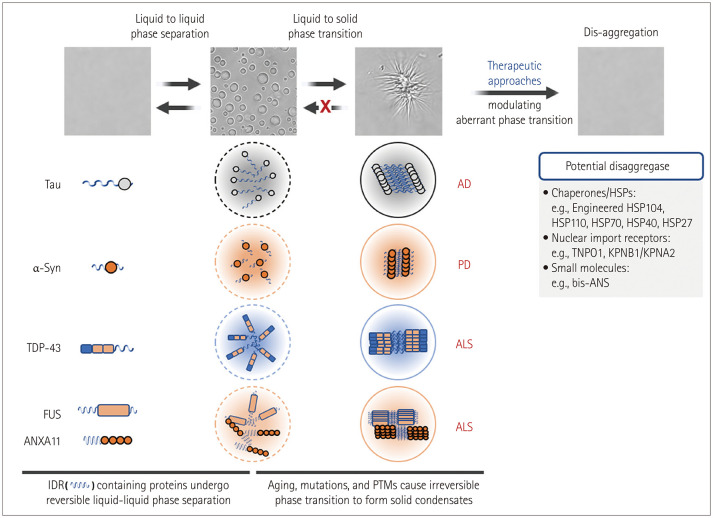
The mechanisms underlying neurotoxicity induced by misfolded and aggregated proteins remain controversial. The deposition of pathogenic aggregates in affected brain regions has been suggested to be linked to the toxic gain-of-function and loss-of-function mechanisms that can contribute to neuronal dysfunction and cell death.134 A therapeutic strategy to restore misfolded/aggregated proteins to their native functional structure and cellular compartments can therefore more effectively alleviate neurotoxicity by simultaneously reversing the gain- and loss-of-toxicity rather than by simply eliminating them. Evidence has accumulated over the past decade that disease-causing proteins, including tau in AD, α-syn in PD, and RBPs (e.g., TDP-43, FUS, and hnRNPA1) in ALS can undergo LLPS.3 LLPS is a reversible process of biomolecules (e.g., proteins and RNAs) that tend to self-assemble to form biological condensates under physiological conditions. Aberrant phase transitions of RNA molecules have also been found to occur in several repeat-RNA-expansion diseases to form toxic nuclear RNA foci (Fig. 4).135 During pathogenic processes such as gene mutations, posttranslational modifications, stress conditions, and aging, aberrant phase separation behavior (from liquid–liquid phase separation to liquid–solid phase transition) may trigger the irreversible and toxic aggregation of target disease proteins. A disaggregase is a therapeutic agent with the ability to control pathological phase transitions, which can therefore have a beneficial effect on neurodegenerative diseases caused by protein misfolding or abnormal aggregation (Fig. 5). HSPs, different proteins that possess molecular chaperone activity, and specific RNAs have been suggested as effective agents for dissolving abnormal protein aggregates by regulating the phase separation of disease-associated proteins with substrate specificity.
Fig. 4. The repeating-nucleotide-expansion disease of LLPS-associated degenerative disease. Repeat-expansion diseases are caused by expansions of DNA repeats of target genes. Repeat expansion can occur in UTRs, coding exons, or introns of target genes. For example, CTG repeats in 3’UTR; DMPK for myotonic DM1, CAG repeats in coding exons; AR for SBMA, HTT for HD, and ATXN2 for spinocerebellar ataxia 2, CCTG repeats in introns; CCHC-type zinc-finger nucleic-acid-binding protein for DM2, GGGGCC repeats in introns; C9ORF72 for ALS/FTD. Transcribed repeats containing RNAs undergo an aberrant phase separation into solid-like structures and form nuclear RNA foci. Furthermore, RNA repeats undergo RAN translation and produce polypeptides that can also form toxic aggregates through irreversible LLPS. These pathogenic structures, RNA foci, and RAN-translated peptide aggregates result in RNA toxicity and repeat-protein toxicity, respectively, via the sequestration of RBPs and/or binding partners. ALS/FTD, amyotrophic lateral sclerosis/frontotemporal dementia; AR, androgen receptor; DM1, myotonic dystrophy type 1; DM2, myotonic dystrophy type 2; DMPK, dystrophia myotonica protein kinase; HD, Huntington’s disease; HTT, huntingtin; LLPS, liquid–liquid phase separation; RAN, repeat-associated non-AUG; SBMA, spinobulbar muscular atrophy; UTRs, untranslated regions.
Fig. 5. Disaggregases may reverse the irreversible transitions of disease-causing proteins that undergo LLPS. IDRs containing proteins in neurodegenerative diseases –tau in AD, α-syn in PD, RBPs (e.g., TDP43, FUS, and ANXA11) in ALS– can undergo LLPS. Differential interference contrast images show that ANXA11 undergoes a reversible LLPS to form liquid droplets, followed by an aberrant phase transition to form a fibrillar structure.6 Disaggregases to reverse the LLPS and aberrant phase transition of disease-causing proteins are a potential therapeutic target. AD, Alzheimer’s disease; ALS, amyotrophic lateral sclerosis; FUS, fused in sarcoma; IDR, intrinsically disordered region; PD, Parkinson’s disease; PTMs, posttranslational modifications; TDP-43, transactive response DNA-binding protein 43; TNPO1, transportin 1; KPNB1/KPNA2, karyopherin-β1/karyopherin-α2; bis-ANS, 4,4′-dianilino-1,1’-binaphthyl-5,5′-disulfonic acid; α-syn, α-synuclein.
HSP104 is one of the most studied AAA+ protein disaggregases found in yeast that maintains proteostasis by remodeling disordered protein aggregates to their original conformation.136 Several studies that used neurodegenerative disease models have found HSP104 to be a potentially effective therapeutic agent. The expression of HSP104 protects dopaminergic neurons by restoring α-syn aggregation in rat and Caenorhabditis elegans models with PD.137,138 The engineered HSP104 variant with enhanced ATPase activity also alleviated the toxicity and aggregation of ALS-associated RBPs in Saccharomyces cerevisiae, including TDP-43, FUS, and TAF15, rather than EWSR1.139 Although HSP104 exhibits a broad spectrum of activity against the pathogenic aggregation of disease-causing proteins, it is crucial to determine endogenous human disaggregase systems, including HSP110, HSP70, HSP40, and sHSP. Similar to HSP104 activity, HSP110, HSP70, and HSP40 cooperate to promote the disaggregation of misfolded and aggregated proteins and the disassembly of SGs.140,141 Furthermore, recent studies have demonstrated that HSP70 and HSP40 may prevent the toxic aggregation of TDP-43 and FUS by regulating their phase separation.142,143 Besides these HSPs acting as disaggregases, karyopherins, which function classically in nucleocytoplasmic transport, were discovered as disaggregases for ALS-associated RBPs to prevent irreversible phase separation and abnormal cytoplasmic aggregation. Transportin 1, a member of the karyopherin family, preferentially binds to the proline–tyrosine NLS of FUS and hnRNPA1, reverses their aberrant phase separation, and mitigates their neurodegeneration, whereas the karyopherin β1–importin-α complex prevents and reverses TDP-43 fibrillization that harbors classical NLSs.144 Moreover, LLPS of RBPs is also regulated by RNA binding, which can be exploited to design novel therapies for neurodegenerative diseases associated with RBP aggregation. Donnelly and colleagues found that TDP-43 targeting oligonucleotides (known as bait RNA) can prevent aberrant TDP-43 phase transitions and alleviate neurotoxicity.145
In conclusion, protein disaggregases that mitigate disease toxicity may yield important advances in the treatment of neurodegenerative diseases driven by abnormal phase transitions in aggregation-prone proteins.
Footnotes
- Conceptualization: all authors.
- Data curation: all authors.
- Visualization: all authors.
- Investigation: all authors.
- Writing—original draft: all authors.
- Writing—review & editing: all authors.
Conflicts of Interest: The authors have no potential conflicts of interest to disclose.
Funding Statement: The research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NRF-2018M3C7A1056512).
Availability of Data and Material
Data sharing not applicable to this article as no datasets were generated or analyzed during the study.
References
- 1.Dugger BN, Dickson DW. Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol. 2017;9:a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hou Y, Dan X, Babbar M, Wei Y, Hasselbalch SG, Croteau DL, et al. Ageing as a risk factor for neurodegenerative disease. Nat Rev Neurol. 2019;15:565–581. doi: 10.1038/s41582-019-0244-7. [DOI] [PubMed] [Google Scholar]
- 3.Zbinden A, Pérez-Berlanga M, De Rossi P, Polymenidou M. Phase separation and neurodegenerative diseases: a disturbance in the force. Dev Cell. 2020;55:45–68. doi: 10.1016/j.devcel.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Feng Z, Chen X, Wu X, Zhang M. Formation of biological condensates via phase separation: characteristics, analytical methods, and physiological implications. J Biol Chem. 2019;294:14823–14835. doi: 10.1074/jbc.REV119.007895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, et al. RNA granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell. 2019;179:147–164.e20. doi: 10.1016/j.cell.2019.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahm M, Lim SM, Kim YE, Park J, Noh MY, Lee S, et al. ANXA11 mutations in ALS cause dysregulation of calcium homeostasis and stress granule dynamics. Sci Transl Med. 2020;12:eaax3993. doi: 10.1126/scitranslmed.aax3993. [DOI] [PubMed] [Google Scholar]
- 7.Vahsen BF, Gray E, Thompson AG, Ansorge O, Anthony DC, Cowley SA, et al. Non-neuronal cells in amyotrophic lateral sclerosis - from pathogenesis to biomarkers. Nat Rev Neurol. 2021;17:333–348. doi: 10.1038/s41582-021-00487-8. [DOI] [PubMed] [Google Scholar]
- 8.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 9.Labbadia J, Morimoto RI. The biology of proteostasis in aging and disease. Annu Rev Biochem. 2015;84:435–464. doi: 10.1146/annurev-biochem-060614-033955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 11.Dai C, Sampson SB. HSF1: guardian of proteostasis in cancer. Trends Cell Biol. 2016;26:17–28. doi: 10.1016/j.tcb.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Labbadia J, Morimoto RI. Rethinking HSF1 in stress, development, and organismal health. Trends Cell Biol. 2017;27:895–905. doi: 10.1016/j.tcb.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- 14.Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS, et al. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. Elife. 2016;5:e18638. doi: 10.7554/eLife.18638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- 16.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 17.Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol. 2005;6:79–87. doi: 10.1038/nrm1552. [DOI] [PubMed] [Google Scholar]
- 19.Murata S, Minami Y, Minami M, Chiba T, Tanaka K. CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep. 2001;2:1133–1138. doi: 10.1093/embo-reports/kve246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esser C, Alberti S, Höhfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 23.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Majeski AE, Dice JF. Mechanisms of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2435–2444. doi: 10.1016/j.biocel.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Jung S, Jeong H, Yu SW. Autophagy as a decisive process for cell death. Exp Mol Med. 2020;52:921–930. doi: 10.1038/s12276-020-0455-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yonekawa T, Thorburn A. Autophagy and cell death. Essays Biochem. 2013;55:105–117. doi: 10.1042/bse0550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eskelinen EL, Saftig P. Autophagy: a lysosomal degradation pathway with a central role in health and disease. Biochim Biophys Acta. 2009;1793:664–673. doi: 10.1016/j.bbamcr.2008.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Cuervo AM, Wong E. Chaperone-mediated autophagy: roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 31.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 32.Hipp MS, Kasturi P, Hartl FU. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 33.Brannan KW, Jin W, Huelga SC, Banks CA, Gilmore JM, Florens L, et al. SONAR discovers RNA-binding proteins from analysis of large-scale protein-protein interactomes. Mol Cell. 2016;64:282–293. doi: 10.1016/j.molcel.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schieweck R, Ninkovic J, Kiebler MA. RNA-binding proteins balance brain function in health and disease. Physiol Rev. 2021;101:1309–1370. doi: 10.1152/physrev.00047.2019. [DOI] [PubMed] [Google Scholar]
- 35.Bryant CD, Yazdani N. RNA-binding proteins, neural development and the addictions. Genes Brain Behav. 2016;15:169–186. doi: 10.1111/gbb.12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 37.Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell. 2020;181:325–345.e28. doi: 10.1016/j.cell.2020.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alami NH, Smith RB, Carrasco MA, Williams LA, Winborn CS, Han SSW, et al. Axonal transport of TDP-43 mRNA granules is impaired by ALS-causing mutations. Neuron. 2014;81:536–543. doi: 10.1016/j.neuron.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheltens P, De Strooper B, Kivipelto M, Holstege H, Chételat G, Teunissen CE, et al. Alzheimer’s disease. Lancet. 2021;397:1577–1590. doi: 10.1016/S0140-6736(20)32205-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano M, Yamashita T, Nagano K, Otani M, Maekura K, Kamada H, et al. Proteomic analysis of the hippocampus in Alzheimer’s disease model mice by using two-dimensional fluorescence difference in gel electrophoresis. Neurosci Lett. 2013;534:85–89. doi: 10.1016/j.neulet.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Magrané J, Smith RC, Walsh K, Querfurth HW. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed beta-amyloid in neurons. J Neurosci. 2004;24:1700–1706. doi: 10.1523/JNEUROSCI.4330-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans CG, Wisén S, Gestwicki JE. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J Biol Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 43.Hoshino T, Murao N, Namba T, Takehara M, Adachi H, Katsuno M, et al. Suppression of Alzheimer’s disease-related phenotypes by expression of heat shock protein 70 in mice. J Neurosci. 2011;31:5225–5234. doi: 10.1523/JNEUROSCI.5478-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, Lallemang M, Hermann B, Wallin C, Loch R, Blanc A, et al. ATP impedes the inhibitory effect of Hsp90 on Aβ40 fibrillation. J Mol Biol. 2021;433:166717. doi: 10.1016/j.jmb.2020.11.016. [DOI] [PubMed] [Google Scholar]
- 45.Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, et al. Macroautophagy--a novel beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu WH, Kumar A, Peterhoff C, Shapiro Kulnane L, Uchiyama Y, Lamb BT, et al. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide over-production and localization in Alzheimer’s disease. Int J Biochem Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 47.Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, et al. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Varo R, Trujillo-Estrada L, Sanchez-Mejias E, Torres M, Baglietto-Vargas D, Moreno-Gonzalez I, et al. Abnormal accumulation of autophagic vesicles correlates with axonal and synaptic pathology in young Alzheimer’s mice hippocampus. Acta Neuropathol. 2012;123:53–70. doi: 10.1007/s00401-011-0896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain. 2011;134(Pt 1):258–277. doi: 10.1093/brain/awq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimura H, Schwartz D, Gygi SP, Kosik KS. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 51.Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, et al. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc Natl Acad Sci U S A. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lackie RE, Maciejewski A, Ostapchenko VG, Marques-Lopes J, Choy WY, Duennwald ML, et al. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front Neurosci. 2017;11:254. doi: 10.3389/fnins.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 54.Lee KY, Clark AW, Rosales JL, Chapman K, Fung T, Johnston RN. Elevated neuronal Cdc2-like kinase activity in the Alzheimer disease brain. Neurosci Res. 1999;34:21–29. doi: 10.1016/s0168-0102(99)00026-7. [DOI] [PubMed] [Google Scholar]
- 55.Stanford PM, Shepherd CE, Halliday GM, Brooks WS, Schofield PW, Brodaty H, et al. Mutations in the tau gene that cause an increase in three repeat tau and frontotemporal dementia. Brain. 2003;126(Pt 4):814–826. doi: 10.1093/brain/awg090. [DOI] [PubMed] [Google Scholar]
- 56.Woollacott IO, Rohrer JD. The clinical spectrum of sporadic and familial forms of frontotemporal dementia. J Neurochem. 2016;138 Suppl 1:6–31. doi: 10.1111/jnc.13654. [DOI] [PubMed] [Google Scholar]
- 57.Ginsberg SD, Crino PB, Lee VM, Eberwine JH, Trojanowski JQ. Sequestration of RNA in Alzheimer’s disease neurofibrillary tangles and senile plaques. Ann Neurol. 1997;41:200–209. doi: 10.1002/ana.410410211. [DOI] [PubMed] [Google Scholar]
- 58.Lester E, Ooi FK, Bakkar N, Ayers J, Woerman AL, Wheeler J, et al. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron. 2021;109:1675–1691.e9. doi: 10.1016/j.neuron.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanaan NM, Hamel C, Grabinski T, Combs B. Liquid-liquid phase separation induces pathogenic tau conformations in vitro. Nat Commun. 2020;11:2809. doi: 10.1038/s41467-020-16580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atlas R, Behar L, Elliott E, Ginzburg I. The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem. 2004;89:613–626. doi: 10.1111/j.1471-4159.2004.02371.x. [DOI] [PubMed] [Google Scholar]
- 61.Maziuk BF, Apicco DJ, Cruz AL, Jiang L, Ash PEA, da Rocha EL, et al. RNA binding proteins co-localize with small tau inclusions in tauopathy. Acta Neuropathol Commun. 2018;6:71. doi: 10.1186/s40478-018-0574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanderweyde T, Apicco DJ, Youmans-Kidder K, Ash PEA, Cook C, Lummertz da Rocha E, et al. Interaction of tau with the RNA-binding protein TIA1 regulates tau pathophysiology and toxicity. Cell Rep. 2016;15:1455–1466. doi: 10.1016/j.celrep.2016.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalia LV, Lang AE. Parkinson’s disease. Lancet. 2015;386:896–912. doi: 10.1016/S0140-6736(14)61393-3. [DOI] [PubMed] [Google Scholar]
- 64.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 65.Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry. 1988;51:745–752. doi: 10.1136/jnnp.51.6.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McLean PJ, Kawamata H, Shariff S, Hewett J, Sharma N, Ueda K, et al. TorsinA and heat shock proteins act as molecular chaperones: suppression of alpha-synuclein aggregation. J Neurochem. 2002;83:846–854. doi: 10.1046/j.1471-4159.2002.01190.x. [DOI] [PubMed] [Google Scholar]
- 67.Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y. Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol. 1988;75:345–353. doi: 10.1007/BF00687787. [DOI] [PubMed] [Google Scholar]
- 68.Shin Y, Klucken J, Patterson C, Hyman BT, McLean PJ. The cochaperone carboxyl terminus of Hsp70-interacting protein (CHIP) mediates alpha-synuclein degradation decisions between proteasomal and lysosomal pathways. J Biol Chem. 2005;280:23727–23734. doi: 10.1074/jbc.M503326200. [DOI] [PubMed] [Google Scholar]
- 69.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 70.Um JW, Im E, Lee HJ, Min B, Yoo L, Yoo J, et al. Parkin directly modulates 26S proteasome activity. J Neurosci. 2010;30:11805–11814. doi: 10.1523/JNEUROSCI.2862-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395:451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 72.Dehay B, Bové J, Rodríguez-Muela N, Perier C, Recasens A, Boya P, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010;30:12535–12544. doi: 10.1523/JNEUROSCI.1920-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanji K, Mori F, Kakita A, Takahashi H, Wakabayashi K. Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol Dis. 2011;43:690–697. doi: 10.1016/j.nbd.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 74.Crews L, Spencer B, Desplats P, Patrick C, Paulino A, Rockenstein E, et al. Selective molecular alterations in the autophagy pathway in patients with Lewy body disease and in models of alpha-synucleinopathy. PLoS One. 2010;5:e9313. doi: 10.1371/journal.pone.0009313. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 75.Higashi S, Moore DJ, Minegishi M, Kasanuki K, Fujishiro H, Kabuta T, et al. Localization of MAP1-LC3 in vulnerable neurons and Lewy bodies in brains of patients with dementia with Lewy bodies. J Neuropathol Exp Neurol. 2011;70:264–280. doi: 10.1097/NEN.0b013e318211c86a. [DOI] [PubMed] [Google Scholar]
- 76.Klucken J, Poehler AM, Ebrahimi-Fakhari D, Schneider J, Nuber S, Rockenstein E, et al. Alpha-synuclein aggregation involves a bafilomycin A 1-sensitive autophagy pathway. Autophagy. 2012;8:754–766. doi: 10.4161/auto.19371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu Y, Dodiya H, Aebischer P, Olanow CW, Kordower JH. Alterations in lysosomal and proteasomal markers in Parkinson’s disease: relationship to alpha-synuclein inclusions. Neurobiol Dis. 2009;35:385–398. doi: 10.1016/j.nbd.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 78.Alvarez-Erviti L, Rodriguez-Oroz MC, Cooper JM, Caballero C, Ferrer I, Obeso JA, et al. Chaperone-mediated autophagy markers in Parkinson disease brains. Arch Neurol. 2010;67:1464–1472. doi: 10.1001/archneurol.2010.198. [DOI] [PubMed] [Google Scholar]
- 79.Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16:394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bento CF, Ashkenazi A, Jimenez-Sanchez M, Rubinsztein DC. The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway. Nat Commun. 2016;7:11803. doi: 10.1038/ncomms11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gan-Or Z, Dion PA, Rouleau GA. Genetic perspective on the role of the autophagy-lysosome pathway in Parkinson disease. Autophagy. 2015;11:1443–1457. doi: 10.1080/15548627.2015.1067364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vilariño-Güell C, Wider C, Ross OA, Dachsel JC, Kachergus JM, Lincoln SJ, et al. VPS35 mutations in Parkinson disease. Am J Hum Genet. 2011;89:162–167. doi: 10.1016/j.ajhg.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zimprich A, Benet-Pagès A, Struhal W, Graf E, Eck SH, Offman MN, et al. A mutation in VPS35, encoding a subunit of the retromer complex, causes late-onset Parkinson disease. Am J Hum Genet. 2011;89:168–175. doi: 10.1016/j.ajhg.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kisos H, Pukaß K, Ben-Hur T, Richter-Landsberg C, Sharon R. Increased neuronal α-synuclein pathology associates with its accumulation in oligodendrocytes in mice modeling α-synucleinopathies. PLoS One. 2012;7:e46817. doi: 10.1371/journal.pone.0046817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schaser AJ, Osterberg VR, Dent SE, Stackhouse TL, Wakeham CM, Boutros SW, et al. Alpha-synuclein is a DNA binding protein that modulates DNA repair with implications for Lewy body disorders. Sci Rep. 2019;9:10919. doi: 10.1038/s41598-019-47227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sawner AS, Ray S, Yadav P, Mukherjee S, Panigrahi R, Poudyal M, et al. Modulating α-synuclein liquid-liquid phase separation. Biochemistry. 2021;60:3676–3696. doi: 10.1021/acs.biochem.1c00434. [DOI] [PubMed] [Google Scholar]
- 90.Brown RH, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 91.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–1446. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 93.Li C, Ji Y, Tang L, Zhang N, He J, Ye S, et al. Optineurin mutations in patients with sporadic amyotrophic lateral sclerosis in China. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16:485–489. doi: 10.3109/21678421.2015.1089909. [DOI] [PubMed] [Google Scholar]
- 94.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and frontotemporal dementia. Nat Neurosci. 2015;18:631–636. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 97.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–1441. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Teyssou E, Takeda T, Lebon V, Boillée S, Doukouré B, Bataillon G, et al. Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathol. 2013;125:511–522. doi: 10.1007/s00401-013-1090-0. [DOI] [PubMed] [Google Scholar]
- 99.Ito H, Nakamura M, Komure O, Ayaki T, Wate R, Maruyama H, et al. Clinicopathologic study on an ALS family with a heterozygous E478G optineurin mutation. Acta Neuropathol. 2011;122:223–229. doi: 10.1007/s00401-011-0842-y. [DOI] [PubMed] [Google Scholar]
- 100.Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–234. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rothenberg C, Monteiro MJ. Ubiquilin at a crossroads in protein degradation pathways. Autophagy. 2010;6:979–980. doi: 10.4161/auto.6.7.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Williams KL, Warraich ST, Yang S, Solski JA, Fernando R, Rouleau GA, et al. UBQLN2/ubiquilin 2 mutation and pathology in familial amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527.e3–2527e10. doi: 10.1016/j.neurobiolaging.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 104.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 105.Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum Mol Genet. 2009;18:3217–3226. doi: 10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Valentine JS, Doucette PA, Zittin Potter S. Copper-zinc superoxide dismutase and amyotrophic lateral sclerosis. Annu Rev Biochem. 2005;74:563–593. doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- 107.Forsberg K, Graffmo K, Pakkenberg B, Weber M, Nielsen M, Marklund S, et al. Misfolded SOD1 inclusions in patients with mutations in C9orf72 and other ALS/FTD-associated genes. J Neurol Neurosurg Psychiatry. 2019;90:861–869. doi: 10.1136/jnnp-2018-319386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- 109.Weisberg SJ, Lyakhovetsky R, Werdiger AC, Gitler AD, Soen Y, Kaganovich D. Compartmentalization of superoxide dismutase 1 (SOD1G93A) aggregates determines their toxicity. Proc Natl Acad Sci U S A. 2012;109:15811–15816. doi: 10.1073/pnas.1205829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kabuta T, Suzuki Y, Wada K. Degradation of amyotrophic lateral sclerosis-linked mutant Cu,Zn-superoxide dismutase proteins by macroautophagy and the proteasome. J Biol Chem. 2006;281:30524–30533. doi: 10.1074/jbc.M603337200. [DOI] [PubMed] [Google Scholar]
- 111.Cheroni C, Marino M, Tortarolo M, Veglianese P, De Biasi S, Fontana E, et al. Functional alterations of the ubiquitin-proteasome system in motor neurons of a mouse model of familial amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:82–96. doi: 10.1093/hmg/ddn319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang F, Ström AL, Fukada K, Lee S, Hayward LJ, Zhu H. Interaction between familial amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants and the dynein complex. J Biol Chem. 2007;282:16691–16699. doi: 10.1074/jbc.M609743200. [DOI] [PubMed] [Google Scholar]
- 113.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 114.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 115.Brown RH, Jr, Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:1602. doi: 10.1056/NEJMc1710379. [DOI] [PubMed] [Google Scholar]
- 116.Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gopal PP, Nirschl JJ, Klinman E, Holzbaur EL. Amyotrophic lateral sclerosis-linked mutations increase the viscosity of liquid-like TDP-43 RNP granules in neurons. Proc Natl Acad Sci U S A. 2017;114:E2466–E2475. doi: 10.1073/pnas.1614462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dang M, Song J. ALS-causing D169G mutation disrupts the ATP-binding capacity of TDP-43 RRM1 domain. Biochem Biophys Res Commun. 2020;524:459–464. doi: 10.1016/j.bbrc.2020.01.122. [DOI] [PubMed] [Google Scholar]
- 119.Hasegawa M, Arai T, Nonaka T, Kametani F, Yoshida M, Hashizume Y, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nonaka T, Suzuki G, Tanaka Y, Kametani F, Hirai S, Okado H, et al. Phosphorylation of TAR DNA-binding protein of 43 kDa (TDP-43) by truncated casein kinase 1δ triggers mislocalization and accumulation of TDP-43. J Biol Chem. 2016;291:5473–5483. doi: 10.1074/jbc.M115.695379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Boyd JD, Lee P, Feiler MS, Zauur N, Liu M, Concannon J, et al. A high-content screen identifies novel compounds that inhibit stress-induced TDP-43 cellular aggregation and associated cytotoxicity. J Biomol Screen. 2014;19:44–56. doi: 10.1177/1087057113501553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liachko NF, McMillan PJ, Guthrie CR, Bird TD, Leverenz JB, Kraemer BC. CDC7 inhibition blocks pathological TDP-43 phosphorylation and neurodegeneration. Ann Neurol. 2013;74:39–52. doi: 10.1002/ana.23870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martínez-González L, Rodríguez-Cueto C, Cabezudo D, Bartolomé F, Andrés-Benito P, Ferrer I, et al. Motor neuron preservation and decrease of in vivo TDP-43 phosphorylation by protein CK-1δ kinase inhibitor treatment. Sci Rep. 2020;10:4449. doi: 10.1038/s41598-020-61265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang P, Wander CM, Yuan CX, Bereman MS, Cohen TJ. Acetylation-induced TDP-43 pathology is suppressed by an HSF1-dependent chaperone program. Nat Commun. 2017;8:82. doi: 10.1038/s41467-017-00088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544:367–371. doi: 10.1038/nature22038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Mackenzie IRA, Neumann M. Fused in sarcoma neuropathology in neurodegenerative disease. Cold Spring Harb Perspect Med. 2017;7:a024299. doi: 10.1101/cshperspect.a024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Martinez FJ, Pratt GA, Van Nostrand EL, Batra R, Huelga SC, Kapeli K, et al. Protein-RNA networks regulated by normal and ALS-associated mutant HNRNPA2B1 in the nervous system. Neuron. 2016;92:780–795. doi: 10.1016/j.neuron.2016.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Doble A. The pharmacology and mechanism of action of riluzole. Neurology. 1996;47(6 Suppl 4):S233–S241. doi: 10.1212/wnl.47.6_suppl_4.233s. [DOI] [PubMed] [Google Scholar]
- 131.Bensimon G, Lacomblez L, Meininger V. A controlled trial of riluzole in amyotrophic lateral sclerosis. ALS/Riluzole Study Group. N Engl J Med. 1994;330:585–591. doi: 10.1056/NEJM199403033300901. [DOI] [PubMed] [Google Scholar]
- 132.Cacabelos R. Donepezil in Alzheimer’s disease: from conventional trials to pharmacogenetics. Neuropsychiatr Dis Treat. 2007;3:303–333. [PMC free article] [PubMed] [Google Scholar]
- 133.Kim SH, Noh MY, Kim HJ, Oh KW, Park J, Lee S, et al. A therapeutic strategy for Alzheimer’s disease focused on immune-inflammatory modulation. Dement Neurocogn Disord. 2019;18:33–46. doi: 10.12779/dnd.2019.18.2.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27:336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Malik I, Kelley CP, Wang ET, Todd PK. Molecular mechanisms underlying nucleotide repeat expansion disorders. Nat Rev Mol Cell Biol. 2021;22:589–607. doi: 10.1038/s41580-021-00382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vashist S, Cushman M, Shorter J. Applying Hsp104 to protein-misfolding disorders. Biochem Cell Biol. 2010;88:1–13. doi: 10.1139/o09-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lo Bianco C, Shorter J, Régulier E, Lashuel H, Iwatsubo T, Lindquist S, et al. Hsp104 antagonizes alpha-synuclein aggregation and reduces dopaminergic degeneration in a rat model of Parkinson disease. J Clin Invest. 2008;118:3087–3097. doi: 10.1172/JCI35781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jackrel ME, DeSantis ME, Martinez BA, Castellano LM, Stewart RM, Caldwell KA, et al. Potentiated Hsp104 variants antagonize diverse proteotoxic misfolding events. Cell. 2014;156:170–182. doi: 10.1016/j.cell.2013.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jackrel ME, Shorter J. Potentiated Hsp104 variants suppress toxicity of diverse neurodegenerative disease-linked proteins. Dis Model Mech. 2014;7:1175–1184. doi: 10.1242/dmm.016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Walters RW, Parker R. Coupling of ribostasis and proteostasis: Hsp70 proteins in mRNA metabolism. Trends Biochem Sci. 2015;40:552–559. doi: 10.1016/j.tibs.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fare CM, Shorter J. (Dis)solving the problem of aberrant protein states. Dis Model Mech. 2021;14:dmm048983. doi: 10.1242/dmm.048983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yu H, Lu S, Gasior K, Singh D, Vazquez-Sanchez S, Tapia O, et al. HSP70 chaperones RNA-free TDP-43 into anisotropic intranuclear liquid spherical shells. Science. 2021;371:eabb4309. doi: 10.1126/science.abb4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gu J, Liu Z, Zhang S, Li Y, Xia W, Wang C, et al. Hsp40 proteins phase separate to chaperone the assembly and maintenance of membraneless organelles. Proc Natl Acad Sci U S A. 2020;117:31123–31133. doi: 10.1073/pnas.2002437117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, et al. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173:677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, et al. RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron. 2019;102:321–338.e8. doi: 10.1016/j.neuron.2019.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the study.