Summary
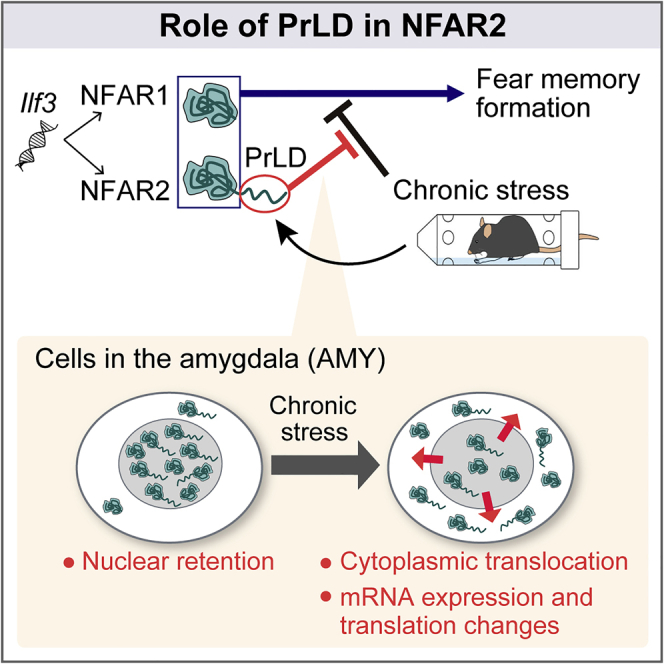
The prion-like domain (PrLD) is a class of intrinsically disordered regions. Although its propensity to form condensates has been studied in the context of neurodegenerative diseases, the physiological role of PrLD remains unclear. Here, we investigated the role of PrLD in the RNA-binding protein NFAR2, generated by a splicing variant of the Ilf3 gene. Removal of the PrLD in mice did not impair the function of NFAR2 required for survival, but did affect the responses to chronic water immersion and restraint stress (WIRS). The PrLD was required for WIRS-sensitive nuclear localization of NFAR2 and WIRS-induced changes in mRNA expression and translation in the amygdala, a fear-related brain region. Consistently, the PrLD conferred resistance to WIRS in fear-associated memory formation. Our study provides insights into the PrLD-dependent role of NFAR2 for chronic stress adaptation in the brain.
Subject areas: Biological sciences, Genetics, Molecular genetics
Graphical abstract
Highlights
-
•
NFAR2 uniquely has a long PrLD, inserted by splicing, among its protein family
-
•
PrLD retains NFAR2 in the nucleus in a stress-sensitive manner in AMY
-
•
NFAR2 PrLD modifies stress-induced changes in mRNA expression and translation in AMY
-
•
NFAR2 PrLD confers chronic stress tolerance in mice in fear memory formation
Biological sciences; Genetics; Molecular genetics
Introduction
Intrinsically disordered regions (IDRs) are protein regions that do not form stable three-dimensional structures. IDRs tend to function as hubs for biomolecular complexes by presenting interaction sites with multiple proteins and RNAs.1,2 Certain IDRs undergo liquid-liquid phase separation (LLPS) to form membraneless organelles (a.k.a. biomolecular condensates) with various proteins and RNAs.3,4 Through these mechanisms, IDR-containing proteins (IDPs) modulate biochemical reactions positively or negatively, depending on the interaction to other molecules, physical property, and the subcellular localization.5,6
A class of IDRs, the prion-like domain (PrLD), has low sequence complexity and is rich in polar amino acids and glycine. This domain has attracted huge attention because its ability to form condensates associated with neurodegenerative diseases, including amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD).7,8,9,10 However, most PrLD-containing proteins including fused in sarcoma (FUS), TAR DNA-binding protein-43 (TDP-43), and heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), whose aggregation causes the diseases listed above, do not form condensates in healthy conditions.11,12
Studies in yeasts and plants have suggested that PrLDs diverge in length among orthologs and alternative splicing variants and that the variety confers evolutional advantages in environmental responses and phenotypic diversities.13,14,15 In general, the length of IDRs encoded in genome increases with the complexity of the organism.16,17 Since IDRs are often encoded in alternatively spliced exons,18,19 the insertion of the IDR may rewire the interaction network of the protein, expanding regulatory layers.20,21 Although, in vertebrates, the pathological aspects of PrLDs have been well recognized, little is known about the physiological relevance of PrLDs.
Interleukin enhancer-binding factor 3 (Ilf3) is a gene conserved in vertebrates and involved in stress responses.22 Alternative splicing of Ilf3 generates transcript variants, nuclear factor associated with dsRNA 1 (NFAR1, a.k.a. NF90) and NFAR2 (a.k.a. NF110 and ILF3), the latter of which possesses a PrLD.23,24 Through the binding to DNA and RNA, both NFAR1 and NFAR2 (NFARs) regulate gene expression in multiple fashions.25 They are predominantly present in the nucleus to promote and repress transcription, while a fraction of them shuttle between the nucleus and cytoplasm, exporting mRNAs for translation.26,27 To respond to virus infection and oxidative stress, NFARs suppress or facilitate translation depending on target mRNAs.22,28,29
In contrast to these common functions, NFAR1 and NFAR2 differ in subcellular and subnuclear localization. NFAR2 is more prone to be in the nucleus than NFAR1, due to the PrLD.26 Within the nucleus, both NFAR1 and NFAR2 are localized in the nucleolus in common, while only NFAR2 is also enriched in the nucleoplasm.25,30,31 In the cytoplasm, NFAR2, but not NFAR1, localizes to the RNP condensate formed by RNG105 (a.k.a. caprin1), a component of neuronal RNA granules and stress granules.32 Given that the potential functional difference of subcellular localization originates from PrLD, further phenotypic studies of NFARs mutants will provide a better understanding of the physiological implications of PrLDs.
For that purpose, we generated Ilf3 mutant mice by deleting the PrLD in NFAR2 (Ilf3ΔPrLD/ΔPrLD mice). In addition, since NFARs have stress-responsive functions, we applied chronic stress to mice, which is known to increase oxidative stress levels in the brain.33,34 Through the investigations by microscopic analysis, ribosome profiling, and behavioral tests, we found that the PrLD of NFAR2 mediates the nuclear retention, regulates chronic stress-dependent changes in mRNA expression and translation, and enables mice to better form conditioned fear memory in a chronic stress environment. These results suggest that the PrLD confers versatility for responding to the environment, by subcellular localization modulation rather than condensate formation.
Results
Prion-like domain insertion in domain associated with zinc finger family proteins
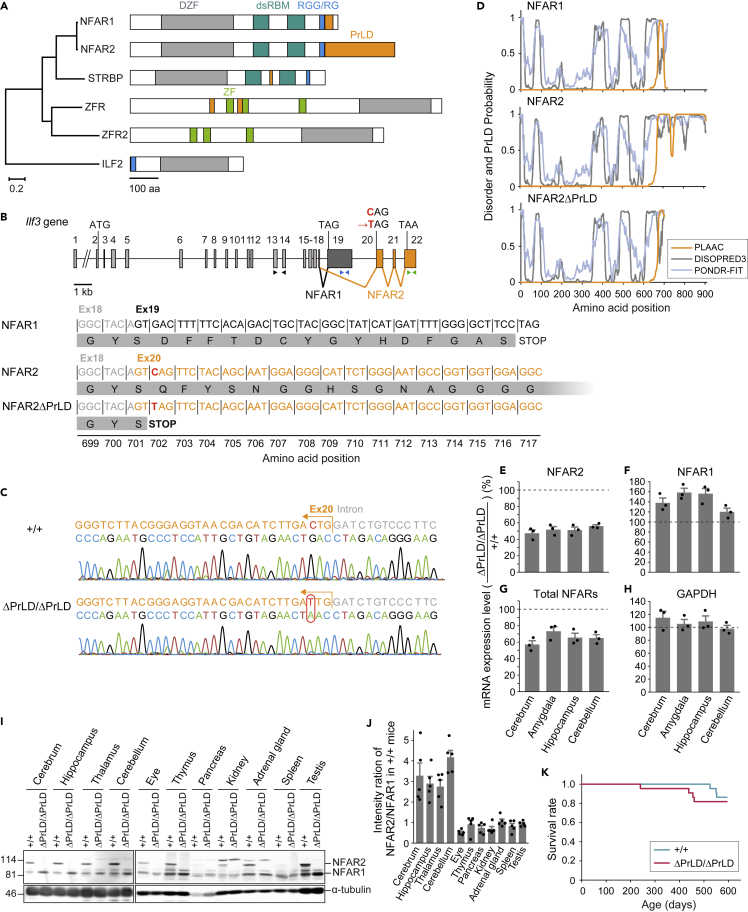
NFARs belong to a metazoan-exclusive protein family containing the domain associated with zinc fingers (DZF domain). This family consists of interleukin enhancer-binding factor 2 (ILF2), zinc-finger protein associated with RNA (ZFR), ZFR2, spermatid perinuclear RNA-binding protein (SPNR, a.k.a. STRBP), and ILF3 (NFARs) (Figure 1A). Of these proteins, ZFR2, STRBP, and NFARs are uniquely found in vertebrates (Figure S1). Among these paralogs, only NFAR2 has a long PrLD (over 200 amino acids) (Figures 1A and S1). Although medium-length PrLDs (100-200 amino acids) are present also in ZFR2 in fish and reptiles, the corresponding region of mammalian ZFR2 is replaced with non-PrLD type IDR (Figure S1). These suggest that the insertion of PrLDs occurred in selective proteins and species. Notably, despite the absence of long PrLDs, STRBP, a closely related paralog of NFARs, plays an important role in brain and sperm function35 as NFARs do. The paralog-wise and isoform-wise comparison led us to study the roles of the uniquely long PrLD in NFAR2.
Figure 1.
Generation of Ilf3ΔPrLD/ΔPrLD mice
(A) Phylogenetic tree of DZF-containing proteins in mice. DZF, double zinc-finger domain; dsRBM, dsRNA-binding motif; RGG/RG, Arg-Gly-Gly/Arg-Gly motif; ZF, C2H2-type zinc-finger domain; PrLD, prion-like domain predicted by PLAAC. Scale bars indicate the number of amino acid substitutions per site (left) and the number of amino acids (right).
(B) Alternative splicing pattern of the mouse Ilf3 gene. The nucleotide and amino acid sequences of NFAR1, NFAR2, and NFAR2ΔPrLD near the splice junction of exon 18 terminal are shown. In NFAR2ΔPrLD, a single nucleotide substitution from C to T (red) introduced a stop codon at the beginning of exon 20. The positions of the primers used for quantitative RT-PCR in E-G are indicated by triangles: black, NFARs; blue, NFAR1; green, NFAR2.
(C) Representative DNA sequencing in mouse genotyping. For simplicity, Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD are denoted as +/+ and ΔPrLD/ΔPrLD, respectively. The antisense strand was sequenced. A red circle indicates the C to T substitution.
(D) IDR and PrLD prediction by DISOPRED3 and PONDR-FIT for IDR and by PLAAC for PrLD.
(E–H) Relative expression levels of mRNAs in the brain of Ilf3ΔPrLD/ΔPrLD mice compared with Ilf3+/+ mice. GAPDH is a control. Data are represented as the mean ± SEM with dot plots of individual values. n = 3.
(I) Western blotting of tissues from the Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice for NFARs and α-tubulin as a control.
(J) Intensity ratio of NFAR2/NFAR1 in the tissues of Ilf3+/+ mice in Western blotting. n = 5, p < 0.0001, one-way ANOVA.
(K) Survival curves of Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice. n = 22, p = 0.6, log rank test.
See also Figure S1.
Generation of Ilf3ΔPrLD/ΔPrLD mice that lack the prion-like domain of NFAR2
To investigate the physiological roles of the PrLD in NFAR2, we deleted the domain in mice (Ilf3ΔPrLD/ΔPrLD mice). Compared to NFAR1, NFAR2 contains more exons at the 3′ end, skipping exon 19 and obtaining exons 20-22 (Figure 1B), and yields NFAR2-specific amino acid residues (702-911) (Figure 1B). The 702-911 region was predicted as IDR32 and as a part of PrLD (671-911) (Figure 1D). In contrast, NFAR1 possesses a short trait of the predicted PrLD (671-698). Given that, we designed to introduce a stop codon at the 5′ end of exon 20, to delete the PrLD from NFAR2. The replacement of c.2104C to T with CRISPR/Cas9 resulted in the truncated NFAR2 (amino acid residues 1-701) (Figures 1B–1D). Since the mutated NFAR2 mRNA has the premature stop codon, it was expected to be degraded by a nonsense-mediated decay (NMD) pathway.36 Consistent with this scenario, NFAR2 mRNA partially reduced in Ilf3ΔPrLD/ΔPrLD mice compared with wild-type (Ilf3+/+) mice (Figure 1E).
However, the remained mRNA may lead to NFAR2ΔPrLD protein translation. Western blotting of mouse tissues with ILF3 antibody detected two proteins in different sizes at 90 kDa and 110 kDa, corresponding to NFAR1 and NFAR2, respectively. The 110 kDa protein disappeared in all tissues from the Ilf3ΔPrLD/ΔPrLD mice (Figure 1I). In contrast, the intensity of the 90 kDa protein was increased 1- to 2-fold in most tissues from the Ilf3ΔPrLD/ΔPrLD mice. This may be due to the similar migration of NFAR2ΔPrLD and NFAR1 on SDS-PAGE and partially due to an increased NFAR1 mRNA (Figure 1F) for unknown reasons.
While Ilf3 knockout mice are perinatal lethal because of neuromuscular respiratory failure,37 the Ilf3ΔPrLD/ΔPrLD mice were viable without a significant decrease in their survival rate (Figure 1K). However, the effect of PrLD deletion could be more prevailing in tissues such as the brain, where NFAR2 expression predominates over NFAR1 (Figure 1J). Therefore, we investigated the effects of PrLD deletion in the brain on the subcellular localization of NFARs, mRNA expression and translational regulation, and mouse behavior, across chronic stress.
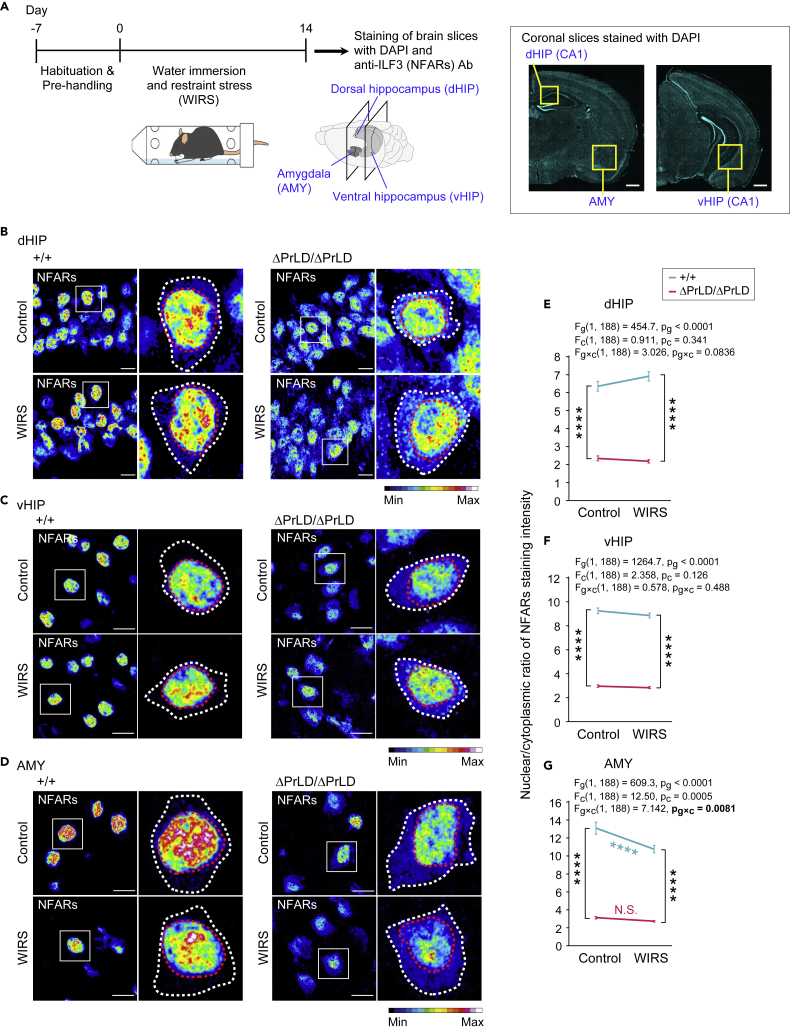
Prion-like domain of NFAR2 is responsible for the chronic stress-sensitive nuclear retention in the amygdala
First, we immunostained NFARs in the brain (Figure 2A). NFARs were predominantly localized in the nucleus of brain regions including the dorsal hippocampus (dHIP), ventral HIP (vHIP), and amygdala (AMY) in the Ilf3+/+ mice (Figures 2B–2D). Although NFARs diffused in the nucleoplasm, a part of them accumulated in nuclear condensates (granules), as previously reported38 (Figure S2A). The Ilf3ΔPrLD/ΔPrLD mice reduced NFARs in the nucleoplasm (Figure S2A) and tended to reduce the nuclear granule formation (Figures S2B–S2L), resulting in the declined nuclear-cytoplasmic ratio of NFARs (Figures 2B–2G). These results suggested that PrLD-deficient forms (NFAR1 and NFAR2ΔPrLD) have a weaker ability to localize in the nucleus. This reduction was primarily due to the loss of NFARs from the nucleoplasm, which is consistent with previous reports that PrLD-containing NFAR2 is more abundant in the nucleoplasm than NFAR1.25,30
Figure 2.
PrLD deletion reduces nuclear retention of NFARs and attenuates amygdala-specific sensitivity of the nuclear retention to chronic stress WIRS
(A) Schematic of NFARs subcellular localization analysis after WIRS. The right panels are DAPI staining of brain slices showing the brain regions. Scale bars, 1 mm.
(B–D) Representative heatmap images of NFARs stained in the dHIP (B), vHIP (C), and AMY (D) of Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice. In the right magnified images, the white and red dotted lines outline the cells and nuclei, respectively. Scale bars, 10 μm.
(E–G) Nuclear-cytoplasmic ratio of NFARs staining intensity in the brain regions. The data are presented as mean ± SEM. n = 48 pictures from three mice in each group. The data were analyzed using two-way ANOVA. The F-values and p values for the main effects of the genotypes (Fg and pg, respectively), conditions (with or without WIRS) (Fc and pc, respectively), and interactions between the genotypes and conditions (Fg×c and pg×c, respectively) are indicated. In E and F, ∗∗∗∗p < 0.0001 in Tukey-Kramer test after the two-way ANOVA. In G, ∗∗∗∗p < 0.0001 in simple effect analysis after significant interaction in the two-way ANOVA. N.S., no significant difference (p ≥ 0.05).
See also Figures S2 and S3.
Next, we stressed those brain regions and tested the localization alteration of NFARs. For this purpose, we harnessed chronic water immersion and restraint stress (WIRS) in mice (Figure 2A), which causes oxidative damage to the most stress-sensitive AMY and HIP in the brain.33,34 Mice were subjected to 3 h WIRS daily for 14 days. This treatment resulted in weight loss in the mice, a hallmark of chronic stress (Figure S11A). Although the impacts on NFARs in the dHIP and vHIP were limited, WIRS reduced the nuclear-cytoplasmic ratio in the AMY of Ilf3+/+ mice (Figures 2B–2G). This WIRS-induced decrease originated from the loss of the diffuse staining in the nucleoplasm, without associated decreases in the number or size of the nuclear granules (Figures S2K and S2L). Remarkably, the Ilf3ΔPrLD/ΔPrLD mice lost this response to WIRS in the AMY (Figures 2D and 2G), whereas no effect in the dHIP or vHIP was observed as in the wild type (Figures 2B, 2C, 2E, and 2F). This phenotype could be attributed to the low nucleoplasm localization of NFARs in Ilf3ΔPrLD/ΔPrLD mice even without stress (Figure S2F). Taken together, these results suggested that the PrLD of NFAR2 is responsible for the nucleoplasmic retention of NFARs, which is sensitive to chronic stress in the AMY.
We also tested the effect of WIRS on the nuclear-cytoplasmic localization of other PrLD-bearing proteins TDP-43 and hnRNP A1 in the AMY. In contrast to NFARs, their localizations were largely unchanged by WIRS (Figures S3A–S3D). TDP-43 and hnRNP A1 have been reported to translocate from the nucleus to the cytoplasm upon osmotic stress, but much less translocation induced by oxidative stress.39,40 Thus, PrLD-containing proteins commonly display stress-sensitive nuclear retention, but the type and intensity of stress that induces translocation to the cytoplasm may differ among them.
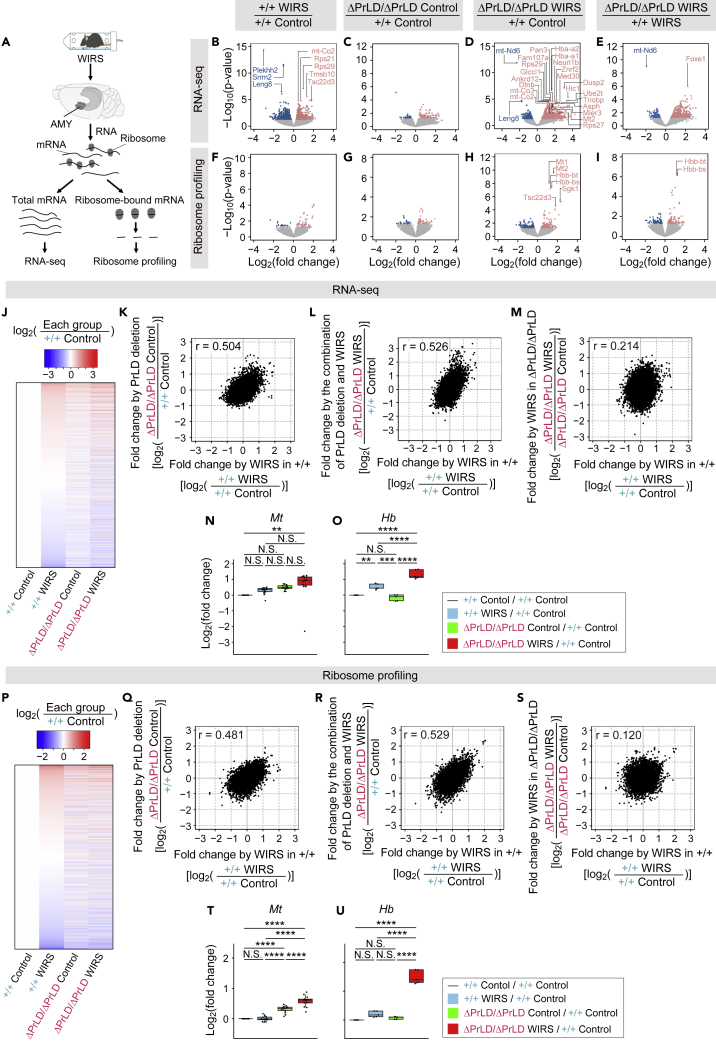
Deletion of NFAR2 prion-like domain alters water immersion and restraint stress-induced changes in mRNA expression and translation in the amygdala
Since NFARs have been reported to participate in gene expression controls in a stress-dependent manner, we explored the role of the PrLD in mRNA expression by RNA-seq in the AMY (Figure 3A). First, we investigated the WIRS-induced mRNA expression changes in Ilf3+/+ mice (Figure 3B; Table S1A). Indeed, our data recapitulated the stress-inducible gene expression reported in earlier studies: glucocorticoid-inducible factor Tsc22d3 mRNA, whose transcriptional activation was reported upon psychological stress,41 and serum and glucocorticoid-regulated kinase 1 (Sgk1), which is known as the gene upregulated by WIRS42 (Figures 3B and S4Q; Table S1A). More broadly, Gene ontology (GO) enrichment analysis revealed that the overrepresented GO terms for up-regulated genes consisted of “Ribosome,” “Nucleosome assembly,” “Oxidative phosphorylation,” and “Parkinson disease” (Figure S4A; Table S2A). These GO terms associated with the ribosome and mitochondrial functions were consistent with the previous study in the prefrontal cortex of chronically stressed mice.43 These data ensured the chronic stress induction by WIRS in our experimental conditions.
Figure 3.
PrLD deletion-caused and WIRS-induced changes in mRNA expression and translation in the AMY
(A) Schematic of RNA-seq and ribosome profiling in the AMY.
(B–I) Volcano plots of differential mRNA expression (B–E) and differential translation (F–I). WIRS-induced changes in the Ilf3+/+ mice (B and F), PrLD deletion-caused changes under control conditions (C and G), changes by the combinational effects of PrLD deletion and WIRS (D and H), and PrLD deletion-caused changes under WIRS (E and I). Blue and red dots indicate transcripts with p < 0.05. The names of transcripts with q < 0.05 are shown.
(J) Heatmap showing the fold changes in mRNA expression by WIRS, PrLD deletion, and the combinational effects of PrLD deletion and WIRS.
(K–M) Scatterplots and Pearson correlation coefficients (r) comparing changes in mRNA expression. WIRS-induced changes in the Ilf3+/+ mice were compared with the changes caused by PrLD deletion (K); with the changes caused by the combinational effects of PrLD deletion and WIRS (L); and with WIRS-induced changes in the Ilf3ΔPrLD/ΔPrLD mice (M).
(N and O) Boxplots showing the changes in mRNA expression of the mitochondrial genome-encoded genes (Mt) (N) and the hemoglobin genes (Hb) (O).
(P) Heatmap showing the fold changes in translation by WIRS, PrLD deletion, and the combinational effects of PrLD deletion and WIRS. Note that the order of transcripts is different from that in J.
(Q–S) Scatterplots and Pearson correlation coefficients (r) comparing changes in translation. The horizontal and vertical axes are the same as in K–M.
(T and U) Boxplots showing the changes in the translation of the Mt (T) and Hb (U) genes.
∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, Welch’s t-test with Bonferroni correction. N.S., no significant difference (p ≥ 0.05).
See also Figure S4 and Tables S1, S2, S3, S4, S5, S6, S7, S8, and S9.
Next, the effects of PrLD deletion on mRNA expression were analyzed (Figure 3C; Table S1B). We noticed that Ilf3ΔPrLD/ΔPrLD mice showed a stress-like gene expression pattern even without WIRS: mRNA expression profile in Ilf3ΔPrLD/ΔPrLD mice was highly correlated with that in the WIRS-treated Ilf3+/+ mice (Figures 3J and 3K; Table S5A). As observed in WIRS-induced mRNA changes in Ilf3+/+ mice (Figure S4A; Table S2A), similar GO terms (such as “Oxidative phosphorylation” and “Parkinson disease”) were also found in mRNAs overexpressed in Ilf3ΔPrLD/ΔPrLD mice (Figure S4B; Table S2B). Thus, NFAR2 PrLD deletion shifted the transcriptome profile to that reminiscent of chronic stress conditions.
WIRS treatment of the PrLD-deficient mice still maintained the stress-related gene induction or even enhanced it (Figures 3D and 3E; Tables S1C and S1D). Overall mRNA expression in the WIRS-treated Ilf3ΔPrLD/ΔPrLD mice was congruent with that in the Ilf3+/+ mice with WIRS (Figures 3J, 3L, and S4C; Tables S2C and S5A). Specifically, WIRS-responsive mRNAs (defined as genes up-regulated in mRNA abundance by WIRS in Ilf3+/+ mice, Figure 3B, red dots) were also upregulated in WIRS-treated Ilf3ΔPrLD/ΔPrLD mice (Figure S4G; Table S7A). Also, mitochondrial genome-encoded genes (Mt) and hemoglobin genes (Hb), whose expressions are considered to be markers of chronic stress in the mouse brain,43,44,45 were upregulated rather more in Ilf3ΔPrLD/ΔPrLD mice than in Ilf3+/+ mice under WIRS (Figures 3N and 3O; Tables S8A and S9A). This, along with the unique downregulation of mt-Nd6 (Figure S4O), was validated by quantitative RT-PCR (Figures S4L–S4N). However, the mRNA expression response to WIRS in the Ilf3ΔPrLD/ΔPrLD mice was markedly altered with little correlation to that in the Ilf3+/+ mice (Figure 3M; Table S6A). These results suggested that the deletion of NFAR2 PrLD altered the mRNA expression dependence on chronic stress in mouse AMY.
To survey the impact of WIRS and PrLD deletion on protein synthesis, we conducted ribosome profiling46,47 from the same materials (Figure 3A). Essentially, we observed similar trends of alteration in translation (Figures 3P–3R; Table S5B) as found in mRNA (Figures 3J–3L; Table S5A): high similarities among WIRS, PrLD deletion, and the combinational effects. We also noticed that, as found in RNA-seq (Figure 3M; Table S6A), the response of the Ilf3ΔPrLD/ΔPrLD mice to WIRS was significantly altered from that of Ilf3+/+ mice (Figure 3S; Table S6B). Interestingly, mRNA expression changes were not always reflected in the protein synthesis of the mRNAs (Figures 3F–3I and S4D–S4F; Tables S3A–S3D and S4A–S4C). Indeed, we noticed that a subset of WIRS-responsive mRNAs was translationally buffered in WIRS in both the genotypes (Figures S4G–S4I; Tables S7A–S7C). Nonetheless, Mt and Hb genes were actively translated upon mRNA accumulation by the combination of PrLD deletion and WIRS (Figures 3T, 3U, S4J, and S4K; Tables S8A–S8C and S9A–S9C), which was reflected in the GO enrichment analysis (Figure S4F; Table S4C). These results suggested that the NFAR2 PrLD deletion altered the translational responsiveness to chronic stress. Taken together, our data suggested that the loss of NFAR2 PrLD mimics stress-induced transcriptome and translatome alterations without WIRS and alters WIRS-responsive control of mRNA expression and translation in mouse AMY (Figure S4R).
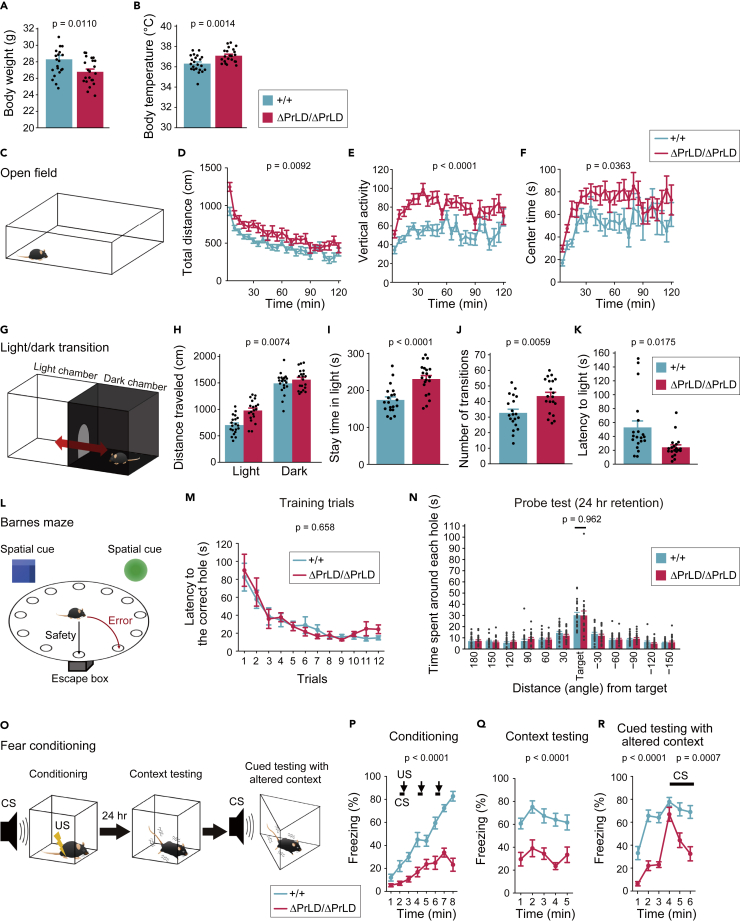
Ilf3ΔPrLD/ΔPrLD mice show increased body temperature and activity, reduced anxiety-like behavior, and impaired conditioned fear memory
Next, we investigated the functional role of NFAR2 PrLD in mouse behavior using the comprehensive battery of behavioral tests48,49 (Table 1). First, Ilf3ΔPrLD/ΔPrLD mice had comparable physical and sensory abilities (Figure S5), but they showed weight loss, increased body temperature, and increased activity compared with wild-type littermates (Figures 4A, 4B, 4D, 4H, S6A, S6C, S7D–S7F, S7J, S7K, S7O, S7P, S7T, S7U, and S7Y; Table 1). Second, the Ilf3ΔPrLD/ΔPrLD mice showed reduced anxiety-like behavior in the open field test (Figure 4F) and the light/dark transition test (Figures 4H–4K). Third, the Ilf3ΔPrLD/ΔPrLD mice and the wild-type controls showed similar performance in the T-maze and the Barnes maze (Figures 4L–4N, S8A, and S8D–S8F). However, the Ilf3ΔPrLD/ΔPrLD mice showed attenuated fear responses in the contextual and cued fear conditioning test (Figures 4O–4R, S8G, and S8H). These changes in emotion- and fear-associated responses suggested that AMY-related neural networks and function were affected.50,51
Table 1.
Comprehensive behavioral test battery of Ilf3ΔPrLD/ΔPrLD mice
| Test | Age (w) | Phenotypes in Ilf3ΔPrLD/ΔPrLD mice | Figures |
|---|---|---|---|
| Body weight and body temperature | 11-12 | Decreased body weight, increased body temperature | 4 |
| Grip strength | 11-12 | N.S. | S5 |
| Wire hang | 11-12 | N.S. | S5 |
| Light/dark transition test | 11-12 | Hyperactivity, decreased anxiety-like behavior | 4 |
| Open field test | 12 | Hyperactivity, decreased anxiety-like behavior | 4 |
| Elevated plus maze | 12 | Hyperactivity | S6 |
| Hot plate test | 12 | N.S. | S5 |
| Social interaction test in a novel environment | 12-13 | Hyperactivity, decreased duration of contact | S7 |
| Rotarod test | 13 | N.S. | S5 |
| Three-chamber social approach test (100 lux) | 15-16 | Hyperactivity | S7 |
| Acoustic startle response/prepulse inhibition test | 16 | Decreased startle | S6 |
| Porsolt forced swim test | 16 | N.S. | S6 |
| Barns maze test | 22-30 | Low task performance in trials after memory retention for a month | 4 and S8 |
| T-maze (spontaneous alternation) | 30-31 | High task performance | S8 |
| Contextual and cued fear conditioning test | 48-53 | Deficits in fear conditioning memory | 4 and S8 |
| Three-chamber social approach test (5 lux) | 65-66 | Hyperactivity | S7 |
| Social interaction test in a home cage | 82-83 | Hyperactivity | S7 |
| Tail suspension | 85 | Depression-like behavior | S6 |
N.S.: no significant differences.
Figure 4.
Ilf3ΔPrLD/ΔPrLD mice show increased body temperature and activity, reduced anxiety, and impaired conditioned fear memory
(A and B) Body weight (A) and temperature (B). n = 22.
(C–F) Open field test. Total distance traveled (D), vertical activity (E), and time spent in the center area of the field (F). n = 22.
(G–K) Light/dark transition test. Total distance traveled (H), time spent in the light chamber (I), number of transitions (J), and latency to first entry into the light chamber (K). n = 20.
(L–N) Barnes maze test. The latency to the correct hole in the training session (M) and the time spent around each hole in the probe trial conducted 24 h after the last training (N). n = 22.
(O–R) Fear conditioning test. CS, conditioned stimuli (white noise); US, unconditioned stimuli (foot shock). Freezing response was measured during conditioning (P), contextual test (Q), and cued test (R). Bars and arrows indicate the time when CS and US were presented, respectively. n = 22.
The data are presented as mean ± SEM. The p values in one-way ANOVA (A, B, H–K, and N) and the genotype effect in two-way repeated measures ANOVA (D–F, M, and P–R) are shown.
See also Table 1 and Figures S5–S8.
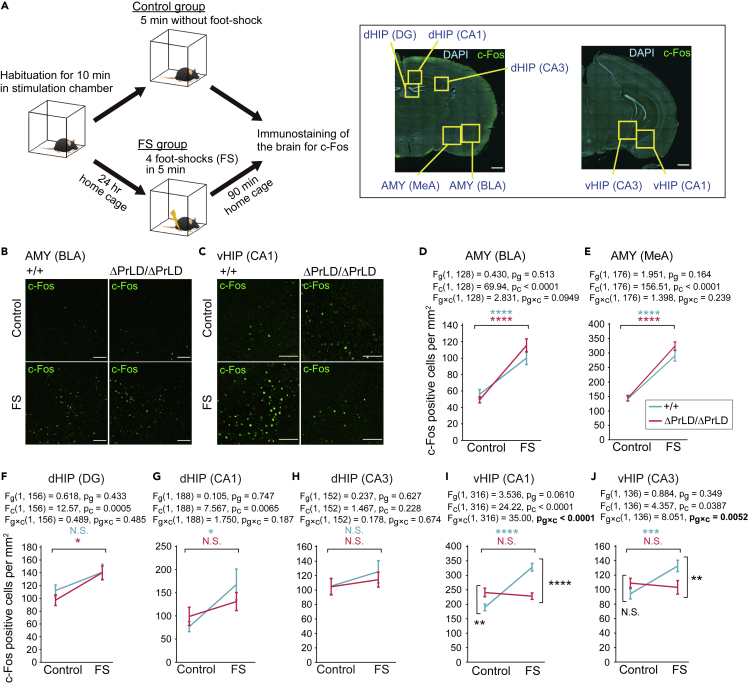
To trace the neural activity in the fear conditioning test, mice were immunostained for c-Fos, a marker of neural activation, in the AMY, dHIP, and vHIP (Figure 5A). In Ilf3+/+ mice, foot shock (FS) caused a mild increase in c-Fos-positive cells in the dHIP and a significant increase in the AMY and vHIP (Figures 5B–5J). Whereas Ilf3ΔPrLD/ΔPrLD mice had similar accumulations of c-Fos-positive cells in the AMY and dHIP (Figures 5B and 5D–5H), vHIP of the mice lost the response (Figures 5C, 5I, and 5J). Since the vHIP and AMY are structurally and functionally connected to encode emotion and fear,52,53 the failure of the vHIP in Ilf3ΔPrLD/ΔPrLD mice to activate in response to FS may be associated with the alterations in the AMY and underlie the attenuated fear responses in those mice. In the context of neural connection, the delayed early development of axons in cultured neurons from the AMY of Ilf3ΔPrLD/ΔPrLD mice might be related to such defects in connectivity (Figure S9).
Figure 5.
Fear-induced upregulation of c-Fos in the vHIP is lost in Ilf3ΔPrLD/ΔPrLD mice
(A) Schematic of immunostaining for c-Fos after foot shock (FS). The right panels are brain slices double-stained with DAPI and c-Fos, showing the brain regions. MeA, medial amygdala; BLA, basolateral amygdala. Scale bars, 1 mm.
(B and C) c-Fos in the AMY (B) and vHIP (C) of Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice with and without FS. Scale bars, 100 μm.
(D–J) Quantification of c-Fos positive cells. The data are presented as mean ± SEM. n = 33 (D), 45 (E), 40 (F), 48 (G), 39 (H), 80 (I), and 35 (J) pictures from four mice in each group. The data were analyzed using two-way ANOVA. The F-values and p values for the main effect of genotypes (Fg and pg, respectively), stimuli (with or without FS) (Fc and pc, respectively), and interaction between genotypes and stimuli (Fg×c and pg×c, respectively) are shown. In D-H, ∗∗∗∗p < 0.0001, ∗p < 0.05 in Tukey-Kramer test after the two-way ANOVA. In I and J, ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01 in simple effect analysis after significant interaction in the two-way ANOVA. N.S., no significant difference (p ≥ 0.05).
See also Figure S9.
Prion-like domain of NFAR2 confers tolerance to chronic stress in forming conditioned fear memory
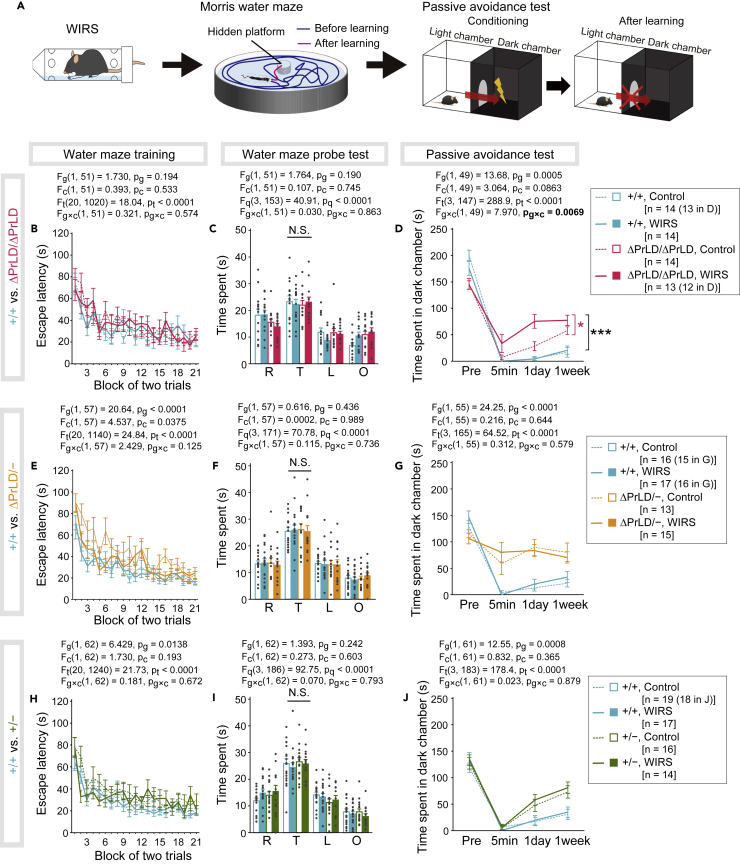
Next, to investigate the behavior outcome associated with WIRS, we applied the Morris water maze and passive avoidance tests to mice with WIRS (Figure 6A). The former test showed that PrLD deletion did not affect spatial learning and memory (Figures 6B and 6C), consistent with the Barnes maze test (Figures 4L–4N). In contrast, the passive avoidance test demonstrated that Ilf3ΔPrLD/ΔPrLD mice spent significantly longer time than Ilf3+/+ mice in a dark chamber where the mice had received the FS (Figure 6D). This indicates that PrLD deletion affected conditioned fear memory, as was the case with the contextual and cued fear conditioning test (Figures 4O–4R). Notably, although the dark chamber avoidance behavior of Ilf3+/+ mice was unaffected by WIRS, WIRS exacerbated the avoidance behavior of Ilf3ΔPrLD/ΔPrLD mice (Figures 6D and S11F). These results suggested that PrLD deletion had an impact on both conditioned fear memory and tolerance to WIRS in the formation of conditioned fear memory.
Figure 6.
Ilf3ΔPrLD/ΔPrLD mice are highly sensitive to WIRS in conditioned fear memory formation
(A) Mice were exposed to WIRS and subsequently underwent the Morris water maze and fear conditioning (passive avoidance) tests.
(B and C) Results for Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice in the Morris water maze. Escape latency to the platform in the training session (B) and time spent in each quadrant in the probe test conducted one day after the last training (C). R, right; T, target; L, left; O, opposite quadrants.
(D) Results for Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice in the passive avoidance test. Time spent in the dark chamber before FS (Pre) and 5 min, 1 day, and 1 week after the conditioning.
(E–G) Results for Ilf3+/+ and Ilf3ΔPrLD/− mice.
(H–J) Results for Ilf3+/+ and Ilf3+/− mice.
The data were analyzed using three-way repeated measures ANOVA. The F-values and p values for the main effects of genotype (Fg and pg, respectively), condition (with or without WIRS) (Fc and pc, respectively), trial (Ft and pt, respectively), and quadrant (Fq and pq, respectively), and the interaction effect between genotype and condition (Fg×c and pg×c, respectively) are indicated. N.S., no significant difference. In D, ∗∗∗p < 0.001, ∗p < 0.05 in simple effect analysis after significant interaction in the three-way repeated measures ANOVA.
See also Figures S10 and S11.
To more precisely identify the PrLD-specific role in conditioned fear memory and stress tolerance, we perturbed the overall expression levels of NFARs and NFAR2ΔPrLD in mice and investigated the behavioral changes. To this end, we generated Ilf3+/− and Ilf3ΔPrLD/− mice using CRISPR/Cas9. The genome editing deleted 14 bp in exon 7, resulting in a frameshift mutation generating a downstream premature stop codon in exon 9 (Figures S10A–S10C). The mutated Ilf3 allele (Ilf3−) encoded amino acid residues 1-224, followed by mutated residues 225-263 (Figure S10C). Since Western blotting using a polyclonal antibody against the N terminus of ILF3 did not detect such low molecular weight proteins (Figure S10G), few proteins were translated and/or accumulated from the Ilf3− allele. As a result, the expression levels of NFARs were reduced by 30-50% in the brains of the Ilf3+/− and Ilf3ΔPrLD/− mice compared with the Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice, respectively (Figures S10H–S10J).
In the Morris water maze, the Ilf3ΔPrLD/− mice required longer escape latency than Ilf3+/+ mice, but the latency decreased with increasing trials, and the performance of the mice in probe tests was equivalent to Ilf3+/+ mice (Figures 6E and 6F). These results showed that neither loss of PrLD nor decreased expression of NFAR2ΔPrLD had a significant effect on spatial learning and memory. In contrast, in the passive avoidance test, the Ilf3ΔPrLD/− mice entered the dark chamber as early as 5 min after FS (Figure 6G). This inability to fully learn the context made it impossible to evaluate the WIRS-caused exacerbations as observed in the Ilf3ΔPrLD/ΔPrLD mice (Figure 6D). This NFAR2ΔPrLD-dose dependence of conditioned fear memory formation suggested that the loss of conditioned fear memory is caused not specifically by the deletion of the PrLD.
Also, the reduced expression of both NFAR1 and NFAR2 in the Ilf3+/− mice did not significantly affect spatial learning and memory in the Morris water maze (Figures 6H and 6I). In contrast, these mice had impaired memory in the passive avoidance test (Figure 6J). This indicated that the loss of conditioned fear memory is caused also by the reduction in the amount of NFARs. However, conditioned fear memory in the Ilf3+/− mice showed no sensitivity to WIRS (Figures 6J and S11L), which was in contrast to the results in Ilf3ΔPrLD/ΔPrLD mice, whose conditioned fear memory was exacerbated by WIRS (Figures 6D and S11F). In summary, comparisons of the Ilf3 mutants and wild-type mice suggested that both NFAR1 and NFAR2 are involved in fear-conditioned memory formation regardless of the PrLD, whereas the presence of the PrLD in NFAR2 is required specifically for tolerance of mice to chronic stress in the formation of conditioned fear memory.
Discussion
Our study demonstrated that PrLD deletion in NFAR2 reduced the tolerance of mice to WIRS in the formation of fear-associated memories. This behavioral outcome was associated with molecular phenotypes in the AMY; the reduction in the WIRS-sensitive nuclear retention of NFARs and the alteration in WIRS-induced gene expression changes. Remarkably, the transcriptional and translational profiles induced by WIRS differed between Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice. Given these results, we hypothesize that the insertion of the PrLD into NFAR2 confers molecular interactions for nuclear retention which may impact WIRS-responsive spatiotemporal regulation of mRNA expression and translation by NFARs. These controls by the PrLD may increase the tolerance of AMY and its associated nervous system to WIRS and, as a result, the tolerance of mice to WIRS in conditioned fear memory formation.
This study showed that PrLD of NFAR2 is required for nuclear retention, rather than condensate formation. Although PrLD has been recognized as aggregation-prone, a recent study challenges this stereotype: the PrLD of TDP-43 is required to prevent, rather than promote, the formation of speckle-like structures in the nucleus.54 Since comprehensive identification of PrLD-containing proteins showed their association with nucleoplasmic localization,55 retention in the nucleoplasm, rather than the formation of condensates, may be a common feature of PrLDs in these proteins.
The reduced nuclear retention of NFARs by PrLD deletion may resemble stress conditions because the export of NFARs from the nucleus to the cytoplasm is facilitated by cellular stress.22,32 In addition, the correlation between the PrLD deletion-caused gene expression changes and the WIRS-induced gene expression changes suggests that the PrLD-deficient mice are in a stress-like state even without WIRS. This notion may be supported by the results showing that Ilf3ΔPrLD/ΔPrLD mice exhibited elevated body temperature and hyperactivity, similar to those caused by chronic stress.56,57 This stress-reminiscent state of PrLD-deficient mice and the misregulation of WIRS-induced gene expression could affect the response to WIRS and stress tolerance. In relation to this model, translation of mRNAs included in the GO terms of “Oxidative phosphorylation” and “Cellular oxidant detoxification” was increased in the AMY of Ilf3ΔPrLD/ΔPrLD mice under WIRS, suggesting that those processes were compromised.
Both Ilf3ΔPrLD/ΔPrLD and Ilf3+/− mice reduced conditioned fear memory compared with Ilf3+/+ mice in the absence of WIRS, suggesting that NFARs are involved in fear memory formation in a PrLD-nonspecific manner. However, exacerbation of conditioned fear memory under WIRS occurred only in Ilf3ΔPrLD/ΔPrLD mice, but not in Ilf3+/− mice, suggesting that PrLD conferred stress tolerance on fear memories. At the cellular level, the effects of PrLD deletion on FS-induced neural activity and WIRS sensitivity were evident in different brain regions, vHIP and AMY, respectively (Figures 2 and 5). These raise the following models: 1) FS-induced neural activity in vHIP may be NFARs-mediated in a PrLD-nonspecific manner and may be NFARs dose-dependent, contributing to fear memory formation, and 2) Chronic stress sensitivity in the AMY is specifically dependent on the PrLD, which regulates the chronic stress tolerance of fear memory. Considering that vHIP and AMY are structurally and functionally connected to regulate fear-associated memory, it is possible that AMY’s stress sensitivity might influence neural circuits including vHIP that control fear memory formation. Nevertheless, RNA-seq in the AMY identified WIRS-induced expression changes not only in stress-related genes, but also in genes related to fear-conditioned memory, such as Lsamp and Bptf (a.k.a. fetal Alzheimer antigen) (Tables S1A, S1C, and S1D).58,59 This suggests that the role of the AMY may be more complex than simply modulating stress sensitivity. At the cellular level, Lsamp is involved in axon outgrowth and targeting,60 which was coincident with the result that cultured neurons from Ilf3ΔPrLD/ΔPrLD AMY showed delayed development of axons. Alternative models may therefore include a more direct role for NFAR2 PrLD in the AMY in conditioned fear memory formation, where the loss of the PrLD could impair the output of neural activity from AMY to vHIP, which may be exacerbated by WIRS.
It is noteworthy that Ilf3ΔPrLD/ΔPrLD and Ilf3+/− mice showed memory impairment in fear conditioning, but not in spatial tasks. Although the reason for this difference was unknown, similar behavioral phenotypes have been reported for RNA granule-associated proteins such as FXR2 and Ataxin-2.61,62 Fear conditioning depends on the AMY and vHIP, whereas spatial memory depends on the dHIP. The AMY and vHIP may be more sensitive than the dHIP to changes in gene expression caused by the misregulation of these RNA-binding proteins and thus leads to behavioral abnormality in fear. Alternatively, fear conditioning can be more stressful to mice than spatial tasks, requiring stress-responsive regulation of gene expression by these RNA-binding proteins. PrLD of NFAR2 may act as an adaptive switch that enables normal AMY-related behaviors even in stressful environments. This role is reminiscent of those of yeast and plant PrLDs in responding to fluctuating environments.13,14,15 Therefore, it is an interesting question of whether other PrLDs in various proteins also contribute to adaptation to the environment.
Like NFARs, alternative splicing of PrLDs occurs in many proteins such as TDP-43 and hnRNP A1.63,64 These splicing isoforms differ in the ability to form pathogenic aggregates,63,64 supporting the idea that the insertion (and deletion) of PrLDs is the basis for the diversification of protein functions. There may be some trade-offs between the physiological advantages and pathological disadvantages. Here, in the case of NFAR2, we have shown the physiological relevance of the PrLD: the NFRA2 PrLD regulates chronic stress-induced gene expression changes and is required for tolerance of conditioned fear memory to chronic stress. However, it is still unclear whether PrLD-deficient NFAR1 also needs to be synthesized. Solving these questions about PrLD-containing proteins will lead to a better understanding of the advantages and disadvantages of the presence of PrLDs and their trade-offs.
Limitations of the study
First, we were unable to detect the subcellular localization of NFAR1 and NFAR2 individually. In addition, we could not distinguish between NFAR1 and NFAR2ΔPrLD in Ilf3ΔPrLD/ΔPrLD mice. These were due to the use of the anti-ILF3 antibody that detects both NFAR1 and NFAR2, as specific antibodies against each of them do not currently exist. One solution is to generate antibodies specific for the C-terminal 15 amino acids of NFAR1 and against PrLD of NFAR2. This enables to analyze the differences in the intracellular behavior of NFAR1 and NFAR2 under WIRS between Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice, allowing us to better understand their functional differences in the presence or absence of PrLD.
Second, we used conventional mutant mice, which limited the analysis of brain region-specific effects of NFAR2 PrLD deletion and NFAR2 deficiency. Future generation and analysis of conditional mutant mice, in which deletions of PrLD and NFAR2 occur at favorable times in targeted cell types, will reveal specific roles for NFAR2 PrLD that may differ in different brain regions such as AMY and vHIP.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-ILF3 | Abcam | Cat#ab92355; RRID: AB_2049804 |
| Rabbit polyclonal anti-ILF3 | Proteintech | Cat#19887-1-AP; RRID: AB_10666431 |
| Rabbit polyclonal anti-TDP-43 | Proteintech | Cat#10782-2-AP; RRID: AB_615042 |
| Rabbit polyclonal anti-hnRNPA1 | Cell Signaling Technology | Cat#8443; RRID: AB_10828725 |
| Rabbit polyclonal anti-α-tubulin | MBL International | Cat#PM054; RRID: AB_10598496 |
| Rabbit monoclonal anti-c-Fos | Cell Signaling Technology | Cat#2250; RRID: AB_2247211 |
| Rabbit polyclonal anti-MAP2 | Abcam | Cat# ab32454; RRID: AB_776174 |
| Mouse monoclonal anti-Tau-1 | Sigma-Aldrich | MAB3420; PRID: AB_94855 |
| Biotin-conjugated anti-rabbit Ig | Cytiva | Cat#RPN1004-2ML; RRID: AB_1062582 |
| Alexa488-conjugated anti-rabbit IgG | Thermo Fisher Scientific | Cat#A-11034; RRID:AB_2576217 |
| Cy3-conjugated anti-mouse IgG | Jackson ImmunoResearch | Code: 715-165-150; PRID: AB_2340813 |
| Chemicals, peptides, and recombinant proteins | ||
| Alkaline phosphatase-conjugated streptavidin | Cytiva | Cat#RPN12234 |
| Turbo DNase | Thermo Fisher Scientific | Cat#AM2238 |
| Opti-MEM1 | Thermo Fisher Scientific | Cat#31985070 |
| Cas9 protein | Integrated DNA Technologies | Cat#1081058 |
| M2 medium | Sigma-Aldrich | Cat#MR-015-D |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat#15140122 |
| ISOGEN | Nippon Gene | Code: 311-02501 |
| M-MLV-Reverse Transcriptase | Thermo Fisher Scientific | Cat#28025013 |
| TB Green Premix Ex Taq II (Tli RNaseH Plus) | Takara Bio | Code: RR820A |
| Tissue-Tek O.C.T. Compound | Sakura Finetek | Code: 45833 |
| 4',6-diamidino-2-phenylindole (DAPI) | FUJIFILM Wako Pure Chemical | Code: 340--7971 |
| Mowiol 4-88 | Sigma-Aldrich | Cat#81381 |
| Neurobasal-A medium | Thermo Fisher Scientific | Cat#10888022 |
| B-27 supplement | Thermo Fisher Scientific | Cat#17504044 |
| L-Glutamine | Thermo Fisher Scientific | Cat#21051024 |
| Neuron Culture Medium | FUJIFILM Wako Pure Chemical | Code: 148-09671 |
| Formaldehyde | FUJIFILM Wako Pure Chemical | Code: 064-00406 |
| TRIzol reagent | Thermo Fisher Scientific | Cat#10296010 |
| RNase I | Lucigen | Code: N6901K |
| SUPERase In | Thermo Fisher Scientific | Cat#AM2694 |
| ProtoScriptII | New England Biolabs | Cat#M0368L |
| CircLigase II | Lucigen | Code: CL9021K |
| Critical commercial assays | ||
| mMESSAGE mMACHINE T7 Transcription Kit | Thermo Fisher Scientific | Cat#AM1344 |
| MEGAclea Transcription Clean-Up Kit | Thermo Fisher Scientific | Cat#AM1908 |
| ExoSAP-IT | Thermo Fisher Scientific | Cat#78201.1.ML |
| MEGAshortscript T7 Transcription Kit | Thermo Fisher Scientific | Cat#AM1354 |
| Qubit RNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q32852 |
| Ribo-Zero Gold rRNA Removal Kit Human/Mouse/Rat) | Illumina | Cat#MRZG12324 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | Cat#20020595 |
| Direct-zol RNA MicroPrep Kit | Zymo Research | Cat#R2060 |
| Deposited data | ||
| RNA-seq and ribosome profiling data | This paper | GEO: GSE209902 |
| Experimental models: Organisms/strains | ||
| Mouse: Ilf3+/ΔPrLD | This paper | N/A |
| Mouse: Ilf3+/− | This paper | N/A |
| Oligonucleotides | ||
| CRISPR/Cas9 targeting sequence: ILF3 ΔPrLD: CCCTGTCTAGGTCAGTTCTACAG | This paper | N/A |
| ssODN: ILF3 ΔPrLD: CGGCTGCG GGCCAGCCTTCAGGGCCCATGACTG CTCCTTCCTCCCCTCCCAGCCCTC ACCCTGGGACCATCCATCTAATC TGCTGCTTCCCTGTCTAGGT TAGTTCTACAGCAATGGAGG GCATTCTGGGAATGCCGGT GGTGGAGGCAGCGGGGGA GGTGGTGGCTCATCCAGCT ACAGCTCCTACTACCAAGGAGACA |
This paper | N/A |
| Primer: ILF3 ΔPrLD genotyping Forward: ATCAGCTGAGGGTGAGTGGAGCTG | This paper | N/A |
| Primer: ILF3 ΔPrLD genotyping Reverse: TGCAGCGGCTTCTTCCCAGCATGC | This paper | N/A |
| CRISPR/Cas9 targeting sequence: ΔILF3: CGTCCCTCCGACACGCCAAG | This paper | N/A |
| Primer: ΔILF3 screening Forward: GAGCTGATGGCACTTTAGCC | This paper | N/A |
| Primer: ΔILF3 screening Reverse: AGTCTCCTCAGGGCTGTCAA | This paper | N/A |
| Primer: ΔILF3 genotyping Forward: TTCTGTGGTTTAAATTTGGGAGAGG | This paper | N/A |
| Primer: ΔILF3 genotyping Reverse control: AAAATGCAAACCTGGAACCACTTG | This paper | N/A |
| Primer: ΔILF3 genotyping Reverse: AAATGCAAACCTTGGCGGAG | This paper | N/A |
| Primer: NFARs qRT-PCR Forward: AGACAGGCCCTGTTCATGCT | This paper | N/A |
| Primer: NFARs qRT-PCR Reverse: TTGGCAAGCCCATGTCCTGT | This paper | N/A |
| Primer: NFAR2 qRT-PCR Forward: ACGCTTTGAGGTGTGGTGGT | This paper | N/A |
| Primer: NFAR2 qRT-PCR Reverse: AACGGCAAGACCTCGGAACA | This paper | N/A |
| Primer: NFAR1 qRT-PCR Forward: TCTACACTGCCTGGCACACA | This paper | N/A |
| Primer: NFAR1 qRT-PCR Reverse: AGGGGCAGTTCTGCAGCAAA | This paper | N/A |
| Primer: GAPDH qRT-PCR Forward: AACGACCCCTTCATTGACCT | This paper | N/A |
| Primer: GAPDH qRT-PCR Reverse: TGGAAGATGGTGATGGGCTT | This paper | N/A |
| Primer: Hba-a1 qRT-PCR Forward: CGATGCTCTGGCCAATGCTG | This paper | N/A |
| Primer: Hba-a1 qRT-PCR Reverse: GCTAGCCAAGGTCACCAGCA | This paper | N/A |
| Primer: mt-Atp8 qRT-PCR Forward: ACATTCCCACTGGCACC |
Long et al.65 | N/A |
| Primer: mt-Atp8 qRT-PCR Forward: GGGGTAATGAATGAGGC |
Long et al.65 | N/A |
| Primer: mt-Nd3 qRT-PCR Forward: GCATTCTGACTCCCCCAAAT |
Weger et al.43 | N/A |
| Primer: mt-Nd3 qRT-PCR Reverse: TGAATTGCTCATGGTAGTGGA |
Weger et al.43 | N/A |
| Primer: mt-Nd6 qRT-PCR Forward: GTATTGGGGGTGATTATAGAG | This paper | N/A |
| Primer: mt-Nd6 qRT-PCR Reverse: CCATCATTCAAGTAGCACAAC | This paper | N/A |
| Primer: Tubb5 qRT-PCR Forward: ACTATGGACTCCGTTCGCTCAG | This paper | N/A |
| Primer: Tubb5 qRT-PCR Reverse: ACAGAGTCAACCAACTCAGCTCC | This paper | N/A |
| Primer: Sgk1 qRT-PCR Forward: GGTGCCAAGGATGACTTTATGGA | This paper | N/A |
| Primer: Sgk1 qRT-PCR Reverse: GGTAAACTCGGGATCGAAGTGC | This paper | N/A |
| Software and algorithms | ||
| MEGA11 | Tamura et al.66 | https://www.megasoftware.net |
| DISOPRED3 | Jones and Cozzetto67 | http://bioinf.cs.ucl.ac.uk/psipred/ |
| PONDR-FIT | Xue et al.68 | http://original.disprot.org/pondr-fit.php |
| PLAAC | Lancaster et al.69 | http://plaac.wi.mit.edu |
| ApE | Davis and Jorgensen70 | https://jorgensen.biology.utah.edu/wayned/ape/ |
| ImageJ | Schneider et al.71 | https://imagej.nih.gov/ij/ |
| Fiji | Schindelin et al.72 | https://fiji.sc |
| fastp | Chen et al.73 | https://github.com/OpenGene/fastp |
| DESeq2 package | Love et al.74 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| DAVID Bioinformatics Resources | Sherman et al.75 | https://david.ncifcrf.gov/home.jsp |
| R | R Core Team76 | https://www.r-project.org |
| Other | ||
| Raw data of behavioral test battery | This paper | http://www.mouse-phenotype.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nobuyuki Shiina (nshiina@nibb.ac.jp).
Material availability
Mouse lines generated in this study are available from the lead contact with a completed Material Transfer Agreement.
Experimental model and subject details
Mice
Mice were maintained on a C57BL/6J genetic background. All animal care and experiments were approved by the Animal Experiment Committee of the National Institutes of Natural Sciences and the University of Toyama and performed in accordance with the guidelines from the National Institutes of Natural Sciences, University of Toyama, and Science Council of Japan.
Method details
Phylogenetic analysis of DZF-containing proteins
Amino acid sequences of DZF-containing proteins were obtained from Ensemble77 and SMART.78 The phylogenetic tree was constructed by the maximum likelihood algorithm using MEGA11.66
IDR and PrLD predictions
IDR predictions were performed using the DISOPRED367 and PONDR-FIT68 predictors. PrLD predictions were conducted with the PLAAC predictor.69
Generation of ILF3 PrLD-deficient mice
ILF3 PrLD-deficient mice were generated following procedures described previously.79 Briefly, gRNA, which targets the exon 20 sequence 5'-CCCTGTCTAGGTCAGTTCTACAG-3' of the Ilf3 gene, and Cas9 mRNA were transcribed in vitro using the mMESSAGE mMACHINE T7 Transcription Kit (Thermo Fisher Scientific, Waltham, MA), and purified with the MEGAclear Transcription Clean-Up Kit (Thermo Fisher Scientific). Single-stranded oligo-DNA (ssODN) was synthesized to introduce a stop codon at the 5' most region of the ILF3 PrLD coding exon. The sequence of ssODN was 5'-CGGCTGCGGGCCAGCCTTCAGGGCCCATGACTGCTCCTTCCTCCCCTCCCAGCCCTCACCCTGGGACCATCCATCTAATCTGCTGCTTCCCTGTCTAGGTTAGTTCTACAGCAATGGAGGGCATTCTGGGAATGCCGGTGGTGGAGGCAGCGGGGGAGGTGGTGGCTCATCCAGCTACAGCTCCTACTACCAAGGAGACA-3'. The RNA and DNA were mixed at concentrations of 25 ng/μl Cas9 mRNA, 12.5 ng/μl gRNA, and 100 ng/μl ssODN, and the injection into fertilized eggs was performed at the RIKEN Research Resource Center.
For genotyping of the ILF3 PrLD-deficient mice, the Ilf3 genomic region containing the substituted nucleotide was amplified using primers 5'-ATCAGCTGAGGGTGAGTGGAGCTG-3' and 5'-TGCAGCGGCTTCTTCCCAGCATGC-3' (PrLD-R). The 1 μl PCR products were diluted with 11.3 μl water and treated with 0.2 μl ExoSAP-IT (Thermo Fisher Scientific) to remove the primers. The purified PCR products were sequenced using the PrLD-R primer. To confirm the single nucleotide substitution in the Ilf3 gene, DNA sequencing chromatograms were visualized with ApE software.70
Generation of ILF3 deficient mice
The ILF3 deficient mice were generated using a CRISPR/Cas9 system with modifications to previous studies.47 The gRNA was generated to target the exon 7 sequence 5'-CGTCCCTCCGACACGCCAAG-3' of the Ilf3 gene. The gRNA template DNA containing the T7 promoter, gRNA spacer (the target sequence), and gRNA scaffold was synthesized using the gBlocks gene fragments service (Integrated DNA Technologies, Coralville, IA) and transcribed to gRNA using the MEGAshortscript T7 Transcription Kit (Thermo Fisher Scientific). The synthesized gRNA was treated with 0.3 U/μl Turbo DNase (Thermo Fisher Scientific) at 37°C for 15 min, purified using phenol-chloroform extraction and ethanol precipitation, and dissolved in Opti-MEM1 (Thermo Fisher Scientific).
In vitro fertilized eggs from C57BL/6J strain mice were washed three times with modified Whitten's medium (mWM) 4 hours after fertilization and then aligned in an electrode gap, which was filled with opti-MEM1 containing 200 ng/μl Cas9 protein (Integrated DNA Technologies) and 200 ng/μl gRNA. Electroporation was performed using a Genome Editor (BEX Co. Ltd., Tokyo, Japan) at 25 V (3 msec ON + 97 msec OFF) three times, alternating the current direction. After electroporation, the eggs were washed three times with M2 medium (Sigma-Aldrich, Burlington, MA) containing 100 units/ml Penicillin-Streptomycin (Thermo Fisher Scientific) and cultured overnight in mWM. On the next day, the surviving 2-cell stage zygotes were transferred to the oviducts of pseudopregnant females.
To identify mutations in the Ilf3 gene that were generated by CRISPR/Cas9, the genomic region containing the gRNA target was amplified by PCR using primers 5'-GAGCTGATGGCACTTTAGCC-3' (exon7-F) and 5'-AGTCTCCTCAGGGCTGTCAA-3'. The 1 μl PCR products were diluted with 11.3 μl water and treated with 0.2 μl ExoSAP-IT (Thermo Fisher Scientific) to remove the primers. The purified PCR products were sequenced using the exon7-F primer. To confirm the insertions and deletions of nucleotides, DNA sequencing chromatograms were visualized using ApE software. Genotyping of the Ilf3+/− mice, which have the Ilf3− allele with 14-bp deletion, was performed by PCR using primers 5'-TTCTGTGGTTTAAATTTGGGAGAGG-3' (common forward primer) and 5'-AAAATGCAAACCTGGAACCACTTG-3' to detect the Ilf3+ allele, and the common forward primer and 5'-AAATGCAAACCTTGGCGGAG-3' to detect the Ilf3− allele.
Water-immersion and restraint stress (WIRS)
Mice were housed in home cages in pairs of identical genotypes (1 or 2 pairs/cage) with a 12-hour light/dark cycle. The mice in the stress group were placed in a 50 mL conical polypropylene centrifuge tube with holes for breathing and soaked in water to a level where the nose was not submerged for 3 hours a day during the light cycle over 14 days. Mice were weighed daily just prior to WIRS (see Figures S11A–S11C). The mice in the control group, which were placed in a home cage separate from the stress group to avoid the effects of interactions between the stressed and control individuals, remained in the home cage during the stress exposure period.
Quantification of mRNA expression
Total RNA was extracted from mouse tissues using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's protocol. Reverse transcription (RT) was performed with M-MLV-Reverse Transcriptase (Thermo Fisher Scientific) and quantitative PCR was performed with TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara Bio, Shiga, Japan) according to the manufacturer's protocols using the 7500 real-time PCR system (Thermo Fisher Scientific). The expression level of each gene was quantified by a standard curve method using the same gene as a standard. The primers used for quantitative RT-PCR were: 5'-AGACAGGCCCTGTTCATGCT-3' and 5'-TTGGCAAGCCCATGTCCTGT-3' for NFARs (both NFAR1 and NFAR2); 5'-ACGCTTTGAGGTGTGGTGGT-3' and 5'-AACGGCAAGACCTCGGAACA-3' for NFAR2; 5'-TCTACACTGCCTGGCACACA-3' and 5'-AGGGGCAGTTCTGCAGCAAA-3' for NFAR1; 5'-AACGACCCCTTCATTGACCT-3' and 5'-TGGAAGATGGTGATGGGCTT-3' for GAPDH; 5'-CGATGCTCTGGCCAATGCTG-3' and 5'-GCTAGCCAAGGTCACCAGCA-3' for Hba-a1; 5’-ACATTCCCACTGGCACC-3’ and 5’-GGGGTAATGAATGAGGC-3’ for mt-Atp8; 5’-GCATTCTGACTCCCCCAAAT-3’ and 5’-TGAATTGCTCATGGTAGTGGA-3’ for mt-Nd3; 5'-GTATTGGGGGTGATTATAGAG-3' and 5'-CCATCATTCAAGTAGCACAAC-3' for mt-Nd6; 5'-ACTATGGACTCCGTTCGCTCAG-3' and 5'-ACAGAGTCAACCAACTCAGCTCC-3' for Tubb5; and 5'-GGTGCCAAGGATGACTTTATGGA-3' and 5'-GGTAAACTCGGGATCGAAGTGC-3' for Sgk1.
Western blotting
Mouse tissue extracts were prepared by homogenization and sonication in Laemmli sample buffer. After boiling for 5 min, the extracts were loaded onto SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Merck KGaA, Darmstadt, Germany). The membranes were probed with anti-ILF3 rabbit monoclonal antibody (ab92355, Abcam, Cambridge, UK), anti-ILF3 rabbit polyclonal antibody (19887-1-AP, Proteintech, Chicago, IL), and anti-α-tubulin rabbit polyclonal antibody (PM054, Medical & Biological Laboratories, Nagoya, Japan). Biotinylated secondary antibody (Cytiva, Marlborough, MA) and alkaline phosphatase-conjugated streptavidin (Cytiva) were used for the detection of protein bands in a bromochloroindolyl phosphate/nitro blue tetrazolium solution. Quantitative comparisons of band intensities between the genotypes were performed as previously described using standard curves generated from the dilution series of each tissue extract from the Ilf3+/+ mice on the same membrane.49 The NFAR2/NFAR1 band intensity ratio was calculated by measuring the NFAR1 and NFAR2 bands in the same lane using ImageJ software.71
Immunostaining
The brains of adult mice were embedded in Tissue-Tek O.C.T. Compound (Sakura Finetek, Tokyo, Japan), frozen in liquid nitrogen, and sliced to a thickness of 12 μm for ILF3 staining and 30 μm for c-Fos staining using a cryostat (Leica CM1950, Leica, Wetzlar, Germany). The sections were mounted on silane-coated coverslips and dried at room temperature. The samples were fixed in 3.7% formaldehyde in phosphate-buffered saline (PBS) for 10 min, washed with PBS, and treated with blocking buffer (10% fetal bovine serum (FBS) and 0.1% Triton X-100 in PBS) for 1 h at room temperature. The samples were then incubated with primary antibodies in blocking buffer at 4°C overnight. The primary antibodies used were the anti-ILF3 rabbit monoclonal antibody (Abcam), anti-TDP-43 rabbit polyclonal antibody (10782-2-AP, Proteintech, Rosemont, IL, USA), anti-hnRNPA1 rabbit monoclonal antibody (#8443, Cell Signaling Technology, Danvers, MA), and anti-c-Fos rabbit monoclonal antibody (#2250, Cell Signaling Technology). After washing with PBS, the samples were stained with Alexa488-conjugated anti-rabbit IgG antibody (Thermo Fisher Scientific) and 4',6-diamidino-2-phenylindole (DAPI) (FUJIFILM Wako Pure Chemical, Osaka, Japan) in blocking buffer at 4°C overnight. The samples were washed with PBS and mounted in Mowiol (Sigma-Aldrich). Fluorescence images were acquired using an A1 confocal laser scanning microscope equipped with a Ti-E inverted microscope (Nikon, Tokyo, Japan) and a 20× or 60× objective.
The fluorescence images were analyzed using ImageJ/Fiji software.72 To calculate the nuclear-cytoplasmic ratio of staining intensity, the images were processed by background subtraction and mean-filtering. The fluorescence intensity of the whole cells and nuclei was measured using the ROI manager, and the intensity of the cytoplasm was calculated by subtracting the intensity of nuclei from the intensity of whole cells. The nuclear-cytoplasmic ratio was calculated by dividing the mean intensity per pixel in the nucleus by that in the cytoplasm. To count the number of c-Fos positive cells, the images were z-stacked (all consecutive images acquired in 1 μm steps) and binarized using the "Triangle" auto threshold function. Particles (c-Fos-positive cells) with a size of 30-200 pixels were counted using the "Analyze particles" function. Multiple cells that were counted as one because they overlapped on the image were manually recounted. Particles that did not overlap with DAPI-stained nuclei were not counted.
Quantification of NFARs-containing granules in the nucleus
Mice were perfused with PBS and subsequently with 3.7% formaldehyde in PBS for fixation using a peristaltic pump (MP-2000, Tokyo Rikakikai, Tokyo, Japan). The brain was removed and post-fixed with 3.7% formaldehyde in PBS overnight at 4°C. The brain was coronally sectioned at a thickness of 50 μm using a vibratome (VT1200S, Leica, Germany). Immunostaining was performed as described above. Fluorescence images were acquired consecutively in 1 μm steps using the A1 confocal laser scanning microscope with a 60× objective. The number and size of NFARs-containing granules in the nucleus were quantified using ImageJ/Fiji software. The quantification used 5 consecutive images with the maximum diameter of the nucleus. Granules that overlapped when the consecutive images were stacked were judged to be the same granule. Granule size was measured in one of the consecutive images with the largest diameter of the overlapping granules.
Analysis of neurite formation in cultured AMY neurons
Dissociated AMY neurons were prepared from Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD littermates at embryonic day 16-18 (E16-18). Neurons were plated at a density of 5.2 × 103 cells/cm2 onto poly-D-lysine-coated coverslips (Matsunami, Osaka, Japan) in Neurobasal-A medium (Thermo Fisher Scientific) containing B-27 supplement (Thermo Fisher Scientific), 0.5 mM glutamine, and 25% Neuron culture medium (FUJIFILM Wako Pure Chemical). Cultured neurons were incubated at 37°C in a 5% CO2 incubator. They were fixed with 3.7% formaldehyde in PBS for 10 min at 3, 5, and 7 days in vitro (DIV).
Immunostaining was performed as described above. The primary antibodies used were anti-MAP2 rabbit polyclonal antibody (ab32454, Abcam) and anti-Tau-1 mouse monoclonal antibody (MAB3420, Sigma-Aldrich). Secondary antibodies were Alexa488-conjugated anti-rabbit IgG antibody (Thermo Fisher Scientific) and Cy3-conjugated anti-mouse IgG antibody (Jackson ImmunoResearch, West Grove, PA). Fluorescence images were acquired using the A1 confocal laser scanning microscope with a 20× or 40× objective. Serial images acquired at 0.5 μm steps were z-stacked. Concentric circles were drawn at 30 μm intervals around the nucleus for Sholl analysis, where the number of intersections was counted. One of the neurites with the strongest tau staining was determined to be the axon.
RNA-seq and ribosome profiling
The Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice in the stress group were exposed to WIRS for 3 h a day for 14 days. The amygdala of control mice without WIRS and stressed mice with WIRS were isolated from the brains and quickly frozen in liquid nitrogen. The samples were lysed in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/ml cycloheximide, 100 μg/ml chloramphenicol, and 1% Triton X-100) using a beads shocker (Yasui Kikai, Osaka, Japan), treated with 25 U/ml Turbo DNase (Thermo Fisher Scientific) on ice for 10 min, and centrifuged at 20,000 g for 10 min at 4°C. The RNA concentration of lysate was measured using a Qubit RNA HS Assay Kit (Thermo Fisher Scientific). The lysate was split into two for the use in RNA-seq and ribosome profiling.
For RNA-seq analysis, the total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) from the lysate containing 500 ng RNA. The rRNA was removed using the Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina, San Diego, CA). The cDNA libraries were prepared using the TruSeq Stranded mRNA Library Prep Kit (Illumina).
Ribosome profiling was conducted as previously reported46,80 with modifications. Lysate containing 1.5 μg RNA was treated with 3 U of RNase I (Lucigen, Middleton, WI) at 25°C for 45 min, followed by the addition of 10 μl SUPERase In (Thermo Fisher Scientific) to stop nuclease digestion. The sample was overlaid on a 1 M sucrose cushion and the ribosomes were pelleted by centrifugation for 1 h at 100,000 rpm at 4°C in a TLA-110 rotor (Beckman Coulter, Brea, CA). Ribosome-bound RNA was isolated using a Direct-zol RNA MicroPrep Kit (Zymo Research, Irvine, CA). After gel electrophoresis, RNA fragments corresponding to 17-34 nt were excised. After ligating the RNA fragments to preadenylated linkers, ribosomal RNAs were removed with a Ribo-Zero Gold rRNA Removal Kit (Human/Mouse/Rat) (Illumina). Reverse transcription was performed by incubation with ProtoScriptII (New England Biolabs, Ipswich, MA) at 50°C for 30 min. The cDNAs were circularized by CircLigase II (Lucigen) and PCR-amplified.
The libraries for RNA-seq and ribosome profiling were sequenced on a HiSeq4000 (Illumina). Read quality filtering and adapter trimming were conducted using fastp.73 After removing noncoding RNA-mapped reads, the remaining reads were aligned to the mouse genome of GRCm38/mm10. For ribosome profiling, the A-site offsets from the 5' end were determined to be 15 for 28-31 nt and 16 for 32-33 nt for cyto-ribosome footprints in the Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD mice; 14 for 21- 28 nt, 15 for 29-33 nt, and 16 for 34 nt for mito-ribosome footprints in the Ilf3+/+ mice; 13 for 22 nt, 14 for 21, 23-29, and 31 nt, and 15 for 30 and 32-34 nt for mito-ribosome footprints in the Ilf3ΔPrLD/ΔPrLD mice. For RNA-seq, an offset of 15 was used for all mRNA fragments. We used the GENCODE VM23 gene annotation and transcripts. The differential expression analysis of RNA-seq and ribosome profiling was performed using the DESeq2 package.74 Translation efficiency, which is the number of reads in ribosome profiling normalized by that in RNA-seq, was also analyzed using DESeq2. Significance was calculated using the likelihood ratio test in a generalized linear model. Reads corresponding to the first and the last 5 codons in the coding sequence were excluded from the analysis. Two replicates from 4 mice were analyzed in each group (each replicate consists of samples from two mice).
Gene ontology enrichment analysis
GO enrichment analysis was performed using the DAVID Bioinformatics Resources.75 The annotation categories used were GOTERM_BP_DIRECT and KEGG_PATHWAY. The significance of the overrepresentation of GO terms was assessed using the Benjamini-Hochberg false discovery rate (FDR) criterion at p < 0.05.
Design of comprehensive behavioral test battery
Male Ilf3+/+ and Ilf3ΔPrLD/ΔPrLD littermates were group-housed (2 pairs/cage) in a room with a 12-hour light/dark cycle. All mice had access to food and water ad libitum. A battery of behavioral tests was conducted in the following sequence starting at 11 weeks of age (Table 1): general health and neurologic screening (body weight, body temperature, grip strength, and wire hang), light/dark transition, open field, elevated plus maze, hot plate, social interaction test, rotarod test, three-chamber social approach test (100 lux), startle response and prepulse inhibition, Porsolt forced swim test, Barnes maze, T-maze (spontaneous alternation), contextual and cued fear conditioning test, three-chamber social approach test (5 lux), social interaction test in a home cage, and tail suspension test. All testing apparatuses were cleaned with diluted hypochlorous solution or 70% ethanol for each test to prevent biases due to olfactory cues.
Body weight and body temperature
Body weight was measured using a weight scale with the mice placed in a stainless bowl. For body temperature, the rectal temperature was measured.
Open field test
Mice were placed in an open field apparatus (40 × 40 × 30 cm; AccuScan Instruments, Columbus, OH) illuminated at 100 lux. The total distance traveled, vertical activity (rearing measured by counting the number of photobeam interruptions), and time spent in the center area of the field were recorded for 120 min using the VeraMax system (AccuScan Instruments).
Light/dark transition test
The light/dark transition test was conducted as previously described.81 Mice were placed in a dark chamber and the door was opened 3 seconds later. The mice were free to move between the two chambers for 10 min with the door open. The total number of transitions between chambers, latency to the first entry to the light chamber, the time spent in each chamber, and the total distance traveled were recorded automatically using the ImageLD program (see section, “Data analysis in behavioral test battery”).
Barnes maze test
The Barnes maze test was conducted as previously described.48 An escape box containing paper bedding was placed under the target hole. Mice were trained in 12 trials. 24 hours after the last training, a probe trial (PT1) was performed without the escape box. 1 month after PT1, another probe trial (PT2) was performed to evaluate memory retention (see Figure S8D). After PT2, mice were trained in 6 additional trials, in which the location of the target hole was not changed from the initial training trials. The mice then underwent a reversal training session. In the reversal training, the location of the target hole was changed to the opposite side of the maze, and the mice were trained in 6 trials. Then, 24 hours after the last reversal training, another probe trial (PT3) was performed (see Figure S8F). The data acquisition and analysis were performed automatically using ImageBM program (see section, “Data analysis in behavioral test battery”).
Contextual and cued fear conditioning test
The contextual and cued fear conditioning test was conducted as previously described.82 Mice were placed in a square test chamber with a metal grid floor. Then, 55 dB of white noise was presented as a conditioned stimulus (CS) for 30 seconds, and a 0.3 mA foot shock was given to the mice as an unconditioned stimulus (US) during the last 2 seconds of the white noise. Each mouse received three CS-US pairings at 2-min intervals in a conditioning session. Then, 24 hours after the conditioning session, a contextual test and a cued test with altered context were performed. In the contextual test, the mice were placed in the same test chamber as the conditioning session for 5 min without CS or US. In the cued test with altered context, the mice were placed in a novel triangular test chamber for 6 min. CS and US were not presented during the first 3 min, and only CS (55 dB white noise) was presented during the last 3 min. Freezing responses during the conditioning, contextual testing, and cued testing sessions were measured automatically using the ImageFZ program (see section, “Data analysis in behavioral test battery”).
Neuromuscular examination
Neuromuscular strength was examined by grip strength and wire hang tests. A grip strength meter (O’Hara & Co., Tokyo, Japan) was used to measure the mouse forelimb grip strength. Mice were held by the tail to allow their forelimbs to grip the wire grid. The mice were then gently pulled backward by the tail until they released the wire grid. The maximum force generated by the mouse forelimbs was recorded. Each mouse was tested three times and the greatest value was used for analysis. In the wire hang test, a box (22 × 22 × 30 cm) with a wire mesh grid (10 × 10 cm) on the top surface (O’Hara & Co.) was used. Mice were placed on the wire mesh grid, and it was inverted gently so that the mice gripped the wire. Latency to fall from the wire was recorded with a cutoff time of 60 sec.
Hot plate test
Mice were placed on a 55.0 ± 0.3°C hot plate (Columbus Instruments, Columbus, OH), and the latency to the first paw response was recorded. The paw response was defined as either paw shaking or licking.
Rotarod test
Motor coordination and balance were tested with the rotarod test. An accelerating rotarod (UGO Basile, Varese, Italy) was used. Mice were placed on a rotating rod (3 cm diameter), and the latency until the mice fell from the rod was measured. The speed of the rotarod accelerated from 4 to 40 rpm in 5 min.
Elevated plus maze test
The elevated plus maze test, which is widely used to assess anxiety-like behavior, was conducted as previously described.83 The apparatus comprised two arms without walls (open arms, 25 × 5 cm), two arms of the same size with transparent walls (15 cm high) (closed arms), and a central square (5 × 5 cm) connecting the arms, which were at 90° to each other (O’Hara & Co., Tokyo, Japan). Arms of the same type were located opposite one another. The arms and the central square were elevated to a height of 55 cm above the floor. Mice were placed in the central square and allowed to freely explore the maze for 10 min. The number of arm entries, the percentage of entries into the open arms, the distance traveled, and the percentage of time spent in the open arms were measured automatically using the ImageEP program (see section, “Data analysis in behavioral test battery”).
Porsolt forced swim test
The Porsolt forced swim test84 was performed to assess depression-related behaviors. Mice were placed in a Plexiglas cylinder (20 cm height × 10 cm diameter, O’Hara & Co.), filled with water (23°C) up to 7.5 cm high, for 10 min a day for two consecutive days. The percentage of time spent immobile was measured automatically using the ImagePS program (see section, “Data analysis in behavioral test battery”).
Tail suspension test
The tail suspension test was performed to assess depression-related behaviors. Mice were suspended by their tails with adhesive tape 30 cm above the floor of a chamber (31 × 41 × 41 cm) (O’Hara & Co.), and the percentage of time spent immobile during a 10-min test period was recorded using the ImageTS program (see section, “Data analysis in behavioral test battery”).
Startle response/prepulse inhibition test
The startle response and prepulse inhibition test was conducted as previously described.85 A startle reflex measurement system (O’Hara & Co.) was used. Mice were placed in a Plexiglas cylinder and left undisturbed for 10 min for habituation just before the test. In the test, 110 or 120 dB of white noise was used for 40 msec as a startle stimulus. The background noise level was 70 dB. The prepulse sound was presented 100 msec before the startle stimulus, and its intensity was 74 or 78 dB. The test session consisted of six consecutive trial types: two types of startle stimulus-only trials (110 and 120 dB) and four types of prepulse inhibition trials (74-110, 78-110, 74-120, and 78-120 dB). The average inter-trial interval was 15 sec (range 10-20 sec). The startle response was recorded for 140 msec, measuring the response every millisecond, starting with the onset of the prepulse stimulus. The peak startle amplitude recorded during the 140-msec sampling window was used as the dependent variable.
Social interaction test in a novel environment
Social interaction was measured in a novel environment as previously described.86 Weight-matched (within 2 g) mice that had been housed in different cages were placed together in a box (40 × 40 × 30 cm) and allowed to explore freely for 10 min. The total number of contacts, total duration of contacts, mean duration per contact, and total distance traveled were measured automatically using the ImageSI program (see section, “Data analysis in behavioral test battery”).
Three-chambered social approach test
The test for sociability and social novelty preference was conducted as previously described.87,88 The apparatus comprised a rectangular, three-chambered box and a lid with a video camera (O’Hara & Co.), and was illuminated at 100 or 5 lux. Each chamber measured 20 × 40 × 47 cm, and the partition walls were made of clear Plexiglas with a small square opening (5 × 3 cm) allowing access to each chamber. First, the test mice were placed in the three-chambered box and allowed to explore for 10 min for habituation, during which an empty wire cage (9 cm in diameter, 11 cm in height, with vertical bars 0.5 cm apart) was placed in the outer corner of each of the left and right chambers. In the test session, an unfamiliar male mouse (stranger 1) that had no prior contact with the test mouse was placed in a wire cage in one of the side chambers. An empty wire cage was placed in the corner of the other side chamber. The location of the stranger mouse in the left vs right chamber was systematically alternated between trials. The test mice were placed in the central chamber and allowed to explore the entire box for 10 min to assess sociability (sociability test). In the next session, another unfamiliar male mouse (stranger 2) was placed in the wire cage that had been empty during the sociability test to evaluate social preference for a new stranger (social novelty preference test). Thus, the test mouse had a choice between the familiar mouse already investigated (stranger 1) and the novel unfamiliar mouse (stranger 2). The time spent around each cage was automatically calculated from video images using the ImageCSI program (see section, “Data analysis in behavioral test battery”).
Social interaction test in a home cage
Monitoring of social behavior between two mice in a familiar environment was conducted as previously described.48 Two mice of the same genotype, which had been housed separately, were placed together in a home cage. Their social interaction and locomotor activity were monitored for seven days. Social interaction was measured by counting the number of particles detected in each frame. In that measurement, two particles indicated that the two mice were separated from each other, and one particle indicated that they were in contact with each other. Data acquisition and analysis were performed automatically using ImageHCSI (see section, “Data analysis in behavioral test battery”).
T-maze (spontaneous alternation)
The T-maze test was conducted as previously described89 using an automated modified T-maze apparatus (O’Hara & Co.). Mice were allowed to freely run 10 laps in a session and subjected to 5 sessions in total. The correct response in each lap means that the mouse has chosen the opposite direction to the last lap. Data acquisition and analysis were performed automatically using ImageTM (see section, “Data analysis in behavioral test battery”).
Data analysis in the behavioral test battery
The behavioral data were acquired automatically using applications (ImageLD, ImageBM, ImageFZ, ImageEP, ImagePS, ImageTS, ImageSI, ImageCSI, ImageHCSI, and ImageTM) based on the public domain NIH Image and ImageJ programs, and modified for each test by Tsuyoshi Miyakawa (O'Hara & Co.). Statistical analysis was conducted using Stat View (SAS Institute, Cary, NC). The data were analyzed using the Student's t-test, one-way ANOVA, or two-way repeated measures ANOVA. The values in the graph are presented as mean ± SEM. In the bar plots, individual values are represented by dot plots.
Morris water maze
The Morris water maze was conducted as previously described.90 A transparent platform (6 cm in diameter) was hidden below the surface of the water in a round water tank with a diameter of 110 cm. The water was clouded with non-toxic white paint (Sakura Color Products, Osaka, Japan). Spatial cues (4 different plane figures) were displayed on the wall of the tank. Mice were placed in the pool and the escape latency to the platform was measured. If the mouse found the platform within a 2-min time limit, then the mouse was allowed to stay on the platform for 30 seconds. If not, then the mouse was guided to the platform before staying on the platform for 30 seconds. The starting position was changed randomly, but the platform position was fixed. In the training session, mice were given 6 trials per day for 7 consecutive days.
The day after the last trial of the training session, the mice underwent a probe test. The platform was removed from the pool and the swimming path of the mouse was tracked for 1 min using a computer-based video tracking system called ANY-Maze (Stoeling, Wood Dale, IL). The time spent in the target quadrant, where the hidden platform had been placed, and in the other quadrants was measured. The data were analyzed in R76 using three-way repeated measures ANOVA.
Contextual fear conditioning test (passive avoidance)
The passive avoidance test was conducted using the LDK-M chamber (Melquest, Toyama, Japan) equipped with the SCANET system (Melquest). The day before the training day, the mice were allowed to freely explore both the light and dark chambers for 10 min with the door open to habituate themselves to the test environment. On the day of the training, the mice were placed in the light chamber, and after the door was opened, the mice were allowed to freely explore the chambers for 10 min. The time spent in the dark chamber was measured in the first 5 min (Pre). Then, when the mouse entered the dark chamber, the door was closed and the mouse received a foot shock (1.5 mA, 2-second duration, 4 times with 1-min intervals). After the foot shock, the mice were returned to their home cage. 5 min, 1day, and 1 week after the foot shock, the mice were replaced into the light chamber, allowing them to freely explore the chambers for 5 min after the door opened. During these 5-min periods, the time spent in the dark chamber was measured. The data were analyzed in R using three-way repeated measures ANOVA. Simple effects were analyzed after interaction was significant in the three-way repeated measures ANOVA.
Quantification and statistical analysis
Sample numbers and experimental repeats are indicated in the figures and figure legends. Statistical significance was calculated using the log-rank test, three-way repeated measures ANOVA, two-way repeated measures ANOVA, two-way ANOVA, one-way ANOVA, simple effect analysis, Welch’s t-test, Tukey-Kramer test, and Student's t-test with R. In the bar plots, individual values were represented as dot plots. In the behavioral tests, mice that died before the end of each experiment or had outlier data were excluded from the data analysis.
Acknowledgments
We thank C. Matsuda for technical assistance; S. Ohsawa for the generation of the ILF3 deficient mice; Dr. H. Koyama for advice on image analysis; Model Organisms, Trans-Omics, and Optics and Imaging Facilities in the Trans-scale Biology Center of the National Institute for Basic Biology for technical supports. This work was supported by a Life Science Research grant from RIKAKEN HOLDINGS CO., LTD. and the Sasakawa Scientific Research Grant from The Japan Science Society (2020-4061) to A.Y.; Grant-in-Aid for Early-Career Scientists (JP21K15023) from the Japan Society for the Promotion of Science (JSPS), Grant-in-Aid for Transformative Research Areas [A] (JP21H05734) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and Special Postdoctoral Researchers and Incentive Research Projects from RIKEN to Y.S.; Grant-in-Aid for Scientific Research (JP19H02959) from the JSPS, Grant-in-Aid for Transformative Research Areas [B] (JP20H05784) from the MEXT, AMED-CREST (JP21gm1410003) from the Japan Agency for Medical Research and Development (AMED), and “Biology of Intracellular Environments” and “Integrated life science research to challenge super aging society” from RIKEN to S.I; Grant-in-Aid for Scientific Research (19H03161, 22H02552 and 16H06276 (AdAMS)) from the JSPS and the Bioscience Research Grant from the Takeda Science Foundation to N.S.
Author contributions
A.Y. designed the research, performed experiments, analyzed the data, and wrote the article. Y.S. and S.I. provided guidance and support in RNA-seq and ribosome profiling and edited the article. M.M. supported RNA-seq and ribosome profiling and generated ILF3 PrLD-deficient mice. S. N. generated ILF3 PrLD-deficient mice and contributed to the writing. K.F., Y.K., M.A., E.S., and K.T. performed the behavioral test battery. N.S. conceived and supervised the research and wrote the article.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: February 19, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106229.
Supplemental information
Data and code availability
-
•
The RNA-Seq and ribosome profiling data used in this study have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. The raw data of behavioral test battery are disclosed in the gene-brain-phenotyping database (http://www.mouse-phenotype.org/), as listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Haynes C., Oldfield C.J., Ji F., Klitgord N., Cusick M.E., Radivojac P., Uversky V.N., Vidal M., Iakoucheva L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput. Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shimizu K., Toh H. Interaction between intrinsically disordered proteins frequently occurs in a human protein-protein interaction network. J. Mol. Biol. 2009;392:1253–1265. doi: 10.1016/j.jmb.2009.07.088. [DOI] [PubMed] [Google Scholar]
- 3.Uversky V.N. Intrinsically disordered proteins in overcrowded milieu: membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 2017;44:18–30. doi: 10.1016/j.sbi.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Gomes E., Shorter J. The molecular language of membraneless organelles. J. Biol. Chem. 2019;294:7115–7127. doi: 10.1074/jbc.TM118.001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boeynaems S., Alberti S., Fawzi N.L., Mittag T., Polymenidou M., Rousseau F., Schymkowitz J., Shorter J., Wolozin B., Van Den Bosch L., et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 2018;28:420–435. doi: 10.1016/j.tcb.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woodruff J.B., Hyman A.A., Boke E. Organization and function of non-dynamic biomolecular condensates. Trends Biochem. Sci. 2018;43:81–94. doi: 10.1016/j.tibs.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Portz B., Lee B.L., Shorter J. FUS and TDP-43 phases in health and disease. Trends Biochem. Sci. 2021;46:550–563. doi: 10.1016/j.tibs.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Choi J.M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M., Maharana S., Lemaitre R., Pozniakovsky A., Drechsel D., et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzmann T.M., Alberti S. Prion-like low-complexity sequences: key regulators of protein solubility and phase behavior. J. Biol. Chem. 2019;294:7128–7136. doi: 10.1074/jbc.TM118.001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maharana S., Wang J., Papadopoulos D.K., Richter D., Pozniakovsky A., Poser I., Bickle M., Rizk S., Guillén-Boixet J., Franzmann T.M., et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halfmann R., Jarosz D.F., Jones S.K., Chang A., Lancaster A.K., Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jung J.H., Barbosa A.D., Hutin S., Kumita J.R., Gao M., Derwort D., Silva C.S., Lai X., Pierre E., Geng F., et al. A prion-like domain in ELF3 functions as a thermosensor in Arabidopsis. Nature. 2020;585:256–260. doi: 10.1038/s41586-020-2644-7. [DOI] [PubMed] [Google Scholar]
- 15.Dorone Y., Boeynaems S., Flores E., Jin B., Hateley S., Bossi F., Lazarus E., Pennington J.G., Michiels E., De Decker M., et al. A prion-like protein regulator of seed germination undergoes hydration-dependent phase separation. Cell. 2021;184:4284–4298.e27. doi: 10.1016/j.cell.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue B., Dunker A.K., Uversky V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]
- 17.Niklas K.J., Dunker A.K., Yruela I. The evolutionary origins of cell type diversification and the role of intrinsically disordered proteins. J. Exp. Bot. 2018;69:1437–1446. doi: 10.1093/jxb/erx493. [DOI] [PubMed] [Google Scholar]
- 18.Romero P.R., Zaidi S., Fang Y.Y., Uversky V.N., Radivojac P., Oldfield C.J., Cortese M.S., Sickmeier M., LeGall T., Obradovic Z., Dunker A.K. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunker A.K., Oldfield C.J., Meng J., Romero P., Yang J.Y., Chen J.W., Vacic V., Obradovic Z., Uversky V.N. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genom. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niklas K.J., Bondos S.E., Dunker A.K., Newman S.A. Rethinking gene regulatory networks in light of alternative splicing, intrinsically disordered protein domains, and post-translational modifications. Front. Cell Dev. Biol. 2015;3:8. doi: 10.3389/fcell.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babu M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016;44:1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harashima A., Guettouche T., Barber G.N. Phosphorylation of the NFAR proteins by the dsRNA-dependent protein kinase PKR constitutes a novel mechanism of translational regulation and cellular defense. Genes Dev. 2010;24:2640–2653. doi: 10.1101/gad.1965010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duchange N., Pidoux J., Camus E., Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–353. doi: 10.1016/s0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- 24.Batlle C., de Groot N.S., Iglesias V., Navarro S., Ventura S. Characterization of soft amyloid cores in human prion-like proteins. Sci. Rep. 2017;7:12134. doi: 10.1038/s41598-017-09714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castella S., Bernard R., Corno M., Fradin A., Larcher J.C. Ilf3 and NF90 functions in RNA biology. Wiley Interdiscip. Rev. RNA. 2015;6:243–256. doi: 10.1002/wrna.1270. [DOI] [PubMed] [Google Scholar]
- 26.Parrott A.M., Walsh M.R., Reichman T.W., Mathews M.B. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J. Mol. Biol. 2005;348:281–293. doi: 10.1016/j.jmb.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 27.Pfeifer I., Elsby R., Fernandez M., Faria P.A., Nussenzveig D.R., Lossos I.S., Fontoura B.M.A., Martin W.D., Barber G.N. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl. Acad. Sci. USA. 2008;105:4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwano Y., Kim H.H., Abdelmohsen K., Pullmann R., Jr., Martindale J.L., Yang X., Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agca C., Boldt K., Gubler A., Meneau I., Corpet A., Samardzija M., Stucki M., Ueffing M., Grimm C. Expression of leukemia inhibitory factor in Müller glia cells is regulated by a redox-dependent mRNA stability mechanism. BMC Biol. 2015;13:30. doi: 10.1186/s12915-015-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viranaicken W., Gasmi L., Chaumet A., Durieux C., Georget V., Denoulet P., Larcher J.C. L-Ilf3 and L-NF90 traffic to the nucleolus granular component: alternatively-spliced exon 3 encodes a nucleolar localization motif. PLoS One. 2011;6:e22296. doi: 10.1371/journal.pone.0022296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reichman T.W., Parrott A.M., Fierro-Monti I., Caron D.J., Kao P.N., Lee C.G., Li H., Mathews M.B. Selective regulation of gene expression by nuclear factor 110, a member of the NF90 family of double-stranded RNA-binding proteins. J. Mol. Biol. 2003;332:85–98. doi: 10.1016/s0022-2836(03)00885-4. [DOI] [PubMed] [Google Scholar]
- 32.Shiina N., Nakayama K. RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45. J. Biol. Chem. 2014;289:21163–21180. doi: 10.1074/jbc.M114.556365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zafir A., Banu N. Modulation of in vivo oxidative status by exogenous corticosterone and restraint stress in rats. Stress. 2009;12:167–177. doi: 10.1080/10253890802234168. [DOI] [PubMed] [Google Scholar]
- 34.Schiavone S., Jaquet V., Trabace L., Krause K.H. Severe life stress and oxidative stress in the brain: from animal models to human pathology. Antioxid. Redox Signaling. 2013;18:1475–1490. doi: 10.1089/ars.2012.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pires-daSilva A., Nayernia K., Engel W., Torres M., Stoykova A., Chowdhury K., Gruss P. Mice deficient for spermatid perinuclear RNA-binding protein show neurologic, spermatogenic, and sperm morphological abnormalities. Dev. Biol. 2001;233:319–328. doi: 10.1006/dbio.2001.0169. [DOI] [PubMed] [Google Scholar]
- 36.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 37.Shi L., Zhao G., Qiu D., Godfrey W.R., Vogel H., Rando T.A., Hu H., Kao P.N. NF90 regulates cell cycle exit and terminal myogenic differentiation by direct binding to the 3'-untranslated region of MyoD and p21WAF1/CIP1 mRNAs. J. Biol. Chem. 2005;280:18981–18989. doi: 10.1074/jbc.M411034200. [DOI] [PubMed] [Google Scholar]
- 38.Jia R., Ajiro M., Yu L., McCoy P., Jr., Zheng Z.M. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation. RNA. 2019;25:630–644. doi: 10.1261/rna.068619.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Y.B., Scotter E.L., Lee D.Y., Troakes C., Mitchell J., Rogelj B., Gallo J.M., Shaw C.E. Cytoplasmic TDP-43 is involved in cell fate during stress recovery. Hum. Mol. Genet. 2021;31:166–175. doi: 10.1093/hmg/ddab227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allemand E., Guil S., Myers M., Moscat J., Cáceres J.F., Krainer A.R. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc. Natl. Acad. Sci. USA. 2005;102:3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang H., Xia L., Chen J., Zhang S., Martin V., Li Q., Lin S., Chen J., Calmette J., Lu M., et al. Stress-glucocorticoid-TSC22D3 axis compromises therapy-induced antitumor immunity. Nat. Med. 2019;25:1428–1441. doi: 10.1038/s41591-019-0566-4. [DOI] [PubMed] [Google Scholar]
- 42.Miyata S., Koyama Y., Takemoto K., Yoshikawa K., Ishikawa T., Taniguchi M., Inoue K., Aoki M., Hori O., Katayama T., Tohyama M. Plasma corticosterone activates SGK1 and induces morphological changes in oligodendrocytes in corpus callosum. PLoS One. 2011;6:e19859. doi: 10.1371/journal.pone.0019859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weger M., Alpern D., Cherix A., Ghosal S., Grosse J., Russeil J., Gruetter R., de Kloet E.R., Deplancke B., Sandi C. Mitochondrial gene signature in the prefrontal cortex for differential susceptibility to chronic stress. Sci. Rep. 2020;10:1018308. doi: 10.1038/s41598-020-75326-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schelshorn D.W., Schneider A., Kuschinsky W., Weber D., Krüger C., Dittgen T., Bürgers H.F., Sabouri F., Gassler N., Bach A., Maurer M.H. Expression of hemoglobin in rodent neurons. J. Cereb. Blood Flow Metab. 2009;29:585–595. doi: 10.1038/jcbfm.2008.152. [DOI] [PubMed] [Google Scholar]
- 45.Stankiewicz A.M., Goscik J., Swiergiel A.H., Majewska A., Wieczorek M., Juszczak G.R., Lisowski P. Social stress increases expression of hemoglobin genes in mouse prefrontal cortex. BMC Neurosci. 2014;15:130. doi: 10.1186/s12868-014-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mito M., Mishima Y., Iwasaki S. Protocol for disome profiling to survey ribosome collision in humans and zebrafish. STAR Protoc. 2020;1:100168. doi: 10.1016/j.xpro.2020.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakazawa K., Shichino Y., Iwasaki S., Shiina N. Implications of RNG140 (caprin2)-mediated translational regulation in eye lens differentiation. J. Biol. Chem. 2020;295:15029–15044. doi: 10.1074/jbc.RA120.012715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takao K., Tanda K., Nakamura K., Kasahara J., Nakao K., Katsuki M., Nakanishi K., Yamasaki N., Toyama K., Adachi M., et al. Comprehensive behavioral analysis of calcium/calmodulin-dependent protein kinase IV knockout mice. PLoS One. 2010;5:e9460. doi: 10.1371/journal.pone.0009460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohashi R., Takao K., Miyakawa T., Shiina N. Comprehensive behavioral analysis of RNG105 (Caprin1) heterozygous mice: reduced social interaction and attenuated response to novelty. Sci. Rep. 2016;6:20775. doi: 10.1038/srep20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phelps E.A., LeDoux J.E. Contributions of the amygdala to emotion processing: from animal models to human behavior. Neuron. 2005;48:175–187. doi: 10.1016/j.neuron.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 51.Babaev O., Piletti Chatain C., Krueger-Burg D. Inhibition in the amygdala anxiety circuitry. Exp. Mol. Med. 2018;50:1–16. doi: 10.1038/s12276-018-0063-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fanselow M.S., Dong H.W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;65:7–19. doi: 10.1016/j.neuron.2009.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim W.B., Cho J.H. Encoding of contextual fear memory in hippocampal-amygdala circuit. Nat. Commun. 2020;11:1382. doi: 10.1038/s41467-020-15121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang I.F., Reddy N.M., Shen C.K.J. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl. Acad. Sci. USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.March Z.M., King O.D., Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res. 2016;1647:9–18. doi: 10.1016/j.brainres.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oka T. Psychogenic fever: how psychological stress affects body temperature in the clinical population. Temperature (Austin) 2015;2:368–378. doi: 10.1080/23328940.2015.1056907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pardon M.C., Kendall D.A., Pérez-Diaz F., Duxon M.S., Marsden C.A. Repeated sensory contact with aggressive mice rapidly leads to an anticipatory increase in core body temperature and physical activity that precedes the onset of aversive responding. Eur. J. Neurosci. 2004;20:1033–1050. doi: 10.1111/j.1460-9568.2004.03549.x. [DOI] [PubMed] [Google Scholar]
- 58.Lamprecht R., Dracheva S., Assoun S., LeDoux J.E. Fear conditioning induces distinct patterns of gene expression in lateral amygdala. Gene Brain Behav. 2009;8:735–743. doi: 10.1111/j.1601-183X.2009.00515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Innos J., Philips M.A., Raud S., Lilleväli K., Kõks S., Vasar E. Deletion of the Lsamp gene lowers sensitivity to stressful environmental manipulations in mice. Behav. Brain Res. 2012;228:74–81. doi: 10.1016/j.bbr.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 60.Pimenta A.F., Zhukareva V., Barbe M.F., Reinoso B.S., Grimley C., Henzel W., Fischer I., Levitt P. The limbic system-associated membrane protein is an Ig superfamily member that mediates selective neuronal growth and axon targeting. Neuron. 1995;15:287–297. doi: 10.1016/0896-6273(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 61.Bontekoe C.J.M., McIlwain K.L., Nieuwenhuizen I.M., Yuva-Paylor L.A., Nellis A., Willemsen R., Fang Z., Kirkpatrick L., Bakker C.E., McAninch R., et al. Knockout mouse model for Fxr2: a model for mental retardation. Hum. Mol. Genet. 2002;11:487–498. doi: 10.1093/hmg/11.5.487. [DOI] [PubMed] [Google Scholar]
- 62.Huynh D.P., Maalouf M., Silva A.J., Schweizer F.E., Pulst S.M. Dissociated fear and spatial learning in mice with deficiency of ataxin-2. PLoS One. 2009;4:e6235. doi: 10.1371/journal.pone.0006235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weskamp K., Tank E.M., Miguez R., McBride J.P., Gómez N.B., White M., Lin Z., Gonzalez C.M., Serio A., Sreedharan J., Barmada S.J. Shortened TDP43 isoforms upregulated by neuronal hyperactivity drive TDP43 pathology in ALS. J. Clin. Invest. 2020;130:1139–1155. doi: 10.1172/JCI130988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deshaies J.E., Shkreta L., Moszczynski A.J., Sidibé H., Semmler S., Fouillen A., Bennett E.R., Bekenstein U., Destroismaisons L., Toutant J., et al. TDP-43 regulates the alternative splicing of hnRNP A1 to yield an aggregation-prone variant in amyotrophic lateral sclerosis. Brain. 2018;141:1320–1333. doi: 10.1093/brain/awy062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Long G., Chen H., Wu M., Li Y., Gao L., Huang S., Zhang Y., Jia Z., Xia W. Antianemia drug roxadustat (FG-4592) protects against doxorubicin-induced cardiotoxicity by targeting antiapoptotic and antioxidative pathways. Front. Pharmacol. 2020;11:1191. doi: 10.3389/fphar.2020.01191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jones D.T., Cozzetto D. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics. 2015;31:857–863. doi: 10.1093/bioinformatics/btu744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim. Biophys. Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lancaster A.K., Nutter-Upham A., Lindquist S., King O.D. PLAAC: a web and command-line application to identify proteins with prion-like amino acid composition. Bioinformatics. 2014;30:2501–2502. doi: 10.1093/bioinformatics/btu310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davis M.W., Jorgensen E.M. ApE, A plasmid editor: a freely available DNA manipulation and visualization program. Front. Bioinform. 2022;2:818619. doi: 10.3389/fbinf.2022.818619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S., Zhou Y., Chen Y., Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.R Core Team . R Foundation for Statistical Computing; 2020. R: A Language and Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 77.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Austine-Orimoloye O., Azov A.G., Barnes I., Bennett R., et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Letunic I., Khedkar S., Bork P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49:D458–D460. doi: 10.1093/nar/gkaa937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Isobe M., Toya H., Mito M., Chiba T., Asahara H., Hirose T., Nakagawa S. Forced isoform switching of Neat1_1 to Neat1_2 leads to the loss of Neat1_1 and the hyperformation of paraspeckles but does not affect the development and growth of mice. RNA. 2020;26:251–264. doi: 10.1261/rna.072587.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McGlincy N.J., Ingolia N.T. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takao K., Miyakawa T. Light/dark transition test for mice. J. Vis. Exp. 2006;1:104. doi: 10.3791/104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shoji H., Takao K., Hattori S., Miyakawa T. Contextual and cued fear conditioning test using a video analyzing system in mice. J. Vis. Exp. 2014;85:50871. doi: 10.3791/50871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Komada M., Takao K., Miyakawa T. Elevated plus maze for mice. J. Vis. Exp. 2008;22:1088. doi: 10.3791/1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Porsolt R.D., Bertin A., Jalfre M. Behavioral despair in mice: a primary screening test for antidepressants. Arch. Int. Pharmacodyn. Ther. 1977;229:327–336. [PubMed] [Google Scholar]
- 85.Matsuo N., Takao K., Nakanishi K., Yamasaki N., Tanda K., Miyakawa T. Behavioral profiles of three C57BL/6 substrains. Front. Behav. Neurosci. 2010;4:29. doi: 10.3389/fnbeh.2010.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shoji H., Takao K., Hattori S., Miyakawa T. Age-related changes in behavior in C57BL/6J mice from young adulthood to middle age. Mol. Brain. 2016;9:11. doi: 10.1186/s13041-016-0191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moy S.S., Nadler J.J., Perez A., Barbaro R.P., Johns J.M., Magnuson T.R., Piven J., Crawley J.N. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Gene Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 88.Fujii K., Koshidaka Y., Adachi M., Takao K. Effects of chronic fentanyl administration on behavioral characteristics of mice. Neuropsychopharmacol. Rep. 2019;39:17–35. doi: 10.1002/npr2.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shoji H., Hagihara H., Takao K., Hattori S., Miyakawa T. T-maze forced alternation and left-right discrimination tasks for assessing working and reference memory in mice. J. Vis. Exp. 2012;26:3300. doi: 10.3791/3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakayama K., Ohashi R., Shinoda Y., Yamazaki M., Abe M., Fujikawa A., Shigenobu S., Futatsugi A., Noda M., Mikoshiba K., et al. RNG105/caprin1, an RNA granule protein for dendritic mRNA localization, is essential for long-term memory formation. Elife. 2017;6:e29677. doi: 10.7554/eLife.29677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-Seq and ribosome profiling data used in this study have been deposited at GEO and are publicly available as of the date of publication. The accession number is listed in the key resources table. The raw data of behavioral test battery are disclosed in the gene-brain-phenotyping database (http://www.mouse-phenotype.org/), as listed in the key resources table. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.