Abstract
The automatic annotation of the protein universe is still an unresolved challenge. Today, there are 229,149,489 entries in the UniProtKB database, but only 0.25% of them have been functionally annotated. This manual process integrates knowledge from the protein families database Pfam, annotating family domains using sequence alignments and hidden Markov models. This approach has grown the Pfam annotations at a low rate in the last years. Recently, deep learning models appeared with the capability of learning evolutionary patterns from unaligned protein sequences. However, this requires large-scale data, while many families contain just a few sequences. Here, we contend this limitation can be overcome by transfer learning, exploiting the full potential of self-supervised learning on large unannotated data and then supervised learning on a small labeled dataset. We show results where errors in protein family prediction can be reduced by 55% with respect to standard methods.
The automatic annotation of the protein universe is still an unresolved challenge. Today, there are 229,149,489 entries in the UniProtKB database, but only 0.25% of them have been functionally annotated. This manual process integrates knowledge from the protein families database Pfam, annotating family domains using sequence alignments and hidden Markov models. This approach has grown the Pfam annotations at a low rate in the last years. Recently, deep learning models appeared with the capability of learning evolutionary patterns from unaligned protein sequences. However, this requires large-scale data, while many families contain just a few sequences. Here, we contend this limitation can be overcome by transfer learning, exploiting the full potential of self-supervised learning on large unannotated data and then supervised learning on a small labeled dataset. We show results where errors in protein family prediction can be reduced by 55% with respect to standard methods.
Main text
Introduction
The protein families database (Pfam) is the most widely used repository of protein families and domains. Pfam uses manually curated “seed” alignments of homologous protein regions (named families) to generate profiles based on hidden Markov models (HMMs). The resulting models are a representation of each profiled family and can be used to classify novel sequences.1 Even though this approach is very successful, there still remain many proteins of UniProtKB2 (≈25%) that have not been annotated yet. Moreover, the number of sequences in this knowledge base grows at a much faster rate than its Pfam coverage, introducing novel sequences that may belong to completely new families.3
Very recently, deep learning (DL) models have emerged4 to potentially provide a powerful alternative to profile-HMMs, which are the dominant technology for protein family classification. DL techniques are capable of inferring patterns shared across the family sequences, allowing autonomous domain annotation on unaligned sequences. This was especially helpful for accelerating the characterization of sequences that do not resemble anything known.5 However, it is well known that DL techniques rely on large-scale data to infer meaningful sequence patterns. This can be a limitation on domain annotation, since many Pfam families comprise few seed sequences. Indeed, it has been an important step toward overcoming this limitation,4 and we show that this issue can be further significantly reduced with transfer learning (TL) by transferring representations of protein sequences already learned without requiring annotations from large-scale protein data.6
Transfer learning for protein representations
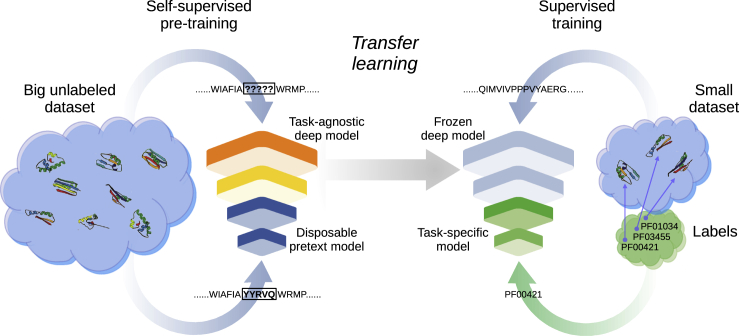
Transfer learning (TL) (Figure 1) is a machine learning technique where one model is first trained with a big unlabeled dataset in a self-supervised way, that is, not using annotations of any specific task but predicting parts of the same data fed as input (e.g., masked small subsequences). This step is also named pre-training, and the result is a task-agnostic deep model and an output model associated with the pretext task for self-supervised learning, which is then discarded. In a second step, the task-agnostic deep model is frozen, and what was learned by it is “transferred” to another deep architecture in order to train a new task-specific model. Here, another model is trained with supervised learning on a small dataset with labeled data for a specific task (e.g., protein family classification). In summary, TL refers to the situation where what has been learned in one setting is exploited to improve generalization in another one.7 For proteins, there are several already available task-agnostic deep models, which integrate in their output different types of protein information in a compact representation usually named embeddings.
Figure 1.
Transfer learning for functional annotation
Transfer learning is a machine learning technique where the knowledge gained by training a model on one general task is transferred to be reused in a second specific task. The first model is trained on a big unlabeled dataset, in a self-supervised way (left). This process is known as pre-training, and the result is a task-agnostic deep model (input layers). Through transfer learning, the first layers are frozen (middle) and transferred to another deep architecture. Then, the last layers of the new model are trained with supervised learning on a small dataset with labeled data for a specific task (right)
Protein embeddings are becoming known and required by the community. So much that UniProtKB now provides embeddings as part of the protein annotations (https://www.uniprot.org/help/embeddings). The available protein embeddings were pre-trained on UniRef50, which provides clustered sets of sequences from the complete UniProtKB.2 A recent review has demonstrated that the evolutionary scale modeling (ESM)5 is one of the most outstanding protein embeddings in terms of representational power.8 ESM was trained using 220 million (unaligned) sequences from UniProtKB. ESM is based on transformers, which have emerged as a powerful general-purpose model architecture for representation learning,9 out-performing deep recurrent and convolutional neural networks. They were originally designed for natural language processing,10 where context within a text is used to predict masked (missing) words. The main hypothesis in this pretext task for self-supervised learning is that the semantics of words can be derived from their contexts. ESM makes an analogy between syllables in text and amino acids in protein sequences: it learns meaningful encodings for each residue in a self-supervised way, by masking some of the residues in the sequence and trying to predict them. This way, ESM builds an embedding per residue position that encodes the “meaning” of the residue in that context. Then, the per residue representation can be collapsed to a per protein embedding. After this, the ESM learnt representation from UniProtKB, already trained and ready-to-use out of the box, can be “transferred” to be used in a specific downstream task.
Transfer learning for annotating protein domains
In order to illustrate how the use of TL can improve a task like protein domain annotation, we trained a new classifier with Pfam data.4 Expertly curated sequences from the 17,929 families of Pfam v.32.0 were used to define a benchmark annotation task. Seed sequences from each family were split into challenging train and test sets by clustering them based on sequence similarity. The clustered split provides a benchmark task for annotation of protein sequences with remote homology, that is, sequences in the test set that have low similarity to the ones in the training set. This is useful as an estimation of how well a model will perform with new sequences that are quite different from the ones in the training data. To this end, single-linkage clustering at 25% similarity within each family was used. The resulting benchmark has a distant held-out test set of 21,293 sequences. For this task, authors proposed ProtCNN and ProtENN.4 ProtCNN receives a one-hot coded sequence and learns to automatically extract features to predict family membership. ProtENN is an ensemble of 19 ProtCNNs using a majority vote strategy, where each model was trained with different random parameter initializations.
For the TL approach, we have obtained the ESM embeddings (ESM-1b) of all the train and test sets for the clustered split (a total of 1,339,083 seed sequences). We used two baseline machine learning classifiers for the supervised downstream task: k-nearest neighbor (kNN) and multilayer perceptron (MLP), both trained with the embeddings collapsed to full sequence, representing each protein domain with a vector in 1,280. After training, these models were tested with the distant held-out test partition for family domain prediction. Finally, we took advantage of TL to improve ProtCNN by training this architecture with the embedding as inputs, instead of the (original) one-hot encoding. The source code is available at https://github.com/sinc-lab/transfer-learning-pfam.
Table 1 shows the results when performance is evaluated by the error rate and the number of errors for classifying the protein domain sequences contained in a held-out clustered test set. The model with the fewest errors is indicated by asterisks. The first four rows reproduce the ProtCNN, ProtENN, TPHMM, and BLASTp results.4 The next rows show the results obtained when TL is used with different classifiers. The first interesting result is that TL with a simple kNN has obtained at least as good results (27.29% error rate) as ProtCNN (27.60%). Similarly, when transferred to the MLP model or an ensemble of 5 MLPs, the error rate is even lower (19.39% and 18.02%, respectively). This is a very remarkable result taking into account that embeddings have not been fine-tuned for this particular downstream task. When TL is used as input to a single ProtCNN, the results improve even further (15.98%), and the best results are achieved when it is used as the input of an ensemble of 10 TL-ProtCNNs (8.35%). All these cases have achieved better performance than ProtCNN with convolutional feature extraction from a one-hot representation. Moreover, when comparing only ensemble models, the TL-ProtCNN ensemble of 10 models has clearly outperformed the ProtENN ensemble of 19 models (8.35% vs. 12.20% error rate, respectively). That is, the error rate has been diminished by 33% thanks to the use of TL for the annotation task. Furthermore, in comparison to the TPHMM, the improvement is an impressive 55%.
Table 1.
Performance on the clustered split of Pfam
| Method | Error rate (%) | Total errors |
|---|---|---|
| ProtCNN | 27.60 | 5,882 |
| ProtENN | 12.20 | 2,590 |
| TPHMM | 18.10 | 3,844 |
| BLASTp | 35.90 | 7,639 |
| TL-kNN | 27.29 | 5,816 |
| TL-MLP | 19.39 | 4,132 |
| TL-MLP-ensemble | 18.02 | 3,840 |
| TL-ProtCNN | 15.98 | 3,405 |
| TL-ProtCNN-ensemble∗ | 8.35∗ | 1,743∗ |
∗Model with the fewest errors.
Closing remarks
In the last few years, several protein representation learning models based on deep learning have appeared, which provide numerical vectors (embeddings) as a unique representation of the protein. With TL, the knowledge encoded in these embeddings can be used in another model to efficiently learn new features of a different downstream prediction task. This TL process allows models to improve their performance by passing knowledge from one task to another, exploiting the information of larger and unlabeled datasets. Protein embeddings has become a new and highly active area of research, with a large number of variants already available in public repositories and easy to use.
The results achieved in a challenging partition of the full Pfam database, with low similarity between train and test sets, have shown superior performance when TL is used in comparison to previous deep learning models. Even in the case of the most simple machine learning classifiers, such as kNN and MLP, the decrease in the error rate was remarkable. Moreover, the best performance is achieved when a convolutional based model is mixed with a pre-trained protein embedding based on transformers. In terms of computational power, even half of ensemble members provided a 33% of improvement in the classification performance.
We hope that this comment will make researchers consider the potential of TL for building better models for protein function prediction. On the practical side, instead of building one’s own embedder for proteins, it is very useful to reuse all the computation time already spent building the available learnt representations. Leveraging TL for new tasks with small sets of annotated sequences is easy to implement and provides significant impact on final performance.
Acknowledgments
This work was supported by Agencia Nacional de Promocion Cientifica y Tecnologica (ANPCyT) (PICT 2018 3384, PICT 2018 2905) and UNL (CAI+D 2020 115). The authors thank and acknowledge Dr. Alex Bateman for his comments on the manuscript.
Author contributions
Conceptualization, G.S. and D.H.M.; Supervision, G.S.; Methodology, D.H.M.; Investigation and Software, L.A.B., E.F., A.A.E., D.H.M. and J.R.; Writing Original Draft, G.S.; Review and Editing, D.H.M., L.A.B., E.F., A.A.E. and J.R.; Visualization, D.H.M.
Declaration of interests
The authors declare no competing interests.
Biographies
About the authors
Diego H. Milone received a bioengineering degree from National University of Entre Rios, Argentina, and a PhD in microelectronics and computer architectures from Granada University, Spain. He is full professor in the Department of Informatics at National University of Litoral (UNL) and was director of the Department and dean for Science and Technology. He is principal research scientist on the National Scientific and Technical Research Council (CONICET). He was founder and first director of the Research Institute for Signals, Systems and Computational Intelligence, sinc(i) (CONICET-UNL). His research interests are machine learning and neural computing, with applications to biosignals and bioinformatics.
G. Stegmayer received an engineering degree in information systems from Facultad Regional Santa Fe (FRSF), Universidad Tecnologica Nacional (UTN), Santa Fe, Argentina, in 2000, and a PhD in engineering from the Politecnico di Torino, Turin, Italy, in 2006. Since 2007, she has been an assistant professor of artificial intelligence at the Universidad Nacional del Litoral (UNL), Santa Fe, Argentina. She is currently principal researcher at the sinc(i) Institute, National Scientific and Technical Research Council (CONICET), Argentina. She is the leader of the Bioinformatics Lab at sinc(i). Her current research interest involves machine learning, data mining, and pattern recognition in bioinformatics.
Contributor Information
Georgina Stegmayer, Email: gstegmayer@sinc.unl.edu.ar.
Diego H. Milone, Email: dmilone@sinc.unl.edu.ar.
Web resources
Source code, https://github.com/sinc-lab/transfer-learning-pfam
References
- 1.Mistry J., Coggill P., Eberhardt R.Y., Deiana A., Giansanti A., Finn R.R., Bateman A., Punta M. The challenge of increasing pfam coverage of the human proteome. Database. 2013;2013 doi: 10.1093/database/bat040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J., et al. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bileschi M.L., Belanger D., Bryant D.H., Sanderson T., Carter B., Sculley D., Bateman A., DePristo M.A., Colwell L.J. Using deep learning to annotate the protein universe. Nat. Biotechnol. 2022;40:932–937. doi: 10.1038/s41587-021-01179-w. [DOI] [PubMed] [Google Scholar]
- 5.Rives A., Meier J., Sercu T., Goyal S., Lin Z., Liu J., Guo D., Ott M., Zitnick C.L., Ma J., Fergus R. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2016239118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Unsal S., Atas H., Albayrak M., Turhan K., Acar A.C., Dogan T. Learning functional properties of proteins with language models. Nat. Mach. Intell. 2022;4:227–245. doi: 10.1038/s42256-022-00457-9. [DOI] [Google Scholar]
- 7.Goodfellow I.J., Bengio Y., Courville A. MIT Press; 2016. Deep Learning.http://www.deeplearningbook.org [Google Scholar]
- 8.Fenoy E., Edera A.A., Stegmayer G. Transfer learning in proteins: evaluating novel protein learned representations for bioinformatics tasks. Brief. Bioinform. 2022;23:4. doi: 10.1093/bib/bbac232. bbac232. [DOI] [PubMed] [Google Scholar]
- 9.Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser L., Polosukhin I. Attention is all you need. Guyon I., Luxburg U.V., Bengio S., Wallach H., Fergus R., Vishwanathan S., Garnett R., editors. Adv. Neural Inf. Process. Syst. 2017;30:5998–6008. [Google Scholar]
- 10.Devlin J., Chang M.W., Lee K., Toutanova. K. BERT: pre-training of deep bidirectional transformers for language understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics. 2019;1:4171–4186. Human Language Technologies. [Google Scholar]