Abstract
Apolipoprotein E4 (APOE4) genotype and sex are significant risk factors for Alzheimer's disease (AD), with females demonstrating increased risk modulated by APOE genotype. APOE is predominantly expressed in astrocytes, however, there is a lack of comprehensive assessments of sex differences in astrocytes stratified by APOE genotype. Here, we examined the response of mixed-sex and sex-specific neonatal APOE3 and APOE4 primary mouse astrocytes (PMA) to a cytokine mix of IL1b, TNFa, and IFNg. Pro-inflammatory and anti-inflammatory cytokine profiles were assessed by qRT-PCR and Meso Scale Discovery multiplex assay. Mixed-sex APOE4 PMA were found to have higher basal messenger RNA expression of several pro-inflammatory cytokines including Il6, Tnfa, Il1b, Mcp1, Mip1a, and Nos2 compared to APOE3 PMA, which was accompanied by increased levels of these secreted cytokines. In sex-specific cultures, basal expression of Il1b, Il6, and Nos2 was 1.5 to 2.5 fold higher in APOE4 female PMA compared to APOE4 males, with both being higher than APOE3 PMA. Similar results were found for secreted levels of these cytokines. Together, these findings indicate that APOE4 genotype and female sex, contribute to a greater inflammatory response in primary astrocytes and these data may provide a framework for investigating the mechanisms contributing to genotype and sex differences in AD-related neuroinflammation.
Keywords: APOE, neuroinflammation, astrocytes, cytokines, sex differences, Alzheimer's disease
Introduction
Neuroinflammation, a complex multicellular response, is a pathological feature in several neurodegenerative disorders. A common feature of neurodegenerative diseases, including Alzheimer's disease (AD), is astrocyte reactivity—a molecular, morphological, and functional change in astrocytes due to noxious influences on the central nervous system (CNS) (Preman et al., 2021). Astrocytes, the most abundant cells in the CNS (Herculano-Houzel, 2014; Keller et al., 2018; Pelvig et al., 2008), play a key role in maintaining the homeostatic balance and participate in crucial processes such as neurotransmitter uptake and recycling, gliotransmitter release, modulation of synaptic activity, maintenance of the blood–brain barrier, ionic balance, and neuroinflammation, among others (Guillamon-Vivancos et al., 2015; Vasile et al., 2017). Depending on the extent of the insult or injury, these reactive astrocytes may undergo resolution or augmentation in their homeostatic, modulatory, and defensive functions. Reactive astrocytes can release several extracellular molecules and secondary messengers, including inflammatory modulators, chemokines, and cytokines that may be deleterious to neurons by inducing neuroinflammation (Sofroniew and Vinters, 2010; Sofroniew, 2009, 2015).
In addition to modulating neuroinflammation, astrocytes are a major source of apolipoprotein E (APOE), a lipoprotein involved in AD pathogenesis in an isoform-dependent manner (Holtzman et al., 2000; Yu et al., 2014). APOE is a polymorphic gene in humans with an allelic frequency in the population in the order APOE3 > APOE4 > APOE2 (Alzheimer's-Association, 2022; Corbo and Scacchi, 1999; Utermann et al., 1980). Genetic studies in patients with late-onset AD (LOAD) demonstrate that APOE is a profound genetic risk factor for developing LOAD, and individuals carrying the ε4 (APOE4) allele are more likely to develop AD at an earlier age (Genin et al., 2011). This association has been reproduced by multiple genome-wide association studies (GWAS), which have confirmed and identified additional independent loci with susceptibility to LOAD (Lambert et al., 2013; Waring and Rosenberg, 2008). In humans, intravenous administration of endotoxin lipopolysaccharide (LPS) in APOE3/APOE4 patients produced higher plasma tumor necrosis factor alpha (TNFa) levels and earlier plasma interleukin 6 (IL6) than in APOE3/APOE3 patients (Gale et al., 2014). LPS-induced inflammation in mice expressing the human APOE4 gene showed greater levels of pro-inflammatory cytokines and neurotoxicity compared to APOE3 (Lynch et al., 2003; Maezawa et al., 2006b, 2006c). Studies on APOE transgenic mice have reported the APOE4 genotype to be more susceptible to inflammation than those expressing APOE3 (Rodriguez et al., 2014; Zhu et al., 2012). While the contribution of APOE4 in inflammation and AD has been largely investigated, few studies have identified the genotype-specific response of astrocytes with conflicting results. Evidence from studies in the targeted replacement (TR) APOE mice indicates increased glial fibrillary acidic protein (GFAP) expression in the APOE4 astrocytes following lesions in the entorhinal cortex (EC) (Belinson and Michaelson, 2009; Blain et al., 2006) as well as after exposure to LPS (Zhu et al., 2012). Gene expression of cytokines such as IL1b, IL6, and C-C motif chemokine ligand 2 was shown to increase from APOE3/APOE3 < APOE3/APOE4 < APOE4/APOE4 in human-induced pluripotent stem cell (hiPSC) astrocytes (Arnaud et al., 2022). Contradictorily, the secretion of cytokines IL1b, IL6, and TNFa from astrocytes has been reported in the order APOE2 > APOE3 > APOE4 after LPS treatment (Maezawa et al., 2006a), while secretion of chemokine CCL3 was induced in the order APOE2 > APOE4 > APOE3 (Cudaback et al., 2015).
Sex differences in gene expression patterns have also been associated with the APOE4 genotype (Hsu et al., 2019). Females are a more susceptible population to the development of AD, and those carrying the APOE4 allele are at a greater risk of developing AD (Altmann et al., 2014; Farrer et al., 1997), have 1% to 1.5% faster atrophy rates in the hippocampus than males, and worsened cognitive abilities (Hua et al., 2010; Koran et al., 2017). Accumulation of Aβ and tau are more pronounced in female APOE4 carriers than their male counterparts and show an accelerated progression of the disease, suggesting a sex-by-genotype interaction of APOE4 in females (Altmann et al., 2014; Buckley et al., 2018; Koran et al., 2017; Lin et al., 2015). Metabolomic analyses in TR-APOE mice indicate a sex and APOE genotype-dependent difference in the plasma and brain, respectively (Shang et al., 2020). Targeted differential gene expression analysis in the hippocampus of these mice revealed an increase in immune-related major histocompatibility complex genes and interferon responses in APOE4 females (Shang et al., 2020).
We have previously shown APOE genotype and sex-dependent effects of LPS on primary microglia from TR-APOE mice (Mhatre-Winters et al., 2022). Here, we sought to determine the genotype-specific effects of an inflammatory stimulus on mixed-specific and sex-specific APOE3 and APOE4 primary astrocytes isolated from TR-APOE3 and APOE4 mice. To induce an inflammatory response, we used a cytokine mix (CYM) containing TNFa, IL1b, and interferon gamma (IFNg), which are present in the pro-inflammatory cytokine milieu in the AD brain (Wang et al., 2015). These cytokines are also secreted by microglia, which are considered the first form of active immune defense (Leng and Edison, 2021; Mhatre-Winters et al., 2022; Sofroniew, 2020). Next, because females are at a higher risk of developing AD earlier in life (Alzheimer's-Association, 2022; Hebert et al., 2013), and sex has been indicated to modulate immune responses (Berkiks et al., 2019; Santos-Galindo et al., 2011), we investigated the sex-specific effects of our CYM on APOE genotypes in astrocytes.
In this study, we report novel sex-specific inflammatory profiles of TR-APOE primary astrocytes, an important factor that has yet to be thoroughly investigated in the literature. These cultures were over 98% pure, and with <2% adulteration with other glial cells, our findings are considerably reflective of astrocytes. Additionally, astrocytes exposed to CYM increased the messenger RNA (mRNA) and protein levels of pro-inflammatory cytokines in the APOE4 genotype, with a greater inflammatory profile observed in females. Transcriptional differences in astrocytes have resulted in the identification of two subtypes—A1 and A2 astrocytes (Clarke et al., 2018; Joshi et al., 2019; Liddelow et al., 2017). The A1 astrocytes assume a neurotoxic characteristic and upregulate several complement cascade genes resulting in the impairment of new synapse formation and decrease the excitatory function of neurons resulting in neuronal damage (Liddelow et al., 2017). Conversely, the A2 astrocytes demonstrate a “protective” attribute by upregulating neurotrophic factors that promote neuronal survival (Liddelow et al., 2017).We further show an increase in all A1, most A2, and pan-inflammation genes in female APOE4 astrocytes. Collectively, these data demonstrate that both APOE genotype and sex contribute to immunological differences, and understanding the multifaceted roles of astrocytes has the potential to contribute to the development of new treatment strategies.
Materials and Methods
Animals
TR-APOE breeders were purchased from Taconic Biosciences (the United States) and bred in-house to obtain mouse pups for primary cultures. These mice are on a C57BL/6 background, with the mouse APOE gene replaced by homozygous human APOE3 or APOE4 gene (Knouff et al., 1999). Breeders were paired at 8 to 10 weeks of age and housed in a 12 h light–dark cycle at 22 ± 2°C and 50 ± 10% of relative humidity. Mice had access to food (standard chow) and water ad libitum. Each litter was obtained from different parents and used as individual isolation for experiments. All procedures were conducted in accordance with the National Institutes of Health's Guide for Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of [anonymized].
Reagents
All cell culture reagents were purchased from Invitrogen. Charcoal-stripped fetal bovine serum (FBS, Cat. No. F6765) was purchased from Sigma-Aldrich (St. Louis, MO). Recombinant mouse TNFa (Cat. No. 410-MT-010), IFNg (Cat. No. 485-MI-100), and IL1b (Cat. No. 401-ML-005) protein were purchased from R&D Systems (Minneapolis, MN).
Primary Mouse Astrocyte Isolation
Mixed-sex and sex-specific mixed glial cultures consisting of microglia and astrocytes were isolated from whole brains of postnatal 0 to 1 day old TR-APOE3 or APOE4 mouse pups as previously described (Neal et al., 2018). The biological sex of the mouse pups was determined by measuring the anogenital distance and the presence or absence of visible pigmentation in the anogenital region (Wolterink-Donselaar et al., 2009). On average, an equal number of male and female brains were used for mixed-sex glial cultures. Briefly, whole brains were collected, washed in 1x phopshate-buffered saline (PBS), trypsinized, thoroughly triturated, and passed through a 70 μm strainer. This mixed glial culture was maintained for 15 days in growth media consisting Dulbecco's Modified Eagle's Medium/F12 (DMEM/F12) supplemented with 10% charcoal-stripped heat-inactivated FBS (HI-FBS), 2 mM L-glutamine, 1 mM sodium pyruvate, 100 µM non-essential amino acids, 1% penicillin/streptomycin, and 2.5 μg/mL Plasmocin™. The addition of charcoal-stripped FBS ensured that no exogenous source of hormones interfered with the experiments. Following the separation of astrocytes and microglial cells by CD11b immunomagnetic positive selection kit (STEMCELL Technologies, Cat. No. 18970), the negative fraction consisting of primary mouse astrocyte (PMA) was cultured in growth media consisting of DMEM supplemented with 10% HI-FBS, 2 mM L-glutamine, 1 mM sodium pyruvate, 1% penicillin/streptomycin, and 2.5 μg/mL Plasmocin™ for 2 to 3 days. After flasks were approximately 80% confluent, astrocytes were seeded for experiments.
Cells were assessed for purity in culture by imaging 3 to 4 random fields per well in a 96-well plate using the 10× objective on the Keyence BZ-X810 microscope. Cells positive for ionized calcium-binding adapter molecule 1 (Iba1) (microglia) or Gfap (astrocytes) were counted against the total number of cells (positive for 4′,6-diamidino-2-phenylindole (DAPI)) per field using the BZ-X800 Analyzer software, and the average was determined as percent purity. Cultures consisting of >98% purity of PMA were consistently obtained. For sex-specific cultures, a subset of cells was examined for sex specificity by examining the presence or absence of the sex-determining region-Y (Sry) gene by quantitative polymerase chain reaction (qPCR).
Treatments
For experiments, PMA were seeded at a density of 30,000 cells/well in a 96-well plate and 70,000 cells/well in a 24-well plate. For each isolation (n), cells were seeded on two separate plates , with 2 to 3 replicates for each treatment group per plate. Treatments were performed in growth media supplemented with 2% HI-FBS. PMA were treated with a CYM of IL1b (5 ng/mL), TNFa (5 ng/mL), and IFNg (10 ng/mL). For all experiments, n = 3 to 4 individual isolations.
RT-qPCR
Primary astrocytes were treated with the CYM for 6 h. RNA was isolated using TRIzol Reagent (Thermo Fisher Scientific, Cat. No. 15-596-018) following the manufacturer's instructions. Complementary DNA (cDNA) was synthesized using the iScript cDNA Synthesis kit (Bio-Rad, Cat. No. 1708891). Primer sets were checked with national center for biotechnology information basic local alignment search tool and ordered from Thermo Fisher Scientific. Subsequent PCR reactions were performed using ItaqTM Universal SYBR green (Bio-Rad, Cat. No. 1725124) following the manufacturer's instructions and previously described (Neal et al., 2020). qPCR was performed in duplicate on the Quantstudio 6 Flex System (Applied Biosystems). A list of genes and primer sequences used in qPCR reactions is listed in Supplemental Table S1. Gene expression was normalized to Rpl13a using the 2−ΔΔCT method (Schmittgen & Livak, 2008). For comparisons between genotypes, data were normalized to the APOE3 control group. For comparisons between the genotypes and sex, data were normalized to the APOE3 male control group since females are at a greater risk of developing AD irrespective of the APOE genotype (Alzheimer's-Association, 2022). The graphs indicate data as fold change compared to the normalized group.
Conditioned Media Collection
Primary astrocytes were treated with CYM for 24 h, washed three times with fresh media, and replaced with new media. Cytokines were allowed to accumulate for 6 h, following which the media was collected and stored in aliquots at −80°C until further use.
Measurement of Secreted Cytokines
Secreted cytokines were measured in the conditioned media using a custom U-PLEX mouse panel for the following pro-inflammatory targets—IL1b, IL6, TNFa, IFNg, monocyte chemoattractant protein 1 (MCP1), macrophage Inflammatory Protein-1 alpha (MIP1a), IL4, and IL10 on the Meso Scale Discovery (MSD) platform following the manufacturer's instructions. Plates were read and analyzed on the MSD QuickPlex SQ 120.
Measurement of Media Nitrite
Nitric oxide (NO) is a signaling molecule that plays a key role in the pathogenesis of neuroinflammation and neurodegeneration (Yuste et al., 2015). Media nitrite is one of the two stable breakdown products of nitric oxide. Quantification of media nitrite was done using the Griess reagent system (Promega, Cat. No. G2930) as previously described (Neal et al., 2018). Following 24 h treatment, media was collected, and nitrite levels were measured at an absorbance of 535 nm along with a nitrite standard reference curve per the manufacturer's protocol.
Immunocytochemistry
Following treatment, cells in a 96 well plates were fixed with 4% paraformaldehyde for 15 min, washed with PBS, and immunocytochemistry was performed as previously described (Neal et al., 2018). Briefly, cells were permeabilized with PBS + 0.5% Triton X-100 and blocked for 1 h in 2% bovine serum albumin (BSA) solution at room temperature, followed by overnight incubation in 2% BSA with the following primary antibodies—Gfap (1:1000, Abcam, Cat. No. ab4674) and Iba1 (1:1000, Wako Chemicals, Cat. No. 019-19741) at 4°C. The following day, cells were washed with PBS and incubated with secondary antibodies from appropriate species (Alexa Fluor® 1:1000, Thermo Fisher Scientific, Cat. No. A11039 and A11037) for 1 h. After washing with PBS, cells were incubated with 10 μg/ mL DAPI (Thermo Fisher Scientific, Cat. No. D1306) for 15 min, washed again, and the nuclei were visualized. The staining specificity was confirmed by omitting the primary or secondary antibody. Cells were imaged using the Keyence BZ-X810 microscope.
Statistics and Software
Statistical analysis was performed using Prism 7.04 (GraphPad Software, San Diego, CA). Data were assessed for normality by D’Agostino–Pearson normality test and Shapiro–Wilk normality test. Basal differences were analyzed in mixed-sex and sex-specific cultures by two-tailed unpaired Student's t-test and two-way analysis of variance (ANOVA), respectively. For comparisons including CYM treated groups, mixed-sex cultures were analyzed using two-way ANOVA, whereas sex-specific cultures were analyzed by three-way ANOVA. Tukey's post hoc test to compare multiple groups was conducted following ANOVA. Statistical significance was considered at p < .05. Heat maps were generated using Displayr software (Pyrmont, Australia).
Results
Inflammatory Gene Expression is Increased in APOE4 Mixed-Sex Primary Astrocytes
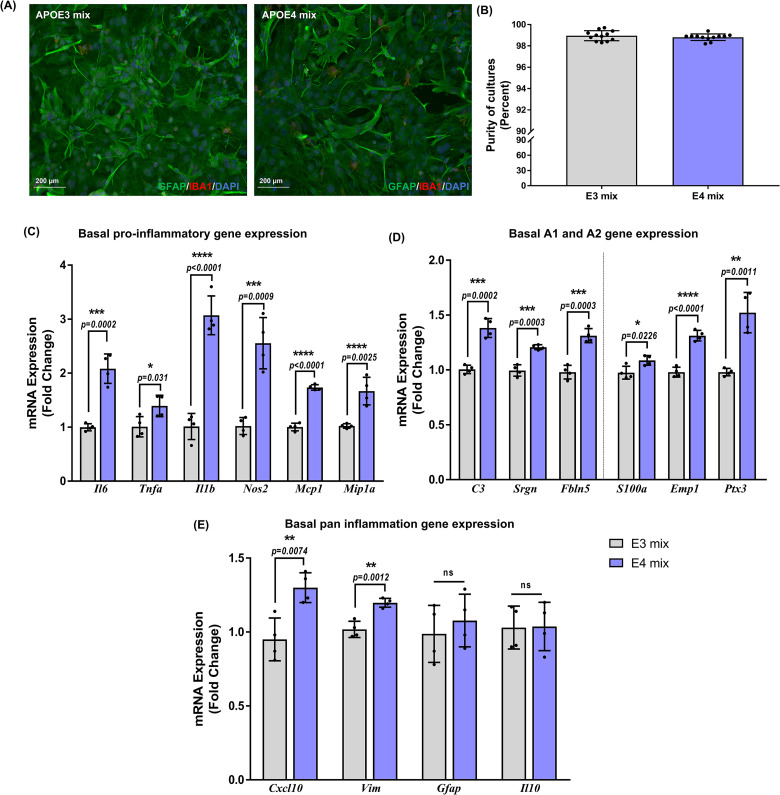
To investigate the differential response of APOE genotype to inflammation, mixed-sex PMA were isolated from whole brains of neonatal APOE3 or APOE4 mice (Figure 1A). Cultures over 98% purity were consistently obtained as assessed by counting Gfap-positive astrocytes against the total number of cells (Figure 1B). We first investigated basal changes between the genotypes in the gene expression of the pro-inflammatory cytokines and chemokines by real-time qPCR (Figure 1C). In the APOE4 astrocytes, baseline expression of Tnfa (p = .02), Mcp1 (p < .0001), and Mip1a (p = .0001) was higher by approximately 1.5-fold, whereas, Il6 (p = .0002), Il1b (p < .0001), and Nos2 (p = .0009) increased by 2, 3, and 2.6-fold, respectively, suggesting that the APOE4 astrocytes inherently have a greater inflammatory profile compared to APOE3. To assess the A1 gene expression, we checked mRNA levels of C3, Srgn, and Fbln5. Basal expression in the APOE4 astrocytes was approximately 1.5-fold higher in C3 (p = .0002), Srgn (p = .0003), and Fbln5 (p = .0003) compared to APOE3 astrocytes (Figure 1D). We then investigated the expression of A2 genes—S100a, Emp1, and Ptx3 in both the genotypes. APOE4 astrocytes showed a baseline increase by 1.1-fold in S100a (p = .03), 1.3-fold in Emp1 (p < .0001) and by 1.5-fold in Ptx3 (p = .0011) (Figure 1D). We further evaluated the expression of other pan-inflammation genes identified from a panel used by Liddelow et al. (2017). Similar to other targets, mRNA expression of Cxcl10 increased by 1.5-fold (p = .007) and Vim by 1.2-fold (p = .0012) in APOE4 astrocytes compared to APOE3 astrocytes (Figure 1E).
Figure 1.
Basal inflammatory gene expression in mixed-sex astrocytes is apolipoprotein E (APOE) genotype dependent: (A) Representative images for purity of mixed-sex astrocyte cultures as determined by staining for Gfap (astrocyte), Iba1 (microglia), and DAPI (nucleus). (B) Purity of mixed-sex astrocyte cultures was >98%. Each data point represents the average percentage of astrocytes frpm 3-4 images per well. Data are represented as mean ± standard deviation(n = 4 isolations). Basal expression of (C) pro-inflammatory, (D) A1–A2, and (E) pan-inflammatory genes was evaluated in nonstimulated mixed-sex astrocytes from APOE3 and APOE4 genotypes. Data were normalized to the APOE3 group for comparisons and indicated as “fold change” on the graphs. Gene expression between genotypes was analyzed using unpaired Student's t-test (n = 4 isolations). Each data point in (C), (D) and (E) denote the average messenger RNA expression from one isolation. Data are represented as mean ± standard deviation; not statistically significant (ns, p > .05).
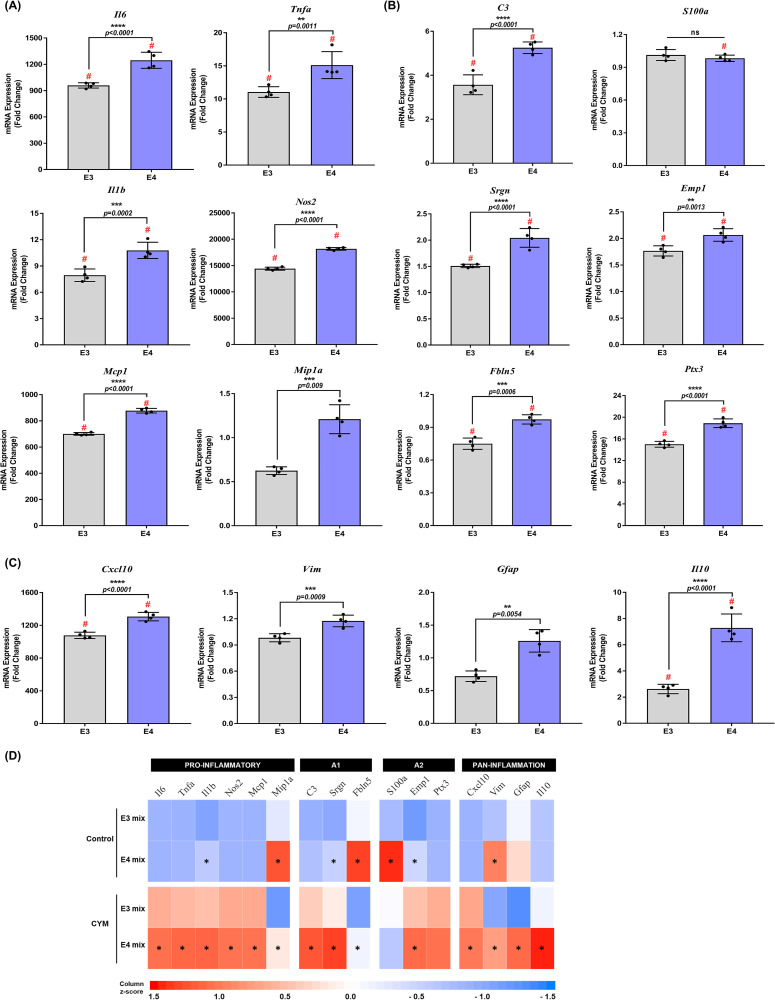
Next, PMA were treated with a CYM of IL1b (5 ng/mL), TNFa (5 ng/mL), and IFNg (10 ng/mL) for 6 h, and gene expression was measured by RT-qPCR. Exposure to CYM increased the mRNA levels of pro-inflammatory cytokines in both APOE3 and APOE4 astrocyte cultures compared to genotype control (Figure 2A). A significant genotype effect was observed for Il6 (F1,12 = 2092, p < .0001), Tnfa (F1,12 = 461.8, p = .0017), Il1b (F1,12 = 551.1, p < .0001), Nos2 (F1,12 = 29023, p < .0001), Mcp1 (F1,12 = 23315, p < .0001) and Mip1a (F1,12 = 30.52, p = .0001) (Supplemental Table S2). A 1.3-fold increase in Il6 (p < .0001), Nos2 (p < .0001), and Mcp1 (p < .0001) and a 1.4-fold increase in Tnfa (p = .0011) and Il1b (p = .0002), was observed in APOE4 PMA after CYM treatment, whereas Mip1a increased by 2-fold (p = .0009).
Figure 2.
Comparison of gene expression between apolipoprotein E3 (APOE3) and APOE4 genotypes: (A) Pro-inflammatory, (B) A1–A2, and (C) pan-inflammatory gene expression were evaluated in mixed-sex astrocytes from APOE3 and APOE4 genotypes following treatment with a cytokine mix (CYM) for 6 h. Data were normalized to the APOE3 control group (data not included in graphs) for comparisons and indicated as “fold change” on the graphs. Gene expression between treatment and genotypes was assessed by two-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test (n = 4 isolations). Each data point represents the average messenger RNA expression from one isolation. # indicates a significant increase compared to the respective genotype control group (data not included in the graphs, p < .05). Data are represented as mean ± standard deviation. (D) Heat map comparing the mean expression of genes in control and CYM-treated APOE3 and APOE4 primary mouse astrocytes (PMA). *indicates a significant (p < .05) increase in APOE4 gene expression compared to APOE3.
Treatment with CYM also affected gene expression of A1 and A2 genes. Two-way ANOVA indicated significant treatment and genotype effect in A1 gene expression (Supplemental Table S2). CYM induced higher mRNA levels in APOE4 astrocytes by approximately 1.5-fold for C3 (p = .0009), Srgn (p = .0026), and Fbln5 (p = .0079) in APOE4 astrocytes (Figure 2B). For the expression of A2 genes, a significant treatment and genotype effect was observed for Emp1 and Ptx3 (Supplemental Table S2). APOE4 astrocytes showed an approximate 1.2-fold increase in Emp1 (p = .0356) and Ptx3 (p = .0001) (Figure 2B). However, CYM was unable to induce S100a expression in either genotype. Finally, gene expression of pan-inflammation targets was assessed following treatment with CYM, and a significant treatment effect was observed (Supplemental Table S2). APOE4 PMA showed an increase in Cxcl10 (p = .0003) and Vim (p = .0087) by 1.2-fold, and Il10 (p < .0001) by 3-fold (Figure 2C). Astrocyte reactivity marker Gfap was significantly increased by 1.8-fold (p = .005) in APOE4 astrocytes after inducing inflammation with CYM (Figure 2C). Together, these data suggest that the APOE4 genotype contributes to greater reactivity and inflammatory response in astrocytes (Figure 2D).
APOE4 Genotype Elevates Secretion of Inflammatory Cytokines in Mixed-Sex Astrocytes
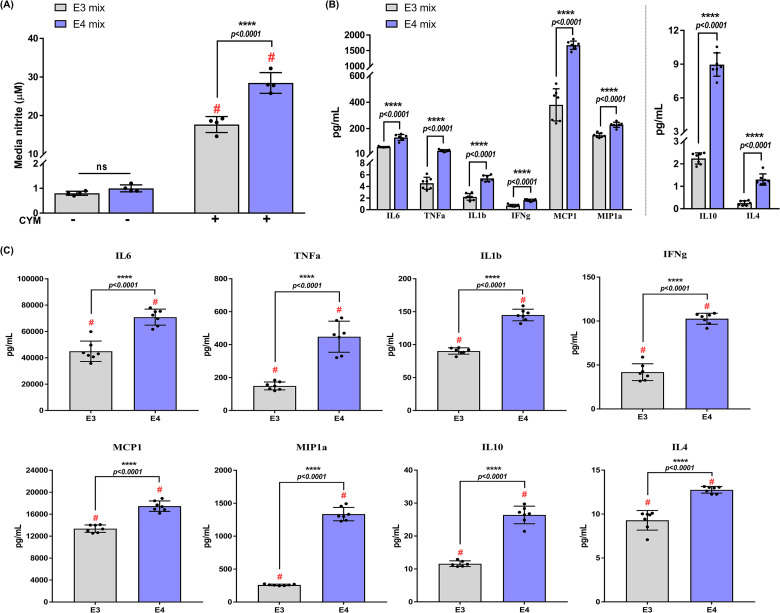
To explore APOE genotype-specific production of inflammatory mediators, astrocytes were stimulated with the CYM for 24 h and the levels of secreted cytokines were measured. While there were no basal changes in media nitrite levels, CYM treatment induced 1.7-fold (p < .0001) increase in media nitrite in the APOE4 astrocytes compared to APOE3 cultures (Figure 3A). In addition to a genotype effect (F1,8 = 41.93, p = .0002), an interaction between genotype and treatment was also observed (F1,8 = 38.84, p = .0003).
Figure 3.
Apolipoprotein E4 (APOE4) genotype exacerbates release of pro-inflammatory and anti-inflammatory cytokines: (A) Average levels of nitrite (± standard deviation) accumulated in the media were measured by Griess assay. Treatment and genotype effects were analyzed using two-way ANOVA (n = 4 isolations). Each data point represents the average media nitrite levels from one isolation. # indicates a significant increase compared to the respective genotype control (p < .05). (B) Basal levels of secreted cytokines in nonstimulated astrocytes after 24 h by Meso Scale Discovery (MSD) multiplex assay. Comparisons between genotypes were made using an unpaired Student's t-test (n = 3–4 isolations). Data points represent technical replicates from 3 to 4 isolations. (C) Cytokine levels in media were measured after treatment with CYM for 24 h. Two-way ANOVA was used to assess cytokine release in mixed-sex astrocytes from APOE3 and APOE4 genotypes (n = 3–4 isolations). Data points represent technical replicates from 3 to 4 isolations. Data in all the panels are represented as mean ± standard deviation. # indicates a significant increase compared to the respective genotype control group (data not included in the graphs, p < .05); not statistically significant (ns, p > .05).
Proteomic studies have demonstrated high levels of inflammatory markers in the CSF of APOE4 carriers (Konijnenberg et al., 2020). Furthermore, hiPSC-derived APOE4 astrocytes have been shown to secrete high levels of chemokines such as C-X-C motif chemokine ligand 1 (CXCL1), CXCL10, CXCL12, and MIP1b, along with other inflammatory proteins, including IL6 and IL8 (Tcw et al., 2022). To assess if the APOE4 astrocytes derived from TR-APOE mice released higher levels of cytokines, conditioned media was analyzed for both pro-inflammatory (IL1b, IL6, TNFa, IFNg, MCP1, and MIP1a) and anti-inflammatory (IL10 and IL4) cytokines on an MSD platform. Basal secretion of IL1b, IL6, TNFa, IFNg, MCP1, MIP1a, IL10, and IL4 was higher in APOE4 astrocytes by 2.5 (p < .0001), 2.2 (p < .0001), 7 (p < .0001), 2.2 (p < .0001), 4.4 (p < .0001), 1.5 (p < .0001), 4 (p < .0001), and 5-fold (p < .0001), respectively (Figure 3B). Two-way ANOVA revealed a significant genotype effect in the secretion of IL1b (F1,23 = 215.3, p < .0001), IL6 (F1,23 = 44.8, p < .0001), TNFa (F1,22 = 64.99, p < .0001), IFNg (F1,22 = 171.4, p < .0001), MCP1 (F1,24 = 146.3, p < .0001), MIP1a (F1,23 = 777.4, p < .0001), IL10 (F1,22 = 308.4, p < .0001), and IL4 (F1,24 = 97.12, p < .0001).
Following CYM treatment, an increase in the secretion of cytokines was detected in both APOE3 and APOE4 astrocytes compared to basal levels; however, APOE4 astrocytes secreted a higher level of cytokines compared to APOE3 astrocyte cultures (Figure 3C). CYM induced significantly higher secretion of pro-inflammatory IL1b (p < .0001), IL6 (p < .0001), and MCP1 (p < .0001) by 1.5-fold, TNFa by 3-fold (p < .0001), IFNg by 2.5-fold (p < .0001), MIP1a by 5-fold (p < .0001). Furthermore, we did not observe a decrease in the anti-inflammatory cytokines. Instead, the secretion of anti-inflammatory IL10 and IL4 increased in the APOE4 astrocytes by 2.3-fold (p < .0001) and 1.4-fold (p < .0001), respectively. A possible explanation for this could be that since inflammation is a dynamic process, the 24 h time point chosen for this investigation highlights the resolving phase of inflammation. Since these astrocytes are obtained from the whole brain, further region-dependent analyses will determine whether these effects are due to the homogeneity of the astrocyte population. Nonetheless, these data verify the increased secretion of cytokines and chemokines observed in the APOE4 hiPSC-derived astrocytes and suggest that the human APOE4 gene contributes to a greater inflammatory response.
APOE-Dependent Basal Inflammatory Gene Expression is Driven by Sex
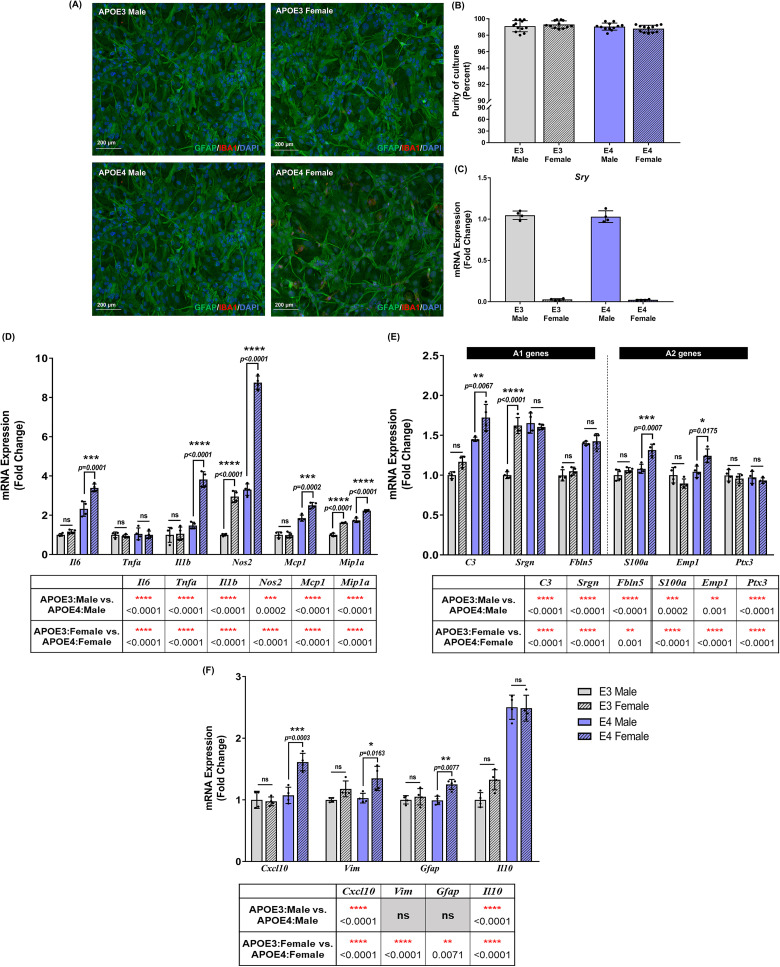
Despite a growing body of literature acknowledging the effect of APOE genotype on AD pathology and inflammation, few studies report the influence of sex on innate immune responses. To assess the contribution of APOE genotype and sex in PMA to an inflammatory stimulus, sex-specific primary cultures were generated from TR-APOE3 and APOE4 mouse pups. Purity of astrocytes in cultures was evaluated to be ≥ 98% (Figure 4A and B), and cells were validated for sex specificity by examining the presence or absence of the sex-determining region-Y (Sry) gene by RT-qPCR (Figure 4C).
Figure 4.
Basal inflammatory gene expression in sex-specific astrocytes is dependent on both apolipoprotein E (APOE) genotype and sex: (A) Representative images for purity of sex-specific astrocyte cultures. (B) Purity >98% was consistently obtained for all sex-specific astrocyte cultures. Each data point in (B) and (D) denotes an average percentage of astrocytes from 3 to 4 images per well. (C) Sex-specificity of cultures was validated by checking for the presence or absence of sex-determining region-Y gene (Sry) by real time quantitative polymerase chain reaction (RT-qPCR). Each data point represents the average messenger RNA (mRNA) expression from one isolation. Data are represented as mean ± standard deviation. Basal expression of (D) pro-inflammatory, (E) A1–A2, and (F) pan-inflammatory genes was evaluated in non-stimulated sex-specific astrocytes from APOE3 and APOE4 genotypes. Data were normalized to the APOE3 male group for comparisons and indicated as “fold change” on the graphs. Gene expression between genotypes and sexes was analyzed using a two-way ANOVA (n = 3–4 isolations). Each data point represents the average mRNA expression from one isolation. Data are represented as mean ± standard deviation; not statistically significant (ns, p > .05).
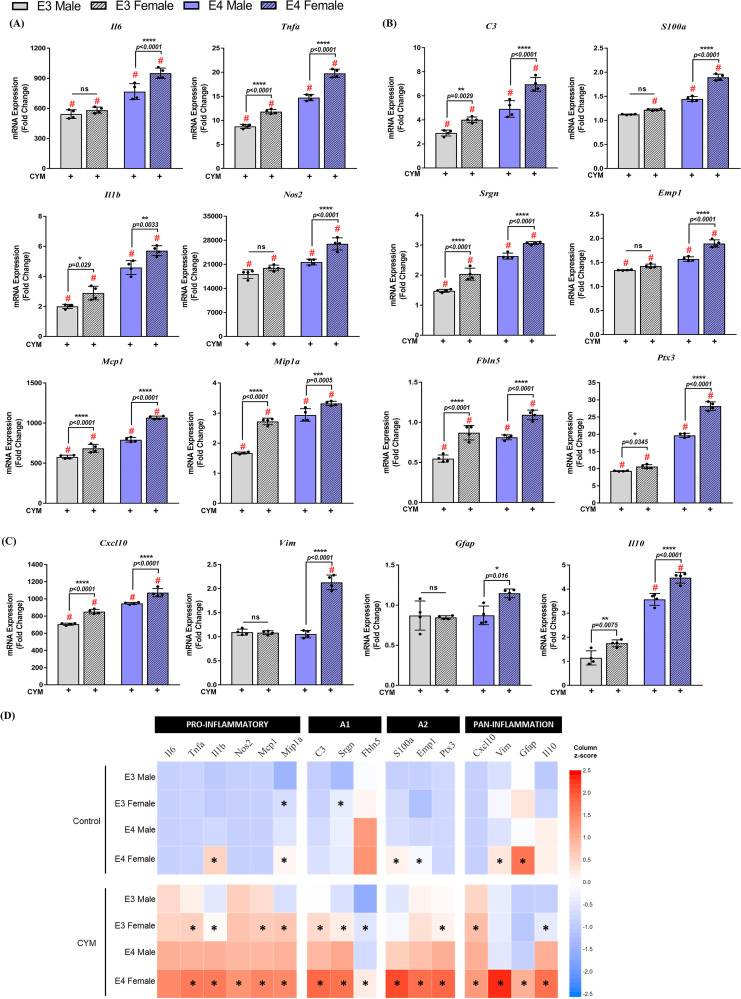
Gene expression in nonstimulated male and female APOE3 and APOE4 astrocytes was measured following 6 h to evaluate basal responses between genotypes and sexes (Figure 4D–F). Results were normalized to the APOE3 male group, and basal mRNA levels were compared by two-way ANOVA with Tukey's posthoc testing. Basal expression of pro-inflammatory genes was increased in APOE4 PMA, with female cultures exhibiting a greater increase (Figure 4D). Il6 increased by 1.5-fold (p = .0001), Il1b and Nos2 increased by 2.6-fold (p < .0001), Mcp1 (p = .0002), and Mip1a (p < .0001) increased by approximately 1.4-fold in female APOE4 PMA compared to male APOE4 cultures. Furthermore, a significant interaction between APOE genotype and sex was detected for Il6 (F1,12 = 16.51, p = .0016), Il1b (F1,12 = 46.12, p < .0001), Nos2 (F1,12 = 168.6, p < .0001), and Mcp1 (F1,12 = 20.31, p = .0007). Compared to PMA derived from female APOE3 mice, APOE4 female cultures showed an approximate 3-fold increase in Il6 (p = .0001), 3.6-fold in Il1b (p < .0001), 3-fold in Nos2 (p < .0001), 2.5-fold in Mcp1 (p < .0001), and 1.5-fold in Mip1a (p < .0001). No baseline differences were detected in Tnfa expression.
We also detected minor variations in basal A1 and A2 gene expression in sex-specific PMA cultures (Figure 4E). Although no changes were observed between male and female APOE3 PMA, APOE4 female PMA showed a modest 1.2-fold increase in C3 (p = .007), S100a (p = .0007), and Emp1 (p = .02) gene expression compared to APOE4 male PMA. Conversely, Srgn expression increased by 1.6-fold (p < .0001) in APOE3 females compared to APOE3 males, however, no basal changes were observed between sexes in the APOE4 genotype. Significant interactions between APOE genotype and sex were noted for Srgn (F1,12 = 61.46, p < .0001), S100a (F1,12 = 8.12, p = .02), and Emp1 (F1,12 = 14.83, p = .002). Expression of A1 genes such as C3 and Fbln5 increased in APOE4 female PMA by approximately 1.5-fold (p < .0001) compared to APOE3 female PMA. Expression of A2 genes S100a and Emp1 genes increased by 1.2 (p = .0004) and 1.4-fold (p = .0002), respectively, in APOE4 females compared to APOE3 females.
For pan-inflammation genes, we did not observe any changes between male and female APOE3 PMA; however, Cxcl10, Vim, and Gfap expression increased by 1.5 (p = .0003), 1.3 (p = .02), and 1.3-fold (p = .008), respectively, in APOE4 female PMA compared to APOE4 male PMA (Figure 4F). Genotype and sex interactions were observed for Cxcl10 (F1,12 = 20.61, p = .0007) and Gfap (F1,12 = 5.32, p = .04). Further comparisons between sex across genotypes revealed increased gene expression of Cxcl10 (1.7-fold, p < .0001), Gfap (1.2-fold, p = .04), and Il10 (1.9-fold, p < .0001) in APOE4 females compared to APOE3 females. Il10 also increased by 2.5-fold (p < .0001) in APOE4 males compared to APOE3 male PMA. These basal differences may, in part attribute to the differences observed in inflammatory gene expression after stimulation with CYM.
Sex Differences in Inflammatory Gene Expression of TR-APOE3 and TR-APOE4 Astrocytes After CYM Treatment
Next, we evaluated the differences in inflammatory gene expression between sex-specific APOE3 and APOE4 PMA cultures stimulated with CYM. We observed an increase in pro-inflammatory cytokine gene expression in the order APOE4 female > APOE4 male > APOE3 female > APOE3 male (Figure 5). Significant interactions between treatment and genotype, treatment and sex, as well as treatment, genotype, and sex, were observed for a majority of cytokines using three-way ANOVA followed by Tukey's multiple comparisons test (Supplemental Table S3). Following CYM treatment, differential response in male and female astrocytes was observed in both genotypes for pro-inflammatory cytokines (Figure 5A). Female APOE4 PMA cultures showed a 1.2 to 1.3-fold increase in Il6 (p < .0001), Tnfa (p < .0001), Il1b (p = .003), Nos2 (p < .0001), Mcp1 (p < .0001) and Mip1a (p = .005) compared to males. Similar increases were also observed in the female APOE3 PMA compared to male cultures, with the exception of Mip1a, which increased by 1.6-fold (p < .0001) in the female PMA. Although a 1.1-fold increase in Il6 and Nos2 gene expression was observed in APOE3 female PMA compared to males, these data were not statistically significant. Furthermore, comparisons between female PMA derived from both genotypes revealed higher levels of Il6 (1.6-fold, p < .0001), Tnfa (1.7-fold, p < .0001), Il1b (2-fold, p < .0001), Nos2 (1.3-fold, p < .0001), Mcp1 (1.6-fold, p < .0001), and Mip1a (1.2-fold, p < .0001) in APOE4 female compared to APOE3 female cultures. This indicates that sex and genotype together modulate inflammation in astrocytes.
Figure 5.
Gene expression in sex-specific astrocytes is dependent on both apolipoprotein E (APOE) genotype and sex: (A) Pro-inflammatory, (B) A1–A2, and (C) pan-inflammatory gene expression was evaluated in sex-specific astrocytes from APOE3 and APOE4 genotypes following a cytokine mix (CYM) treatment for 6 h. Data were normalized to the APOE3 male control group (data not included in graphs) for comparisons and indicated as “fold change” on the graphs. Gene expression between treatment, genotypes, and sexes was analyzed using a three-way analysis of variance (n = 3–4 isolations) followed by Tukey's multiple comparisons test. Each data point represents the average messenger RNA expression from one isolation. # indicates a significant increase compared to the respective genotype and sex control group (data not included in the graphs, p < .05). Data are represented as mean ± standard deviation; not statistically significant (ns, p > .05). (D) Heat map comparing the mean expression of genes in sex-specific APOE3 and APOE4 primary mouse astrocytes (PMA) treated with CYM. * indicates a significant (p < .05) increase in fold changes of gene expression compared to genotype-matched male PMA.
Investigation of selective A1 and A2 genes indicated that CYM treatment increased mRNA levels of both A1 and A2 genes in all the groups compared to vehicle control. A 1.2 to 1.4-fold increase in A1 genes C3 (p < .0001), Srgn (p < .0001), and Fbln5 (p < .0001) was observed in APOE4 females compared to male APOE4 PMA (Figure 5B). Similarly, A1 gene expression was also increased in APOE3 by approximately 1.5-fold in female PMA compared to APOE3 males. Comparisons between female PMA of both genotypes revealed an increase in C3, Srgn, and Fbln5 by 1.7 (p < .0001), 1.5 (p < .0001), and 1.3-fold (p = .01), respectively. Parallel increases were also observed in male PMA from both genotypes. Subsequent evaluation of A2 genes, including S100a, Emp1, and Ptx3, also indicated sex-specific responses, albeit mostly in the APOE4 genotype (Figure 5B). S100a and Emp1 increased by approximately 1.2-fold (p < .0001) in female APOE4 PMA compared to males. However, no statistical significance was observed between the male and female APOE3 PMA. Ptx3, a TNF inducible gene, increased in the female cultures of both APOE3 and APOE4 genotype by 1.1 (p < .0001) and 1.4-fold (p < .0001), respectively, compared to their counterpart male cultures. Furthermore, comparisons between females across both genotypes showed a 1.6 (p < .0001), 1.3 (p < .0001), and 2.7-fold (p < .0001) increase in S100a, Emp1, and Ptx3, respectively. Similar observations were noted for male PMA from both genotypes.
Lastly, we assessed the differential genotype and sex responses in pan-inflammation genes to the CYM inflammatory stimulus (Figure 5C). We observed sex differences in both APOE3 and APOE4 genotypes for Cxcl10 and Il10. mRNA levels increased by 1.1 (p < .0001) and 1.3-fold (p < .0001) in female APOE4 PMA compared to parallel male cultures, whereas an increase of 1.2 (p < .0001) and 1.5-fold (p = .008) in Cxcl10 and Il10, respectively, was noted in APOE3 females compared to counterpart male cultures. While no sex differences were observed in the gene expression of Gfap and Vim in the APOE3 genotype, a 1.3-fold (p = .02) increase in Gfap and a 2-fold (p < .0001) increase in Vim were observed in the APOE4 females compared to APOE4 male PMA. Comparisons between the genotypes of the same sex revealed greater differences between the female PMA cultures for Gfap and Vim expression, by approximately 1.5 (p = .008) and 2-fold (p < .0001), respectively, compared to male PMA cultures. Conversely, Il10 expression increased by 2.6-fold (p < .0001) in APOE4 females and 3.1-fold (p < .0001) in APOE4 males compared to same-sex APOE3 cultures. Cxcl10 expression increased equally in males and females across genotypes. Together, these data indicate that an inflammatory stimulus increases the upregulation of pro-inflammatory and pan-inflammatory genes in the female APOE4 astrocytes compared to APOE4 male PMA and APOE3 cells (Figure 5D).
Female APOE4 Astrocyte Cultures Have Increased Secretion of Inflammatory Cytokines
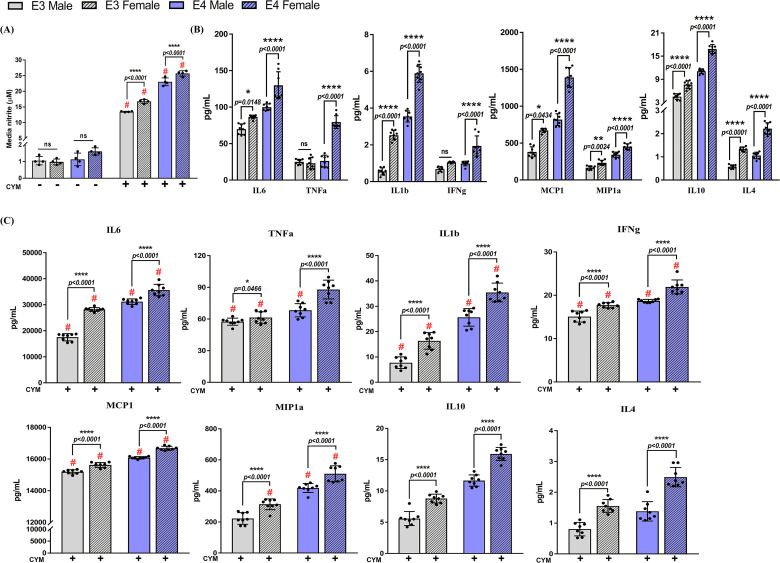
Nitric oxide levels were measured indirectly by quantifying levels of media nitrite in media collected from APOE3 and APOE4 male and female astrocyte cultures (Figure 6A). No basal differences were observed across groups; however, CYM treatment increased media nitrite levels significantly in both APOE3 and APOE4 male and female cultures compared to the respective vehicle control. Data analysis using three-way ANOVA indicated a treatment effect on genotype (F1,24 = 422.8, p < .0001) and sex (F1,24 = 41.59, p < .0001). Furthermore, an equivalent increase in media nitrite was observed between male and female PMA from both genotypes. Comparisons between the same-sex across genotypes revealed an approximately 1.5-fold increase in the APOE4 genotype.
Figure 6.
Increased secretion of inflammatory cytokines depends on both apolipoprotein E (APOE) genotype and sex: (A) Nitrite levels accumulated in the media were measured by Griess assay. Three-way ANOVA followed by Tukey's multiple comparisons test revealed a significant treatment-by-genotype-by-sex effect (n = 4 isolations). Each data point represents the average media nitrite levels from one isolation. # indicates a significant increase compared to the respective genotype control (p < .05). (B) Basal levels of secreted cytokines in nonstimulated astrocytes by Meso Scale Discovery (MSD) multiplex assay. Comparisons between sex and genotype were made using two-way ANOVA followed by Tukey's multiple comparisons test (n = 3–4 isolations). Data points represent technical replicates from 3 to 4 isolations. (C) Secretion of cytokines was measured post treatment with a cytokine mix (CYM) by a three-way ANOVA followed by Tukey's multiple comparisons test (n = 3–4 isolations). Data points represent technical replicates from 3 to 4 isolations. # indicates a significant increase compared to the respective genotype and sex control group (data not included in the graphs, p < .05). Data in all panels are represented as mean ± standard deviation; not statistically significant (ns, p > .05).
We continued to assess secreted pro-inflammatory and anti-inflammatory cytokines in conditioned media collected from sex-specific astrocyte cultures. As expected, basal secretion of cytokines including IL6, TNFa, MCP1, MIP1, IL1b, IFNg, IL10, and IL4 was higher in the APOE4 PMA (Figure 6B). In addition, female APOE4 PMA cultures secreted 1.3 to 3-fold higher levels of cytokines compared to male APOE4 PMA. Similarly, increased levels of other cytokines were observed in conditioned media from female APOE3 PMA compared to the male APOE3 conditioned media, except TNFa and IFNg. These data suggest that both APOE3 and APOE4 female astrocyte cultures basally secrete higher cytokine levels than males. Following treatment with CYM, a significant increase was observed in secreted pro-inflammatory cytokines (Figure 6C).
A treatment by genotype effect was observed for pro-inflammatory IL6 (F1,56 = 418.7, p < .0001), IL1b (F1,56 = 177.6, p < .0001), TNFa (F1,56 = 8.847, p = .0043), IFNg (F1,56 = 69.37, p < .0001), and MCP1 (F1,56 = 54.42, p < .0001) as well as anti-inflammatory IL10 (F1,56 = 7.803, p = .0071). A treatment by sex effect was also observed in pro-inflammatory IL6 (F1,56 = 216.3, p < .0001), TNFa (F1,56 = 37.82, p < .0001), TNFa (F1,56 = 16.87, p = .0001), and IFNg (F1,56 = 32.78, p < .0001) but no interaction was observed in anti-inflammatory cytokines. Pro-inflammatory cytokines increased by 1.2 to 1.4-fold in female APOE4 cultures whereas, in female APOE3 cultures, an increase in secreted cytokines ranging from 1.2 to 2-fold was observed. Anti-inflammatory IL10 and IL4 were also found to be approximately 1.5 to 2-fold higher in the female cultures from both genotypes. This indicates that male and female astrocyte cultures distinctively respond to an inflammatory stimulus. Furthermore, treatment by genotype by sex effect was only observed in IL6 (F1,56 = 36.87, p < .0001) and TNFa (F1,56 = 33.68, p < .0001). Collectively, measurement of secreted cytokines from sex-specific astrocyte cultures from both APOE3 and APOE4 genotypes demonstrate the divergent potential of both sex and genotype to modulate inflammation.
Discussion
While there are numerous data focusing on the effects of APOE genotype on inflammatory profiles, the vast majority derive data from blood, serum, brain, or microglia, few studies have investigated the role of astrocytes in contributing to APOE genotype-driven inflammation. The current study revealed novel sex-specific inflammatory profiles of primary astrocytes generated from TR-APOE3 and TR-APOE4 neonatal mice. We confirmed that these cultures consisted of over 98% astrocytes with minimal contamination from other cell types. Our findings also extend the observations made by previous reports in mixed-sex cultures. Here, we first evaluated the inflammatory response of mixed-sex TR-APOE3 and TR-APOE4 astrocytes. Previous studies in rat primary mixed-glial cultures consisting of astrocytes and microglia showed an increase in Il1b mRNA levels when exposed to recombinant APOE4 (Guo et al., 2004). In our studies, basal gene expression of pro-inflammatory cytokines revealed that APOE4 PMA innately expressed higher mRNA levels. This finding in astrocytes is congruent with evidence suggesting that the presence of APOE4, either endogenously present or recombinantly treated, increases the production of pro-inflammatory cytokines in rodents and humans.
Microglial secreted cytokines, such as IL1b, IFNg, and TNFa that are known to activate astrocytes, were used to induce and compare the inflammatory profile in PMA (Leng & Edison, 2021). We observed an increase in both mRNA and secreted protein levels of pro-inflammatory cytokines in APOE4 PMA cultures compared to APOE3 PMA. In contrast, previous studies in TR-APOE mice have demonstrated that primary astrocytes treated with a toll-like receptor 4 (TLR4) agonist such as LPS resulted in increased secretion of pro-inflammatory cytokines in the order APOE2 > APOE3 > APOE4 (Maezawa et al., 2006a). The differences in our findings may be attributed to astrocyte purity in the cultures. It has been previously proposed that the increased cytokine expression or secretion observed in astrocytes could be due to microglial contamination of cultures (Saura, 2007). While the PMA cultures used in our current study were at least 98% enriched in astrocytes, the cultures used by Maezwa and colleagues utilized a different technique of PMA preparation which yielded ≥93% pure astrocytes. The presence of microglia in an astrocyte culture has been suggested to enhance the inflammatory outcome, possibly due to crosstalk between astrocytes and microglia (Linnerbauer et al., 2020; Saura, 2007; Sola et al., 2002). Moreover, while mouse astrocytes have TLR4 on their surface, the extent of its expression has been controversial (Hasel & Liddelow, 2021). TLRs bind to adaptor proteins such as MyD88 and TIR-domain-containing adapter-inducing interferon-β (TRIF) that subsequently activate the intracellular signaling cascades ensuing secretion of inflammatory cytokines (Takeuchi & Akira, 2010). RNA sequencing studies in mice indicate that expression of TLR4, MYD88, and TRIF through which early activation occurs is approximately 10 to 14 times lower in astrocytes compared to microglia (Zhang et al., 2014).
Astrocytes undergo a change in reactivity under noxious environmental cues, which is reflected on their gene expression profile (Escartin et al., 2021; Perez-Nievas & Serrano-Pozo, 2018). APOE4 astrocytes also have lower levels of brain-derived neurotrophic factors (BDNF) and have been shown to disrupt neuronal support and synaptic transmission during aging (Sen et al., 2017; Verkerke et al., 2021). APOE4 carriers without AD pathology have altered levels of complement proteins and inflammatory cytokines, including reactive astrocyte markers CCL4, S100b, and GFAP, compared to age-matched noncarriers (Bussy et al., 2019; Dai et al., 2018; Ji et al., 2003). Although emerging literature shows that the A1–A2 binary polarization of astrocytes is not universal and similar to microglia, heterogeneity in the morphology, cell signaling, reactivity, transcriptome, and proteome of astrocytes exist depending on the brain region, sex, age, and the disease/disorder stage (Anderson et al., 2014; Escartin et al., 2021; Hasel et al., 2021; Matias et al., 2019; Sofroniew, 2020), we used this distinction as a preliminary tool to screen the CYM induced alterations in APOE genotype-specific astrocytes. Extensive studies by Liddelow et al. (2017) demonstrated that A1 astrocytes make up a large proportion of astrocytes in AD. A1 reactive astrocytes overexpress many classical complement cascade genes in AD mice which may alter calcium homeostasis and synaptic responses (Orre et al., 2014). Conversely, A2 astrocytes upregulate several neurotrophic factors when activated in cases such as ischemia and promote neuronal survival and repair (Liddelow & Barres, 2017). We integrated selective A1, A2, and pan reactive astrocyte markers into our study to identify if their gene expression was modulated by APOE genotype. Indeed, we observed a basal increase in A1, A2, and in some, but not all, pan reactive genes in APOE4 PMA compared to the APOE3 genotype. Upon stimulating the PMA cultures with CYM, we observed a significant increase in all A1 genes, most A2 genes, and pan-inflammation genes in the order APOE4 > APOE3. These data suggest primarily that APOE4 PMA are basally more activated than APOE3. Secondly, the process of neuroinflammation is not a clear dichotomous process; instead, as supported by previous and continuously emerging evidence, it is dynamic. The increase in both A1 and A2 gene expression, although at different magnitudes, indicates that the 6 h time point may be reflective of a mixed pattern of competing pro-inflammatory and anti-inflammatory pathways. Investigating earlier and later time points may aid in understanding the pattern of these genes expressed over time. Additionally, the cytokine mix used in this study included IL1b (5 ng/mL), TNFa (5 ng/mL), and IFNg (10 ng/mL), whereas other studies have used a different combination of cytokines, including LPS, IL-1a, TNFa, transforming growth factor beta-1, IL1b, C1q, or fibroblast growth factor (FGF) for immune stimulation (Liddelow et al., 2017; Zhang et al., 2020). Thus the variation observed in the A1/A2 gene expression in our study compared to others may be attributed to the use of different stimulators or cocktail of cytokines.
Astrocytes produce NO in response to detrimental stimuli, including inflammation, through NOS2 enzyme activity (Yuste et al., 2015). We observed a significant increase in APOE4 media nitrite levels post-treatment with CYM. These data are in line with previous evidence from TR-APOE4 astrocytes secreting higher levels of media nitrate plus nitrite levels 72 h after LPS treatment compared to TR-APOE3 astrocytes (Maezawa et al., 2006a). These changes were not observed at 24 and 48 h which could possibly be due to the reported lack of LPS receptors and response elements TLR4 and MYD88 in astrocytes (Zhang et al., 2014), suggesting that the observed increase in NO at 72 h may be the result of an indirect effect. We also noted basal differences in the secretion of other pro-inflammatory and anti-inflammatory cytokines, with APOE4 releasing significantly higher levels of cytokines. This increase was consistent after CYM treatment suggesting that APOE4 PMA are potentially primed and thus secrete higher levels of cytokines after an inflammatory insult. These findings are consistent with evidence from previous studies indicating increased TLR-mediated secretion and mRNA levels of chemokine CCL3 in APOE4 PMA (Cudaback et al., 2015). Contradictorily, evidence from another study in PMA derived from TR-APOE mice indicates IL1b, IL6, and TNFa secretion following LPS treatment in the order APOE2 > APOE3 > APOE4 (Maezawa et al., 2006a). In the study, Maezawa et al. further demonstrated decreased nuclear factor (NF)-kB p50 subunit activity in APOE3 and APOE4 PMA compared to APOE2 PMA, suggesting that the increased secretion of cytokines in APOE2 PMA is associated with NF-kB signaling. While we did not investigate signaling pathways, the opposing findings may be attributed to the origin of PMA. While their study used PMA derived from the cerebral cortex, we generated PMA cultures from the whole brain. Future region-specific studies with a highly enriched astrocyte culture are warranted to determine the inflammation profile of astrocytes across brain regions. A multimodal approach involving technologies such as single-cell RNA sequencing, spatial transcriptomics and glial gene profiling will provide valuable insight into the heterogeneity in the inflammatory profile of astrocyte subtypes across brain regions.
We further identified sex-specific responses of APOE3 and APOE4 PMA. Basal differences in pro-inflammatory mRNA levels were mainly limited to the APOE4 genotype, with increased expression in females. For basal A1 gene expression, C3 increased in female APOE4 PMA compared to APOE4 males. No sex-specific basal differences were observed in A2 and pan-inflammation genes of APOE3 cultures. Post-CYM exposure, inflammatory gene expression increased in a genotype and sex-specific manner. While APOE4 PMA expressed a higher inflammatory profile, female PMA from both APOE3 and APOE4 genotypes demonstrated higher mRNA levels than males. This increase was representative of most cytokines and A1–A2 genes in APOE3 females and all genes in APOE4 females. These sex-specific effects also extend to secreted cytokines detected in the order APOE4 female > APOE4 male > APOE3 female > APOE3 male. Basally, most cytokines were elevated in the media of female cultures, with an additional increase in APOE4 female secreted cytokine levels.
While the specific mechanism linking the APOE genotype to inflammation is not clearly understood, several hypotheses have been proposed. ApoE is an immunomodulatory protein involved in various functions, including lipid transport, blood brain barrier integrity, neuronal plasticity, regulation and clearance of Aβ, and neuroimmune modulation of astrocytes and microglia (Flowers & Rebeck, 2020; Liao et al., 2017; Yamazaki et al., 2020). Previous studies have shown that mice lacking the Apoe gene exhibit an increase in the CNS pro-inflammatory cytokines, indicating a link between APOE and the production of inflammatory cytokines (Liu et al., 2015). Furthermore, ApoE has been demonstrated to modulate the activation of microglia and mixed glial cultures (Barger & Harmon, 1997). The addition of ApoE or ApoE-mimetic peptides reduced levels of pro-inflammatory cytokines in microglia (Laskowitz et al., 2001; Lynch et al., 2003; Vitek et al., 2009), in TR-APOE mice (Lynch et al., 2003; Vitek et al., 2009), mouse models of traumatic brain injury (Laskowitz et al., 2017), sepsis (Wang et al., 2009), and ischemic stroke (Tu et al., 2017). Although there is no evidence establishing this link in astrocytes, varying levels of cellular and secreted ApoE from astrocytes might be a step in the direction. Previous studies have shown that TR-APOE astrocytes secrete ApoE in a genotype-dependent manner APOE2 > APOE3 > APOE4 (Cudaback et al., 2015; Lanfranco et al., 2021), whereas reports for basal levels of cellular APOE from astrocytes are variable, with some studies showing no genotype-specific change (Lanfranco et al., 2021) while others demonstrated a decrease in the order APOE2 > APOE3 > APOE4 (Cudaback et al., 2015). Interestingly, activation of astrocytes with TNFa reduced both secreted and cellular ApoE in the APOE4 PMA, which was reversed by IL10 (Lanfranco et al., 2021). Thus, targeting ApoE and recovering levels in the APOE4 genotype could be a valuable tool to address a mechanistic link in astrocytes.
Lipidated Apoe is a primary cholesterol carrier, which neurons rely heavily upon. In humans and mice, Apoe is primarily produced by astrocytes in the brain. This shuttling of cholesterol is APOE genotype-dependent, with the APOE4 genotype implicated in the dysregulation of cholesterol homeostasis in astrocytes (Lin et al., 2018). A recent study reported elevated levels of chemokines and cytokines including IL6, CXCL1, CXCL10, and others, in isogenic APOE4 human astrocytes (Tcw et al., 2022). Compared to the APOE3 genotype, APOE4 astrocytes treated with pro-inflammatory factors like IFNg and TNFa upregulated cholesterol biosynthesis genes but downregulated cholesterol efflux genes leading to lipid dysregulation. These findings indicate an adverse loop in the APOE4 astrocytes where increased pro-inflammatory cytokines promote altered cholesterol trafficking, thus leading to functional deficits in other cell types by assuming an AD-like pathology (Jeong et al., 2019; Lee et al., 2021; Lin et al., 2018).
In vivo and postmortem evidence in AD has implicated three main signaling pathways in astrocyte reactivity—the Janus kinase/signal transducer and activator of transcription (JAK/STAT3) pathway, the NF-kB pathway, and mitogen-activated protein kinases (MAPK) pathway (Ben Haim et al., 2015; Perez-Nievas & Serrano-Pozo, 2018). These pathways are activated by pro-inflammatory agents such as LPS and molecular triggers such as cytokines and growth factors. This, in turn, is responsible for inducing astrocyte reactivity by activating various intracellular signaling cascades. Although these pathways have been extensively studied in microglia and neurons, much remains unknown in association with astrocytes. The NF-kB pathway is activated by pro-inflammatory cytokines such as TNFa, IL6, and IL1b. Among NF-kB targets is complement protein C3 which is an A1 astrocyte marker. In addition, astrocytic deletion of STAT3 resulted in a phenotypic switch of astrocytes from A1 to A2 (Reichenbach et al., 2019). APOE genotype-specific modulation of the MAPK pathway has also been implicated in Aβ and Aβ-induced neuroinflammation (Huang et al., 2017). Increased brain inflammation in APOE4 mice has been linked to dysregulation of the NF-kB signaling pathway (Ophir et al., 2005). In a recent study, Arnaud et al. identified an APOE-transgelin (TAGLN3)-NF-κB axis that is dysregulated in astrocytes derived from APOE4-carrying AD patients (Arnaud et al., 2022). Their findings indicated that TAGLN3 downregulation in APOE4 carriers exacerbated inflammation through NF-κB activation. Pharmacological interventions rescued the hiPSC APOE4 astrocytes from an altered neuroinflammatory phenotype, thus suggesting that TAGLN3 could be used as a potential biomarker. While several of these posited pathways are linked to APOE, it remains to be seen how they are modified by sex. To date, few studies exist elucidating the role of astrocytes in APOE genotype-mediated inflammation (Maezawa et al., 2006a, 2006b; Cudaback et al., 2012, 2015). To our knowledge, these data are the first highlighting sex-specific effects of inflammation in APOE genotype-specific astrocytes and are consistent with largely recognized studies in humans and TR-APOE mice indicating sex and APOE genotype differences in the inflammatory pathways (Shang et al., 2020; Hsu et al., 2019; Zhao et al., 2020). In summary, our most important finding indicates that, similar to microglia, astrocytes are equally involved in complex neuroinflammatory mechanisms, and these effects are present in an APOE genotype-dependent as well as sex-dependent manner. However, our study is not immune to caveats associated with experiments reproduced in an ex vivo culture system. Although these experiments are pertinent to understanding the underlying mechanisms involved in a cell-type-specific manner, culturing primary cells disrupts the normal cellular interactions and may result in slight alterations of activity. Nevertheless, these in vitro systems are valuable in recognizing cell-type-specific inflammatory profiles in a controlled environment. Furthermore, the heterogeneity of reactive PMA at regional, subregional, and cellular levels needs to be thoroughly investigated by taking advantage of modern techniques such as two-photon microscopy and single-cell RNA sequencing (Ben Haim et al., 2015). Ongoing and future studies will assess an elaborate panel of inflammatory genes in the hippocampus and frontal cortex, regions vulnerable to AD.
Supplemental Material
Supplemental material, sj-xlsx-1-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-2-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-3-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-4-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Footnotes
Authors’ Contributions: IM-W, AE, KT, and JRR contributed to the conceptualization and design of the research. IM-W and YH generated cultures and acquired the data. JRR and IM-W interpreted the data. IM-W drafted and revised the manuscript. KT and JRR provided revisions. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement: No large datasets were generated from this study. All data supporting the findings of this study are available from the corresponding author on reasonable request.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health (grant number R01-ES033892, R01ES021800, R01ES024288-03S1, R01ES026057, R35ES030523).
ORCID iD: Isha Mhatre-Winters https://orcid.org/0000-0003-3498-8355
Supplemental Material: Supplemental material for this article is available online.
References
- Altmann A., Tian L., Henderson V. W., et al. (2014). Sex modifies the APOE-related risk of developing Alzheimer disease. Annals of Neurology, 75(4), 563–573. 10.1002/ana.24135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer's-Association (2022). 2022 Alzheimer’s disease facts and figures. Alzheimer's Dementia, 2022(4), 700–789. 10.1002/alz.12638 [DOI] [PubMed] [Google Scholar]
- Anderson M. A., Ao Y., Sofroniew M. V. (2014). Heterogeneity of reactive astrocytes. Neuroscience Letters, 565, 23–29. 10.1016/j.neulet.2013.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud L., Benech P., Greetham L., et al. (2022). APOE4 Drives inflammation in human astrocytes via TAGLN3 repression and NF-kappaB activation. Cell Reports, 40(7), 111200. 10.1016/j.celrep.2022.111200 [DOI] [PubMed] [Google Scholar]
- Barger S. W., Harmon A. D. (1997). Microglial activation by Alzheimer amyloid precursor protein and modulation by apolipoprotein E. Nature, 388(6645), 878–881. 10.1038/42257 [DOI] [PubMed] [Google Scholar]
- Belinson H., Michaelson D. M. (2009). ApoE4-dependent Abeta-mediated neurodegeneration is associated with inflammatory activation in the hippocampus but not the septum. Journal of Neural Transmission (Vienna), 116(11), 1427–1434. 10.1007/s00702-009-0218-9 [DOI] [PubMed] [Google Scholar]
- Ben Haim L., Carrillo-de Sauvage M. A., Ceyzeriat K., et al. (2015). Elusive roles for reactive astrocytes in neurodegenerative diseases. Frontiers in Cellular Neuroscience, 9, 278. 10.3389/fncel.2015.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkiks I., Garcia-Segura L. M., Nassiri A., et al. (2019). The sex differences of the behavior response to early life immune stimulation: Microglia and astrocytes involvement. Physiology & Behavior, 199, 386–394. 10.1016/j.physbeh.2018.11.037 [DOI] [PubMed] [Google Scholar]
- Blain J. F., Sullivan P. M., Poirier J. (2006). A deficit in astroglial organization causes the impaired reactive sprouting in human apolipoprotein E4 targeted replacement mice. Neurobiology of Disease, 21(3), 505–514. 10.1016/j.nbd.2005.08.010 [DOI] [PubMed] [Google Scholar]
- Buckley R. F., Mormino E. C., Amariglio R. E., et al. (2018). Sex, amyloid, and APOE epsilon4 and risk of cognitive decline in preclinical Alzheimer's disease: Findings from three well-characterized cohorts. Alzheimer's & Dementia, 14(9), 1193–1203. 10.1016/j.jalz.2018.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussy A., Snider B. J., Coble D., et al. (2019). Effect of apolipoprotein E4 on clinical, neuroimaging, and biomarker measures in noncarrier participants in the dominantly inherited Alzheimer network. Neurobiology of Aging, 75, 42–50. 10.1016/j.neurobiolaging.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L. E., Liddelow S. A., Chakraborty C., et al. (2018). Normal aging induces A1-like astrocyte reactivity. Proceedings of the National Academy of Sciences of the United States of America, 115(8), E1896–E1905. 10.1073/pnas.1800165115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo R. M., Scacchi R. (1999). Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Annals of Human Genetics, 63(Pt 4), 301–310. 10.1046/j.1469-1809.1999.6340301.x [DOI] [PubMed] [Google Scholar]
- Cudaback E., Li X., Yang Y., et al. (2012). Apolipoprotein C-I is an APOE genotype-dependent suppressor of glial activation. Journal of Neuroinflammation, 9, 192. 10.1186/1742-2094-9-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudaback E., Yang Y., Montine T. J., et al. (2015). APOE genotype-dependent modulation of astrocyte chemokine CCL3 production. Glia, 63(1), 51–65. 10.1002/glia.22732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Johnson E. C. B., Dammer E. B., et al. (2018). Effects of APOE genotype on brain proteomic network and cell type changes in Alzheimer's disease. Frontiers in Molecular Neuroscience, 11, 454. 10.3389/fnmol.2018.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escartin C., Galea E., Lakatos A., et al. (2021). Reactive astrocyte nomenclature, definitions, and future directions. Nature Neuroscience, 24(3), 312–325. 10.1038/s41593-020-00783-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer L. A., Cupples L. A., Haines J. L., et al. (1997). Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer disease meta analysis consortium. JAMA, 278(16), 1349–1356. 10.1001/jama.1997.03550160069041 [DOI] [PubMed] [Google Scholar]
- Flowers S. A., Rebeck G. W. (2020). APOE in the normal brain. Neurobiology of Disease, 136, 104724. 10.1016/j.nbd.2019.104724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale S. C., Gao L., Mikacenic C., et al. (2014). APOepsilon4 is associated with enhanced in vivo innate immune responses in human subjects. Journal of Allergy and Clinical Immunology, 134(1), 127–134.e9. 10.1016/j.jaci.2014.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genin E., Hannequin D., Wallon D., et al. (2011). APOE and Alzheimer disease: A major gene with semi-dominant inheritance. Molecular Psychiatry, 16(9), 903–907. 10.1038/mp.2011.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillamon-Vivancos T., Gomez-Pinedo U., Matias-Guiu J. (2015). Astrocytes in neurodegenerative diseases (I): Function and molecular description. Neurologia (Barcelona, Spain), 30(2), 119–129. 10.1016/j.nrl.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Guo L., LaDu M. J., Van Eldik L. J. (2004). A dual role for apolipoprotein e in neuroinflammation: Anti- and pro-inflammatory activity. Journal of Molecular Neuroscience, 23(3), 205–212. 10.1385/JMN:23:3:205 [DOI] [PubMed] [Google Scholar]
- Hasel P., Liddelow S. A. (2021). Isoform-dependent APOE secretion modulates neuroinflammation. Nature Reviews. Neurology, 17(5), 265–266. 10.1038/s41582-021-00483-y [DOI] [PubMed] [Google Scholar]
- Hasel P., Rose I. V. L., Sadick J. S., et al. (2021). Neuroinflammatory astrocyte subtypes in the mouse brain. Nature Neuroscience, 24(10). 10.1038/s41593-021-00905-6 [DOI] [PubMed] [Google Scholar]
- Hebert L. E., Weuve J., Scherr P. A., et al. (2013). Alzheimer disease in the United States (2010-2050) estimated using the 2010 census. Neurology, 80(19), 1778–1783. 10.1212/WNL.0b013e31828726f5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herculano-Houzel S. (2014). The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia, 62(9), 1377–1391. 10.1002/glia.22683 [DOI] [PubMed] [Google Scholar]
- Holtzman D. M., Bales K. R., Tenkova T., et al. (2000). Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America, 97(6), 2892–2897. 10.1073/pnas.050004797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu M., Dedhia M., Crusio W. E., et al. (2019). Sex differences in gene expression patterns associated with the APOE4 allele. F1000research, 8, 387. 10.12688/f1000research.18671.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Hibar D. P., Lee S., et al. (2010). Sex and age differences in atrophic rates: An ADNI study with n = 1368 MRI scans. Neurobiology of Aging, 31(8), 1463–1480. 10.1016/j.neurobiolaging.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. A., Zhou B., Wernig M., et al. (2017). Apoe2, ApoE3, and ApoE4 differentially stimulate APP transcription and Abeta secretion. Cell, 168(3), 427–441 e421. 10.1016/j.cell.2016.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong W., Lee H., Cho S., et al. (2019). ApoE4-induced cholesterol dysregulation and its brain cell type-specific implications in the pathogenesis of Alzheimer's disease. Molecules and Cells, 42(11), 739–746. 10.14348/molcells.2019.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Gong Y., Gan W., et al. (2003). Apolipoprotein E isoform-specific regulation of dendritic spine morphology in apolipoprotein E transgenic mice and Alzheimer's disease patients. Neuroscience, 122(2), 305–315. 10.1016/j.neuroscience.2003.08.007 [DOI] [PubMed] [Google Scholar]
- Joshi A. U., Minhas P. S., Liddelow S. A., et al. (2019). Fragmented mitochondria released from microglia trigger A1 astrocytic response and propagate inflammatory neurodegeneration. Nature Neuroscience, 22(10), 1635–1648. 10.1038/s41593-019-0486-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D., Ero C., Markram H. (2018). Cell densities in the mouse brain: A systematic review. Frontiers in Neuroanatomy, 12, 83. 10.3389/fnana.2018.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouff C., Hinsdale M. E., Mezdour H., et al. (1999). Apo E structure determines VLDL clearance and atherosclerosis risk in mice. Journal of Clinical Investigation, 103(11), 1579–1586. 10.1172/JCI6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konijnenberg E., Tijms B. M., Gobom J., et al. (2020). APOE Epsilon4 genotype-dependent cerebrospinal fluid proteomic signatures in Alzheimer's disease. Alzheimer's Research & Therapy, 12(1), 65. 10.1186/s13195-020-00628-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koran M. E. I., Wagener M., Hohman T. J., et al. (2017). Sex differences in the association between AD biomarkers and cognitive decline. Brain Imaging and Behavior, 11(1), 205–213. 10.1007/s11682-016-9523-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J. C., Ibrahim-Verbaas C. A., Harold D., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature Genetics, 45(12), 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco M. F., Sepulveda J., Kopetsky G., et al. (2021). Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia, 69(6), 1478–1493. 10.1002/glia.23974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowitz D. T., Thekdi A. D., Thekdi S. D., et al. (2001). Downregulation of microglial activation by apolipoprotein E and apoE-mimetic peptides. Experimental Neurology, 167(1), 74–85. 10.1006/exnr.2001.7541 [DOI] [PubMed] [Google Scholar]
- Laskowitz D. T., Wang H., Chen T., et al. (2017). Neuroprotective pentapeptide CN-105 is associated with reduced sterile inflammation and improved functional outcomes in a traumatic brain injury murine model. Scientific Reports, 7, 46461. 10.1038/srep46461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. I., Jeong W., Lim H., et al. (2021). APOE4-carrying human astrocytes oversupply cholesterol to promote neuronal lipid raft expansion and Abeta generation. Stem Cell Reports, 16(9), 2128–2137. 10.1016/j.stemcr.2021.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng F., Edison P. (2021). Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nature Reviews. Neurology, 17(3), 157–172. 10.1038/s41582-020-00435-y [DOI] [PubMed] [Google Scholar]
- Liao F., Yoon H., Kim J. (2017). Apolipoprotein E metabolism and functions in brain and its role in Alzheimer's disease. Current Opinion in Lipidology, 28(1), 60–67. 10.1097/MOL.0000000000000383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S. A., Barres B. A. (2017). Reactive astrocytes: Production, function, and therapeutic potential. Immunity, 46(6), 957–967. 10.1016/j.immuni.2017.06.006 [DOI] [PubMed] [Google Scholar]
- Liddelow S. A., Guttenplan K. A., Clarke L. E., et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541(3), 481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K. A., Choudhury K. R., Rathakrishnan B. G., et al. (2015). Marked gender differences in progression of mild cognitive impairment over 8 years. Alzheimer's & Dementia, 1(2), 103–110. 10.1016/j.trci.2015.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Seo J., Gao F., et al. (2018). APOE4 causes widespread molecular and cellular alterations associated with Alzheimer's disease phenotypes in human iPSC-derived brain cell types. Neuron, 98(6), 1294. 10.1016/j.neuron.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnerbauer M., Wheeler M. A., Quintana F. J. (2020). Astrocyte crosstalk in CNS inflammation. Neuron, 108(4), 608–622. 10.1016/j.neuron.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Xu X., Dou H., et al. (2015). Apolipoprotein E knockout induced inflammatory responses related to microglia in neonatal mice brain via astrocytes. International Journal of Clinical and Experimental Medicine, 8(1), 737–743. [PMC free article] [PubMed] [Google Scholar]
- Lynch J. R., Tang W., Wang H., et al. (2003). APOE Genotype and an ApoE-mimetic peptide modify the systemic and central nervous system inflammatory response. Journal of Biological Chemistry, 278(49), 48529–48533. 10.1074/jbc.M306923200 [DOI] [PubMed] [Google Scholar]
- Maezawa I., Maeda N., Montine T. J., et al. (2006a). Apolipoprotein E-specific innate immune response in astrocytes from targeted replacement mice. Journal of Neuroinflammation, 3, 10. 10.1186/1742-2094-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maezawa I., Nivison M., Montine K. S., et al. (2006b). Neurotoxicity from innate immune response is greatest with targeted replacement of E4 allele of apolipoprotein E gene and is mediated by microglial p38MAPK. FASEB Journal, 20(6), 797–799. 10.1096/fj.05-5423fje [DOI] [PubMed] [Google Scholar]
- Maezawa I., Zaja-Milatovic S., Milatovic D., et al. (2006c). Apolipoprotein E isoform-dependent dendritic recovery of hippocampal neurons following activation of innate immunity. Journal of Neuroinflammation, 3, 21. 10.1186/1742-2094-3-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias I., Morgado J., Gomes F. C. A. (2019). Astrocyte heterogeneity: Impact to brain aging and disease. Frontiers in Aging Neuroscience, 11, 59. 10.3389/fnagi.2019.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhatre-Winters I., Eid A., Han Y., et al. (2022). Sex and APOE genotype alter the basal and induced inflammatory states of primary microglia from APOE targeted replacement mice. International Journal of Molecular Sciences, 23(17). 10.3390/ijms23179829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M. L., Boyle A. M., Budge K. M., et al. (2018). The glycoprotein GPNMB attenuates astrocyte inflammatory responses through the CD44 receptor. Journal of Neuroinflammation, 15(1), 73. 10.1186/s12974-018-1100-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal M. L., Fleming S. M., Budge K. M., et al. (2020). Pharmacological inhibition of CSF1R by GW2580 reduces microglial proliferation and is protective against neuroinflammation and dopaminergic neurodegeneration. FASEB Journal, 34(1), 1679–1694. 10.1096/fj.201900567RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ophir G., Amariglio N., Jacob-Hirsch J., et al. (2005). Apolipoprotein E4 enhances brain inflammation by modulation of the NF-kappaB signaling cascade. Neurobiology of Disease, 20(3), 709–718. 10.1016/j.nbd.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Orre M., Kamphuis W., Osborn L. M., et al. (2014). Isolation of glia from Alzheimer's mice reveals inflammation and dysfunction. Neurobiology of Aging, 35(12), 2746–2760. 10.1016/j.neurobiolaging.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Pelvig D. P., Pakkenberg H., Stark A. K., et al. (2008). Neocortical glial cell numbers in human brains. Neurobiology of Aging, 29(11), 1754–1762. 10.1016/j.neurobiolaging.2007.04.013 [DOI] [PubMed] [Google Scholar]
- Perez-Nievas B. G., Serrano-Pozo A. (2018). Deciphering the astrocyte reaction in Alzheimer's disease. Frontiers in Aging Neuroscience, 10, 114. 10.3389/fnagi.2018.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preman P., Alfonso-Triguero M., Alberdi E., et al. (2021). Astrocytes in Alzheimer's disease: Pathological significance and molecular pathways. Cells, 10(3). 10.3390/cells10030540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichenbach N., Delekate A., Plescher M., et al. (2019). Inhibition of Stat3-mediated astrogliosis ameliorates pathology in an Alzheimer's disease model. Embo Molecular Medicine, 11(2). 10.15252/emmm.201809665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez G. A., Tai L. M., LaDu M. J., et al. (2014). Human APOE4 increases microglia reactivity at Abeta plaques in a mouse model of Abeta deposition. Journal of Neuroinflammation, 11, 111. 10.1186/1742-2094-11-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Galindo M., Acaz-Fonseca E., Bellini M. J., et al. (2011). Sex differences in the inflammatory response of primary astrocytes to lipopolysaccharide. Biology of Sex Differences, 2, 7. 10.1186/2042-6410-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saura J. (2007). Microglial cells in astroglial cultures: A cautionary note. Journal of Neuroinflammation, 4, 26. 10.1186/1742-2094-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T. D., Livak K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nature Protocols, 3(6), 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Sen A., Nelson T. J., Alkon D. L. (2017). Apoe isoforms differentially regulates cleavage and secretion of BDNF. Molecular Brain, 10(1), 19. 10.1186/s13041-017-0301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Mishra A., Wang T., et al. (2020). Evidence in support of chromosomal sex influencing plasma based metabolome vs APOE genotype influencing brain metabolome profile in humanized APOE male and female mice. PLoS ONE, 15(1), e0225392. 10.1371/journal.pone.0225392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. (2009). Molecular dissection of reactive astrogliosis and glial scar formation. Trends in Neurosciences, 32(12), 638–647. 10.1016/j.tins.2009.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. (2015). Astrocyte barriers to neurotoxic inflammation. Nature Reviews Neuroscience, 16(5), 249–263. 10.1038/nrn3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V. (2020). Astrocyte reactivity: Subtypes, states, and functions in CNS innate immunity. Trends in Immunology, 41(9), 758–770. 10.1016/j.it.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew M. V., Vinters H. V. (2010). Astrocytes: Biology and pathology. Acta Neuropathologica, 119(1), 7–35. 10.1007/s00401-009-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola C., Casal C., Tusell J. M., et al. (2002). Astrocytes enhance lipopolysaccharide-induced nitric oxide production by microglial cells. European Journal of Neuroscience, 16(7), 1275–1283. 10.1046/j.1460-9568.2002.02199.x [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell, 140(6), 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tcw J., Qian L., Pipalia N. H., et al. (2022). Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell, 185, 2213–2233 e2225. 13). 10.1016/j.cell.2022.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu T. M., Kolls B. J., Soderblom E. J., et al. (2017). Apolipoprotein E mimetic peptide, CN-105, improves outcomes in ischemic stroke. Annals of Clinical and Translational Neurology, 4(4), 246–265. 10.1002/acn3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utermann G., Langenbeck U., Beisiegel U., et al. (1980). Genetics of the apolipoprotein E system in man. American Journal of Human Genetics, 32(3), 339–347. [PMC free article] [PubMed] [Google Scholar]
- Vasile F., Dossi E., Rouach N. (2017). Human astrocytes: Structure and functions in the healthy brain. Brain Structure & Function, 222(5), 2017–2029. 10.1007/s00429-017-1383-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerke M., Hol E. M., Middeldorp J. (2021). Physiological and pathological ageing of astrocytes in the human brain. Neurochemical Research, 46(10). 10.1007/s11064-021-03256-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitek M. P., Brown C. M., Colton C. A. (2009). APOE genotype-specific differences in the innate immune response. Neurobiology of Aging, 30(3), 1350–1360. 10.1016/j.neurobiolaging.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Christensen D. J., Vitek M. P., et al. (2009). APOE genotype affects outcome in a murine model of sepsis: Implications for a new treatment strategy. Anaesthesia and Intensive Care, 37(1), 38–45. 10.1177/0310057X0903700111 [DOI] [PubMed] [Google Scholar]
- Wang W. Y., Tan M. S., Yu J. T., et al. (2015). Role of pro-inflammatory cytokines released from microglia in Alzheimer's disease. Annals of Translational Medicine, 3(10), 136. 10.3978/j.issn.2305-5839.2015.03.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring S. C., Rosenberg R. N. (2008). Genome-wide association studies in Alzheimer disease. Archives of Neurology, 65(3), 329–334. 10.1001/archneur.65.3.329 [DOI] [PubMed] [Google Scholar]
- Wolterink-Donselaar Inge G, Meerding Jennifer M., Fernandes Cathy. (2009). A method for gender determination in newborn dark pigmented mice. Lab Animal, 38(1), 35–38. 10.1038/laban0109-35 [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Shinohara M., Yamazaki A., et al. (2020). Apoe (apolipoprotein E) in brain pericytes regulates endothelial function in an isoform-dependent manner by modulating basement membrane components. Arteriosclerosis Thrombosis and Vascular Biology, 40(1), 128–144. 10.1161/ATVBAHA.119.313169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. T., Tan L., Hardy J. (2014). Apolipoprotein E in Alzheimer's disease: An update. Annual Review of Neuroscience, 37, 79–100. 10.1146/annurev-neuro-071013-014300 [DOI] [PubMed] [Google Scholar]
- Yuste J. E., Tarragon E., Campuzano C. M., et al. (2015). Implications of glial nitric oxide in neurodegenerative diseases. Frontiers in Cellular Neuroscience, 9, 322. 10.3389/fncel.2015.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Y., Wang Y., He Y., et al. (2020). A1 astrocytes contribute to murine depression-like behavior and cognitive dysfunction, which can be alleviated by IL-10 or fluorocitrate treatment. Journal of Neuroinflammation, 17(1), 200. 10.1186/s12974-020-01871-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S. A., et al. (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. Journal of Neuroscience, 34(36), 11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Ren Y., Yamazaki Y., et al. (2020). Alzheimer's risk factors age, APOE genotype, and sex drive distinct molecular pathways. Neuron, 106(5), 727–742 e726. 10.1016/j.neuron.2020.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nwabuisi-Heath E., Dumanis S. B., et al. (2012). APOE genotype alters glial activation and loss of synaptic markers in mice. Glia, 60(4), 559–569. 10.1002/glia.22289 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-2-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-3-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro
Supplemental material, sj-xlsx-4-asn-10.1177_17590914221144549 for Sex and APOE Genotype Alter the Basal and Induced Inflammatory States of Primary Astrocytes from Humanized Targeted Replacement Mice by Isha Mhatre-Winters, Aseel Eid, Yoonhee Han, Kim Tieu and Jason R. Richardson in ASN Neuro