Abstract
Gene expression in plant mitochondria is still inadequately analyzed. To learn more about transcription and RNA processing in plant mitochondria, the 5′- and 3′-RNA extremities and the promoters of the cytochrome oxidase gene (cox2) were analyzed in pea. Both 5′ and 3′ ends of cox2 transcripts were examined by RT–PCR across the ligation site of circularized mitochondrial RNA as template. This approach identified 5′ ends a few nucleotides shorter than three major 5′ ends mapped by primer extension analysis. Presumably, only monophosphate 5′ ends derived from processing can be ligated. In vitro transcription assays using a homologous mitochondrial protein extract from pea strongly suggest the major 5′ ends to derive from transcription initiation. The cDNA analysis of the head-to-tail ligated cox2 mRNA identified 3′ ends within a thymidine stretch ∼300 nt downstream of the reading frame in a sequence segment that was not present in the previous investigation of this gene. Nuclease S1 protection experiments confirmed this newly identified 3′ terminus and corroborated the validity of this technique in mRNA end analysis. The general use of the circularized RNA (CR)–RT–PCR approach for the simultaneous analysis of the 5′ and 3′ extremities of mRNA molecules is discussed.
INTRODUCTION
The mitochondrial DNAs (mtDNAs) of higher plants encode approximately 50–60 genes storing the genetic information for structural RNAs and proteins indispensable for the function of these organelles (1,2). To maintain and implement this genetic information a complete enzymatic equipment for DNA replication, transcription, RNA processing and translation within this compartment is required. Despite the progress that has been achieved in recent years, the majority of cis elements, trans-acting factors and mechanisms of plant mitochondrial gene expression are still unknown (3). For instance, the detailed analysis of transcription initiation sites in maize and pea revealed the promoter requirements, but the number of promoters analyzed or even definitely identified is still very limited and the overall picture of plant mitochondrial promoters remains patchy (4–8). Even less is known about the processes at the opposite end of the mRNA. Whereas transcription termination has not been investigated at all to date, our understanding of post-transcriptional processing of mitochondrial RNAs is likewise very limited. Beside RNA splicing and RNA editing altering the primary structure of an RNA molecule, various RNA processing steps in plant mitochondria promote or inhibit degradation of an RNA and are thus important determinants of RNA stability (3). This includes the alteration of the 3′ ends either by post-transcriptional trimming or the addition of non-encoded nucleotides (mostly adenosines) to the 3′ ends of at least some RNA species. This polyadenylation of transcripts was found in mitochondria from sunflower and maize and was suggested to accelerate degradation of these RNAs (9,10). Poly(A) tails were also found downstream of double stem–loop structures in Oenothera atp1 and pea atp9 transcripts. However, in the atp9 mRNAs only the post-transcriptional 3′-end formation is substantially accelerated, whereas in contrast to mRNAs without stem–loop structure the complete degradation of the mRNA seems only slightly stimulated (11). Short extensions primarily composed of adenosines and cytidines were observed at the 3′ ends of pea atp9 and maize rps12, cox2 and atp9 transcripts, but their function is unknown at present (11,12). The number of identified and thoroughly analyzed 3′-transcript termini, an absolute prerequisite for the investigation of both transcription termination events and mechanisms for post-transcriptional 3′-end formation, is still insufficient.
In the present work transcripts of the mitochondrially encoded gene cox2 from pea are investigated. The circularized RNA (CR)–RT–PCR method employed simultaneously and rapidly identifies the 5′ and 3′ ends of the pea cox2 mRNA. Controls with primer extension, nuclease S1 protection and in vitro transcription analyses calibrate and confirm the validity of this technique.
MATERIALS AND METHODS
RT–PCR analysis of circularized RNA
Total mitochondrial RNA was extracted from isolated and purified mitochondria as described by Binder et al. (13). Between 5 and 9 µg of this mtRNA was self-ligated with T4 RNA ligase in a total volume of 10 µl under conditions recommended by the manufacturer of the enzyme (Roche Molecular Biochemicals). After an adjustment of the total volume to 400 µl, proteins were removed by phenol–chloroform extraction following standard protocols and the total volume was reduced to 10 µl. First-strand synthesis was then carried out with primer PC2 (105 to 125) and Superscript reverse transcriptase on 5–9 µg ligated RNA following the instructions of the respective manual (Life Science Technologies). DNA fragments comprising the 5′–3′-ligated mRNA ends were subsequently amplified with primer pairs C4/PC8 (648–669) followed by a second PCR with nested primers PC3/PC9 (670–691) using 0.5–0.75 kb amplification products of the first PCR as template. The products were finally cloned into an XcmI linearized pBluescript vector according to standard procedures followed by sequence analysis of 15 different cox2 cDNA clones (14). The locations of the primers are given in respect to the translation initiation codon (+1).
Nucleic acids analysis
Primer extension analysis was carried out with primer PC3 (–201 to –218) as described by Marchfelder et al. (15). The 5′ ends were identified by electrophoresis of the primer extension products alongside sequencing fragments obtained with the same primer (PC3). Nuclease S1 protection experiments were essentially done as described by Hoffman et al. (16) with the following modifications: a 0.5 kb HindIII–EcoRI fragment covering nucleotides 200–700 downstream of the reading frame was labeled with [α-32P]dATP by the Klenow fragment of DNA polymerase I and used as antisense probe.
In vitro transcription analysis
Pea mitochondria were isolated, purified and processed to transcriptionally active lysates as described by Binder et al. (7). To generate the pea cox2 genomic clone pcox2HE75 a respective DNA fragment was amplified by PCR with primers PC1 (–628 to –608) and PC2 digested with HindIII–EcoRV and cloned into pBluescript vectors digested with the same enzymes. Template pcox2HC468 was obtained after amplification with primers PC1 and C4 (48 to 31), digestion of the fragment with HindIII and cloning into a HindIII–SmaI digested pBluescript vector. PCR was performed using KlenTaq polymerase according to the conditions suggested by the manufacturer with pea mtDNA as template. Promoter activity was tested with ∼100 ng of the DNA templates linearized with NcoI (pcox2HE75) and BstXI (pcox2HC468) as described by Binder et al. (7).
Miscellaneous methods
Filters (Hybond N) carrying HindIII and EcoRI pea mtDNA libraries were prepared and hybridized according to a protocol given by the manufacturer (Amersham Pharmacia Biotech). Oligonucleotide pcox2INS-1 (1063–1045) was used as probe for the isolation of clones representing the cox2 gene and the respective genomic downstream region.
Sequence analyses of the genomic and cDNA clones including those used as DNA templates for in vitro transcription were done with a T7 sequencing kit and [35S]dATP or with a Thermo Sequenase fluorescent labeling kit according to the instructions of the manufacturer (Amersham Pharmacia Biotech). Sequencing products were either separated by conventional polyacrylamide gel electrophoresis or on an ALF express sequencer.
RESULTS
An RT–PCR analysis of 5′–3′ self-ligated cox2 mRNA reveals a new 3′ end within a oligothymidine stretch
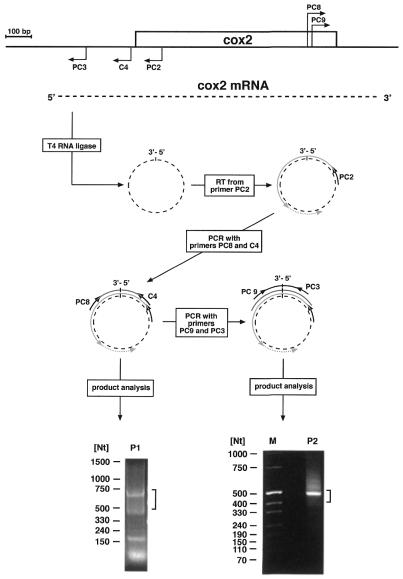
A previous analysis of the pea cox2 gene and its expression had revealed this gene to be transcribed into monocistronic transcripts, with two 5′ ends located at nucleotide positions –302 and –285 upstream of the reading frame and a single 3′ end 193–195 nt downstream of the translation stop codon (17). Thus, this gene represents a good candidate to test a method employing RT–PCR across the 5′–3′-end-ligated mRNAs, which would allow to simultaneously investigate both extremities of the transcripts and which has successfully been used for the analysis of land snail mitochondrial tRNAs (18). To this end, total RNAs from pea mitochondria were self-ligated with T4 RNA ligase and specific first-strand cDNA synthesis was initiated with primer PC2 followed by a PCR with primers PC8 and C4 (Fig. 1). Separation of the amplification products revealed three major molecule species of ∼650, 420 and 170 bp, and a smaller one most likely representing non-incorporated primers (Fig. 1, bottom part). A PCR product of ∼650–670 nt was expected if the 5′ and 3′ termini of the steady state 1.2 kb transcripts are correctly ligated. Accordingly, DNA fragments with sizes between 0.5 and 0.75 kb were recovered from the gel and used as templates in a second PCR with nested primers PC3/PC9 (Fig. 1). This generated a predominant cDNA fragment of ∼500 nt, slightly larger than the 450 nt expected for ligation at the previously reported mRNA extremities (17). To cover these sizes cDNA fragments between ∼420 and 530 nt were cloned and 15 clones were sequenced. No PCR fragment could be obtained by the same experimental strategy without RNA ligase indicating that 5′–3′-end-ligated cox2 mRNAs are not detectable in the steady state RNA, which, however, does not necessarily exclude the existence of RNA molecules with such a structure.
Figure 1.
CR–RT–PCR analysis of cox2 transcripts. Location and orientation of the primers (bent arrows) used on the cox2 transcripts (dashed line) are indicated in a cartoon of the pea cox2 locus (upper part). The 5′ and 3′ ends of the transcripts are joined using T4 RNA ligase. The circular form of the RNA is then used as template for first-strand synthesis initiated from primer PC2 using reverse transcriptase (RT) followed by two consecutive amplification reactions with primer pairs C4/PC8 and PC3/PC9, each enclosing the ligation site. Products obtained in the CR–RT–PCR analysis are separated on agarose gels (bottom part). Sizes of coelectrophoresed marker molecules are given in nucleotides. Angular brackets indicate the range of excised fragments.
The 5′ end in a single clone maps 6 nt upstream of position –285, whereas the 5′ termini of the residual 14 clones map 5–24 nt downstream of this position (Fig. 2). In contrast to this congruent assignment of the 5′ sequences, the 3′-terminal nucleotides do not match the previously published sequence (17). The cDNA sequences could be aligned with genomic sequences at primer PC9 from nucleotide 109 upstream of the translation stop codon to 237 nt downstream of the reading frame, ∼44 nt beyond the originally mapped 3′ end (+193 to +195). This sequence is followed by 61–64 nt, depending on the clone, not found in the published genomic sequence (17). To test whether the newly identified sequence segment is present at the 5′ or the 3′ ends, corresponding RT–PCR analyses were carried out. Whereas no cDNA fragment was observed with an RT–PCR strategy consistent with an attachment of the newly identified sequence at the 5′ end of cox2 transcripts, cDNA fragments were generated in two amplification reactions corresponding to an mRNA containing the new sequence at the 3′ end. This suggests these nucleotides to be an integral part of the 3′-non-translated region of the cox2 mRNA (data not shown).
Figure 2.
5′ and 3′ ends detected by the CR–RT–PCR. Sequences of the cDNA covering the 5′–3′-ligation sites of pea cox2 transcripts. The numbering of the 5′ end refers to the translation start codon and the 3′ numbers are relative to the stop codon. The numbers on the left indicate the frequency with which the distinct 5′ and 3′ ends are found in individual clones. The ligation site is indicated by a horizontal dash. Sequences corresponding to the 5′-terminal part are given in white letters in fading black boxes.
Mapping three major 5′-transcript termini upstream of the pea cox2 gene
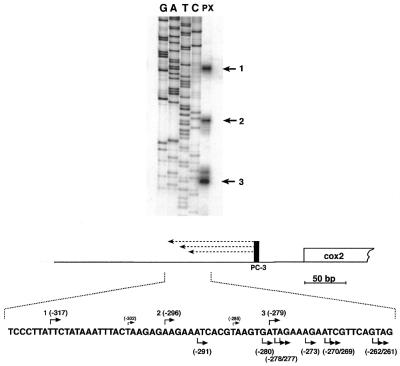
Since the 5′ ends detected in the CR–RT–PCR analysis did not match the previously mapped termini these were re-examined by primer extension analysis from primer PC3, which anneals to sequences 201–218 nt upstream of the translation start codon. This analysis revealed three major ends in repeated experiments in the initially described region, but all differing from the previously mapped sites, and also from the 5′ ends detected in the cDNA derived from the CR–RT–PCR analysis (Figs 2 and 3). The 5′ ends (sites 1–3) were found at positions –316, –296 and –279 with respect to the ATG.
Figure 3.
Primer extension analysis of pea cox2 transcripts. Three major 5′-transcript termini (indicated by numbered arrows on the right) were detected by running the extension products (lane PX, illustrated as dashed lines in the bottom part) alongside a sequencing ladder (lanes G, A, T, C). Both primer extension and sequencing analysis were carried out with primer PC3 (vertical black bar) annealing to the mRNA upstream of the cox2 reading frame (open box). The sequence surrounding the detected 5′ ends (numbered bent arrows above the sequence) is shown in the bottom part. Nucleotide positions relative to the ATG are given in brackets. Previously mapped 5′ termini are also given above the sequence (17). 5′ Extremities found by the CR–RT–PCR analysis are given below. All numberings are in respect to the translation start codon (+1).
Two of the newly identified 5′ ends (–296 and –279) are associated with sequences showing strong similarity with the consensus promoter motif identified in dicot plant species (8). To test the functional significance of this similarity, the entire region was further analyzed.
In vitro transcription analysis confirms promoters upstream of the cox2 reading frame
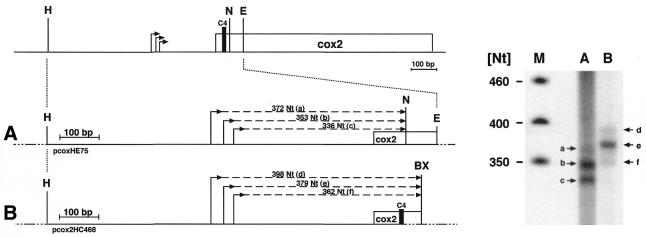
In order to investigate whether the identified ends indeed derive from transcription initiation, two different DNA templates comprising the detected 5′ ends were incubated with a transcriptionally active mitochondrial protein lysate. The sizes of the strongest signals (Fig. 4, signals b and e in lanes A and B) correlate well with de novo transcription initiation at site 2 (corresponding to position –296), strongly suggesting the surrounding sequences to function as a bona fide mitochondrial promoter. Signals of slightly lower intensities correspond to transcription initiation events at site 3 (–279), supporting the respective 5′ end to be derived from a transcription start site (Fig. 4, signals c and f in lanes A and B). Two products with much lower abundance correspond to run-off transcripts expected upon transcription initiation at site 1 (–317). This suggests low but active transcription initiation at this site, even though the surrounding sequences do not resemble any known promoter sequence from dicot plant mitochondria (Fig. 4, signals a and d in lanes A and B).
Figure 4.
In vitro transcription analysis of pea cox2 DNA templates. A diagram of the pea cox2 genomic region and the respective DNA templates used for in vitro transcription analysis are shown in the left part. Both DNA templates (A and B) contain the 5′ region of the cox2 reading frame (indicated by the open box) including the three 5′ ends (bent arrows) mapped as detailed above. Expected sizes of the run-off products (dashed lines) of the NcoI and BstXI linearized templates (A) pcox2HE75 and (B) pcox2HC468, respectively, are indicated. Restriction sites are given for HindIII (H), EcoRV (E) and NcoI (N). The BstXI site (BX) used for the linearization of pcox2HC468 is located in vector sequences (lines interrupted by dots). The location of primer C4 determining the downstream end of the cloned mitochondrial sequences is indicated (C4, vertical black bar). The in vitro transcription analysis is shown in the right-hand part. The designation of the lanes corresponds to the templates used in the in vitro assays. The numbering of the detected in vitro run-off products (a–g, indicated by numbered arrows) corresponds to the one given for the expected products.
The new sequence segment is present in the genomic DNA downstream of the cox2 gene
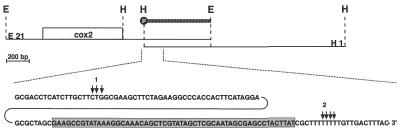
Although the presence of 3′-end non-encoded nucleotides of different identities has been reported for several plant mitochondrial transcripts, the attachment of non-homopolymeric sequences of a length of >60 nt as found here (see above) seems highly unlikely (11,12). Therefore, we examined the 3′ region of the pea cox2 gene on the level of the genomic DNA. Using oligonucleotide pcox2INS-1, corresponding to part of the newly identified sequence, a 1.8 kb EcoRI and a 1.7 kb HindIII clone were isolated from pea mtDNA libraries. Sequence analysis of the former showed this to carry the complete cox2 reading frame and ∼350 nt as well as 650 nt 5′- and 3′-flanking sequences, respectively (accession no. AJ414385). This clone corresponds to the EcoRI clone used for the previously published sequence analysis (17). The HindIII clone represents the cox2 3′ region and overlaps with the EcoRI clone by 600 nt. This clone also contains the newly identified sequence confirming its presence in the genomic sequence (Fig. 5). Thus, the CR–RT–PCR of cox2 transcripts locates the 3′-transcript terminus ∼100 nt downstream of the previously mapped end (17). To confirm this by an independent experimental approach, an S1 protection analysis covering the 3′-non-translated region 200–700 nt downstream of the cox2 reading frame was performed (Fig. 5). The protected fragment of ∼100 nt corresponds to a 3′ end ∼300 nt downstream of the translation stop codon, consistent with the 3′-transcript termini detected in the CR–RT–PCR clones (Fig. 6). These results confirm the validity of the CR–RT–PCR method for 3′-end mapping of mitochondrial transcripts. Comparison of the cDNA sequences derived from the CR–RT–PCR analysis with genomic sequences revealed no non-encoded nucleotides at either end of the cox2 mRNA.
Figure 5.
Sequence context of the pea cox2 3′ region. Two pea mtDNA clones designated E21 (EcoRI 1.8 kb) and H1 (HindIII, 1.7 kb) (indicated by solid lines) representing the cox2 genomic environment were used for the genomic sequence analysis of the respective cox2 region. The new sequence segment identified in the CR–RT–PCR analysis is highlighted by a gray box. The previously mapped (1) and the newly identified 3′-transcript termini (2) are indicated by vertical arrows above the sequence. Restriction sites are given by dashed vertical lines for EcoRI (E) and HindIII (H). The probe used in the nuclease S1 protection assays (Fig. 6) is given as a hatched bar. Position of the radioactive label is indicated by a white P in a black circle.
Figure 6.
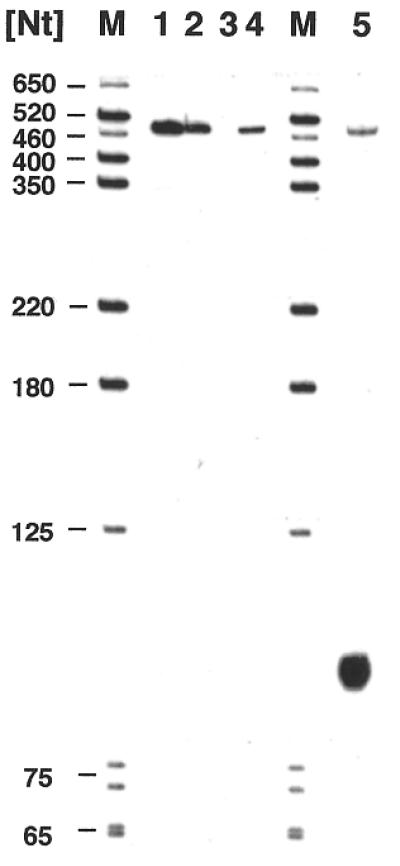
Nuclease S1 protection analysis confirms the new mRNA 3′ end detected by the CR–RT–PCR. Protected products (lane 5) were coelectrophoresed with products obtained in control experiments and with DNA marker fragments (lanes M), whose sizes are given in the left margin. The control reactions are the antisense probe without any further treatments (lane 1), incubated under S1 conditions without nuclease and RNA (lane 2), incubated with 75 U S1 and without RNA (lane 3) or 40 µg of wheat germ tRNA products (lane 4). The size of the protected fragment of ∼100 nt corresponds to a 3′ end ∼300 bp downstream of the translation stop codon, which is consistent with the 3′ terminus mapped by the CR–RT–PCR analysis.
DISCUSSION
Pea cox2 mitochondrial transcripts: test object for the CR–RT–PCR analysis of mRNA extremities
Previous analysis of the cox2 gene encoded in pea mitochondria had indicated a relatively simple transcript pattern, which suggested this gene as an ideal candidate for a detailed functional investigation of its expression. Two 5′-transcript termini had been mapped in a region containing sequence elements with substantial similarity to dicot plant mitochondrial promoters, and a nuclease S1 protection analysis suggested a 3′ end ∼193–195 nt downstream of the reading frame (17). Later analysis of cDNAs originating from this gene indicated several nucleotide differences compared with genomic sequences recommending a detailed descriptive re-examination (19).
Re-examination of the cox2 gene encoded in pea mitochondria
We started the re-investigation of the transcript extremities followed by a complete sequence analysis of the gene and its 5′ and 3′ genomic environments. The latter revealed several nucleotide differences with respect to the previously published sequences with the additional 54 nt in the 3′-terminal part of the mRNA as the most striking discrepancy (see accession no. AJ414385 and Fig. 5). All non-RNA editing DNA/RNA differences between the previously published genomic (17) and the cDNA sequences (19) were found to be correct in the latter. From the 14 RNA editing DNA/RNA differences identified by the cDNA analysis, 12 were confirmed in our genomic sequence, whereas the editing events at 44 nt upstream and 412 nt downstream of the ATG indicated by C/T differences have to be corrected. At these sites thymidines are already present in our genomic pea sequence (19). Comparison of the new cDNA sequences obtained from the CR–RT–PCR covering the complete 3′- and partially the 5′-non-translated parts of the mRNA does not indicate any further editing site.
Similarity between the pea cox2 promoter sequences and conserved nonanucleotide promoters from dicot plants
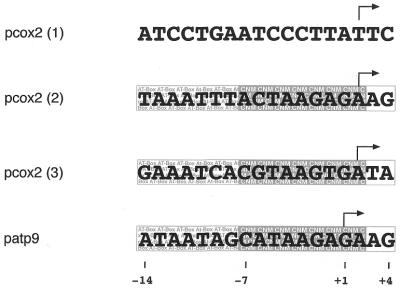
In contrast to the two cox2 mRNA 5′ ends previously mapped by nuclease S1 protection experiments, the primer extension technique applied here revealed three 5′ ends each differing by several nucleotides from the previously mapped ends. These discrepancies are probably caused by the different techniques used, an often observed phenomenon. Comparison of the sequences at sites 2 and 3 shows substantial similarities to the pea atp9 promoter, which has been investigated by detailed functional in vitro transcription studies (7,8). As shown in Figure 7, well conserved nonanucleotide motifs (CNM) are present at both sites 2 (–296) and 3 (–279) with minor variations at the first two positions of this motif at site 2. In addition, the presence of upstream-located AT-rich regions and the presence of at least one purine at positions +3 and +4 fulfill the requirements of a typical dicot plant mitochondrial promoter (8). Consequently, run-off transcripts consistent with transcription initiation at these sites are detected in the functional in vitro studies confirming the function of these sequences as mitochondrial promoters. In contrast, a completely different sequence surrounds the most upstream 5′ end at site 1 (–317), with no similarity to either CNM or to other non-CNM-type transcription initiation sites identified in mitochondria of various dicot plant species (20–23). Nevertheless, weak signals that could correspond to run-off products initiated at this site are detected in the in vitro analysis, suggesting promoter activity of this sequence, the first observation of a non-CNM promoter activity in vitro.
Figure 7.
Sequences of pea cox2 promoters. The sequences surrounding the three different 5′ ends of pea cox2 transcripts [pcox2 (1–3)] are aligned with the –14 to +4 promoter sequence located upstream of the pea atp9 (patp9) gene. Whereas sequences at sites 2 and 3 were aligned for best nucleotide consistence with the pea atp9 promoter, the sequence at site 1 is aligned with respect to the detected 5′ ends (indicated by bent arrows). Nucleotide positions are given for the pea atp9 promoter referring to the transcription start site (+1). The AT-rich region (AT-Box, –14 to –8) and the CNM (–7 to +2) as well as purine-rich nucleotide positions +3/+4, all hallmarks of the CNM-type dicot plant mitochondrial promoters, are boxed and highlighted.
Primary and secondary structures at the 3′ end of the pea cox2 transcripts
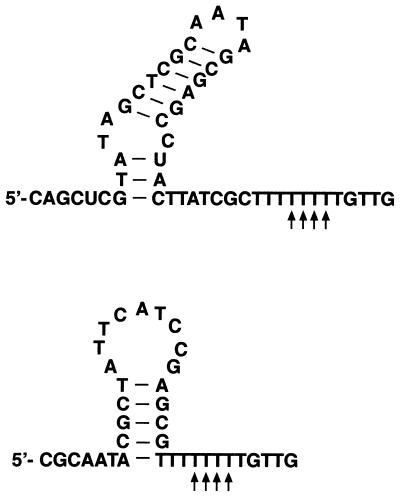
At present little is known about the generation of 3′ termini in higher plant mitochondria. Generally, 3′-end formation can be achieved either directly by transcription termination or by subsequent post-transcriptional RNA processing. The modes and structures involved in the former are completely unknown, and the double stem–loop structures located immediately upstream of several characterized 3′ ends are not able to terminate transcription in vitro. These structures are instead processing signals impeding the progression of 3′–5′ exonucleolytic degradation in post-transcriptional 3′ trimming (24,25). How the cox2 3′ mRNA ends investigated here in pea mitochondria are formed remains unclear at present. Two alternative stem–loop structures can be folded a few nucleotides upstream of the 3′ termini. These structures and the location of the 3′ ends within a polythymidine stretch of 9 nt resemble prokaryotic transcription terminators and may suggest this mRNA terminus to be derived by transcription termination (Fig. 8) (26). Thus, detailed in vitro studies are required to learn more about 3′-end formation of pea cox2 transcripts and plant mitochondrial 3′-end formation in general.
Figure 8.
Potential secondary structures at the 3′ end of the pea cox2 transcripts identified here. The 3′ ends detected by nuclease S1 and CR–RT–PCR analysis are indicated by vertical arrows beneath the sequence.
Non-encoded nucleotides at the 3′ end of pea cox2 transcripts
Besides the mapping of RNA 5′ and 3′ extremities the CR–RT–PCR analysis further allows the simultaneous detection of non-encoded nucleotides. However, in the case of pea cox2 transcripts non-encoded nucleotides could not be detected by this approach. Although the total number of 15 clones analyzed might be too low, an alternative strategy using a 3′-RACE experiment initiated with oligo(dT) adapter primer revealed only a single clone deriving from an annealing of the oligo(dT) primer to an endogenous oligoadenosine stretch, but failed to detect 3′-non-encoded nucleotides (A.Dospil, J.Kuhn and S.Binder, unpublished data). Since this latter approach has been successfully used in the analysis of other mitochondrial transcripts (9–11), the presence of 3′-non-encoded nucleotides at pea cox2 transcripts is highly unlikely.
CR–RT–PCR, a convenient method for the investigation of 5′ and 3′ extremities of plant mitochondrial RNA
The mapping of 5′ ends by an RT–PCR analysis of circularized mRNA revealed different termini, most of them located up to 20 nt downstream of site 3. Whereas one clone ends between sites 2 and 3, the 5′ ends of three clones coincide with a guanidine at position –280, 1 nt upstream of the major product at site 3 at position –279. This distribution of the 5′ ends is in contrast to the primer extension experiments, which clearly indicate three major 5′ ends. The exact reason for this discrepancy is unclear at present. We assume that 5′ ends with di- or triphosphate groups present at primary transcripts whose 5′ ends derive from de novo initiation events, are ligated with much lower efficiency (or not at all) compared with termini with 5′-monophosphate groups at transcripts originating from processing events. This bias is strongly suggested by the mechanism of the ATP-dependent T4 RNA ligase reaction. In the first step, AMP, which is formed by the release of pyrophosphate from ATP, is bound to the 5′-phosphoryl-terminated nucleic acid donor via a pyrophosphate bond yielding an adenylated donor [A5′pp-5′Np(Np)x]. In the next step, AMP is released from the adenylated donor and replaced by a phosphodiester bond between the donor and a 3′-hydroxyl-terminated acceptor. The latter ligation step, which is independent from ATP, can be performed by T4 RNA ligase with various adenylated donor molecules and 3′ hydroxyl acceptor, but not with an A5′ppp5′G, which would theoretically result from the first reaction step from a 5′ diphosphate donor molecule (27,28). Concluding, it is thus highly unlikely that the 5′ di- or triphosphate ends found at 5′ termini from transcription initiation events can be accepted as substrates of the RNA ligase reactions.
The cox2 mRNAs slightly shortened at the 5′ end are possibly products of processing or degradation events. These transcripts are most likely shortened at the 5′ end by endo- or exonucleolytic activities, rather than resulting from initiation events downstream of the major initiation sites and thus carry monophosphate groups at the 5′ ends. This hypothesis is further supported by the sequence analysis of 21 cDNA clones obtained from CR–RT–PCR investigation of self-ligated Oenothera atp1 transcripts, which revealed no 5′ end identical with the originally mapped primary 5′ end (J.Kuhn and S.Binder, manuscript in preparation). Instead, an alternative 5′ terminus was found in 20 of these clones, which is located ∼76–79 nt downstream of the transcription initiation site as confirmed by primer extension experiments and which is most likely derived from a processing event. Only a single clone contained a 5′ end close to the initiation site, but is still lacking the terminal 5 nt.
If the RNA ligase indeed prefers monophosphate over di- or triphosphate 5′ ends, which has of course to be investigated experimentally, the CR–RT–PCR method will give a straightforward assignment of 5′ ends originating from processing or degradation events without any pretreatment. On the other hand, primary 5′ termini could only be ligated once di- or triphosphate groups have been converted to monophosphate groups. Such a pretreatment, e.g. by consecutive phosphatase and kinase reactions, would also make these termini accessible for the RNA ligase. Such a differential pretreatment of the RNA could allow the assignment of the origin of any 5′ end from either de novo transcription initiation or from processing events.
General suitability of the method for mapping of 5′- and 3′-RNA extremities
The application of the CR–RT–PCR for the transcript analysis of the pea cox2 as well as pea atp9 and Oenothera atp1 genes (J.Kuhn and S.Binder, manuscript in preparation) revealed the 5′ and 3′ extremities mapped by this technique to be generally consistent with respective ends determined by other methods. For instance the cox2 3′-transcript termini detected here in the CR–RT–PCR analysis could be clearly confirmed by a nuclease S1 experiment (Fig. 6). The 3′ ends mapped previously most likely originate from a misleading nuclease S1 protection strategy using a fragment that is entirely complementary to internal sequences of the mRNA and should thus have been entirely protected. The observed shortening by a few nucleotides is perhaps caused by the use of too stringent nuclease S1 digestion conditions.
The application of the CR–RT–PCR technique presented here for the analysis of the above mentioned transcripts is facilitated by the knowledge of both ends, which allowed to set up the appropriate PCR strategy and to preselect cDNA fragments obtained in the amplification reactions. But will this technique also be suitable for 5′- and 3′-end mapping of totally uncharacterized transcripts? This will probably be rather difficult and at least the rough size of the transcript(s), e.g. determined by northern blot experiments, will help to estimate the extent of the 5′- and 3′-untranslated regions. This in turn will allow a prediction of the size(s) of the expected RT–PCR fragment(s). Such a northern blot analysis will also reveal the overall transcript pattern, which for plant mitochondrial genes can be very complex and will complicate the 5′- and 3′-end mapping by any method.
The requirement of monophosphate 5′ termini defines the types of RNA that can be investigated by CR–RT–PCR. The analysis of RNA species such as those derived from processed mRNAs in any system like mitochondria, plastids or bacteria will be straightforward. For capped RNAs this method has previously been successfully used to characterize the 5′- and 3′-terminal regions of non-polyadenylated linear RNA genomes from several tick-borne flaviviruses. In this case, the 5′-terminal cap had to be removed by tobacco acid pyrophosphatase to allow the 5′–3′-end-ligation by T4 RNA ligase (29).
However, the same strategy should also allow the analysis of polyadenylated mRNAs, whose 5′ ends are presently often mapped by ligation of an anchor primer to the 5′ end instead of circularization of the mRNA (30).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bärbel Weber for excellent technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to S.B. and a fellowship of the Studienstiftung des Deutschen Volkes to J.K.
DDBJ/EMBL/GenBank accession no. AJ414385
REFERENCES
- 1.Unseld M., Marienfeld,J.R., Brandt,P. and Brennicke,A. (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nature Genet., 15, 57–61. [DOI] [PubMed] [Google Scholar]
- 2.Kubo T., Nishizawa,S., Sugawara,A., Itchoda,N., Estiati,A. and Mikami,T. (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNACys (GCA). Nucleic Acids Res., 28, 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kuhn,J., Däschner,K. and Binder,S. (2001) The RNA world of plant mitochondria. In Moldave,K. (ed), Progress in Nucleic Acid Research and Molecular Biology. Academic Press, San Diego, Vol. 70, pp. 119–154. [DOI] [PubMed]
- 4.Rapp W.D. and Stern,D.B. (1992) A conserved 11 nucleotide sequence contains an essential promoter element of the maize mitochondrial atp1 gene. EMBO J., 11, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapp W.D., Lupold,D.S., Mack,S. and Stern,D.B. (1993) Architecture of the maize mitochondrial atp1 promoter as determined by linker-scanning and point mutagenesis. Mol. Cell. Biol., 13, 7232–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caoile A.G. and Stern,D.B. (1997) A conserved core element is functionally important for maize mitochondrial promoter activity in vitro. Nucleic Acids Res., 25, 4055–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder S., Hatzack,F. and Brennicke,A. (1995) A novel pea mitochondrial in vitro transcription system recognizes homologous and heterologous mRNA and tRNA promoters. J. Biol. Chem., 270, 22182–22189. [DOI] [PubMed] [Google Scholar]
- 8.Dombrowski S., Hoffmann,M., Guha,C. and Binder,S. (1999) Continuous primary sequence requirements in the 18-nucleotide promoter of dicot plant mitochondria. J. Biol. Chem., 274, 10094–10099. [DOI] [PubMed] [Google Scholar]
- 9.Gagliardi D. and Leaver,C.J. (1999) Polyadenylation accelerates the degradation of the mitochondrial mRNA associated with cytoplasmic male sterility in sunflower. EMBO J., 18, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupold D.S., Caoile,A.G. and Stern,D.B. (1999) Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell, 11, 1565–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn J., Tengler,U. and Binder,S. (2001) Transcript lifetime is balanced between stabilizing stem–loop structures and degradation-promoting polyadenylation in plant mitochondria. Mol. Cell. Biol., 21, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams M.A., Johzuka,Y. and Mulligan,R.M. (2000) Addition of non-genomically encoded nucleotides to the 3′-terminus of maize mitochondrial mRNAs: truncated rps12 mRNAs frequently terminate with CCA. Nucleic Acids Res., 28, 4444–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Binder S., Marchfelder,A. and Brennicke,A. (1994) RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet., 244, 67–74. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY.
- 15.Marchfelder A., Brennicke,A. and Binder,S. (1996) RNA editing is required for efficient excision of tRNAPhe from precursors in plant mitochondria. J. Biol. Chem., 271, 1898–1903. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann M., Dombrowski,S., Guha,C. and Binder,S. (1999) Cotranscription of the rpl5–rps14–cob gene cluster in pea mitochondria. Mol. Gen. Genet., 261, 537–545. [DOI] [PubMed] [Google Scholar]
- 17.Moon E., Kao,T.H. and Wu,R. (1985) Pea cytochrome oxidase subunit II gene has no intron and generates two mRNA transcripts with different 5′-termini. Nucleic Acids Res., 13, 3195–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokobori S. and Paabo,S. (1995) Transfer RNA editing in land snail mitochondria. Proc. Natl Acad. Sci. USA, 92, 10432–10435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covello P.S. and Gray,M.W. (1990) Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res., 18, 5189–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown G.G., Auchincloss,A.H., Covello,P.S., Gray,M.W., Menassa,R. and Singh,M. (1991) Characterization of transcription initiation sites on the soybean mitochondrial genome allows identification of a transcription-associated sequence motif. Mol. Gen. Genet., 228, 345–355. [DOI] [PubMed] [Google Scholar]
- 21.Fey J. and Marechal-Drouard,L. (1999) Expression of the two chloroplast-like tRNAAsn genes in potato mitochondria: mapping of transcription initiation sites present in the trnN1–trnY–nad2 cluster and upstream of trnN2. Curr. Genet., 36, 49–54. [DOI] [PubMed] [Google Scholar]
- 22.Remacle C. and Marechal-Drouard,L. (1996) Characterization of the potato mitochondrial transcription unit containing ‘native’ trnS (GCU), trnF (GAA) and trnP (UGG). Plant Mol. Biol., 30, 553–563. [DOI] [PubMed] [Google Scholar]
- 23.Binder S., Thalheim,C. and Brennicke,A. (1994) Transcription of potato mitochondrial 26S rRNA is initiated at its mature 5′ end. Curr. Genet., 26, 519–523. [DOI] [PubMed] [Google Scholar]
- 24.Dombrowski S., Brennicke,A. and Binder,S. (1997) 3′-Inverted repeats in plant mitochondrial mRNAs are processing signals rather than transcription terminators. EMBO J., 16, 5069–5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bellaoui M., Pelletier,G. and Budar,F. (1997) The steady-state level of mRNA from the Ogura cytoplasmic male sterility locus in Brassica cybrids is determined post-transcriptionally by its 3′ region. EMBO J., 16, 5057–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lesnik E.A., Sampath,R., Levene,H.B., Henderson,T.J., McNeil,J.A. and Ecker,D.J. (2001) Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res., 29, 3583–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugino A., Snopek,T.J. and Cozzarelli,N.R. (1977) Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem., 252, 1732–1738. [PubMed] [Google Scholar]
- 28.England T.E., Gumport,R.I. and Uhlenbeck,O.C. (1977) Dinucleoside pyrophosphate are substrates for T4-induced RNA ligase. Proc. Natl Acad. Sci. USA, 74, 4839–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandl C.W., Heinz,F.X., Puchhammer-Stockl,E. and Kunz,C. (1991) Sequencing the termini of capped viral RNA by 5′–3′ ligation and PCR. Biotechniques, 10, 484–486. [PubMed] [Google Scholar]
- 30.Fromont-Racine M., Bertrand,E., Pictet,R. and Grange,T. (1993) A highly sensitive method for mapping the 5′ termini of mRNAs. Nucleic Acids Res., 21, 1683–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]