Abstract
During sexual reproduction, Euplotes crassus precisely fragments its micronuclear chromosomes and synthesizes new telomeres onto the resulting DNA ends to generate functional macronuclear minichromosomes. In the micronuclear chromosomes, the macronuclear-destined sequences are typically separated from each other by spacer DNA segments, which are eliminated following chromosome fragmentation. Recently, in vivo chromosome fragmentation intermediates that had not yet undergone telomere addition have been characterized. The ends of both the macronuclear-destined and eliminated spacers were found to consist of six-base, 3′ overhangs. As this terminal structure on the macronuclear-destined sequences serves as the substrate for de novo telomere addition, we sought to determine if the spacer DNAs might also undergo telomere addition prior to their elimination. Using a polymerase chain reaction approach, we found that at least some spacer DNAs undergo de novo telomere addition. In contrast to macronuclear-destined sequences, heterogeneity could be observed in the position of telomeric repeat addition. The observation of spacer DNAs with telomeric repeats makes it unlikely that differential telomere addition is responsible for differentiating between retained and eliminated DNA. The heterogeneity in telomere addition sites for spacer DNA also resembles the situation found for telomeric repeat addition to macronuclear-destined sequences in other ciliate species.
INTRODUCTION
In most organisms, double-stranded breaks in DNA are mended by cellular DNA repair pathways. Chromosome breaks that escape such repair are recombinogenic and often result in deleterious changes to the genome. In contrast, organisms such as Ascaris and the ciliated protozoa undergo programmed chromosome fragmentation as part of their normal developmental programs. In these cases the broken chromosome ends are stabilized by the de novo addition of telomeric repeat sequences. In Ascaris, the chromosome fragmentation/telomere addition process occurs in somatic, but not germline cells (reviewed in 1). An analogous situation exists for the unicellular ciliated protozoa (reviewed in 2–4). These organisms are binucleate, containing both a transcriptionally inactive ‘germline’ micronucleus (MIC) with conventional chromosomes, and a transcriptionally active ‘somatic’ macronucleus (MAC) with minichromosomes. Both the MIC and MAC replicate their DNA and divide during asexual (vegetative) growth, but the MAC is destroyed following sexual reproduction (conjugation) and a new one is generated from a copy of the MIC. MAC development involves extensive rearrangement of the MIC genome, and includes steps of DNA splicing, gene amplification, DNA elimination, the specific fragmentation of the MIC chromosomes and addition of simple telomeric repeats to DNA ends.
The degree of rearrangement varies among different ciliate species. For example, Tetrahymena thermophila fragments its five MIC chromosomes to yield approximately 200 MAC chromosomes, and eliminates ∼15% of the MIC genome during MAC development. In contrast, the spirotrichous ciliates (e.g. Oxytricha, Stylonychia and Euplotes; formerly termed hypotrichs) undergo an extreme rearrangement process. Greater than 90% of the MIC genome is eliminated in these organisms, and the MIC chromosomes are fragmented at >10 000 positions to generate short MAC DNA molecules that typically contain single genes. Whereas the chromosome fragmentation/telomere addition process is quite reproducible during independent cycles of MAC development, the process is not entirely precise in most ciliates. The positions of chromosome fragmentation/telomere addition typically vary by tens of base pairs, but in Paramecium they can vary by hundreds of base pairs (reviewed in 2,4). In addition, some organisms utilize a subset of chromosome fragmentation/telomere addition sites in an alternative manner, giving rise to related MAC chromosomes that differ in size by kilobases (5–7).
The proteins that carry out chromosome fragmentation are unknown, but information on the cis-acting sequence elements that specify the process is available for some ciliates. The best understood system is T.thermophila, where a 15 bp sequence element (5′-AAAGAGGTTGGTTTA-3′) termed the chromosome breakage sequence (Cbs) has been demonstrated to be necessary and sufficient for chromosome breakage (8). The Cbs element resides between what will become the ends of two separate MAC chromosomes, and telomeres are ultimately added 4–22 bp from the ends of the Cbs. This has led to suggestions that Cbs-specified cleavage of the chromosome is followed by exonucleolytic degradation of the ends prior to de novo telomere formation (2). De novo telomere formation is tightly coupled to chromosome fragmentation and is known to be mediated by the telomerase enzyme (9). Information on the sequences specifying fragmentation in other ciliates is generally lacking, but it is clear that strong matches to the Tetrahymena Cbs are not present at most fragmentation sites. Some recent work on Stylonychia lemnae has provided evidence that sequences near the ends of the MAC-destined sequences are required for fragmentation, but the precise sequence element(s) responsible has not yet been defined (10).
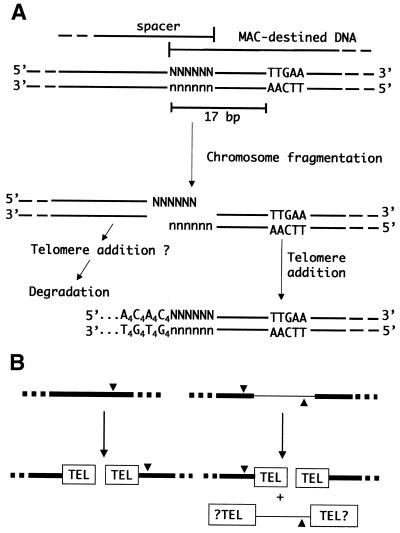
The subject of this study, Euplotes crassus, displays a number of differences in the chromosome fragmentation/telomere addition process relative to the other ciliates. First, chromosome fragmentation/telomere addition is precise and reproducible in this organism (11). Secondly, the likely cis-acting sequence specifying fragmentation can reside either near the end of the MAC-destined sequence, or in the flanking DNA. A 10 bp consensus sequence (5′-HATTGAAaHH-3′) termed the Euplotes Cbs (E-Cbs) has been identified in E.crassus (11,12). When the E-Cbs resides within the MAC-destined DNA, the highly conserved core TTGAA sequence is located 17 bp from the site of telomere addition; when the E-Cbs is located in the flanking DNA, it is in inverted orientation with the core TTGAA positioned 11 bp from the telomere addition site. Based on this difference in spacing, a model of chromosome fragmentation/telomere addition was proposed (11,12) in which the E-Cbs directs a double-stranded cut in the DNA upstream of itself (i.e. in the 5′ direction) so as to generate DNA ends with six-base, 3′ overhangs (Fig. 1A). Telomerase would then initiate de novo telomere synthesis by adding 5′-GGGGTTTT-3′ telomeric repeats (G4T4 repeats) to the 3′ end of the fragmentation intermediate. The C4A4 strand of the telomere then appears to be synthesized by a conventional DNA polymerase (13), and this would also serve to fill in the initial six-base overhang. As in Tetrahymena, chromosome fragmentation and de novo telomere addition appear to be tightly coupled during MAC development (14). Nonetheless, using a sensitive polymerase chain reaction (PCR)-based procedure, a recent study has provided evidence for the existence of the predicted chromosome fragmentation intermediates with six-base, 3′ overhangs in vivo at the time of chromosome fragmentation, providing strong support for the model (15).
Figure 1.
Chromosome fragmentation/telomere addition in E.crassus. (A) Current model of chromosome fragmentation and de novo telomere formation (15). A segment of MIC DNA with an E-Cbs core (TTGAA) is shown at the top. The E-Cbs directs a double-stranded break in the DNA to generate fragmentation intermediates with six-base, 3′ overhangs. Telomerase then adds G4T4 telomeric repeats to the 3′ end of the MAC-destined sequence, and a DNA polymerase synthesizes the complementary C4A4 strand. The spacer DNA segment is degraded soon after chromosome fragmentation. (B) The two possible arrangements of adjacent MAC-destined sequences (black rectangles) in the MIC chromosomes are shown. Adjacent MAC-destined sequences may overlap by 6 bp, such that a single E-Cbs (triangle) specifies chromosome fragmentation and both ends receive telomeres (TEL). More typically, adjacent MAC-destined sequences are separated by eliminated spacer DNA (thin line), such that two E-Cbs elements are required to specify chromosome fragmentation at the two MAC-destined DNA ends. The E-Cbs can reside either within the MAC-destined DNA end, or in the flanking spacer DNA.
In the present work, we examine the question of whether the developmentally eliminated spacer DNA might also undergo de novo telomere addition during MAC development. Adjacent MAC-destined sequences sometimes overlap by 6 bp in E.crassus (11,12), in which case a single E-Cbs specifies chromosome fragmentation and both of the resulting ends undergo de novo telomere formation (Fig. 1B). The more common situation is for adjacent MAC-destined sequences to be separated by a short spacer segment that is eliminated from the genome soon after chromosome fragmentation (Fig. 1B). In these cases, an E-Cbs is positioned adjacent to each MAC-destined sequence end.
The basis for retention of the MAC-destined DNA and the elimination of the spacers is not known. However, one of the functions of telomeres, in concert with telomere-binding proteins, is to protect free chromosome ends from degradation and recombination (reviewed in 16). As a result, the spacers might be degraded because they fail to undergo de novo telomere addition. The in vivo characterization of chromosome fragmentation intermediates suggests that this notion may not be correct. Specifically, in the case of overlapping MAC-destined sequences, both DNA ends generated by chromosome fragmentation undergo de novo telomere addition (Fig. 1B). Moreover, the spacer DNA ends appear to have the same terminal sequence organization as the MAC-destined sequence following fragmentation (15). As a result, if the terminal sequence organization of the DNA alone is important for recognition by telomerase, one would expect that spacer DNAs would undergo de novo telomere addition. In the present work, we characterized three spacer intermediates after fragmentation using a PCR-based procedure. In all cases we observed spacer DNAs with telomeres prior to their degradation during MAC development. In contrast to MAC-destined sequences, where telomere addition is precise to the base (11), the spacers have multiple telomere addition sites. This heterogeneity resembles the type of microheterogeneity in telomere addition to MAC-destined sequences reported for other spirotrichs, and oligohymenophoran ciliates such as Tetrahymena and Paramecium (reviewed in 2,4).
MATERIALS AND METHODS
Growth of cells and DNA isolation
All studies employed E.crassus strains X1 and X2 (17). Cells were grown in artificial seawater prepared using Reef Crystals (Aquarium Systems, Mentor, OH) and supplemented as previously described by Roth et al. (18). The algae Dunaliella salina was used as the food source. Matings of strains X1 and X2 were carried out at 24°C as described previously (18,19).
Developing macronuclei were isolated and DNA was purified using published procedures (18,19), except that 3 mM CaCl2 and 50 mM NaCl were used in place of spermidine in nuclear isolation buffers.
Oligonucleotides and linkers
All oligonucleotides were purchased from GIBCO-BRL Life Technologies (Gaithersburg, MD). The names and sequences of the oligonucleotides used in studies (see Fig. 2C) of the addition of telomeres to spacer sequences were: 2FMacC, 5′-CTAGAGGCTTAGATGGAGAG-3′; FMacC, 5′-TGTCAAGCAGCTTTAAGGAC-3′; LMacC-3, 5′-CTATCAAGGAACTGGGATTC-3′; LMacC-4, 5′-TCCATCCTCTAATATCCAGG-3′; RFr29-1, 5′-TCTTTCCTAGTATTTGGATG-3′; RFr29-3, 5′-AGCTTTGAAAGTTAATATTC-3′.
Figure 2.
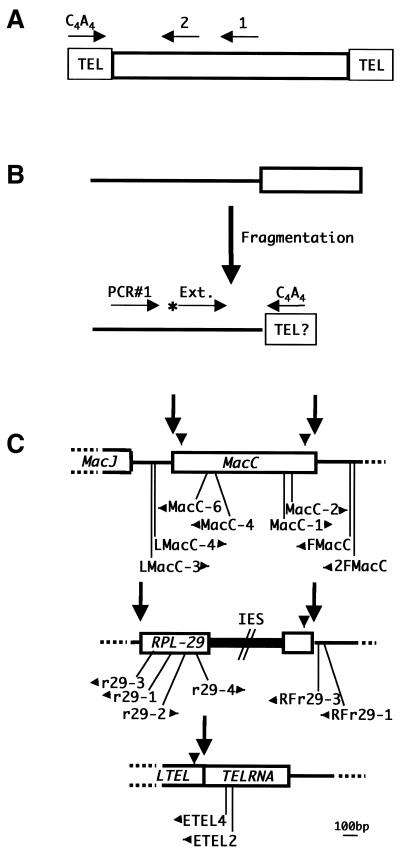
PCR strategies to amplify DNA segments capped by telomeric repeats. (A) The MAC-end PCR strategy is illustrated. PCR is first performed using a synthetic oligonucleotide (primer 1) complementary to an internal segment of a MAC DNA molecule (open rectangle) in combination with an oligonucleotide complementary to the telomeric repeat sequence (C4A4 primer). Specificity is increased by performing a second PCR reaction with a nested oligonucleotide (primer 2) and the C4A4 primer. (B) In spacer-end PCR, an oligonucleotide (PCR#1) complementary to the spacer (thin line) is first used in combination with the C4A4 primer. The products of this reaction are then extended with a radioactively labeled nested primer (Ext.) and displayed on an acrylamide–urea gel. (C) MIC maps of the three loci (MacC, RPL-29 and TelRNA) analyzed in this study are shown. MAC-destined sequences are indicated as open rectangles, spacers as thin lines and an IES as a filled rectangle. Arrows indicate the fragmentation sites analyzed and arrowheads indicate the positions of E-Cbs. The positions of the PCR primers used in the analyses are shown below the maps, with the arrows denoting the directions in which they prime DNA synthesis. The maps are based on previously published data (12), as well as unpublished results (C.Pearson, A.Sanchez-Blanco and L.A.Klobutcher, unpublished data). GenBank accession numbers for the sequences of these loci are: MacJ, AF072708 and AF072707; MacC, AF072708 and AF072706; RPL-29, AF061111, AF061112 and U132070; and LTEL/TelRNA, AF061110 and AF061108.
The following oligonucleotides were used in MAC-end PCR analyses: MacC-4, 5′-CCAAGTTCTGAGCCGTTCTA-3′; MacC-6, 5′-ATCTCTATCACAAAATTATGGC-3′; MacC-1, 5′-ACCCACACAAATGTTCTAGG-3′; MacC-2, 5′-TAGGGCTTACGAAGAATCTG-3′; r29-1, 5′-ATGTCTCATACCGTGCTTTCC-3′; r29-3, 5′-GTGACCTCTGAGTTTCCTCG-3′; r29-2, 5′-AGTTCCTCTCGTTGTCAAGG-3′; r29-4, 5′-AGGTGCCTGTGTATTGAGAG-3′; ETEL2, 5′-CTAAACGCACCTATTGATCG-3′; ETEL4, 5′-TTGTTTGACAGATTTGACGG-3′.
The telomere specific primer (C4A4) was 5′-CCCCAAAACCCCAAAACCCCAAAACCCC-3′.
When noted, oligonucleotides were 5′-end-labeled using [γ-32P]ATP and T4 polynucleotide kinase as described by Ausubel et al. (20).
PCR procedures
The analyses of telomere addition to spacer DNAs involved a first round of PCR employing 50 ng of total cellular DNA from either vegetative cells or from cells at different developing stages as the substrate, and an oligonucleotide complementary to the spacer plus the C4A4 oligonucleotide as the primers. PCR reactions were carried out in a volume of 50 µl using KlenTaq DNA polymerase (Sigma, St Louis, MO), with other components as specified by the manufacturer. Twenty cycles of PCR were carried out, with a cycle consisting of a 95°C denaturation step for 1 min and an annealing/elongation step of 1 min. The temperature for the annealing/elongation step was adjusted based on the GC content of the primers. Five microliters of these PCR reactions were then used as the substrate in subsequent elongation reactions, which were prepared as for PCR, except with a final volume of 25 µl. The primer for the elongation reactions consisted of an oligonucleotide that was 5′-end-labeled with 32P, and that was complementary to the spacer and nested relative to the spacer-specific primer used in the initial PCR reaction. Fifteen cycles of elongation were carried out, and the elongation products were then analyzed by electrophoresis on 20 cm, 8–12% Explorer (J.T. Baker, Phillipsburg, NJ)–urea gels prepared and run in 0.6× TBE. The size standard was the 1 kb Plus DNA Ladder (GIBCO-BRL Life Technologies) 5′-end-labeled with 32P. Following electrophoresis, gels were vacuum-dried and exposed to X-ray film along with an intensifying screen.
To generate sufficient material for sequencing, aliquots of the PCR reactions that generated products were subjected to another round of PCR. The products of the first round of PCR were run on a 0.8–1.2% low melting point (LMP) agarose gel, and the regions of the gel containing products of the expected size, plus 50 bp above as well as below, were cut out. The gel slice was melted by boiling, and 5 µl of the melted gel slice was used as the template for 25 cycles of PCR using the nested spacer oligonucleotide and the C4A4 oligonucleotide as primers. The resulting products were then gel-purified as described by Qian and Wilkinson (21), and either sequenced directly or cloned for further analysis.
MAC-end PCRs were performed with whole-cell DNA from vegetative cells (strains X1 and X2) and developing MAC DNA isolated from cells 48 and/or 60 h after mixing the cells for mating. To increase the specificity of the procedure, ∼2 µg of each DNA was run on a 0.8% LMP agarose gel and the region of the gel containing the target MAC DNA molecules was cut out. The gel slice was melted and used as a template for a first round of PCR (25 cycles) using a MAC-specific primer in combination with the C4A4 oligonucleotide and the conditions described above. The PCR products were then run on a 0.8–1.2% LMP agarose gel and the region of the gel containing DNA of the expected size of the product ±50 bp was cut out. The gel slice DNA was used as the substrate in a second PCR (25 cycles) using a nested MAC-specific oligonucleotide in combination with the C4A4 oligonucleotide. The resulting products were then run on a LMP agarose gel, excised, purified (21) and sequenced.
DNA cloning and sequencing
PCR products were cloned into the pCR®t-TOPO® vector using the TOPO TA Cloning® Kit for Sequencing (Invitrogen, Carlsbad, CA). Sequencing of cloned PCR products, as well as direct sequencing of total PCR products, was carried out by the University of Connecticut Health Center Molecular Core facility using the Taq Dideoxy Termination Cycle Sequencing Kit (Perkin Elmer Cetus, Norwalk, CT). The T3 sequencing primer was used for sequencing of clones, while the nested oligonucleotides were used for bulk sequencing of MAC-end and spacer-end PCR products.
RESULTS
Detection of telomere repeat addition to spacer DNA
To address the question of whether telomeric repeats are added to spacer DNAs prior to their elimination during MAC development, we used an approach similar to the ‘MAC-end PCR’ procedure (Fig. 2A) that has been used to isolate the ends of MAC DNA molecules (12,22). In this method, PCR is performed on cellular DNA using one primer that is complementary to the specific MAC DNA molecule to be analyzed (primer 1 in Fig. 2A) in combination with a primer that is complementary to the telomeric repeat sequence (C4A4 primer). Since all MAC DNA molecules are bounded by telomeres, the telomeric repeats essentially serve as a universal primer-binding site, allowing the ends of any MAC DNA molecule to be amplified if a small segment of internal sequence is known. The specificity of the procedure is greatly enhanced by carrying out a second round of PCR using a nested primer (primer 2) in combination with the telomeric repeat primer.
The analysis of spacer ends employed a similar approach, which we will refer to as ‘spacer-end PCR’, on DNA isolated from cells at various stages of MAC development (Fig. 2B). A first round of PCR was performed using a primer complementary to the spacer (PCR#1 in Fig. 2B) in combination with the C4A4 primer. An aliquot of this reaction was then used as the substrate for a multi-cycle extension reaction employing a nested oligonucleotide that was labeled at its 5′ end with 32P (primer Ext.), and the products were displayed on acrylamide–urea gels. PCR products of the expected size (i.e. based on the position of the nested primer relative to the end of the fragmentation intermediate predicted by the position of the E-Cbs sequence) are expected only if telomeric repeats are added to the ends of the spacers following chromosome fragmentation.
The three spacers that were analyzed by this approach are shown in Figure 2C, along with the names of the spacer-specific oligonucleotides employed in the analyses. Both spacer ends flanking the MIC copy of the MacC MAC DNA molecule (12,23), which contains a gene of unknown function, were analyzed, along with the spacer end abutting the right end of the MAC-destined sequence that contains the gene for ribosomal protein L29 (RPL-29) (12,24).
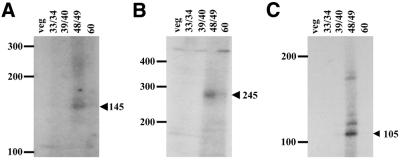
The extension products from the spacer-end PCRs of these three spacers are shown in Figure 3. For each spacer, we observed no products in the expected size range when vegetative cell DNA or DNAs prepared from cells at 33/34 or 39/40 h of development (times represent hours after mixing cells of complementary mating types to initiate conjugation) were used as the substrate. However, diffuse bands of products of approximately the expected size were observed with DNA isolated from cells at 48/49 h of development. This represents the end of the polytene chromosome stage of MAC development and the period during which chromosome fragmentation/telomere addition is initiated (15). For both of the MacC spacers (Fig. 3A and B), weaker smears of products are also visible at 60 h of development. These signals are likely due to DNA from a subpopulation of cells that mated late in the cultures used to purify developing macronuclei, and thus proceed through development asynchronously from the bulk of the population. Overall, these results provide evidence that some spacer sequences undergo de novo telomere addition transiently, and in a developmental-specific manner, during MAC development.
Figure 3.
Spacer-end PCR analyses. Autoradiographs of urea–acrylamide gels containing the extension reaction products from spacer-end PCRs of the left spacer of MacC (A), the right spacer of MacC (B) and the right spacer of RPL-29 (C) are shown. The positions of size standards (in bases) are indicated on the left. The sizes and positions of the expected spacer-end PCR products are indicated by the arrowheads on the right. In each case, the spacer end PCR procedure was performed on total DNA from vegetative cells (lane veg), as well as on total DNA from cells undergoing MAC development at the times (h) indicated (lanes 33/34, 39/40, 48/49 and 60). Images were obtained using a Kaiser RA1 digital camera and AlphaImager 2200 version 5.1 software, and the brightness of images was adjusted using Microsoft PowerPoint software (version 8.0).
Analysis of telomere addition sites for spacer DNAs
For each of the spacer ends analyzed, a smear of products was observed, rather than a distinctly sized band. For example, in the case of the left spacer of MacC, the expected size of a spacer-end PCR product that contains 28 bases of telomeric repeats at its end (the length of the C4A4 oligonucleotide) was 145 bases, while a smear of products ranging from approximately 140–162 bases is observed (Fig. 3A). In addition, for the right spacer of RPL-29 (Fig. 3C), there is not only a smear of products of about the expected size of 105 bases, but also some discretely sized larger products. There are two potential explanations for these observations. First, telomere addition to spacer DNA may occur at multiple sites. Secondly, some variability in products likely results from the spacer-end PCR strategy. Given that the telomeres are tandem arrays of an eight-base sequence, the C4A4 primer can hybridize with telomeric ends at multiple positions. Moreover, the newly synthesized telomeres of MAC-destined sequences are longer (∼80 bp) than in vegetative cells (14,25), and this may also be the case for spacer telomeres.
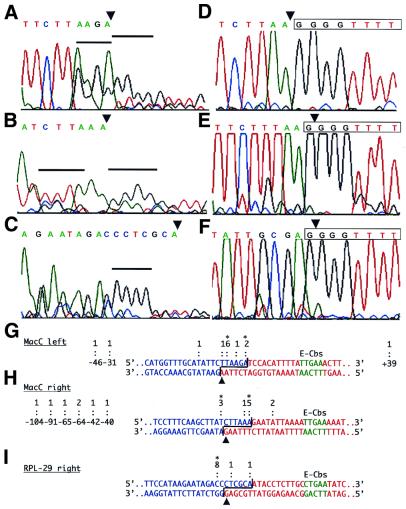
Two approaches were employed to determine if there were multiple telomere addition sites for spacer DNAs. The first was direct or bulk sequencing of PCR products. To obtain sufficient DNA for direct sequencing, the first round spacer-end PCR products were resolved on a LMP agarose gel, and a section of the gel containing DNA in the size range of the expected first round PCR product ±50 bp was excised. This DNA was then used in a second round of PCR utilizing the nested oligonucleotide in combination with the C4A4 oligonucleotide. The resulting products were then sequenced. For each of the spacer ends, a relatively clear pattern of peaks was seen on the sequencing electropherogram that corresponded to the expected spacer sequence until near the predicted end of the fragmentation intermediate (Fig. 4A–C). At this point the quality of the sequences degenerated, as one would expect for mixtures of spacers with telomeres added at different positions. Indeed, runs of the telomeric sequence G4T4 are evident. Close inspection of the sequencing electropherograms, and comparison with the MIC sequences of the fragmentation regions, led us to interpret the results as indicating that there were at least two predominant positions of telomeric repeat addition for each of the MacC spacer ends, and a single predominant position for the RPL-29 right-end spacer (Fig. 4A–C and G–I). For the spacer ends at the left and right sides of MacC, one of the predominant positions appears to be at the end of the predicted fragmentation intermediate (i.e. at the end of the six-base, 3′ overhang). The second major position for the MacC right spacer end occurs at position –7 (the position corresponding to the 3′ end of the spacer fragmentation intermediate is designated –1, with negative numbering into the spacer), which corresponds to the base just prior to the start of the six-base, 3′ overhang of the predicted fragmentation intermediate. The second position for the MacC left spacer is at position –5, within the predicted 3′ overhang of the fragmentation intermediate. In the case of the RPL-29 right spacer, the single discernible telomere addition site is at position –8, which would be just internal to the 3′ overhang of the fragmentation intermediate.
Figure 4.
Sequence analyses of spacer-end and MAC-end PCR products. (A–C) Electropherograms from the direct sequencing of the spacer-end PCR products from 48/49 h DNA from the left spacer of MacC (A), the right spacer of MacC (B) and the right spacer of RPL-29 (C) are shown. The sequence above each electropherogram represents that of each spacer up to the 3′ end of the fragmentation intermediate. In each case, the observed sequence degenerates in quality approaching the end of the predicted fragmentation intermediate, suggesting that the PCR products represent a mixture with telomeres added at different positions. Blocks of four consecutive G residues that we interpret as the beginning of the telomeric repeats for a major subset(s) of the PCR products are indicated by the black horizontal bars. (D–F) Electropherograms from the sequencing of the MAC-end PCR products obtained from 48 h developing MAC DNA for the left and right ends of MacC and the right end of the RPL-29 MAC DNAs, respectively. The sequence of the region is given above the electropherogram, with the first telomeric repeat boxed. Note that for the MacC right end and the RPL-29 right end, the fragmentation intermediates end with a G residue, and this becomes part of the first telomeric repeat (12). (G–I) Summaries of the results from the bulk sequencing of PCR products and cloned PCR products. The MIC DNA sequences at the left and right ends of the MacC MAC-destined sequence and the right end of the RPL-29 MAC-destined sequence are shown. Based on the position of the E-Cbs core (green), and previous analyses (15), bases forming the spacer segment after fragmentation are shown in blue, whereas bases that form the MAC-destined sequence are shown in red. The brackets denote the six-base, 3′ overhangs of the fragmentation intermediates. Arrowheads below the sequences denote the positions of telomere addition (i.e. the nucleotide to which telomeric repeats are added) for the MAC-destined sequences based on bulk sequencing of MAC-end PCR products. Telomere addition sites for the spacers are indicated above each sequence: colons denote telomere addition sites inferred from sequencing cloned spacer-end PCR products, with the numbers above each colon indicating the number of clones obtained for each observed site; and asterisks denote the major telomere addition sites inferred from the bulk sequencing of PCR products. Also indicated to the left and right of the sequences are more distal positions of telomere addition observed in the clones. Numbering (e.g. –46) is based on designating the 3′ base of the spacer fragmentation intermediate as –1, with negative numbering extending into the spacer, and positive numbering into the MAC-destined sequence.
To more precisely determine sites of telomere addition, we also cloned the second-round PCR products. For each spacer end, at least 10 clones whose insert sizes were approximately the size expected for a spacer fragmentation intermediate with telomeric repeats were individually sequenced. The sequences revealed that all of the cloned inserts corresponded to spacer DNA with telomeric repeats at their ends, but multiple telomere addition sites were observed among the clones for each spacer examined. The results are summarized in Figure 4G–I. In each case we observed at least one clone with telomeres added to the 3′ end of the predicted fragmentation intermediate, as well as multiple clones for each of the other major telomere addition sites that we deduced from the bulk sequencing analysis. However, additional telomere addition sites were also observed, most of which correspond to positions within the 3′ overhangs of the predicted fragmentation intermediates. In addition, for the right end spacer of MacC we obtained two clones that had telomeres added at position +5, a position that we would not normally expect to be present in the spacer (Fig. 4H). Intriguingly, the +5 position is 17 bp away from a perfect match to the 5′-TTGAA-3′ core of the E-Cbs sequence, suggesting that these clones could represent telomere addition to the 3′ ends of a more rarely used fragmentation site.
In addition to clones with inserts of approximately the expected size, we also observed some spacer-end PCR product clones for the MacC spacer ends with much smaller inserts, and, in one case, a significantly larger insert. A number of these were selected and sequenced, and all again contained the expected spacer sequences with telomeric repeats at their ends. For the MacC left spacer end, three clones were analyzed and they had telomere addition sites at positions –46, –31 and +39 (Fig. 4G). For the MacC right spacer end, seven clones were analyzed and they all showed telomere addition at internal positions in the spacer ranging from –104 to –40 (Fig. 4H).
Overall, the sequencing results indicate that the predominant positions for telomere addition exist in the vicinity of the overhang of the fragmentation intermediates, but that telomere addition can also occur at more internal regions of the spacers and more rarely at positions that would normally be expected to form part of the MAC DNA molecule. These results suggest to us that the spacers are capable of serving as the substrates for de novo telomere addition, but that they are often subjected to nuclease degradation before the telomere addition process, and that the 3′ overhangs of the spacer fragmentation intermediates are particularly susceptible to degradation. The one spacer end that is somewhat at odds with this interpretation is the one at the right end of RPL-29, where the bulk of telomere addition appears to occur just prior to the 3′ overhang (Fig. 4I). We suspect that this spacer terminus may be particularly susceptible to nuclease degradation prior to telomeric repeat addition.
Telomere addition to MAC-destined sequences
A number of aspects of the above analyses led us to re-address the issue of the fidelity of telomere addition to MAC-destined sequences. The conclusion that telomere addition to MAC-destined sequences is precise rests on two types of analyses. First, a hybrid selection procedure was initially used to isolate and characterize MAC DNA termini (11). For five different MAC DNA ends, the results indicated unique sites of chromosome fragmentation/telomere addition and that these sites were reproducibly used in independent episodes of MAC development. Secondly, in both our laboratory and other laboratories, multiple independent recombinant clones of particular MAC DNA molecules have often been sequenced, and no variability in the position of telomere addition has ever been reported. Nonetheless, the observation of heterogeneity in telomeric addition sites for the spacer DNA, including some cases where short segments of the MAC-destined DNA were present (e.g. the two MacC right spacer clones with telomere addition sites at the +5 position; Fig. 4H), led us to ask whether the MAC-destined DNA ends adjacent to the spacer ends might also display a corresponding level of variability in telomere addition sites.
To address this we performed MAC-end PCR on both ends of MacC and the RPL-29 right end using the primers indicated in Figure 2C. Bulk sequencing of the PCR products obtained using E.crassus strains X1 and X2 vegetative cell DNA each produced a clear sequence with telomeric repeats beginning at a position corresponding to the 3′ end of the fragmentation intermediate (data not shown; see Fig. 4G–I). While we obtained no evidence for shortened MAC DNA ends that would be the complement of the abnormally long spacer DNAs with telomeres in vegetative cell DNA, it remained possible that such forms are generated during MAC development, but for some reason are not retained in the mature MAC. To investigate this we carried out the same MAC-end PCR analyses on DNA isolated from purified developing macronuclei at 48 and 60 h after mating, the period when new MAC DNA molecules are forming. Again we observed no evidence for a significant number of MAC DNA molecules with telomeres at positions other than the expected site (Fig. 4D–I and data not shown). These results indicate that if any variant MAC DNA termini exist, they must be quite rare.
Role of the E-Cbs in precise telomere addition
The tight coupling of chromosome fragmentation and de novo telomere formation has led to suggestions that the enzymatic activities necessary to carry out these processes may reside in a complex (15,26). One possible explanation for our results is that such a hypothetical complex acts asymmetrically. That is, a complex containing both telomerase and the fragmentation activity would be recruited to the sites of chromosome fragmentation via interaction with the E-Cbs sequence. Following DNA cleavage, the terminus containing the E-Cbs would retain the complex and telomerase would be able to directly initiate telomeric repeat addition onto this end. The complementary end, lacking the bound complex, would be susceptible to the action of nucleases, until a free telomerase recognizes the end to initiate telomere synthesis. Since the E-Cbs sequence resides within the MAC-destined sequence ∼75% of the time (12), one would expect the majority of MAC DNA ends to display unique positions of telomere addition under this model, while most spacer ends would display multiple telomere addition sites. Indeed, for the three spacer ends analyzed in this study, the E-Cbs is located within the MAC-destined sequence (Fig. 2C).
To test the notion that the E-Cbs sequence is correlated with precise telomere addition, we used the MAC-end PCR procedure to analyze ends of two MAC DNA molecules where the E-Cbs is located in the flanking spacer DNA (Fig. 2C): the left end of the MAC DNA molecule containing the gene for the RNA component of telomerase (TelRNA) (12,27) and the left end of the RPL-29 MAC DNA molecule (12,24). MAC-end PCR was performed on vegetative DNA from both E.crassus strains X1 and X2, as well as on developing MAC DNA isolated 48 h after mating these two strains. Direct sequencing of the products from these three MAC-end PCR reactions for the left end of the TelRNA MAC DNA molecule are shown in Figure 5. In each case we observed a clear sequence with no evidence of variability in the position of telomeric repeat addition. The same result was obtained in the analysis of the left end of the RPL-29 MAC DNA molecule (data not shown). Moreover, the end of an additional MAC DNA molecule lacking an E-Cbs was previously analyzed by a hybrid selection procedure, and it too was found to display precise telomere addition (11). Thus, it appears that the presence of an E-Cbs on a MAC-destined fragmentation intermediate is not required for precise telomere addition.
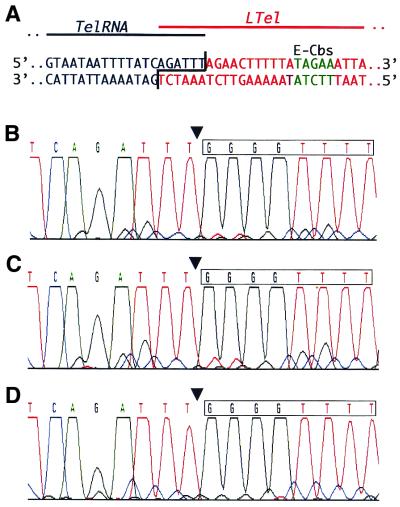
Figure 5.
MAC-end PCR analyses of ends lacking an E-Cbs. (A) The MIC sequence of the overlapping TelRNA and LTel Mac-destined sequences are shown. Bases that will form MAC TelRNA are shown in black, those that will form MAC LTel are shown in red, and the E-Cbs core is shown in green. (B–D) Electropherograms from the bulk sequencing of MAC-end PCR products for the left end of the TelRNA MAC DNA molecule from 48 h developing MAC DNA (B), E.crassus strain X1 whole cell DNA (C) and E. crassus strain X2 whole cell DNA (D) are shown. The sequence is given above the electropherogram, with the position of telomere addition indicated by the arrowhead and the first telomeric repeat boxed.
DISCUSSION
In this study we provide evidence that the developmentally eliminated spacers that separate MAC-destined DNA sequences can undergo de novo telomere addition during the chromosome fragmentation stage of MAC development. However, analysis of the spacers with telomeres indicates that the position of telomere addition is variable. While telomeres were sometimes added to positions corresponding to the ends of the six-base, 3′ overhangs generated by chromosome fragmentation, addition of telomeres to positions ‘within’ the 3′ overhangs were also frequently observed. Moreover, we also identified numerous situations in which telomeric repeats were added to positions within the spacer DNA that were quite distant from the site of chromosome fragmentation. The variability of telomere addition sites for spacers is in stark contrast to the situation observed for the ends of MAC-destined sequences. The MAC-end PCR analyses reported here, along with previous analyses (11), provide a clear indication that telomere addition to MAC-destined DNA sequences is precise. This level of fidelity of telomere addition to MAC-destined sequences is unique to Euplotes, but the observed variability in spacer DNA telomere addition sites is very reminiscent of the situation for de novo telomere addition to MAC-destined sequences in other ciliates, including Tetrahymena, Paramecium, and other non-euplotid spirotrichs (reviewed in 2,4).
Telomere addition to spacer DNA segments was detected via a sensitive PCR procedure. While it is possible that the observed products represent some form of PCR artifact, at least two observations argue against this possibility. First, the products are observed in a developmental-specific manner. They are seen primarily at the time of chromosome fragmentation, the same period when MAC-destined sequences are undergoing de novo telomere formation, but not at earlier stages of development or from DNA isolated from vegetatively growing cells. Secondly, sequencing of the PCR products provides no evidence that they arise from ‘mis-priming’ events by the C4A4 telomeric repeat primer on unfragmented DNA forms. This primer has four C residues at its 3′ end, so that if mis-priming were occurring, we would expect to often observe multiple G residues in the template strand at positions adjacent to those where we observe telomeric repeats in our clones. In fact, of the 21 telomere adddition sites defined through the analysis of clones, there are only two cases where even a single G residue was observed.
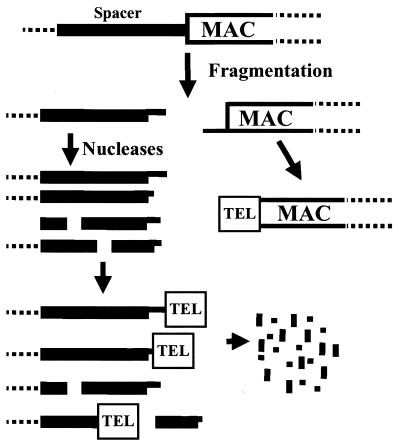
We note that the PCR approach is not a quantitative one, and that it would indeed be difficult to directly address the abundance of spacer DNAs with telomeres due to the fact that the genome complexity is in flux during MAC development and because the spacers are rapidly degraded following chromosome fragmentation. As a result, the analyses presented must be viewed as providing evidence that at least some spacer DNAs, but not necessarily all, undergo de novo telomere addition during MAC development. Taking this into consideration, Figure 6 provides a model of the fate of spacer DNA segments that is consistent with our results. Following chromosome fragmentation, both spacer DNA and MAC-destined DNA have the same terminal structure: six-base, 3′ overhangs. The MAC-destined DNA is either protected from degradation or quickly becomes a substrate for telomerase, resulting in the de novo addition of telomeric repeats to the terminal base of the overhang. In contrast, spacer segments are more readily attacked by nucleases prior to interacting with telomerase. The six-base, 3′ overhang is particularly susceptible to nuclease attack, but internal regions of the spacer are susceptible as well. Either an exonuclease or an endonuclease, or a combination of both, could be responsible for the degradation of spacer DNA. During this process, at least some of the spacers are recognized by telomerase, but all spacer DNA is ultimately degraded by nucleases. Finally, we note that the addition of telomeres to developmentally eliminated DNA likely extends beyond Euplotes. Muller and Tobler (1) refer to unpublished data indicating a similar phenomenon during the chromatin diminuition process in the nematode Ascaris.
Figure 6.
Model of telomere addition to MAC-destined and spacer DNA. Following chromosome fragmentation, MAC-destined sequences undergo de novo telomere addition to the 3′ end of the fragmentation intermediate. In contrast, both the 3′ overhang and internal regions of the spacer DNA are attacked by exonucleases, with some of the resulting ends undergoing de novo telomere addition. Ultimately, the spacer DNAs are completely degraded.
Implications for de novo telomere formation
It is clear that telomerase is responsible for de novo telomere formation during MAC development in Tetrahymena (9), and this is likely to be true of other ciliates as well. Nonetheless, it has been difficult to completely reconstruct de novo telomere formation in vitro. For both T.thermophila and E.crassus, de novo addition of telomeric repeats to single-stranded oligonucleotides with non-telomeric repeat termini has been demonstrated in vitro (26,28,29). In E.crassus, only telomerase from developing cells, which exists in higher molecular weight complexes compared with vegetative cell telomerase, is capable of this form of de novo telomere synthesis. However, it has not been possible to reconstruct de novo telomere formation using double-stranded DNA, the likely substrate for this reaction in vivo.
It was previously suggested that the specific terminal structure of the fragmentation intermediate (i.e. the six-base, 3′ overhang) might be required for recognition by telomerase for de novo telomere formation (15). The current results suggest this may not be the case. As discussed, the observed multiple positions of telomere addition for spacer DNA is most easily explained by nucleases that partially degrade spacers. It is unlikely that this partial degradation maintains six-base, 3′ overhangs at all ends, particularly for the preferred positions of addition within the fragmentation intermediate overhangs (Fig. 4G–I). As a result, our results suggest that the de novo telomere addition activity is quite promiscuous, initiating telomere repeat addition on both retained and eliminated sequences with a variety of terminal structures.
Differentiating MAC-destined and spacer DNAs
The observation of spacer DNAs with telomeric repeats makes it unlikely that differential de novo telomere addition is responsible for discriminating between MAC-destined and eliminated DNA sequences. Another way discrimination might be accomplished is through chromatin structure. Changes in chromatin structure have been implicated in other DNA rearrangement processes, such as mating type switching in yeast (30) and the V(D)J recombination process that occurs in cells of the vertebrate immune system (31). In the case of ciliates, the role of chromatin structure in chromosome fragmentation has not been examined, but evidence is accumulating that indicates chromatin structure is involved in the DNA splicing, or internal eliminated sequence (IES) excision, aspect of MAC development. In T.thermophila, excised DNA segments localize to heterochromatin-like structures in the developing MAC following DNA rearrangement (32). Moreover, two developmental-specific proteins (Pdd1p and Pdd3p) co-localize with the eliminated DNA (32,33), and these proteins possess chromodomains, motifs that have been associated with heterochromatin-associated proteins in other organisms. While these proteins have not been identified in E.crassus, there are other indications that specialized chromatin may be involved in IES excision in this organism. First, variant histone proteins have been identified that are expressed specifically during development, and that are targeted to the developing MAC (34,35). Secondly, large transposon IESs in E.crassus have been shown to assume an alternative chromatin structure in which the spacing of nucleosomes is altered and the chromatin is highly resistant to nuclease digestion, whereas the MAC-destined sequences do not display these properties (34,36).
Changes in chromatin structure could also help to explain the different fates of MAC-destined and spacer DNAs. Specifically, we suggest that the MAC-destined sequences exist in a chromatin structure that protects them from degradation by endogenous nucleases, whereas the spacer DNA exists in an alternative chromatin conformation that renders them accessible to nuclease degradation. This type of model is attractive because it could help explain at least two puzzling aspects of ciliate chromosome fragmentation. First, in E.crassus, one can find good matches to the E-Cbs within MAC-destined sequences, yet these regions are not subject to chromosome fragmentation. It may be that the proposed protective chromatin structure shields these sites from recognition by the fragmentation machinery.
Secondly, the hypothesis can be extended to provide an explanation for the differences in the fidelity of chromosome fragmentation/telomere addition process among ciliates. In an organism like E.crassus, it may be that there is a close correspondence between the boundaries of the protective chromatin structure for MAC-destined sequences and the positions of the functional E-Cbs, such that essentially no degradation occurs following chromosome fragmentation. In other organisms, the boundaries of the protective MAC-destined sequence chromatin structure may not closely abut the sequence element(s) that specify chromosome fragmentation. As a result, variable amounts of nuclease-mediated erosion of the ends might occur prior to the initiation of de novo telomere formation. We note that in the spirotrichs, the gene-sized MAC DNA molecules typically have short 5′- and 3′-non-coding regions, but that the Euplotes species have the shortest non-coding regions within this group (4). This may be a reflection of an extremely pared-down E.crassus genome, in which the proposed chromatin-structure boundaries have come to reside in close proximity to the fragmentation signals. Further studies assessing chromatin structure during the period of chromosome fragmentation in different ciliates would provide information critical for assessing the validity of this model.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mary Ellen Jacobs for many helpful discussions. This work was supported by National Science Foundation grant MCB-9816765 to L.A.K.
REFERENCES
- 1.Muller F. and Tobler,H. (2000) Chromatin diminution in the parasitic nematodes Ascaris suum and Parascaris univalens. Int. J. Parisitol., 30, 391–399. [DOI] [PubMed] [Google Scholar]
- 2.Coyne R.S., Chalker,D.L. and Yao,M.-C. (1996) Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet., 30, 557–578. [DOI] [PubMed] [Google Scholar]
- 3.Klobutcher L.A. and Herrick,G. (1997) Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol., 56, 1–62. [DOI] [PubMed] [Google Scholar]
- 4.Prescott D.M. (1994) The DNA of ciliated protozoa. Microbiol. Rev., 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baroin A., Prat,A. and Caron,F. (1987) Telomeric site position heterogeneity in macronuclear DNA of Paramecium primaurelia. Nucleic Acids Res., 15, 1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forney J.D. and Blackburn,E.H. (1988) Developmentally controlled telomere addition in wild-type and mutant Paramecia. Mol. Cell. Biol., 8, 251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrick G., Hunter,D., Williams,K. and Kotter,K. (1987) Alternative processing during development of a macronuclear chromosome family in Oxytricha fallax. Genes Dev., 1, 1047–1058. [DOI] [PubMed] [Google Scholar]
- 8.Yao M.-C., Yao,C.-H. and Monks,B. (1990) The controlling sequence for site-specific chromosome breakage in Tetrahymena. Cell, 63, 763–772. [DOI] [PubMed] [Google Scholar]
- 9.Yu G.L. and Blackburn,E.H. (1991) Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell, 67, 823–832. [DOI] [PubMed] [Google Scholar]
- 10.Jonsson F., Steinbruck,G. and Lipps,H.J. (2001) Both subtelomeric regions are required and sufficient for specific DNA-fragmentation during macronuclear development in Stylonychia lemnae. Genome Biol., 2, research0005.1–0005.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baird S.E. and Klobutcher,L.A. (1989) Characterization of chromosome fragmentation in two protozoans and identification of a candidate fragmentation sequence in Euplotes crassus. Genes Dev., 3, 585–597. [DOI] [PubMed] [Google Scholar]
- 12.Klobutcher L.A., Gygax,S.E., Podoloff,J.D., Vermeesch,J.R., Price,C.M., Tebeau,C.M. and Jahn,C.L. (1998) Conserved DNA sequences adjacent to chromosome fragmentation sites in Euplotes crassus. Nucleic Acids Res., 26, 4230–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan X. and Price,C.M. (1997) Coordinate regulation of G- and C-strand length during new telomere synthesis. Mol. Biol. Cell, 8, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roth M. and Prescott,D.M. (1985) DNA intermediates and telomere addition during genome reorganization in Euplotes crassus. Cell, 41, 411–417. [DOI] [PubMed] [Google Scholar]
- 15.Klobutcher L.A. (1999) Characterization of in vivo developmental chromosome fragmentation intermediates in Euplotes crassus. Mol. Cell, 4, 695–704. [DOI] [PubMed] [Google Scholar]
- 16.Blackburn E.H. and Greider,C.W. (eds) (1995) Telomeres. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Frels J.S. and Jahn,C.L. (1995) DNA rearrangements in Euplotes crassus coincide with discrete periods of DNA replication during the polytene chromosome stage of macronuclear development. Mol. Cell. Biol., 15, 6488–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roth M., Lin,M. and Prescott,D.M. (1985) Large scale synchronous mating and the study of macronuclear development in Euplotes crassus. J. Cell Biol., 101, 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tausta S.L. and Klobutcher,L.A. (1990) Internal eliminated sequences are removed prior to chromosome fragmentation during development in Euplotes crassus. Nucleic Acids Res., 18, 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (2001) Current Protocols in Molecular Biology. John Wiley and Sons, New York, NY.
- 21.Qian L. and Wilkinson,M. (1991) DNA fragment purification. Biotechniques, 10, 736–737. [PubMed] [Google Scholar]
- 22.Seegmiller A., Williams,K.R., Hammersmith,R.L., Doak,T.G., Messick,T., Witherspoon,D., Storjohann,L.L. and Herrick,G. (1996) Internal eliminated sequences interrupting the Oxytricha 81 locus: allelic divergence, conservation, conversions and possible transposon origins. Mol. Biol. Evol., 13, 1351–1362. [DOI] [PubMed] [Google Scholar]
- 23.Jahn C.L., Nilles,L.A. and Krikau,M.F. (1988) Organization of the Euplotes crassus micronuclear genome. J. Protozool., 35, 590–601. [DOI] [PubMed] [Google Scholar]
- 24.Jahn C.L., Erbeznik,M., Jaraczewski,J.W., Melek,M. and Shippen,D.E. (1994) Sequence of the macronuclear DNA encoding large subunit ribosomal protein 29 (L29) in Euplotes crassus and cycloheximide sensitivity. Gene, 151, 231–235. [DOI] [PubMed] [Google Scholar]
- 25.Vermeesch J.R. and Price,C.M. (1993) Telomeric DNA sequence and structure following de novo telomere synthesis in Euplotes crassus. Mol. Cell. Biol., 14, 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melek M., Greene,E.C. and Shippen,D.E. (1996) Processing of nontelomeric 3′ ends by telomerase: default template alignment and endonucleolytic cleavage. Mol. Cell. Biol., 16, 3437–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shippen L.D. and Blackburn,E.H. (1990) Functional evidence for an RNA template in telomerase. Science, 247, 546–552. [DOI] [PubMed] [Google Scholar]
- 28.Wang H. and Blackburn,E.H. (1997) De novo telomere addition by Tetrahymena telomerase in vitro. EMBO J., 16, 866–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bednenko J., Melek,M., Greene,E.C. and Shippen,D.E. (1997) Developmentally regulated initiation of DNA synthesis by telomerase: evidence for factor-assisted de novo telomere formation. EMBO J., 16, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasmyth K. (1982) The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell, 30, 567–578. [DOI] [PubMed] [Google Scholar]
- 31.Stanhope-Baker P., Hudson,K.M., Shaffer,A.L., Constantinescu,A. and Schlissel,M.S. (1996) Cell type-specific chromatin structure determines the targeting of V(D)J recombination activity in vitro. Cell, 85, 887–897. [DOI] [PubMed] [Google Scholar]
- 32.Madireddi M.T., Coyne,R.S., Smothers,J.F., Mickey,K.M., Yao,M.-C. and Allis,C.D. (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell, 87, 75–84. [DOI] [PubMed] [Google Scholar]
- 33.Nikiforov M.A., Gorovsky,M.A. and Allis,C.D. (2000) A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol., 20, 4128–4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jahn C.L., Ling,Z., Tebeau,C.M. and Klobutcher,L.A. (1997) An unusual histone H3 specific for early macronuclear development in Euplotes crassus. Proc. Natl Acad. Sci. USA, 94, 1332–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghosh S. and Klobutcher,L.A. (2000) A developmental-specific histone H3 localizes to the developing macronucleus of Euplotes.Genesis, 26, 179–188. [DOI] [PubMed] [Google Scholar]
- 36.Jahn C.L. (1999) Differentiation of chromatin during DNA elimination in Euplotes crassus. Mol. Biol. Cell, 10, 4217–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]