Abstract
Class 1 release factor in eukaryotes (eRF1) recognizes stop codons and promotes peptide release from the ribosome. The ‘molecular mimicry’ hypothesis suggests that domain 1 of eRF1 is analogous to the tRNA anticodon stem–loop. Recent studies strongly support this hypothesis and several models for specific interactions between stop codons and residues in domain 1 have been proposed. In this study we have sequenced and identified novel eRF1 sequences across a wide diversity of eukaryotes and re-evaluated the codon-binding site by bioinformatic analyses of a large eRF1 dataset. Analyses of the eRF1 structure combined with estimates of evolutionary rates at amino acid sites allow us to define the residues that are under structural (i.e. those involved in intramolecular interactions) versus non-structural selective constraints. Furthermore, we have re-assessed convergent substitutions in the ciliate variant code eRF1s using maximum likelihood-based phylogenetic approaches. Our results favor the model proposed by Bertram et al. that stop codons bind to three ‘cavities’ on the protein surface, although we suggest that the stop codon may bind in the opposite orientation to the original model. We assess the feasibility of this alternative binding orientation with a triplet stop codon and the eRF1 domain 1 structures using molecular modeling techniques.
INTRODUCTION
In the standard genetic code, 61 amino acid codons are recognized by tRNAs via the codon–anticodon interaction. The remaining three codons, UAA, UAG and UGA, are thought to be recognized by proteins called class 1 release factors (RFs) that terminate nascent protein synthesis (1–4). Bacteria use two class 1 RFs, RF1 and RF2, that likely result from an ancient gene duplication in the bacterial lineage. RF1 binds to UAA and UAG codons, while RF2 binds to UAA and UGA codons (1–4). Most eukaryotes and archaea each have single RF proteins, called eRF1 and aRF1, respectively, which recognize all three stop codons (2,5,6). aRF1 and eRF1 show obvious sequence similarity and thus the eukaryotic termination system is thought to have originated from an archaeal-like version in the common ancestor of eukaryotes and archaea (6). This view is strongly supported by the fact that aRF1 can catalyze the release of nascent peptides from eukaryotic ribosomes in response to UAA, UAG and UGA stop codons in vitro (5). Interestingly, sequence similarity between bacterial RFs and the eRF1/aRF1 family is difficult to detect; it is unclear whether the two RF families evolved from a single ancestral protein (2,6).
Extensive mutational studies using Escherichia coli have clearly indicated that three consecutive residues in RF1 and RF2, Pro-Ala-Thr and Ser-Pro-Phe, respectively, govern their codon specificities (7). Molecular studies of the eukaryotic and archaeal termination systems are less advanced and it is still unclear how eRF1 and aRF1 distinguish stop codons from codons specifying amino acids. Presumably, the stop codon-binding sites (and the catalytic centre promoting peptidyl-tRNA hydrolysis) are highly conserved, since accurate eRF1 activities are obviously essential for cell viability. An attractive hypothesis has been formulated in which class 1 RFs mimic tRNAs in terms of their functions and tertiary structures (1,3,8). Indeed, the recently determined human eRF1 structure resembles that of tRNA to a striking degree (9). This structural resemblance implies that eRF1 domains 1 and 2, which are highly conserved among standard code eRF1s (SC-eRF1s), correspond to the anticodon stem–loop and acceptor stem of tRNA, respectively (9).
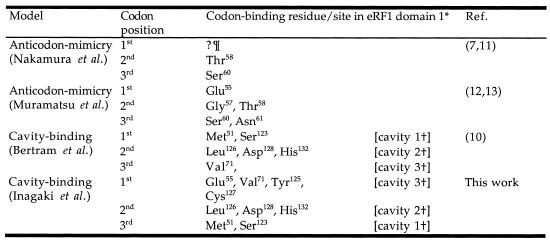
The analogy between the anticodon stem–loop of tRNA and eRF1 domain 1 is further supported by genetic studies using yeast (10). Mutations in yeast eRF1 that abolish recognition of only one of the three stop codons (so called ‘unipotent’ suppressors) are located in domain 1. Molecular modeling by Bertram et al. of the binding of the trinucleotide UAA to human eRF1 has shown that the unipotent suppressor sites were located near surface cavities that could potentially bind the stop codon (10). Others propose, however, that it is the conserved residues in the α2-helix–loop–α3-helix region (henceforth referred to as α2–loop–α3) that form the stop codon-binding site (11–13). Either hypothesis is consistent with the structural similarity shared between eRF1 and tRNA and the fact that these conserved residues occupy a position on eRF1 that is analogous to the tRNA anticodon (11–13). Putative models for the interaction between eRF1 and stop codons are summarized in Table 1.
Table 1. Alternative models for stop codon binding by eRF1.
Domain 2 of eRF1 is believed to promote peptidyl-tRNA hydrolysis at the peptidyltransferase center in ribosomes. At the tip of domain 2 there is a strictly conserved motif, Gly-Gly-Gln (GGQ) (positions 183–185 in human eRF1). Genetic studies in yeast eRF1 showed that only Gln185 is essential for cell viability (9). On the other hand, human proteins with substitutions at positions 183 and 184 could not catalyze peptide release from ribosomes in vitro (14), while the release activity was not lost completely by substitutions at position 185 (15). Although these results from yeast and human eRF1s are incongruent in detail, the GGQ motif clearly plays a critical role in promoting peptidyl-tRNA hydrolysis. Interestingly, the mutations in human eRF1 affected neither ribosome binding nor its interaction with the GTPase subunit eRF3 (14). Domain 3, the least conserved of the three eRF1 domains, is thought to bind to eRF3, since deletions of this domain abolished the eRF1–eRF3 interaction (16–20).
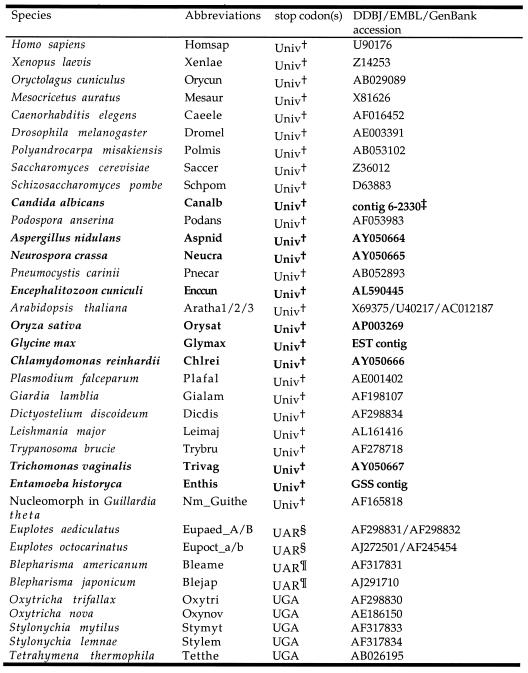
Table 2. eRF1 sequences used in this study.
†The standard genetic code, where UAA, UAG and UGA serve as termination signals.
§R for A or G and UGA for Cys.
¶R for A or G and UGA for Trp.
‡Refer to the Stanford Genome Technology Center website (http://www-sequence.stanford.edu/group/candida). The DNA sequences of the E.histolytica GSS and G.max EST contigs and the putative amino acid sequences are available on request from Y.I. The novel eRF1 sequences determined/identified in this study are shown in bold.
Although translation must be accurately terminated at stop codons, it is well known that stop codons in the standard code have been re-assigned to code for several different amino acids in some bacterial and eukaryotic lineages (reviewed in 21). The ciliates (Ciliophora) are a unicellular eukaryotic group that includes several lineages with variant genetic codes. Although broad-scale ciliate phylogeny remains contentious, evidence suggests that ciliate lineages converted the standard genetic code to variant codes multiple times in evolution (22,23). Species of the class Oligohymenophorea (e.g. Tetrahymena) assigns the UAA and UAG (UAR, R = A or G) codons to Gln, while UGA is reserved as the sole termination signal. However, two different variant codes, UAR for Gln/UGA for stop (UGA = *) and UGA for Cys/UAR for stop (UAR = *), have been reported in the class Spirotrichea (e.g. Oxytricha and Euplotes), which is distantly related to the Oligohymenophorea (22,23). Furthermore, an extensive survey of ciliate gene sequences revealed that the classes Colpodea (e.g. Colpoda) and Heterotrichea (e.g. Blepharisma) utilize the UGA codon for Trp, while UAR codons are retained as termination signals (UAR = *) (23).
In ciliate groups with variant codes, the stop codon-binding site on eRF1 in domain 1 must have been altered to accommodate the variant stop codons. We have previously shown that the domain 1 sequences in variant code eRF1s (VC-eRF1s) from ciliates are indeed significantly diverged from those of SC-eRF1s (24). On the other hand, little sequence divergence was detected in domains 2 and 3 (24). Based on similar sequence comparisons between SC- and VC-eRF1s, conserved residues in the α2–loop–α3 region of domain 1 were independently nominated for the stop codon-binding site by two groups. The Thr-Ala-Ser tripeptide (TAS, positions 58–60 in human eRF1) was proposed by Nakamura and collaborators to be the discriminator region based on analogy to the ‘peptide anticodons’ in bacterial RFs (11). Muramatsu and collaborators, in contrast, hypothesized that a set of residues, Glu55, Gly57, Thr58, Ser60 and Asn61, discriminates stop codons from amino acid codons (13). While the two candidates for the codon-binding site overlap, the stop codon–eRF1 interactions proposed were different in detail. Furthermore, Lozupone et al. found that substitutions at still a third site in ciliate eRF1 domain 1 are shared by variant codes to the exclusion of other SC-eRF1 homologs (23). Interestingly some of these sites were identical to residues forming the putative stop codon-binding cavities proposed by Bertram et al. (10), but completely different from those proposed by Muramatsu et al. or Nakamura et al. (11,13).
Evolutionary comparisons like these should provide important clues to the molecular mechanism of eRF1 codon recognition, by identifying sites that are highly conserved in SC-eRF1s but different in VC-eRF1s. However, ‘highly conserved’ sites have so far been identified by considering a SC-eRF1 dataset that is phylogenetically restricted to relatively few eukaryotic groups (e.g. animals, fungi and plants). Sequences conserved amongst these organisms may in fact be variable when a more diverse set of eukaryotes is considered. Therefore, to determine which sites are absolutely conserved among SC-eRF1s, it is crucial that sampling of the gene be extended to encompass the phylogenetically diverse protists. Data from a variety of protistan SC-eRF1s should permit a more precise definition of functional residues in eRF1, including the stop codon-binding site. Furthermore, it is important to compare VC-eRF1s that have independently evolved similar recognition characteristics. Amino acid assignments that are specifically and universally shared between such VC-eRF1s (and different from those of their standard code ancestors) comprise what we here call ‘converged unvaried sites’ and will be key in identifying amino acid substitutions specifically involved in altered codon recognition.
We sequenced or identified the genes encoding SC-eRF1 from three amitochondrial protists [Trichomonas vaginalis (Parabasalia), Entamoeba histolytica (Entamoebea) and Encephalitozoon cuniculi (Microsporidia)], three fungi (Candida albicans, Neurospora crassa and Aspergillus nidulans), a green alga (Chlamydomonas reinhardii) and two plants (Oryza sativa and Glycine max). Using an eRF1 dataset that included diverse protistan eRF1s, we re-assessed sites that substituted convergently in the eRF1s from ciliates with the UGA = * code (Oxytricha, Stylonychia and Tetrahymena) and the UAR = * code (Euplotes and Blepharisma). Combining evolutionary computational analyses of our eRF1 dataset and detailed analyses of the tertiary structure of human eRF1, we examined candidates proposed for the stop codon-binding site in eRF1 domain 1. Although our observations favor the location of the stop codon-binding site in eRF1 suggested by Bertram et al. (10), we suggest that the stop codon triplet associates with the codon-binding ‘cavities’ in the reverse orientation to their model.
MATERIALS AND METHODS
Novel eRF1 genes
The Neurospora and Apergillus eRF1 cDNA clones (a4b11 and y4b01, respectively) were provided by the Fungal Genetic Stock Center (University of Kansas Medical Center, Kansas). A cDNA clone (TV-0925) of Trichomonas eRF1 was provided by T. M. Embley (Natural History Museum, London, UK). Chlamydomonas cDNA clones CM016g10_r, CM021b05_r and HCL030f11_r were provided by the Kazusa DNA Research Institute (Kisarazu, Chiba, Japan) (25). All clones were sequenced completely on both strands.
The entire Oryza eRF1 gene in clone P0504E02 (GenBank accession no. AP003269) and the Encephalitozoon eRF1 gene on chromosome V (GenBank accession no. AL590445) were identified with TBLASTN (26) against the GenBank non-redundant (nr) database. We also found five genome survey sequences (GSS) from Entamoeba that show strong similarity to eRF1s with TBLASTN in the Genome Survey Database at GenBank. The GSSs were assembled to form a contig covering almost the entire coding region of eRF1.
The putative amino acid sequence of the Glycine eRF1 was obtained from a contig assembled from more than 20 expressed sequence tag (EST) sequences. The eRF1 gene of Candida was identified with TBLASTN (26) and the DNA sequence was obtained from the Stanford DNA Sequencing and Technology Center website (http://www-sequencing.stanford.edu/group/candida). Sequencing of C.albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund.
Phylogenetic analyses
The putative amino acid sequences of the eRF1s that were newly determined or identified in this study were manually added to our previous eRF1 alignment (24). All sequences available as of June 2001 were used for the analyses. Ambiguously aligned sites and sites with a gap(s) were excluded from the initial alignment. The final dataset included 39 eRF1s with 339 aligned amino acid sites [henceforth referred to as the full-length (FL) dataset]. A maximum likelihood (ML) distance matrix was calculated from the FL dataset using TREE-PUZZLE v.4.0.2 (27) employing the JTT amino acid substitution model incorporating among-site rate variation [modeled by a gamma (Γ) distribution approximated with 16 discrete rate categories]. A phylogenetic tree based on this FL dataset was reconstructed from the JTT + Γ distance matrix using FITCH with global rearrangements implemented in PHYLIP v.3.573 (28). Branch lengths for optimal distance trees were estimated by the ML method using the JTT + Γ model implemented in TREE-PUZZLE v.4.0.2 (16 discrete Γ distribution categories). Each node in the distance tree was examined by bootstrap analyses. 500 resampled datasets were generated from the FL dataset using SEQBOOT in PHYLIP v.3.573 and subsequently JTT + Γ ML distance matrices were calculated using TREE-PUZZLE v.4.0.2 and PUZZLEBOOT v.1.02 (distributed by A. J. Roger and M. E. Holder; http://members.tripod.de/korbi/puzzle/) with parameter settings as above. Bootstrap proportions (BPs) were obtained using SEQBOOT, FITCH and CONSENSE in PHYLIP v.3.573. We also examined the tree topology by percent occurrence in 10 000 quartet puzzling trees (QP score), calculated using TREE-PUZZLE v.4.0.2. For all of the analyses above, a shape parameter α = 0.95 was used, obtained by ML estimation on a neighbor-joining tree using TREE-PUZZLE v.4.0.2.
We generated a data subset that included 103 sites in eRF1 domain 1 from the FL dataset. Since four vertebrate, Podospora and Neurospora and two Stylonychia, eRF1 domain 1 sequences are identical, redundant sequences were removed to save computational time. The partial eRF1 sequence of Blepharisma japonicum (GenBank accession no. AJ291710) was included, yielding a final dataset of 35 eRF1s [the domain-1 (D1) dataset]. The FITCH (Γ-FITCH) tree based on a JTT + Γ ML distance matrix for the D1 dataset was estimated with branch lengths optimized as described above. A shape parameter of α = 1.01 was re-estimated from the D1 dataset for this series of analyses.
A SC-eRF1 domain 1 dataset was generated from the D1 dataset by exclusion of 11 ciliate eRF1s. From this SC-eRF1 dataset, we used the conditional mode relative substitution rates of each site (site rates) derived using TREE-PUZZLE v.4.0.2 (27) with a discrete Γ distribution approximated with 16 categories (with shape parameter α = 0.89 estimated from the data). The calculated relative site rates were categorized into five classes and mapped on the tertiary structure of human eRF1 domain 1 (PDB accession no. 1DT9), with a color gradient from blue to red; the slowest evolving (blue; site rate < 0.2000), the second slowest evolving (light blue; 0.2000 < site rate < 0.5000), moderately evolving (white; 0.5000 < site rate < 0.8000), the second fastest evolving (pink; 0.8000 < site rate < 1.3000) and the fastest evolving (red; site rate > 1.3000).
Based on detailed observations of the human eRF1 domain 1 considering relative site rates, we classified the residues corresponding to the slowest evolving sites, where possible, into two functional groups: structural and non-structural. The structural residues included all slowest evolving residues involved in intramolecular interactions with other slowest evolving residues that most likely stabilize the backbone fold of eRF1 domain 1. Slowest evolving residues failing to meet this criterion were deemed non-structural and are potentially responsible for RNA binding and/or codon specificity. All tertiary structures were visualized using VMD v.1.6.1 (29).
Identification of converged unvaried sites using site log-likelihood comparison
Since the tree topologies inferred from the FL and D1 datasets (FL and D1 topologies) were incongruent (see Results and Discussion), log-likelihood (lnL) scores for individual amino acid alignment positions (site lnLs) were calculated from the D1 dataset under the FL and D1 topologies using the JTT + Γ model with eight discrete Γ rate categories (shape parameter α = 1.01) using CODEML implemented in PAML v.3.0 (30). The FL topology was modified by adding B.japonicum to form a clade with Blepharisma americanum and excluding redundant sequences (three vertebrates, Podospora and Stylonychia mytilus). By subtracting the site lnL scores under the FL topology from those under the D1 topology, a number of sites that favored the D1 topology were identified (i.e. those sites with positive ΔlnL scores).
We postulate that some fraction of these had undergone convergent evolution in the ciliate lineages with the same codon specificity (UGA = * or UAR = *). Here we define a subset of such sites we call ‘converged unvaried’ sites to refer to those sites in which an identical residue is shared among the eRF1s with the same codon specificity (i.e. UGA = * or UAR = *) but a different residue is present in their common ancestral sequence. Ancestral sequences of each node in the FL topology were reconstructed using the AAML option of CODEML implemented in PAML v.3.0 (30). The details of the analysis are described in the previous section. Our analyses indicated that seven sites were converged unvaried sites in Oxytricha + Stylonychia and Tetrahymena (UGA = *) or in Euplotes and Blepharisma (UAR = *) (see Results and Discussion). To test whether the presence of these seven converged unvaried sites in the alignment are sufficient to yield the anomalous relationship among ciliates in the D1 tree, Γ-FITCH trees were reconstructed from the D1 dataset: (i) excluding the seven converged unvaried sites; or (ii) excluding the seven sites with negative ΔlnL scores. Phylogenetic analyses were carried out as described above. For these calculations, shape parameters (α) were re-estimated for each new dataset.
Molecular modeling
Stop codon-binding was simulated using residues 28–132 of the human eRF1 structure and a trinucleotide 5′-UAA-3′. The unipotent suppressor data (10) and the converged sites data (above and 23) suggested that the trinucleotide in the domain 1 structure can be modeled in the opposite orientation to that proposed by Bertram et al. (10) (see detailed explanation below). Therefore, the first, second and third bases of the trinucleotide were roughly positioned within cavities 3, 2 and 1, respectively [Table 1; cavity numbering as per Bertram et al. (10)]. The binding conformational search was performed using AFFINITY (MSI, San Diego, CA) using a random docking complex search on a flexible ligand. The best complex was refined with a 50 step (0.1 ps/step) simulated annealing (500–300 K), using the force field cff91. Finally, 1000 iterations of minimization were performed on the bound complex.
RESULTS AND DISCUSSION
Characteristics of novel SC-eRF1s
Neurospora cDNA clone a4b11 is 2002 bp long and contains the entire eRF1 coding region. Aspergillus clone y4b01 (1426 bp) lacks a part of eRF1 domain 1, but, by assembly with EST sequence AA965980, we succeeded in recovering almost the entire coding region of the eRF1. Trichomonas cDNA clone TV-0925 appeared to contain almost the entire coding region of eRF1 (1291 bp long, 429 residues) with a 3′-untranslated region and poly(A) tail. Comparison of the putative amino acid sequence to other complete eRF1 sequences suggested that this cDNA probably lacks a very small portion of the N-terminus (data not shown). We sequenced three Chlamydomonas cDNA clones, but only HCL030f11_r covered the entire eRF1 coding region. The HCL030f11_r clone is 2568 bp long and includes a 1317 bp open reading frame (ORF). In the 3′-untranslated regions, four polymorphic sites were detected among the three clones (data not shown).
The Entamoeba eRF1 gene occupies nearly two-thirds of the contig comprising five GSSs (AZ537755, AZ534941, AZ533867, AZ534936 and AZ540149; total 1762 bp). The identified coding region is 1241 bp long (413 residues) and the region corresponding to the C-terminal end of eRF1 (corresponding to the β15 sheet and the acidic amino acid tail in human eRF1) was truncated (data not shown). No obvious introns were identified in the Entamoeba eRF1 gene. The consensus DNA sequence from the Glycine eRF1 EST contig has an ORF of 1314 bp (437 residues). The Oryza eRF1 gene was identified in a BAC clone of chromosome I (GenBank accession no. AP003269; positions 132930–134231). The gene is 1311 bp long (436 residues) and no introns were found. The Encephalitozoon eRF1 gene (1161 bp long; 386 residues), without any introns, was identified between positions 49544 and 50704 in the chromosome V sequence (GenBank accession no. AL590445). The Candida eRF1 gene was found in contig 6-2330 (positions 12329–13639). The gene is 1311 bp long and encodes 346 residues without any introns.
eRF1 phylogeny
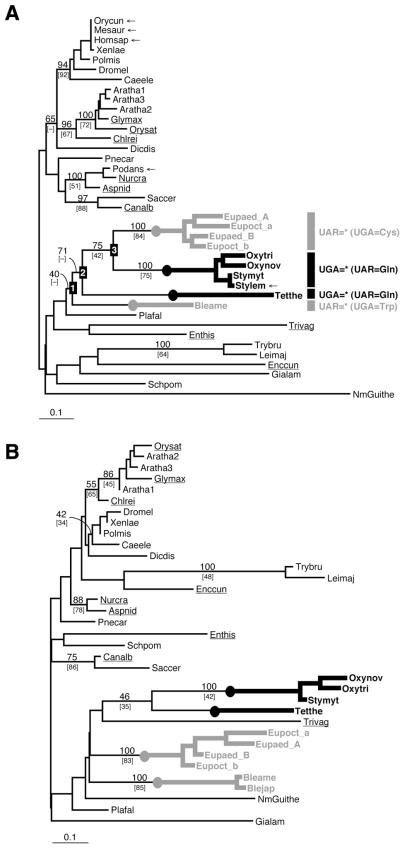
Phylogenetic relationships among eukaryotic groups were poorly resolved by the eRF1 tree based on the FL dataset. Only seven major nodes were supported by a BP >70% (Fig. 1A): (i) an animal clade (BP = 94%); (ii) a plant + Chlamydomonas clade (BP = 96%); (iii) a kinetoplastid (Trypanosoma + Leishmania) clade (BP = 100%); (iv) an Aspergillus + Neurospora + Podospora clade (BP = 100%); (v) a Saccharomyces + Candida clade (BP = 97%); (vi) a Spirotrichea clade (Oxytricha, Stylonychia and Euplotes) (BP = 75%); (vii) a Spirotrichea + Tetrahymena clade (BP = 71%).
Figure 1.
(A) An unrooted eRF1 tree based on the FL dataset using the Γ-FITCH protein distance method. Abbreviations of the sequence names are listed in Table 2. BP are listed for major nodes. Percent occurrence in 10 000 quartet puzzling trees are given in brackets. Dashes indicate that given nodes were not supported by bootstrap or quartet puzzling analyses. The novel sequences determined or identified in this study are underlined. The redundant sequences removed from the analyses using the D1 dataset are marked by arrows. The ciliate lineages that independently converted the standard code to variant codes [UGA = * (UAR = Gln) and UAR = * (UGA = Cys or Trp)] are highlighted by thick black and gray lines, respectively. (B) An unrooted eRF1 tree based on the D1 dataset using the Γ-FITCH protein distance method.
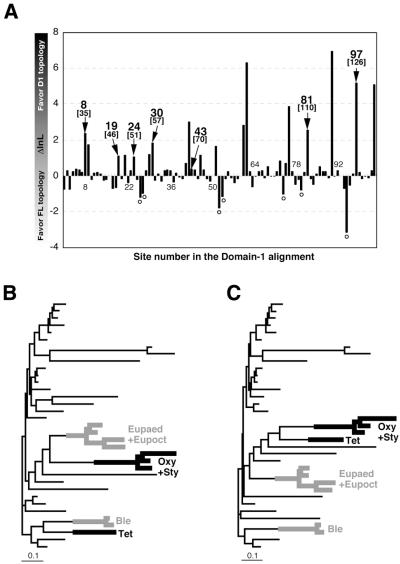
Although the ciliates as a whole were recovered as monophyletic in the optimal Γ-FITCH tree, this clade received low statistical support (BP = 40%; Fig. 1A). In contrast, a single clade for the Spirotrichea was not recovered in the tree based on the D1 dataset. Instead, the ciliate lineages with UGA = * codes (Oxytrich + Stylonychia and Tetrahymena) were clustered together (BP = 46%; Fig. 1B). In general, the resolution of this tree was worse than the FL dataset tree. We suspected that the aberrant relationships among ciliates in the D1 tree could result from sites in the D1 dataset that had independently converged on the same amino acid residues, conferring the same variant codon specificities of eRF1 on the independent ciliate lineages, such as Oxytricha + Stylonychia and Tetrahymena (UGA = *) or Euplotes and Blepharisma (UAR = *). To test this, we compared the lnL scores of domain 1 sites under the FL and D1 topologies by subtracting the former from the latter (Fig. 2A). This comparison indicates that 55 of 103 sites in domain 1 had higher lnL scores under the D1 topology than the FL topology.
Figure 2.
(A) Comparison of site 1nL calculated under the FL and D1 tree topologies. lnL scores calculated from the D1 dataset under the D1 and FL topologies (see Fig. 1A and B) were compared on a site-by-site basis. The sites that were inferred to have evolved convergently in the eRF1s from Oxytricha + Stylonychia and Tetrahymena (UGA = *) and Euplotes and Blepharisma (UAR = *) are highlighted by arrows with site numbers in the alignment (the human eRF1 numbering is given in parentheses). A Γ-FITCH tree based on the D1 dataset excluding the seven convergent sites (B) and a tree based on the D1 dataset excluding the seven sites with negative ΔlnL scores (marked with open circles in Fig. 2A) (C). Details of the analyses are described in the text. Sequence names except for ciliates are omitted. Oxy, Oxytricha; Sty, Stylonychia; Tet, Tetrahymena; Eupaed, Euplotes aediculatus; Eupoct, Euplotes octocarinatus; Ble, Blepharisma. The ciliate lineages that independently converted the standard code to variant codes [UGA = * (UAR = Gln) and UAR = * (UGA = Cys or Trp)] are highlighted by thick black and gray lines, respectively.
It is important to note that the FL phylogeny for ciliates, although poorly supported, is in agreement with those based on sequences of small subunit and large subunit rRNA and features of cell ultrastructure (reviewed in 31), as well as macronuclear genome organization (32,33). Thus, the conclusion that, in ciliates, VC-eRF1s have arisen four separate times from SC-eRF1s (UGA = * code twice independently in Oxytricha + Stylonychia and Tetrahymena and UAR = * code twice independently in Euplotes and Blepharisma) rests on much more than the apparent phylogeny of the eRF1s themselves (22,23). The fact that the D1 dataset supports a topology that unites the two UAR = * code groups therefore likely reflects convergence in the eRF1 primary sequences, constrained by similarly altered codon recognition requirements. Amino acid position assignments that are identical within either the ciliate groups with the UAR = * or UGA = * code, and different from inferred ancestral assignments for ciliates (based on our expanded SC-eRF1 dataset), are therefore good candidates for involvement in codon recognition. We call these ‘variant code-specific converged and then unvaried sites (converged unvaried sites)’.
Variant code-specific converged unvaried sites in ciliate eRF1
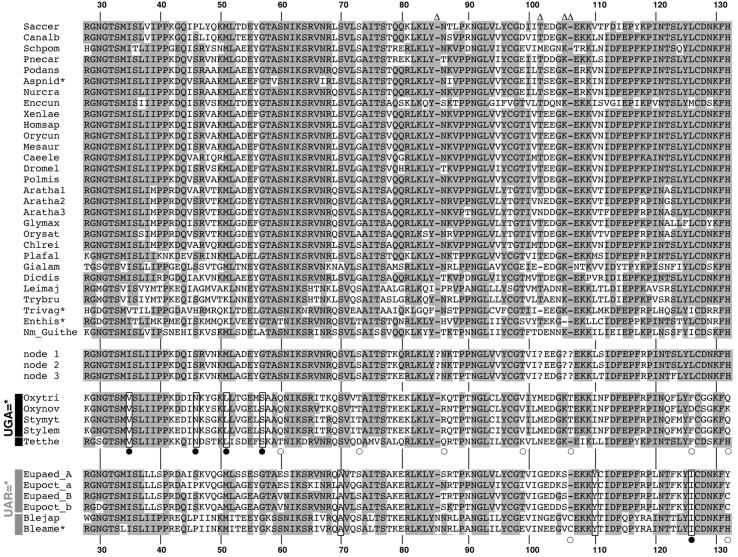
Since the FL and D1 trees differed in the placement of SC-eRF1s as well the branching order of ciliate eRF1s (Fig. 2A and B), only a subset of the 55 ‘D1 topology-favoring’ sites correspond to ciliate variant code-specific converged unvaried sites. To identify the latter sites, we filtered out 48 positions where identical residues were not conserved within the eRF1s corresponding to the UGA = * or UAR = * code. This left seven sites, four of which were identical among the UGA = * code ciliates (Oxytricha + Stylonychia and Tetrahymena) and three that were shared among UAR = * code ciliates (Euplotes and Blepharisma) (Fig. 3). We used ML reconstruction of ancestral sequences of the ciliate eRF1s at nodes 1–3 in the FL topology (Fig. 1A) to confirm that these seven sites were indeed lineage-specific convergent substitutions within the UGA = * or UAR = * code groups (Fig. 3). Five of the sites we identify as convergent were previously identified by Lozupone et al. (23) (boxed and marked with closed circles, Fig. 3). However, our method identifies two novel converged unvaried sites in the UAR = * eRF1s (boxed, Fig. 3). In no cases were the converged unvaried sites in the two variant code groups located at the same alignment position (Fig. 3).
Figure 3.
An amino acid alignment of eRF1 D1. Abbreviations of the sequence names are listed in Table 2. Residues conserved among >70% of standard code eRF1s are shaded. Converged unvaried sites in ciliate eRF1s that are identified both in this study and the study by Lozupone et al. (23) are boxed with filled circles. Converged unvaried sites newly identified in this study are boxed. ‘Convergent’ sites identified only in the study of Lozupone et al. (23) are marked with open circles. The sequences labeled as nodes 1–3 are the ancestral ciliate sequences inferred for these nodes on the full-length topology (see Fig. 1A). The residue numbers are based on human eRF1 (GenBank accession no. U90176). Asterisks indicate partial sequences. Sites that were excluded from the computational analyses are indicated by open triangles. The amino acid residues in the ancestral ciliate eRF1s (nodes 1–3 in Fig. 1A) for these excluded sites are not available and are indicated by question marks.
A Γ-FITCH tree based on the D1 dataset with the seven converged unvaried sites removed no longer grouped the UGA = * code ciliates together, instead recovering a monophyletic Spirotrichea clade (Fig. 2B). In contrast, a tree from the D1 dataset with the seven negative ΔlnL scores removed (marked by open circles in Fig. 2A) once again recovered the UGA = * code ciliates as a group (Fig. 2C). These results indicate that the seven sites identified by our site lnL comparisons and ancestral sequence reconstructions as convergently substituted among the UGA = * or UAR = * code lineages are sufficient to cause the phylogenetically anomalous relationships among ciliates in the D1 tree.
Nine of the ‘code-specific’ substitutions detected by Lozupone et al. (23) (positions 60, 73, 86, 99, 106–107, 126 and 132) are not identified as converged unvaried sites by our method (marked with open circles in Fig. 3). For instance, positions 60 and 86 had negative ΔlnL scores, indicating that they favor the FL topology and disqualifying them from consideration by our criteria. At position 60 the Tetrahymena sequence has a Thr, whereas the Oxytricha and Stylonychia sequences have a Gln. Position 86 is not conserved between UGA = * code ciliate sequences and is unlikely to be necessary for UGA = * code specificity, since the Gln in the Tetrahymena sequence is chemically more similar to the ancestral Asn, found in the SC-eRF1s, than the Lys and Arg residues with long and basic side chains found in the Oxytricha and Stylonychia sequences.
Residues at positions 73, 99, 126 and 132 have positive ΔlnL scores (Fig. 2A) but can be excluded from consideration for other reasons. At position 99, the residue found in the Tetrahymena eRF1 (Lys) is chemically and structurally dissimilar to the amino acid found in the Oxytricha and Stylonychia sequences (Val) (Fig. 3). At position 73, the Thr with a polar side chain in the Oxytricha and Stylonychia sequences is not similar to the negatively charged Asp in the Tetrahymena eRF1 (Fig. 3). Position 126 in Tetrahymena and the Oxytricha + Stylonychia group appears to represent a convergent substitution in these two lineages, as Lozupone et al. argued (23). However, the Oxytricha nova sequence has an Asp at this position, which is chemically and structurally dissimilar to the Phe in the Tetrahymena, Oxytricha trifallax and Stylonychia sequences, questioning its absolute necessity for conferring the UGA = * code specificity (Fig. 3). At position 132, where a His is strictly conserved among SC-eRF1s, the Oxytricha, Stylonychia and Euplotes sequences (Spirotrichea) have various residues. However, the Tetrahymena and Blepharisma sequences retain the His residue from the ancestral sequences (Fig. 3). Thus the substitutions found at this position in the Spirotrichea eRF1s are unlikely to contribute to their variant code specificities.
Finally, the insertion of a single amino acid (Thr or Cys) between positions 106 and 107 is found only in the Oxytricha, Stylonychia and Blepharisma eRF1s, whereas neither the Tetrahymena nor Euplotes proteins share this insertion (Fig. 3). Clearly, such insertions cannot be required for variant code specificity in the UGA = * or UAR = * eRF1s (a ΔlnL score is not available, as the missing data mandated its exclusion from the site lnL analyses).
Mapping the converged unvaried sites on the human eRF1 domain 1 structure
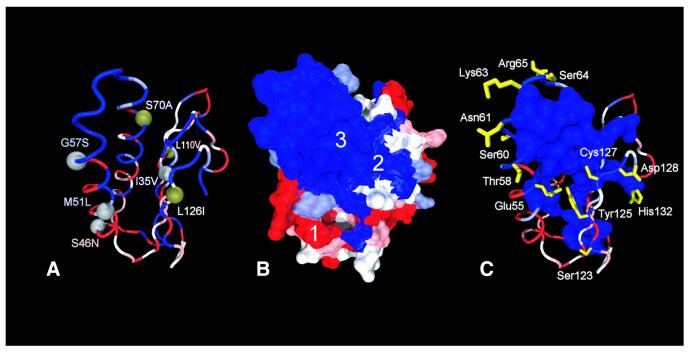
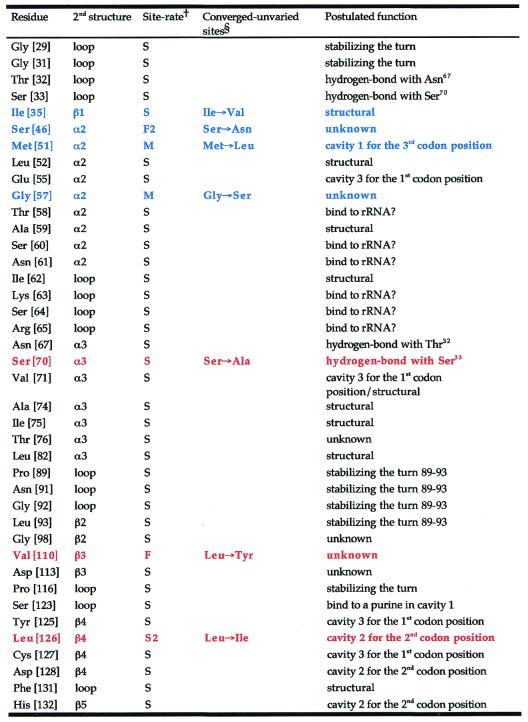
We postulated that some or all of the convergent sites we identified could account for the alteration of codon specificities in the ciliate eRF1s, responding to the UGA = * or UAR = * code. However, mapping these sites onto the tertiary structure reveals that they are widely spread across eRF1 domain 1 (Fig. 4A). There appears to be no general correlation between the convergent substitutions and the estimated site rates at a given site, although positions 35, 51, 57, 70 and 126 are located within or near the cluster of slowest evolving residues (Fig. 4B and C). Of most interest are positions 51 (Met→Leu in the UGA = * eRF1s) and 126 (Met→Ile in the UAR = * eRF1s); these sites map to cavities 1 and 2 on the protein surface, respectively, identified by Bertram et al. as potential stop codon-binding ‘cavities’ (10). Postulated functions of the slowest evolving and converged unvaried sites are summarized in Table 3.
Figure 4.
Distribution of site rates in eRF1 D1 and converged unvaried sites in the UGA = * and UAR = * eRF1s mapped on the human eRF1 D1 structure (PDB accession no. 1DT9). The five site rates categories are color coded from blue (slowest) to red (fastest). (A) Distribution of the converged unvaried sites mapped on the human eRF1 D1 structure. Gray spheres indicate a convergence in the ciliate VC-eRF1s with the UAR = * code (Euplotes and Blepharisma) and green spheres a convergence in the VC-eRF1s with the UGA = * code (Oxytricha, Stylonychia and Tetrahymena). The labels indicate the identity of the ancestral residue on the left of the number and the convergent change on the right of the number. Numbering is based on human eRF1 (GenBank accession no. U90176). (B) Site rates of D1 of eRF1 mapped on the solvent-accessible surface. The surface cavities are labeled as per Bertram et al. (10). (C) Distribution of the residues included in the slowest category. All side chains potentially performing a structural function (see criteria in the text) are represented as a molecular surface. Polar residues for which no structural function could be assigned are displayed in yellow.
Table 3. Putative function of the slowest evolving and converged unvaried residues in eRF1 domain 1.
Residue numbers based on human eRF1 (GenBank accession no. U90176) are given in brackets.
†S, slowest evolving; S2, second slowest evolving; M, moderately evolving; F, fastest evolving; F2, second fastest evolving residue.
§Substitutions shown in blue and red display the convergent site in the UGA = * eRF1s from Oxytricha, Stylonychia and Tetrahymena and those in the UAR = * eRF1s from Euplotes and Blepharisma, respectively. Cavity numbering as per Bertram et al. (10).
Another interesting convergent substitution is Ser→Ala at position 70 in the UAR = * eRF1s (Figs 3 and 4A). In the human structure, the two slowest evolving residues, Ser70 and Ser33, are predicted to form a potential hydrogen bond (Table 3). The ciliate VC-eRF1s with this substitution retain a Ser at position 33 (Fig. 3) and so would not be able to form a hydrogen bond. This substitution may not have a direct mechanical impact on codon binding, but it could alter the codon specificity of eRF1 by indirectly affecting the flexibility of the stop codon-binding site.
Indeed, the structural rationale behind many of the converged unvaried sites is likely to extend beyond the postulated function of codon recognition. As discussed above, only a few residues are directly involved in the cavity-binding model proposed by Bertram et al. (10). However, many of the convergent changes (including that at position 70) are more likely to reflect group-specific constraints on what residues are involved in stabilizing the structure. This is consistent with the observation that many of the converged unvaried changes are occasionally observed in SC-eRF1 homologs from other organisms (see for example positions 35, 70 and 126 in Fig. 3).
Revised cavity-binding model
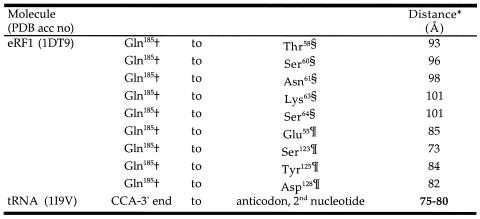
The pattern of site rates mapped on the human eRF1 structure (Fig. 4B) sensibly corresponds to the general area of binding of the stop codon in the model proposed by Bertram et al. (10). This large number of slowest evolving residues comprises a structural (and hydrophobic) core that is likely to position the side chains whose function it is to recognize the stop codons (Fig. 4B). Four of the solvent-accessible (and slowest evolving) residues, Glu55, Tyr125, Cys127 and Asp128, have the added property that they are located near conserved cavities on the protein surface (Fig. 4C) suggested to specifically bind the nucleotides of the stop codons on mRNA in the cavity-binding model proposed by Bertram et al. (10). Assuming that eRF1 interacts with the ribosome in a similar fashion to a tRNA molecule, the distance between the codon and the elongation peptide chain should be similar to the preceding tRNA already in the ribosome. Thus we measured the distances between the anticodon and the CCA end based on the Saccharomyces cerevisiae phenylalanine tRNA (PDB accession no. 1I9V) and between Gln185 in eRF1 domain 2 (which is thought to be analogous to the 3′-end of tRNAs) and various slowest evolving residues in domain 1 (Table 4). Interestingly, the distance between Gln185 and Tyr125 in cavity 2 (domain 1) is 84 Å, roughly matching the distance between the anticodon and the CCA end of 75–80 Å in the tRNA (Table 4).
Table 4. Dimensional comparisons between tRNA and eRF1.
Residue numbering is based on human eRF1 (GenBank accession no. U90176).
*Measured from the Cα of Gln185 to the tip of the target side chain.
†Part of the GGQ motif in eRF1 domain 2.
§In the α2-helix–loop–α3-helix region in domain 1.
¶In cavity 2 in domain 1. Cavity numbering as per Bertram et al. (10).
In the cavity-binding model of Bertram et al. (10), residue Met51 is part of cavity 1, where it is thought to bind the first nucleotide of stop codons (Table 1). However, it is hard to explain how the Met→Leu convergence found in the UGA = * eRF1s (Fig. 3), and Met→Ile in a UAG unipotent suppressor mutant (10), selectively forbid the binding of a G at the third position of the stop codon. Furthermore, cavity 3, which was proposed to originally bind with the third nucleotide (10), is lined with absolutely conserved residues among SC-eRF1s and ciliate VC-eRF1s (Glu55, Tyr125 and Cys127; Fig. 3). These results suggest to us that the three nucleotides of the stop codon and the three cavities on the eRF1 surface could interact in an opposite orientation to that originally proposed (10); cavities 1 and 3 could account for discrimination of the third and first nucleotides of stop codons, respectively [Table 1; cavity numbering as per Bertram et al. (10)].
To test this, we modeled binding of the UAA trinucleotide such that the first nucleotide is inserted in cavity 3, the third nucleotide in cavity 1 and the second nucleotide still interacts with cavity 2. In this conformation, the mRNA backbone is extended along a surface groove in which the proposed binding site is located (Fig. 5). The first nucleotide, U, is held in place by absolutely conserved Glu55 and Tyr125 residues, while its sugar moiety interacts with Cys127. The second position is tucked into cavity 2, with matching Asp128 and His132. Finally, the third position is located in the least evolutionarily constrained cavity, nearby Met51 in the human structure. The only slowest evolving residue in cavity 1 is Ser123, which can donate a hydrogen bond to the purine ring of either A or G in the third codon position (Fig. 5).
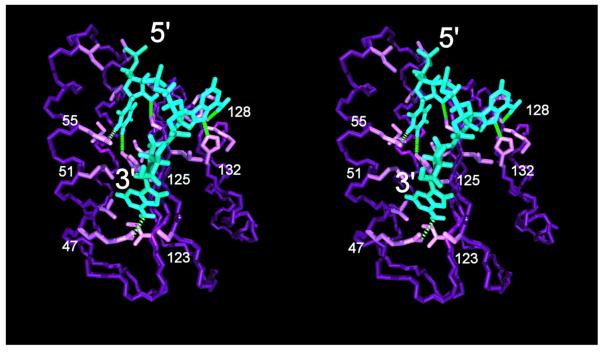
Figure 5.
Stereoview of stop codon 5′-UAA-3′ (blue) bound to the human release factor eRF1 N-terminal domain (purple) in the reverse orientation to the previously proposed model (10). Only the backbone atoms of eRF1 and the side chain interacting with the trinucleotide are displayed. Green dotted lines indicate possible dipolar interactions conferring binding specificity to the stop codon.
Our cavity-binding model shown in Figure 5 also rationalizes why a higher aliphatic branching at position 51 (i.e. the Met→Leu convergence in UGA = * ciliates and the Met→Ile change in the unipotent suppressor mutation) would prevent binding of a G by steric hindrance of the amino group at C2 of the purine ring. It provides a structural basis for binding of the first nucleotide (which apparently never changes amongst SC or VC organisms) in a patch of absolutely conserved residues and positions the three nucleotides within the surface cavities without imposing a strain on the mRNA backbone. However, it is currently impossible to definitively decide whether the model presented in Figure 5 or that proposed by Bertram et al. (10) has the correct codon-binding orientation. Based on what is known about mRNA–tRNA interaction in the ribosome (34), the binding groove is located in the face of domain 1 that is distal to the 5′-end of the mRNA. Thus, the mRNA has to wrap around the release factor in either cavity-binding model; only the direction of the wrap would differ.
Is the α2-helix–loop–α3-helix region directly involved in stop codon recognition?
Although currently available data (this study; 10,23) fit well with the cavity-binding models, a set of residues in the α2–loop–α3 region, Glu55, Thr58, Ser60, Lys63, Ser64 and Arg65, are slowest evolving and are not classified as ‘structural’ by our criteria (Fig. 4C). These residues are probably not interacting with other slowest evolving residues and so could, in principle, bind to mRNA. These results also support the two models proposed by Muramatsu et al. (13) and Nakamura et al. (11), which suggest that the side chains of these residues could interact with stop codons directly (Table 1). However, the distances between Gln185 in eRF1 domain 2 and the slowest evolving residues in the α2–loop–α3 region are ∼100 Å, much larger than the estimated distance based on the tRNA structure (∼80 Å) or the cavity-binding models (∼84 Å) (Table 4). Further crystallographic studies are needed to examine whether the tertiary structure of eRF1 bound with the GTPase subunit, eRF3, or in the A site of the ribosome might be different from that determined by Song et al. (9).
Other lines of evidence also cast doubt on involvement of the α2–loop–α3 region in stop codon recognition. For instance, a recent in vitro study showed that the Methanococcus jannaschii aRF1 with Gln at position 58 (rather than the Thr conserved amongst eukaryotes) is capable of terminating protein synthesis on human ribosomes responding to UAA, UAG and UGA codons (5). Similarly, Ser60 has changed to Thr in the Entamoeba SC-eRF1 (Fig. 3). These results indicate that positions 58 and 60 are changeable (at least to similar amino acids) without generating a concomitant change in the codon specificity of SC-eRF1. No substitution at position 59 has been found in SC-eRF1s before this study and Ala59 is a part of the potential ‘peptide anticodon’ in the model proposed by Nakamura et al. (11). However, this residue is substituted by Val in the SC-eRF1 from Aspergillus (Fig. 3). Our examination of the human eRF1 structure indicates that the Ala59 side chain points into and forms part of the slowest evolving domain 1 core (Fig. 4C). Thus it is likely that the high degree of conservation of Ala at this position is not necessary for eRF1 codon recognition. Instead, the residue is probably constrained to maintain the conformation of domain 1.
The combination of site rate analysis and structural observations of eRF1 domain 1 defined Ile62 as one of the slowest evolving residues in the domain 1 core (Fig. 4B). Other slowest evolving residues, Asn61, Lys63 and Ser64, adjacent to Ile62, are all polar or positively charged and solvent exposed (Fig. 4C), suggestive of a potential role as a ‘peptide anticodon’. In fact, position 64 was once suggested to be the eRF1 discriminator residue, since the Tetrahymena sequence, the only VC-eRF1 available at the time, possessed Asp (35). Yet this hypothesis was rejected, because other ciliate VC-eRF1s have the regular Ser at this position (Fig. 3) (23). Additionally, our novel SC-eRF1 from Trichomonas has the substitution, Ser64→Asn (Fig. 3), suggesting that Ser at position 64 is unlikely to be involved in eRF1 codon specificity.
Given the novel eRF1s determined/identified in this study and the in vitro experiments with the M.jannaschii aRF1 (5), it appears that at least one of the residues at positions 58–60 and 64 is substitutable to UAA, UAG or UGA without loss of codon specificity. It is intriguing how divergent this suite of residues can be, since most aRF1s have more than one substitution at these positions as compared with the sequence conserved in most SC-eRF1s (TASNIKS) (24). Particularly, the aRF1s from Aeropyrum pernix (GenBank accession no. AP000063, ITDNIKL; versus the standard TASNIKS motif), Sulfolobus solfataricus (GenBank accession no. AE006836, IAQNIKL) and Methanobacterium thermautotrophicum (GenBank accession no. AE000666, QSANIKS) are extremely divergent in this region.
Yet, the high degree of conservation of the α2–loop–α3 region amongst eukaryotic class 1 RFs suggests that this region has an important function. Interestingly, we have noted that yeast ribosomal protein L30 (RPL30; PDB accession no. 1CK8) appears to have an analogous tertiary structure to human eRF1 domain 1 (despite having a different connectivity of secondary structural elements) by direct comparison of three-dimensional protein structure with the VAST algorithm (36,37). In this case, residues in the helices of RPL30 that are analogous to the eRF1 α2 and α3 helices interact with, and bind to, the rRNA molecule (38). eRF1 works at the A site of ribosomes and it is likely to interact with rRNA and ribosomal proteins adjacent to the A site during the translation termination process. Extrapolating the rRNA–RPL30 interaction to human eRF1 domain 1, one may speculate that the side chains of slowest evolving residues in the α2–loop–α3 region may interact with a part of small subunit rRNA (Table 3). Further in vitro experiments combining various aRF1s and eukaryotic ribosomes as well as large-scale sequencing of eRF1 genes will help elucidate the function of the residues in the α2–loop–α3 region.
CONCLUSION
Are there multiple ways to change the codon specificity of eRF1?
As discussed above, both cavity-binding models, regardless of the trinucleotide orientation, appear to explain the data better than the anticodon mimicry models between the tRNA anticodon loop and the eRF1 α2–loop–α3 region. If either of the cavity-binding models were true, variant codon specificity would be modulated by (i) overall shapes of the codon-binding cavities and/or (ii) interactions between stop codon nucleotides and residues that are adjacent to the cavities. It is known that diplomonads and dasycladacean green algae have converted the standard code to the UGA = * code independently (39–41). Furthermore, independent code conversions (from the standard to UGA = * code) from those in either the Oligohymenophrea or Spirotichea have been reported in other ciliate lineages [i.e. the classes Nassophorea and Karyorelictea and Condylostoma (Heterotrichea)] (23). Some VC-eRF1s from these lineages may alter their codon specificities via substitutions that have occurred in different positions to the VC ciliates tested here (e.g. positions 51 and 126). Thus, sequencing of additional SC-eRF1s and VC-eRF1s and structural analyses are required to further elucidate the mechanism of the translation termination system at the molecular level.
Acknowledgments
ACKNOWLEDGEMENTS
We thank J. M. Archibald and J. O. Andersson (Dalhousie University, Halifax, Canada) for critical reading of this manuscript. We also thank T. M. Embley (Natural History Museum, London, UK) for providing Trichomonas cDNA clone TV-0925. The Neurospora and Aspergillus cDNA clones a4b11 and y4b01 were kindly provided by the Fungal Genetic Stock Center (University of Kansas Medical Center, KS). Chlamydomonas cDNA clones CM016g10_r, CM021b05_r and HCL030f11_r were provided by Kazusa the DNA Research Institute (Kisarazu, Chiba, Japan). Sequence data for C.albicans were obtained from the Stanford Genome Technology Center (http://www-sequence.stanford.edu/group/candida). Sequencing of C.albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. Y.I. was supported by a post-doctoral research fellowship awarded to A.J.R. from the CIAR and grant MT4467 from the Canadian Institutes for Health Research awarded to W.F.D. This work was supported by Genomics grant 228253-99 awarded to A.J.R. by the Natural Sciences and Engineering Research Council of Canada.
DDBJ/EMBL/GenBank accession nos+ To whom correspondence should be addressed. Tel: +1 902 494 2968; Fax: +1 902 494 1355; Email: yinagai@is.dal.ca AY050664–AY050667
REFERENCES
- 1.Nakamura Y. and Ito,K. (1998) How proteins read the stop codon and terminate translation. Genes Cells, 3, 265–278. [DOI] [PubMed] [Google Scholar]
- 2.Kisselev L.L. and Buckingham,R.H. (2000) Translational termination comes of age. Trends Biochem. Sci., 25, 561–566. [DOI] [PubMed] [Google Scholar]
- 3.Poole E. and Tate,W. (2000) Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim. Biophys. Acta, 1493, 1–11. [DOI] [PubMed] [Google Scholar]
- 4.Bertram G., Innes,S., Minella,O., Richardson,J. and Stansfield,I. (2001) Endless possibilities: translation termination and stop codon recognition. Microbiology, 147, 255–269. [DOI] [PubMed] [Google Scholar]
- 5.Dontsova M., Frolova,L., Vassilieva,J., Piendl,W., Kisselev,L. and Garber,M. (2000) Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett., 472, 213–216. [DOI] [PubMed] [Google Scholar]
- 6.Inagaki Y. and Doolittle,W.F. (2000) Evolution of the eukaryotic translation termination system: origins of release factors. Mol. Biol. Evol., 17, 882–889. [DOI] [PubMed] [Google Scholar]
- 7.Ito K., Uno,M. and Nakamura,Y. (2000) A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature, 403, 680–684. [DOI] [PubMed] [Google Scholar]
- 8.Ito K., Ebihara,K., Uno,M. and Nakamura,Y. (1996) Conserved motifs in prokaryotic and eukaryotic polypeptide release factors: tRNA-protein mimicry hypothesis. Proc. Natl Acad. Sci. USA, 93, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H., Mugnier,P., Das,A.K., Webb,H.M., Evans,D.R., Tuite,M.F., Hemmings,B.A. and Barford,D. (2000) The crystal structure of human eukaryotic release factor eRF1—mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell, 100, 311–321. [DOI] [PubMed] [Google Scholar]
- 10.Bertram G., Bell,H.A., Ritchie,D.W., Fullerton,G. and Stansfield,I. (2000) Terminationg eukaryotic translation: domain 1 of release factor eRF1 functions in stop codon recongnition. RNA, 6, 1236–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y., Ito,K. and Ehrenberg,M. (2000) Mimicry grasps reality in translation termination. Cell, 101, 349–352. [DOI] [PubMed] [Google Scholar]
- 12.Liang A., Brunen-Nieweler,C., Muramatsu,T., Kuchino,Y., Beier,H. and Heckmann,K. (2001) The ciliate Euplotes octocarinatus expresses two polypeptide release factors of the type eRF1. Gene, 262, 161–168. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu T., Heckmann,K., Kitanaka,C. and Kuchino,Y. (2001) Molecular mechanism of stop codon recognition by eRF1: a wobble hypothesis for peptide anticodons. FEBS Lett., 488, 105–109. [DOI] [PubMed] [Google Scholar]
- 14.Frolova L.Y., Tsivkovskii,R.Y., Sivolobova,G.F., Oparina,N.Y., Serpinsky,O.I., Blinov,V.M., Tatkov,S.I. and Kisselev,L.L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA, 5, 1014–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seit Nebi A., Frolova,L., Ivanova,N., Poltaraus,A. and Kisselev,L. (2000) Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity. Mol. Biol. (Mosk.), 34, 899–900. [PubMed] [Google Scholar]
- 16.Ito K., Ebihara,K. and Nakamura,Y. (1998) The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA, 4, 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mugnier P. and Tuite,M.F. (1999) Translation termination and its regulation in eukaryotes: recent insights provided by studies in yeast. Biokhimiia, 64, 1360–1366. [PubMed] [Google Scholar]
- 18.Ebihara K. and Nakamura,Y. (1999) C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA, 5, 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eurwilaichitr L., Graves,F.M., Stansfield,I. and Tuite,M.F. (1999) The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol. Microbiol., 32, 485–496. [DOI] [PubMed] [Google Scholar]
- 20.Frolova L.Y., Merkulova,T.I. and Kisselev,L.L. (2000) Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA, 6, 381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight R.D., Freeland,S.J. and Landweber,L.F. (2001) Rewiring the keyboard: evolvability of the genetic code. Nature Rev. Genet., 2, 49–58. [DOI] [PubMed] [Google Scholar]
- 22.Tourancheau A.B., Tsao,N., Klobutcher,L.A., Pearlman,R.E. and Adoutte,A. (1995) Genetic code deviations in the ciliates: evidence for multiple and independent events. EMBO J., 14, 3262–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozupone C.A., Knight,R.D. and Landweber,L.F. (2001) The molecular basis of nuclear genetic code change in ciliates. Curr. Biol., 11, 65–74. [DOI] [PubMed] [Google Scholar]
- 24.Inagaki Y. and Doolittle,W.F. (2001) Class I release factors in ciliates with variant genetic codes. Nucleic Acids Res., 29, 921–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Asamizu E., Nakamura,Y., Sato,S., Fukuzawa,H. and Tabata,S. (1999) A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii. I. Generation of 3433 non-redundant expressed sequence tags. DNA Res., 6, 369–373. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strimmer K. and von Haeseler,A. (1996) Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol., l3, 964–969. [Google Scholar]
- 28.Felsenstein J. (1993) PHYLIP v.3.573. University of Washington, Seattle, WA.
- 29.Humphrey W., Dalke,A. and Schulten,K. (1996) VMD: visual molecular dynamics. J. Mol. Graph., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z. (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci., 13, 555–556. [DOI] [PubMed] [Google Scholar]
- 31.Katz L.A. (2001) Evolution of nuclear dualism in ciliates: a reanalysis in light of recent molecular data. Int. J. Syst. Evol. Microbiol., 51, 1587–1592. [DOI] [PubMed] [Google Scholar]
- 32.Prescott D.M. (1994) The DNA of ciliated protozoa. Microbiol. Rev., 68, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley J.L. and Katz,L.A. (2001) Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol. Biol. Evol., 18, 1372–1377. [DOI] [PubMed] [Google Scholar]
- 34.Yusupov M.M., Yusupova,G.Z., Baucom,A., Lieberman,K., Earnest,T.N., Cate,J.H. and Noller,H.F. (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science, 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 35.Knight R.D. and Landweber,L.F. (2000) The early evolution of the genetic code. Cell, 101, 569–572. [DOI] [PubMed] [Google Scholar]
- 36.Madej T., Gibrat,J.F. and Bryant,S.H. (1995) Threading a database of protein cores. Proteins, 23, 356–369. [DOI] [PubMed] [Google Scholar]
- 37.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Surprising similarities in structure comparison. Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 38.Mao H., White,S.A. and Williamson,J.R. (1999) A novel loop-loop recognition motif in the yeast ribosomal protein L30 autoregulatory RNA complex. Nature Struct. Biol., 6, 1139–1147. [DOI] [PubMed] [Google Scholar]
- 39.Schneider P.R., Leiblé,M.B. and Yang,X.-P. (1989) Strong homology between the small subunit of ribulose-1,5-biphosphate carboxylase/oxygenase of two species of Acetabularia and the occurrence of unusual codon usage. Mol. Gen. Genet., 218, 445–452. [DOI] [PubMed] [Google Scholar]
- 40.Keeling P.J. and Doolittle,W.F. (1996) A non-canonical genetic code in an early diverging eukaryotic lineage. EMBO J., 15, 2285–2290. [PMC free article] [PubMed] [Google Scholar]
- 41.Keeling P.J. and Doolittle,W.F. (1997) Widespread and ancient distribution of a noncanonical genetic code in diplomonads. Mol. Biol. Evol., 14, 895–901. [DOI] [PubMed] [Google Scholar]