Abstract
Surface-enhanced Raman spectroscopy (SERS) is a powerful tool for molecule identification. However, profiling complex samples remains a challenge because SERS peaks are likely to overlap, confounding features when multiple analytes are present in a single sample. In addition, SERS often suffers from high variability in signal enhancement due to nonuniform SERS substrate. The machine learning classification techniques widely used for facial recognition are excellent tools to overcome the complexity of SERS data interpretation. Herein, we reported a sensor for classifying coffee beverages by integrating SERS, feature extractions, and machine learning classifiers. A versatile and low-cost SERS substrate, called nanopaper, was used to enhance Raman signals of dilute compounds in coffee beverages. Two classic multivariate analysis techniques, Principal Component Analysis (PCA) and Discriminant Analysis of Principal Components (DAPC), were used to extract the significant spectral features, and the performance of various machine learning classifiers was evaluated. The combination of DAPC with Support Vector Machine (SVM) or K-Nearest Neighbor (KNN) shows the best performance for classifying coffee beverages. This user-friendly and versatile sensor has the potential to be a practical quality-control tool for the food industry.
Keywords: Surface-enhanced Raman spectroscopy (SERS), Machine learning, Feature extraction, Coffee, Classification
1. Introduction
Raman spectroscopy is a valuable tool for chemical identification in its ability to provide fingerprint information of molecules. A significant challenge of Raman spectroscopy is the inherently weak Raman scattering signal; surface-enhanced Raman spectroscopy (SERS) is an excellent approach to overcome this challenge. SERS amplifies the local electromagnetic field near nanostructured metal surfaces, providing magnitudes of enhancement of Raman signals for molecules adsorbed on metal surfaces (Sharma et al., 2012). The large signal enhancement makes SERS an effective tool for the detection of dilute analytes.
However, two prominent factors inhibit the interpretation of spectral data in SERS sensing applications. First, SERS signals often suffer from high variability (Fornasaro et al., 2020). The signal enhancement of SERS is primarily caused by localized electric field enhancement and is particularly significant in hot spots, such as nanoscale gaps between metal particles and highly structured metal surfaces (Weatherston et al., 2016). A tiny variation of SERS substrates could lead to a high variability of SERS signal. Second, many applications of Raman spectroscopy, such as food quality analysis and disease diagnosis, intend to compare samples that contain similar types of chemicals. The sample states are determined by the relative concentrations of multiple chemicals (Kuhar et al., 2018). Although Raman spectroscopy provides molecular fingerprint spectra, Raman peaks are likely to overlap when a large number of analytes are present in a sample. Thus, sample states cannot be simply determined by a few prominent Raman bands. Instead, the whole Raman spectra that include the covariate features of multiple molecules should be considered (Liu et al., 2020). Therefore, advanced data analysis techniques like machine learning are needed to uncover patterns of Raman spectra.
To address complex Raman data interpretation, we applied multivariate analysis and machine learning classifiers widely used in pattern recognition applications (e.g., facial recognition). Similar to SERS spectra, image datasets often suffer from high variability caused by facial expression, illumination, blocking, resolution, and noise, leading to practical challenges in computer vision applications (Wright et al., 2009). Multivariate analysis has been applied to extract the relevant features and minimize data variations (Jade et al., 2003, Kim et al., 2010, Seena and Yomas, 2014). The dimension reduction of the high dimension image dataset is achieved through feature extraction and feature selection. The removal of the data unrelated to classification improves the dataset quality and classification performance (Khalid et al., 2014). Similarly, SERS suffers from high variability in signals; therefore, the combination of feature extractions and machine learning classifiers could serve as a powerful tool to identify the SERS spectra of complex samples (Lussier et al., 2020). In the past decade, multivariate analysis, such as Principal Component Analysis (PCA) and Discriminant Analysis of Principal Components (DAPC), have already been utilized to analyze Raman and SERS spectra for different applications, including medical diagnosis (Chen et al., 2013; Liu et al., 2016; Muratore, 2013; Senger et al., 2020; Sigurdsson et al., 2004), explosive compound detection (Hwang et al., 2013), material research (Mao et al., 2020), biological samples identification (Jamieson et al., 2018; Liu et al., 2020; Lu et al., 2018; Prakash et al., 2020; Senger and Scherr, 2020), and food industry (de Toledo et al., 2017; Jiang et al., 2021).
In this study, we developed a sensor for classifying coffee using Raman spectroscopy. Flavors of coffee beverages are determined by a large number of flavor and aroma compounds. The compositions of these compounds are highly dependent on the growth of coffee beans, the fermentation and roasting processes, and the drying and storage conditions (Poltronieri and Rossi, 2016). Although quality assurance is typically performed by experienced tasters, analytical methods that provide rapid, robust, and precise quality evaluations are desired (Wermelinger et al., 2011). Raman spectroscopy has been used to discriminate Arabica and Robusta coffee samples (El-Abassy et al., 2011; Rubayiza and Meurens, 2005; Wermelinger et al., 2011). For example, Wermelinger et al. quantified the fraction of the Robusta beans in the coffee blend by measuring the intensity ratio of 1570/1665 cm−1 and 1570/1460 cm−1 (Wermelinger et al., 2011). The Raman peak at 1570 cm−1 is associated with kahweol in Arabica coffee and is absent in Robusta coffee. When the composition of Robusta coffee increased, the peak position at 1665 cm−1 shifted towards a lower wavenumber because Robusta coffee contains a higher amount of unsaturated fatty acids. In another study, Rubayiza et al. observed the absence of 1567 and 1487 cm−1 peaks in Robusta coffee due to the trace kahweol content in Robusta (Rubayiza and Meurens, 2005). Moreover, El-Abassy et al. discriminated between Arabica and Robusta coffee beans based on chlorogenic acid (CGA) and lipid contents (El-Abassy et al., 2011). These tests only selected a few molecules as indicators to differentiate Arabica and Robusta beans. Such analysis, however, cannot distinguish the same types of coffee beans treated by distinct roasting, fermentation, and aging processes. Caprioli et al. had a review on coffee aroma profiles and found there are more than 40 types of potent odorants of roast coffee with many more still not being identified (Caprioli et al., 2015). All of these flavor and aroma compounds are essential for quality control (Cruz et al., 2012; Semmelroch and Grosch, 1996).
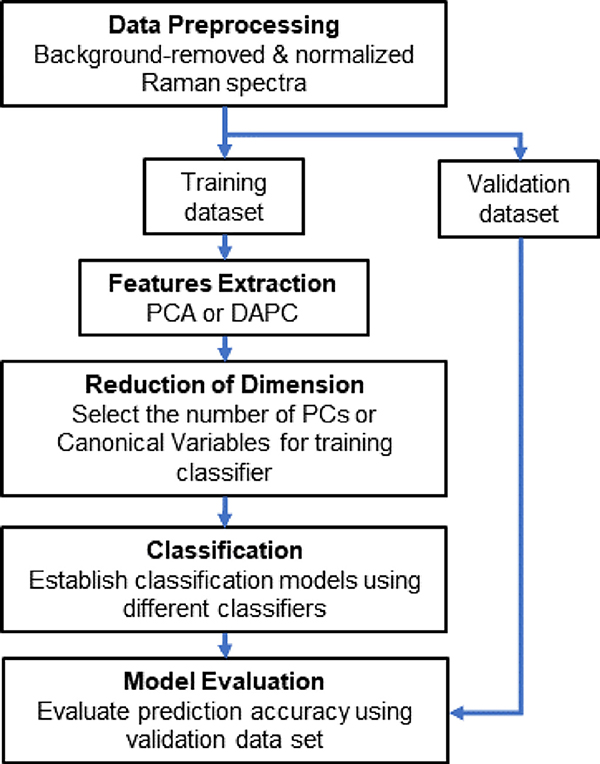
Herein, we integrate SERS, feature extractions, and machine learning classifiers to overcome the challenges in coffee beverage analysis. First, to enhance Raman signals of dilute compounds, we used a versatile and inexpensive SERS substrate, called nanopaper (i.e., a glass fiber paper decorated with silver nanoparticles) as shown in Fig. 1 (Weatherston et al., 2018; Weatherston et al., 2019). Second, we integrated feature extraction algorithms and machine learning classifiers to address the high variability in SERS signals with spectral overlap in high fluorescing coffee samples. The workflow is shown in Fig. 2. We selected two multivariate analysis techniques for feature extraction, including Principal Component Analysis (PCA) and Discriminant Analysis of Principal Components (DAPC), and four machine learning classifiers, including Naïve Bayes, Decision Tree, Support Vector Machine (SVM), and K-Nearest Neighbor (KNN) (Kotsiantis et al., 2007; Yuan et al., 2019; Yuan et al., 2020). Feature extraction tools, such as PCA, have already been utilized for Raman analysis (Lussier et al., 2020). DAPC applies PCA then discriminant analysis to further explore variances between sample groups (Jombart et al., 2010). Our results show that DAPC has a better performance than PCA for all classifiers. In addition, the combination of DAPC with SVM or KNN delivers the best performance for classifying coffee beverages. The results also show that the machine learning classifiers can detect trivial differences among similar SERS spectra.
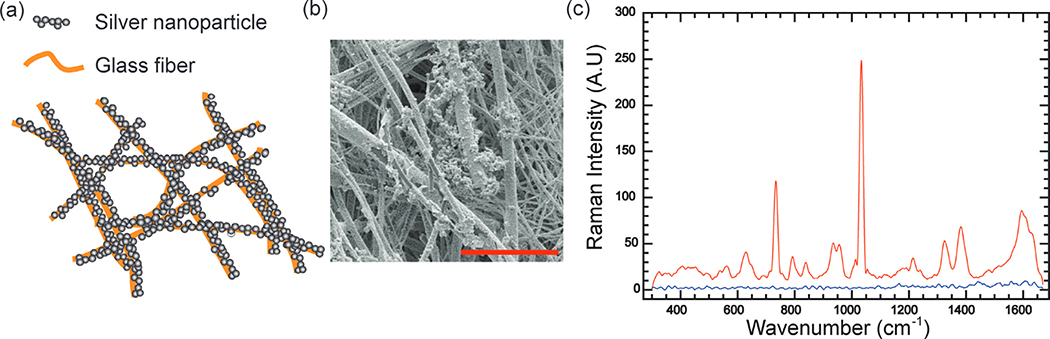
Fig. 1. SERS measurement of the coffee.
(a) Schematic illustration of nanopaper, a glass fiber paper coated with the silver nanoparticle. (b) SEM image of nanopaper. The scale bar is 10 μm. (c) Averaged Raman spectra (N = 80) of coffee Sample 1 on nanopaper (red), and on the bare glass fiber paper (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 2.
Flow chart for the classification algorithm.
2. Experimental methods
2.1. Materials
Double deionized (DDI) water was produced with a Milli-Q system from Millipore. Five different types of ground coffee samples were purchased at a local grocery store. Four of them were Arabica beans from the Colombia Region with varying degrees of roasting, and one of them was the Robusta bean (The detailed information about coffee products is listed in Supplementary Information). Filter paper (Qualitative P8 – Fluted, Fisher Brand), binder-free glass microfiber filters (Whatman grade 934-AH, 110 mm circles), sodium hydroxide, methanol, silver nitrate (99.9995% (metals basis), Alfa Aesar), and 2-propanol were obtained from Fisher Scientific. Sodium citrate dihydrate, potassium hydroxide, ammonia hydroxide solution (28%–30%) were purchased from Sigma-Aldrich. Citric acid and D-glucose anhydrous were purchased from VWR International. All chemicals were ACS grade or better and used without further purification.
2.2. Nanopaper fabrication and characterization
Nanopapers were fabricated as previously reported (Weatherston et al., 2018). In brief, a typical synthesis consisted of mixing aqueous solution of ammonia and silver nitrate with potassium hydroxide; 40 mL of 35% aqueous D-glucose solution was added to a 2L glass beaker and shaken vigorously by hand. Glass microfiber papers were immersed into the solution, and the container was shaken for 5 min by hand. The container was covered with aluminum foil to minimize the exposure to light and left at room temperature for 1 h. The filter papers were then rinsed thoroughly with DDI water and 2-propanol. The final products, i.e., the nanopapers, were stored in 2-propanol and the container was covered with aluminum foil and stored in drawers to protect from light exposure. Before the Raman measurement, the nanopaper was dried in a hot air oven and cut into a 1 cm × 1 cm square shape.
2.3. Coffee beverage preparation
Ground coffee sample (1.2 g) was added to 10 mL of deionized water. The solution was vigorously vortexed and kept at 4 °C for 15 h. The solution was then centrifuged by a Thermofisher Megafuge Centrifuge at 2500 RPM for 15 mins, and the resulting liquid was stored in a 4 °C fridge and covered with aluminum foil.
2.4. Raman measurements
Nanopapers were washed with 0.1 M citrate buffer (pH = 3.6) and immersed afterwards into the coffee extract solution for one minute. Then, the paper was dried in a hot air oven at 70 °C and transferred to the Raman microscope for SERS measurement. The Raman spectra were collected from a 780 nm diode laser with a 10x objective lens, a Rayleigh rejection filter, a diffraction grating (4.7–8.7 cm−1 resolution), a 25 μm pinhole, and a black-illuminated charged-coupled device (CCD) detector using a Thermo Scientific DXR Raman microscope (Thermo Fisher Scientific, Inc.). Each spectrum was acquired at 1 mW laser power with 10 accumulations of 90 s integration time each, 15 min in total. To investigate the reproducibility of spectra, the same experiment protocol was repeated on five different days. A total number of 400 spectra were collected, with 80 spectra per sample.
2.5. Data processing and multivariate data analysis
Data processing, multivariate analysis, and classification algorithms were conducted using Matlab 2021 (Mathworks Inc., Natick, US) Fig. 2. shows the data process flow chart. The spectra were first processed using Asymmetric Least Square (ALS) baseline correction with OriginPro Software (OriginLab Corp., Northampton, 2021). Then, baselined spectra were vector normalized and Savitzky–Golay smoothed (4th order polynomial, with a frame size of 37) using Matlab 2021. Finally, multivariate analysis techniques and classification algorithms were performed in the spectral range, 300–1670 cm−1.
Before applying classifiers, the normalized and smoothed spectra were processed using multivariate statistical analysis to reduce the complexity of Raman spectra and extract the significant features that explain the most variance. The two multivariate analysis methods chosen to process the spectra were namely Principal Component Analysis (PCA) and Discriminant Analysis of Principal Components (DAPC) (Jombart et al., 2010; Yang and Yang, 2003). PCA is a multivariate statistical analysis that calculates the orthogonal combinations of the original variables and summarizes the variances in data. The dimension reduction of data was achieved by considering the number of significant principal components (PCs) that explain the most variance. For DAPC, PCA was applied to reduce the complexity of the spectra dataset; then, a supervised multivariate analysis, Discriminant Analysis, was used to further discriminate the dataset by correlating the variation in the data with the coffee information for each sample.
After feature extraction, common machine learning classifiers were used to classify coffee beverages. The classification classifiers in the Statistics and Machine Learning Toolbox of Matlab, including Decision Tree, Support Vector Machine (SVM), K-Nearest Neighbors (KNN), and Naïve Bayes, were selected. The number of the principal components (PCs) or canonical variables from the PCA or DAPC was varied from 1 to 80 in order to observe its influence on classification performance. The influences of the classifier hyperparameters on the classification accuracy were also evaluated. For SVM, linear and polynomial kernel functions were selected (Kancherla et al., 2019). For KNN, we assessed the impact of the number of nearest neighbors and the distance metrics, including Euclidean, Cosine, Chebyshev, Cityblock, Correlation, Mahalanobis, and Spearman (Abu Alfeilat et al., 2019; Chomboon et al., 2015). For Naïve Bayes, the classic Gaussian model was used. The classification model parameters from the training dataset were used to predict the validation dataset in each repetition. A 5-fold cross-validation was performed to assess the suitability of each classification algorithm, avoiding overfitting (Berrar, 2019). In brief, the training and the validation sets were established by randomly selecting from the Raman spectra data. The training dataset was used to generate a classification model, and the model predicted the validation dataset to evaluate the performance. The cross-validation approach was repeated five times, wherein the validation set consisted of 80 randomly selected Raman spectra in each repetition. The performance of the model was measured by classification accuracies, sensitivity, and selectivity. The accuracy, sensitivity, and the selectivity are defined as (Trevethan, 2017):
An additional two packages of Sample 3 were purchased to demonstrate the potential of the sensor for quality control. The SERS spectra from the additional coffee batches were collected using the same protocol described above, and 40 spectra were collected for each batch. The original 400 SERS spectra were used to train the classification models, and the established models were used to predict the class label of 80 spectra from the two new coffee batches. The performance was evaluated by the prediction accuracy.
3. Results and discussion
3.1. SERS spectra of coffee beverages
To evaluate the flavors of coffee beverages, we prepared aqueous coffee samples using the classic cold brew method (Baggenstoss et al., 2008; Rao et al., 2020; Semmelroch and Grosch, 1996). Although water extraction is better for evaluating flavor and aroma compounds in coffee beverages than organic extraction methods, strong fluorescent signals and extremely dilute analytes in coffee beverages impede aqueous coffee analysis using standard Raman spectroscopy (Dias and Yeretzian, 2016; El-Abassy et al., 2015). This issue could be resolved by SERS. A home-made SERS substrate, called nanopaper, was used to enhance Raman signals of coffee samples. The nanopaper is made by decorating a dense layer of silver nanoparticles on glass fiber paper, which possesses significant SERS enhancement (Fig. 1. (a) and Fig. 1. (b)). This low-cost SERS substrate was made by the well-known silver mirror reaction in a batch reactor. Our prior study has shown that the enhancement factor can reach 1.15 × 105, sufficient to detect dilute molecules in coffee beverages (Weatherston et al., 2018). In Fig. 1. (c), the SERS spectral features of the coffee beverage are much more significant than those in the Raman spectrum (Fig. 1. (c)).
SERS enhancement is highly dependent on hot spots, such as nanoscale gaps between nanoparticles or highly structured surfaces. A SERS substrate with uniform signal enhancement is almost impossible to make; thus, SERS often suffers from high variability in Raman signals Fig. 3. (a) shows five individual SERS spectra from the same coffee sample collected on a single SERS substrate at different locations; it is apparent that the uneven distributions and size variations of silver particles led to high variability in SERS signals. Moreover, SERS is a near-field electromagnetic effect, and signal enhancement decays exponentially due to distance from nanoparticle surfaces. Therefore, uneven adsorption of analytes on SERS substrates could also introduce signal variations.
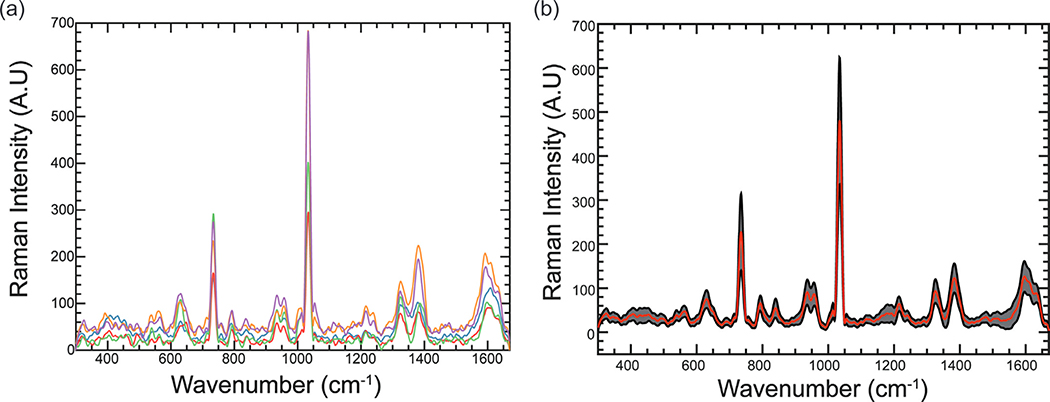
Fig. 3.
SERS spectra with high variations. (a) 5 individual SERS spectra of Sample 1 on a single SERS substrate at five random locations. (b) The average (red curve) and one standard deviation (grey area) of Sample 1 SERS spectra collected on 5 different days. (N = 80). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The inter-day and inter-substrate variations were also evaluated Fig. 3. (b) shows the average and standard deviation of SERS spectra (n = 80) collected on 5 different days using different SERS substrates. Although SERS signals fluctuate significantly, the overall morphologies of SERS spectra are relatively similar. Therefore, multivariate analysis can be applied to extract the significant features of SERS spectra.
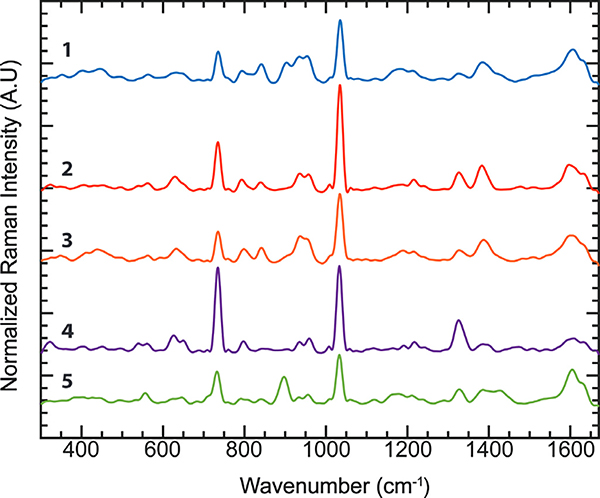
To evaluate the performance of this coffee classifier, we purchased five commercial coffees from local stores. Four of them (Sample 1 to Sample 4) were 100% Arabica beans from the Colombia Region with various degrees of roasting. The fifth sample (Sample 5) was the Robusta bean. The brand information is available in the Supplementary Materials. The averaged SERS spectra of these five samples are shown in Fig. 4. The peaks identified in the average SERS spectra were compared with the peaks reported in the literature. The positions of SERS peaks were shifted slightly from the positions of typical Raman spectra due to the interaction between chemical molecules and SERS substrates (Blum et al., 2012). We assigned those Raman bands to the closest peaks reported in the literatures and books (Table 1) with some of the peaks that have not been identified yet. Most observed SERS peaks are associated with aroma and flavor molecules in coffee beverages, including aromatic compounds (e.g., furan, kahweol, etc.), saccharides, organic acids, and caffeine (Caprioli et al., 2015). Although SERS provides rich structural information, the spectral difference among coffee samples is subtle. Therefore, multivariate analysis is used to extract significant spectral features such as the trivial difference between peaks, which are required to distinguish between the coffee samples properly.
Fig. 4.
Averaged Vector Normalized and Smoothed SERS spectra of coffee (1) Sample 1, (2) Sample 2, (3) Sample 3, (4) Sample 4, (5) Sample 5.
Table 1.
Tentative Peak Assignment Table.
| SERS Position (cm−1) | Band Assignment | Reference Position (cm−1) | Ref. |
|---|---|---|---|
|
| |||
| 311 | “Chain expansion” n-Alkanes | 150–425 | (Lin-Vien et al., 1991) |
| 325 | “Chain expansion” n-Alkanes | 150–425 | (Lin-Vien et al., 1991) |
| 348 | Skeletal deformation Monoalkyl acetylenes | 335–355 | (Lin-Vien et al., 1991) |
| 411 | “Chain expansion” n-Alkanes | 150–425 | (Lin-Vien et al., 1991) |
| 440 | Unknown | ||
| 495 | C-N-C | 482 | (El-Abassy et al., 2015) |
| 544 | Unknown | ||
| 564 | Unknown | ||
| 686 | ring breathing hydrothiophene | 688 | (Lin-Vien et al., 1991) |
| 709 | Ring Vibration Alkyl cyclohexanes | 700–785 | (El-Abassy et al., 2011, Lin-Vien et al., 1991) |
| 734 | O=C-C | 742 | (El-Abassy et al., 2015) |
| 760 | Ring Vibration Alkyl cyclohexanes | 700–785 | (Lin-Vien et al., 1991) |
| 799 | N-C-H | 801 | (El-Abassy et al., 2015) |
| 842 | Polysaccharides | 835 | (Rubayiza and Meurens, 2005) |
| 903 | Polysaccharides | 911 | (El-Abassy et al., 2011, El-Abassy et al., 2015, Wermelinger et al., 2011) |
| 937 | ring breathing 1,3-Dioxolane | 939 | (Lin-Vien et al., 1991) |
| 952 | Symmetric COC stretch from Aliphatic ethers | 830–930 | (Lin-Vien et al., 1991) |
| 1011 | Trigonal ring breathing from Pyridines, CGA | 1010–1030 | (El-Abassy et al., 2011, Lin-Vien et al., 1991, Zhang et al., 2016) |
| 1033 | asymmetric stretching of N-CH3 | 1029 | (El-Abassy et al., 2015, Zhang et al., 2016) |
| 1060 | Polysaccharides | 1062 | (Zhang et al., 2016) |
| 1077 | deformation of C-C | 1072 | (El-Abassy et al., 2015, Zhang et al., 2016) |
| 1098 | CC stretch n-alkynes, CGA | 950–1150 | (El-Abassy et al., 2011, Lin-Vien et al., 1991) |
| 1120 | cyclohexane (eye) CH, COH bending | 1120 | (Rubayiza and Meurens, 2005) |
| 1190 | phenyl ring CH, COH bending | 1193 | (Wermelinger et al., 2011) |
| 1215 | ring vibrations from Para-disubstituted benzenes | 1200–1230 | (Lin-Vien et al., 1991, Luna et al., 2019) |
| 1240 | deformation vibration of CH-N | 1250 | (Wermelinger et al., 2011) |
| 1290 | Stretching Vibration Mode of C-N | 1291 | (Figueiredo et al., 2019, Rubayiza and Meurens, 2005, Zhang et al., 2016) |
| 1327 | Unknown | (Wermelinger et al., 2011) | |
| 1387 | Ring stretch from Anthracenes, CGA | 1385–1415 | (El-Abassy et al., 2011, Lin-Vien et al., 1991) |
| 1484 | Kahweol C=C Furan | 1485 | (Rubayiza and Meurens, 2005, Wermelinger et al., 2011) |
| 1509 | 16-methyl-O-cafesto | 1507 | (El-Abassy et al., 2011, Figueiredo et al., 2019) |
| 1602 | phenyl ring stretching/CGA | 1605/1606 | (El-Abassy et al., 2011, Figueiredo et al., 2019) |
| 1634 | C=C ethylenic stretch vibration | 1630 | (El-Abassy et al., 2011) |
| 1662 | C=C cyclohexane | 1657 | (Figueiredo et al., 2019, Rubayiza and Meurens, 2005) |
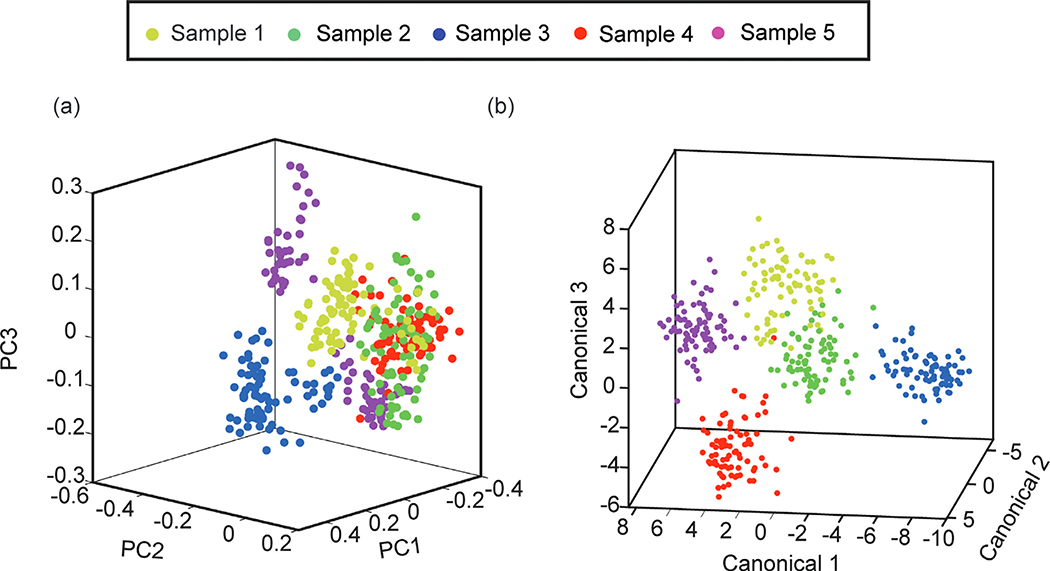
In this study, the performance of PCA and DAPC was evaluated. PCA was used to extract the features that explain the most difference among spectral data Fig. 5. (a) shows that the scatter plots of the top three PCA scores accounted for 74.6% (PCI, 44.4%; PC2, 20.0%; PC3, 10.1%) of the total Raman variations for the whole spectra. The loading plots of the top 3 PCs show the critical peaks explaining the variances among the coffee samples (Fig. S1). The major variations of Raman peaks are correlated to aroma and flavor molecules. Because PCA did not include group information, the coffee samples were not separated well Fig. 5. (b) shows the scatter plot of DAPC analysis fed by the top 27 PCs that accounted for 95.1% of the total variations. Since the spectral difference among the coffee samples is subtle, a higher number of PCs is required for catching trivial differences. The DAPC plot shows a better separation between coffee samples than the PCA plot.
Fig. 5.
Scatter plots of PCA and the DAPC scores. (a) Top three PCA scores. (b) Top three DAPC canonical scores for 27 PCs.
Before moving to classification, we investigated whether the background signals interfere with the SERS spectra from the coffee (Fig. S2). No significant overlapping were observed in coffee SERS spectra. To ensure the background interference is minimal, we repeated the experiments on different days using nanopapers from different synthesis batches.
3.2. Machine learning classifiers
After feature extraction, the principal components (PCs) from PCA or canonical variables from DAPC were used as input variables for training machine learning classifiers. The prior studies often combined probabilistic classifiers with DAPC or DA for classification (Fisher et al., 2018; Liu et al., 2016). In this study, the performance of a few common classifiers was evaluated, including Naïve Bayes, Decision Tree, K-Nearest Neighbor (KNN), Support Vector Machine (SVM). The Naïve Bayes classifier is a straightforward probabilistic classifier with the independence assumption based on Bayes’ theorem. However, the efficiency of the Naïve Bayes classifier depends on the size of the dataset and the probability models (Archana and Sachin, 2015). The Decision tree classifier is a standard machine learning algorithm and has been applied for facial recognition (Huang et al., 1996).
Both SVM and KNN have shown superior performance in the fields of computer vision and Raman analysis (Archana and Sachin, 2015; Bouzalmat et al., 2014; Li et al., 2009; Rebrošová et al., 2017). SVM finds a hyperplane to enhance the separation between classes, and according to the types of kernel functions, SVM could be applied to classify linear and nonlinear tasks (Kancherla et al., 2019; Savas and Dovis, 2019). In this study, we selected the linear and 3rd polynomial functions to evaluate the performances (Kancherla et al., 2019).
KNN is a non-parametric classifier that relies on distance metrics for classification. Therefore, the number of nearest neighbors and the choice of the distance measures could affect the performance of KNN (Abu Alfeilat et al., 2019). To select the best hyperparameters, we first found the optimal number of nearest number (k). Then, we evaluated the effect of distance metrics on classification accuracy.
3.3. Comparison of feature extraction and classification algorithms
We evaluated the performances of feature extraction (PCA and DAPC) and classification (Naïve Bayes, Decision Tree, KNN, SVM) algorithms using the 5-fold cross-validation. In the feature extraction step, SERS spectra from training data are first processed with PCA to reduce the dimensionality of the data and obtain the PC scores. Then, the desired number of PCs from PCA is used in the following process. If PCA is chosen for feature extraction, PCs will be directly used to train classifiers. For DAPC, PCs are further processed by discriminant analysis (DA) to obtain the canonical variables, and these canonical variables are then used to train classifiers. The 5-fold cross-validation accuracies of classification with different feature extractions and classifiers are reported in Table S1 and S2.
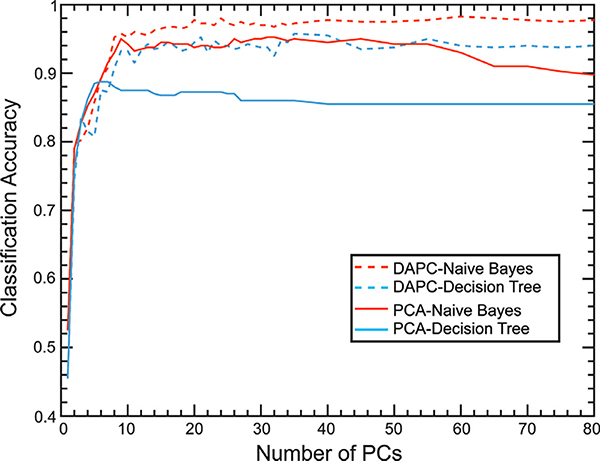
We first compared the combination of the Naïve Bayes classifier with PCA or DAPC (Fig. 6). The DAPC method gives better accuracies regardless of the number of PCs. The best classification accuracies of PCA and DAPC were 95.25% and 98.25%, respectively. The number of PCs used to train classifiers influences the classification accuracies, the maximum accuracies for PCA and DAPC were observed at 31 and 60 PCs, respectively. The confusion matrix, sensitivity, and selectivity of the best classification case are reported in Figure S3. In the PCA case with 31 PCs (95.7% of total variance explained), the classification accuracy reached 95.25%, and the sensitivity and selectivity of all samples were above 90%. In the DAPC case with 60 PCs (98.1% of total variance explained), the classification accuracy was improved to 98.25%, and the sensitivity and selectivity of all samples were over 93%.
Fig. 6.
Classification accuracies for Naïve Bayes classifier (Red) and Decision Tree classifier (Blue) with different numbers of PCs from spectra processed by PCA (Solid) or DAPC (Dash). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The Decision Tree classifier also shows a similar trend (Fig. 6). The best accuracies of the decision tree were 88.75% for PCA and 95.75% for DAPC. The maximum accuracies for PCA and DAPC were observed at 6 and 35 PCs, respectively. DAPC improved the classification performances for both classifiers. However, the overall classification performances of the Decision Tree were worse than the Naïve Bayes classifier (Fig. S4). Beyond these maximum points, we observed that the increase of PCs provides no additional benefits in improving classification accuracies. This trend is similar to data reported in the literature (Rebrošová et al., 2017). Overfitting of classifiers with noise in the training dataset could be the cause of this phenomenon (Ying, 2019).
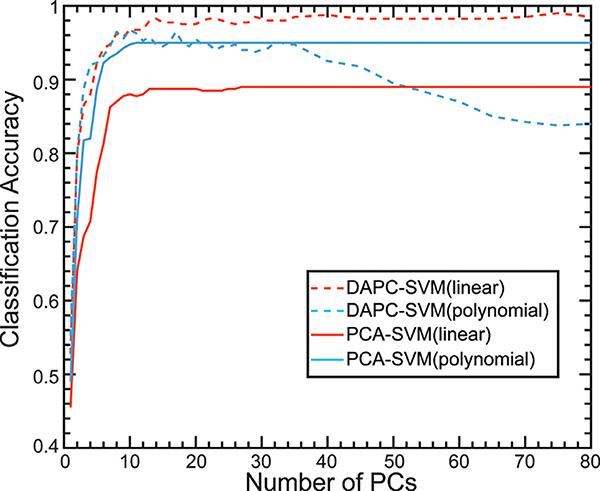
Fig. 7 shows the classification accuracies of SVM with PCA or DAPC. It has been reported that the kernel functions influence the performance of SVM (Kancherla et al., 2019; Savas and Dovis, 2019). Here, we studied the impact of the linear and the third-order polynomial kernel functions on classification performance. For PCA-SVM, the classification accuracies of linear kernel are worse than that of the polynomial kernel; however, for DAPC-SVM, the linear kernel offers better accuracy. Kancherla et al. has provided a general guideline regarding the use of SVM kernel functions based on the boundaries (Kancherla et al., 2019). If the class boundaries are nonlinear or overlapping, the nonlinear kernels are the more appropriate choices for SVM. In contrast, the linear kernel is preferable for a dataset with linear boundaries. Since PCA is a linear method, the polynomial kernel should provide better classification performance. For DAPC, the class boundaries are well defined (Fig. 5), so the linear kernel is a superb choice. In general, DAPC-SVM offers superior performance when compared to PCA-SVM. In addition, the maximum accuracy of SVM reached 99% with 75 PC (98.74% of total variables explained), which is superior to the Decision Tree classifiers and comparable to the Naïve Bayes classifier. The corresponding sensitivity and selectivity of each sample are close to 99% (Fig. S5).
Fig. 7.
Classification accuracies for SVM classifier with the linear kernel (Blue) or 3rd order polynomial kernel (Red) with different number of PCs from spectra processed by PCA (Solid) or DAPC (Dash). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
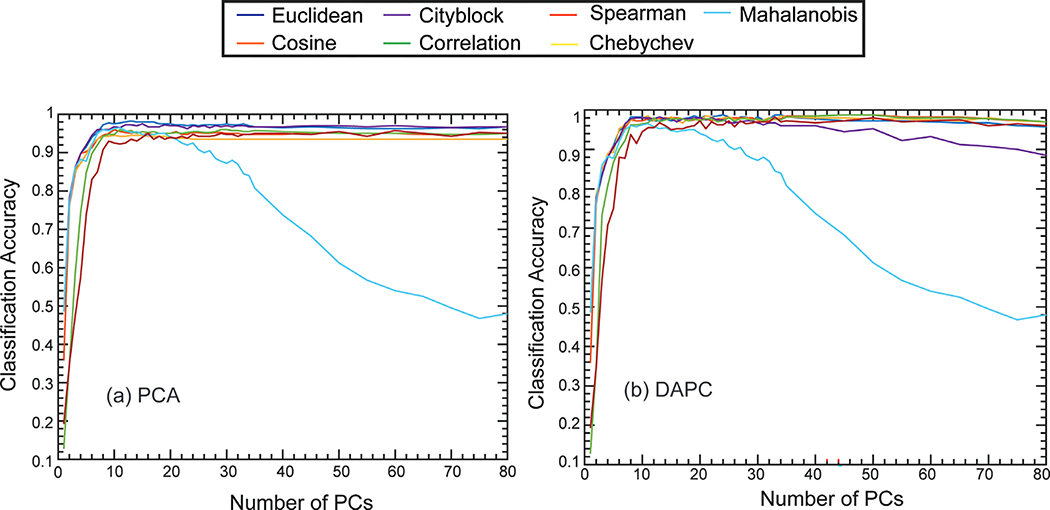
The performance of KNN depends on the hyperparameters, such as the number of nearest neighbors (k) and the distance metrics. Here, we selected seven typical distance metrics, including Minkowski distances (i.e., Euclidean, Cityblock, and Chebyshev), inner product distance (i.e., Cosine), and other distance measures (i.e., Correlation, Mahalanobis, and Spearman) (Abu Alfeilat et al., 2019; Chomboon et al., 2015). Before comparing the effects of distance metrics, we first searched the optimal number of nearest neighbors (k). Fig. S7 shows the average accuracy of KNN with different k values. The optimal accuracy appeared at k = 9; thus, we conducted the 9-nearest neighbor algorithm to evaluate the performance of the distance metrics.
The accuracies of PCA-KNN with different distance metrics are reported in Fig. 8. (a). Euclidean distance offers the best classification accuracy among seven metrics. For the PCA case, the maximum accuracy of the Euclidean distance reached 98.25% when 13 PCs (91.6% of total variance explained) were used to train the classifier. The Mahalanobis distance is observed to be sensitive to the number of PCs. The accuracy dropped when the number of PCs increased. In general, DAPC improves the classification accuracy of KNN, regardless of the selection of the distance metrics (Fig. 8. (b)). The maximum accuracies of different distance metrics except Mahalanobis are similar (98~09%). Still, the Euclidean Distance, Cosine Distance, and Correlation Distance offered the best accuracies (98.75%) using 24 PCs (94.6% of total variance explained) and 45 PCs (97.1% of total variance explained), respectively. The corresponding sensitivity and selectivity of each coffee sample were near 99%. In summary, the overall performance of KNN is comparable to the SVM linear kernel in DAPC.
Fig. 8.
Classification accuracies for KNN classifier using different distance metrics (Euclidean, Chebyshev, Correlation, Spearman, Cosine, City block, and Mahalanobis) with varying numbers of PCs from spectra processed by (a) PCA or (b) DAPC.
To demonstrate the potential application as a quality control tool, we analyzed the same coffee product from different packages. Coffee Sample 3 was selected for this multi-batch test because this coffee product was purchased from a source with better quality management. A total of 80 spectra from the two new coffee packages were collected (Fig. S8). The intensities of the spectra vary slightly, likely due to the variations of SERS enhancement and the content variations among coffee batches, but the overall morphologies are similar. The original 400 SERS spectra were used to build the classification model; then, the established model was used to predict the class label of the 80 new SERS spectra. The data points of these three different batches cluster together in the DAPC loading plot (Fig. S9). A 100% prediction accuracy was achieved by Naïve Bayes and SVM classifiers.
4. Conclusion
We demonstrated a coffee classifier that integrates SERS, feature extraction, and machine learning classifiers. The signal enhancement of SERS substrates enables the detection of the dilute molecules in coffee beverages. Although we observed high variability in SERS signals, the combinations of feature extraction tools and machine learning classifiers could successfully classify coffee beverages. Compared with PCA, DAPC improved the accuracies of all the classifiers tested in this study. SVM and KNN are superior to the Naïve Bayes and the Decision Tree classifiers. The performances of DAPC-SVM and DAPC-KNN are comparable (99% for SVM vs. 98.75% for KNN). The prior classification studies often combined DAPC or DA with probabilistic classifiers, such as Naïve Bayes classifier (Fisher et al., 2018; Liu et al., 2016). Thus, SVM and KNN could serve as an alternative choice for Raman spectra classification. In conclusion, this platform successfully classified coffee beverages with high accuracy using our machine learning tool and the inexpensive, versatile nanopaper.
Supplementary Material
Acknowledgment
This work was supported by the funds from National Science Foundation (award number: CHE-1904784), National Institutes of Health (award number: R03A1139650 and R21A1149383), and the AggiE-Challenge fund from the College of Engineering at Texas A&M University.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dche.2022.100020.
References
- Abu Alfeilat HA, Hassanat ABA, Lasassmeh O, Tarawneh AS, Al Hasanat MB, Eyal Salman HS, Prasath VBS, 2019. Effects of distance measure choice on K-nearest neighbor classifier performance: a review. Big Data 7, 221–248. [DOI] [PubMed] [Google Scholar]
- Archana H, Sachin D, 2015. Dimensionality reduction and classification through PCA and LDA. Int. J. Comput. Appl. 122, 4–8. [Google Scholar]
- Baggenstoss J, Poisson L, Kaegi R, Perren R, Escher F, 2008. Coffee roasting and aroma formation: application of different time–temperature conditions. J. Agric. Food Chem. 56, 5836–5846. [DOI] [PubMed] [Google Scholar]
- Berrar D, 2019. Cross-validation. In: Ranganathan S, Gribskov M, Nakai K, Schonbach C (Eds.), Encyclopedia of Bioinformatics and Computational Biology. Academic Press, Oxford. [Google Scholar]
- Blum C, Schmid T, Opilik L, Weidmann S, Fagerer SR, Zenobi R, 2012. Understanding tip-enhanced Raman spectra of biological molecules: a combined Raman, SERS and TERS study. J. Raman Spectrosc. 43, 1895–1904. [Google Scholar]
- Bouzalmat A, Kharroubi J & Zarghili A 2014. Comparative study of PCA, ICA, LDA using SVM classifier. [Google Scholar]
- Caprioli G, Cortese M, Sagratini G, Vittori S, 2015. The influence of different types of preparation (espresso and brew) on coffee aroma and main bioactive constituents. Int. J. Food Sci. Nutr. 66, 505–513. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang Y, Liu NR, Lin D, weng CC, Zhang J.x., Zhu LH, Chen WS, Chen R, Feng SY, 2013. Near-infrared confocal micro-Raman spectroscopy combined with PCA-LDA multivariate analysis for detection of esophageal cancer. Laser Physics 23. [Google Scholar]
- Chomboon K, Chujai P, Teerarassamee P, Kerdprasop K, Kerdprasop N, 2015. An empirical study of distance metrics for k-nearest neighbor algorithm. In: Proceedings of the 3rd international conference on industrial application engineering, pp. 280–285. [Google Scholar]
- Cruz R, Cardoso MM, Fernandes L, Oliveira M, Mendes E, Baptista P, Morais S, Casal S, 2012. Espresso coffee residues: a valuable source of unextracted compounds. J. Agric. Food Chem. 60, 7777–7784. [DOI] [PubMed] [Google Scholar]
- De Toledo PRAB, De Melo MMR, Pezza HR, Toci AT, Pezza L, Silva c.M., 2017. Discriminant analysis for unveiling the origin of roasted coffee samples: a tool for quality control of coffee related products. Food Control 73, 164–174. [Google Scholar]
- Dias RCE, Yeretzian C, 2016. Investigating coffee samples by Raman spectroscopy for quality control - preliminary study. Int. J. Exp. Spectrosc. Tech. 1, 1–5. [Google Scholar]
- El-Abassy RM, Donfack P, Materny A, 2011. Discrimination between Arabica and Robusta green coffee using visible micro Raman spectroscopy and chemometric analysis. Food Chem. 126, 1443–1448. [Google Scholar]
- El-Abassy RM, Von der Kammer B, Materny A, 2015. UV Raman spectroscopy for the characterization of strongly fluorescing beverages. LWT - Food Sci. Technol. 64, 56–60. [Google Scholar]
- Figueiredo LP, Borem FM, Almeida MR, Oliveira LFC, Alves APC, santos CMD, Rios PA, 2019. Raman spectroscopy for the differentiation of Arabic coffee genotypes. Food Chem. 288, 262–267. [DOI] [PubMed] [Google Scholar]
- Fisher AK, Carswell WF, Athamneh AIM, Sullivan MC, Robertson JL, Bevan DR, Senger R.s., 2018. The Rametrix™ LITE toolbox vi.o for MATLAB®. J. Raman Spectrosc. 49, 885–896. [Google Scholar]
- Fornasaro S, Al Samad F, Baia M, Batista De Carvalho LAE, Beleites C, Byrne HJ, Chiado A, Chis M, Chisanga M, Daniel A, Dybas J, Eppe G, Falgayrac G, Faulds K, Gebavi H, Giorgis F, Goodacre R, Graham D, La Manna P, Laing S, Litti L, Lyng FM, Malek K, Malherbe C, Marques MPM, Meneghetti M, Mitri E, Mohacek-Groöev V, Morasso C, Muhamadali H, Musto P, Novara C, Pannico M, Penel G, Piot O, Rindzevicius T, Rusu EA, Schmidt MS, Sergo V, Sockalingum GD, Untereiner V, Vanna R, Wiercigroch E, Bonifacio A, 2020. Surface enhanced raman spectroscopy for quantitative analysis: results of a large-scale European multi-instrument interlaboratory study. Anal. Chem. 92, 4053–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Gutta S, Wechsler H, 1996. Detection of human faces using decision trees. In: Proceedings of the Second International Conference on Automatic Face and Gesture Recognition, 14 Quarterly-16 Oct. 1996, pp. 248–252. [Google Scholar]
- Hwang J, Choi N, Park A, Park J-Q, Chung JH, Baek S, Cho SG, Baek S-J, Choo J, 2013. Fast and sensitive recognition of various explosive compounds using Raman spectroscopy and principal component analysis. J. Mol. Struct. 1039, 130–136. [Google Scholar]
- Jade AM, Srikanth B, Jayaraman v.K., Kulkarni BD, Jog JP, Priya L, 2003. Feature extraction and denoising using kernel PCA. Chem. Eng. Sci. 58, 4441–4448. [Google Scholar]
- Jamieson LE, Li A, Faulds K, Graham D, 2018. Ratiometric analysis using Raman spectroscopy as a powerful predictor of structural properties of fatty acids. R. Soc. open Sci. 5, 181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Hassan MM, Ali S, Li H, Sheng R, Chen Q, 2021. Evolving trends in SERS-based techniques for food quality and safety: a review. Trends Food Sci. Technol. 112, 225–240. [Google Scholar]
- Jombart T, Devillard S, Balloux F, 2010. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 11, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kancherla D, Bodapati JD, Veeranjaneyulu N, 2019. Effect of different kernels on the performance of an SVM based classification. Int. J. Recent. Technol. Eng. 1–6. [Google Scholar]
- Khalid S, Khalil T, Nasreen S, 2014. A survey of feature selection and feature extraction techniques in machine learning. 2014 Science and Information Conference, 2014-08-01. IEEE. [Google Scholar]
- Kim S-K, Park YJ, Toh K-A, Lee S, 2010. SVM-based feature extraction for face recognition. Pattern Recognit. 43, 2871–2881. [Google Scholar]
- Kotsiantis SB, Zaharakis I, Pintelas P, 2007. Supervised machine learning: a review of classification techniques. Emerg. Artif. Intell. Appl. Comput. Eng. 160, 3–24. [Google Scholar]
- Kuhar N, Sil S, Verma T, Umapathy S, 2018. Challenges in application of Raman spectroscopy to biology and materials. RSC Adv. 8, 25888–25908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao B, Zhang H, Jiao J, 2009. Face recognition system using SVM classifier and feature extraction by PCA and LDA combination. 2009 International Conference on Computational Intelligence and Software Engineering, 2009-12-01. IEEE. [Google Scholar]
- Lin-Vien D, Colthup NB, Fateley WG, Grasselli JG, 1991. The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules,. Elsevier Science & Technology, Saint Louis, United States. [Google Scholar]
- Liu W, Sun Z, Chen J, Jing C, 2016. Raman spectroscopy in colorectal cancer diagnostics: comparison of PCA-LDA and PLS-DA models. J. Spectrosc. 1–6 2016. [Google Scholar]
- Liu Y-J, Kyne M, Wang C, Yu X-Y, 2020. Data mining in Raman imaging in a cellular biological system. Comput. Struct. Biotechnol. J. 18, 2920–2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Lin Y, Zheng Z, Tang X, Lin J, Liu X, Liu M, Chen G, Qiu S, Zhou T, Lin Y, Feng S, 2018. Label free hepatitis B detection based on serum derivative surface enhanced Raman spectroscopy combined with multivariate analysis. Biomed. opt. Exp. 9, 4755–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A.s., Da Silva AP, Da Silva CS, Lima ICA, De Gois J.s., 2019. Chemometric methods for classification of clonal varieties of green coffee using Raman spectroscopy and direct sample analysis. J. Food Compos. Anal. 76, 44–50. [Google Scholar]
- Lussier F, Thibault V, Charron B, Wallace GQ, Masson J-F, 2020. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. Trends Anal. Chem. 124, 115796. [Google Scholar]
- Mao Y, Dong N, Wang L, Chen X, Wang H, Wang Z, Kislyakov IM, Wang J, 2020. Machine learning analysis of raman spectra of MoS2. Nanomaterials 10, 2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore M, 2013. Raman spectroscopy and partial least squares analysis in discrimination of peripheral cells affected by Huntington’s disease. Anal. Chim. Acta 793, 1–10. [DOI] [PubMed] [Google Scholar]
- Poltronieri P, Rossi F, 2016. Challenges in specialty coffee processing and quality assurance. Challenges 7, 19. [Google Scholar]
- Prakash O, Sil S, Verma T, Umapathy S, 2020. Direct detection of bacteria using positively charged ag/au bimetallic nanoparticles: a label-free surface-enhanced Raman scattering study coupled with multivariate analysis. J. Phys. Chem. C 124, 861–869. [Google Scholar]
- Rao NZ, Fuller M, Grim MD, 2020. Physiochemical characteristics of hot and cold brew coffee chemistry: the effects of roast level and brewing temperature on compound extraction. Foods (Basel, Switzerland) 9, 902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebrošová K, Šiler M, Samek 0, Růžička F, Bernatová S, Holá V, JEŽEK J, Zemánek P, Sokolová J, Petráš P, 2017. Rapid identification of staphylococci by Raman spectroscopy. Sci. Rep. 7, 14846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubayiza AB., Meurens M, 2005. Chemical discrimination of arabica and robusta coffees by fourier transform Raman spectroscopy. J. Agric. Food Chem. 53, 4654–4659. [DOI] [PubMed] [Google Scholar]
- Savas C, Dovis F, 2019. The impact of different kernel functions on the performance of scintillation detection based on support vector machines. Sensors 19, 5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seena V, Yomas J, 2014. A review on feature extraction and denoising of ECG signal using wavelet transform. 2014 2nd International Conference on Devices, Circuits and Systems (ICDCS), 2014-03-01. IEEE. [Google Scholar]
- Semmelroch P, Grosch W, 1996. Studies on Character Impact Odorants of Coffee Brews. J. Agric. Food Chem. 44, 537–543. [Google Scholar]
- Senger RS, Scherr D, 2020. Resolving complex phenotypes with Raman spectroscopy and chemometrics. Curr. Opin. Biotechnol. 66, 277–282. [DOI] [PubMed] [Google Scholar]
- Senger RS, Sullivan M, Gouldin A, Lundgren S, Merrifield K, Steen C, Baker E, Vu T, Agnor B, Martinez G, Coogan H, Carswell W, Kavuru V, Karageorge L, Dev D, Du P, Sklar A, Pirkle J, Guelich S, Orlando G, Robertson JL, 2020. Spectral characteristics of urine from patients with end-stage kidney disease analyzed using Raman Chemometric Urinalysis (Rametrix). PLOS One 15, e0227281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma B, Frontiera RR, Henry A. l., Ringe E, van Duyne RP, 2012. SERS: materials, applications, and the future. Mater. Today 15, 16–25. [Google Scholar]
- Sigurdsson S, Philipsen PA, Hansen LK, Larsen J, Gniadecka M, Wulf HC, 2004. Detection of skin cancer by classification of raman spectra. IEEE Trans. Biomed. Eng. 51, 1784–1793. [DOI] [PubMed] [Google Scholar]
- Trevethan R, 2017. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front. Public Health 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherston JD, Seguban RKO, Hunt D, wu H-J, 2018. Low-Cost and Simple Fabrication of Nanoplasmonic Paper for Coupled Chromatography Separation and Surface Enhanced Raman Detection. ACS Sensors 3, 852–857. [DOI] [PubMed] [Google Scholar]
- Weatherston JD, Worstell NC, Wu H-J, 2016. Quantitative surface-enhanced Raman spectroscopy for kinetic analysis of aldol condensation using Ag–Au core–shell nanocubes. Analyst 141, 6051–6060. [DOI] [PubMed] [Google Scholar]
- Weatherston JD, Yuan S, Mashuga CV, wu H-J, 2019. Multi-functional SERS substrate: collection, separation, and identification of airborne chemical powders on a single device. sens. Actuat. B 297, 126765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermelinger T, D’ambrosio L, Klopprogge B, Yeretzian C, 2011. Quantification of the Robusta Fraction in a coffee blend via Raman spectroscopy: proof of principle. J. Agric. Food Chem. 59, 9074–9079. [DOI] [PubMed] [Google Scholar]
- Wright J, Yang AY, Ganesh A, Sastry SS, Ma Y, 2009. Robust face recognition via sparse representation. IEEE Trans. Pattern Anal. Mach. Intell. 31, 210–227. [DOI] [PubMed] [Google Scholar]
- Yang J, Yang J-Y, 2003. Why can LDA be performed in PCA transformed space? Pattern Recognit. 36, 563–566. [Google Scholar]
- Ying X, 2019. An overview of overfitting and its solutions. J. Phys. Conf. Ser. 1168, 022022. [Google Scholar]
- Yuan S, Jiao Z, Quddus N, Kwon JS-I, Mashuga CV, 2019. Developing quantitative structure–property relationship models to predict the upper flammability limit using machine learning. Ind. Eng. Chem. Res. 58, 3531–3537. [Google Scholar]
- Yuan S, Zhang Z, Sun Y, Kwon JS-I, Mashuga CV, 2020. Liquid flammability ratings predicted by machine learning considering aerosolization. J. Hazard. Mater. 386, 121640. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang C, Liu F, He Y, 2016. Mid-infrared spectroscopy for coffee variety identification: comparison of pattern recognition methods. J. Spectrosc. 1–7 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.