Abstract
Telomerase is a ribonucleoprotein responsible for maintaining telomeres in nearly all eukaryotic cells. The enzyme is able to utilize a short segment of its RNA subunit as the template for the reverse transcription of d(TTAGGG) repeats onto the ends of human chromosomes. Transfection with telomerase was shown to confer immortality on several types of human cells. Moreover, telomerase activation appears to be one of the key events required for malignant transformation of normal cells. Inhibition of telomerase activity in transformed cells results in the cessation of cell proliferation in cultures and provides the rationale for the selection of telomerase as a target for anticancer therapy. Using oligonucleotide N3′→P5′ phosphoramidates (NPs) we have identified a region of the human telomerase RNA subunit (hTR) ∼100 nt downstream from the template region whose structural integrity appears crucial for telomerase enzymatic activity. The oligonucleotides targeted to this segment of hTR are potent and specific inhibitors of telomerase activity in biochemical assays. Mutant telomerase, in which 3 nt of hTR were not complementary to a 15 nt NP, was found to be refractory to inhibition by that oligonucleotide. We also demonstrated that the binding of NP, oligonucleotides to this hTR allosteric site results in a marked decrease in the affinity of a telomerase substrate (single-stranded DNA primer) for the enzyme.
INTRODUCTION
Telomerase is the enzyme responsible for maintaining telomeres in nearly all eukaryotic cells. It is a ribonucleoprotein that utilizes a short sequence within its RNA subunit as the template for reverse transcription, synthesizing d(TG)-rich repeats, which in vertebrates comprises the hexanucleotide d(TTAGGG) (1). In human tissues, telomerase activity can be detected in germ and stem cells, but not in most somatic or differentiated cells (2). The absence of telomerase activity leads to telomere erosion due to the inability of the conventional replication machinery to duplicate the ends of linear chromosomes (3). Cells cultured in vitro may undergo senescence when their telomeres shorten (4). The expression of various viral oncoproteins in cells allows them to bypass this state of growth arrest and re-enter the cell cycle (5). However, at some critical point, continued cellular proliferation in the absence of telomerase activity results in chromosomal instability—a phenomenon known as cell crisis (6,7). Although crisis normally refers to cells grown in culture, data from telomerase RNA gene knockouts in mice strongly suggest that analogous events may also occur in vivo (8,9). During crisis, most cells in a population undergo apoptotic death. In rare instances cells can emerge from crisis, which is nearly always concomitant with the re-acquisition of telomerase activity (10). Cells expressing telomerase have the potential to be immortal, which in fibroblasts, retinal pigment epithelial cells and endothelial cells also appears to be sufficient for proliferative immortality (11–13). Moreover, telomerase expression is one of several key events required for the malignant transformation of cells in vitro and in vivo (14), which serves to explain the earlier correlation linking telomerase activity with the vast majority of malignant tumors (15). This, as well as the finding that over-expression of a dominant negative form of the enzyme could drive tumor cell lines into crisis (16), provided the rationale for selecting telomerase as an attractive target for anticancer therapy.
In vitro, telomerase catalyzes the addition of d(TTAGGG) repeats onto a single-stranded DNA substrate. Two telomerase components are necessary and sufficient for activity in vitro: an RNA, part of which serves as the template for the synthesis of hexanucleotide repeats, and a protein that comprises the catalytic subunit. Vertebrate telomerase RNAs are similar in length and recently a secondary structure has been proposed based on phylogenetic comparisons (17). Several conserved domains emerged from this comparison. In the 5′-half of the RNA, a putative pseudoknot structure was identified in addition to the template region. Three conserved domains could be also found in the 3′-half of the molecule. One of these, the box H/ACA domain, was recognized previously in human telomerase RNA (hTR) and found to be essential for RNA processing and stability (18). The core H/ACA SnoRNA proteins have been shown to interact with hTR (19–21) and are probably part of the telomerase holoenzyme.
The telomerase protein components (TERTs) that have been identified in yeast, ciliates and mammals are similar in size (∼100–130 kDa), but their region of sequence homology is limited to a small portion of the gene product. While there is a small stretch of amino acids that represents a unique motif among the TERTs (22), the majority of the homology is shared with other reverse transcriptases (RTs). These RT motifs are critical structural elements, as single amino acid substitutions have been shown to abolish telomerase activity (23–26). This information has limited utility, though, in aiding the design of inhibitors for the catalytic subunit. Apart from nucleoside analogs, in the absence of additional structural information, it is difficult to consider a rational approach for choosing inhibitors of the active site of telomerase. The RNA component, on the other hand, appears more amenable to such a rational approach and may be an ideal candidate for an oligonucleotide-based approach to inhibition. Indeed, there have been several reports describing oligonucleotides complementary to the template region as potent inhibitors of the enzyme (27–30).
We have designed modified oligonucleotides complementary to the template region of hTR, and have found these compounds to inhibit telomerase with high potency in vitro (IC50 values of ∼1 nM; R.Pruzan and S.Gryaznov, unpublished data). These are N3′→P5′ phosphoramidate (NP) oligonucleotides, where a 3′-amino group is substituted for the 3′-oxygen in the 2′-deoxyribose ring. These compounds have been shown to form very stable duplexes with single-stranded RNA, are resistant to nuclease degradation and display high specificity for RNA and DNA targets, with a relatively low affinity for proteins (31). While NP oligonucleotides targeted against the template region of hTR are potent telomerase inhibitors, we were interested in finding additional regions of the RNA that might be sensitive to inhibition by NP oligonucleotides. We have identified a segment of hTR nearly 100 nt downstream from the template region that is susceptible to inhibition by NP oligonucleotides complementary to this region. Upon binding of an NP oligonucleotide to that region, we have found a marked decrease in affinity of single-stranded DNA primer for the enzyme.
MATERIALS AND METHODS
Telomerase extracts and activity assay
Telomerase was prepared from 293 suspension cells that over-expressed the hTERT gene, using a myeloproliferative sarcoma virus promoter (MPSV). Whole cell extracts were prepared from frozen cell pellets. The cell pellets were resuspended in one packed cell volume of ‘H’ buffer [10 mM HEPES pH 7.9, 1 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.5 µg/ml leupeptin) and then lyzed with a dounce homogenizer. The concentration of salt in the lysate was adjusted to 0.3 M NaCl, which was stirred for 15 min and then centrifuged at 100 000 g. Solid ammonium sulfate was added to the supernatant (42% saturation) and insoluble proteins were pelleted. The pellets were resuspended in one-fifth of their original volume and dialyzed against buffer ‘A’ [20 mM HEPES–KOH pH 7.9, 1 mM MgCl2, 1 mM DTT, 1 mM ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 10% glycerol] containing 0.1 M NaCl. Following dialysis the extract was spun at 25 000 g to remove insoluble material. Telomerase was purified by antisense affinity chromatography similar to methods described previously, using a 2′-O-Me RNA complementary to the template region of hTR (32,33). Affinity purified extracts were further purified by gel filtration chromatography (G5000PW, Tosoh Biosep LLC). Extracts contained between 0.1 and 0.2 pmol of telomerase per ml of extract, as determined by quantitative northern blot analysis of hTR and by measuring activity using conditions that allowed only a single nucleotide addition to a pre-bound primer. Activity was determined by incubating telomerase extract with 100 nM primer [d(TTAGGG)3], 200 µM dTTP, 50 µM dGTP, 10 µM dATP, 10 µCi [α-33P]dATP (2000–4000 Ci/mmol) in a buffer containing 50 mM (N-2-hydroxyethyl piperazine-N′-3 propanesulfonic acid)–NaOH pH 8.5, 1 mM MgCl2, 1 mM DTT, 5% glycerol, 0.5 mM EGTA and 100 mM KOAc in a final reaction volume of 40 µl. Incubations were routinely done for 90 min at 37°C. Reactions were terminated with SDS (0.1%), EDTA (2.5 mM) and NaCl (100 mM) in a final volume of 200 µl. Single-stranded DNA (50 µg) was added along with sodium pyrophosphate (5 mM); the reactions were adjusted to 5% trichloroacetic acid (TCA) (w/v) and allowed to set on ice for 30 min. Following centrifugation, the pellets were suspended in water and re-precipitated with cold 5% TCA (w/v). The precipitate was collected on a glass fiber filter (no. 31; Schleicher and Schuell), washed with 1% cold TCA (w/v) followed by ethanol. The filters were air dried and counted following the addition of scintillation fluid. Background counts, which were derived from reactions carried out without the addition of primer (or in the presence of 5 mM EDTA) were subtracted from the total counts.
Enzyme inhibition was followed by plotting enzyme activity as a function of the log of the inhibitor concentration. IC50 values were derived by fitting the following equation to the data using the KaleidaGraph software package (Synergy): y = b + (a – b)/[1 + 10 exp (log c – x)], where b and a represented the bottom and top values of the sigmoidal curve, respectively, and c represents the concentration of inhibitor where 50% activity is observed (IC50).
Binding of NP oligonucleotides using native gels
NP oligonucleotides were labeled using [γ-32P]ATP and polynucleotide kinase. Specific activities of 2000–4000 c.p.m./fmol of oligonucleotides were obtained. Affinity purified telomerase (1 fmol) was incubated with the labeled oligonucleotide (2 nM) for 30 min at room temperature in a volume of 20 µl. Half of each sample was treated with proteinase K (100 µg/ml, 0.2% SDS) for 10 min at 37°C. Samples were analyzed by electrophoresis on a native gel. The gel (140 × 170 × 1.5 mm) contained 3.5% acrylamide (1:60 bisacrylamide), 0.5% agarose, 75 mM Tris and 75 mM glycine. The gel was pre-run for 30 min, and then electrophoresed at 200 V until the xylene cyanole dye migrated 10 cm. Gels were dried and analyzed with a PhosphorImager (Molecular Dynamics).
Determination of dissociation rates of primer
Telomerase-containing extract (18 µl, G5000PW pool) was incubated with d(TTAGGG)3 (100 nM) at 30°C for 30 min in an initial reaction volume of 180 µl. d(TTAGGG)4 was added (20 µM) and aliquots (18 µl) were removed at times indicated in the figure legend (see Fig. 5). A nucleotide mix (2 µl) containing 2 mM dTTP and 10 µCi [α-32P]dATP (3000 Ci/mmol) was added and the labeling reaction was allowed to proceed for 5 min. The reaction was terminated with SDS (0.1%), EDTA (2.5 mM), NH4OAc (750 mM) and pellet paint (2 µl; Novagen) in a final volume of 200 µl. The terminated reactions were extracted with an equal volume of phenol–chloroform (1:1) and precipitated with 2.5 vol of ethanol. Following centrifugation, the pellets were washed with 80% ethanol and resuspended in 90% formamide (10 µl) containing bromophenol blue and xylene cylanole. The samples were applied to an 18% urea (7 M) polyacrylamide gel containing 115 mM Tris, 15 mM borate and 0.6 mM EDTA. The gel (140 × 170 × 0.8 mm) was run at 21 W until the bromophenol blue ran off the gel. The gel was fixed in 5% TCA (w/v), rinsed in water, dried and analyzed with a PhosphorImager (Molecular Dynamics). The rate constants for disappearance of the +3 nt product were determined by fitting the data to the equation: y = A exp (–kt) (where A represents the amount of enzyme in complex at time 0, k is the rate constant and t is the time), using the Kaleidagraph software package (Synergy).
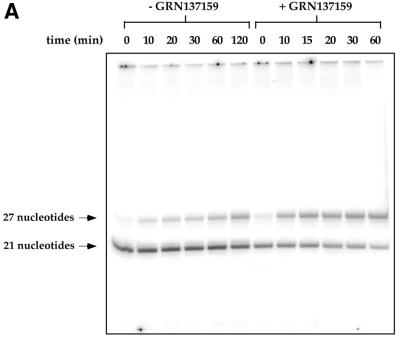
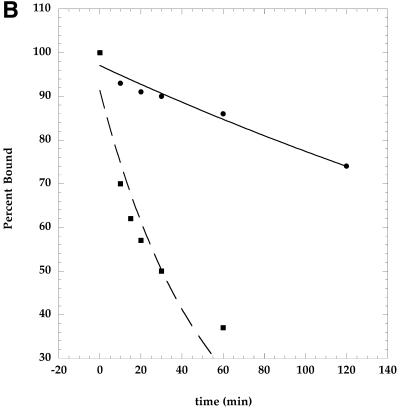
Figure 5.
Dissociation rate of primer from telomerase is increased in the presence of phosphoramidate oligonucleotide, GRN137159. Purified telomerase (G5000PW pool) was incubated with or without the phosphoramidate oligonucleotide for 15 min, followed by the addition of primer d(TTAGGG)3 (100 nM) at 30°C for 30 min. d(TTAGGG)4 was added (20 µM) and aliquots (18 µl) were removed at the times indicated above. A nucleotide mix (2 µl) containing 2 mM dTTP and 10 µCi [α-32P]dATP (3000 Ci/mmol) was added and the labeling was allowed to proceed for 5 min. Labeling will extend each primer by 3 nt. The samples were separated on an 18% poylacrylamide gel containing 7 M urea. (A) The gel was dried and analyzed by a PhosphoImager. (B) The bands in (A) were quantified and expressed as a percentage of the initial amount of ennzyme–primer complex. The amount of the primer labeled immediately following the challenge with the longer primer was assigned the value of 100%. The data were fit to the equation for exponential decay as described in Materials and Methods. The t1/2 for the primer’s dissociation from telomerase is ∼300 min without the presence of the NP oligonucleotide GRN137159, versus ∼35 min in the presence of GRN137159.
Expression of recombinant telomerase in mammalian cells
The SV40-transformed monkey cell line COS-7 (34) was transiently transfected with expression constructs for hTERT and hTR. Plasmid pGRN335 expresses hTERT from the MPSV promoter. Plasmid pGRN33 (35) expresses hTR under the control of its native genomic promoter. Cells at ∼50% confluence were transfected with 8 µg pGRN335 and 22 µg pGRN33, and mixed with 100 µl Lipofectamine reagent (Life Technologies, Inc.), according to the manufacturer’s instructions. At 48 h post-transfection, cultures were harvested and extracted by CHAPS detergent lysis (0.5%) and clarified by 100 000 g centrifugation. This extract was purified using Toyopearl Super Q-650 M (Tosoh Biosep LLC) in batch mode. The extract was bound to the resin in buffer ‘A’ containing 0.1 M NaCl, washed with buffer ‘A’ containing 0.18 M NaCl and eluted in the same buffer containing 0.3 M NaCl. Telomerase activity was found in the 0.3 M fraction.
RESULTS
The template region of hTR is an attractive target for telomerase inhibition with oligonucleotides. hTR has a stretch of 11 nt partially complementary to telomeric DNA. While six of the nucleotides serve as a template for reverse transcription, others may be involved in the enzyme’s substrate recognition or stabilization of its complex with telomeres. Several types of oligonucleotides complementary to this region (27–30) have already been described as potent telomerase inhibitors in biochemical assays and in vitro cell assays. We were interested in finding additional regions of hTR that might be susceptible to oligonucleotide-mediated inhibition. We initially targeted a region of hTR (nucleotides 137–179) that we believed contained a single-stranded RNA region, and which also appeared essential for telomerase activity (R.Pruzan and J.Kealey, unpublished observations; 36).
In order to assess the effects of various oligonucleotide inhibitors, we converted the conventional gel-based telomerase assay to one that followed the incorporation of radiolabeled nucleotide triphosphates into DNA, monitored as acid insoluble material. This assay allowed us to measure enzymatic activity directly, and to quantify the results in a simple manner (Materials and Methods). Performing this assay with highly purified telomerase extract allowed us to obtain a signal-to-background ratio of greater than 20:1.
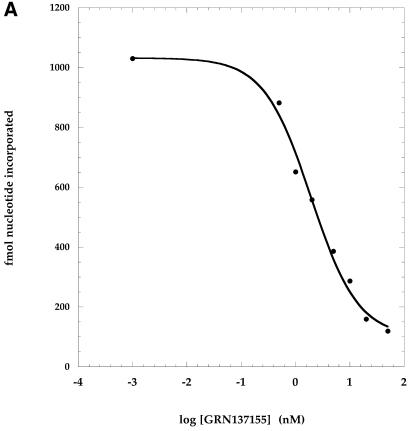
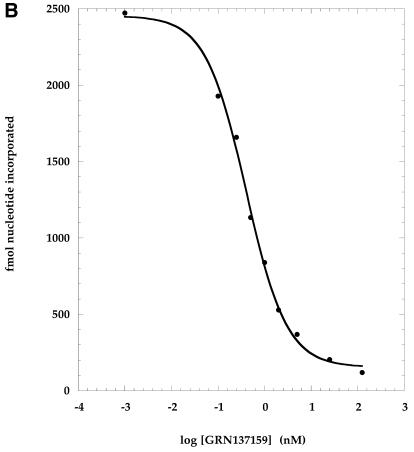
Inhibition of telomerase activity was determined following incubation of the enzyme with varying concentrations of NP oligonucleotides prior to the addition of DNA primer and deoxynucleotide triphosphates (dNTPs). The enzymatic activity was measured and expressed as femtomoles of nucleotides added to primer. The addition of nucleotides to primer was linear over the period of time used and can thus be considered a rate. Nucleotide addition rates were plotted as a function of the inhibitor–NP oligonucleotide concentration (Fig. 1). These graphs in turn were used to derive the IC50 values (as described in the legend to Fig. 1), which are summarized in Table 1.
Figure 1.
Dose–response inhibition of telomerase by phosphoramidate oligonucleotides directed against a non-template region of hTR. Phosphoramidate oligonucleotides were pre-incubated with purified telomerase extract at concentrations ranging from 0.2–125 nM. Primer [100 nM d(TTAGGG)3] and nucleotides [10 µM dATP (50 c.p.m./fmol [α-33P]dATP), 50 µM dGTP, 200 µM dTTP] were added and the reactions were incubated for 90 min at 37°C. Enzymatic activity was measured by determining the rate of nucleotide addition to primer as assessed by the incorporation of [α-33P]dATP into acid insoluble counts (described in Materials and Methods). Activity was plotted as a function of the log of the oligonucleotide concentration. These curves were used to derive the IC50 values shown in Table 1 (Materials and Methods). (A) GRN137155 (ACATTTTTTGTTTGCTCTAG); (B) GRN137159 (GTGGAAGGCGGCAGG).
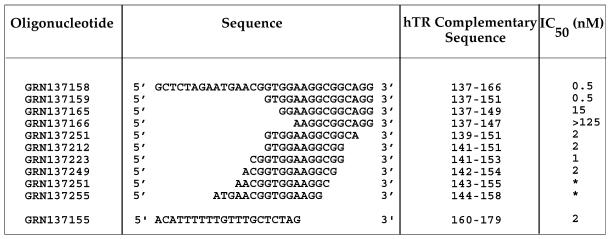
Table 1. Mapping non-template antisense region for the inhibition of telomerase activity.
Phosphoramidate oligonucleotides were pre-incubated with purified telomerase extract at concentrations ranging from 0.2–125 nM. Primer [100 nM d(TTAGGG)3] and nucleotides [10 µM dATP (50 c.p.m./fmol [α-33P]dATP), 50 µM dGTP, 200 µM dTTP] were added and the reactions were incubated for 90 min at 37°C. Enzymatic activity was measured by determining the rate of nucleotide addition to primer as assessed by the incorporation of [α-33P]dATP into acid insoluble counts. Dose-response curves as shown in Figure 1 were used to derive the concentration where 50% inhibition occurred (IC50).
*Compete inhibition was not reached even at concentrations over 100 nM; partial inhibition (20–50%) was observed in the low nanomolar range.
Inhibition of telomerase activity by non-template addressed oligonucleotide NPs
Several NP oligonucleotides were synthesized that were complementary to the hTR region mentioned above (nucleotides 137–179; Table 1). One of these, GRN137155, complementary to nucleotides 160–179, was found to inhibit telomerase activity with an IC50 value of ∼2 nM (Fig. 1; Table 1). A second oligonucleotide, GRN137158, complementary to nucleotides 137–166 and overlapping with the 3′-end of GRN137155 had an IC50 value of ∼0.5 nM (Table 1). We focused our attention on the latter oligonucleotide because a shortened version of it retained potency, whereas attempts to shorten GRN137155 without loss of activity were unsuccessful (data not shown). We found that the 3′-half of GRN137158 (compound GRN137159) was as active as the parental 30-nt molecule (Fig. 1B; Table 1).
In order to further define the active region, we synthesized several additional NP oligonucleotides. An 11mer NP oligonucleotide (GRN137212, Table 1) was nearly as active as the 15mer (GRN137159, Table 1). While removal of 4 nt from the 3′-end of GRN137159 had little effect on its potency, elimination of 4 nt from the 5′-end completely abolished the oligonucleotide’s activity (GRN137166, Table 1). Both of these 11mers were able to form duplexes with similar stabilities when mixed with a segment of telomerase RNA from this region (nucleotides 137–166) (Tm of >70°C in phosphate-buffered saline, data not shown). Shortening GRN137159 by even 2 nt from the 5′-end resulted in a >10-fold reduction in the compound’s activity (GRN137165, Table 1). Nucleotides complementary to 148–151 on hTR therefore appear critical for activity of these oligonucleotides. Additional nucleotides at the 5′-end of the oligonucleotide, complementary to nucleotides 152–153 of hTR, do not increase the potency of the compound (GRN137223, Table 1).
In contrast to the 5′-end of GRN137159, where removal of 4 nt results in complete inactivation of the compound, deletions of 5 nt from the 3′-end of GRN137159 are well tolerated (GRN137249, Table 1). Any further shortening from the 3′-end of GRN137249, however, affected the potency of the compounds. Eliminating 1 additional nt at the 3′-end (GRN137251, Table 1) or 2 additional nt from the 3′-end (GRN137255, Table 1) resulted in only a partial inhibition (50 and 20%, respectively) of activity (Table 1 and data not shown). This may be explained by partial inhibition of all the enzyme molecules, or the existence of two discrete enzyme populations—one sensitive and the other refractory to the inhibition by the oligonucleotides. Our observations are consistent with the first possibility, as partial inhibition with compound GRN137251 was by ∼50%, whereas inhibition with GRN137255 was only by ∼20%. Furthermore, this partial inhibition revealed an inelastic response to inhibitor concentrations over a 100-fold range of concentrations (data not shown), suggesting that hTR binding with the NP compounds can occur, but the resultant complexes retain activity. Taken together, the experimental data indicate the presence of 10 nt complementary to positions 142–151 on hTR are necessary to render the NP oligonucleotide an effective telomerase inhibitor. As a result of the high stability of an RNA–NP duplex, even an 11-nt long NP oligonucleotide is quite a potent telomerase inhibitor when targeted to a functionally important and accessible hTR region (GRN137212, Table 1). The dramatic decrease in potency observed with deletions from the 5′-end of GRN137159 suggest that activity is lost due to the inability of that NP oligonucleotide to bind to telomerase. The partial loss of activity resulting from 3′-end deletions may suggest the compound’s inability to perturb a structure necessary for enzymatic activity.
A mutant hTR, having a substitution within the critical region, is refractory to inhibition by GRN137159
Identification of the minimal effective NP oligonucleotide sufficient for inhibition of telomerase activity indicated the indispensability of 4 nt 5′-(GTGG) at the 5′-end of NP GRN137159, which may form potential base pairs with nucleotides 148–151 of hTR (Table 1 and see Fig. 6). The removal of these nucleotides resulted in a >250-fold reduction in potency as was seen with GRN137166 (Table 1). In order to unequivocally demonstrate that the 5′-end of GRN137159 potentiates the compound’s activity because of its ability to form a duplex with complementary sequences of hTR, we tested the ability of GRN137159 to inhibit a mutant telomerase enzyme that had an altered hTR sequence at positions 149–152. This mutant hTR would not allow the formation of 3 bp with the 3 5′-terminal nt of GRN137159, which were already shown to be critical for activity. If inhibition of telomerase by the NP oligonuceotide were a consequence of duplex formation with hTR, then the absence of activity with GRN137166 would predict that the mutant enzyme should also be refractory to inhibition by GRN137159.
Figure 6.
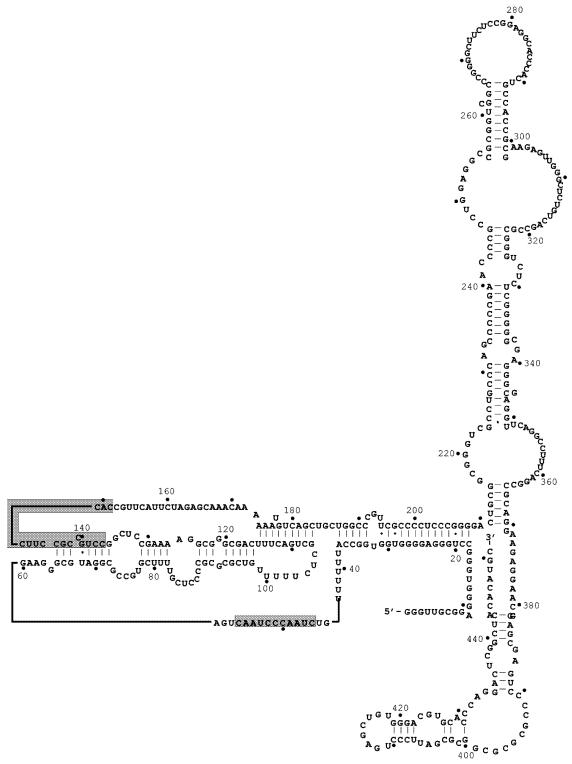
Proposed secondary structure of human hTR (17). The structure of human hTR was derived from the phylogenic comparison of 35 different vertebrate telomerase RNAs. Co-variation of potential base pairs in different organisms is used as an indication that a particular stem–loop structure may indeed exist. The template region (nucleotides 46–56) as well as the region complementary to the phosphoramidate oligonucleotide GRN137159 (137–151) have been shaded.
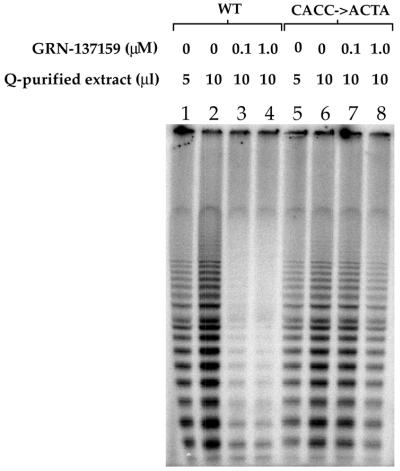
A system was designed that facilitated the overproduction of telomerase comprising mutant hTR (nucleotides 149–152 were changed from 5′-CACC to 5′-ACTA), or wild-type hTR, along with hTERT. Two plasmids containing SV40 origins of replication were used to transfect COS-7 cells, one expressing hTERT and the other expressing hTR. The COS-7 cells, which express SV40 large T antigen, allow the plasmids to replicate to a high copy number. The cells were harvested following transfection with the plasmids, and telomerase-containing extracts were prepared. The extracts were purified (see Materials and Methods) and assayed for the presence or absence of telomerase activity. It is important to note that the enzymatic activity in this system was completely dependent upon transfection with both the hTR and the hTERT bearing plasmids (data not shown). The hTR 5′-CACC to 5′-ACTA mutation of nucleotides 149–152 appeared to have a minimal effect on telomerase activity (Fig. 2, compare lanes 1 and 2 with 5 and 6).
Figure 2.
Recombinant telomerase containing a mutant hTR with nucleotide substitutions at positions 149–152 (CACC→ACTA) is refractory to inhibition by GRN137159. Wild-type or mutant hTR was transiently co-expressed with hTERT (as described in Materials and Methods). The resulting recombinant enzymes were isolated by detergent lysis and partial purification on Super Q-650 M (Tosoh Biosep). Telomerase activities of wild-type and mutant enzymes were measured (described in Materials and Methods) after a 30 min pre-incubation with GRN137159 at the concentrations indicated. Telomerase products were analyzed on an 8% denaturing polyacrylamide gel.
Two different concentrations of extract were used to measure telomerase activity in order to demonstrate the linearity of the assay (Fig. 2, lanes 1 and 2, and 5 and 6). Addition of 100 nM of the oligonucleotide (GRN137159) to the assay mixture abolished the activity of the wild-type enzyme (Fig. 2, compare lanes 2 and 3), whereas telomerase comprising the mutant hTR was refractory to this treatment, and only a modest effect can be observed at 1 µM (compare lanes 6–8). The latter concentration is 1000-fold above the IC50 that we have determined for this NP oligonucleotide (Table 1). The data support the notion described above, that the 5′-end of GRN137159 is critical for its inhibitory effect on telomerase, and its role may be to facilitate the binding of the oligonucleotide to telomerase.
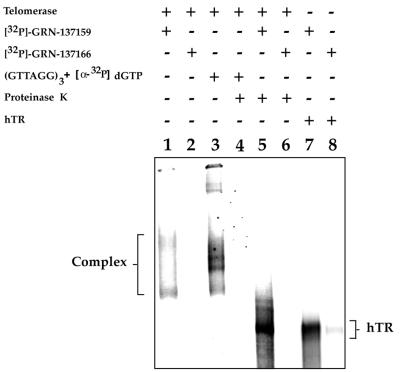
Although unlikely, there remained the formal possibility that the difference in potency between GRN137159 and GRN137166 was not a manifestation of their ability to bind to telomerase. In order to eliminate this possibility, we carried out an experiment to directly examine the potential of these labeled NP oligonucleotides to bind telomerase. Affinity purified telomerase was incubated with either 32P end-labeled GRN137159 or GRN137166. Following incubation the samples were analyzed by native gel electrophoresis (Fig. 3). In the sample that contained labeled GRN137159, a complex can be visualized that migrates in the region of the gel where active telomerase is found (Fig. 3, lanes 1 and 3). This is in sharp contrast to the absence of a visible complex when labeled GRN137166 is used (Fig. 3, lane 2). The position of telomerase on this native gel was determined by the auto-labeling of a pre-bound telomeric primer, having a 3′-terminal sequence of d(AGG), following the addition of [α-32P]dGTP (Fig. 3, lane 3). When the GRN137159-labeled complex is treated with proteinase K, the complex is converted into a discrete band that migrates in the position of hTR (Fig. 3, lanes 5–8). Once again nothing can be seen in the GRN137166 sample treated with proteinase K.
Figure 3.
GRN137159 binds to telomerase by association with hTR. Affinity purified telomerase was incubated with either 32P-labeled GRN137159 (4000 c.p.m./fmol) or GRN137166 (2000 c.p.m./fmol), as indicated above, followed by analysis on a native gel (described in Materials and Methods). The position of the active telomerase RNP complex (lane 3) was used as a reference and obtained by incubating the enzyme with (GTTAGG)3 and [α-32P]dGTP. Duplicate samples were treated with proteinase K, where indicated. The labeled oligonucleotides were also allowed to bind in vitro synthesized hTR (lanes 7 and 8), which serves as a marker for free telomerase RNA.
This demonstrates that labeled GRN137159 binds to telomerase and that binding occurs via a stable interaction with hTR. The tight interaction of telomerase with primer, in comparison, is destroyed by proteinase K treatment (Fig. 3, lanes 4 and 5), which indicates that the primer–enzyme complex is primarily stabilized by contacts with the protein.
The auto-labeled telomerase complex reveals the presence of four distinct telomerase-specific bands (Fig. 3, lane 3). It is unclear if they represent telomerase complexes differing in protein content, conformation or multimers of the enzyme. A similar pattern, however, is observed following blotting and probing with an anti-sense hTR (R.Pruzan, unpublished data). Interestingly, only two bands are illuminated following incubation with labeled GRN137159, which may indicate that the compound is able to partially disrupt the complex. We also tested the ability of the labeled oligonucleotides to bind in vitro synthesized hTR. GRN137166 is also severely limited in its ability to bind hTR (Fig. 3, lanes 7 and 8), which would suggest that the intrinsic structure of the RNA is the determining factor in binding.
Mixed mode of inhibition with non-template addressed NP oligonucleotide
In an effort to uncover the mode of action of GRN137159, we measured the inhibition of telomerase activity by this oligonucleotide as a function of the DNA primer concentration. If the compound acts as a competitive inhibitor with the DNA primer substrate of telomerase, then its inhibitory effect should be overcome by higher primer concentrations. This inhibitory mechanism would result in a higher Km value for the primer while the Vmax of the enzyme would not be affected. If, on the other hand, inhibition was non-competitive with respect to the primer concentration, then the Km values should not change and only the Vmax should be lower.
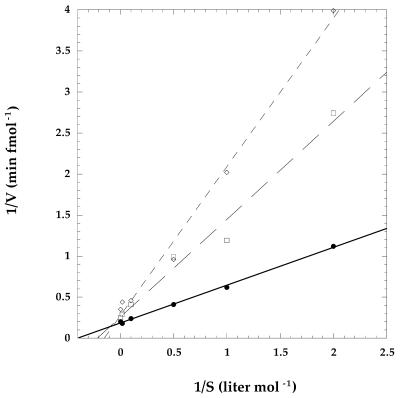
Telomerase activity was measured at primer concentrations ranging from 0.5 to 250 nM. The primer titration was done without the inhibitor oligonucleotide and in the presence of 1 or 2 nM of GRN137159. The resulting data were used to generate double-reciprocal plots: 1/rate of nucleotide addition as a function of 1/concentration of primer (Fig. 4). The Vmax and Km at each concentration of the inhibitor were derived from the y and x intercepts, respectively (Table 2). Both the Km and Vmax are affected by GRN137159 (Table 2). This indicates a mixed mode of telomerase inhibition, which might be the result of the inhibitor directly affecting both the catalytic rate of the enzyme and its interaction with primer, or solely affecting the latter. A decreased enzyme affinity for the primer would result in a less processive enzyme and, thus, appear as an overall lower rate of nucleotide addition, if one assumes that processive elongation occurs faster than substrate turnover.
Figure 4.
The double-reciprocal Lineweaver–Burke plot of telomerase inhibition by phosphoramidate oligonucleotide GRN137159. Telomerase activity was measured by following the incorporation of [α-33P]dATP (50 c.p.m./fmol dATP) onto an 18 nt primer d(TTAGGG)3 (described in Materials and Methods), over a range of primer concentrations (0.5–250 nM) with either 0 (circles), 1 (squares) or 2 (diamonds) nM inhibitor oligonucleotide present. For each inhibitor concentration, a double-reciprocal plot of the rate of nucleotide addition versus the primer concentration was performed. Linear regression was applied to each data set and the resulting equations were used to derive the Vmax and Km from the respective x and y intercepts. The correlation coefficients (r) of the equations for their respective data were ≥0.99.
Table 2. The Vmax of the enzyme and the Km of the primer are affected by the presence of phosphoramidate oligonucleotide GRN137159.
| [GRN137159] (nM) | Km (nM) | Vmax (fmol nucleotide min–1) |
|---|---|---|
| 0 | 2.5 | 5.5 |
| 1 | 4.8 | 3.9 |
| 2 | 6.7 | 3.7 |
A more rapid dissociation rate (koff) is observed for a primer in the presence of GRN137159
One possible explanation for the observed higher Km value for the primer in the presence of GRN137159 would be a reduction of affinity for that primer by telomerase in the presence of the NP oligonucleotide. The estimation of stability of the enzyme–primer complex can be derived from either a determination of dissociation equilibrium constant (KD), or alternatively through measuring the dissociation rate of the primer from the enzyme (koff). Experimentally, the latter approach was the more tractable one. Dissociation rates were determined by a gel-based electrophoresis method using two primers of different lengths (Materials and Methods). We measured the koff values in the absence or in presence of the NP oligonucleotide inhibitor (GRN137159). The enzyme was equilibrated with a saturating amount of a short telomeric primer [100 nM d(TTAGGG)3]. This incubation was followed by challenge with vast molar excess of a longer primer [20 µM d(TTAGGG)4]. At various times following the addition of the longer primer, aliquots were taken from the reaction mixture and pulse labeled (5 min) with dTTP and α-32P-labeled dATP. Only the primer associated with the enzyme will be labeled by dTTP and radioactive dATP using this experimental design. A labeled product of either a 21mer or a 27mer oligonucleotide product should result. The disappearance of the 21-nt oligonucleotide product and the concomitant appearance of the 27mer with passage of time were monitored by a denaturing polyacrylamide gel. The obtained comparison of the dissociation rates of the primer from telomerase with or without the addition of the inhibitory NP oligonucleotide is presented in Figure 5. The bands corresponding to the two possible products seen in Figure 5A were quantified and presented graphically in Figure 5B. The half-life (t1/2) of an 18-nt long telomeric primer d(TTAGGG)3 is ∼300 min at 30°C, whereas it is reduced to ∼35 min in the presence of the NP oligonucleotide (GRN137159). Moreover, we observed formation of a more labile complex between the enzyme and primer irrespective of whether GRN137159 was added prior or subsequent to the addition of primer to the reaction mixture (data not shown).
DISCUSSION
We have identified a segment of hTR, ∼100 nt downstream of the hTR template region, whose structural integrity is crucial for telomerase enzymatic activity. An NP oligonucleotide complementary to nucleotides 137–151 of hTR inhibits the enzyme at sub-nanomolar concentrations. Elimination of 4 nt at the 5′-end of this compound, complementary to nucleotides 148–151 of hTR, render it inactive. Conversely, with modified telomerase, where 3 of the 4 nt of hTR were mutated to prevent formation of proper Watson–Crick base pairs with the 5′-end of GRN137159, this NP oligonucleotide has a dramatic reduction in its inhibitory activity. Furthermore, we have shown directly that GRN137159 can bind to telomerase, while GRN137166, which is shortened by 4 nt at the 5′-end, has a limited ability to bind to telomerase.
Taken together, our results are consistent with a proposed mechanism whereby GRN137159 inhibits telomerase activity via binding to hTR and by doing so disrupts its functionally important secondary structure. The inability of a 5′-shortened oligonucleotide GRN137166 to inhibit telomerase, as well as the observation that an enzyme containing the mutated hTR sequence (at nucleotides 148–151) becomes refractory to inhibition by GRN137159, suggest that these 4 nt may be part of a single-stranded RNA region. This region may serve as an ‘entry point’ for the oligonucleotide GRN137159 facilitating its hybridization with hTR. This conclusion is supported by the direct binding data, which demonstrate that indeed the 4 nt complementary to 148–151 in hTR facilitates that binding to telomerase. The obtained data also suggest that GRN137159 probably disrupts the active hTR structure by forming a more stable heteroduplex with the RNA segment. It was reported earlier that NP oligonucleotide duplexes with RNA are up to ∼3°C more stable (per base pair) than for those with the corresponding native phosphodiester oligonucleotides (31). Indeed, a phosphodiester oligonucleotide, isosequential to GRN137159, does not inhibit telomerase activity (data not shown).
Recently, a secondary structure for hTR has been proposed, which is based on phylogenetic conservation among other vertebrate telomerase RNAs (17). This structure suggests that hTR nucleotides 136–144 are involved in an intramolecular duplex with nucleotides 65–72 (Fig. 6). This proposed secondary structure of hTR is consistent with the suggestion that hTR nucleotides 148–151 may provide a single-stranded ‘entry point’ for the hybridization of NP oligonucleotide (GRN137159) to telomerase. Given the stability of a NP–RNA duplex, it is reasonable to propose that this partial NP–RNA duplex can proceed further and invade the RNA helix just beyond the template region (Fig. 6, nucleotides 65–72 paired with 136–144, or P2a.1; 17).
One of the conserved features that emerged from the previously mentioned phylogenetic comparison of mammalian telomerase RNAs was a pseudoknot structure, which in hTR would predict base pairing between nucleotides 107–115 with nucleotides 174–183 (17; Fig. 6). While we did not pursue further experiments with GRN137155, it is interesting to note that this oligonucleotide overlaps extensively with this proposed structure and it is reasonable to hypothesize that GRN137155 may inhibit telomerase by its ability to disrupt this putative structure. This compound may prove to be a useful reagent for further studies involving this region of hTR.
While binding of the NP oligonucleotide (GRN137159) to its complementary hTR sequence appears to be crucial for its inhibitory effect, the interesting question arises as to this oligonucleotide’s mechanism of action. We observed a mixed mode of telomerase inhibition by GRN137159, both competitive and non-competitive with a DNA primer. We measured telomerase enzymatic activity as a rate of nucleotides incorporated into DNA, where this number represents the activity of an aggregate enzyme population. A lower Vmax could represent a reduced rate of nucleotide addition or possibly a lower percentage of active enzymes in the total population. Thus, the allosteric effect of GRN137159 binding to hTR, which is manifested in a reduced telomerase affinity for the primer, may either result in less efficient catalysis or, for example, in a less stable telomerase ribonucleoprotein complex. Presently we are unable to distinguish between these alternatives. The inhibition can be explained in part by the determined reduction (nearly 10-fold) in affinity of the telomere sequence based primer d(TTAGGG)3 for the enzyme. Reduced affinity for the primer was observed irrespective of the order of the inhibitors addition to the reaction mixtures. It is yet unclear how perturbation of the structure of hTR distal to the template region might affect primer binding, which must await further elucidation of the hTR structure.
Further work will elucidate the extent to which GRN137159 acts as an effective inhibitor in cells. In a preliminary experiment we have not observed inhibition in ACHN cells by GRN137159. However an NP oligonucleotide directed to the template region of hTR inhibited enzyme activity with an IC50 of 0.3 µM under similar experimental conditions (data not shown; 37). Lack of telomerase inhibition in cells by GRN137159 may either reflect the inaccessibility of the targeted hTR sequence in cells, or may reflect the differences in affinity to the natural substrate in cells versus a synthetic primer used in the purified system. The exact structure of DNA–protein complex substrate for telomerase, as well as potential important telomerase accessory factors in cells are still unknown.
We have, however, discovered an allosteric oligonucleotide-based reagent that apparently perturbs the secondary structure of the telomerase, resulting in the enzyme inhibition in a cell-free system. Further study will be required to define the relationship between the targeted region of hTR and the catalytic and template sites of telomerase.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to acknowledge data from Dr E. M. Atkinson and Dr S. L. Weinrich that prompted our initial examination into this region of hTR. We would also like to acknowledge Dr R. Carlos for providing us with whole cell extracts, and Dr D. J. Earp and Dr C. B. Harley for critically reading the manuscript.
REFERENCES
- 1.O’Reilly M., Teichmann,S.A. and Rhodes,D. (1999) Telomerases. Curr. Opin. Struct. Biol., 9, 56–65. [DOI] [PubMed] [Google Scholar]
- 2.Wright W.E., Piatyszek,M.A., Rainey,W.E., Byrd,W. and Shay,J.W. (1996) Telomerase activity in human germline and embryonic tissues and cells. Dev. Genet., 18, 173–179. [DOI] [PubMed] [Google Scholar]
- 3.Olovnikov A.M. (1996) Telomeres, telomerase and aging: origin of the theory. Exp. Gerontol., 4, 443–448. [DOI] [PubMed] [Google Scholar]
- 4.Levy M.Z., Allsopp,R.C., Futcher,A.B., Greider,C.W. and Harley,C.B. (1992) Telomere end-replication problem and cell aging. J. Mol. Biol., 225, 951–960. [DOI] [PubMed] [Google Scholar]
- 5.Shay J.W., Wright,W.E. and Werbin,H. (1991) Defining the molecular mechanisms of human cell immortalization. Biochim. Biophys. Acta, 1072, 1–7. [DOI] [PubMed] [Google Scholar]
- 6.Girardi A.J., Jensen,F.C. and Koprowski,H. (1965) SV40-induced transformation of human diploid cells: crisis and recovery. J. Cell. Comp. Physiol., 65, 69–84. [DOI] [PubMed] [Google Scholar]
- 7.Counter C.M., Avilion,A.A., LeFeuvre,C.E., Stewart,N.G., Greider,C.W., Harley,C.B. and Bacchetti,S. (1992) Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J., 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herrera E., Samper,E., Martin-Caballero,J., Flores,J.M., Lee,H.W. and Blasco,M.A. (1999) Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. EMBO J., 18, 2950–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudolph K.L., Chang,S., Lee,H.W., Blasco,M., Gottlieb,G.J., Greider,C. and DePinho,R.A. (1999) Longevity, stress response and cancer in aging telomerase-deficient mice. Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]
- 10.Kim N.W., Piatyszek,M.A., Prowse,K.R., Harley,C.B., West,M.D., Ho,P.L., Coviello,G.M., Wright,W.E., Weinrich,S.L. and Shay,J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 11.Bodnar A.G., Ouellette,M., Frolkis,M., Holt,S.E., Chiu,C.P., Morin,G.B., Harley,C.B., Shay,J.W., Lichtsteiner,S. and Wright,W.E. (1998) Extension of life-span by introduction of telomerase into normal human cells. Science, 279, 349–352. [DOI] [PubMed] [Google Scholar]
- 12.Counter C.M., Hahn,W.C., Wei,W., Caddle,S.D., Beijersbergen,R.L., Lansdorp,P.M., Sedivy,J.M. and Weinberg,R.A. (1999) Dissociation among in vitro telomerase activity, telomere maintenance and cellular immortalization. Proc. Natl Acad. Sci. USA, 96, 3339–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J., Chang,E., Cherry,A.M., Bangs,C.D., Oei,Y., Bodnar,A., Bronstein,A., Chiu,C.P. and Herron,G.S. (1999) Human endothelial cell life extension by telomerase expression. J. Biol. Chem., 274, 26141–26148. [DOI] [PubMed] [Google Scholar]
- 14.Hahn W.C., Counter,C.M., Lundberg,A.S., Beijersbergen,R.L., Brooks,M.W. and Weinberg,R.A. (1999) Creation of human tumour cells with defined genetic elements. Nature, 400, 401–402. [DOI] [PubMed] [Google Scholar]
- 15.Shay J.W. and Bacchetti,S. (1997) A survey of telomerase activity in human cancer. Eur. J. Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- 16.Hahn W.C., Stewart,S.A., Brooks,M.W., York,S.G., Eaton,E., Kurachi,A., Beijersbergen,R.L., Knoll,J.H., Meyerson,M. and Weinberg,R.A. (1999) Inhibition of telomerase limits the growth of human cancer cells. Nature Med., 10, 1164–1170. [DOI] [PubMed] [Google Scholar]
- 17.Chen J.L., Blasco,M.A. and Greider,C.W. (2000) Secondary structure of vertebrate telomerase RNA. Cell, 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell J.R. and Collins,K. (2000) Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell, 2, 361–371. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell J.R., Wood,E. and Collins,K. (1999) A telomerase component is defective in the human disease dyskeratosis congenita. Nature, 402, 551–555. [DOI] [PubMed] [Google Scholar]
- 20.Dragon F., Pogacic,V. and Filipowicz,W. (2000) In vitro assembly of human H/ACA small nucleolar RNPs reveals unique features of U17 and telomerase RNAs. Mol. Cell. Biol., 9, 3037–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pogacic V., Dragon,F. and Filipowicz,W. (2000) Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol., 20, 9028–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryan T.M., Sperger,J.M., Chapman,K.B. and Cech,T.R. (1998) Telomerase reverse transcriptase genes identified in Tetrahymena thermophila and Oxytricha trifallax. Proc. Natl Acad. Sci. USA, 95, 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lingner J., Hughes,T.R., Shevchenko,A., Mann,M., Lundblad,V. and Cech,T.R. (1997) Reverse transcriptase motifs in the catalytic subunit of telomerase. Science, 276, 561–567. [DOI] [PubMed] [Google Scholar]
- 24.Haering C.H., Nakamura,T.M., Baumann,P. and Cech,T.R. (2000) Analysis of telomerase catalytic subunit mutants in vivo and in vitro in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 97, 6367–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinrich S.L., Pruzan,R., Ma,L., Ouellette,M., Tesmer,V.M., Holt,S.E., Bodnar,A.G., Lichtsteiner,S., Kim,N.W., Trager,J.B. et al. (1997) Reconstitution of human telomerase with the template RNA component hTR and the catalytic protein subunit hTRT. Nature Genet., 17, 498–502. [DOI] [PubMed] [Google Scholar]
- 26.Collins K. and Gandhi,L. (1998) The reverse transcriptase component of the Tetrahymena telomerase ribonucleoprotein complex. Proc. Natl Acad. Sci. USA, 95, 8485–8490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukai S., Kondo,Y., Koga,S., Komata,T., Barna,B.P. and Kondo,S. (2000) 2–5A antisense telomerase RNA therapy for intracranial malignant gliomas. Cancer Res., 60, 4461–4467. [PubMed] [Google Scholar]
- 28.Herbert B., Pitts,A.E., Baker,S.I., Hamilton,S.E., Wright,W.E., Shay,J.W. and Corey,D.R. (1999) Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc. Natl Acad. Sci. USA, 96, 14276–14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glukhov A.I., Zimnik,O.V., Gordeev,S.A. and Severin,S.E. (1998) Inhibition of telomerase activity of melanoma cells in vitro by antisense oligonucleotides. Biochem. Biophys. Res. Commun., 248, 368–371. [DOI] [PubMed] [Google Scholar]
- 30.Norton J.C., Piatyszek,M.A., Wright,W.E., Shay,J.W. and Corey,D.R. (1996) Inhibition of human telomerase activity by peptide nucleic acids. Nat. Biotechnol., 5, 615–619. [DOI] [PubMed] [Google Scholar]
- 31.Gryaznov S.M. (1999) Oligonucleotide N3′→P5′ phosphoramidates as potential therapeutic agents. Biochim. Biophys. Acta, 1489, 131–140. [DOI] [PubMed] [Google Scholar]
- 32.Schnapp G., Rodi,H.P., Rettig,W.J., Schnapp,A. and Damm,K. (1998) One-step affinity purification protocol for human telomerase. Nucleic Acids Res., 26, 3311–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingner J. and Cech,T.R. (1996) Purification of telomerase from Euplotes aediculatus: requirement of a primer 3′ overhang. Proc. Natl Acad. Sci. USA, 93, 10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gluzman Y. (1981) SV40-transformed simian cells support the replication of early SV40 mutants. Cell, 1, 175–182. [DOI] [PubMed] [Google Scholar]
- 35.Feng J., Funk,W.D., Wang,S.S., Weinrich,S.L., Avilion,A.A., Chiu,C.P., Adams,R.R., Chang,E., Allsopp,R.C., Yu,J. et al. (1995) The RNA component of human telomerase. Science, 269, 1236–1241. [DOI] [PubMed] [Google Scholar]
- 36.Autexier C., Pruzan,R., Funk,W.D. and Greider,C.W. (1996) Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J., 15, 5928–5935. [PMC free article] [PubMed] [Google Scholar]
- 37.Gryaznov S., Pongracz,K., Matray,T., Schultz,R., Pruzan,R., Aimi,J., Chin,A., Harley,C., Shea-Herbert,B., Shay,J., Oshima,Y., Asai,A. and Yamashita,Y. (2001) Telomerase inhibitors-oligonucleotide phosphoramidates as potential therapeutic agents. Nucleosides Nucleotides Nucleic Acids, 20, 401–410. [DOI] [PubMed] [Google Scholar]