Abstract
The polypyrimidine tract binding protein (PTB), a homodimer that contains four RRM-type RNA binding domains per monomer, plays important roles in both the regulation of alternative splicing and the stimulation of translation initiation as directed by the internal ribosome entry sites of certain picornaviruses. We have used chemical shift mapping experiments to probe the interactions between PTB-34, a recombinant fragment that contains the third and fourth RRM domains of the protein, and a number of short pyrimidine-rich RNA oligonucleotides. The results confirm that the RNAs interact primarily with the β-sheet surface of PTB-34, but also reveal roles for the two long flexible linkers within the protein fragment, a result that is supported by mutagenesis experiments. The mapping indicates distinct binding preferences for RRM3 and RRM4 with the former making a particularly specific interaction with the sequence UCUUC.
INTRODUCTION
The polypyrimidine tract binding protein (PTB) is an RNA binding protein that functions to regulate the utilisation of messenger and pre-messenger RNA in a number of different contexts. While the principal physiological role of PTB appears to be tissue-specific regulation of alternative splicing of a large number of genes, such as α-actinin, α- and β-tropomyosin, c-src, fibroblast growth factor receptors and the γ2 subunit of the GABAA receptor (1–11), the protein has also been implicated in the control of polyadenylation (12,13) and mRNA localisation (14). Moreover, PTB is recruited by a number of picornaviruses as a stimulator of translation initiation driven by internal ribosome entry sites (IRES) (15–20). Recent work suggests that the protein may also regulate the translation of hepatitis C virus by interacting with both the 5′ and 3′ ends of the viral RNA (21,22) and that it can stimulate the activity of the cellular IRES of APAF-1 (23).
PTB functions in all these systems by interacting with RNA and other accessory proteins (splicing and translation initiation factors), although the mechanisms of action remain obscure. PTB binding sites within intron and IRES RNA are observed to contain repeats of short pyrimidine motifs (e.g. UCUU, UCUUC, UUCUCU, CUCUCU) (1,3,8,11,24), which are often, but not exclusively, contained within a pyrimidine-rich background. These observations are supported by results from in vitro selection (8,25) and binding experiments (26).
PTB is a homodimer (26) and contains four RRM-type RNA binding domains per monomer (27). Deletion mutagenesis studies have mapped the primary RNA binding activity to the third and fourth RRM domains of each monomer, although the contribution of RRM1 and RRM2 may also be significant (17,26,28–30). RRM2 appears to play an important role in the dimerisation of PTB (26,28). The solution structure of PTB-34, a monomeric fragment that contains the third and fourth RRM domains of PTB, revealed that RRM3 has an atypical structure for this type of module, as the four-stranded β-sheet, which acts as the RNA binding surface has been extended by one strand (29). The structure of PTB-34 is also dominated by two extended and flexible linker polypeptides, one (17 amino acids) connecting β-strands 4 and 5 of RRM3 and the other (25 amino acids) joining the two RRM domains, indicating that the protein has a high degree of conformational variability, at least prior to its interaction with RNA.
To extend our structural studies of the interactions of PTB with RNA targets, we have performed chemical shift mapping experiments on complexes between PTB-34 and a number of short, synthetic, pyrimidine-rich RNA molecules. This experimental approach reveals the amino acids that are in contact with the RNA ligand. Comparison of the different distributions of amino acid contacts observed with different RNA oligonucleotides reveals new insights regarding the sequence specificity of PTB.
MATERIALS AND METHODS
NMR spectroscopy
15N-labelled PTB-34, which contains residues 335–531 of human PTB-1, was expressed in Escherichia coli and purified as described previously (29). Synthetic RNA oligonucleotides were prepared and gel-purified by Dharmacon Research, Inc. For NMR mapping experiments, RNA oligonucleotides were titrated into solutions containing 0.5 mM 15N-labelled PTB-34 in 20 mM sodium acetate, pH 5.4. 15N-1H HSQC spectra (31) were recorded at RNA:PTB-34 mole ratios of 0.5:1, 1:1 and 2:1 to facilitate tracking and thus assignment of resonances perturbed by RNA binding to the protein. All the NMR spectra were acquired at 302 K using a four-channel Bruker DRX500 equipped with a z-shielded gradient and triple resonance probe. NMR data were processed using NMRPipe/NMRDraw (32) and analysed using NMRView (33).
Mutagenesis and RNA binding experiments
Mutants of hisPTB-34a (residues 324–531 of human PTB-1) were prepared by overlap PCR using established protocols and expressed in E.coli (29). All mutations were confirmed by cDNA sequencing. PTB–RNA binding experiments were performed using α-32P-labelled RNA transcripts of domain 1 from the EMCV IRES (17) in nitrocellulose filter binding assays as described (29). Briefly, PTB–RNA binding reactions (75 µl) were prepared and incubated for at 24°C at least 15 min in 10 mM HEPES (pH 7.25), 100 mM KCl, 3 mM MgCl2, 5% glycerol, 1 mM DTT, 50 µg/ml yeast tRNA (Boehringer Mannheim), 50 µg/ml human serum albumin (Delta Biotechnology). The RNA concentration was typically fixed at 4 nM. Assays were performed using the protein binding Protran BA-85 nitrocellulose membrane (Schleicher and Schuell). The membrane was washed extensively in 10 mM HEPES (pH 7.25), 3 mM MgCl2, 5% glycerol, 1 mM DTT and mounted on a 96-well dot-blotter (Bio-Rad). Before and after application of 65 µl of the binding reaction, the membrane was washed with 180 µl of wash buffer. Following the experiment, the membrane was dried and the quantity of bound PTB–RNA complex determined by scintillation counting of Cerenkov radiation. Except where stated otherwise, all reagents were purchased from Sigma-Aldrich.
RESULTS
Selection of RNA oligonucleotides
Naturally occurring PTB binding sites consist of multiple repeats of short pyrimidine sequences embedded in a stretch of intron or IRES RNA that is typically >100 nt in length. Such large targets are not readily amenable to analysis by NMR so our studies focused on the binding of short motifs. The sequences selected ranged in length from 4 to 10 nt and included motifs identified previously in PTB binding sites or slight variations of these (Table 1).
Table 1. RNA oligonucleotides used in chemical shift mapping experiments.
| No. | RNA motif | Occurrence in splice site sequences |
|---|---|---|
| 1 | UCUUa | Caspase-9 (4); clathrin light chain B (11); calcitonin/calcitoning gene related peptide (13); fibroblast growth receptor-1 (51); GABA γ2 subunit (1); N-methyl-d-aspartate receptor (11); α-actinin (5); α-tropomyosin (8); β-tropomyosin (6) |
| 2 | CUUC | |
| 3 | UCUUC | |
| 4 | UCUUUa | Fibroblast growth receptor-2 (2) |
| 5 | UCUCU | Caspase-9 (4); clathrin light chain B (11); c-src (52); GABA γ2 subunit (1); α-tropomyosin (8); β-tropomyosin (6) |
| 6 | CUCUCU | c-src (52); α-tropomyosin (8) |
| 7 | UCCUCUUC | As above for UCUUC |
| 8 | UCUUCUCU | bCaspase-9 (4); clathrin light chain B (11); c-src (52); GABA γ2 subunit (1); α-tropomyosin (8); β-tropomyosin (6) |
| 9 | UCUUCUCUUC |
Mapping the RNA binding site on PTB-34
15N-1H HSQC experiments (31) were used to monitor the backbone amide chemical shift changes in PTB-34 upon addition of RNA oligonucleotides (see Materials and Methods). This sensitive method detects perturbations in the environment of backbone amides due to RNA binding. Amide chemical shifts are usually also sensitive to the perturbations of connected side-chains that interact with the ligand, but they may also be affected by conformational changes in the protein induced by ligand binding, and this can complicate the location of the binding site. To facilitate proper delineation of the binding surface, emphasis was placed on large chemical shift titrations and perturbed amides that fell into concerted, surface-exposed patches. Among the RNAs studied in this report, the two RNA tetramers (UCUU and CUUC) showed no amide perturbations under our experimental conditions, indicating that there was no significant binding to PTB-34. However, addition of the pentameric, hexameric or octameric RNA oligonucleotides (oligonucleotides 3–8 in Table 1) in each case resulted in appreciable chemical shift changes for many PTB-34 resonances without causing significant increases in relaxation rates. The binding of these RNA oligonucleotides to PTB-34 were generally in the fast exchange limit on the NMR timescales, and approached saturation of binding at a 1:1 ratio of RNA:PTB-34.
Figure 1 shows overlays of HSQC spectra of uniformly 15N-labelled PTB-34 with and without an equal molar amount of ligand RNA for oligonucleotides 3–8. It should be noted that several resonances belonging to the highly flexible loop connecting β-strands 4 and 5 in RRM3 or the long flexible inter-domain linker were not assignable in the apo-protein due to severe overlap in the centre of the spectrum (29). Many of these show chemical shift changes upon binding to RNA, but it was not possible to record three-dimensional NMR data for the complexes (to facilitate sequence-specific assignment) because the RNA oligonucleotides induced degradation of the protein sample within 8 h.
Figure 1.
Overlay of 15N-1H HSQC spectra of 15N-PTB-34 ± RNA recorded at pH 5.38, 302 K. The spectra of apo-PTB-34 are shown in black and those of holo-PTB-34 in magenta.
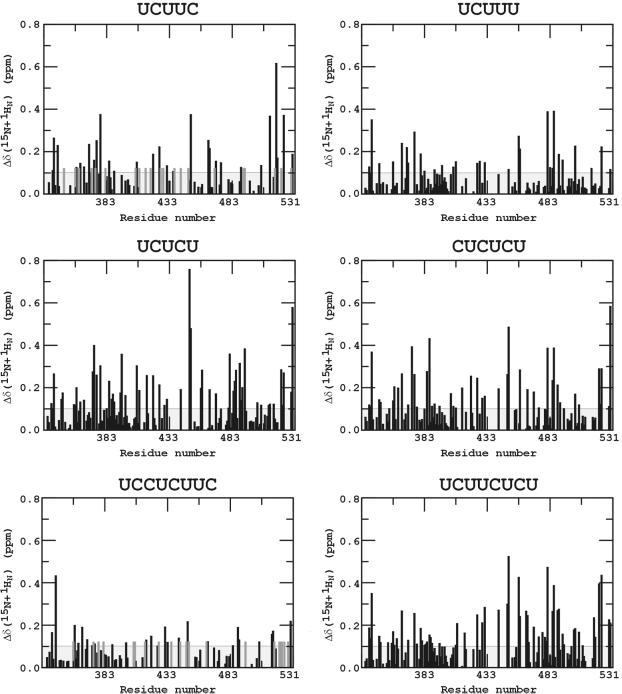
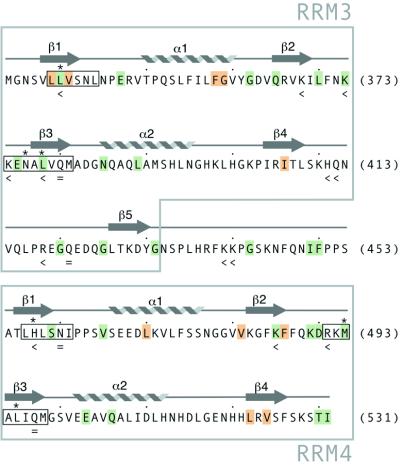
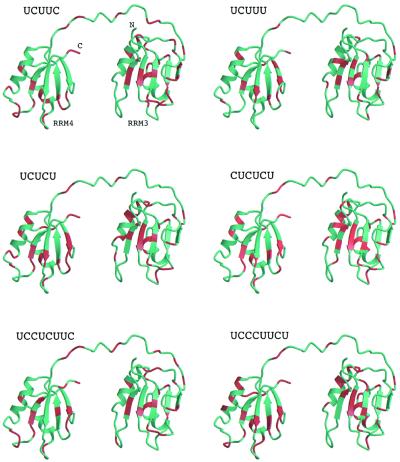
The combined backbone amide chemical shift changes in PTB-34 due to the binding of RNA oligonucleotides 3–8 (Table 1) are plotted as histograms in Figure 2. To give a general impression of residues involved in interactions with RNA ligands, Figure 3 indicates those residues with weighted backbone amide chemical shift perturbation ≥0.1 p.p.m. upon interaction with at least four out of the six RNA oligonucleotides used in our study. Residues with a weighted chemical shift perturbation ≥0.1 p.p.m. in PTB-34 upon binding to each of these six RNAs are indicated on the structure of PTB-34 in Figure 4. Collectively, the data for these six oligonucleotides show that the majority of residues perturbed by RNA binding occur on the surfaces of the β-sheets in the two RRMs, as expected for this type of module. The chemical shift data confirm and extend the characterisation of the RNA binding surface obtained from site-directed mutagenesis and binding experiments (29; and this work). Notably, β-strand 5 of RRM3, which is a novel extension of the canonical RRM, is also perturbed, thus confirming its importance in RNA recognition. The β1–α1 and β2–β3 loops in both RRM3 and RRM4 are also frequently perturbed by different oligonucleotides, consistent with the demonstrated role of these features in RNA binding by PTB (29) and other RRM proteins (34). Other residues in the core of the RRM domain and on the ‘back-side’ are also perturbed but these changes are most likely due to small conformational alterations in the protein induced by RNA binding (Fig. 4). In addition to the β-sheet surface, residues in the β4–β5 loop and the inter-domain linker are clearly affected by RNA binding (Figs 3 and 4). Previous mutagenesis data had already implicated the inter-domain linker in RNA recognition (29). The β4–β5 loop, however, is a feature that so far is unique to PTB. In support of the chemical shift data, mutations in this loop were found to significantly depress the affinity of PTB-34 for EMCV IRES domain 1 in binding assays (Table 2). The double mutation H411A/Q412A and the single mutation R418A both reduced RNA binding affinity by ∼2-fold, similar to the magnitude of the binding defect caused by mutations on the β-sheet binding surface (29).
Figure 2.
Combined chemical shift perturbation (Δδ) of the backbone 15N and 1HN resonances of PTB-34 upon binding of RNA. The absolute values of 15N and 1HN chemical shift changes are combined using this equation: Δδ(15N + 1HN) = │Δδ 15N│/4.69 + │Δδ 1HN│. The correction factor for 15N chemical shift changes is calculated as the ratio of the spectral width of the 15N versus that of the 1H (46,47). The 0.1 p.p.m. threshold is indicated by a horizontal line.
Figure 3.
Overview of amino acids residues in PTB-34 with significant chemical shift changes upon binding of RNA. Residues with a combined shift of perturbation of ≥0.1 p.p.m. for at least four out of the six RNA oligonucleotides which bound to the protein are shaded green or orange. Green shading indicates that the amino acid side-chain is solvent-exposed; buried side-chains are shaded orange. The hexameric RNP-2 and octameric RNP-1 motifs in both RRMs are boxed. Asterisks mark the positions of residues that are aromatic in most RRM sequences (48). Mutation of residues marked with ‘<’ reduced the RNA binding affinity; residues marked with ‘=’ had no effect on RNA binding affinity (29).
Figure 4.
Structure mapping of the chemical shift patterns obtained with the different RNA oligonucleotides used in this study. The relative disposition shown for RRM3 and RRM4 is arbitrary since the two domains appear to tumble independently in solution. Residues with a combined shift of perturbation of ≥0.1 p.p.m. are shaded red. The figure was prepared using Molscript (49) and Raster3D (50).
Table 2. Dissociation constants for binding of hisPTB-34s mutants to EMCV IRES domain 1 RNA.
| Mutation | Kd (µM) | Structural location |
|---|---|---|
| Wild-type | 0.083 ± 0.009 | – |
| H411A/Q412A | 0.14 ± 0.01 | β4– β5 loop |
| R418A | 0.20 ± 0.01 | β4– β5 loop |
| Q424A | 0.09 ± 0.01 | β4– β5 loop |
Data were fitted to a single-site binding model. Values quoted are the average of three experiments (± standard deviation).
As well as demonstrating that the unusual features of PTB (β5, the β4–β5 loop and the long inter-domain linker) are all involved in RNA binding, the data reveal that a collection of hydrophobic, polar and positively charged amino acids are involved in RNA recognition (Fig. 3). PTB is notable for the absence of a triad of aromatic residues that is found in most other RRM proteins and makes important stacking interactions with RNA bases. However, several of the hydrophobic residues that in PTB occupy these generally conserved aromatic positions (Leu 340 and Leu 378 in RRM3, Met 493 in RRM4) are found to have significant chemical shift alterations indicating that they maintain a role in RNA binding. These aliphatic side-chains in PTB may make similar interactions to the hydrophobic Ile-base contacts observed in the crystal structures of sex-lethal and HuD (35,36). Although the three remaining ‘aromatic’ positions in PTB (Asn 376 in RRM3, His 457 and Leu 495 in RRM4) show only small or negligible chemical shift changes in the presence of RNA, this does not preclude their involvement in RNA interactions. Indeed, mutation of His 457 has been observed to reduce RNA binding (29).
Comparison of the patterns of chemical shifts associated with the different RNA oligonucleotides reveals a number of intriguing differences. The sequence UCUUC induces chemical shifts that are almost exclusively clustered on the binding surface of RRM3 indicating that this oligonucleotide may be specifically bound to RRM3 (Fig. 4). This interpretation is supported by the observation that the RNA decamer (UCUUCUCUUC, oligonucleotide 9), comprising two copies of the UCUUC sequence, yields significant line broadening across the whole of the spectrum (data not shown), indicative of the formation of a high molecular weight complex, probably with a protein:RNA stoichiometry greater than 1:1. A simple explanation for this would be that two molecules of PTB-34 bind to the oligonucleotide primarily via their RRM3 modules. The oligonucleotide UCUUCUCU which differs from oligonucleotide 9 only in that it lacks 2 nt at the 3′ end, does not cause the same dimerisation, reinforcing the inference that two copies of UCUUC are required for the binding of more than one PTB-34 molecule to the oligonucleotide.
Minor modifications of the UCUUC sequence, to give UCUUU and UCUCU, dramatically alter the distribution of chemical shifts, yielding a larger perturbation surface that extends to cover much of the β-sheet surface in RRM4 (Fig. 4). The crystal structure of the N-terminal pair of RRM domains of poly(A)-binding protein (PABP) complexed with poly(A) revealed that the RNA binds in a highly extended configuration such that 6 nt were sufficient to span the RRM pair (37). The UCUUU and UCUCU pentamers may adopt similar extended configurations when bound to PTB-34, an interpretation supported by the observation that the chemical shift changes were close to maximal at a 1:1 RNA:PTB-34 mole ratio. Nevertheless, it is possible that the distributed patterns of shifts for these two oligonucleotides are due to shuttling of the ligands across the β-sheet surfaces within the two RRM domains, so that the observed perturbation surface represents an average of different binding conformations. A pattern of shifts that extends across RRM3 and RRM4 was also observed for the hexamer (CUCUCU), which shows a very similar perturbation surface to that of UCUCU (see Figs 1 and 4). The extra pyrimidine (cytosine) at the 5′ end does not seem to have affected the binding specificity.
The two octamers UCCUCUUC and UCUUCUCU each contain the UCUUC motif but have three additional pyrimidine residues at the 5′ and 3′ ends respectively. The magnitude of chemical shift perturbations for UCCUCUUC are significantly smaller than for UCUUC (Fig. 2), implying that UCCUCUUC induces smaller conformational changes in the protein or causes further chemical shift averaging over several conformations. Thus, the binding specificity of the sequence UCUUC for RRM3 seems to have been compromised in UCCUCUUC. For the other octamer UCUUCUCU, the magnitude of conformational perturbation to RRM3 is similar to that for UCUUC; but in addition, it also causes significant changes in RRM4 of a similar magnitude and specificity to that of the UCUCU pentamer (Fig. 4), which matches the five residues at the 3′ end of UCUUCUCU.
DISCUSSION
The RRM is an extremely versatile module which binds RNA in a single-stranded configuration but in a diverse range of sequence and structural contexts, as revealed by structural analyses of U1A, U2B″, sex-lethal, PABP and nucleolin (36–40). PTB remains something of an outlier in the extended family of RRM proteins, as it possesses unusual RNP-1 and RNP-2 sequence motifs and has a novel fifth strand in RRM3 (29). The presence of this fifth strand alters the topology of RRM-3 and raises the possibility that the arrangement of the tandem RRM domains in PTB-34 may not conform to that observed for other proteins (in which the C-terminal RRM binds upstream of the N-terminal RRM on the RNA target). However, our results are consistent with a model in which the pair of RRMs in PTB-34 are required to bind 5–6 nt sequence motifs, most likely in an extended configuration similar to that observed for poly(A) bound to PABP (although our present data give no indication that the RNA ligands help to lock a particular configuration of PTB-34 and the actual binding configuration will require further work). Given that PTB acts as a dimer (with a total of eight RRMs), our data also support the idea that PTB can make multiple contacts with intron and IRES RNA targets (3,24), and needs to do so to make a specific, functional interaction (e.g. to stabilise an active configuration). In particular, our findings suggest that a single PTB dimer could bind pairs of UCUUC or CUCUCU motifs, consistent with cross-linking models of PTB–RNA interactions in which the PTB dimer bridges two RNA motifs that are widely separated in the primary sequence (3,13).
A universal PTB recognition sequence common to all PTB binding sites identified to date has not been found (Table 1). Our mutagenesis results tend to highlight the relatively non-specific nature of the interaction of PTB with RNA targets, given that single or double amino acid substitutions in the protein reduce the binding affinity by a factor of four at most (Table 2) (29). In contrast, the chemical shift mapping reveals clear differences in the binding interactions of oligonucleotides that only differ by a single nucleotide. In this regard the observation of specific binding of UCUUC to RRM3 is very striking, particularly in comparison with the lack of binding of UCUU and the very different pattern of chemical shift changes observed with UCUUU. The specific binding of UCUUC to RRM3 is intriguing as the same RNA sequence motif was obtained by in vitro selection experiments even though these were done with the full-length protein (8). However, it should be borne in mind that our studies were carried out using purified PTB-34. The binding of the protein to pyrimidine-rich motifs may be modulated by RNA sequence and structural contexts and by the presence of protein co-factors (e.g. Nova, hnRNP-L, unr, ITAF45) that interact with PTB and/or with RNA sequences proximal to PTB binding sites (23,41–45). While our chemical shift data give a clear indication of the modulation of PTB–RNA interactions by sequence context variations (compare UCUUC, UCUUCUCU and UCCUCUUC), further work towards the determination of the structure of a PTB–RNA complex will be required to reveal the nature of the protein–RNA interaction in greater detail.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Philip Sharp (MIT, USA), Richard Jackson and Ann Kaminski (Cambridge University, UK), Graham Belsham (IAH, Pirbright, UK) and Doug Black (UCLA, USA) for reagents and discussions. This work is funded by grant support from the Wellcome Trust and the Biotechnology and Biological Sciences Research Council, UK.
REFERENCES
- 1.Ashiya M. and Grabowski,P.J. (1997) A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA, 3, 996–1015. [PMC free article] [PubMed] [Google Scholar]
- 2.Carstens R.P., Wagner,E.J. and Garcia-Blanco,M.A. (2000) An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol., 20, 7388–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chou M.Y., Underwood,J.G., Nikolic,J., Luu,M.H. and Black,D.L. (2000) Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol. Cell, 5, 949–957. [DOI] [PubMed] [Google Scholar]
- 4.Coté J., Dupuis,S. and Wu,J.Y. (2001) Polypyrimidine track-binding protein binding downstream of caspase-2 alternative exon 9 represses its inclusion. J. Biol. Chem., 276, 8535–8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gooding C., Roberts,G.C. and Smith,C.W. (1998) Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA, 4, 85–100. [PMC free article] [PubMed] [Google Scholar]
- 6.Mulligan G.J., Guo,W., Wormsley,S. and Helfmant,D.M. (1992) Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem., 267, 25480–25487. [PubMed] [Google Scholar]
- 7.Norton P.A. (1994) Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res., 22, 3854–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez I., Lin,C.-H., McAfee,J.G. and Patton,J.G. (1997) Mutation of PTB binding sites causes misregulation of alternative 3′ splice sites selection in vivo. RNA, 3, 764–778. [PMC free article] [PubMed] [Google Scholar]
- 9.Southby J., Gooding,C. and Smith,C.W.J. (1999) Polypyrimidine tract binding protein functions as a repressor to regulate alternative splcing of α-actinin mutually exclusive exons. Mol. Cell. Biol., 19, 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner E.J. and Garcia-Blanco,M.A. (2001) Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol., 21, 3281–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang L., Liu,W. and Grabowski,P.J. (1999) Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA, 5, 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreira A., Takagaki,Y., Brackenridge,S., Wollerton,M., Manley,J.L. and Proudfoot,N.J. (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev., 12, 2522–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lou H., Helfman,D.M., Gagel,R.F. and Berget,S.M. (1999) Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol. Cell. Biol., 19, 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cote C.A., Gautreau,D., Denegre,J.M., Kress,T.L., Terry,N.A. and Mowry,K.L. (1999) A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell, 4, 431–437. [DOI] [PubMed] [Google Scholar]
- 15.Jang S.J. and Wimmer,E. (1990) Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev., 4, 1560–1572. [DOI] [PubMed] [Google Scholar]
- 16.Niepmann M., Petersen,A., Meyer,K. and Beck,E. (1997) Functional involvement of polypyrimidine tract binding protein in translation initiation complexes with the internal ribosome entry site of foot-and-mouth disease virus. J. Virol., 71, 8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminski A., Hunt,S.L., Patton,J.G. and Jackson,R.J. (1995) Direct evidence that polypyrimidine tract binding protein (PTB) is essential for internal initiation of translation of encephalomyocarditis virus RNA. RNA, 1, 928–938. [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski A. and Jackson,R.J. (1998) The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA, 4, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt S.L. and Jackson,R.J. (1999) Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA, 5, 344–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert R., Chang,K.H., Rijnbrand,R., Yi,M., Sangar,D.V. and Lemon,S.M. (2000) Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol., 20, 1583–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito T. and Lai,M.M. (1999) An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology, 254, 288–296. [DOI] [PubMed] [Google Scholar]
- 22.Anwar A., Ali,N., Tanveer,R. and Siddiqui,A. (2000) Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem., 275, 34231–34235. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell S.A., Brown,E.C., Coldwell,M.J., Jackson,R.J. and Willis,A.E. (2001) Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol. Cell. Biol., 21, 3364–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolupaeva V.G., Hellen,C.U.T. and Shatsky,I.N. (1996) Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA, 2, 1199–1212. [PMC free article] [PubMed] [Google Scholar]
- 25.Singh R., Valcárcel,J. and Green,M.R. (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science, 268, 1173–1176. [DOI] [PubMed] [Google Scholar]
- 26.Pérez I., McAfee,J.G. and Patton,J.G. (1997) Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry, 36, 11881–11890. [DOI] [PubMed] [Google Scholar]
- 27.Ghetti A., Pinol-Roma,S., Michael,W.M., Morandi,C. and Dreyfuss,G. (1992) hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res., 20, 3671–3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh Y.L., Hahm,B., Kim,Y.K., Lee,H.K., Lee,J.W., Song,O.-K., Tsukiyama-Kohara,K., Kohara,M., Nomoto,A. and Jang,S.K. (1998) Determination of functional domains in polypyrimidine-tract binding protein. Biochem. J., 331, 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conte M.R., Grüne,T., Ghuman,J., Kelly,G., Ladas,A., Matthews,S. and Curry,S. (2000) Structure of tandem RNA recognition motifs from polypyrimidine tract binding protein reveals novel features of the RRM fold. EMBO J., 19, 3132–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S., Deerinck,T.J., Ellisman,M.H. and Spector,D.L. (1997) The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol., 137, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenhausen G. and Ruben,D.J. (1980) Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett., 69, 185–189. [Google Scholar]
- 32.Delaglio F., Grzesiek,S., Vuister,G.W., Zhu,G., Pfeifer,J. and Bax,A. (1995) NMRPipe – a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 33.Johnson B.A. and Blevins,R.A. (1994) NMRView – a computer-program for the visualization and analysis of NMR data. J. Biomol. NMR, 4, 603–614. [DOI] [PubMed] [Google Scholar]
- 34.Nagai K., Oubridge,C., Jessen,T.H., Li,J. and Evans,P.R. (1990) Crystal structure of the RNA-binding domains of the U1 small nuclear ribonucleoprotein A. Nature, 348, 515–520. [DOI] [PubMed] [Google Scholar]
- 35.Wang X. and Tanaka Hall,T.M. (2001) Structural basis for recognition of AU-rich element RNA by the HuD protein. Nature Struct. Biol., 8, 141–145. [DOI] [PubMed] [Google Scholar]
- 36.Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- 37.Deo R.C., Bonanno,J.B., Sonenberg,N. and Burley,S.K. (1999) Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell, 98, 835–845. [DOI] [PubMed] [Google Scholar]
- 38.Oubridge C., Ito,N., Evans,P.R., Teo,C.-H. and Nagai,K. (1995) Crystal structure of the RNA-binding domains of the U1A spliceosomal protein complexed with an RNA hairpin. Nature, 372, 432–438. [DOI] [PubMed] [Google Scholar]
- 39.Price S.P., Evans,P.R. and Nagai,K. (1998) Crystal structure of the spliceosomal U2B′′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature, 394, 645–650. [DOI] [PubMed] [Google Scholar]
- 40.Allain F.H., Bouvet,P., Dieckmann,T. and Feigon,J. (2000) Molecular basis of sequence-specific recognition of pre-ribosomal RNA by nucleolin. EMBO J., 19, 6870–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahm B., Cho,O.H., Kim,J.-E., Kim,Y.K., Kim,J.H., Oh,Y.L. and Jang,S.K. (1998) Polypyrimidine tract-binding protein interacts with hnRNP-L. FEBS Lett., 425, 401–406. [DOI] [PubMed] [Google Scholar]
- 42.Hunt S.L., Hsuan,J.J., Totty,N. and Jackson,R.J. (1999) unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev., 13, 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polydorides A.D., Okano,H.J., Yang,Y.Y., Stefani,G. and Darnell,R.B. (2000) A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl Acad. Sci. USA, 97, 6350–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilipenko E.V., Pestova,T.V., Kolupaeva,V.G., Khitrina,E.V., Poperechnaya,A.N., Agol,V.I. and Hellen,C.U. (2000) A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev., 14, 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 45.Markovtsov V., Nikolic,J.M., Goldman,J.A., Turck,C.W., Chou,M.Y. and Black,D.L. (2000) Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol., 20, 7463–7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wishart,D.S., Sykes,B.D. and Richards,F.M. (1991) Relationship between nuclear-magnetic-resonance chemical-shift and protein secondary structure. Mol. Cell. Biol., 222, 311–333. [DOI] [PubMed]
- 47.Williamson R.A., Carr,M.D., Frenkiel,T.A., Feeney,J. and Freedman,R.B. (1997) Mapping the binding site for matrix metalloproteinase on the N-terminal domain of the tissue inhibitor of metalloproteinases-2 by NMR chemical shift perturbation. Biochemistry, 36, 13882–13889. [DOI] [PubMed] [Google Scholar]
- 48.Kenan D.J., Query,C.C. and Keene,J.D. (1991) RNA recognition: towards identifying determinants of specificity. Trends Biochem. Sci., 16, 214–220. [DOI] [PubMed] [Google Scholar]
- 49.Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 50.Merrit E.A. and Murphy,M.E.P. (1994) Raster3D Version 2.0 – a program for photorealistic molecular graphics. Acta Crystallogr., D50, 869–873. [DOI] [PubMed] [Google Scholar]
- 51.Jin W., McCutcheon,I.E., Fuller,G.N., Huang,E.S. and Cote,G.J. (2000) Fibroblast growth factor receptor-1 alpha-exon exclusion and polypyrimidine tract-binding protein in glioblastoma multiforme tumors. Cancer Res., 60, 1221–1224. [PubMed] [Google Scholar]
- 52.Chan R.C. and Black,D.L. (1995) Conserved intron elements repress splicing of a neuron-specific c-src exon in vitro. Mol. Cell. Biol., 15, 6377–6385. [DOI] [PMC free article] [PubMed] [Google Scholar]