Abstract
The chicken lysozyme (cLys) locus has been shown to contain all of the cis-elements necessary for position-independent and tissue-specific expression entirely within a 24-kb region defined by general DNase I sensitivity and flanked by matrix attachment regions. As such, it has been viewed as an example of a functional chromatin domain, which is structurally and functionally isolated from neighbouring chromatin. We report here the identification and characterisation of the chicken glioma-amplified sequence (cGas41) locus, which though widely expressed, is contained entirely within the lysozyme chromatin domain. The cGas41 transcript encodes a putative transcription factor, starts 207 bp downstream of the cLys polyadenylation site and is preceded by a CpG island with proposed dual promoter/origin function. The location and differential expression of cGas41 compels re-evaluation of the accumulated literature on the lysozyme domain, and represents an example of two unrelated, differentially expressed vertebrate genes coexisting in the same functional chromatin domain.
INTRODUCTION
Functional chromatin domains in vertebrates have been defined as extended regions of ‘open’, DNase I-sensitive chromatin that contain a gene or a related gene cluster with all the cis-elements necessary for their appropriate expression as transgenes. This definition also includes that such regions of general DNase I-sensitive chromatin are flanked by domain boundary/insulator elements, which act to segregate or protect against chromosomal position effects (1). Three prominent examples of such functional domains are the chicken lysozyme locus, and the chicken and human β-globin loci (reviewed in 1,2). However, the genome-wide relevance of this model has been questioned, as a number of characterised loci are not organised into structurally confined regulatory units (reviewed in 3). Evidence is mounting that a functional gene domain can be equally well defined by the gene-specific interaction of cis-elements (reviewed in 4).
Chicken lysozyme is expressed in the oviduct and in myeloid cells, where expression progressively increases during macrophage differentiation. In these tissues, lysozyme is located in a 24-kb domain of general DNase I sensitivity, whose 5′ and 3′ boundaries are 14 kb upstream and 6 kb downstream, respectively, of the lysozyme transcription start (1,5). The transition from increased to decreased DNase I sensitivity at each domain boundary coincides with matrix attachment regions (MARs) (6). Consequently, the lysozyme chromatin domain was thought to be structurally isolated. It was hypothesised that this separation from the chromosomal environment is required for the appropriate expression of the lysozyme gene (1,6). This hypothesis was supported by the finding that a reporter gene was buffered from position-effects when flanked by the 5′ MAR and stably transfected into cultured cells (7). Several DNase I hypersensitive sites (DHS) have been identified around the lysozyme coding region, most of which correspond to discrete cis-elements that regulate the spatial and temporal expression of lysozyme, such as enhancers, a silencer, hormone response element, a promoter (reviewed in 8) and a replication origin (9).
Bonifer et al. (10) demonstrated that transgenic mice carrying multiple copies of a 21.4-kb domain fragment randomly integrated into the genome, expressed lysozyme in a copy number-dependent manner, leading to the conclusion that the chicken lysozyme chromatin domain acted as a functional regulatory unit. Deletion analysis experiments aimed at localising the elements responsible for this feature revealed that position independence was conferred by the combined effect of several upstream tissue-specific cis-elements and the promoter (11). However, questions remain about how the tissue-specific activity of the lysozyme locus is assured at the molecular level. Subsequent studies have, for that reason, focused on the molecular interactions at various cis-elements, particularly at the level of chromatin fine structure (12) and non-histone protein–DNA associations (reviewed in 13).
We report here that a BLASTN search of the complete genomic sequence of the chicken lysozyme domain resulted in the discovery of a widely expressed second gene, cGas41, that is highly homologous to a ubiquitously expressed human gene of unknown function, but which is often amplified in gliomal tumors (glioma-amplified sequence; GAS41). We also report the preliminary characterisation of cGas41 and discuss the implications of its proximity to cLys, as well as its general location within the lysozyme chromatin domain.
MATERIALS AND METHODS
Cell lines, tissues and RNA preparation
Cell lines MEP, HD37, DT40, MSB1, BM2 and HD11 were all grown in Iscove’s modified Dulbecco’s medium containing l-glutamine (Gibco BRL), 8% foetal calf serum, 2% chicken serum, 100 U/ml penicillin, 100 mg/ml streptomycin and 0.15 mM monothioglycerol. Mouse macrophages carrying cLys/cGas41 were obtained by the differentiation in vitro of bone marrow progenitor cells obtained from adult transgenic mice as described previously (10). Total RNA was prepared from cultured cells and tissues with Trizol Reagent (Gibco BRL), according to the manufacturer’s instructions.
RT–PCR
A 5–10 µg sample of intact total RNA was treated with 1 U/µg RQ1 RNase-free DNase (Promega) in a total volume of 50–100 µl for 10 min at 37°C followed by heat inactivation of the enzyme at 65°C for 10 min. cDNA was prepared from the RNA pellets with M-MLV reverse transcriptase (Gibco BRL) as recommended by the manufacturer, using 250 ng of random primers (Gibco BRL). As a result of its GC-richness, PCR of cGas41 was performed with HotStarTaq DNA polymerase and Q-solution (Qiagen). cLys and control (chicken β-actin, mouse GAPDH) PCR reactions were all performed as described previously (12). All PCR reactions, containing ∼5% of each cDNA mixture in a total volume of 30 µl, were amplified for 30 (cGas41), 30 (cLys), 25 (chicken β-actin) or 20 cycles (mouse GAPDH). The cGas41 PCR primers are identical to the gene-specific, forward inner primer and a reverse outer primer used for 5′ RACE below. RT–PCR products were resolved on 1.5–2.5% agarose gels.
5′ and 3′ RNA ligase-mediated rapid amplification of cDNA ends
RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) was performed with Ambion’s FirstChoice RLM-RACE kit using either DT40 (5′ RACE) or MSB1 (3′ RACE) total RNA according to the manufacturer’s instructions, except that the 5′ RACE inner adapter primer was not used due to mispriming problems. Additional gene-specific PCR primers used for 5′ RACE included a reverse outer primer, 5′-ACCGTCCACTGATGCGTGTG-3′, as well as an inner primer set, forward 5′-ATGTTCAAGAGAATGGCTGAG-3′ and reverse 5′-ATTGGTACCTACACTACCGGCTTCACGAT-3′. Similarly, gene-specific primers designed for 3′ RACE included a forward outer primer, 5′-AAACTCGAAGAGGATGATCAGTC-3′, and inner primer, 5′-CGGGGTACCAAAGATATGTGATGAGTGTTG-3′. KpnI restriction sites incorporated into PCR primers are underlined. RACE products were either sequenced directly (5′ RACE) or subcloned into pBluescript II KS+ and then sequenced (3′ RACE).
RESULTS
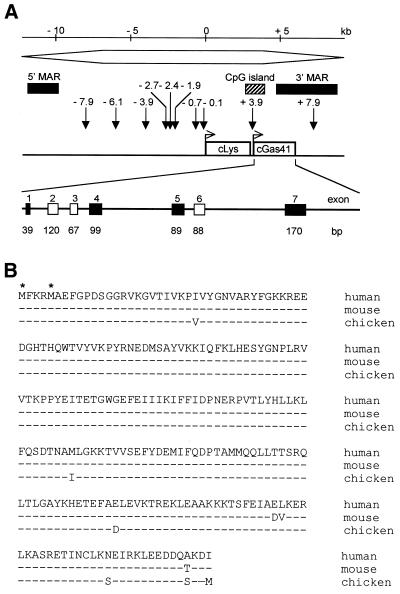
We recently completed the sequence of a fragment carrying the chicken lysozyme chromatin domain (10; submitted to the GenBank database under accession no. AF410481). This sequence was used in a BLASTN search of the National Center for Biotechnology Information (NCBI) non-redundant nucleotide database. To our surprise, high percentage identity to human glioma-amplified sequence (GAS41, GenBank accession no. NM_006530) was observed at several discrete sequences in the 3′ half of the lysozyme domain. The finding that each region of homology corresponded to individual human GAS41 exons suggested that we had identified the chicken orthologue of this gene. As can be seen in Figure 1A, the first sequence with homology to human GAS41 (exon 1, 92% identity over 38 bp) is located immediately 3′ to the chicken lysozyme coding region, in a CpG island reported previously to function as an origin of replication (9). Other sequences in the lysozyme domain exhibiting homology to human GAS41 are located further downstream (exon 4, 88% identity over 50 bp; exon 5, 86% identity over 93 bp; exon 7, 84% identity over 151 bp) and extend into the 3′ MAR (Fig. 1A).
Figure 1.
(A) Summary of important features in the cLys chromatin domain. The open sharp-ended box at top represents the region of general DNase I sensitivity. Below this, MARs and the CpG island are indicated by black and hatched boxes, respectively. Vertical arrows indicate sites of DNase I hypersensitivity, and the orientation of cLys and cGas41 are also shown. At the bottom, cGas41 has been expanded to show individual exons and introns; filled boxes represent those exons initially identified by homology to human GAS41 in the BLASTN search. (B) Multiple alignments of GAS41 amino acid sequences. The complete amino acid sequence of human GAS41 is shown, with asterisks designating the two possible translational start sites. The mouse and chicken gas41 sequences are shown below. A consensus mouse gas41 amino acid sequence was generated from multiple entries in the GenBank database (accession nos BE448250, BF022066, AW210316, AW412092, AW476549 and BF011985). Only those amino acids that differed from the human are shown, those that are identical to human are marked by a dash (–).
Human GAS41 was initially identified as part of a multigene region at 12q13.15 that was amplified and expressed in gliomas (14). As a result of its homology to human AF-9 and ENL genes, it is believed to be a transcription factor (15,16). The chicken Gas41 genomic locus spans >2.8 kb and consists of seven exons and six introns. All of the exon/intron junctions except one, at the 3′ end of intron 4, conform to the GT/AG rule. cGas41 encodes either a 223 or 227 amino acid protein, depending on which of two possible translation start sites are used, and a multiple sequence alignment using the Pileup and Pretty programs from the Genetics Computer Group sequence analysis software package illustrates that GAS41 is highly conserved, with cGas41 exhibiting 97.4 and 96.5% identity to human and mouse Gas41, respectively, at the amino acid level (Fig. 1B). Interestingly, the proximity of lysozyme and GAS41 is conserved in humans, where they are located 5.53 kb apart on 12q14.3.
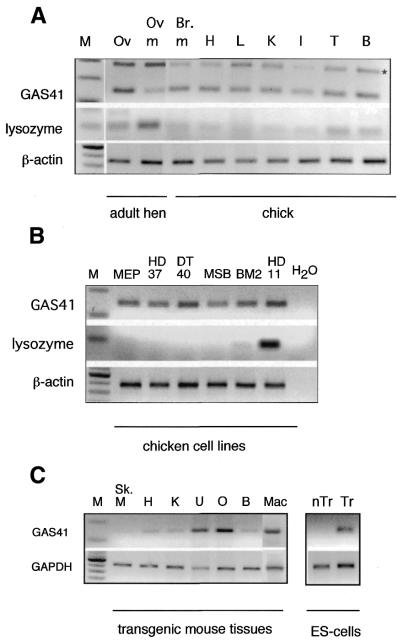
Figure 2 details the results of RT–PCR analyses of endogenous cGas41 expression in various chicken tissues and cell lines. The forward and reverse PCR primers were designed to anneal to exons 1 and 2, respectively, so that PCR products containing the intervening intron 1 region could be easily identified. In Figure 2A, several chick and adult hen tissue samples were tested, all of which were positive for cGas41 expression. The two bands obtained for cGas41 correspond to spliced and unspliced transcripts. No such PCR products were obtained in control reactions done without reverse transcriptase (data not shown). In contrast, cLys is only expressed in adult hen oviduct tissues. Faint bands corresponding to lysozyme expression in all the chick tissues were most likely due to the inevitable contamination of tissues with peripheral blood (and therefore macrophages) at harvest. This was confirmed by analysis of six chicken cell lines grown as pure cultures, corresponding to multipotent myeloid progenitor cells (HD50 MEP), erythroblasts (HD37), B cells (DT40), T cells (MSB1), monocytes (BM2) and macrophages (HD11) (Fig. 2B). cGas41 is clearly expressed to a similar extent in all the chicken cell lines tested, whilst cLys exhibits its characteristic tissue and developmental stage-specific expression profile, with a low level of expression in monocytes and a high level of expression in macrophages. In summary, endogenous cGas41 is widely expressed and, whilst the range of tissues tested was not exhaustive, a cGas41 non-expressing tissue was not identified. Expression of chicken Gas41 thus seems to be similar to the ubiquitous expression seen for human GAS41.
Figure 2.
Expression analysis of endogenous chicken gas41 and cLys in (A) a range of chicken tissues, (B) cell lines and (C) tissues of transgenic mice as well as in transfected (Tr) or non-transfected (nTr) mouse embryonic stem (ES) cells. Tissues: Ov, Oviduct; Ov.m, oviduct magnum; Br.m, breast muscle; H, heart; L, liver; K, kidney; I, intestine; T, testis; B, brain; Sk.M, skeletal muscle; U, uterus; O, ovar; Mac, macrophages. Cell lines: MEP, HD50 MEP multipotent precursor cells; HD37, erythroblasts; DT40, B cells; MSB, T cells; BM2, monocytes; HD11, macrophages. RT–PCR was carried out described in Materials and Methods. M, markers; *, unspliced transcript. Unspliced cGas41 was not detected in chicken cell lines. Chicken β-actin or mouse GAPDH, was used as a control, respectively.
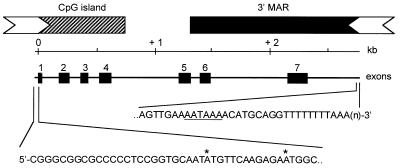
RLM-RACE was used to determine the 5′ and 3′ ends of the cGas41 transcript and the results are shown in Figure 3. The full-length cGas41 transcript is comprised of 1125 nt, and is contained entirely within the chicken lysozyme chromatin domain. More specifically, cGas41 transcription is in the same orientation as cLys and starts 207 bp downstream of the lysozyme polyadenylation site, in a previously characterized unmethylated CpG island (9). As such, cGas41 appears to have a CpG island promoter typical of constitutively expressed genes, including a 26-bp GC-box with perfect dyad symmetry (17). At the 3′ end, cGas41 exons 5–7 and the 3′ untranslated region are all either partially or fully localized within a previously characterized MAR (6). In accordance with its widespread expression, the CpG island promoter of cGas41 displays a DHS in all tissues tested so far (18,19). These earlier DHS mapping studies as well as experiments performed in our laboratory studies also demonstrated that probes covering most of cGas41 do not cross-hybridise with any other gene in the chicken genome (M.Huber and C.Bonifer, unpublished observation).
Figure 3.
Summary of results from 5′ and 3′ RLM-RACE. The 2.8-kb cGas41 transcript is shown at top, with each of the seven exons indicated by a black box. Expanded sections of the 5′ and 3′ ends are shown below. Asterisks designate the two possible ATG initiation codons at the 5′ end. At the 3′ end, a region matching the consensus polyadenylation signal (AATAAA) is underlined, followed by A(n) at the polyadenylation site. The novel sequence data reported in this manuscript have been submitted to the GenBank database under accession no. AF410481.
A transgenic mouse line, TgH(cLys)3, carrying a single copy of the 21.4-kb cLys/cGas41 chromatin domain targeted to the X-linked mouse Hprt locus, and created to further examine chicken lysozyme regulation, was used to determine whether cGas41 expression could be reproduced in the mouse genome (S.Chong, J.Kontaraki, C.Bonifer and A.Riggs, manuscript in preparation). Previous studies revealed expression of the chicken lysozyme transgene only in the brain and macrophages (10,11; data not shown). In contrast, the results in Figure 2C show that the expression of the cGas41 transgene is detectable in all tested mouse tissues. Control experiments with the embryonic stem (ES) cell lines used to generate the transgenic mice and non-transgenic ES cells demonstrate that cGas41 expression originates exclusively from the transgene.
DISCUSSION
cGas41 was identified when a BLASTN comparison of the 21.4-kb chicken lysozyme chromatin domain to the NCBI nucleotide database revealed high homology to human GAS41 cDNA. The cGas41 genomic locus is at least 2.8 kb and consists of seven exons. A 1.1-kb cGas41 mRNA is produced in the same orientation as cLys transcription, and encodes a 223–227 amino acid protein, which exhibits at least 96% identity to human and mouse Gas41 at the amino acid level. Accordingly, an avian example can be added to the growing list of highly conserved GAS41 proteins of fungal, yeast, plant and mammalian origin (16). Human GAS41 is a nuclear protein that has a proposed role as a transcription factor, but no apparent DNA-binding domain (15,16).
Human GAS41 produces a single transcript of ∼1.7 kb, which is present in a wide range of human tissues (16). Likewise, RT–PCR analysis of endogenous cGas41 revealed its constitutive expression in a range of adult hen and chick tissues as well as chicken cell lines. Initiation of cGas41 transcription occurs 207 bp downstream of the cLys polyadenylation site. This is intriguing for a number of reasons. First, the 5′ end of cGas41 coincides with a region characterised previously both as a CpG island and replication origin (9). Unmethylated CpG islands are commonly associated with the promoters of housekeeping genes (20), and this report establishes that the CpG island at the 3′ end of cLys is, similarly, associated with the promoter of the widely expressed cGas41. It has also been shown that many CpG islands are origins of replication (21,22). Correspondingly, the GC-rich region between cLys and cGas41 represents a specific example of a CpG island with dual promoter/origin function. Moreover, as it was reported that replication at this site is bidirectional and initiates early in S phase in both lysozyme-expressing and non-expressing cells (9), the question arises whether in this case a common replication origin is used for both a constitutive gene and a tissue-specific gene.
The most important result from this study is our finding that a highly expressed, tissue-specific gene and a widely expressed gene with a housekeeping promoter coexist in close proximity on the same structurally defined chromatin domain. Moreover, the expression in several tissues of a cLys/cGas41 transgene in the mouse genome suggests that all of the required cis-regulatory elements may be present on the transgene and thus in the chromatin domain. Support for this notion comes from earlier DHS mapping experiments (18,19). A second, constitutive DHS is located 4 kb downstream of the cGas41 transcription start site and is contained within the transgene. No other DHS is found within the next 15 kb downstream of this site. Our results also invite a re-evaluation of the literature regarding the definition of a domain of general DNase I sensitivity. The example presented here suggests a strong link between gene expression levels and the degree of nuclease sensitivity across the entire chromatin domain, as in cLys/cGas41 expressing cells a significant difference in nuclease sensitivity could be detected as compared with cells expressing cGas41 only (1,5). A similar link between promoter activity and general DNase I sensitivity is also apparent in the β-globin locus where intergenic transcripts were identified that may be implicated in the generation of open chromatin in this locus (23).
Our data provide an example of two unrelated, differentially expressed vertebrate genes coexisting in the same functional chromatin domain. The implications to chicken lysozyme regulation, in particular, and functional chromatin domains, in general, illustrate the power of genomics to both alter established view points and to open up new avenues of research.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Gerd Pfeiffer for valuable insight. The authors also thank Dr Helen Sang, Roslin Institute, for preparation of chicken and hen tissues. This work was funded by a grant from the National Insitutes of Health (GM50575) to A.D.R. and grants from the Wellcome Trust, the BBSRC and the Leukaemia Research Fund to C.B.
DDBJ/EMBL/GenBank accession no. AF410481
REFERENCES
- 1.Sippel A.E., Schäfer,G., Faust,N., Saueressig,H., Hecht,A. and Bonifer,C. (1993) Chromatin domains constitute regulatory units for the control of eukaryotic genes. Cold Spring Harb. Symp. Quant. Biol., 58, 37–44. [DOI] [PubMed] [Google Scholar]
- 2.Bell A.C. and Felsenfeld,G. (1999) Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev., 9, 191–198. [DOI] [PubMed] [Google Scholar]
- 3.Bonifer C. (2000) The cis-regulatory information required for the correct developmental regulation of eukaryotic gene loci. Trends Genet., 16, 310–315. [DOI] [PubMed] [Google Scholar]
- 4.Dillon N. and Sabbattini,P. (2000) Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays, 22, 657–665. [DOI] [PubMed] [Google Scholar]
- 5.Jantzen K., Fritton,H.P. and Igo-Kemenes,T. (1986) The DNase I sensitive domain of the chicken lysozyme gene spans 24 kb. Nucleic Acids Res., 14, 6085–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phi-van L. and Strätling,W.H. (1988) The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J., 7, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stief A., Winter,D.M., Strätling,W.H. and Sippel,A.E. (1989) A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature, 341, 343–345. [DOI] [PubMed] [Google Scholar]
- 8.Bonifer C., Jägle,U. and Huber,M.C. (1997) The chicken lysozyme locus as a paradigm for the complex developmental regulation of eukaryotic gene loci. J. Biol. Chem., 272, 26075–26078. [DOI] [PubMed] [Google Scholar]
- 9.Phi-van L. and Strätling,W.H. (1999) An origin of bidirectional DNA replication is located within a CpG island at the 3′ end of the chicken lysozyme gene. Nucleic Acids Res., 27, 3009–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonifer C., Vidal,M., Grosveld,F. and Sippel,A.E. (1990) Tissue specific and position independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J., 9, 2843–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonifer C., Yannoutsos,N., Krüger,G., Grosveld,F. and Sippel,A.E. (1994) Dissection of the locus control function located on the chicken lysozyme gene domain in transgenic mice. Nucleic Acids Res., 22, 4202–4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kontaraki J., Chen,H.-H., Riggs,A. and Bonifer,C. (2000) Chromatin fine structure profiles for a developmentally regulated gene: reorganisation of the lysozyme locus before trans-activator binding and gene expression. Genes Dev., 14, 2106–2122. [PMC free article] [PubMed] [Google Scholar]
- 13.Sippel A.E., Borgmeyer,U., Püschel,A.W., Rupp,R.A.W., Stief,A., Strech-Jurk,U. and Theisen,M. (1987). Multiple nonhistone protein-DNA complexes in chromatin regulate the cell- and stage-specific activity of an eukaryotic gene. In Hennig,W. (ed.), Results and Problems in Cell Differentiation 14. Structure and Function of Eukaryotic Chromosomes. Springer, Heidelberg, Germany, pp. 255–269. [DOI] [PubMed]
- 14.Fischer U., Meltzer,P. and Meese,E. (1996) Twelve amplified and expressed genes localized in a single domain in glioma. Hum. Genet., 98, 625–628. [DOI] [PubMed] [Google Scholar]
- 15.Fischer U., Heckel,D., Michel,A., Janka,M., Hulsebos,T. and Meese,E. (1997) Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum. Mol. Genet., 6, 1817–1822. [DOI] [PubMed] [Google Scholar]
- 16.Harborth J., Weber,K. and Osborn,M. (2000) GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. J. Biol. Chem., 275, 31979–31985. [DOI] [PubMed] [Google Scholar]
- 17.Hauber J., Nelbock,P. and Jantzen,K. (1988) A remarkable nucleotide sequence on the 3′ border of the chicken lysozyme gene that possibly creates a constitutively DNase I hypersensitive site. Nucleic Acids Res., 16, 4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritton H.P., Sippel,A.E. and Igo-Kemenes,T. (1983) Nuclease-hypersensitive sites in the chromatin domain of the chicken lysozyme gene. Nucleic Acids Res., 11, 3467–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fritton H.P., Igo-Kemenes,T., Nowock,J., Strech-Jurk,U., Theisen,M. and Sippel,A.E. (1987) DNase I-hypersensitive sites in the chromatin structure of the lysozyme gene in steroid hormone target and non-target cells. Biol. Chem. Hoppe Seyler, 368, 111–119. [DOI] [PubMed] [Google Scholar]
- 20.Gardiner-Garden M. and Frommer,M. (1987) CpG islands in vertebrate genomes. J. Mol. Biol., 196, 261–282. [DOI] [PubMed] [Google Scholar]
- 21.Delgado S., Gómez,M., Bird,A. and Antequera,F. (1998) Initiation of DNA replication at CpG islands in mammalian genomes. EMBO J., 17, 2426–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antequera F. and Bird,A. (1999) CpG islands as genomic footprints of promoters that are associated with replication origins. Curr. Biol., 9, R661–R667. [DOI] [PubMed] [Google Scholar]
- 23.Gribnau J., Diderich,K., Pruzina,S., Calzolari,R. and Fraser,P. (2000). Intergenic transcription and developmental remodelling of chromatin sub-domains in the human β-globin locus. Mol. Cell, 5, 377–386. [DOI] [PubMed] [Google Scholar]