Abstract
Purpose
Psoriasis and atherosclerosis are immunometabolic diseases. This study aimed to integrate bioinformatics and updated public resources to find potential biological markers associated with atherosclerosis that can cause psoriasis.
Patients and Methods
Microarray datasets were downloaded from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) were screened, and functional enrichment analysis was performed. We identified psoriasis and atherosclerosis common immune-related genes (PA-IRGs) by overlapping immune-related genes (IRGs) with genes in the module most associated with psoriasis and atherosclerosis obtained by weighted gene co-expression network analysis (WGCNAs). Receiver operating characteristic (ROC) was conducted to evaluate the predictive ability. The skin expression levels of diagnostic biomarkers were further verified by immunohistochemical staining. CIBERSORT, single-sample gene set enrichment analysis (ssGSEA), and Pearson’s correlation analysis were applied to evaluate immune and lipid metabolism relationships in psoriatic tissues. In addition, a lincRNA-miRNA-mRNA network was constructed to find the pathogenesis in which diagnostic markers may be involved.
Results
Four PA-IRGs (SELP, CD93, IL2RG, and VAV1) demonstrated the optimal diagnostic value, with an AUC above 0.8. The immune cell infiltration analysis showed that dendritic resting cells, NK cell activation, neutrophils, macrophages M2, macrophages M0, and B-cell memory were highly abundant in psoriasis. Immune response analysis showed that TNF family members, chemokine receptors, interferons, natural killer cells, and TGF-β family members might be involved in psoriasis. Diagnostic biomarkers are strongly associated with various infiltrating immune cells, immune responses, and lipid metabolism. A lincRNA-miRNA-mRNA regulatory network consisting of 31 lincRNAs and 23 miRNAs was constructed. LINC00662 is involved in modulating four diagnostic biomarkers.
Conclusion
This study identified atherosclerosis-related genes SELP, CD93, VAV1, and IL2RG as potential psoriasis diagnostic markers. Provide novel insights into the possible regulatory mechanisms involved in psoriasis.
Keywords: psoriasis, atherosclerosis, biomarker, immune, lipid metabolism, ceRNA network
Introduction
Psoriasis is an immune-mediated systemic chronic inflammatory genetic disease associated with immune abnormalities, dyslipidemia, and cardiometabolic disorders. Psoriasis is linked to multiple complications, including cardiometabolic diseases and metabolic syndrome (diabetes, obesity, hypertension, and atherogenic dyslipidemia), and they share genes. Psoriasis is an independent risk factor for cardiovascular disease, with a significant association with the incidence of major adverse cardiovascular events.1,2 Atherosclerosis is a hallmark of cardiovascular disease. Accumulating evidence supports that psoriasis increases the development and prevalence of atherosclerosis. Disturbed lipid metabolism and immune abnormalities play a role in both disorders. Exploring biomarkers that make early diagnosis possible and molecular mechanisms of co-morbidity can help early detection of subclinical atherosclerosis and reduce cardiovascular morbidity and mortality in patients with psoriasis.
Psoriasis is a chronic inflammatory skin disease characterized by hyperproliferative keratinocytes, immune cell infiltration, and aberrant innate and adaptive immune responses. It has been shown that psoriasis and atherosclerosis exhibit common features of infiltrating T cells, monocytes/macrophages, neutrophils, dendritic cells, and mast cells.2 The development of atherosclerosis is an active process. Inflammatory mechanisms may play a significant role in triggering plaque rupture. Studies have suggested that lipid disorder is an essential part of the pathogenesis of psoriasis and is related to immune abnormalities.3,4 Psoriasis patients are more frequently hyperlipidemic and prone to atherosclerosis.5 An imbalance in lipid metabolism contributes to accelerated inflammatory responses and promotes atherogenesis. Early detection of the pathogenesis of inflammatory and lipid disorders in psoriasis is essential for preventing and treating atherosclerosis. In recent years, the role of competitive endogenous RNA (ceRNA) networks has attracted much attention in diagnostic and prognosis markers and therapeutic targets. The lincRNA-miRNA-mRNA ceRNA network can provide new insights into the development of psoriasis.
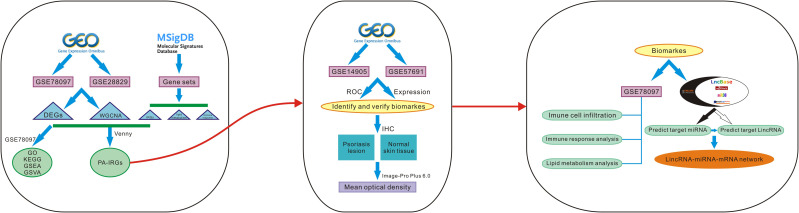
Comprehensive bioinformatics approaches can help to identify potential molecular biomarkers, explore disease mechanisms, and predict molecular targets for precision therapy. This study focused on finding shared genes for psoriasis and atherosclerosis and searching for diagnostic markers. Investigating the interaction of diagnostic biomarkers with immune cells, immune responses, and lipid metabolism provide new insights into the pathogenesis of cardiometabolic disorders in psoriasis. The construction of lincRNA-miRNA-mRNA networks for diagnostic markers provides a new theoretical basis for the underlying regulatory mechanisms involved in diagnostic marker genes. Study procedures are displayed in the study flow chart (Figure 1).
Figure 1.
The flow chart of the analysis procedure on bioinformatics data.
Materials and Methods
Microarray Data Acquisition
Four expression profile data sets, GSE78097, GSE28829, GSE14905, and GSE57691, were downloaded from the GEO (http://www.ncbi.nlm.nih.gov/geo) database by using the GEO query package of R software (version 4.1.2, https://www.r-project.org). All data were preprocessed using robust multi-array average analysis, which included background correction, quantile normalization, and summarization. The GPL570 datasets GSE78097 (n=27 psoriasis, n=6 normal) and GSE28829, including 16 chips at an advanced stage (advanced atherosclerosis, thin or thick fibrous cap atherosclerosis with stenosis greater than 70%) and 13 chips at an early stage (early atherosclerosis, pathological, intimal thickening and intimal xanthoma), were selected as test sets. The psoriatic skin biopsies were obtained from patients 18 years of age and older with active psoriatic vulgaris lesions divided into 14 mild skin psoriasis and 13 severe psoriasis skin according to Psoriasis Area and Severity Index (PASI) scores. Mild psoriatic skin has been shown to have the same histological features as severe psoriatic skin. The GPL570 datasets GSE14905 (n=33 psoriasis, n=21 normal) and the GPL10558 datasets GSE57691 (n=9 atherosclerosis, n=10 normal) were selected as the validation set (Table 1).
Table 1.
Data Cohort Characteristics
| Series | Platform | Platform Name | Sample Information |
|---|---|---|---|
| GSE78097 | GPL570 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | Psoriasis (N=27), Normal (N=6) |
| GSE14905 | GPL570 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | Psoriasis (N=33), Normal (N=21) |
| GSE28829 | GPL570 | [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array | Advanced Atherosclerotic (N=16), Early Atherosclerotic (N=13) |
| GSE57691 | GPL10558 | Illumina HumanHT-12 V4.0 expression beadchip | Aortic occlusive disease (N=9), Control aortic specimen (N=10) |
DEGs Identification and Functional Enrichment Analysis
The DEGs in the GSE78097 and GSE28829 microarray were identified using the Sangerbox tools (http://vip.sangerbox.com/), a free online platform for data analysis. Log-fold change (log2FC) >1 or < −1 and p-value < 0.05 were defined as statistically significant for the DEGs. Volcano plots were made to visualize these DEGs better. With functional enrichment analysis tools: Funrich (http://www.funrich.org/), Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs in GSE78097 were performed and visualized with histograms.
GSEA and GSVA in GSE78097
Gene Set Enrichment Analysis (GSEA) is a gene set-based enrichment analysis method used to determine whether gene sets have statistically significant, consistent differences between two biological states. Gene Set Variation Analysis (GSVA) is a non-parametric unsupervised analysis method to assess whether different metabolic pathways are enriched among different samples. In this study, GSEA analysis was performed with the “GSEA” R package for all genes in GSE78097. GSVA was performed on GSE78097 using the “GSVA” R package, and pathways with significant differences between samples were identified using the R package “Limma” Pathways with P < 0.05 were considered significant signaling pathways.
Weight Gene Co-Expression Network Analysis and PA-IRGs Identification
The WGCNA was constructed by ImageGP (an easy-to-use data visualization web server for scientific, http://www.ehbio.com) to screen hub modules significantly related to clinical characters. First, a Pearson correlation matrix was performed for all paired genes to construct a co-expression similarity matrix. Then soft threshold power values and appropriate parameters β were calculated and screened to construct a scale-free network to transform the similarity matrix into an adjacency matrix. The degree of dissimilarity between nodes was calculated to transform the adjacency matrix into the topology matrix (TOM). The dynamic shearing tree algorithm identifies gene network modules. Finally, the correlation between module signature genes and clinical features is analyzed to identify modules of interest significantly associated with clinical features. The most relevant module genes in the two diseases were taken to intersect, and key genes were selected by |log2FC| > 0.6 and p-value < 0.05 criteria. PA-IRGs were obtained by overlapping key genes and 3615 IRGs downloaded from the the Molecular Characterization Database (MSigDB) (14 gene sets in total, Supplementary Table 1).
Screening and Validation of Diagnostic Markers
The expression levels and the ROC analysis of the PA-IRGs were performed with the validation sets GSE14905 and GSE57691. P-values < 0.05 were considered statistically significant. Generally, an area under the curve (AUC) > 0.9 represents a high diagnostic value, 0.7 < AUC <= 0.9 represents a moderate diagnostic value, and 0.5 < AUC <= 0.7 represents a low diagnostic value.
Samples of Psoriasis and Normal Human Skin Tissue
The samples included 15 psoriatic lesions and 5 normal human skin tissues. This study was obtained from the Department of Pathology, Hospital for Dermatology, Southern Medical University, from December 2018 to December 2019. All the samples were histologically confirmed as psoriatic and normal human tissues by pathologists. Paraffin embedding was used for immunohistochemical staining analysis.
Immunohistochemistry and Image Analysis of Diagnostic Markers
The paraffin-embedded tissue blocks were sectioned at 3-μm thickness. IHC was performed using a DAB kit (GK600710, Shanghai, China) following the manufacturer’s protocol. The anti-VAV1 antibody (ab97574, 1:100), anti-IL2RG antibody (ab273023, 1:50), anti-CD93 antibody (ab198854, 1:100), and anti-CD62P antibody (ab182135, 1:300) used for IHC was purchased from Abcam (Shanghai, China). Goat anti-rabbit (ZF-0513, Zhongshan Golden Bridge, Beijing, China) as a secondary antibody. The NanoZoomer S210 Digital slide scanner scanned the image. Five randomly selected fields per section per sample at 400x magnification. Image-Pro Plus 6.0 was used to determine integrated optical density (IOD) values, and the IOD per unit area (mean density) represented the specific protein relative expression level. Statistical analyses were performed using GraphPad Prism (version 8.0.1).
Evaluation of Immune Cell Infiltrates and Immune Response
CIBERSORT is an analytical tool that uses deconvolution methods to estimate the composition and abundance of immune cells. Using the online tool sangerbox (http://vip.sangerbox.com/), CIBERSORT was used to assess the relationship between diagnostic marker genes and immune cell infiltrates in GSE78097. Psoriatic gene expression profiling was used to evaluate cell populations, with adequate analysis and visualization of the correlated heatmap, proportion profiles, and differential expression between immune cells and samples. The Pearson correlation analysis explored potential associations between diagnostic marker genes and immune cell infiltration. Immune response gene sets were downloaded from the MsigDB database (Supplementary Table 2). The expression differences between immune responses in psoriasis and normal tissues were analyzed, and the correlation between immune responses in psoriasis was analyzed using the ssGSEA method in GSVA. Pearson correlation analysis was performed to investigate the correlation between diagnostic marker genes and immune response. Results associated with p< 0.05 were considered significant.
Correlation Analysis of Lipid Metabolic
Lipid metabolism, cholesterol, and plasma lipoprotein-related genes were downloaded from the MSigDB database (72 gene sets in total, Supplementary Table 3 and Supplementary Table 4). Psoriatic lipometabolism-related gene was obtained by intersecting with lipid metabolism genes (Supplementary Table 5) and the key module genes of psoriasis. SsGSEA analysis using the online tool sangerbox (http://vip.sangerbox.com/) was performed to assess the enrichment fraction of lipid metabolism, cholesterol, and plasma lipoproteins in psoriasis samples. Pearson correlation analysis was performed to analyze the correlation between diagnostic marker genes and lipids.
Construction of a lincRNA-mRNA-miRNA Network
StarBase (https://starbase.sysu.edu.cn/), Lncbase (http://carolina.imis.athena-innovation.gr/diana tools/web/index.php?r=lncbasev2%2Findex-predicted), Miwalk (http://mirwalk.umm.uni-heidelberg.de/), MiRDB (http://mirdb.org/), and TargetScan (https://www.targetscan.org/vert_72/) are online databases that predict mRNA-miRNA and miRNA-lncRNA interactions. mRNA-miRNA interactions were predicted by Starbase, Miwalk, MiRDB, and TargetScan databases, and miRNA-lincRNA interactions were predicted by Starbase and Lncbase databases. They are constructing a lincRNA-miRNA-mRNA ceRNA network with Sangerbox tools (http://vip.sangerbox.com/).
Statistical Analysis
GraphPad Prism 8.0.1 software was used to perform a Mann–Whitney test to analyze mean optical density (MOD) differences between psoriasis and normal skin tissues. P < 0.05 was regarded as significant.
Results
DEGs Identification and Functional Enrichment Analysis in GSE78097
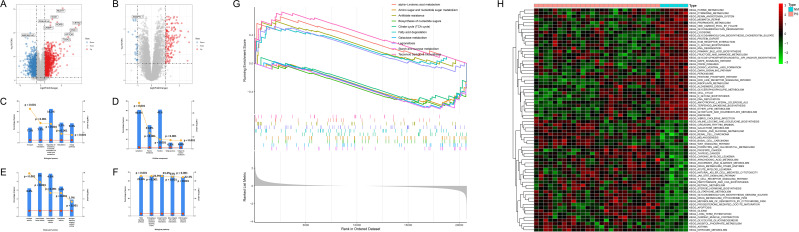
We used |log2FC| > 1 and p-value < 0.05 as criteria for screening DEGs. 1220 up-regulated and 1020 down-regulated genes were identified in GSE78097 (Figure 2A). 148 up-regulated and 28 down-regulated genes were identified between advanced and early atherosclerosis in GSE28829 (Figure 2B). The functional enrichment analysis was performed to further investigate the characteristics of genes in psoriasis. As a result, the GO term showed that the nucleic acid metabolic process (16.2%, P<0.001) (Figure 2C), nucleus (41%, P < 0.001) (Figure 2D), and transcription factor activity (5%, P < 0.001) (Figure 2E) were significantly enriched in biological progress (BP), cellular components (CC), and molecular functions (MF), respectively. And the biological pathway was mainly enriched to integrin family cell surface interactions (23.4%, P<0.001), β1 integrin cell Surface interactions (23.0%, P<0.001), proteoglycan-syndecan mediated signal events (22.9%, P<0.001) (Figure 2F). The GSEA results of GSE78097 showed that the KEGG pathways were enriched in alpha−linolenic acid metabolism, amino sugar and nucleotide sugar metabolism, antifolate resistance, biosynthesis of nucleotide sugars, citrate cycle (TCA cycle), fatty acid degradation, galactose metabolism, legionellosis, starch and sucrose metabolism, terpenoid backbone biosynthesis (Figure 2G). The GSVA result showed that multiple pathways and biological processes, including inflammatory response and lipid metabolic pathways, were activated in psoriasis (Figure 2H). It was consistent with the GSEA analysis.
Figure 2.
Identification of DEGs and functional enrichment analysis in GSE78097. (A and B) Volcano map of DEGs for GSE78097 and GSE28829. Red represents up-regulated differential genes, grey represents no significant difference, and blue represents down-regulated differential genes. (C–F) Column chart of GO functional and KEGG pathway analysis of DEGs in GSE78097. (G) The top five up and down-regulated signaling pathways of GSEA in GSE78097. (H) Heatmap showing the GSVA scores of potential pathways and biological processes in GSE78097.
WGCNA Analysis Identified Key Genes and Screening IRGs
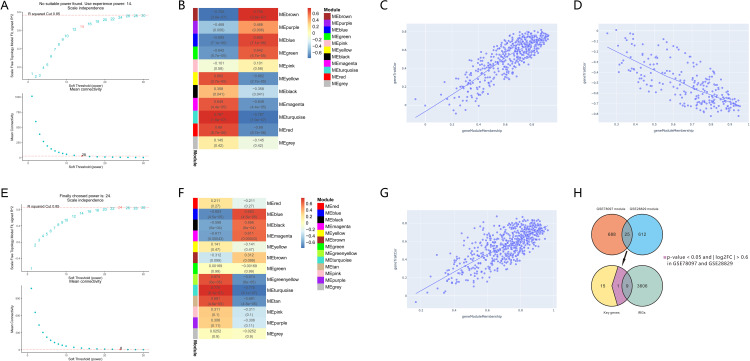
A soft-threshold power was introduced into the network topology, which affected the network’s scale independence and means of connectivity. A soft threshold of 14 in the GSE78097 dataset and 24 in the GSE28829 dataset was used to obtain the approximate scale-free topology with a scale-free topology fit index >0.85 and the lowest power (Figure 3A and E). Module-trait correlation analyses showed that 11 and 13 modules were related to psoriasis and atherosclerotic. The gene modules most significantly associated with psoriasis and atherosclerosis were selected by WGCNA analysis with selection criteria of correlation coefficient > 0.7 and P < 0.05. The turquoise module (correlation coefficient = 0.767, P = 1.9e-07) and brown module (correlation coefficient = 0.755, P = 3.8e-07) were significantly associated with psoriasis (Figure 3B), while the turquoise module (correlation coefficient = 0.775, P = 8.1e-07) was significantly related to atherosclerotic (Figure 3F). Scatter plot showing linear correlation of genes in significantly associated modules for psoriasis and atherosclerosis (Figure 3C, D, and G). 25 key genes were obtained by overlapping the significantly associated modules in psoriasis and atherosclerosis via the Venn diagram (Figure 3H). 10 of 25 key genes had p-values < 0.05 and | log2FC | > 0.6 in psoriasis and atherosclerosis (Table 2). A total of 9 (SELP, CD93, CHIT1, CLEC2B, PSMB9, IL2RG, ITGAL, SELPLG, and VAV1) PA-IRGs were obtained by taking the intersection of 10 key genes and 3615 IRGs downloaded from MSigDB database (Figure 3H).
Figure 3.
Identify key immune-related genes associated with psoriasis and atherosclerosis. (A and E) Soft threshold power map of GSE78097 and GSE28829. (B and F) Weighted gene co-expression network modules of GSE78097 and GSE28829. Each row represents one module. Each column represents one trait attribute. Each cell contains the correlation coefficients corresponding to the cell color. (C and D) Scatter plot of correlations between the significance of turquoise and brown module genes and module members in GSE78097. (G) Scatter plot of correlations between the significance of turquoise module genes and module members in GSE28829. (H) The overlapped immune-related genes and most significant module genes in GSE78097 and GSE28829 with p-values < 0.05 and | log2FC | > 0.6.
Table 2.
| Gene | GSE28829 | GSE78097 | ||
|---|---|---|---|---|
| logFC | P.Value | logFC | P.Value | |
| VAMP8 | 1.825385282 | 8.06362E-09 | 0.203679227 | 0.472787023 |
| RAMP3 | 0.696952452 | 0.001610804 | −0.53167484 | 0.065273301 |
| CA2 | 0.420668724 | 0.119512375 | 1.724556011 | 4.56994E-09 |
| CD93 | 0.891338719 | 2.26795E-05 | −0.64299386 | 0.048000587 |
| SELP | 0.749599531 | 0.003251036 | 0.908797137 | 0.00028967 |
| RGS2 | 0.38638396 | 0.092757205 | 2.092202153 | 2.38606E-08 |
| NR2F1 | 0.391996014 | 0.132185195 | 0.352657691 | 0.312375166 |
| BTN3A2 | 0.144066634 | 0.489958067 | 0.724773779 | 0.049465993 |
| SERPINF1 | 0.545130957 | 0.009835755 | 2.178993489 | 1.22702E-08 |
| CHIT1 | 0.771168825 | 0.015316339 | 0.655586953 | 0.047471415 |
| CLEC2B | 0.670727519 | 0.002816242 | 1.108542065 | 0.006154346 |
| PSMB9 | 0.64668543 | 0.004368308 | 2.64450084 | 4.95238E-09 |
| ITGA4 | 0.604361727 | 1.59957E-05 | 0.057918401 | 0.76693857 |
| IL2RG | 0.90531887 | 4.65795E-06 | 1.188662026 | 4.13288E-07 |
| COL15A1 | 0.405691379 | 0.077338158 | 1.557504909 | 0.000838612 |
| SLN | 0.405138825 | 0.251574205 | 1.019646052 | 0.004029667 |
| ITGAL | 0.667317774 | 4.39303E-08 | 0.950917934 | 0.00396419 |
| HK3 | 0.584703237 | 3.81198E-05 | 7.789277812 | 9.47856E-14 |
| SELPLG | 0.645274716 | 4.89998E-07 | 3.228598273 | 3.36721E-14 |
| CD8A | 0.403669024 | 0.018690373 | −0.68168786 | 0.002472851 |
| TMEM51 | 0.585396895 | 0.000437638 | 0.437601941 | 0.314962022 |
| OXSM | 0.264948994 | 0.057457685 | 1.489394533 | 8.82478E-05 |
| VAV1 | 2.307365828 | 9.50509E-11 | 2.307365828 | 9.50509E-11 |
| EGFL6 | 0.666860274 | 0.012482715 | 1.410981972 | 1.64257E-06 |
| PSENEN | 0.296561977 | 0.069801198 | 0.993352781 | 0.003170017 |
Screening and Validating Potential Diagnostic Markers
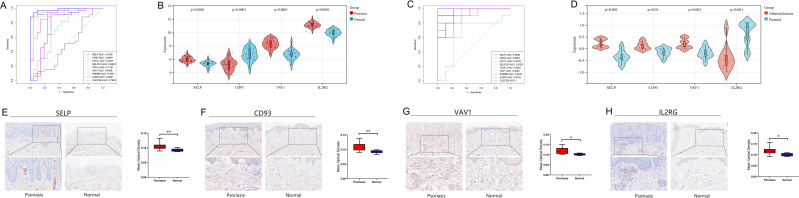
In this study, ROC analysis was performed to evaluate the diagnostic validity of the 9 PA-IRGs (SELP, CD93, CHIT1, CLEC2B, PSMB9, IL2RG, ITGAL, SELPLG, and VAV1). We used the GSE14905 and GSE57691 datasets as a validation set for psoriasis and atherosclerotic. An AUC > 0.8 was selected as the diagnostic marker. The AUCs of SELP, CD93, VAV1, and IL2RG were 0.9105, 0.8846, 0.9408, and 0.9827 in psoriasis (Figure 4A) and 0.9556, 0.8667, 0.9222, and 0.9333 in atherosclerosis (Figure 4C), respectively. Further, the four genes have significant differential expression in the validation sets of psoriasis and atherosclerosis (Figure 4B and D). Therefore, the four genes were selected for further analysis.
Figure 4.
Identify and validate diagnostic efficacy and expression in psoriasis of diagnostic biomarkers. (A and C) ROC analysis of PA-IRGs in validation sets GSE14905 and GSE57691. The greater the AUC, the higher the diagnostic value. (B and D) The expression level of diagnostic biomarkers in GSE14905 and GSE57691. P-value < 0.05 was considered statistically significant. (E–H) IHC staining (x200) and histogram of mean optical density of diagnostic biomarkers in psoriatic lesions and healthy skin tissue. * p < 0.05, **P<0.01, the difference was statistically significant.
Expression of Diagnostic Markers in Skin Lesions of Psoriasis
The IHC was carried out to verify the relative protein expression levels of four diagnostic markers in psoriatic lesional and healthy skin tissues. The positive expression of SELP was mainly found in the endothelial cells of superficial dermal vessels in psoriasis (Figure 4E), and the positive expression of IL2RG, CD93, and VAV1 was mainly found around infiltrating inflammatory cells and vascular endothelial cells in the superficial dermis in psoriasis (Figure 4F–H). SELP (p-value = 0.0077), CD93 (p-value = 0.0054), VAV1 (p-value = 0.0193), and IL2RG (p-value = 0.0054) were significantly different compared to normal skin tissue.
Immune Cell Infiltration of Diagnostic Markers in Psoriasis
We analyzed the correlation between immune cells and each sample’s score and proportion of immune cells. The results showed that the composition of Neutrophils, Dendritic cells resting, T cells CD4 memory resting, T cells CD4 naive, B cells memory, Mast cells activated, and NK cells activated was dominant in psoriasis tissues, while Dendritic cells activated, Macrophages M2, Macrophages M1, Macrophages M0, NK cells resting, T cells gamma delta, T cell regulatory (Tregs), and Plasma cells were rarely expressed in psoriasis tissues (Figure 5A). The correlation heatmap of 13 immune cells showed that NK cells activated and Dendritic cells resting, Macrophages M2 and Dendritic cells activated, B cells memory and Dendritic cells activated showed a significant positive correlation. NK cells activated and Macrophages M2, Macrophages M2 and Dendritic cells resting, NK cells activated and Neutrophils, NK cells activated and Dendritic cells activated, Dendritic resting cells and Dendritic cells activated, B cells and Dendritic cells activated, B cells and NK cells activated memory were significantly negatively correlated (Figure 5B). The result of the scores of 13 types of immune cells showed that the abundance of Dendritic resting cells and NK cells activated were significantly higher in psoriasis than in normal samples, while the abundance of Neutrophils, Macrophages M2, Macrophages M0, Dendritic cells activated, and B cells memory were significantly lower in psoriasis than in normal samples (Figure 5C).
Figure 5.
Evaluation of immune cell infiltration and correlation analysis between immune cells and diagnostic markers in GSE78097. (A) Histogram of immune cell ratio in psoriasis dataset GSE78097. (B) Correlation heat map of 13 types of immune infiltrating cells. (C) Differential expression plot of 13 immune infiltrating cells. (D–G) Lollipop diagram showing the relationship between diagnostic markers expression and 13 immune cell infiltrates in psoriasis. The size of the dots represents the strength of the correlation. Positive numbers represent positive correlations, and negative numbers represent negative correlations.
We analyzed the relationship between diagnostic markers and psoriatic immune infiltrating cells. Correlation analysis showed that SELP was positively correlated with T cells regulatory (Tregs) (r = 0.39, p = 0.0251) and Dendritic resting cells (r = 0.36, p = 0.0369) and negatively correlated with Macrophages M2 (r = - 0.58, p = 0.0004) (Figure 5D). CD93 was positively correlated with Dendritic cells activated (r = 0.43, p = 0.0136) and B cells memory (r = 0.39, p = 0.0233) and negatively correlated with NK cells activated (r = - 0.42, p = 0.0139) (Figure 5E). VAV1 was positively correlated with NK cells activated (r = 0.61, p = 0.0002), and negatively correlated with Neutrophils (r = - 0.57, p = 0.0006) and B cells activated (r = - 0.55, p = 0.0001) (Figure 5F). IL2RG was positively correlated with Dendritic cells resting (r = 0.46, p = 0.0076) and NK cells activated (r = 0.45, p = 0.0091) and negatively correlated with Dendritic cells activated (r = - 0.59, p = 0.0003) and T cell CD4 naïve (r = - 0.36, p = 0.0383) (Figure 5G).
These results indicated that Dendritic resting cells and NK cells activated account for a large proportion of psoriatic immune cell infiltration. The abundance of the two categories is significantly higher in psoriasis than in normal samples. SELP was positively correlated with dendritic resting cells, VAV1 was positively correlated with NK cell activation, IL2RG was positively correlated with dendritic resting cells and NK cell activation, and CD93 was negatively correlated with NK cell activation. Therefore, SELP, CD93, VAV1, and IL2RG may play an important role in the immune infiltration of psoriasis patients.
Immune Responses of Diagnostic Markers in Psoriasis
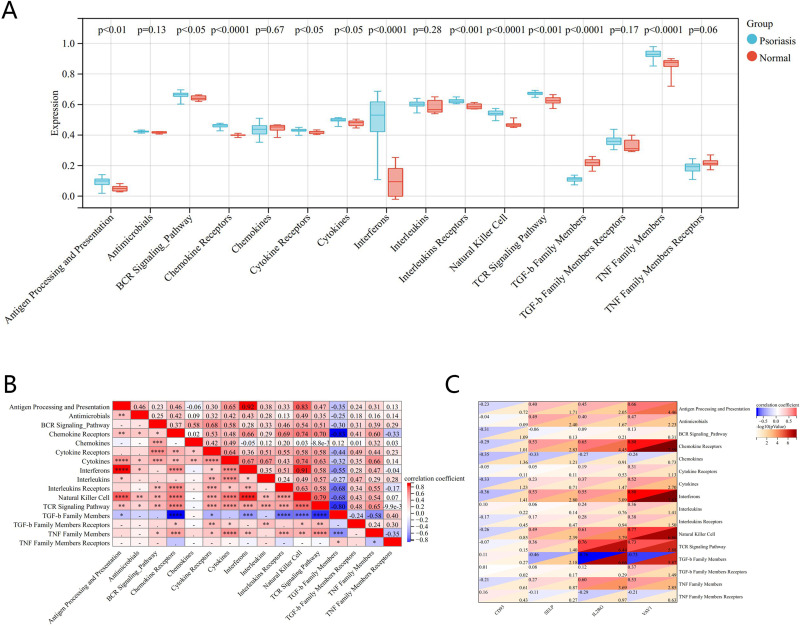
The differences in expression in 16 immune responses in psoriasis showed that tumor necrosis factor (TNF) family members, chemokine receptors, interferons, natural killer (NK) cells, and transforming growth factor-β (TGF-β) family members had significant differences (P<0.0001) in expression between psoriasis and normal tissue (Figure 6A). The heat map of the correlation between immune responses showed an extremely strong positive correlation between antigen processing and presentation and interferons (r = 0.92, P < 0.0001), NK cell (r = 0.83, P < 0.0001), interferons and NK cell (r = 0.91, P < 0.0001); and an extremely strong negative correlation between TGF-β family members and chemokine receptors (r = - 0.83, P < 0.0001), T cell receptor (TCR) signaling pathway (r = - 0.80, P < 0.0001) (Figure 6B). Correlation analysis between four genes and the immune response showed that VAV1 was extremely positively correlated with chemokine receptors and interferons, strongly positively correlated with NK cell and TCR signaling pathway members, moderately positively correlated with TNF Family Members, and extremely negatively correlated with TGF-β family members; IL2RG was strongly positively correlated with TCR signaling pathway, chemokine receptors, NK cell, and TNF family members, moderately positively correlated with interferons, and strongly negatively correlated with TGF-β family members; SELP was moderately positively correlated with chemokine receptors, NK cell, and interferons, moderately negatively correlated with TGF-β family members, and weakly negatively correlated with interferons; CD93 was weakly negatively correlated with interferons and chemokines (Figure 6C).
Figure 6.
Correlation analysis between diagnostic markers and immune responses in GSE78097. (A) Differences in expression of 16 immune responses. (B) Correlation heatmap of 16 immune responses. (C) Correlation between diagnostic biomarkers and different immune responses.
These results show that VAV1, CD93, IL2RG, and SELP may all be involved in the biological process of psoriasis through immune pathways such as chemokine receptors, natural killer cells, interferons, TGF-β family members, and interferons.
Lipid Metabolism Analysis of Diagnostic Markers in Psoriasis
Interacting lipid metabolism genes with genes from the psoriasis key module identified 18 lipid metabolism-related genes (Figure 7A). Pearson correlation analysis was performed between 18 lipid metabolism-related genes and diagnostic marker genes. The results showed that OXCT2, CPNE7, ABCD1, ACER2, and ACOT6 were a strongly positive correlation with SELP, IL2RG, and VAV1. CD93 showed a moderate negative correlation with OXCT2 and CPNE7 (Figure 7B). Subsequently, we explored the relationship between four diagnostic marker genes and lipid metabolism in psoriasis. SsGSEA analysis showed that the metabolism of fatty acid, sphingolipid, glycosphingolipid, cholesterol, lipid, phospholipid, ketone body, triglyceride (TG), low-density lipoprotein (LDL) lipoprotein, and high-density lipoprotein (HDL) lipoprotein were consistent correlation with SELP, IL2RG, and VAV1 (Figure 7C and D). The results suggested that these four genes may be involved in the lipid metabolic process of psoriasis.
Figure 7.
Correlation analysis between diagnostic biomarkers and lipid metabolism in GSE78097. (A) Venn diagram of lipid genes and most significant module genes in GSE78097. (B) Correlation heat map of diagnostic biomarkers and most significant lipid genes in GSE78097. (C) Correlation between diagnostic biomarkers and lipid metabolism. (D) Correlation between diagnostic biomarkers and TC, TG, LDL cholesterol, and HDL cholesterol.
LincRNA-miRNA-mRNA Network of Diagnostic Markers
A lincRNA-miRNA-mRNA network was built to investigate further the regulatory mechanism of the four diagnostic marker genes (Figure 8). We analyzed the upstream regulation of SELP, CD93, IL2RG, and VAV1. MiRNAs or lincRNAs targeting these four genes were screened. Upstream miRNAs predicted by more than three databases in miRWalk, Starbase, TargetScan Human, and miRDB were extracted. 24, 13, 9, and 7 upstream miRNAs target SELP, CD93, VAV1, and IL2RG, respectively (Supplementary Table 6). Furthermore, overlapping the StarBase and Lncbase, the predicted lincRNA-miRNA relationships of related miRNAs for the four genes were obtained. SELP, CD93, IL2RG, and VAV1 miRNAs were targeted by 11, 16, 9, and 2 lincRNAs (Supplementary Table 7), respectively. LINC00662 had the highest degree with miRNAs in the ceRNA network. Therefore, we speculated that LINC00662 might affect the development of psoriasis by regulating SELP, CD93, VAV1, and IL2RG.
Figure 8.
Sankey diagram of the lincRNAs-miRNAs-mRNAs network. Each rectangle represents a gene, and the size indicates each gene’s degree of connectivity.
Discussion
Psoriasis is an immune-mediated systemic inflammatory genetic disorder that may provide additional independent cardiovascular risk factors and increase the incidence and mortality of the cardiovascular disease.6 Immune disorders and dyslipidemia increase the risk of psoriasis and cardiovascular disease. Atherosclerosis is a hallmark of cardiovascular disease. Psoriasis and atherosclerosis share common immune-inflammatory pathogenic mechanisms and pathophysiology, and specific hub genes may control common pathogenic mechanisms.7 Psoriasis may be involved in the development and progression of cardiovascular events through elevated systemic inflammation, immune cells, lipoprotein dysfunction, and metabolic dysregulation.8 Adipose tissue is an integral part of the innate immune system and contains immune cells that regulate adipocyte function and lead to metabolic disorders.9 Lipoprotein dysfunction and metabolic dysregulation can alter immune cell and adipokine profiles and exacerbate endothelial dysfunction. The GSEA results showed that lipid metabolism, such as α-linolenic acid metabolism and fatty acid degradation, was associated with psoriasis. It suggests that there are abundant lipid metabolic responses in psoriasis.
In this study, atherosclerosis-related genes SELP, CD93, VAV1, and IL2RG were identified as potential diagnostic markers of psoriasis in combination with the MSigDB database and WGCNA and ROC analysis methods. SELP (P-selectin) is an adhesion protein belonging to the selection, which is expressed on the surface of activated endothelial cells and is a cell surface adhesion molecule involved in leukocyte rolling and attachment. SELP is associated with autoimmune diseases, particularly rheumatoid arthritis (RA), and is critical for inflammatory cell recruitment, adhesion, and migration to joints and correlates with disease severity.10,11 SELP exposure can trigger various thrombotic and inflammatory responses,12 and variants in the SELP gene are risk factors for Systemic lupus erythematosus (SLE).13 It has been shown that SELP is upregulated in mice with lupus nephritis, and applying SELP-blocking antibodies improves renal prognosis in lupus mice.14 The critical role of SELP during recruitment of cerebrovascular CD8 cells during acute attacks in multiple sclerosis patients.15 Human P-selectin on platelets and endothelial cells is required for enhanced atherogenesis.16 High soluble SELP predicts future cardiovascular events.17,18 IHC results showed that SELP was highly expressed in vascular endothelial cells of the papillary dermis in psoriasis than in normal skin tissue. CD93 is a type 1 transmembrane glycoprotein that promotes adhesion, penetration, and diapedesis of inflammatory cells to the endothelium.19 CD93 is expressed in various inflammatory diseases and on various inflammatory cells such as neutrophils, monocytes, and endothelial cells.20,21 Chen Su showed that CD93 might be a potential new reporter for noninvasive atherosclerotic plaque imaging.22 CD93 levels were significantly higher in asthmatics than in healthy controls, and inhibition of CD93 increased levels of the inflammatory cytokine IL-6. IL-6 and its receptor are present in human coronary atherosclerotic plaques.23 CD93 is a regulator and indicator of inflammation in many inflammatory diseases such as SLE and rheumatic joints. Soluble CD93 (sCD93) primarily regulates angiogenesis. The expression of sCD93 is inversely correlated with prednisone dose in SLE patients.24 SCD93 expression is increased in the synovial fluid of RA patients and may be a potential biomarker of RA.25 There is currently no research evidence that CD93 is associated with multiple sclerosis. Therefore, SELP and CD93 are closely related to cardiovascular disease and may play a role in the formation of atherosclerosis in psoriasis, which needs further investigation. VAV1 is a Rac/Rho guanine nucleotide exchange factor required for foam cell formation and c-Jun N-terminal (JNK) kinase activation.26 The JNK signaling pathway mediates cell proliferation, apoptosis, differentiation, and inflammatory responses.27 JNK is involved in keratinocyte production and cytokine release, which recruits immune cells, stimulates skin cell proliferation, and amplifies the inflammatory response.28 VAV1 plays a critical role in developing and activating T and B cells and is associated with several immune-mediated diseases, including RA, SLE, and multiple sclerosis. VAV1 is involved in the regulation of T-cell signaling and is related to anti-CCP-negative RA.29 A study based on a comprehensive Bayesian network approach highlighting key drivers in SLE patients showed downregulation of expression levels of VAV1 in SLE patients.30 VAV1 expression is elevated in individuals with multiple sclerosis and correlates with tumor necrosis factor and interferon-γ expression.31 IL2RG (interleukin-2 receptor subunit gamma), which encodes the IL2γ receptor Il2Rγ, mediates proliferative signaling via the Janus Kinase (JAK) / signal transducer and activator of transcription (STAT) pathway. In vivo, IL2RG overexpression promotes the growth of pancreatic cancer cells.32 According to Lei-Lei Zhang, JAK phosphorylation is dependent on IL2RG expression and is associated with IL-2 and IL-21 secretion.33 Silencing IL2RG resulted in the inactivation of JAK3 and downstream STAT5, IL-2RG activates signal transducer and activator of STAT5 via JAK3.34 STAT3 and STAT5 regulate Th17 cells via anti-inflammatory effects. STAT5 deficiency enhances autoimmune diseases.35 IL2RG may contribute to the development of psoriasis through the JAK3-STAT5 regulatory pathway. IL2RG is an essential component of T cell proliferation and mediating immune responses, and it is upregulated in RA synovial tissue and SLE.36,37 IL2RG may activate the primary immunodeficiency pathway to increase the risk of RA development.38 Multiple sclerosis is an immune-mediated demyelinating disease of the central nervous system caused primarily by abnormal T-cell responses. There is no evidence that multiple sclerosis is associated with IL2RG.
CD93, VAV1, and IL2RG were mostly expressed surrounding inflammatory cells and vascular endothelial cells in the superficial dermis of psoriasis, according to IHC results. Four diagnostic markers are associated with atherosclerosis and may play a role in biological processes such as immune cells and immune responses. These specific mechanisms warrant further investigation.
Psoriatic arthritis is a chronic inflammatory arthritis associated with psoriasis. It has general features of chronic inflammation, such as intimal lining hyperplasia, inflammatory cell influx, and neovascularization with RA and other arthritides. SELP, CD93, VAV1, and IL2RG can participate in the pathogenesis of RA by participating in cell adhesion,11 inducing monocyte differentiation,25 signal transduction,39 and immune cell infiltration,40 respectively. Psoriatic arthritis and RA share common physiological and pathological mechanisms and immune pathways. There is no evidence that these genes are involved in the pathogenesis of psoriatic arthritis, and further studies are needed.
Aberrant activation of immune cells is an immunopathological process in psoriasis. Immune infiltration is a feature of psoriasis, with increased dendritic cells and macrophages in the dermis and neutrophils in the epidermis.41 Dendritic resting cells are involved in the activation and differentiation of T lymphocytes and the production of cytokines and chemokines.42 Macrophages play a key role in lymphocyte activation and proliferation, promoting the inflammatory process in psoriasis.43 Neutrophils are a significant source of proinflammatory mediators, such as IL-17, that are involved in the progression of psoriasis.42 In psoriasis, NK cell infiltration is increased, and cytokines released by NK cells are implicated in the control of immunological responses.44 Inflammation caused by innate and adaptive immune cell activation contributes to the development of atherosclerosis in psoriasis patients.45 Unstable atherosclerotic plaques usually contain infiltrates of activated macrophages and activated T cells. Dendritic cells may participate in the immune process of atherosclerosis by activating T cells, leading to plaque instability.46 NK cells also influence the formation and progression of atherosclerosis, and dysfunctional NK cells observed in atherosclerosis may be attributed to abnormal lipid metabolism, which is also a major cause of plaque formation.47 In this study, we analyzed the correlation between diagnostic markers and immune cells in psoriasis. We found that SELP, IL2RG, CD93 may be involved in the inflammatory process of psoriasis by affecting dendritic resting cell infiltration, and VAV1, IL2RG, CD93 may be involved in regulating immune responses by affecting activated NK cell infiltration.
The immune response is overactive in psoriasis.48 Chemokines are involved in the migration and localization of various immune cells.49,50 Abnormalities in interferon in plasmacytoid dendritic cells can trigger T cell activation.51 NK cells stimulate keratinocytes by inducing the expression of adhesion molecules and chemokines in keratinocytes leading to intraepidermal infiltration of inflammatory cells.52,53 Psoriasiform skin inflammation is closely related to TGF-β1 expression in the skin.54 TGF-β1 is required for Th17 cell differentiation.55 There are several critical immune factors important in psoriasis that overlap with atherosclerosis. Cell-mediated immunity has been shown to enhance the atherosclerotic process. We found that the diagnostic biomarkers were closely associated with immune cells and responses, consistent with the abundant immune cell infiltration and immune response results in psoriasis.
Abnormal metabolism is an essential factor in the pathogenesis of psoriasis.56,57 Studies have shown that serum of TC, LDL cholesterol, and TG levels are significantly increased, and HDL cholesterol is decreased in psoriasis.3 Lowering the LDL cholesterol, TG, and TC reduces atherosclerosis-related cardiovascular events. Plasma HDL cholesterol concentrations are negatively associated with atherosclerosis. Lipids are involved in cell proliferation, apoptosis, angiogenesis, inflammatory conditions, or immune responses.58 Precise targeting of genes involved in lipoprotein metabolism is an emerging approach for atherosclerosis prevention and treatment. This study preliminarily explored the correlation between four genes and lipid metabolism. The results show that the metabolism of fatty acid, sphingolipid, glycosphingolipid, cholesterol, lipid, phospholipid, ketone body, TG, LDL, and HDL were consistently correlated with SELP, IL2RG, and VAV1. Lipid metabolism genes of OXCT2, CPNE7, ABCD1, ACER2, and ACOT6 were positively correlated with SELP, IL2RG, and VAV1. While CD93 was moderate negative correlated with OXCT2 and CPNE7. OXCT2 is involved in ketone body catabolism, and increased OXCT2 expression may be associated with diabetic chronic kidney disease.59 These four genes may interact with lipid metabolism genes and influence lipid metabolism pathways, potentially affecting psoriasis pathogenesis. Further investigation into the role of lipid metabolism and its associated genes in psoriasis will help to provide a complete risk assessment and complement known classical atherosclerotic risk factors.
The lncRNA-miRNA-mRNA axis plays a crucial role in the pathogenesis of cardiovascular, inflammatory, and immune-related diseases.60–63 lincRNA is a subgroup of lncRNA that can act as ceRNA by competing with mRNA by binding to miRNA, and an increasing number of lincRNAs have been demonstrated to act as modulators of the immune system. To further investigate the four genes’ potential regulatory mechanism and lincRNAs’ role in disease progression. Based on ceRNA regulatory network theory, we predicted miRNAs and lincRNAs for diagnostic markers. A lincRNA-miRNA-mRNA network was constructed, including 31 lincRNAs, 23 miRNAs, and four mRNAs. We found that LINC00662 had the highest degree with miRNAs in the ceRNA network. lncRNAs play crucial roles in the cell cycle, differentiation, apoptosis, immune responses, cardiovascular disease, and cancer progression.64 LINC00662 is considered a lncRNA that promotes tumors, such as hepatocellular carcinoma and Non-small cell lung cancer.65,66 The lncRNA-miRNA-mRNA ceRNA network plays an essential role in immune and inflammatory pathways.67 Further studies are required to validate the regulatory mechanism of LINC00662.
Conclusion
The immune genes SELP, CD93, VAV1, and IL2RG linked to atherosclerosis were screened and validated as potential diagnostic biomarkers for psoriasis and showed a strong correlation with psoriasis. Four immune genes were highly correlated with immune cell infiltration and immunological response and consistently associated with multiple lipid metabolic processes, indicating that they may contribute to the development of psoriasis by influencing immune mechanisms and lipid metabolisms. LINC00662 may be involved in the pathogenesis of these four genes in psoriasis. The role of four diagnostic marker genes in immune and lipid metabolism may reveal the underlying molecular mechanisms of psoriasis and atherosclerosis.
Acknowledgments
We thank the GEO database for providing the platform and the contributors to the GSE78097, GSE28829, GSE14905, and GSE57691 datasets for uploading their meaningful datasets; we thank Sangerbox tools, Omicstudio platforms, venny tools, ImageGP tools, and Funrich software for providing online data analyses for free; we thank Starbase, Lncbase, Miwalk, MiRDB, and TargetScan databases for providing free online platforms for gene interaction searches; we thank our researchers for their hard work and the reviewers for their valuable advice.
Abbreviations
GEO, Gene Expression Omnibus; DEGs, differentially expressed genes; WGCNA, weighted gene co-expression network analysis; PA-IRGs, psoriasis and atherosclerosis immune-related genes; MSigDB, molecular characterization database; ROC, receiver operating characteristic; IHC, immunohistochemical; ssGSEA, single-sample gene set enrichment analysis; ceRNA, competitive endogenous RNA; GO, gene ontology; KEGG, Kyoto encyclopedia of genes and genomes; GSEA, gene set enrichment analysis; GSVA, gene set variation analysis; AUC, area under the curve; IOD, integrated optical density; MOD, mean optical density; BP, biological progress; CC, cellular components; MF, molecular functions.
Data Sharing Statement
The datasets and R code analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval and Informed Consent
The samples of psoriasis and normal skin tissues used in the study were approved by the Ethics Committee of the Hospital of Dermatology, Southern Medical University (Approval No: 2022040). Informed consent was waived due to the anonymized processing of patient data. Our study complies with the Declaration of Helsinki.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Gelfand JM, Neimann AL, Shin DB, Wang X, Margolis DJ, Troxel AB. Risk of myocardial infarction in patients with psoriasis. JAMA. 2006;296(14):1735–1741. doi: 10.1001/jama.296.14.1735 [DOI] [PubMed] [Google Scholar]
- 2.Armstrong EJ, Harskamp CT, Armstrong AW. Psoriasis and major adverse cardiovascular events: a systematic review and meta-analysis of observational studies. J Am Heart Assoc. 2013;2(2):e000062. doi: 10.1161/jaha.113.000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pietrzak A, Michalak-Stoma A, Chodorowska G, Szepietowski JC. Lipid disturbances in psoriasis: an update. Mediators Inflamm. 2010;2010:1–13. doi: 10.1155/2010/535612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miao C, Li J, Li Y, Zhang X. Obesity and dyslipidemia in patients with psoriasis: a case-control study. Medicine. 2019;98(31):e16323. doi: 10.1097/md.0000000000016323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu H, Daugherty A. Atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35(3):485–491. doi: 10.1161/atvbaha.115.305380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masson W, Lobo M, Molinero G. Psoriasis and cardiovascular risk: a comprehensive review. Adv Ther. 2020;37(5):2017–2033. doi: 10.1007/s12325-020-01346-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Zang J, Liu C, Yan Z, Shi D. Interleukin-17 links inflammatory cross-talks between comorbid psoriasis and atherosclerosis. Front Immunol. 2022;13:835671. doi: 10.3389/fimmu.2022.835671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aksentijevich M, Lateef SS, Anzenberg P, Dey AK, Mehta NN. Chronic inflammation, cardiometabolic diseases and effects of treatment: psoriasis as a human model. Trends Cardiovasc Med. 2020;30(8):472–478. doi: 10.1016/j.tcm.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose S, Stansky E, Dagur PK, et al. Characterization of immune cells in psoriatic adipose tissue. J Transl Med. 2014;12:258. doi: 10.1186/s12967-014-0258-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ertenli I, Kiraz S, Arici M, et al. P-selectin as a circulating molecular marker in rheumatoid arthritis with thrombocytosis. J Rheumatol. 1998;25(6):1054–1058. [PubMed] [Google Scholar]
- 11.Burkhardt J, Blume M, Petit-Teixeira E, et al. Cellular adhesion gene SELP is associated with rheumatoid arthritis and displays differential allelic expression. PLoS One. 2014;9(8):e103872. doi: 10.1371/journal.pone.0103872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George R, Bhatt A, Narayani J, Thulaseedharan JV, Sivadasanpillai H, Tharakan JA. Enhanced P-selectin expression on platelet-A marker of platelet activation, in young patients with angiographically proven coronary artery disease. Mol Cell Biochem. 2016;419(1–2):125–133. doi: 10.1007/s11010-016-2756-4 [DOI] [PubMed] [Google Scholar]
- 13.Morris DL, Graham RR, Erwig LP, et al. Variation in the upstream region of P-Selectin (SELP) is a risk factor for SLE. Genes Immun. 2009;10(5):404–413. doi: 10.1038/gene.2009.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Chen S, Liu Y, et al. P-selectin blockade ameliorates lupus nephritis in MRL/lpr mice through improving renal hypoxia and evaluation using BOLD-MRI. J Transl Med. 2020;18(1):116. doi: 10.1186/s12967-020-02284-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Battistini L, Piccio L, Rossi B, et al. CD8+ T cells from patients with acute multiple sclerosis display selective increase of adhesiveness in brain venules: a critical role for P-selectin glycoprotein ligand-1. Blood. 2003;101(12):4775–4782. doi: 10.1182/blood-2002-10-3309 [DOI] [PubMed] [Google Scholar]
- 16.Zhang N, Liu Z, Yao L, Mehta-D’souza P, McEver RP. P-selectin expressed by a human SELP transgene is atherogenic in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2016;36(6):1114–1121. doi: 10.1161/atvbaha.116.307437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ridker PM, Buring JE, Rifai N. Soluble P-selectin and the risk of future cardiovascular events. Circulation. 2001;103(4):491–495. doi: 10.1161/01.cir.103.4.491 [DOI] [PubMed] [Google Scholar]
- 18.Hillis GS, Terregino C, Taggart P, et al. Elevated soluble P-selectin levels are associated with an increased risk of early adverse events in patients with presumed myocardial ischemia. Am Heart J. 2002;143(2):235–241. doi: 10.1067/mhj.2002.120303 [DOI] [PubMed] [Google Scholar]
- 19.Greenlee MC, Sullivan SA, Bohlson SS. Detection and characterization of soluble CD93 released during inflammation. Inflamm Res. 2009;58(12):909–919. doi: 10.1007/s00011-009-0064-0 [DOI] [PubMed] [Google Scholar]
- 20.Greenlee-Wacker MC, Briseño C, Galvan M, Moriel G, Velázquez P, Bohlson SS. Membrane-associated CD93 regulates leukocyte migration and C1q-hemolytic activity during murine peritonitis. J Immunol. 2011;187(6):3353–3361. doi: 10.4049/jimmunol.1100803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenlee MC, Sullivan SA, Bohlson SS. CD93 and related family members: their role in innate immunity. Curr Drug Targets. 2008;9(2):130–138. doi: 10.2174/138945008783502421 [DOI] [PubMed] [Google Scholar]
- 22.Su C, Han Y, Qu B, et al. CD93 in macrophages: a novel target for atherosclerotic plaque imaging? J Cell Mol Med. 2022;26(8):2152–2162. doi: 10.1111/jcmm.17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Golden JB, Fritz Y, et al. Interleukin 6 regulates psoriasiform inflammation-associated thrombosis. JCI Insight. 2016;1(20):e89384. doi: 10.1172/jci.insight.89384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HJ, Han H, Lee SC, et al. Soluble CD93 in serum as a marker of allergic inflammation. Yonsei Med J. 2017;58(3):598–603. doi: 10.3349/ymj.2017.58.3.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeon JW, Jung JG, Shin EC, et al. Soluble CD93 induces differentiation of monocytes and enhances TLR responses. J Immunol. 2010;185(8):4921–4927. doi: 10.4049/jimmunol.0904011 [DOI] [PubMed] [Google Scholar]
- 26.Rahaman SO, Swat W, Febbraio M, Silverstein RL. Vav family Rho guanine nucleotide exchange factors regulate CD36-mediated macrophage foam cell formation. J Biol Chem. 2011;286(9):7010–7017. doi: 10.1074/jbc.M110.192450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bode AM, Dong Z. The functional contrariety of JNK. Mol Carcinog. 2007;46(8):591–598. doi: 10.1002/mc.20348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammouda MB, Ford AE, Liu Y, Zhang JY. The JNK signaling pathway in inflammatory skin disorders and cancer. Cells. 2020;9(4):857. doi: 10.3390/cells9040857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerreiro-Cacais AO, Norin U, Gyllenberg A, et al. VAV1 regulates experimental autoimmune arthritis and is associated with anti-CCP negative rheumatoid arthritis. Genes Immun. 2017;18(1):48–56. doi: 10.1038/gene.2016.49 [DOI] [PubMed] [Google Scholar]
- 30.Maleknia S, Salehi Z, Rezaei Tabar V, Sharifi-Zarchi A, Kavousi K. An integrative Bayesian network approach to highlight key drivers in systemic lupus erythematosus. Arthritis Res Ther. 2020;22(1):156. doi: 10.1186/s13075-020-02239-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jagodic M, Colacios C, Nohra R, et al. A role for VAV1 in experimental autoimmune encephalomyelitis and multiple sclerosis. Sci Transl Med. 2009;1(10):10ra21. doi: 10.1126/scitranslmed.3000278 [DOI] [PubMed] [Google Scholar]
- 32.Ayars M, O’Sullivan E, Macgregor-Das A, et al. IL2RG, identified as overexpressed by RNA-seq profiling of pancreatic intraepithelial neoplasia, mediates pancreatic cancer growth. Oncotarget. 2017;8(48):83370–83383. doi: 10.18632/oncotarget.19848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang LL, Pan HX, Wang YX, Guo T, Liu L. Genome profiling revealed the activation of IL2RG/JAK3/STAT5 in peripheral T‑cell lymphoma expressing the ITK‑SYK fusion gene. Int J Oncol. 2019;55(5):1077–1089. doi: 10.3892/ijo.2019.4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirken RA, Rui H, Malabarba MG, et al. Activation of JAK3, but not JAK1, is critical for IL-2-induced proliferation and STAT5 recruitment by a COOH-terminal region of the IL-2 receptor beta-chain. Cytokine. 1995;7(7):689–700. doi: 10.1006/cyto.1995.0081 [DOI] [PubMed] [Google Scholar]
- 35.Rujimongkon K, Ampawong S, Reamtong O, Buaban T, Aramwit P. The therapeutic effects of Bombyx mori sericin on rat skin psoriasis through modulated epidermal immunity and attenuated cell proliferation. J Tradit Complement Med. 2021;11(6):587–597. doi: 10.1016/j.jtcme.2021.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kyogoku C, Smiljanovic B, Grün JR, et al. Cell-specific type I IFN signatures in autoimmunity and viral infection: what makes the difference? PLoS One. 2013;8(12):e83776. doi: 10.1371/journal.pone.0083776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang X, Yue L, Liu W, et al. CD38 and E2F transcription factor 2 have uniquely increased expression in rheumatoid arthritis synovial tissues. Clin Exp Immunol. 2014;176(2):222–231. doi: 10.1111/cei.12268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang R, Yang X, Wang J, et al. Identification of potential biomarkers for differential diagnosis between rheumatoid arthritis and osteoarthritis via integrative genome‑wide gene expression profiling analysis. Mol Med Rep. 2019;19(1):30–40. doi: 10.3892/mmr.2018.9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pawlik A, Malinowski D, Paradowska-Gorycka A, Safranow K, Dziedziejko V. VAV1 gene polymorphisms in patients with rheumatoid arthritis. Int J Environ Res Public Health. 2020;17(9):3214. doi: 10.3390/ijerph17093214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng ZW, Tang YC, Sheng XY, et al. Screening and identification of potential hub genes and immune cell infiltration in the synovial tissue of rheumatoid arthritis by bioinformatic approach. Heliyon. 2023;9(1):e12799. doi: 10.1016/j.heliyon.2023.e12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowes MA, Suárez-Fariñas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–255. doi: 10.1146/annurev-immunol-032713-120225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiricozzi A, Romanelli P, Volpe E, Borsellino G, Romanelli M. Scanning the Immunopathogenesis of Psoriasis. Int J Mol Sci. 2018;19(1):179. doi: 10.3390/ijms19010179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorthois I, Asselineau D, Seyler N, Pouliot R. Contribution of in vivo and organotypic 3D models to understanding the role of macrophages and neutrophils in the pathogenesis of psoriasis. Mediators Inflamm. 2017;2017:7215072. doi: 10.1155/2017/7215072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucuksezer UC, Aktas Cetin E, Esen F, et al. The role of natural killer cells in autoimmune diseases. Front Immunol. 2021;12:622306. doi: 10.3389/fimmu.2021.622306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sajja AP, Joshi AA, Teague HL, Dey AK, Mehta NN. Potential immunological links between psoriasis and cardiovascular disease. Front Immunol. 2018;9:1234. doi: 10.3389/fimmu.2018.01234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yilmaz A, Lochno M, Traeg F, et al. Emergence of dendritic cells in rupture-prone regions of vulnerable carotid plaques. Atherosclerosis. 2004;176(1):101–110. doi: 10.1016/j.atherosclerosis.2004.04.027 [DOI] [PubMed] [Google Scholar]
- 47.Li Y, Wang F, Imani S, Tao L, Deng Y, Cai Y. Natural killer cells: friend or foe in metabolic diseases? Front Immunol. 2021;12:614429. doi: 10.3389/fimmu.2021.614429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harden JL, Krueger JG, Bowcock AM. The immunogenetics of Psoriasis: a comprehensive review. J Autoimmun. 2015;64:66–73. doi: 10.1016/j.jaut.2015.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mabuchi T, Chang TW, Quinter S, Hwang ST. Chemokine receptors in the pathogenesis and therapy of psoriasis. J Dermatol Sci. 2012;65(1):4–11. doi: 10.1016/j.jdermsci.2011.11.007 [DOI] [PubMed] [Google Scholar]
- 50.Zdanowska N, Kasprowicz-Furmańczyk M, Placek W, Owczarczyk-Saczonek A. the role of chemokines in psoriasis-an overview. Medicina. 2021;57(8):1. doi: 10.3390/medicina57080754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang C, Yang M. The role and potential application of antimicrobial peptides in autoimmune diseases. Front Immunol. 2020;11:859. doi: 10.3389/fimmu.2020.00859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sato Y, Ogawa E, Okuyama R. Role of innate immune cells in psoriasis. Int J Mol Sci. 2020;21(18):6604. doi: 10.3390/ijms21186604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ottaviani C, Nasorri F, Bedini C, de Pità O, Girolomoni G, Cavani A. CD56brightCD16(-) NK cells accumulate in psoriatic skin in response to CXCL10 and CCL5 and exacerbate skin inflammation. Eur J Immunol. 2006;36(1):118–128. doi: 10.1002/eji.200535243 [DOI] [PubMed] [Google Scholar]
- 54.Han G, Williams CA, Salter K, Garl PJ, Li AG, Wang XJ. A role for TGFbeta signaling in the pathogenesis of psoriasis. J Invest Dermatol. 2010;130(2):371–377. doi: 10.1038/jid.2009.252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441(7090):231–234. doi: 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- 56.Pietrzak A, Chodorowska G, Szepietowski J, Zalewska-Janowska A, Krasowska D, Hercogová J. Psoriasis and serum lipid abnormalities. Dermatol Ther. 2010;23(2):160–173. doi: 10.1111/j.1529-8019.2010.01311.x [DOI] [PubMed] [Google Scholar]
- 57.Nowowiejska J, Baran A, Flisiak I. Aberrations in lipid expression and metabolism in psoriasis. Int J Mol Sci. 2021;22(12):6561. doi: 10.3390/ijms22126561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng C, Wen B, Hou G, et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. GigaScience. 2017;6(10):1–11. doi: 10.1093/gigascience/gix087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harada F, Uehara O, Morikawa T, et al. Effect of systemic administration of lipopolysaccharides derived from Porphyromonas gingivalis on gene expression in mice kidney. Med Mol Morphol. 2018;51(3):156–165. doi: 10.1007/s00795-018-0181-3 [DOI] [PubMed] [Google Scholar]
- 60.Marques-Rocha JL, Samblas M, Milagro FI, Bressan J, Martínez JA, Marti A. Noncoding RNAs, cytokines, and inflammation-related diseases. FASEB J. 2015;29(9):3595–3611. doi: 10.1096/fj.14-260323 [DOI] [PubMed] [Google Scholar]
- 61.Li M, Duan L, Li Y, Liu B. Long noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in cardiovascular diseases. Life Sci. 2019;233:116440. doi: 10.1016/j.lfs.2019.04.066 [DOI] [PubMed] [Google Scholar]
- 62.Haemmig S, Simion V, Yang D, Deng Y, Feinberg MW. Long noncoding RNAs in cardiovascular disease, diagnosis, and therapy. Curr Opin Cardiol. 2017;32(6):776–783. doi: 10.1097/hco.0000000000000454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972. doi: 10.1038/ni.3771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian X, Wu Y, Yang Y, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol Oncol. 2020;14(2):462–483. doi: 10.1002/1878-0261.12606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lv X, Lian Y, Liu Z, Xiao J, Zhang D, Yin X. Exosomal long non-coding RNA LINC00662 promotes non-small cell lung cancer progression by miR-320d/E2F1 axis. Aging. 2021;13(4):6010–6024. doi: 10.18632/aging.202522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu GC, Pan HF, Leng RX, et al. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun Rev. 2015;14(9):798–805. doi: 10.1016/j.autrev.2015.05.004 [DOI] [PubMed] [Google Scholar]